Abstract
UNC-89 is a giant sarcomeric M-line protein required for sarcomere organization and optimal muscle function. UNC-89 contains two protein kinase domains, PK1 and PK2, separated by an elastic region. Here we show that PK2 is a canonical kinase expected to be catalytically active. C. elegans expressing UNC-89 with a lysine to alanine (KtoA) mutation to inactivate PK2 have normally organized sarcomeres and SR, and normal muscle function. PK2 KtoA mutants have fragmented mitochondria, correlated with more mitochondrially-associated DRP-1. PK2 KtoA mutants have increased ATP levels, increased glycolysis and altered levels of electron transport chain complexes. Muscle mitochondria show increased complex I and decreased complex II basal respiration, each of which cannot be uncoupled. This suggests that mutant mitochondria are already uncoupled, possibly resulting from an increased level of the uncoupling protein, UCP-4. Our results suggest signaling from sarcomeres to mitochondria, to help match energy requirements with energy production.
Subject terms: Cytoskeleton, Cell signalling, Mitochondria
UNC-89, a giant protein localized to muscle sarcomeres, contains 2 protein kinase domains, one of which, PK2 is a canonical kinase. A missense mutation expected to inactivate PK2 results in worms with an unexpected phenotype suggesting signaling from sarcomeres to mitochondria.
Introduction
Muscle sarcomeres contain extraordinarily large proteins, >700,000 Da1. Examples include twitchin, UNC-89, and TTN-1 in C. elegans2; projectin, obscurin, and Sallimus in Drosophila3, and titin and obscurin in vertebrates4,5. These proteins contain multiple immunoglobulin (Ig) and fibronectin type III domains (Fn), one or two protein kinase domains, and often highly elastic intrinsically disordered regions.
Loss-of-function mutations in the C. elegans gene unc-89 result in slow-moving worms with disorganized muscle sarcomeres, including a lack of M-lines6–9. unc-89 is a complex gene, which utilizes three promoters and alternative splicing to generate at least 16 polypeptides, ranging in size from 156,000 to 900,000 Da8,10,11 (WormBase). Overlapping sets of isoforms are expressed in body wall muscle, pharyngeal muscle (for pumping in and grinding up bacterial food), egg-laying and intestinal muscles, and several gonadal epithelial cells. The largest of these isoforms, UNC-89-B, consists of 53 Ig domains, two Fn3 domains, a triplet of SH3, DH, and PH domains near their N-termini, and two protein kinase domains (PK1 and PK2) near their C termini. Antibodies localize UNC-89 to the M-line8,10. To learn how UNC-89 is localized and performs its functions, we have screened a yeast two-hybrid library with segments of the largest UNC-89 isoform and identified at least 7 interacting partners with 6 out of 7 having orthologs or homologs in humans2. Interactions were verified by a combination of in vitro binding assays using purified recombinant proteins, co-immunolocalization, and analysis of muscle mutant phenotypes. Two types of protein phosphatases interact with the protein kinase region of UNC-89. Segments containing either of the kinase domains, Fn1-Ig52-PK1 and Ig53-Fn2-PK2, interact with SCPL-112, a CTD-type protein phosphatase. The segment, Ig53-Fn2, which lies just the N-terminal of PK2, interacts with PPTR-2, a regulatory subunit of protein phosphatase 2 A (PP2A)13. Although we do not know the substrates for UNC-89 kinases, or SCPL-1 and PP2A phosphatases, it could be envisioned that some of the substrates might be shared.
The Drosophila genome contains a single unc-89 gene called obsc. Mutant and RNAi knockdown flies are unable to fly and their indirect flight muscles have abnormal sarcomeric structures with missing M-lines and misplaced or absent H-zones resulting in abnormally long or short thin filaments14. Thus, the role of UNC-89 in sarcomere assembly is conserved in worms and flies.
Mammalian striated muscle expresses three UNC-89-like proteins from separate genes: obscurin (OBSC)15,16, obscurin-like 1 (obsl1)17, and striated muscle preferentially expressed gene (SPEG)18. The most homologous protein to UNC-89 is obscurin, which also exists as many isoforms, including giant isoforms (obscurin A), and shorter isoforms with 2 protein kinase domains5. While obscurin A contains all the same domains as UNC-89, the SH3, DH, and PH domains are located near the C-terminus rather than near the N-terminus as they are in UNC-89. Although UNC-89 is located only at the M-line, various obscurin isoforms are located at either the M-line or the Z-disk16,19.
Polymorphisms or mutations in the human OBSC gene are implicated, but not yet proven, to cause various types of cardiomyopathies, systolic heart failure, and distal skeletal myopathy20. In addition to serving as a platform for the assembly of M-line proteins, obscurin links myofibrils to the sarcoplasmic reticulum (SR). The C-terminus of obscurin interacts with an ankyrin protein localized to the SR21,22. The obscurin KO mouse has normal myofibril organization but disorganized SR23. This myofibril to SR linkage is conserved in nematode UNC-8924.
Obsl1 is an ~200 kDa protein consisting solely of 20 tandem Ig domains, and in heart muscle is localized to M-lines, Z-disks, and intercalated disks17. Several portions interact with titin and myomesin at the M-line25. However, obsl1 is ubiquitously expressed, the global KO is embryonic lethal, and mutations in the human Obsl1 gene result in a growth defect (3M-growth syndrome)26.
Substrates for UNC-89 PK2 have not yet been identified and an understanding of the in vivo substrates for the kinase domains of obscurin is in its infancy. However, sequence analysis suggests that human obscurin PK1 and PK2 are both likely to be active kinases27, and each has been shown to undergo autophosphorylation in vitro28. Obscurin PK2 can phosphorylate N-cadherin in vitro28. Obscurin PK1 has been shown to autophosphorylate several serines in the interkinase region immediately C-terminal of PK129. Deletion of both OBSC and Obsl1 in the skeletal muscle of mice results in changes in proteins implicated in metabolic pathways and the mitochondrial electron transport chain (ETC)30. Specifically, nearly all mitochondrial electron transport chain proteins were downregulated. Analysis of the proteome data suggested downregulation of ETC proteins was more associated with loss of obscurin than with Obsl1.
PK1 from Drosophila UNC-89 has been shown to be an inactive pseudokinase, with inactivity features being strictly conserved across arthropods31. Drosophila UNC-89 PK1 has also been shown to interact with PK2, suggesting that both kinases form a functional dual kinase hub, where the inactive PK1 likely serves as regulator of PK231.
So far, the experiments on obscurin have utilized a knockout of the entire protein, not specifically eliminating the activity of one or more kinase domains. Taking advantage of the genetic manipulability of C. elegans, we used CRISPR/Cas9 to create a worm that expresses UNC-89 having the established kinase inactivating mutation, lysine-to-alanine, in the ATP pocket of the PK2 catalytic domain. These mutants have normal sarcomere structure, normal force generation, even better than normal muscle function, and yet display fragmented mitochondria that have increased basal respiration that cannot be uncoupled further. We gained insight into molecules and processes that lead to mitochondrial fragmentation, uncoupling, and higher ATP. Our results suggest signaling from sarcomere to mitochondria which may function to match energy needs to energy production, and that PK2 of UNC-89 (obscurin) is key to this communication.
Results
Bioinformatic analysis indicates that PK2 is an active kinase and that a mutation expected to abolish activity is not expected to result in instability
A sequence analysis of PK2 from C. elegans UNC-89 using BLAST confirmed that it belongs to the DMT class of CAMK kinases (Ca2+/Calmodulin-dependent kinases). The DMT class takes its name from their representative Death-associated-protein kinases (DAPKs), Myosin light-chain-kinases (MLCKs), and Triple functional domain protein-related kinases (TRIO) and contains, in addition, various muscle kinases including human titin kinase, nematode twitchin kinase (TwcK) and mammalian obscurin and SPEG kinases32. The BLAST search revealed that PK2 shares the highest sequence identity levels with DAPKs, TwcK, and MLCK (Supplementary Fig. 1).
DAPK, TwcK, and MLCK kinases are proven to be catalytically active and their phosphotransfer has been characterized33–35. Referencing the UNC-89-PK2 sequence to these other kinases reveals a high conservation of catalytically relevant motifs in PK2. Specifically, canonical kinases are characterized by a glycine-rich GxGxxG loop motif involved in ATP-binding, an AAK motif in β-strand β3 containing an essential lysine (that optimally positions the non-transferable α- and β-phosphate groups of ATP), a conserved glutamate residue in helix C that forms a salt bridge with the essential lysine during the catalytically committed state of the kinase, a DFG signature motif (where the aspartate residue chelates the magnesium ion required for catalysis), a catalytic aspartate and a conserved asparagine residue (also involved in cation coordination), both in the catalytic loop motif36,37. These groups are conserved in UNC-89-PK2 (Fig. 1c and Supplementary Fig. 1). A minor variation is present only in the glycine-rich motif, which in UNC-89-PK2 adopts the sequence FxGxxS. In fact, this motif is commonly diversified across obscurin and UNC-89 PK2 kinases across phyla (Supplementary Fig. 2). Degeneration of the first and last positions of this motif is common among kinases and is known not to compromise ATP-binding or phosphotransfer. For example, the active kinases CDK1138, TP53RK39, and mouse obscurin PK228 contain ExGxxG, KxGxxA, and RxGxxS motif variants, respectively.
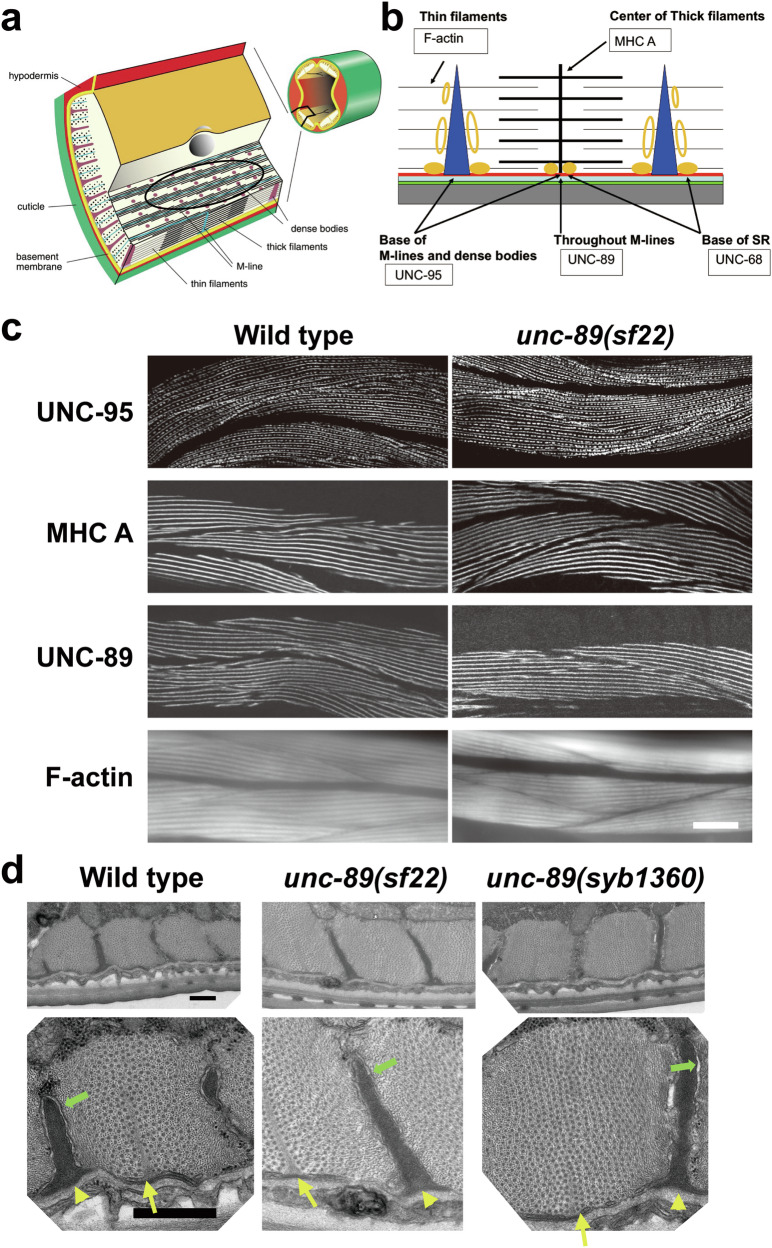
Fig. 1. Three-dimensional model of UNC-89 PK2, and CRISPR/Cas9 generation of a nematode expressing PK2 with an inactivating KtoA mutation.
