Abstract
Slow waves are a distinguishing feature of non-rapid-eye-movement (NREM) sleep, an evolutionarily conserved process critical for brain function. Non-human studies suggest that the claustrum, a small subcortical nucleus, coordinates slow waves. We show that, in contrast to neurons from other brain regions, claustrum neurons in the human brain increase their spiking activity and track slow waves during NREM sleep, suggesting that the claustrum plays a role in coordinating human sleep architecture.
Subject terms: Slow-wave sleep, Non-REM sleep, Sleep
The role of the human claustrum during slow wave sleep is unknown. Here the authors characterize the spiking activity of claustrum neurons in humans and demonstrate that claustrum neurons track slow waves during NREM sleep.
Introduction
Slow waves are low-frequency (0.3–1.5 Hz) electrographic brain oscillations present across the N2 and N3 stages of non-rapid-eye-movement (NREM) sleep and are a defining feature of the deeper N3 stage (known as slow-wave sleep, SWS)1–4. NREM sleep is important for brain homeostasis and memory consolidation, and its disruption has been linked to a variety of neurocognitive and sleep disorders5,6. At the single neuron level, slow waves are characterized by alternating periods of synchronous silence (hyperpolarized DOWN states) and firing (depolarized UP states) in the cortex7. This synchrony is intrinsically generated in the cortex but highly regulated by a complex network of subcortical regions, including the thalamus8–11. Despite great interest in the contribution of slow waves to normal sleep physiology, the source of their regulation is poorly understood in humans.1,2
The claustrum is an evolutionarily conserved subcortical sheet of neurons with a high density of reciprocal connections with the cortex12–14. Its ability to modulate many cortical regions simultaneously has motivated inquiry into its contribution to diverse functions, including perception, salience detection, and memory15–21. Recent work in reptiles and rodents has shown that the claustrum coordinates slow waves in the cortex during sleep20,22–24. Specifically, the claustrum is thought to regulate slow waves by activating a network of cortical parvalbumin-expressing interneurons, suppressing activity across broad regions of the cortex through feedforward inhibition16,20,21. On a longer timescale, claustrum neurons in rodents increase spiking activity during NREM sleep compared to rapid-eye-movement (REM) sleep or wakeful states24. While this function of the claustrum appears to be conserved across reptiles and rodents, it has not yet been examined in humans.
We, therefore, sought to examine the relationship of the human claustrum with slow waves during sleep. We recorded overnight sleep sessions from epilepsy patients who underwent intracranial electrode placement into the middle insula for seizure localization, which gave us the rare opportunity to sample single units from the human claustrum. Using this paradigm, we show that claustrum neurons increase spiking activity during NREM sleep and periods of slow waves while other brain regions behave in an opposite manner. We conclude that the claustrum plays an important role in the regulatory network that coordinates slow waves during NREM sleep in humans.
Results
Microwire sampling of claustrum single units in humans
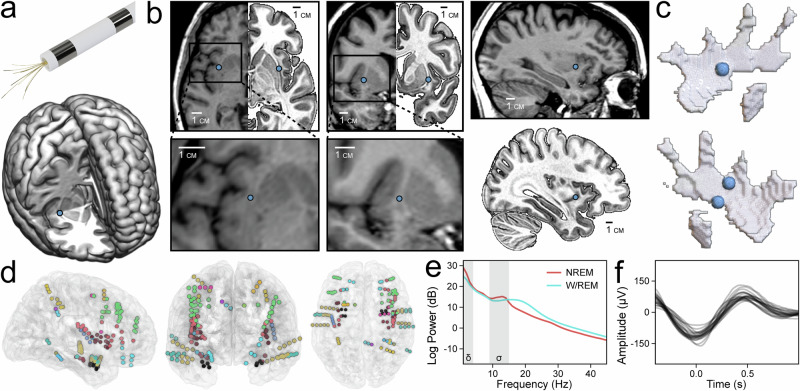
The small axial cross-section of the claustrum and its proximity to the insula and striatum have made human lesion studies difficult to interpret, as lesions frequently entail damage to adjacent structures25. Similarly, electrical stimulation using depth electrodes may not be confined to the claustrum alone, limiting its ability to define claustrum function26,27. We inserted 40 μm microwires into the claustrum through insular electrodes placed for clinical seizure localization in two human subjects, allowing us to overcome these technical challenges. Accurate placement was verified by fusion of pre-operative and post-operative imaging, which projected microwire locations onto subject-specific anatomy (Fig. 1a–c, Supplementary Table 1, Supplementary Figs. 1, 2).
Fig. 1. Claustrum single units sampled with microwires.
a Model of a Behnke–Fried depth electrode with protruding microwires for sampling the claustrum (top). Location of right claustrum microwires (blue dot) in Subject A on the MNI152 template (bottom). b Axial, coronal, and sagittal T1 MR images of Subject A with corresponding mirrored MNI152 templates in the axial and coronal views (top row) marking the location of right claustrum microwires and accompanied by magnified inserts and a sagittal MNI152 template (bottom row). c Locations of distal microwires when superimposed onto MNI152 models of the right and left claustrum, respectively (left top and bottom). Sagittal T1 MR image of Subject A (top right) marking the location of the right claustrum microwires. d Electrode locations for all subjects superimposed onto the N27 template in axial, coronal, and sagittal views (left, middle, right). e Power spectral density of NREM and W/REM sleep across all channels in Subject B, Night 03. Delta and sigma frequency bands (indicating SWA and sleep spindles, respectively) are shaded. f Average waveforms of detected slow waves in each channel of Subject B, Night 03.
After validating microwire placement, we tracked claustrum single unit activity during sleep using established spike-sorting methods (Supplementary Fig. 3). Over four nights and 33 h of sleep, we recorded 49 single units in the claustrum (CLA) along with 73 units in cortical (anterior cingulate cortex, ACC) and subcortical (amygdala, AMY) regions (Supplementary Tables 2, 3). Simultaneously, we recorded slow-wave activity (SWA, 0.3–4 Hz) in 150 intracranial macroelectrode contacts across 77 unique brain regions (Fig. 1d, e, Supplementary Table 4, Supplementary Fig. 4). SWA was defined as the z-score of the log-normalized 0.3–4 Hz band power. To identify morphological slow waves (SWs) for complementary analyses, we adapted an established slow-wave detection algorithm (Fig. 1f, Supplementary Fig. 4). Briefly, it applies a 0.3–1.5 Hz bandpass filter before pairing negative and positive peaks to produce candidate SWs, which are then subjected to numerous quality control parameters before acceptance.
Neurons typically decrease spiking activity during NREM sleep compared to REM sleep or wakefulness (except for neurons with very low firing rates <1 Hz)28–30. However, studies in mice have found the opposite pattern in the claustrum20,24. We, therefore, hypothesized that single units in the human CLA would increase spiking activity during periods of SWA in NREM sleep and that neurons in other brain regions (the ACC and AMY) would concomitantly decrease spiking activity.