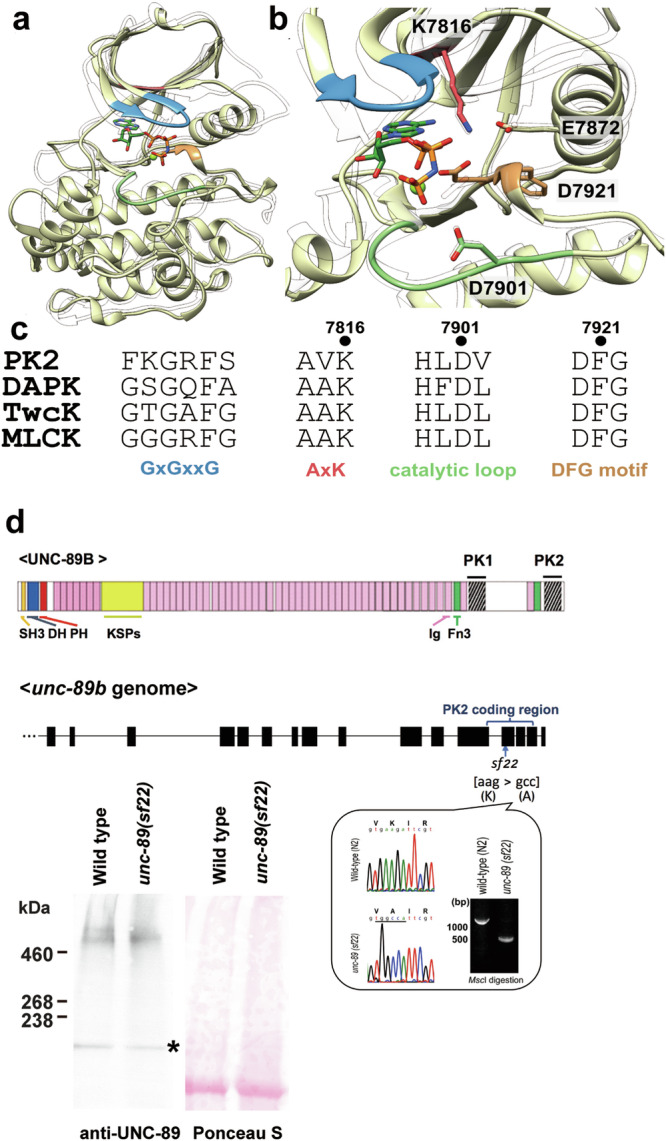
a Cartoon representation (yellow) of the 3D model generated by AlphaFold68. Catalytically relevant segments are color-coded as in (c). As a transparent ribbon trace, the crystal structure of the active, canonical kinase DAPK1 in complex with the ATP-analog AMPPNP and Mg2+ and in the catalytically committed state is shown (PDB code 3F5U). b Close-up of the active site of UNC-89-PK2. Color code is as in (a). Catalytically relevant residues described in the main text are displayed and annotated. c Alignment of catalytically relevant sequence motifs in UNC-89-PK2 with those of representative DMT kinases: UNC-89-PK2 (PK2), human death-associated-protein kinase (DAPK1), twitchin kinase from C. elegans (TwcK), human myosin light-chain kinase (MLCK). d On top is a schematic representation of domains within the largest isoform of UNC-89. Ig domains, purple; Fn3 domains, green; KSPs, a region with tandem repeats and 44 KSPs; other domains as indicated. Note the two protein kinase domains, PK1 and PK2, at the C-terminus. Below is representation of part of the exon/intron organization of part of the unc-89 gene, with the location of the PK2 coding region. Bottom right: confirmation of the CRISPR/Cas9 generated aag to gcc sequence change in unc-89(sf22) which specifies a K7816A mutation in PK2. Bottom, left: western blot comparing the levels of UNC-89 giant isoforms in wild type vs. unc-89(sf22). The giant isoforms of UNC-89 are ~800 kDa. The asterisk indicates a protein of nonspecific reactivity detected by the anti-UNC-89 antibodies46.
During the course of this study, the recombinant production of UNC-89-PK2 samples compatible with viable phosphotransfer experimentation could not be established. Thus, in order to further investigate whether a phosphor-transfer productive conformation is attainable by UNC-89-PK2, we calculated its 3D model using AlphaFold and tested in silico its capability of accommodating ATP/Mg2+ (Fig. 1a, b and Supplementary Fig. 3a). In kinases, the mutual arrangement of N- and C-terminal fold lobes, the position of the ATP-binding glycine-rich loop, the orientation of the catalytic helix αC hosting the conserved glutamate residue that coordinates the magnesium counterion, the conformation of the activation loop that governs access to the catalytic aspartate at the peptide binding site, the conformation of the conserved DFG-motif, and the assembly state of two dynamic structural networks of hydrophobic residues termed spines, is dynamically altered to achieve catalytic readiness36. Nucleotide binding to the kinase normally induces the formation of the catalytically committed state. Distinctly, DMT kinases exist in a catalytically-primed state in the absence of ATP-binding, which does not induce a further conformational rearrangement31,32. In agreement with this existent knowledge, the 3D model of UNC-89-PK2 displayed a regular kinase fold with features typical of the catalytically-primed conformational state of a kinase (Fig. 1a, b). At the residue level, the ATP-binding lysine K7816 is modeled forming a typical salt bridge to the conserved glutamic acid E7827 in the catalytic helix αC. The canonical DFG-motif (residues 7921-7923) is modeled in the “in”-conformation and the activation loop is in an open state. Both the regulatory (R)-spine (residues L7831, L7842, H7899 and F7922) and the catalytic (C-)spine (I7799, A7814, V7863, I7907, M7908, F7909, V7965 and L7969) are assembled. The modeled structure of UNC-89-PK2 closely resembles that of human DAPK1 and was also compatible with its ATP/Mg2+ binding mode (Fig. 1a, b and Supplementary Fig. 3b). Thus, we conclude that UNC-89-PK2 shares the sequence and conformational features of established canonical kinases.
Mutation of the K in the AAK motif to A or several other residues has been shown to inactivate most known kinases40. To assess whether the introduction of a KtoA mutation in PK2 could render the kinase unstable, we tested the effects of this mutation on the 3D model of PK2 using FoldX v441. This yielded a calculated comparative protein unfolding energy, ΔΔG, of −0.15 kcal/mol and indicated that no significant stability change arises from the introduction of the mutation.
To summarize, UNC-89-PK2 is modeled by Alphafold in a catalytically-primed conformational state compatible with kinase activity, the active site comprises the essential functional groups necessary for phosphor-transfer and the introduced mutation is not expected to cause structural instability. Taken together, bioinformatics analysis indicates that UNC-89-PK2 is a canonical, active kinase and that the observed nematode phenotype in this work results from hindered phosphor-transfer by PK2 resulting from the inactivating KtoA mutation.
Nematodes expressing UNC-89 with PK2 KtoA have normally organized sarcomeres, SR, and microtubules, and move faster than wild-type nematodes
We used CRISPR/Cas9 to create a worm (Fig. 1d), unc-89(sf22), in which PK2 has a K7816A mutation. The strain was outcrossed 4 times to wild type to remove most of the potential off-target mutations induced by CRISPR. By western blot, this KtoA mutation does not detectably affect the expression or stability of full-length UNC-89 isoforms (Fig. 1d) using an established antibody10. Surprisingly, unlike all other unc-89 mutants, unc-89(sf22) worms have normal sarcomere structure as assessed by immunostaining using antibodies to various sarcomeric proteins, including UNC-95 for the base of M-lines and dense bodies (Z-disks), MHC A myosin heavy chain for A-bands, UNC-89 for M-lines, and F-actin for I-bands (Fig. 2a–c). Detection and localization of the PK2 KtoA mutant UNC-89 to M-lines, the normal localization of wild-type UNC-89 in sarcomeres8,10, is consistent with the western blot showing no major loss of the mutant UNC-89 protein. To examine the myofilament lattice at higher resolution we used EM. As shown in Fig. 2d, the ultrastructure of the sarcomeres in unc-89(sf22) and in unc-89(syb1360), a second, independently isolated PK2 KtoA mutant allele (see below), appear as in wild type; there are normally organized A- and I-bands, normal numbers of thick and thin filaments, and normally organized dense bodies, M-lines and membranous sacs of the SR.
Fig. 2. Nematodes expressing UNC-89 with the KtoA mutation in PK2 have normal muscle structure.
a The drawing on the right shows a cross-section through an adult worm indicating that the body wall muscle is organized into four quadrants. Each quadrant is composed of interlocking pairs of mononuclear fusiform-shaped cells (23 or 24 per quadrant). The enlargement emphasizes that the myofilament lattice is restricted to one side of the cell. The drawing depicts several planes of section; one plane emphasizes that this is striated muscle with typical A-bands containing thick filaments organized around M-lines, and overlapping thin filaments attached to dense bodies (Z-disk analogs). The black oval indicates the plane viewed when an animal, lying on a glass slide, is examined by light microscopy as shown in part (c). This drawing is a slight modification of a published drawing2, under terms of a Creative Commons Attribution License. b Idealized sarcomere and SR with the location of proteins localized in part (c). MHC A: myosin heavy chain A. Red: muscle cell membrane; blue-green: ECM; green: hypodermis; and grey: cuticle. Orange ovals: location of SR determined by electron microscopy; solid orange ovals: location of UNC-68 (Ryanodine receptor). c Wild type (left column), and unc-89(sf22) (right column) animals immunostained with various antibodies to proteins indicated in part (b). Phalloidin was used to image F-actin in thin filaments. In each panel, several spindle-shaped body wall muscle cells are shown. Note that there is no difference in the pattern of localization of these sarcomeric markers between wild type and unc-89(sf22). Scale bars, 10 μm. d Transmission EM of body wall muscle from wild type, unc-89(sf22) and a second independently-generated PK2 KtoA mutant, unc-89(syb1360). Bottom row displays the enlargement of one portion of the image above it. Yellow arrows, M-lines; yellow arrowheads, dense bodies, green arrows, SR. There is no discernible difference between the wild type and the two mutants in terms of numbers and organization of thick and thin filaments, M-lines, dense bodies, and SR. Scale bars, 0.5 μm.
Because deficiency of mouse obscurin23 and worm UNC-8924 result in disorganized SR, we examined SR organization using the marker UNC-68 (Ryanodine receptor). However, HA-tagged UNC-68 endogenously expressed from a CRISPR/Cas9 strain (Supplementary Fig. 4a), or myc-tagged UNC-68 overexpressed from a transgene (Supplementary Fig. 4b) show normal localization in unc-89(sf22).
Obscurin is also required for normal microtubule organization42. However, we found no difference in microtubule organization comparing wild type vs. unc-89(sf22), using a transgenic line expressing TBA-1 (α-tubulin) tagged with HA (Supplementary Fig. 5).
unc-89 is also expressed in the single sarcomere muscle cells of the pharynx. UNC-89 is normally localized to the middle of the A-bands of pharyngeal sarcomeres8,10. All previously characterized unc-89 mutants, except for unc-89(tm752), have abnormally organized filaments as observed by polarized light or immunostaining6,9,11, with characteristic arc-like thick filaments at the periphery of the terminal bulb. As shown in Supplementary Fig. 6, both unc-89(sf22) and unc-89(syb1360) display normal pharyngeal muscle as assessed by staining with anti-UNC-89 and the pharyngeal muscle-specific MHC C.
Nearly all previously characterized unc-89 mutants have reduced whole animal locomotion and generate reduced maximal force9, most likely because sarcomere disorganization results in reduced muscle activity. The one known exception is unc-89(ad539), but this mutant only affects the organization of the pharyngeal muscle. Two independently derived UNC-89 KtoA mutants, unc-89(sf22) and unc-89(syb1360), crawl faster than wild-type worms on an agar surface (Fig. 3a), and generate the same amount of maximal force43 as wild type (Fig. 3b).
Fig. 3. UNC-89 PK2 KtoA mutants move faster than wild type but exert the same amount of force as wild type.
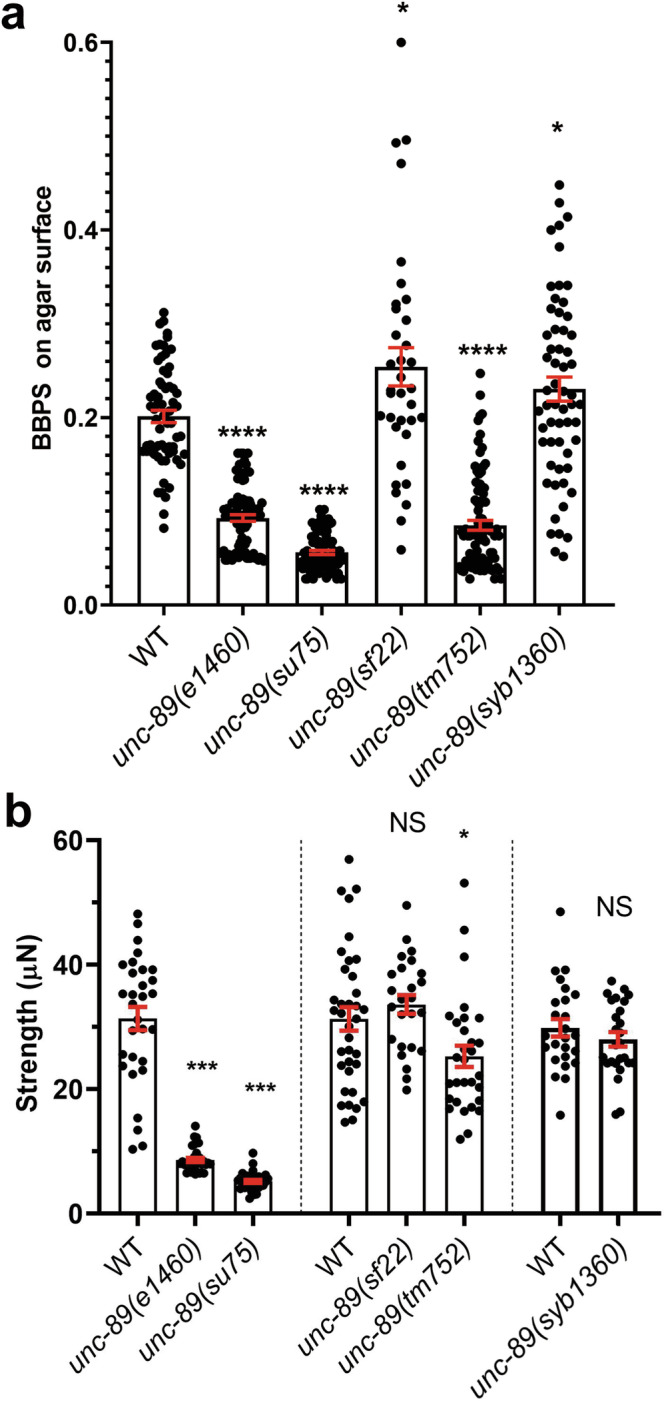
a Crawling assays on wild type (WT), several unc-89 loss of function alleles (e1460, su75, tm752), and two independently derived PK2 KtoA mutants (sf22 and syb1360). Body bends per second (BBPS) are quantified for individual animals of each strain. In the graphs, each dot represents the result from an independently selected animal. Welch’s t-test was used for significance. Error bars (red) indicate SEMs, *p ≤ 0.05, ****p ≤ 0.0001. b NemaFlex force measurements of individual worms of the same strains indicated in part (a). Statistical analysis was performed using the Wilcoxon rank-sum test. N = 25–36; error bars (red) indicate SEMs, *p ≤ 0.05, ***p ≤ 0.001.
Nematodes expressing UNC-89 PK2 KtoA, or an unc-89 mutant lacking expression of all kinase-containing isoforms, have fragmented mitochondria
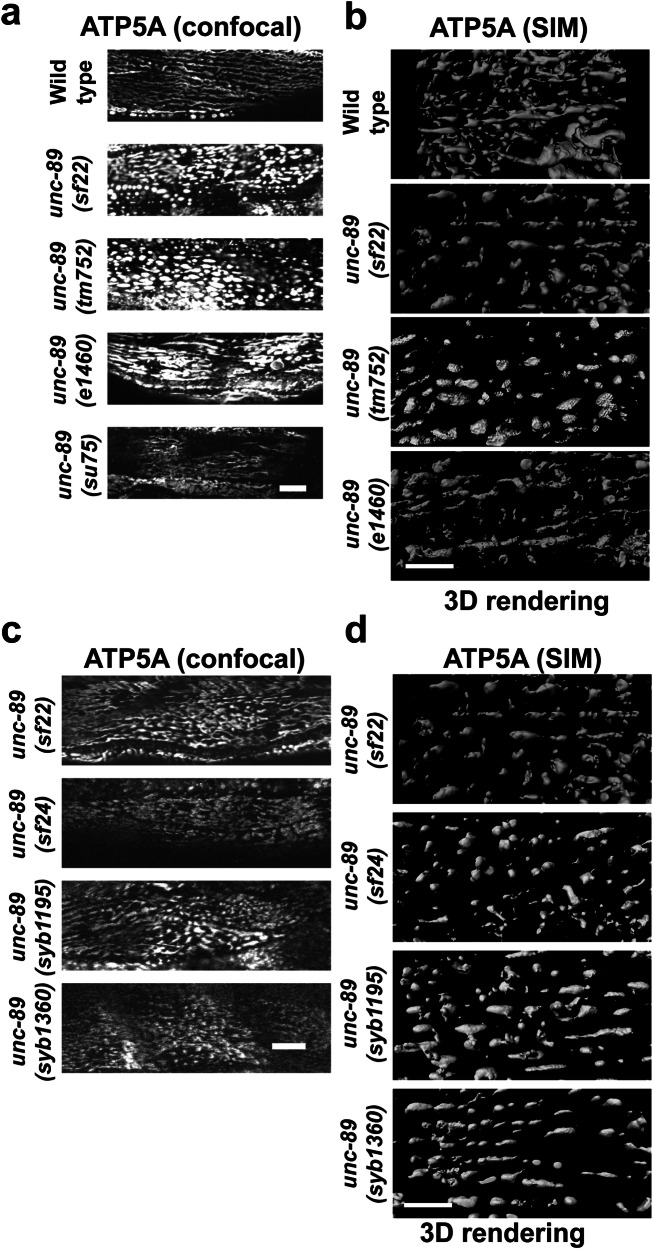
Blondelle et al.30 created a mouse doubly knocked out for both obscurin and obsl1 and demonstrated that by proteome analysis, these mice have changes in metabolic pathways, including decreased levels of nearly all mitochondrial electron transport chain complexes. This report prompted us to examine the morphology and function of mitochondria in UNC-89 KtoA mutants. Using an antibody to the mitochondrial inner membrane protein ATP5A, and identifying the body wall muscle cells using antibodies to UNC-95, we found that mitochondria in wild-type muscle are organized into long wavy chains, parallel to the sarcomeric A- and I-bands (Fig. 4a), as reported by others44. In comparison, mitochondria in unc-89(sf22) appear fragmented. This can be seen by confocal (Fig. 4a) and structured illumination microscopy (SIM) (Fig. 4b). In contrast, nematodes expressing the related giant protein twitchin with a catalytically dead protein kinase domain, unc-22(sf21), or twitchin with a missense mutation in Ig7, unc-22(e105)45 have normally organized mitochondria (Supplementary Fig. 7). This suggests that effects on mitochondrial morphology are specific for UNC-89 PK2 and not a function shared by protein kinases of the other giant muscle proteins in the worm. We find similar abnormal fragmented morphology of the mitochondria in unc-89(tm752) animals, which lack expression of all kinase-containing isoforms, but do express other isoforms8,11(Fig. 4a). In contrast, a more normal (wild type) morphology of mitochondria is seen in two other unc-89 mutants: unc-89(e1460)—a stop codon in an alternatively spliced exon for Ig20-Ig2624,46, which expresses kinase-containing isoforms; and in unc-89(su75)—a frameshift mutation in the exon for Ig3946, that lacks expression of all giant isoforms but expresses the small kinase-containing isoforms (Fig. 4a, b). Our impressions of mitochondrial morphology are supported by a quantitative analysis of the SIM images (Supplementary Fig. 8): binning the mitochondria for length indicates that unc-89(sf22) and unc-89(tm752) have no mitochondria >10 μm, and fewer mitochondria between 5 and 10 μm as compared to either wild type or unc-89(e1460). Therefore, either inactivation of PK2 kinase or lack of expression of PK2-containing isoforms results in fragmented mitochondria.
Fig. 4. Body wall muscle cells of UNC-89 PK2 KtoA mutants have fragmented mitochondria.
a Confocal images of body wall muscle stained with antibodies to ATP5A to reveal the morphology of mitochondria. Note that wild type has mitochondria organized as long wavy lines running parallel to the A- and I-bands, described as “tubular”. In contrast, unc-89(sf22) which carries the PK2 KtoA mutation, and unc-89(tm752) which lacks expression of all kinase-containing isoforms have mitochondria that appear as dots or can be described as fragmented. By comparison, unc-89(e1460) which maintains expression of all kinase-containing isoforms, and unc-89(su75) which lacks expression of all giant isoforms but still expresses small kinase-containing isoforms have mitochondria that appear more tubular or more like wild type. b 3D rendering of SIM images of muscle mitochondria of the indicated strains. Note the presence of linear or tubular mitochondria in wild type and unc-89(e1460) and fragmented mitochondria in unc-89(sf22) and unc-89(tm752). c, d Three additional independently isolated PK2 KtoA mutants (sf24, syb1195, and syb1360) show fragmented mitochondria. Scale bars, 10 μm in (a and c); 5 μm in (b and d).
Because CRISPR/Cas9 can result in off-target effects, we generated 3 more CRISPR alleles (sf24, syb1195, and syb1360) resulting in the same KtoA amino acid change. sf22 and sf24 are independent alleles generated from the same sgRNA, and syb1195 and syb1360 are independent alleles generated using a different sgRNA. The mutant alleles were outcrossed 3X to wild type. The new mutants displayed the same mitochondrial fragmentation as unc-89(sf22) worms (Fig. 4c, d), suggesting that the KtoA mutation in UNC-89 PK2 adversely affects mitochondrial morphology.
In PK2 KtoA mutants, mitochondrial fragmentation results from greater association of DRP-1 with mitochondria
In C. elegans, mitochondrial fusion and fission are primarily controlled by 3 proteins44 (Fig. 5a): fusion is mediated by FZO-1 (ortholog of mitofusin 1 (Mfn1) and mitofusin 2 (Mfn2)) on the outer membranes of opposing mitochondria, and by the inner membrane protein EAT-3 (OPA1); fission is mediated by DRP-1 (Drp1), which is mostly cytosolic but can be recruited to mitochondria where it forms a large spiral that constricts mitochondrial membranes. We find that the drp-1(tm1108) mutant produces large clumped mitochondria as reported previously47(Fig. 5b), and that an unc-89(sf22); drp-1(tm1108) double mutant shows the Drp-1 phenotype, i.e., drp-1 is epistatic to unc-89(sf22). The epistasis of drp-1 over sf22 indicates that the fragmentation of mitochondria in unc-89(sf22) requires DRP-1 protein. One possibility is that UNC-89 PK2 may be an inhibitor of DRP-1 activity, and DRP-1 may be more active when PK2 has no catalytic activity. This further suggests that DRP-1 protein could be a target of the protein kinase activity of UNC-89 PK2, directly or indirectly through a kinase cascade.
Fig. 5. drp-1 is epistatic to unc-89(sf22).