Claustrum single units track slow-wave activity over hours of sleep
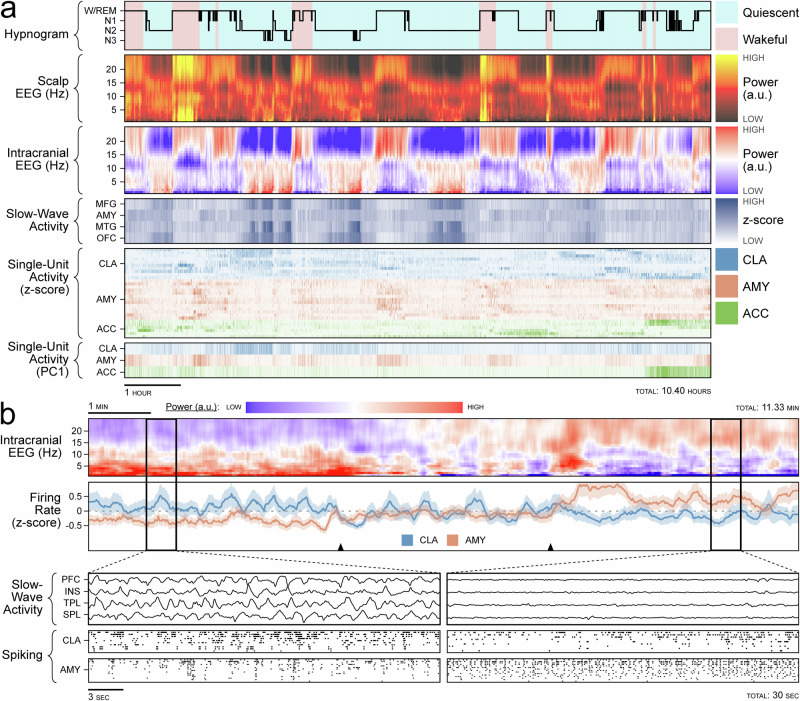
On a timescale of hours, we observed that CLA neurons increase their spiking activity during NREM sleep. This was in contrast to ACC and AMY neurons, which decreased their spiking activity over the same periods (Fig. 2a, Supplementary Fig. 5). On a timescale of minutes, this pattern was most prominent during transitions out of NREM sleep, in which CLA spiking activity decreased and AMY population activity increased (Fig. 2b, Supplementary Fig. 6). These initial qualitative findings were consistent with observations of claustrum activity, relative to cortical activity, during NREM sleep in rodents20,24.
Fig. 2. Claustrum single units and slow-wave activity tracking over hours of sleep.
a Sleep recording from Subject B, Night 03. Hypnogram is colored by behavioral state observed on avEEG: red indicates wakefulness and blue indicates behavioral quiescence (first row). Power spectrogram from the C4 scalp electrode (second row). Illustrative power spectrogram from right middle frontal gyrus (third row). Binned z-score of slow-wave activity from four illustrative regions: middle frontal gyrus, amygdala, middle temporal gyrus, and orbitofrontal cortex (fourth row). Binned z-score of the firing rate for claustrum (blue), anterior cingulate cortex (green), and amygdala (orange) single units (fifth row). First principal component of the firing rate for single units in the above regions (sixth row). b Transition out of NREM sleep in Subject A. Power spectrogram of the left middle frontal electrode (first row). z-scored population firing rates in the claustrum (blue) and amygdala (orange) with black triangles indicating the transition period (second row). Magnified windows before and after sleep transition (bottom left and bottom right panels) showing SWA in the prefrontal cortex, insula, temporal lobe, and parietal lobe (first rows of bottom panels) and raster plots of spiking activity for the claustrum (second rows of bottom panels) and amygdala (third rows of bottom panels).
Claustrum single units increase spiking with slow-wave activity (SWA) and non-rapid-eye-movement (NREM) sleep
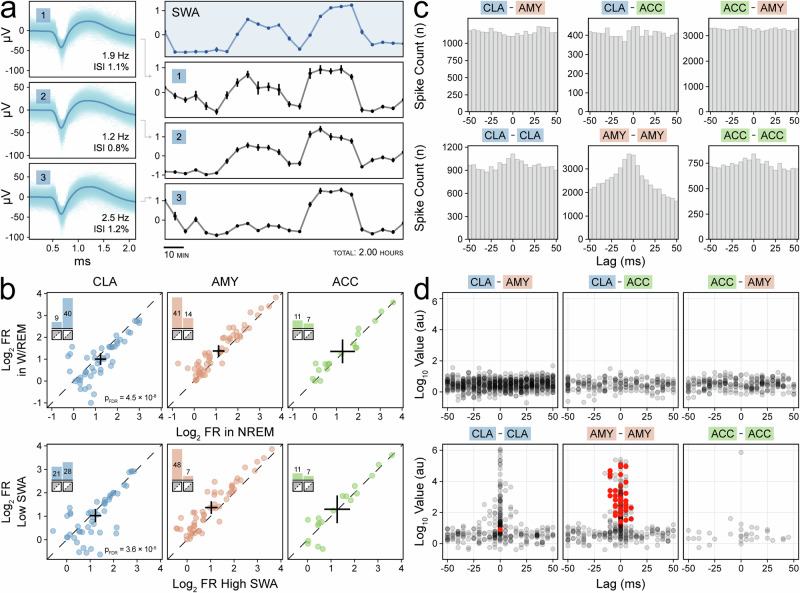
We confirmed the initial observation that CLA neurons increase their spiking activity during NREM sleep by inspecting CLA units individually (Fig. 3a, Supplementary Fig. 7). First, we divided sleep recordings into epochs classified by indirect measures of SWs: sleep stage and SWA. We found that a majority (40/49) of CLA neurons increased spiking activity during NREM sleep relative to wakefulness and REM sleep (combined into W/REM). This was significantly different than the pattern seen in pooled control AMY and ACC units (χ2: pFDR = 4.5 × 10−8). A similar pattern was observed when we compared periods of high SWA to periods of low SWA (χ2: pFDR = 3.6 × 10−5) (Fig. 3b). Cross-correlograms between single units were generated to assess correlated spiking behavior across regions (Fig. 3c). However, while strong cross-correlation existed between some units in the same region, especially in the amygdala, no significant interactions across regions were found (Fig. 3d).
Fig. 3. Claustrum single units increase spiking with slow-wave activity (SWA) and non-rapid-eye-movement (NREM) sleep.
a Average waveforms of three claustrum units with inset firing rate and inter-spike interval violations (left panels) with corresponding mean z-scored firing rate (black panels) aligned to the illustrative z-scored slow-wave activity (SWA) in the right orbitofrontal cortex from Subject A (blue panels) (right panels). Standard error for each 5-min interval (30 epochs of 10 s each) is indicated by vertical bars. b Scatterplots of log2 firing rate of claustrum (blue), amygdala (orange), and anterior cingulate (green) units stratified by sleep stage (not NREM sleep vs. NREM sleep, left) and SWA (25th vs. 75th percentiles, middle). Crosses indicate population averages with 95% confidence intervals. Units in the lower triangle favor SWs. Inset bar plots indicate the number of units in the upper and lower triangles, and their sum indicates the sample size. c Example cross-correlograms during NREM sleep between single unit pairs with p < 0.05 prior to FDR correction on threshold-free cluster enhancement (TFCE) permutation testing (except for ACC–ACC). d Scatterplots stratified by region pairs showing the maximum TFCE value of each unit pair’s cross-correlogram (in log10) and the time lag at which it occurred; red indicates unit pairs with p < 0.05 after FDR correction on TFCE permutation testing.