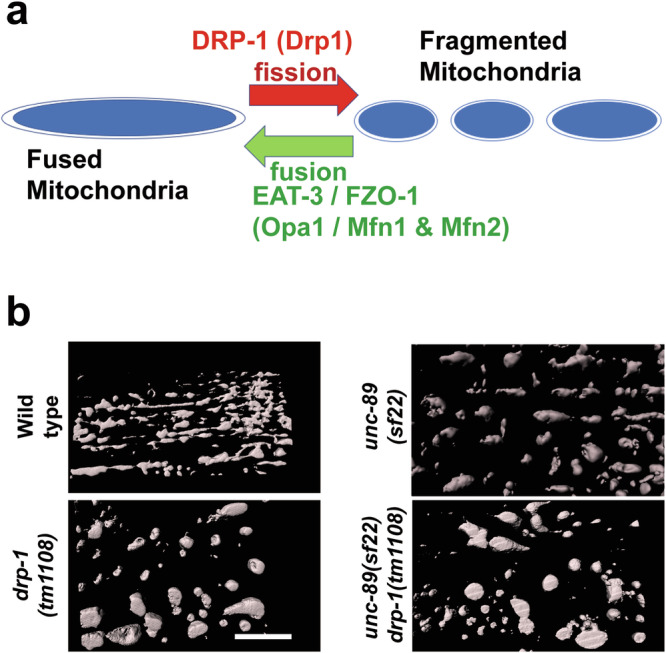
a Mitochondria are constantly undergoing fusion and fission, through the actions of several conserved GTPases of the dynamin family. The names of the nematode proteins are indicated with their mammalian orthologs shown in parentheses. b 3D rendering of SIM images of muscle mitochondria stained with anti-ATP5A. The drp-1 null mutant shows abnormally clumped mitochondria, whereas unc-89(sf22) shows fragmented (smaller) mitochondria. The double mutant, unc-89(sf22); drp-1(tm1108) shows the Drp-1 single mutant phenotype. Scale bar, 5 μm.
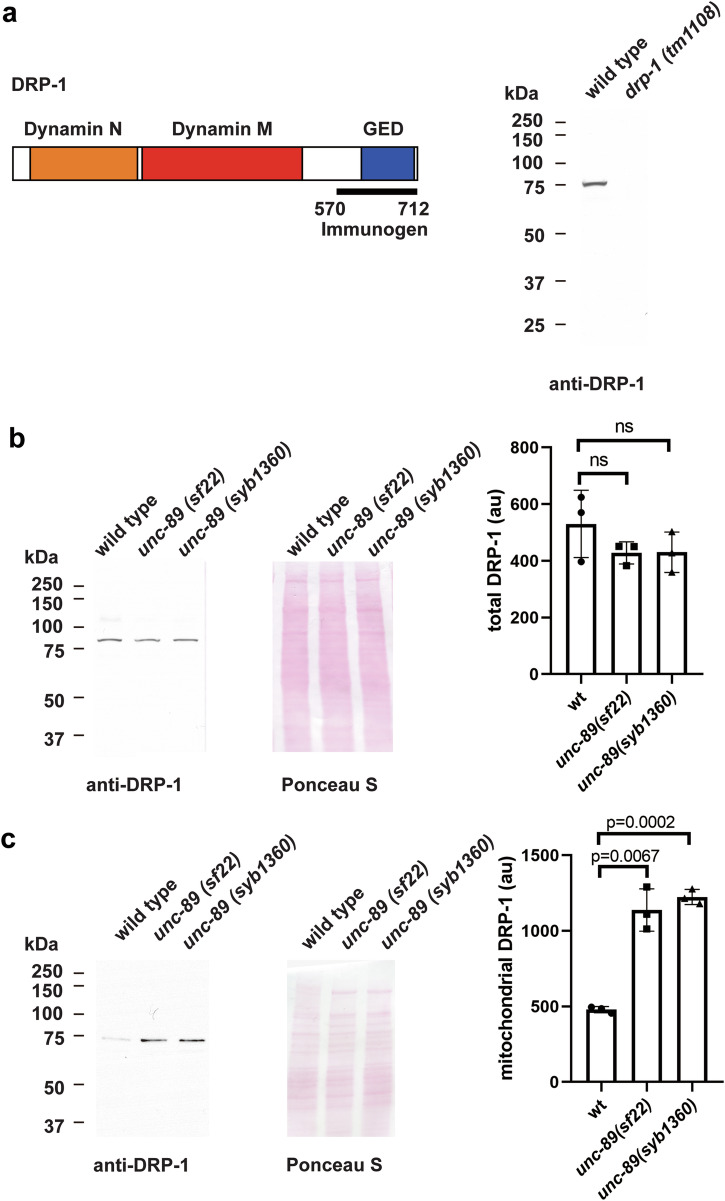
In mammals, Drp-1 shuttles between the cytosol and mitochondria, and upon association with mitochondria, fission is promoted48. We tested the hypothesis that part of the mechanism by which unc-89 KtoA mutants have more mitochondrial fragmentation is that more DRP-1 is associated with their mitochondria. We developed an antibody to DRP-1 and validated the antibody by showing that it detects a protein of expected size from wild type (~80 kDa), but no protein is detectable from a drp-1 deletion mutant (Fig. 6a). We observed no differences in the total levels of DRP-1 between wild type and either of two unc-89 PK2 KtoA mutants (unc-89(sf22) or unc-89(syb1360)) (Fig. 6b). However, we found an approximately two-fold higher level of DRP-1 associated with purified mitochondria from either of the PK2 mutants compared to wild type (Fig. 6c). Therefore, the greater association of DRP-1 with mitochondria at least partially explains how the mitochondria in PK2 KtoA mutants are more fragmented.
Fig. 6. More DRP-1 is associated with UNC-89 PK2 KtoA mutant mitochondria as compared to wild-type mitochondria.
a Left: Schematic representation of domains in DRP-1 as predicted by PFAM, and location of the immunogen used to raise antibodies to DRP-1. Right: Western blot showing the reaction of affinity-purified anti-DRP-1 to a protein of expected size for DRP-1 from wild type, and absence of detectable DRP-1 protein from the drp-1 null mutant. b Left: A representative western blot showing the reaction of anti-DRP-1 against total protein extracts from wild type, unc-89(sf22), and unc-89(syb1360). Right: quantitation of results indicating no significant difference in total DRP-1 levels in wild type vs. the two unc-89 mutants. au: arbitrary units; ns: not significant. Statistical significance was assessed using an unpaired t-test with Welch’s correction. c Left: A representative western blot showing the reaction of anti-DRP-1 against extracts of mitochondrial preparations from wild type, unc-89(sf22), and unc-89(syb1360). Right: quantitation of results indicating that there is more DRP-1 associated with mitochondria in the two mutants vs. wild type. au: arbitrary units. Statistical significance was assessed using an unpaired t-test with Welch’s correction.
Increased levels of ATP in PK2 KtoA mutants
Because mitochondrial fragmentation is usually associated with decreased respiration and ATP production49,50, we compared levels of ATP in wild type vs. each of the 4 PK2 KtoA mutant alleles. Using a luciferase assay on whole worms, we were surprised to find that each mutant shows at least a 40% increase of ATP compared with wild type (Fig. 7). However, unc-89 is expressed only in the body wall, pharyngeal, vulval and intestinal muscle, and a few myoepithelial cells of the oviduct. Thus, we utilized a method to assess ATP levels primarily in the body wall muscle, the major site of UNC-89 expression. The ATP:ADP ratio, is a sensitive indicator of cellular energy status51. The fluorescent biosensor PercevalHR can measure this ratio in living cells52,53. The ATP:ADP ratio is proportional to fluorescence at 529 nm at two excitation wavelengths (405 and 488 nm)54. The myo-3 gene, which encodes myosin heavy chain A (MHC A), is primarily expressed in body wall muscle but is also expressed in a few single sarcomere muscle cells and in a few myoepithelial cells of the gonadal sheath. We created transgenic worms that express PercevalHR primarily in body wall muscle using the myo-3 promoter. We first validated the technique by showing: (i) the ATP:ADP ratio is decreased in animals treated with antimycin A (inhibits complex III) (Supplementary Fig. 9a, c), and (ii) the ATP/ADP ratio is decreased in drp-1(tm1108) (Supplementary Fig. 9b, d), consistent with a report that drp-1 mutants have decreased ATP levels55. The ATP/ADP ratio is increased in unc-89(sf22) (Supplementary Fig. 9b, d), consistent with the increased levels of ATP in the whole animals (Fig. 7). Although most of these results do not reach the level of statistical significance, they do show trends consistent with our tentative conclusions.
Fig. 7. PK2 KtoA mutants have elevated ATP levels.
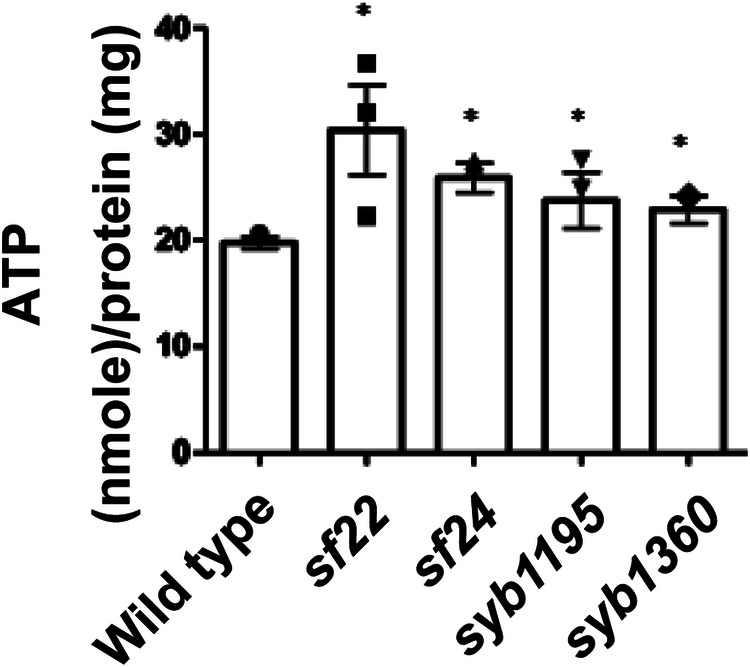
Results of luciferase assays on extracts of whole worms from the indicated strains. For each strain, n = 3, and *: p < 0.05 by Dunnett’s test.
Complex I respiration is increased, complex II respiration is decreased, and each cannot be uncoupled in unc-89 PK2 KtoA mutants
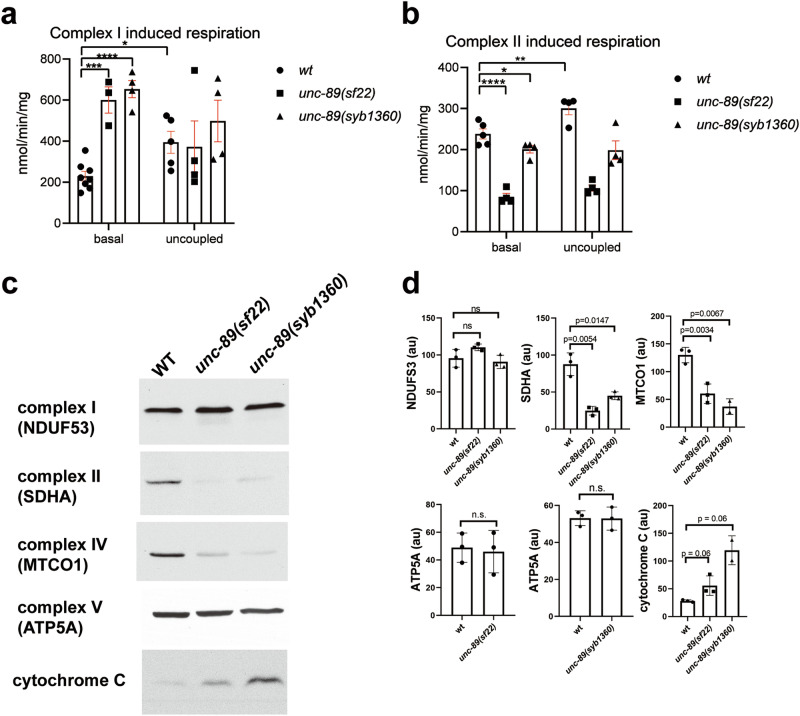
We next tested the hypothesis that the increase in ATP levels in PK2 KtoA mutants is due to enhanced mitochondrial function. We utilized a method to isolate mitochondria from any cell type in C. elegans56 to isolate mitochondria primarily from body wall muscle cells. We used a strain in which the fusion protein tomm-20-mKate2-HA was expressed from the myo-3 promoter, total mitochondria were purified, and then mostly muscle-specific mitochondria were pulled out using anti-HA-magnetic beads. Mitochondrial respiration in muscle mitochondria was assayed using an oxygen electrode. We investigated both complex I and complex II-based respiration as electrons can take two paths through the electron transport chain: either I to III to IV or II to III to IV. When assessed for complex I induced respiration (addition of substrates malate and glutamate), muscle mitochondria from unc-89(sf22) and unc-89(syb1360) showed significantly higher rates of basal respiration, and the respiration could not be uncoupled by adding FCCP (Fig. 8a). When assessed for complex II-induced respiration (addition of the substrate succinate), the UNC-89 KtoA mutants showed lower rates of basal respiration, which could also not be uncoupled (Fig. 8b).