Claustrum single units positively correlate with measures of slow-wave sleep (SWS)
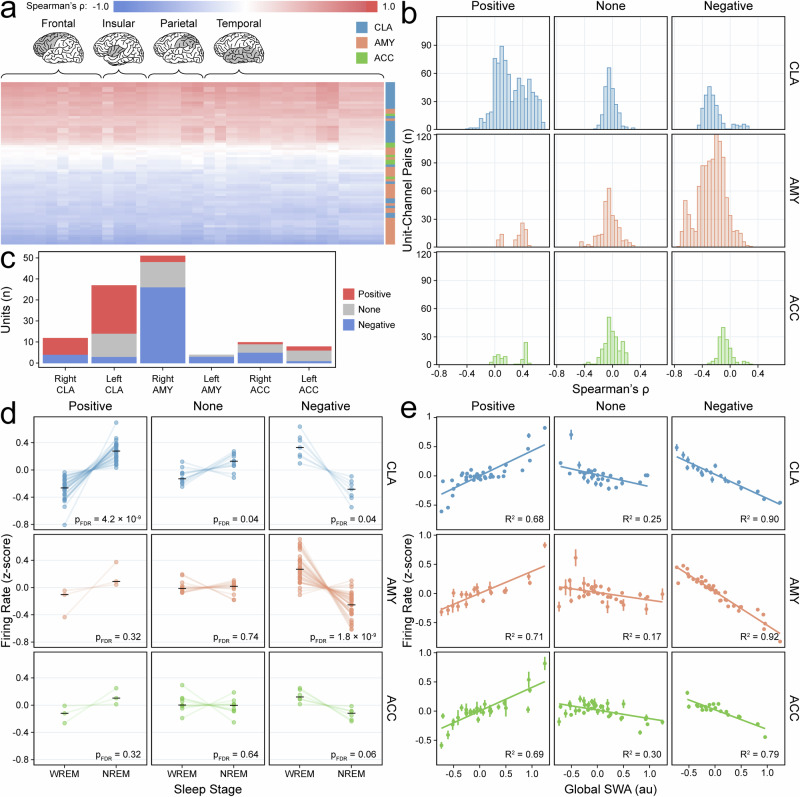
Based on these observations, we quantified the heterogeneity of responses among neurons: Single units were classified into positive-, negative-, and non-responding units based on Spearman’s ρ correlations with SWA and SWs in ipsilateral brain regions (Fig. 4a, b, Supplementary Fig. 8). This analysis showed that 63% of CLA single units were positive responders compared to 8% of units in other regions (17% ACC, 5% AMY); conversely, only 14% of CLA single units were negative responders compared to 62% of units in other regions (33% ACC, 71% AMY) (Fig. 4c). The assigned response types were further validated by assessing population spiking behavior across sleep stages and deciles of SWA stratified by unit response type (Fig. 4d,e). Thus, the majority of CLA neurons displayed a preference for spiking during periods of SWs compared to neurons in other regions.
Fig. 4. Claustrum single units positively correlate with measures of slow-wave sleep (SWS).
a Heatmap of Spearman’s ρ correlations between units (rows) and SWA across channels (columns) for Subject A. Colors indicate unit region. Highlighted lobes indicate channel locations. b Histogram of Spearman’s ρ values for unit-channel pairs (122 units and 64 unique channels across four recordings) stratified by unit response type and unit region. Colors indicate unit region. c Bar plot showing distribution of single units across all subjects with positive (red), negative (blue), or no correlation (gray) with ipsilateral SWA and SWs. d Paired dot plots of z-scored firing rates for units (n = 122) in WREM versus NREM sleep stratified by unit response type and unit region; black horizontal lines indicate group medians, and p-values are derived from FDR-corrected Wilcoxon signed-rank tests (two-sided). Colors indicate unit region. e Decile scatterplots stratified by unit response type and unit region show the average z-scored firing rate for each decile of SWA. Deciles are plotted separately for each of four sleep recordings and are derived from 10 s epochs (underlying sample size varies based on recording length and the number of units in each response-region subset); vertical lines indicate the standard error; fitted lines and R2 values are based on linear models. Colors indicate unit region.
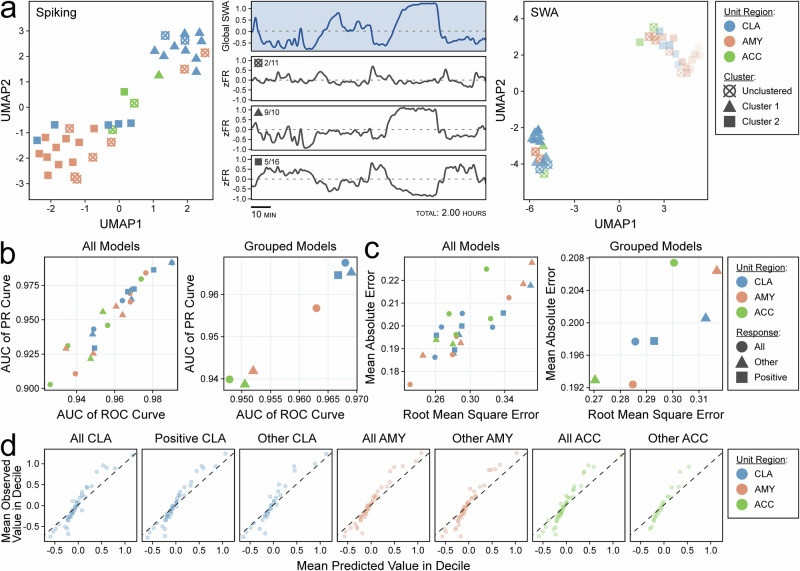
Claustrum population activity predicts slow-wave activity
To compare the population activities of the CLA, ACC, and AMY across full sleep recordings, we used uniform manifold approximation and projection (UMAP) to collapse single unit activity across time31. Single units were segregated into clusters in native space using hierarchical density-based spatial clustering of applications with noise (HDBSCAN) prior to UMAP32. Clusters positively correlated with SWA were enriched in CLA units, while negatively correlated with SWA were enriched in ACC and AMY units (Fig. 5a, Supplementary Fig. 9). We then collapsed single unit activity into population activity using the temporal potential of heat-diffusion for affinity-based transition embedding (T-PHATE), which is a dimensionality reduction technique designed to preserve time33. These new dimensions were used as model features for the prediction of sleep stage and SWA. The T-PHATE population activity from all regions, including CLA population activity, was highly accurate in classification of sleep stage (mean AUC: CLA = 0.97, AMY = 0.96, ACC = 0.95) and regression of SWA (mean RMSE: CLA = 0.29, AMY = 0.28, ACC = 0.30), reflecting that population activity in all sampled regions undergoes major changes during SWS, even though such changes may be opposite in direction (Fig. 5b–d).
Fig. 5. Claustrum population activity predicts slow-wave activity (SWA).
a Scatterplot of UMAP dimensions for Subject A’s single-unit spiking activity demonstrating self-segregation of single units into two clusters (and one unclustered group) indicated by shapes (left). Global slow-wave activity (middle, top panel) with aligned z-scored population firing rates for UMAP clusters with fractions indicating the proportion of claustrum units in each cluster (middle). Scatterplot of two UMAP dimensions for the same units after dimensionality reduction of correlation with slow wave presence across channels (right). b Scatterplot displaying the area under the curve (AUC) for the receiver operating characteristic (ROC) curve and the precision-recall (PR) curve for support vector machine (SVM) models classifying sleep stage using dimensionally reduced population activity from single unit groups stratified by microwire region, response type, and sleep recording (left panel). The same results are shown after averaging across sleep recordings (right panel). Colors indicate unit region, and shapes indicate unit response type. c Scatterplot displaying the root mean square error (RMSE) and mean absolute error (MAE) of SVM models regressing global SWA using dimensionally reduced population activity from single unit groups stratified by microwire region, response type, and sleep recording (left panel). The same results are shown after averaging across sleep recordings (right panel). Colors indicate unit region, and shapes indicate unit response type. d Decile-wise calibration plots for the models in (c) stratified by response type and unit region. Values are arbitrary units (z-scored log10 of global SWA power). Colors indicate unit region.
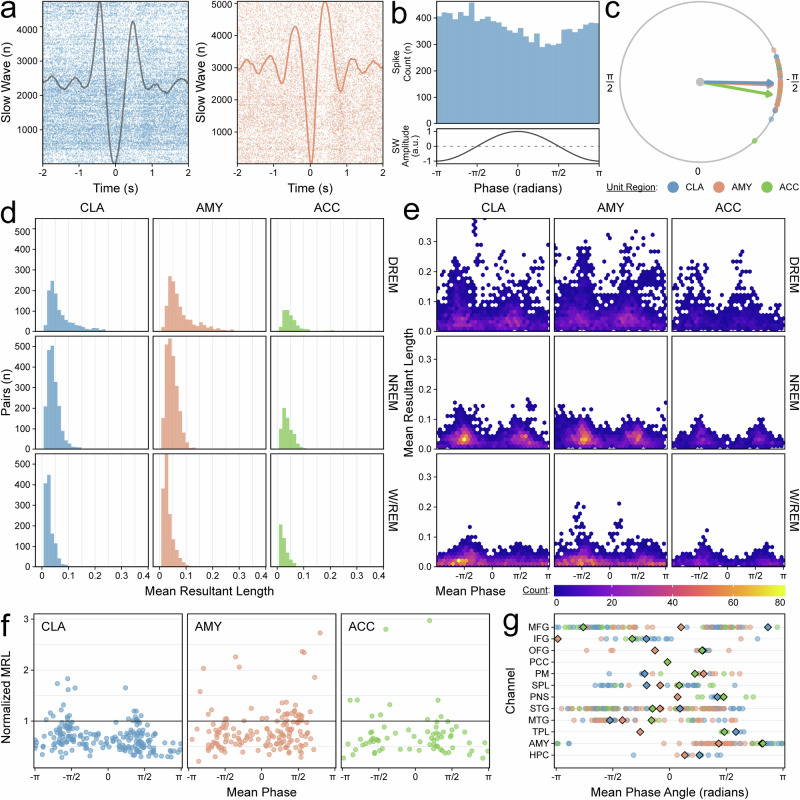
Claustrum single units phase-lock with the slow-wave band
We then examined the relationship between the CLA activity and the slow-wave band (SWB, 0.3–1.5 Hz), which more narrowly captures morphological slow waves, during NREM sleep, using spike-phase coupling (SPC) (Fig. 6a–c). The strength of SPC, as measured by the distributions of mean resultant lengths (MRLs, a measure of the concentration of phase angles), increased from W/REM to NREM (Fig. 6d). The MRLis further increased when comparing NREM sleep with high delta-band power (DREM) to all NREM sleep to periods of NREM sleep with high delta-band power (DREM) (Fig. 6e). The strength of coupling between AMY single units and AMY SWB phase was designated as the positive control given that it was the only region with simultaneous sampling of single units and macroelectrode field potentials (Fig. 6a, f). Strong SPC relationships in CLA clustered around phases of −π/2 and π/2; a similar effect was seen in the AMY and ACC, although less markedly (Fig. 6f). Significant SPC of CLA units with SWB phase was present in nearly all sampled macroelectrode regions (Fig. 6g, Supplementary Table 5)31.
Fig. 6. Claustrum single units phase-lock with the slow-wave band.
a Raster plot demonstrating phase-locking of a claustrum unit with slow waves (SW) in an illustrative orbitofrontal channel (top) compared to phase-locking in an amygdala unit with its adjacent amygdala channel (bottom) in Subject B, Night 03. The average slow-wave waveforms are superimposed. b Phase histogram for the same claustrum-orbitofrontal pair (top); the amplitude of an idealized slow wave for each phase is shown (bottom). c Polar plot for the same claustrum-orbitofrontal pair indicating the preferred phase angle for every unit-orbitofrontal pair with a phase distribution significantly different from uniform. Color indicates unit region, and arrows indicate the average preferred phase angle among all pairs of the same unit region. d Histograms of mean resultant lengths (MRLs) for unit-channel pairs stratified by unit region and sleep stage. High delta NREM (DREM) indicates NREM sleep ≥75th percentile of NREM SWA power. Colors indicate unit region. e Heatmaps of the phases at which MRLs occurred for each unit-channel pair, stratified by unit region and sleep stage. f Scatterplot of phases at which MRLs occurred during DREM sleep. Only pairs with a significant relationship in DREM sleep but not in WREM sleep are displayed. The MRLs have been normalized to the mean MRL of AMY-AMY pairs, which is indicated by the horizontal black line. Colors indicate unit region. g Region-wise preferred phase angles for all unit-channel pairs with a phase distribution significantly different from uniform in DREM sleep but not W/REM sleep (combined across all subjects). Colors indicate unit region. Diamonds indicate the average preferred phase angles for each unit region. See Supplementary Table 5 for abbreviations.
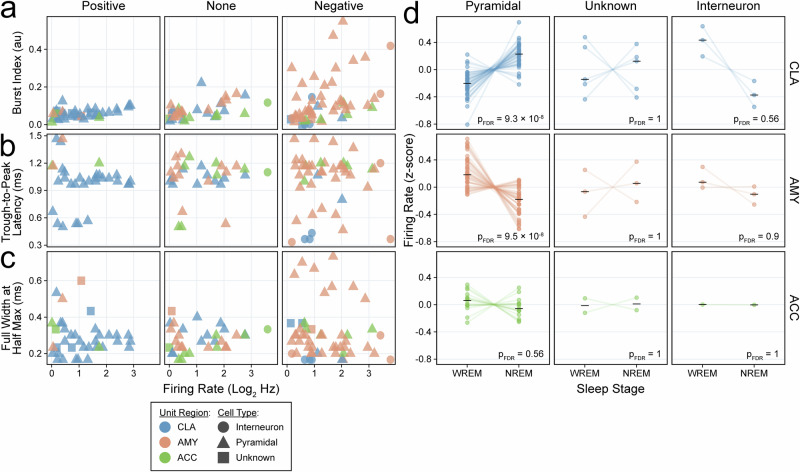
Pyramidal neurons positively correlate with slow-wave activity in the human claustrum
Finally, we inferred the cell types of our 122 single neurons using established metrics (Fig. 7a–c)34. Most CLA units were classified as pyramidal cells (84%) with a similar proportion in control region units (AMY 89%, ACC 83%); the remaining cells were classified as interneurons or did not meet criteria for either cell type. Notably, nearly all CLA units classified as pyramidal cells exhibited increased spiking activity during NREM sleep (p = 9.3 × 10−8), whereas all CLA units classified as interneurons and about half of CLA units classified as unknown displayed a decrease in spiking activity during NREM sleep (Fig. 7d).
Fig. 7. Claustrum units that increase spiking with slow-wave activity are enriched with pyramidal cells.
a–c Scatterplots of firing rate (log2, Hz) versus a burst index (au), b trough-to-peak latency (negative-spiking units only, ms), and c full-width at half-maximum (ms) stratified by unit response type. Colors indicate unit region and shapes indicate cell type classification. d Paired dot plots of z-scored firing rates for units in WREM versus NREM sleep stratified by cell type classification and unit region; black horizontal lines indicate group medians, and p-values are derived from FDR-corrected Wilcoxon signed-rank tests (two-sided). Colors indicate unit region.
Discussion
We recorded single neurons in the human claustrum. In contrast to neurons in other brain regions, CLA neuronal activity tracked SWA in NREM sleep, at multiple timescales and over multiple nights of sleep: Spiking activity increased with SWA at NREM sleep onset, maintained heightened levels throughout NREM sleep periods, and decreased with the transition to REM sleep or wakefulness. These correlations were robust for SWA measured in many brain regions, including the frontal lobe, which is known to have strong connections with the human claustrum14,20,35. We also found that CLA units displayed broadly significant phase locking with the cortical SWB during NREM sleep, which is consistent with recent animal findings that CLA neuronal stimulation aligns SWs across the cortex24. CLA neurons were mostly classified as pyramidal cells; this result may be consistent with the theory of CLA feedforward inhibition, as CLA pyramidal cells are connected to the SW-generating cortex in animals16,20,21. We believe that our results support previous work in animals and argue for a conserved role of the claustrum in the coordination of human SWs.