Fig. 8. PK2 KtoA mutants display altered mitochondrial respiration and altered levels of some electron transport chain complexes.
a, b Muscle mitochondria from PK2 KtoA mutants have increased complex I and decreased complex II basal respiration and each cannot be uncoupled. a Complex I (glutamate and malate) stimulated basal and uncoupled mitochondrial oxygen consumption rates measured in isolated muscle mitochondria from wild type (wt), unc-89(sf22), and unc-89 (syb1360) mutant strains (n = 3–8 per strain). b Complex II (succinate) stimulated basal and uncoupled mitochondrial oxygen consumption rates measured in isolated muscle mitochondria from wild type (wt), unc-89(sf22), and unc-89 (syb1360) mutant strains (n = 4–5 per strain). Means and standard errors of the mean are represented. Significance was tested by one-way ANOVA followed by Tukey’s post hoc test. *p ≤ 0.05, **p ≤ 0.005, ***p ≤ 0.0005, ****p ≤ 0.00005. c, d PK2 KtoA mutants have altered levels of electron transport chain complexes. c Representative western blots reacted against antibodies to the indicated components of complexes I, II, IV, V, and cytochrome C. d Quantitation of these results with n = 3. au: arbitrary units. Statistical significance was assessed using an unpaired t-test with Welch’s correction. Note that as compared to wild type, complexes I and V are normal, but complexes II and IV are reduced, and cytochrome C is elevated in the PK2 KtoA mutants.
To gain insight into why complex I respiration is increased and complex II respiration is decreased, we assessed the protein levels of one component of each electron chain complex by western blots. Unfortunately, we could not identify a commercially available antibody that reacted against a complex III component, so complex III was not assessed. As shown in Fig. 8c, d, the levels of complexes I and V are normal, complexes II and IV are decreased, and cytochrome C (which shunts electrons from III to IV) is increased. These results only partially explain why complex II-based respiration is decreased.
Increased glycolysis in PK2 KtoA mutants
Although the increase in complex I-based respiration might partially explain the increased level of ATP in the PK2 KtoA mutants, we explored the additional possibility that glycolysis was increased. There is precedence for a compensatory increase in glycolysis when mitochondria do not perform well57. We measured the activity of the rate-limiting enzyme in glycolysis, 6-phosphofructose kinase. The activity of this enzyme is significantly increased in all 4 PK2 KtoA mutants (Fig. 9). Thus, increased glycolysis at least partially explains why there is an increase in total ATP levels in PK2 KtoA mutants, however, the mechanism of this increase remains elusive.
Fig. 9. PK2 KtoA mutants display increased activity of the rate-limiting enzyme of glycolysis, 6-phosphofructose kinase.
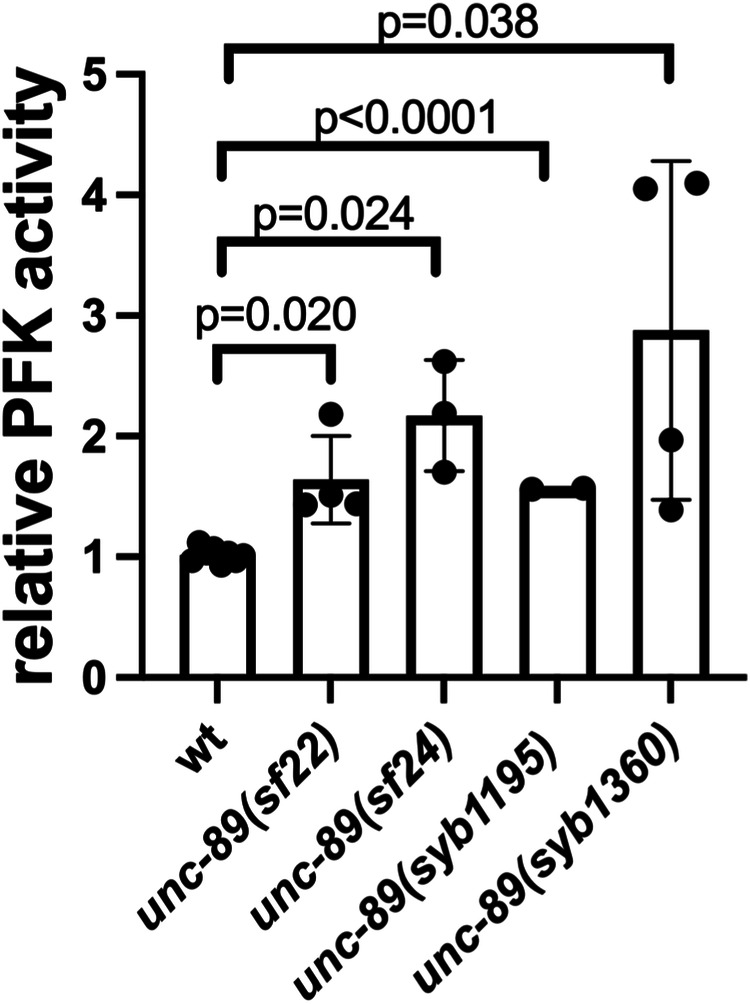
Statistical significance was assessed using an unpaired t-test with Welch’s correction.
Uncoupling might result from increased levels of the uncoupling protein UCP-4 in PK2 KtoA mutants
That both ETC I and ETC II-based respiration cannot be further increased by uncoupling reagents (Fig. 8a, b), suggests that the mutant mitochondria are already uncoupled. The proton gradient across the intermembrane space is generated by electron transfer, but the gradient can be dissipated by uncoupling proteins (UCPs)58. UCPs are a family of transporters that permit proton movement back into the mitochondrial matrix, and some UCPs are involved in generating heat. Thus, uncoupling should result in less ATP production, and the observed increase in ATP is likely due to increased glycolysis (Fig. 9). Mammals express 5 UCP proteins, but C. elegans express only a single UCP, called UCP-4. Nematode UCP-4 is primarily expressed in the body wall and pharyngeal muscles59. We wished to explore the possibility that the level of UCP-4 is altered in the PK2 KtoA mutants. We created a CRISPR/Cas9 strain in which UCP-4 has an HA tag at its C-terminus, and this strain was crossed into unc-89(sf22) and unc-89(syb1360). As shown in Fig. 10, the level of UCP-4 protein, but not mRNA, is increased in PK2 KtoA mutants. The higher level of UCP-4 likely explains the uncoupled state of PK2 KtoA mutants, but why UCP-4 levels are increased awaits further study.
Fig. 10. PK2 KtoA mutants have elevated levels of UCP-4 protein but not mRNA.
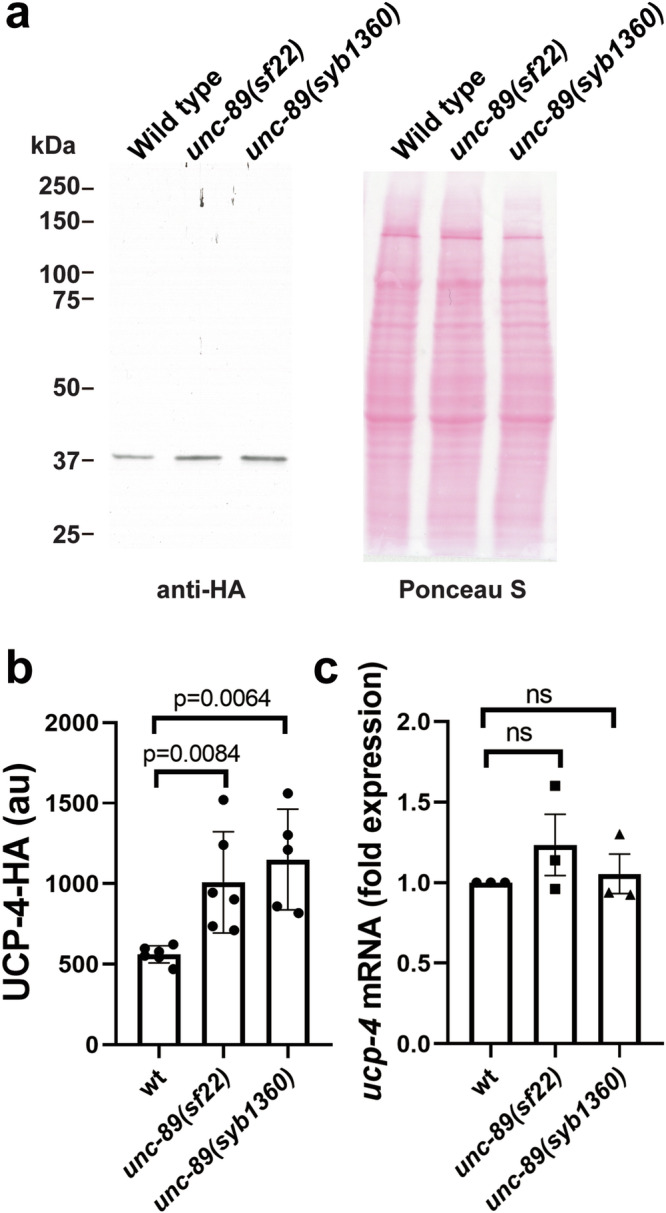
a Representative western blot reacted with anti-HA to detect UCP-4-HA. An HA tag was inserted at the C-terminus of UCP-4 by CRISPR/Cas9 in a wild-type background and crossed into the unc-89 mutants. b Quantitation of western blot results with n = 3–5. Statistical significance was assessed using an unpaired t-test with Welch’s correction. c Results of quantitative real-time PCR comparing levels of ucp-4 mRNA in wild type vs. the unc-89 mutants. Statistical significance was assessed using an unpaired t-test with Welch’s correction.
Increasing the level of PK2 increases mitochondrial clumping and decreases DRP-1 associated with mitochondria
Since the presumed inactivation of PK2 leads to mitochondrial fragmentation we wondered what would happen if the activity of PK2 was increased. We tested this by overexpressing a portion of UNC-89 containing wild-type PK2. We reported that a segment consisting of the C-terminal third of the interkinase region followed by Ig53, Fn2, and PK2 (1/3 IK-Ig-Fn-Pk2), when transgenically expressed from a heat shock promoter localizes to the M-line, the normal location of UNC-8946. This strain and wild type, as day 1 adult animals, were heat-shocked at 30° for 2 h, and then immunostained using anti-ATP5A to image the mitochondria. As shown in Supplementary Fig. 10a, although mitochondria from the wild type showed the expected linear/tubular organization, mitochondria from the strain expressing the PK2 fragment showed more clumping. As shown in Supplementary Fig. 10b, heat shock of this extra PK2 did not change the total level of DRP-1, but as shown in Supplementary Fig. 10c, it decreased the level of DRP-1 associated with mitochondria.
Discussion
We first performed bioinformatics analysis indicating that UNC-89 PK2 is a canonical active protein kinase and that a particular missense mutation (KtoA) is expected to abolish activity but not the stability of the protein. We used CRISPR/Cas9 to create worms that express UNC-89 isoforms with catalytically inactive PK2 protein kinase domains and we observed an unexpected and specific phenotype. In contrast to all previously described unc-89 loss of function mutants, these worms have normal sarcomere and SR structures and move faster than the wild type. In addition, PK2 KtoA mutants have fragmented mitochondria. Although mitochondrial fragmentation is usually associated with decreased mitochondrial function, we found that ATP levels were increased and complex I basal respiration was increased and was already uncoupled, and there was no obvious increase in complex I proteins. Nevertheless, complex II basal respiration was decreased and the level of complex II was decreased. Thus, it seems that the effect of PK2 KtoA mutation both increases and decreases mitochondrial function. The observed increase in glycolysis might be a compensatory response to altered mitochondrial respiration.