A broader question remains: how is the claustrum integrated into the known circuitry of NREM sleep regulation? Many subcortical nuclei participate in the NREM regulatory network36. Specifically, the thalamus has been shown to orchestrate neocortical slow waves during NREM sleep, via projections to parvalbumin-positive interneurons37,38. It is possible that the claustrum plays a complementary role in the regulation of SWS by coordinating SWs across brain regions via trans-claustral circuits39. For example, slow waves in humans have been shown to predominantly originate from the anterior-inferior regions of the brain (e.g. prefrontal cortex, temporal lobe, insula) and propagate in a broadly posterior-superior direction40,41. Trans-claustral circuits may be a mechanism that facilitates this propagation. Alternatively, the claustrum may serve to suppress the cortex through slow-wave activity in NREM sleep—as opposed to thalamic sleep spindles that may be associated with excitatory post-synaptic potentials, faster cortical frequencies, and cortical sleep spindles42,43. Recent evidence suggests that the claustrum is also involved in the regulation of other sleep stages, such as REM23. This, however, was outside the scope of the current work.
Our study has several limitations. First, we studied claustrum activity in subjects with epilepsy undergoing inpatient seizure monitoring, which may reduce the generalizability of our results to the broader population. Furthermore, while emerging evidence suggests that the CLA may be organized into functional modules, all of our microwires sampled the mid-claustrum, as dictated by clinical planning44,45. This prevented us from generalizing our results to the entire claustrum. Additionally, claustrum subregions are known to have anatomically distinct efferent projections. However, our analyses did not directly identify the efferent targets of our sampled neurons46,47. Thus, we considered all possible efferent connections, reducing the specificity of our claims. Future studies combining high-resolution tractography to trace efferent claustrum projections to the cortex combined with higher-density macroelectrode intracranial sampling may overcome this limitation and allow for the testing of the hypothesis that trans-claustral circuits facilitate SW propagation across the cortex. Other technical limitations of our study include the unbalanced contribution of subjects to the pool of single units and the possibility that some units may be counted across sleep sessions if they persist across days of recording.
Despite these limitations, we found that a population of human claustrum neurons increased their activity with SWs during NREM sleep. This relationship was consistent across multiple timescales and several measures of SWs and is in accordance with animal literature. Taken together, our observations build on existing causal studies in animals to converge on the theory that the human claustrum plays a key role in coordinating SWs during NREM sleep. These results advance our understanding of the normal physiology of human NREM sleep and suggest that claustrum disruption may lead to sleep-related neuropathology.
Methods
Participants
Two right-hand dominant female subjects (Subject A was 47 years old; Subject B was 27 years old) with medication-refractory epilepsy undergoing intracranial EEG (icEEG) electrode implantation for clinical seizure localization were enrolled in the study after providing informed consent. Plans for the icEEG studies were made exclusively for clinical purposes; both subjects had macroelectrodes placed into the middle insula, and microwires extending from those electrodes sampled the claustrum (bilaterally in Subject A, left in Subject B). Both subjects also remained on their home anti-seizure medications for the duration of the study, and sleep sessions were recorded in the epilepsy monitoring unit. One sleep session was recorded for Subject A (2 h), and three sleep sessions were recorded for Subject B (9.7, 10.6, and 10.4 h). The study was approved by the Institutional Review Board at Yale University.
Electrophysiology system
Subdermal scalp electrodes were placed according to the 10–20 system. Spencer Probe and Behnke-Fried depth electrodes (Ad-Tech Medical Instrument Corp., Oak Creek, WI, USA) were placed using a ROSA surgical robot (Zimmer Biomet, Warsaw, IN, USA) to bilaterally sample field potentials from numerous brain regions48. Scalp and macroelectrode data were recorded with a NeuroPort Neural Signal Processor (Blackrock Neurotech, Salt Lake City, UT, USA) at a sampling rate of 2048 Hz with a 0.3–500 Hz hardware bandpass filter. Extracellular microwire recordings were sampled at 30 kHz with a 250 Hz–7.5 kHz bandpass filter in Subject A and a 0.3 Hz–7.5 kHz bandpass filter in Subject B. The online reference electrodes were screwed into the outer cortex of the frontal bone, and left orbitofrontal contacts were selected as the ground electrodes in both subjects.
Electrode localization
Pre-operative axial non-contrast T1-weighted magnetic resonance (MR) sequences with 1 mm slices were co-registered with post-operative axial non-contrast CT scans with 0.625 mm slices using Statistical Parametric Mapping (SPM) 1249. Macro- and microelectrodes were automatically localized on the fused image and manually adjusted using LeGUI software (Supplementary Fig. 1)50. Gray and white matter were automatically segmented, and macroelectrodes in white matter were later excluded from analysis after re-referencing. The fused image and electrode positions were warped into Montreal Neurological Institute (MNI) 152 space for the assignment of electrodes to the nearest region of interest (ROI) in the Yale Brain Atlas (YBA)51. Electrode coordinates were then transformed into MNI305 and then tkrRAS space for visualization in MRIcroGL and the threeBrain package52,53. A total of 76 contacts in Subject A and 74 contacts in Subject B were selected for analysis (Supplementary Table 4). Perpendicular distances from the tips of the claustrum microwires to the axial centerline of the claustrum were calculated to quantify placement accuracy (Supplementary Table 1). Claustrum microwire locations were projected onto a three-dimensional claustrum model derived from segmentation of the MNI 152 template using an algorithm that has been previously described54. The position of microwires in the claustrum was also confirmed using Lead-DBS, which uses Advanced Normalization Tools for CT-MRI co-registration and transformation into MNI 152 space prior to manual localization of the microwires (Supplementary Fig. 2)55,56.
Pre-processing
The Blackrock Neural Processing Matlab Kit (NPMK) was used to concatenate data and repair segments of packet loss (https://github.com/BlackrockNeurotech/NPMK). Data were then cropped to sleep intervals, notch filtered for 60 Hz electrical line noise (and its harmonics), lowpass filtered at 128 Hz, and decimated to 256 Hz using the MNE software library57. Subdermal scalp and intracranial macroelectrodes were separately re-referenced to the common average reference (CAR) for their electrode type.
Sleep staging
Sleep recordings were divided into 30-second epochs, and a previously validated algorithm with integral artifact rejection was used to automatically classify sleep stages based on scalp EEG (C4 in both subjects)58. Automatic sleep staging was internally validated using manual sleep stage classification by a board-certified neurologist. Sleep staging yielded 65 min of N2/N3 sleep in Subject A and 240, 319, and 321 min of N2/N3 sleep in Subject B’s three recordings (Supplementary Table 3).