We gained molecular insight into how the mitochondria are fragmented. The conserved GTPase Drp1 is required for mitochondrial fragmentation, and in mammals, this occurs by recruitment of Drp1 to the outer membrane by adaptors, then oligomerization, recruitment of dynamin 2, and membrane scission60. We found that there is more DRP-1 associated with mitochondria of PK2 KtoA mutants, thus explaining the higher degree of fragmentation. Conversely, increasing the level of wild-type PK2 by transgenic overexpression increases mitochondrial clumping and decreases DRP-1 associated with mitochondria. The association of Drp1 is known to be regulated by phosphorylation in a complex manner, some phosphorylation sites promoting association, others inhibiting it48. Our finding that genetically drp-1 is epistatic to unc-89(sf22) is consistent with the DRP-1 protein being a direct or indirect target of UNC-89 PK2 protein kinase activity.
That both complex I and complex II respiration could not be further uncoupled led us to postulate that the mitochondria are already uncoupled. The proton gradient across the intermembrane space can be dissipated by uncoupling proteins which transport protons back into the mitochondrial matrix. Although mammals express 5 UCP proteins, C. elegans expresses only a single UCP, UCP-4. This allowed us to easily explore whether this mechanism of uncoupling is abnormal in PK2 KtoA mutants. We found that the level of UCP-4 protein is elevated in PK2 KtoA mutants as compared to wild type. The higher level of UCP-4 likely explains the uncoupled state of PK2 KtoA mutants; however, how this occurs awaits further experiments. One possibility is that the expression, stability, or activity of UCP-4 is regulated by phosphorylation, which also may occur by PK2 or a PK2 kinase cascade. Moreover, more uncoupling means that mitochondria produce less ATP, and thus elevated ATP levels in PK2 KtoA mutants are most likely due to increased glycolysis.
Further insight into the mechanisms that generate the phenotype of PK2 KtoA mutants awaits determining the in vivo substrates for PK2. Unfortunately, during the course of this study, we were unsuccessful in producing recombinant PK2 samples suitable for in vitro phosphotransfer experimentation. Nonetheless, the phenotype of PK2 KtoA mutants suggests the existence of a signaling pathway between the sarcomere, where UNC-89 is located, and mitochondria. Such communication would help match the energy requirements of muscle contraction with energy production by mitochondria. In mammalian striated muscle, large quantities of ATP are required to fuel sarcomere contraction and the ionic pumps that drive muscle contraction, and most of this ATP is supplied by numerous mitochondria. There is very close packing of organelles in striated muscle and the spaces between adjacent myofibril/SR/T-tubule cylinders are filled with mitochondria. There is a complex 3D network of mitochondria in mammalian skeletal muscle, mostly in the form of fused or elongated mitochondria61. This organization likely ensures rapid and efficient transfer of ATP to the contractile apparatus62. It is believed that during muscle contraction, calcium ions, and ATP are released into a privileged microdomain and taken up into myofibrils before they have time to diffuse beyond the myofibrils. Because of the close proximity of mitochondria to sarcomeres, even in C. elegans muscle, and the need to match energetic output to contractile demands, we speculate that there is signaling from sarcomeres to mitochondria that coordinate the structural organization and metabolic activity of mitochondria, and this is essential for optimal muscle performance.
Although our studies provide a possible molecular mechanism by which sarcomeres communicate with mitochondria, our study is not the first to provide evidence of communication between these organelles. In a remarkably sophisticated study using Drosophila, Avellaneda et al.63 demonstrated that there is communication between mitochondria and myofibrils during muscle development. Drosophila have two types of muscle, fibrillar, exemplified by indirect flight muscle in which individual myofibrils are not laterally aligned with their neighbors, and body muscles such as in the legs in which myofibrils are aligned laterally to form a tube. The morphology and organization of mitochondria are different in these muscle types. In indirect flight muscle, the mitochondria are elongated and lie between the myofibrils, whereas in leg muscle the mitochondria lie mostly within the tube and peripherally. When Avellaneda et al. overexpressed a mitofusin homolog required for mitochondrial fusion or expressed a dominant negative Drp1 required for fission, the morphology of indirect flight muscle resembled leg muscle with myofibrils aligned laterally and often forming tubes and mitochondria located inside.
Increased mitochondrial fragmentation has been linked to a reduction in mitochondrial function49,64. However, some studies suggest that mitochondrial fragmentation is not always associated with decreased function and may be beneficial. For example, in rat gastrocnemius muscle, resistance exercise resulted in an increase in activated Drp1 (Drp1 phosphorylated at serine 616), suggesting activation of mitochondrial fission65. Feng et al.66 reported that in rats during early exercise, there was increased expression of the fission mediator Fis1. A more definitive study was reported on heart muscle67. After exercising mice for 1 h, the authors found that mitochondria were fragmented, total Drp1 and activated Drp1 were increased, oxidative stress was not increased, mitochondrial membrane potential was maintained, pro-mitophagy proteins decreased, and mitochondrial respiration was increased. Their conclusion is that mitochondrial fragmentation associated with increased mitochondrial function is part of the normal cardiac response to increased energetic demand encountered during normal levels of exercise. Our findings in C. elegans striated muscle—that a kinase-dead version of UNC-89 results in mitochondrial fragmentation but increased respiration and ATP levels—are consistent with this report on exercised mouse heart.
Materials and methods
Bioinformatics analysis of PK2
Three-dimensional structural models of UNC-89-PK2 (UniProtKB O01761) were calculated using AlphaFold v2.3.168 as implemented in the ColabFold 1.5.5 server (https://colab.research.google.com/github/sokrypton/ColabFold/blob/main/AlphaFold2.ipynb).
In order to estimate the effect of the KtoA mutation (K7816A in UniProtKB O01761) on kinase stability, we calculated a comparative protein unfolding energy, ΔΔG, between wild type and in silico mutated PK2 models, where ΔΔG = ΔGKtoA – ΔGwt. To calculate the ΔΔG value, the best-ranking AlphaFold model was used as template in FoldX v441, applying the RepairPDB function for gentle model equilibration, followed by the introduction of the KtoA mutation using the PositionScan function, as described in ref. 69.
C. elegans strains
N2 (wild type, Bristol)
GB360 unc-89(sf22), outcrossed 4× to wild type
GB361 unc-89(sf24), outcrossed 3× to wild type
GB362 unc-89(syb1195), outcrossed 3× to wild type
GB363 unc-89(syb1360), outcrossed 3× to wild type
unc-89(tm752)
CB1460 unc-89(e1460)
HE75 unc-89(su75)
GB359 unc-22(sf21), outcrossed 4× to wild type
unc-22(e105)
CU6372 drp-1(tm1108)
GB364 unc-89(sf22); drp-1(tm1108)
ZW129 zwIs108 [myo-3p::myc::unc-68; myo-3p::GFP]
GB369 unc-89(sf22); zwIs108
SU1002 unc-68(jc78[HA::unc-68]) cash-1(jc60[TagRFP::myc::cash-1]V
GB366 unc-89(sf22); unc-68(jc78[3Xha::unc-68]) cash-1(jc60[TagRFP::myc::cash-1]
GB357 sfEx[myo-3p::TBA-1::HA; sur-5::GFP]
GB358 unc-89(sf22); sfEx[myo-3p::TBA-1::HA; sur-5::GFP]
SJZ884 foxSi127 [myo-3p::tomm20::mKate2::HA::tbb-2 3′UTR]
GB353 unc-89(sf22) I; foxSi127 [myo-3p ::tomm20::mKate2::HA::tbb-2 3′UTR]
GB354 unc-89(syb1360); foxSi127 [myo-3p ::tomm20::mKate2::HA::tbb-2 3′UTR]
GB296 sfIs21[myo-3p::PercevalHR; lin-44p::GFP]
GB355 unc-89(sf22); sfIs21[myo-3p ::PercevalHR; lin-44p::GFP]
GB356 drp-1(tm1108); sfIs21[myo-3p ::PercevalHR; lin-44p::GFP]
PHX6586 ucp-4(syb6586)
GB367 unc-89(sf22); ucp-4(syb6586)
GB368 unc-89(syb1360); ucp-4(syb6586)
GB267 sfIs13[hsp ::HA ::UNC-89 (1/3 IK-Ig-Fn-PK2 ; sur-5 ::GFP)]
CRISPR/Cas9 generation of UNC-89 PK2 KtoA mutants, and HA tagging of UCP-4
The UNC-89 PK2 KtoA mutants, unc-89(sf22) and unc-89(sf24) were generated using the following strategy.
Vector: pPD162 (Peft-3::Cas9 + Empty sgRNA)
Target sequence in pPD162: ACGAATCTTCACAGCGCAA
Template sequence (100 nt ssDNA):
3′ATTCATACTCTGAGATCGCTTCAGACGATGGATGACGAATggcCACtGCGCAgTGtGCGTGACCTTCTGTTGTGGAATCGACGGCGTCGCGGATCACAGA5′
ggc: KtoA point mutation
under lined: MscI site used to detect heterozygotes
Lower case g: silent mutation
Primers for insertion of sgRNA target sequence:
Fw: 5′ACGAATCTTCACAGCGCAAgttttagagctagaaatagcaagt3′
Rv: 5′caagacatctcgcaataggaggtg3′
PK2 coding region (length 968 bp) from genomic DNA:
ATGATCCAACAGGACGGTTCCAGgtaagggacacttcaggaaataaaaagaaagtcatgtgaaattttccctttatactgggaataattaggcaaaatccctactaacaatatccatattttggtacactgaaagtaattctaaacctgtattttataacgagacatctataaattttctagcagaaatttgcccccaagaaaattatttttttgaataatttttaaaagagttttttttggtgttttggggtgagtttcataatacatatataattaatacaattttgcactcacaagtaaaaaatagttcttgtggcattttaaaatttaaatttatatcattttttcatttcactattccacataaaattgttcagATCGGTGGTCTCAAGTTCAAGGGACGTTTCTCTGTGATCCGCGACGCCGTCGATTCCACAACAGAAGGTCACGCCCATTGCGCTGTGAAGATTCGTCATCCATCGTCTGAAGCGATCTCAGAGTATGAATCGCTTCGTGATGGTCAGCATGAAAATGTTCAACGCCTTATCGCCGCATTCAATAACTCCAATTTCTTGTATCTATTATCGGAAAGACTCTACGAAGATGTGTTTTCTCGTTTTGTGTTCAACGATTATTATACAGAAGAACAAGTTGCATTGACAATGAGACAAGTCACTTCGGCACTTCATTTCTTGCATTTCAAAGGgtgagctttaacagttattaacagttattaattcctaattctaatattttgattttagAATTGCCCATCTTGATGTGAATCCACACAACATAATGTTCCAATCAAAACGTAGTTGGGTCGTGAAACTAGTTGATTTTGGAAGAGCACAAAAAGTGTCGGGAGCTGTGAAACCAGTTGATTTTGATACTAAATGGGCTTCACCAGAATTCCATATTCCGGAAACTCCGGTTACCGTTCAAAGTGACATGTGGGGTATGGGA
Under lined: PAM sequence (GN19NGG)
Red Upper case: AAG (K) in PK2
MscI digestion: (968 bp -> 504 bp/464 bp)
Primers for examination of the point mutations:
Fw: 5′ATGATCCAACAGGACGGTTCCAG3′
Rv: 5′TCCCATACCCCACATGTCACTTTG3′
The UNC-89 PK2 KtoA mutants, unc-89(syb1195) and unc-89(syb1360), and placement of an HA tag at C-terminus of UCP-4 was carried out by SunyBiotech (http://www.sunybiotech.com). Details about the sgRNAs and repair templates used are given in Supplementary Fig. 11.