Artifact rejection
For automatic intracranial macroelectrode channel rejection, the raw time series was converted into a low-frequency time series (0–50 Hz), high-frequency time series (50–120 Hz), and first derivative time series. The local outlier factor (LOF) algorithm (10 neighbors) was then computed for each time series, and any channel with a LOF score less than −2 for any time series was rejected. The power spectral density across the entire sleep recording was computed for every channel using Welch’s method, and outliers were manually identified and cross-referenced to confirm that all channels contaminated by artifact had been removed. Next, interictal epileptiform discharges (IEDs) were detected using a previously developed algorithm, and time intervals within one second of an IED were excluded from subsequent slow-wave detection in a channel-wise manner (https://github.com/Kleen-Lab/Linelength-spike-detector-PYTHON). A total of 83, 796, 1407, and 1130 IEDs were detected in at least one channel across the four sleep recordings of 2, 9.7, 10.6, and 10.4 h, respectively. Finally, sleep intervals classified as N2 or N3 sleep were divided into 3-s epochs for further artifact rejection. Numerous features were extracted for each epoch (mean, variance, standard deviation, peak-to-peak amplitude, skewness, kurtosis, root-mean-squared value, quantile, zero crossings, Hurst exponent, band-wise average power for eight frequency bands covering 0.5–128 Hz, and slope of the power spectral density)59. The LOF score was then computed on the extracted features for each epoch in a channel-wise manner, and epochs with a LOF score of less than −2 in more than three channels were excluded from slow-wave detection (with an additional one-second interval before and after the epoch time interval).
Audiovisual review
Audiovisual recordings of candidate sleep sessions were manually reviewed at five-minute intervals on a Natus Neuroworks clinical EEG system (Natus Medical Inc., Middleton, WI, USA). Behavioral state was classified as either wakeful or quiescent, and these results were manually compared to the hypnogram generated by the sleep staging algorithm for each candidate sleep session.
Time–frequency processing
Time-frequency power spectrograms were generated with the Morlet wavelet transform (cycles = 6) for 1–25 Hz and log-ratio normalized by frequency. Spectrograms for whole-sleep recordings were averaged into 30-s time bins and smoothed with a Gaussian window. Slow-wave activity (SWA) was extracted from the envelope of the Hilbert Transform on the band-passed time series (0.3–4 Hz). Sleep transitions were identified via manual review of whole-recording intracranial spectrograms.
Slow-wave detection
A previously validated algorithm for slow-wave detection on scalp EEG was adapted for use on intracranial EEG58. Only artifact-free intracranial macroelectrode contacts were selected for use. By default, data is 0.3–1.5 Hz bandpass filtered, both negative and positive peaks are labeled and sequentially paired, feature thresholding is applied, and outliers are removed using the Isolation Forest algorithm. For adaptation to icEEG, raw time series were transformed into channel-wise robust z-scores (rZS) before slow-wave detection. Feature thresholds were set for the duration of negative deflection (0.3–1.5 s), duration of positive deflection (0.1–1.5 s), negative peak amplitude (>1 rZS), positive peak amplitude (>1 rZS), and peak-to-peak amplitude (>4 rZS). Channels with <3 detected slow waves per minute across time intervals classified as N2 or N3 sleep were excluded from the analysis. After exclusion, we detected a median of 8.0 SWs per contact per minute of N2/3 sleep in Subject A and 6.3, 7.0, and 6.8 in the three Subject B recordings.
Spike sorting
The Combinato algorithm was used to detect spikes, reject artifacts, and cluster spikes into units using a validated superparamagnetic clustering algorithm (maximum cluster distance for grouping = 2.5, minimum spikes for cluster selection = 25)60. Candidate units were manually reviewed for artifacts and split or merged as appropriate. In addition to the quality control steps in Combinato, unit stability was ensured throughout the recording sessions by assessment of the cumulative spike distribution and five metrics of waveform stability calculated with randomly sampled spikes (n = 1000) from every single unit, including the l-ratio and the coefficients of variation of the maximum amplitude, full-width half-maximum, the area under the curve, and Euclidean distance from the mean waveform (Supplementary Fig. 3). Candidate units with an average firing rate of <1 Hz or an inter-spike interval <3 ms in more than 5% of spikes were excluded from the analysis. Spike sorting yielded 49 claustrum single units (37 left, 12 right), 18 anterior cingulate single units (8 left, 10 right), and 55 amygdala single units (4 left, 51 right) across all recordings (Supplementary Table 2).
Cross-correlation
Raw cross-correlograms (21 bins, 5 ms bin width) were computed for the spike trains of all possible pairs of single units (excluding unit pairs from the same region) during NREM sleep. For each raw cross-correlogram, surrogate cross-correlograms (n = 1000) were computed by jittering spike times (20 ms, uniform random distribution) in the spike train of the second unit in each pair61,62. The means and standard deviations of the surrogate cross-correlogram bins were then used to transform the raw cross-correlograms into z-scored cross-correlograms in a bin-wise fashion. The z-scored cross-correlograms then underwent a threshold-free cluster enhancement (TFCE) procedure to produce a TFCE-valued cross-correlogram63. Surrogates were generated by randomly shuffling the z-scored cross-correlogram bins and then performing the TFCE procedure (n = 1000). The maximum bin values of the TFCE-valued cross-correlogram and its surrogates were extracted (the lag time of the maximum bin value of the TFCE-valued cross-correlogram was also extracted), and the p-value for each unit pair was defined as the percent of maximum surrogate TFCE bin values greater than the true TFCE bin value. The p-values were corrected for the false discovery rate (FDR) using the Benjamini–Hochberg method.
SW measures
Data were divided into 10-s epochs, and for each epoch, three SW measures were calculated: sleep stage (assigned based on 30-s epochs from sleep staging), z-score of the average log-normalized delta power (SWA), and slow-wave presence (percent of the epoch that a slow wave was present). Adjacent slow waves within one second were merged when calculating slow-wave presence. For each SW measure scatterplot, the χ2 test was used to compare the proportion of CLA units in the upper and lower triangles of the scatterplot to those of other regions (ACC and AMY).
Unit responsiveness
For each unit, the average firing rate was separately compared epoch-wise to each of the three SW measures using a Spearman’s ρ correlation; this comparison was channel-wise in the case of SWA and SW presence. Spearman’s ρ correlation p-values were FDR-corrected using the Benjamini–Hochberg method. Units were defined as positive responders if they had a positive Spearman’s ρ with FDR-corrected p-values < 0.01 across at least half of the ipsilateral intracranial channels for both SWA and SW presence. Negative responders were defined conversely, and units failing to meet either criterion were defined as non-responders. Two auxiliary analyses were performed for the validation of unit responsiveness. First, FDR-corrected paired Wilcoxon signed-rank tests were used to assess differences in z-scored firing rate between periods of W/REM sleep and periods of NREM sleep for single units after stratification by unit responsiveness and microelectrode region. Second, sleep recordings were divided into deciles of normalized global SWA, and the average z-scored firing rate for each stratum was calculated per decile. A linear regression was then fit with normalized global SWA as the dependent variable and z-scored firing rate as the independent variable, and the R2 values were compared between strata.
Population modeling
Population modeling of unit spiking was undertaken for the classification of sleep stage (NREM versus non-NREM) and regression of global SWA power, respectively. Features were extracted from 10-s epochs of single-unit spiking activity by collapsing units into three dimensions using temporal potential of heat-diffusion for affinity-based transition embedding (T-PHATE)33. Support vector machines (SVMs) were trained on these features using 10-fold cross-validation of the training set (70% random split), and performance was evaluated on the testing set (30% random split). Classification models were evaluated using the area under the curves (AUC) of the receiver operating characteristic (ROC) curve and the precision-recall (PR) curve. Regression models were evaluated using the mean absolute error (MAE) and the root mean squared error (RMSE). Modeling was performed independently for all strata as defined by sleep recording, unit response type (positive, negative, other), and microwire region (CLA, AMY, ACC). Strata with <5 single units were excluded from the analysis.
Spike-phase coupling
The Hilbert transform was used to extract the phase of the 0.3–1.5 Hz frequency band for every macroelectrode channel. Spike times of single units during N2 or N3 sleep were then intersected with the nearest sampled phase, and these phases were pooled for every unit-channel pair. The set of phases for each unit-channel pair was tested for a significant difference from the uniform distribution using a permutation-based Rayleigh’s test with FDR correction. Briefly, z-statistics were extracted from Rayleigh’s tests for each unit-channel pair. The same procedure was applied to surrogate data (n = 1000), which were generated by randomly jittering phase angles by up to a quarter wavelength in either direction62. The p-value was then defined as the percent of surrogate z-statistics that were equal to or larger than the original z-statistic. p-values for all unit-channel pairs underwent FDR correction. Mean phase angles and mean resultant lengths were extracted for all unit-channel pairs. This analysis was performed independently for different sleep stages of the data: W/REM, NREM, and high delta power NREM (DREM). DREM was defined as periods of NREM sleep where the SWA was at or above the 75th percentile of NREM SWA power.
Dimensionality reduction
The spiking activity of single units was binned into 10-s epochs across sleep recordings and z-scored. This data was collapsed from an n × t matrix (where t represents time bins) into an n × 2 matrix using uniform manifold approximation and projection (UMAP)31. Correlation matrices of the Spearman’s ρ values between single-unit activity and either the SWA or SW presence on macroelectrode channels (after binning into 10-s epochs) were also reduced from n × c matrices into n × 2 matrices (where c represents the macroelectrode channels) with UMAP. Units were clustered using the hierarchical density-based spatial clustering of applications with Noise (HDBSCAN) algorithm for further analysis (the minimum points per cluster were manually optimized for each sleep recording due to differing numbers of units)32.
Cell type classification
Several established metrics (firing rate, burst index, and trough-to-peak latency) for inferring cell type were calculated for each single unit64. Burst index was defined as the number of spikes occurring within 10 ms of each other divided by the total number of spikes. Trough-to-peak latency was not calculated for positive-spiking units. Thresholds favoring classification as a pyramidal cell were firing rate <10 Hz, trough-to-peak latency >0.5 ms, and burst index >0.2. Single units were classified as pyramidal cells if the majority of criteria favored classification as pyramidal; they were classified as interneurons if the converse was true. In positive-spiking units where trough-to-peak latency could not be calculated, single units with ties in the number of criteria were classified as an unknown cell type. Full width at half maximum (FWHM) was also computed to quantify spike width across both positive- and negative-spiking units.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We thank the members of the Yale Comprehensive Epilepsy Center for their excellent patient care, the patients who participated in this study, Ayman Aljishi for his assistance in early data collection, and Dr. Pue Farooque for her assistance in the internal validation of automatic sleep stage classification. This study was supported by grants from the National Institutes of Health (KL2TR001862, ECD; TR001864, BG; K24MH121571, CP) and the Hypothesis Fund Seed Grant (ECD).
Author contributions
Conceptualization, E.C.D.; Methodology, E.C.D., L.L., M.L., B.G., G.S., and K.J.K.; Software, L.L., M.L., B.G., and G.S.; Validation, L.L., M.L., B.G., and G.S.; Formal analysis, E.C.D., L.L., M.L., B.G.; Investigation, E.C.D., L.L., M.L., B.G., and G.S.; Resources, E.C.D.; Data curation, L.L., M.L., B.G., and G.S.; Writing—original draft, E.C.D. and L.L.; Writing—review and editing, E.C.D., L.L., B.G., L.J.H., C.P., A.P.K., K.N.S.; J.H.K.; Visualization, L.L. and G.S.; Supervision, E.C.D.; Project administration, E.C.D., L.L., M.L.; Funding acquisition E.C.D.
Peer review
Peer review information
Nature Communications thanks Edward Chang, Shuo Wang, Ziv Williams and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The data generated in this study are freely available without stipulation or restrictions and can be obtained by request to the corresponding author via email.
Code availability
The code for this project is available at https://github.com/damisahlab/sleep_2.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-53477-x.
References
- 1.Nir, Y. et al. Regional slow waves and spindles in human sleep. Neuron70, 153–169 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Staresina, B. P., Niediek, J., Borger, V., Surges, R. & Mormann, F. How coupled slow oscillations, spindles and ripples coordinate neuronal processing and communication during human sleep. Nat. Neurosci.26, 1429–1437 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.von Ellenrieder, N. et al. How the human brain sleeps: direct cortical recordings of normal brain activity. Ann. Neurol.87, 289–301 (2020). [DOI] [PubMed] [Google Scholar]
- 4.McConnell, B. V. et al. The aging slow wave: a shifting amalgam of distinct slow wave and spindle coupling subtypes define slow wave sleep across the human lifespan. Sleep44, zsab125 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Léger, D. et al. Slow-wave sleep: from the cell to the clinic. Sleep. Med. Rev.41, 113–132 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Brodt, S., Inostroza, M., Niethard, N. & Born, J. Sleep—a brain-state serving systems memory consolidation. Neuron111, 1050–1075 (2023). [DOI] [PubMed] [Google Scholar]
- 7.Adamantidis, A. R., Gutierrez Herrera, C. & Gent, T. C. Oscillating circuitries in the sleeping brain. Nat. Rev. Neurosci.20, 746–762 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Lemieux, M., Chen, J.-Y., Lonjers, P., Bazhenov, M. & Timofeev, I. The impact of cortical deafferentation on the neocortical slow oscillation. J. Neurosci.34, 5689–5703 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lőrincz, M. L. et al. A distinct class of slow (∼0.2–2 Hz) intrinsically bursting layer 5 pyramidal neurons determines UP/DOWN state dynamics in the neocortex. J. Neurosci.35, 5442–5458 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weber, F. & Dan, Y. Circuit-based interrogation of sleep control. Nature538, 51–59 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Scammell, T. E., Arrigoni, E. & Lipton, J. O. Neural circuitry of wakefulness and sleep. Neuron93, 747–765 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernández-Miranda, J. C., Rhoton, A. L., Kakizawa, Y., Choi, C. & Álvarez-Linera, J. The claustrum and its projection system in the human brain: a microsurgical and tractographic anatomical study—laboratory investigation. J. Neurosurg.108, 764–774 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Bruguier, H. et al. In search of common developmental and evolutionary origin of the claustrum and subplate. J. Comp. Neurol.528, 2956–2977 (2020). [DOI] [PubMed] [Google Scholar]
- 14.Torgerson, C. M., Irimia, A., Goh, S. Y. M. & Van Horn, J. D. The DTI connectivity of the human claustrum. Hum. Brain Mapp.36, 827–838 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crick, F. C. & Koch, C. What is the function of the claustrum? Philos. Trans. R. Soc. B: Biol. Sci.360, 1271–1279 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jackson, J., Karnani, M. M., Zemelman, B. V., Burdakov, D. & Lee, A. K. Inhibitory control of prefrontal cortex by the claustrum. Neuron99, 1029–1039.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McBride, E. G. et al. Influence of claustrum on cortex varies by area, layer, and cell type. Neuron111, 275–290.e5 (2023). [DOI] [PubMed] [Google Scholar]
- 18.Madden, M. B. et al. A role for the claustrum in cognitive control. Trends Cogn. Sci.26, 1133–1152 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Atilgan, H. et al. Human lesions and animal studies link the claustrum to perception, salience, sleep and pain. Brain145, 1610–1623 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Narikiyo, K. et al. The claustrum coordinates cortical slow-wave activity. Nat. Neurosci.23, 741–753 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Timofeev, I. & Chauvette, S. Global control of sleep slow wave activity. Nat. Neurosci.23, 693–695 (2020). [DOI] [PubMed] [Google Scholar]