Western blots
The method of Hannak et al.70 was used to prepare total protein lysates from wild type, unc-89(sf22), unc-89(syb1360), drp-1(tm1108), PHX6586 ucp-4(syb6586), GB367 unc-89(sf22); ucp-4(syb6586), GB368 unc-89(syb1360); ucp-4(syb6586), and GB267 sfIs13[hsp::HA::UNC-89 (1/3 IK-Ig-Fn-PK2; sur-5::GFP)] worms. To detect UNC-89 proteins, we separated the protein lysate on a 3% stacking and 5% separating Laemmli SDS-PAGE gel, transferred to nitrocellulose membrane for 2 h, and reacted with anti-UNC-89 EU30 affinity-purified rabbit polyclonal antibodies at 1:400 dilution10, followed by reaction with anti-rabbit Ig-horseradish peroxidase and ECL. Western blots to detect DRP-1 were used in three types of experiments. First, to check the specificity of anti-DRP-1 antibodies, equal quantities of total protein lysates from wild type and drp-1(tm1108) were separated on a 10% Laemmli SDS-PAGE gel, transferred to nitrocellulose, and reacted against affinity-purified anti-DRP-1 at 1:200 dilution which had been pre-absorbed against an acetone powder of E. coli OP50 to reduce or eliminate reaction to bacterial proteins. Second, we used a similar approach to compare the levels of DRP-1 in total protein lysates from wild type, unc-89(sf22) and unc-89(syb1360). Third, we compared the level of DRP-1 from total worm mitochondria from wild type, unc-89(sf22) and unc-89(syb1360). Preparation of total worm mitochondria by differential centrifugation was conducted as described below in the section on purifying muscle mitochondria. We also performed western blots to compare the levels of representative components of electron chain complexes from total protein lysates. Antibodies and concentrations were: complex I, mouse monoclonal to NDUFS3 (Abcam cat. no. ab14711) at 1:2,500 dilution; complex II, rabbit anti-SDHA (Cell Signaling, cat. no. 11998 P) at 1:500 dilution; complex IV, mouse monoclonal to MTCO1 (Abcam cat. no. ab14705) at 1:2000 dilution; complex V, mouse monoclonal to ATP5A (Abcam cat. no. ab147481) at 1:5000 dilution; and mouse monoclonal to cytochrome C (Abcam cat. no. ab110325) at 1:1000 dilution. Finally, we performed western blots to measure the level of UCP-4-HA in wild type, unc-89(sf22) and unc-89(syb1360) backgrounds using anti-HA (rabbit monoclonal cat. no. C29F4 from Cell Signaling Technology) at 1:1000 dilution. Before use, the optimal dilutions of each antibody were determined to yield linear detection of the proteins. Protein bands were quantitated by comparing to scans of Ponceau S staining of a gel lane on the blot. The uncropped and unedited blot images used to prepare Figs. 1d, 6a–c, 8c, 10a, and Supplementary Fig. 10b, c, are shown respectively in Supplementary Figs. 12–16.
Immunostaining and confocal microscopy of body wall muscle myofilament lattice and mitochondria, and pharyngeal myofilament lattice
Day 0 or day 1 adult worms were fixed and immunostained using the method described by Nonet et al.71. The following antibodies were used to stain parts of the myofilament lattice, all at 1:200 dilution: anti-UNC-95 (rabbit polyclonal Benian-13)72, anti-MHC A (mouse monoclonal 5–6)73, and anti-UNC-89(rabbit polyclonal EU30)10. Fixation and phalloidin-rhodamine staining were conducted as described in ref. 74. To visualize mitochondria, we used mouse monoclonal antibody (15H4C4) also at 1:200 dilution (Abcam cat. no. ab14748). For pharyngeal muscle imaging, the fixed adult day 1 worms were incubated with anti-UNC-89 (EU30) at 1:100, and anti-myosin heavy chain C (MHC C; mouse monoclonal 9.2.175) at 1:100. Secondary antibodies, used at 1:200 dilution, included anti-rabbit Alexa 488, and anti-mouse Alexa 594, all purchased from Invitrogen. Confocal images were captured at room temperature with a Zeiss confocal system (LSM510) equipped with an Axiovert 100 M microscope and an Apochromat x63/1.4 numerical aperture oil immersion objective, in 1× and 2.5× zoom mode. For the confocal images, the color balances were adjusted by using Adobe Photoshop (Adobe, San Jose, CA).
Super-resolution microscopy was performed using a Nikon N-SIM system in 3D structured illumination mode on an Eclipse Ti-E microscope equipped with a 100×/1.49 NA oil immersion objective, 561-nm solid-state laser, and an EM-CCD camera (DU-897, Andor Technology). Super-resolution images were reconstructed using the N-SIM module in NIS-Elements software. The color balances of the images were adjusted by using Adobe Photoshop (Adobe, San Jose, CA). A Z-series every 0.2 μm was taken from the outer muscle cell membrane deeper into the muscle cell. This Z stack was then reconstructed into a 3D image using software on the N-SIM microscope. The reconstructed 3D z-stacks were analyzed using Imaris 8.6 (Bitplane), to visualize the mitochondria.
Measurement of mitochondrial length in μm from the 3D reconstructions of SIM images was performed using Imaris software on a combination of 2 images from each wild type and mutant strain. The longest axis of each anti-ATP5A-staining object (mitochondrion) was determined using the Imaris parameter called “BoundingBoxOOLength”. After eliminating objects <1 μm, the number of objects was placed and counted in 3 bins: 1 ≤ × < 5 μm, 5 ≤ × < 10 μm, and x ≥ 10 μm. The Y axis of the graph in Supplementary Fig. 8 represents the number of objects in each bin divided by the total number objects.
Electron microscopy
Electron microscopy of body wall muscle was performed as described in Matsunaga et al.45. From each strain, approximately 200 days 2 adults were fixed and processed, and approximately 10 randomly selected ultrathin sections were examined. Approximately 30 low-powered images were captured that revealed all of the sarcomeres of single body wall muscle cells, and from these 10–20 images at higher magnification were captured that revealed 1–2 sarcomeres each. All these images were examined by two individuals both on a computer screen and by printouts, looking for any differences between the wild type and the two mutant alleles, sf22 and syb1360. Representative images are shown in Fig. 2d.
Crawling assays and NemaFlex force measurement
For crawling assays, day two adults were harvested from one 6 cm NGM OP50 seeded plate with M9 buffer, and a final wash in M9 containing 0.2 g/L gelatin. Five microliters of worm suspension were added to the center of a 6 cm unseeded NGM plate and then the excess liquid was removed using a twisted KimWipe. After a five-minute adaptation time, worm crawling movement was recorded using a dissecting stereoscopic microscope fitted with a CMOS camera (Thorlabs). For all strains a total of fifteen, 10-s. videos were recorded from various sections of the plate with each video tracking an average of 8 individual animals. Video data was analyzed by Image J WrmTracker software plug-in to obtain body bends per second (BBPS) for individual animals. Worms that moved out of frame and outliers were removed during data analysis and an average of 20 animals were analyzed for each strain. The resulting BBPS values for each mutant strain were compared to the wild type and further tested for statistically significant differences using Welch’s T-test.
The maximum exertable muscle force by C. elegans strains was measured using the NemaFlex method43. The technique is based on the deflection of soft micropillars as the animals crawl through an array of micropillars. The pillars are arranged in a square lattice with a pillar diameter of 44 μm and height of 87 μm. The gap between the pillars is 71 μm. Day 1 adult animals were loaded individually in each chamber76, and a 1-min video was captured for each animal in a food-free environment at a temperature of 20 ± 1 °C. Imaging was performed in brightfield using a Nikon Ti-E microscope and an Andor Zyla sCMOS 5.5 camera. Images were acquired at 5 frames per second with a 4× objective at a pixel resolution of 1.63 μm per pixel. Movies were processed and analyzed for force values using in-house-built image processing software (MATLAB, R2016a). Animal muscle strength measurements were calculated by identifying the maximal force exerted in each frame for an individual animal. We define the maximum exertable force f95 corresponding to the 95th percentile of these maximal forces. Statistical analysis was performed using the Wilcoxon rank-sum test.
Generation of antibodies to DRP-1
Rabbit polyclonal antibodies were generated to the C-terminal 143 residues of DRP-1b, amino acids 570-712. The coding sequence of this segment was amplified by PCR from a cDNA pool (RB2 library) and cloned into vectors pGEX-KK1 and pMAL-KK, to express glutathione S-transferase (GST) and MBP fusion proteins in E. coli77. In brief, to express each fusion protein, the plasmids were transformed into E. coli Rosetta 2 (DE3) (Millipore Corporation, cat. no. 71397-4), and grown in Luria-Bertani broth containing 10 µg/ml ampicillin at 37° to OD600 = 0.6–0.8, followed by induction of expression with 0.5 mM isopropyl β-D-1-thiogalactopyranoside at 20° for 5 h. Cells were pelleted by centrifugation and resuspended in 50 mM Tris-HCl pH 8.0 containing 100 mM phenylmethylsulfonyl fluoride (PMSF) and cOmplete Mini protease inhibitor cocktail (Roche, Inc., cat. no. 11836170001), and broken in a French pressure cell at 1000 pounds inch−2, the addition of Triton X-100 to 1%, followed by spinning out debris by centrifugation at 12,000 × g for 20 min. The lysates were added to either a slurry of glutathione-agarose beads (Sigma, cat. no. G-4510) or amylose resin (New England BioLabs, cat. no. E8021L) and incubated with mixing for 30 min at 4°. The beads were washed 5X with 50 mM Tris-HCl pH 8.0 containing 1% Triton X-100 and 100 mM PMSF, and then 4× with 50 mM Tris-HCl pH 8.0 containing 100 mM PMSF. The beads were placed into minicolumns and eluted with either 10 mM free glutathione or 10 mM maltose in 50 mM Tris-HCl pH 8.0. The GST fusions were shipped to Noble Life Sciences (Sykesville, Maryland) for the production of rabbit antibodies. Antibodies were affinity-purified using Affi-gel (Bio-Rad)-conjugated MBP fusions, as described78. Briefly, the MBP fusion proteins were dialyzed against 100 mM 3-morpholinopropane-1-sulfonic acid (MOPS) pH 7.0, and then 3–10 mg were covalently coupled to 1 ml of a 50:50 mixture of Affi-Gel 10 and Affi-Gel 15 beads (Bio-Rad Laboratories, cat. nos. 1536099 and 153-6051). These columns were then used to affinity purify ~2 ml of antiserum and the antibodies eluted with low and high pH and concentrated with a centrifugal filter (Centriprep 10, Millipore, cat. no. 4304).
Measurement of ATP levels in whole worms
100 day 1 adults were collected and washed free from bacteria, pelleted in a 1.5 ml Eppendorf tube, frozen in liquid nitrogen, and then placed in a boiling water bath for 15 min. After placing on ice for 5 min and vortexing for 5 s, debris was pelleted by centrifugation at maximum speed in a microfuge for 10 min at 4 °C. Supernatants were transferred to fresh tubes and diluted 5–10 fold with deionized water and kept on ice. Using this material, the total protein content was determined by the BCA protein assay kit (Pierce, cat. no. 23225), and the total ATP determined using a CellTiter-Glo Luminescent Cell Viability Assay kit from Promega Corporation. Results were expressed as nmoles of ATP per milligram of protein.