- 22.Norimoto, H. et al. A claustrum in reptiles and its role in slow-wave sleep. Nature578, 413–418 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Fenk, L. A., Riquelme, J. L. & Laurent, G. Interhemispheric competition during sleep. Nature616, 312–318 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marriott, B. A. et al. Brain-state-dependent constraints on claustrocortical communication and function. Cell Rep.43, 113620 (2024). [DOI] [PubMed] [Google Scholar]
- 25.Duffau, H., Mandonnet, E., Gatignol, P. & Capelle, L. Functional compensation of the claustrum: lessons from low-grade glioma surgery. J. Neurooncol.81, 327–329 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Bickel, S. & Parvizi, J. Electrical stimulation of the human claustrum. Epilepsy Behav.97, 296–303 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Kurada, L., Bayat, A., Joshi, S. & Koubeissi, M. Z. The claustrum in relation to seizures and electrical stimulation. Front. Neuroanat.13, 10.3389/fnana.2019.00008 (2019). [DOI] [PMC free article] [PubMed]
- 28.Vyazovskiy, V. V. et al. Cortical firing and sleep homeostasis. Neuron63, 865–878 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ravagnati, L., Halgren, E., Babb, T. L. & Crandall, P. H. Activity of human hippocampal formation and amygdala neurons during sleep. Sleep2, 161–173 (1979). [DOI] [PubMed] [Google Scholar]
- 30.Watson, B. O., Levenstein, D., Greene, J. P., Gelinas, J. N. & Buzsáki, G. Network homeostasis and state dynamics of neocortical sleep. Neuron90, 839–852 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McInnes, L., Healy, J., Saul, N. & Großberger, L. UMAP: uniform manifold approximation and projection. J. Open Source Softw.3, 861 (2018). [Google Scholar]
- 32.McInnes, L., Healy, J. & Astels, S. hdbscan: hierarchical density based clustering. J. Open Source Softw.2, 205 (2017). [Google Scholar]
- 33.Busch, E. L. et al. Multi-view manifold learning of human brain-state trajectories. Nat. Comput. Sci.3, 240–253 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee, E. K. et al. Non-linear dimensionality reduction on extracellular waveforms reveals cell type diversity in premotor cortex. Elife10, e67490 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Milardi, D. et al. Cortical and subcortical connections of the human claustrum revealed in vivo by constrained spherical deconvolution tractography. Cereb. Cortex25, 406–414 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Sulaman, B. A., Wang, S., Tyan, J. & Eban-Rothschild, A. Neuro-orchestration of sleep and wakefulness. Nat. Neurosci.26, 196–212 (2023). [DOI] [PubMed]
- 37.Zucca, S., Pasquale, V., Lagomarsino de Leon Roig, P., Panzeri, S. & Fellin, T. Thalamic drive of cortical parvalbumin-positive interneurons during down states in anesthetized mice. Curr. Biol.29, 1481–1490.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schreiner, T., Kaufmann, E., Noachtar, S., Mehrkens, J. H. & Staudigl, T. The human thalamus orchestrates neocortical oscillations during NREM sleep. Nat. Commun.13, 5231 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qadir, H. et al. The mouse claustrum synaptically connects cortical network motifs. Cell Rep.41, 111860 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murphy, M. et al. Source modeling sleep slow waves. Proc. Natl Acad. Sci. USA106, 1608–1613 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brancaccio, A., Tabarelli, D., Bigica, M. & Baldauf, D. Cortical source localization of sleep-stage specific oscillatory activity. Sci. Rep.10, 6976 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steriade, M., McCormick, D. A. & Sejnowski, T. J. Thalamocortical oscillations in the sleeping and aroused brain. Science (1979)262, 679–685 (1993). [DOI] [PubMed] [Google Scholar]
- 43.Mak-McCully, R. A. et al. Coordination of cortical and thalamic activity during non-REM sleep in humans. Nat. Commun.8, 15499 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chia, Z., Augustine, G. J. & Silberberg, G. Synaptic connectivity between the cortex and claustrum is organized into functional modules. Curr. Biol.30, 2777–2790.e4 (2020). [DOI] [PubMed] [Google Scholar]
- 45.Ham, G. X. & Augustine, G. J. Topologically organized networks in the claustrum reflect functional modularization. Front. Neuroanat.16, 901807 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jackson, J., Smith, J. B. & Lee, A. K. The anatomy and physiology of claustrum–cortex interactions. Annu Rev. Neurosci.43, 231–247 (2020). [DOI] [PubMed] [Google Scholar]
- 47.Wang, Q. et al. Regional and cell-type-specific afferent and efferent projections of the mouse claustrum. Cell Rep.42, 112118 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Minxha, J., Mamelak, A. N. & Rutishauser, U. Surgical and electrophysiological techniques for single-neuron recordings in human epilepsy patients. Extracell. Rec. Approaches134, 267–293 (2018). [Google Scholar]
- 49.Friston, K. J. et al. Statistical parametric maps in functional imaging: a general linear approach. Hum. Brain Mapp.2, 189–210 (1994). [Google Scholar]
- 50.Davis, T. S. et al. LeGUI: a fast and accurate graphical user interface for automated detection and anatomical localization of intracranial electrodes. Front. Neurosci.15, 769872 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McGrath, H. et al. High-resolution cortical parcellation based on conserved brain landmarks for localization of multimodal data to the nearest centimeter. Sci. Rep.12, 18778 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rorden, C. & Brett, M. Stereotaxic display of brain lesions. Behav. Neurol.12, 191–200 (2000). [DOI] [PubMed] [Google Scholar]
- 53.Magnotti, J. F., Wang, Z. & Beauchamp, M. S. RAVE: comprehensive open-source software for reproducible analysis and visualization of intracranial EEG data. Neuroimage223, 117341 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li, H. et al. Automated claustrum segmentation in human brain MRI using deep learning. Hum. Brain Mapp.42, 5862–5872 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Neudorfer, C. et al. Lead-DBS v3.0: mapping deep brain stimulation effects to local anatomy and global networks. Neuroimage268, 119862 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Avants, B. B. et al. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage54, 2033–2044 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gramfort, A. MEG and EEG data analysis with MNE-Python. Front. Neurosci.7, 267 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vallat, R. & Walker, M. P. An open-source, high-performance tool for automated sleep staging. Elife10, 70092 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schiratti, J. B., Le Douget, J. E., Le Van Quyen, M., Essid, S. & Gramfort, A. An ensemble learning approach to detect epileptic seizures from long intracranial EEG recordings. In ICASSP, IEEE International Conference on Acoustics, Speech and Signal Processing— Proceedings 2018 April 856–860 (Institute of Electrical and Electronics Engineers Inc., 2018).
- 60.Niediek, J., Boström, J., Elger, C. E. & Mormann, F. Reliable analysis of single-unit recordings from the human brain under noisy conditions: tracking neurons over hours. PLoS ONE11, e0166598 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Denker, Mi., Yegenoglu, A. & Grun, S. Collaborative HPC-enabled workflows on the HBP Collaboratory using the Elephant framework. Neuroinformatics 19 (2018) 10.12751/incf.ni2018.0019.
- 62.Louis, S., Gerstein, G. L., Grun, S. & Diesmann, M. Surrogate spike train generation through dithering in operational time. Front. Comput. Neurosci.4, 127 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smith, S. M. & Nichols, T. E. Threshold-free cluster enhancement: addressing problems of smoothing, threshold dependence and localisation in cluster inference. Neuroimage44, 83–98 (2009). [DOI] [PubMed] [Google Scholar]
- 64.Valero, M. et al. Sleep down state-active ID2/Nkx2.1 interneurons in the neocortex. Nat. Neurosci.24, 401–411 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are freely available without stipulation or restrictions and can be obtained by request to the corresponding author via email.
The code for this project is available at https://github.com/damisahlab/sleep_2.