Use of PercevalHR to measure ATP/ADP ratio
The plasmid pRsetB-PercevalHR was purchased from Addgene (Cambridge, MA), and The XbaI/XhoI fragment of pRsetB-PercevalHR was inserted into the vector pPD95.86 via its XbaI/SalI sites to allow expression of PercevalHR from the muscle-specific promoter for the myo-3 gene. The resulting plasmid, myo-3p::PercevalHR at 10 ng/ml, was co-injected into worms together with the transformation marker plasmid lin-44p::GFP at 90 ng/ml, into wild-type worms, and transgenic animals were selected on the basis of GFP expression in the worm tail. The extrachromosomal array was then integrated randomly into the genome by ultraviolet irradiation79 with modifications (Peter Barrett, personal communication), to create strain GB296, sfIs21[myo-3p::PercevalHR; lin-44p::GFP]. sfIs21 was crossed into unc-89(sf22) and into drp-1(tm1108), to create strains GB355 and GB356, respectively. PercevalHR is based on GlnK1 which undergoes a major conformational change depending on whether ATP or ADP is bound. Coupling of GlnK1 to mVenus creates a fluorescent read-out of this conformation. An ATP to ADP ratio is proportional to emission at 533 nm upon excitation at 405 nm and 488 nm. Day 1 transgenic adults were immobilized for imaging without drugs using a combination of a PDMS microfluidic device with channels just large enough to accommodate adult worms and Pluronic F12780. The microfluidic devices were kindly manufactured and provided by Gongchen Sun from the Hang Lu laboratory at the Georgia Institute of Technology. Aqueous solutions of Pluronic F127 are fluid at 4°, and solid at room temperature. Body wall muscle was excited with a laser at 405 and at 488 nm using an Olympus FV1000 inverted confocal microscope. Ratiometric images of body wall muscle were created by dividing the emissions, Em 488/Em 405. Using Fiji software on these images, the total number of pixels with intensity greater than 100 were counted, and the means and standard deviations were calculated from multiple body wall muscle cells from 5 animals of each strain.
Purification of muscle mitochondria and respirometry
Body wall muscle-specific mitochondria were purified from strains expressing tomm20-mKate2-HA from the promoter of the muscle-specific gene myo-3, using a modified version of the procedure developed by Ahier et al.56. Briefly, for each strain, we harvested a mixed-stage population of nematodes from 2, 15 cm, high peptone plates seeded with E. coli strain NA22. Worms were washed free from bacteria by at least 3 washes of M9 buffer, followed by 2 washes with deionized water. The worm pellets were resuspended in 3.5 mls of ice-cold Mitochondrial Isolation Buffer (MIB; 50 mM KCl, 110 mM mannitol, 70 mM sucrose, 0.1 mM EDTA, 5 mM Tris pH 7.4 without protease inhibitors) and transferred to a 7 ml Dounce homogenizer. While on ice, we brought the pestle up and down 25 times. The homogenate was transferred to 3, 1.5 ml Eppendorf tubes, and spun in a microfuge at 4° for 5 min at 200 × g. The resulting supernatant was transferred to 3 fresh tubes, and centrifuged for 10 min at 800 × g. This supernatant was then centrifuged for 10 min at 12,000 × g to pellet the mitochondria. The pellets from all 3 Eppendorf tubes were resuspended in a final volume of 600 μl of ice-cold PEB (PBS, 2 mM EDTA, 1% bovine serum albumin, without protease inhibitors). To this suspension was added 30 μl of a suspension of anti-HA magnetic beads (Pierce, cat. no. 88836), and the tube placed on a rotating wheel at 4° for one hour. The beads were then pulled out of the solution using a tube stand containing neodymium magnets (Millipore Corporation), and then the beads were washed twice with 1 ml of PEB, and then resuspended in 50–100 μl of MIB and kept on ice.
Mitochondrial respiration was measured using a Clark electrode (Oxytherm System, Hansatech) as described81. Briefly, isolated muscle mitochondria were suspended in respiration buffer containing 120 mM KCl, 3 mM HEPES, 1 mM EGTA, 5 mM KH2PO4, and 0.3% (w/v) fat-free BSA. Complex I substrates (10 mM glutamate, and 2 mM malate) or complex II substrates (7 mM succinate) were added in addition to 1.2 mM ADP to stimulate basal respiration. Uncoupled respiration was measured with the addition of 80 nM FCCP (Carbonyl cyanide-p-trifluoromethoxyphenylhydrazone), and non-mitochondrial oxygen consumption was assessed with the addition of potassium cyanide (500 μM). To assess mitochondrial respiration, the non-mitochondrial oxygen consumption rate was subtracted from the total oxygen consumption rate.
Glycolysis assay
We used Abcam’s “6-Phosphofructokinase Activity Assay Kit (colorimetric)” (ab155898) essentially according to the manufacturer’s instructions. Approximately 50 milligrams of packed worms were homogenized in 200 μl of PFK Assay Buffer, using a Pellet Pestle Motor (DWK Life Sciences Kimble Kontes). After determining the protein concentrations using a Bradford assay, approximately 1 μg of total protein extract was used per assay. PFK activity was recorded as mU/mg protein.
Statistics and reproducibility
GraphPad Prism9 was used to plot and analyze numerical data. Figure legends explain the n values, whether standard deviations or standard errors of the mean were plotted, which statistical tests were used, and the definition of * values. Statistical significance was assessed using an unpaired t-test with Welch’s correction unless otherwise specified. Each experiment was conducted several times independently and only if the same results or tendencies were obtained, were figures and conclusions made. Many experiments involved imaging and evaluation of the images was conducted by at least two individuals but in most cases, no quantitation was involved, so statistical analysis was not used.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary File
Acknowledgements
These studies were primarily supported by grant no. 2050009 from the National Science Foundation to G.M.B and J.Q.K. Additional partial support includes the following: NIH grant R01HL160693 to G.M.B.; NIH grant R01GM144729 to J.Q.K.; AAF grant 06/16 from the University of Konstanz to O.M.; a Stafford Fox Research Foundation Fellowship and a National Health and Medical Research Council (NHMRC) grant GNT2010813 to S.Z.; and NASA grant NNX15AL16G to S.A.V. The authors thank SunyBiotech Corporation for generating CRISPR/Cas9 lines PHX1195, PHX1360, and PHX6586; Jeannette Taylor of Emory University’s Integrated Electron Microscopy Core for EM images; Gongcheng Sun and Hang Lu (Georgia Institute of Technology) for the microfluidic device used to immobilize worms for the Perceval imaging; Laura Fox-Goharioon and April Reedy of Emory University’s Integrated Cellular Imaging (ICI) Core for help in using an N-SIM microscope, 3D rendering, and ratiometric imaging; Nathaniel Szewczyk (Ohio University) for advice on which antibodies to use to detect mitochondrial complex proteins; Kenneth Norman (Albany Medical School) for nematode strain ZW129; the Caenorhabditis Genetics Center, which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440) for multiple nematode strains; and Robert Barstead (Oklahoma Medical Research Foundation) for the cDNA library RB2.
Author contributions
G.M.B. contributed by conceiving the project, designed and interpreted most experiments, carried out some experiments, and wrote most of the manuscript. J.Q.K. designed and interpreted some experiments carried out some experiments, and wrote some parts of the manuscript. O.M. and T.D. performed the bioinformatics analysis, and O.M. wrote some sections of the manuscript and edited it. Y.M., H.Q., N.G., L.L., J.C.M., A.A., and C.J.M. performed and interpreted experiments, with Y.M., H.Q., and N.G. doing most of them. These same 7 authors also wrote small parts of the manuscript and edited it. S.Z. and S.A.V. contributed by interpreting the results and edited the manuscript.
Peer review
Peer review information
Communications Biology thanks Belinda Bullard and the other, anonymous, reviewers for their contribution to the peer review of this work. Primary Handling Editors: Martina Rauner and Joao Valente. A peer review file is available.
Data availability
The authors declare that the data supporting the results of this study are displayed in the Figures and the Supplementary Figs. The numerical source data for the graphs presented in the figures are available in the Supplementary Data.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-024-07042-3.
References
- 1.Kontrogianni-Konstantopoulos, A., Ackermann, M. A., Bowman, A. L., Yap, S. V. & Bloch, R. J. Muscle giants: molecular scaffolds in sarcomerogenesis. Physiol. Rev.89, 1217–1267 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gieseler K, Qadota H, Benian GM. Development, structure, and maintenance of C. elegans body wall muscle. (2017). WormBook, ed. The C. elegans Research Community, WormBook, 10.1895/wormbook.1.173.1. [DOI] [PMC free article] [PubMed]
- 3.Schueder, F. et al. Nanobodies combined with DNA-PAINT super-resolution reveal a staggered titin nanoarchitecture in flight muscles. Elife12, e79344 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loescher, C. M., Hobbach, A. J. & Linke, W. A. Titin (TTN): from molecule to modifications, mechanis, and medical significance. Cardiovasc. Res.118, 2903–2918 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perry, N. A., Ackermann, M. A., Shriver, M., Hu, L. Y. & Kontrogianni-Konstantopoulos, A. Obscurins: unassuming giants enter the spotlight. IUBMB Life65, 479–486 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Waterston, R. H., Thomson, J. N. & Brenner, S. Mutants with altered muscle structure in C. elegans. Dev. Biol.77, 271–302 (1980). [DOI] [PubMed] [Google Scholar]
- 7.Benian, G. M., Ayme-Southgate, A. & Tinley, T. L. The genetics and molecular biology of the titin/connectin-like proteins of invertebrates. Rev. Physiol. Biochem. Pharmacol.138, 235–268 (1999). [DOI] [PubMed] [Google Scholar]
- 8.Small, T. M. et al. Three new isoforms of C. elegans UNC-89 containing MLCK-like protein kinase domains. J. Mol. Biol.342, 91–108 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Qadota, H. et al. A region of UNC-89 (obscurin) lying between two protein kinase domains is a highly elastic spring required for proper sarcomere organization. J. Mol. Biol.432, 4799–4814 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benian, G. M., Tinley, T. L., Tang, X. & Borodovsky, M. The Caenorhabditis elegans gene unc-89, required for muscle M-line assembly, encodes a giant modular protein composed of Ig and signal transduction domains. J. Cell Biol.132, 835–848 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferrara, T. M., Flaherty, D. B. & Benian, G. M. Titin / connectin-related proteins in C. elegans: a review and new findings. J. Musc. Res. Cell Motil.26, 435–447 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Qadota, H. et al. A novel protein phosphatase is a binding partner for the protein kinase domains of UNC-89 (obscurin) in C. elegans. Mol. Biol. Cell19, 2424–2432 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qadota, H. et al. Protein phosphatase 2A is crucial for sarcomere organization in Caenorhabditis elegans striated muscle. Mol. Biol. Cell29, 2084–2097 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katzemich, A. et al. The function of the M-line protein obscurin in controlling the symmetry of the sarcomere in the flight muscle of Drosophila. J. Cell Sci.125, 3367–3379 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bang, M. L. et al. The complete gene sequence of titin, expression of an unusual approximately 700-kDa titin isoform, and its interaction with obscurin identify a novel Z-line to I-band linking system. Circ. Res.89, 1065–1072 (2001). [DOI] [PubMed] [Google Scholar]
- 16.Young, P., Ehler, E. & Gautel, M. Obscurin, a giant sarcomeric Rho guanine nucleotide exchange factor protein involved in sarcomere assembly. J. Cell Biol.154, 123–136 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geisler, S. B. et al. Obscurin-like 1, OBSL1, is a novel cytoskeletal protein related to obscurin. Genomics89, 521–531 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsieh, C. M. et al. Striated muscle preferentially expressed genes alpha and beta are two serine/threonine protein kinases derived form the same gene as the aortic preferentially expressed gene-1. J. Biol. Chem.275, 36966–36973 (2000). [DOI] [PubMed] [Google Scholar]
- 19.Bowman, A. L. et al. Different obscurin isoforms localize to distinct sites at sarcomeres. FEBS Lett.581, 1549–1554 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grogan, A. & Kontrogianni-Konstantopoulos, A. Unraveling obscurins in heart disease. Pflug. Arch.471, 735–743 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kontrogianni-Konstantopoulos, A., Jones, E. M., Van Rossum, D. B. & Bloch, R. J. Obscurin is a ligand for small ankyrin 1 in skeletal muscle. Mol. Biol. Cell14, 1138–1148 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bagnato, P., Barone, V., Giacomello, E., Rossi, D. & Sorrentino, V. Binding of an ankyrin-1 isoform to obscurin suggests a molecular link between the sarcoplasmic reticulum and myofibrils in striated muscles. J. Cell Biol.160, 245–253 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lange, S. et al. Obscurin determines the architecture of the longitudinal sarcoplasmic reticulum. J. Cell Sci.122, 2640–2650 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spooner, P. M., Bonner, J., Maricq, A. V., Benian, G. M. & Norman, K. R. Large isoforms of UNC-89 (obscurin) are required for muscle cell architecture and optimal calcium release in Caenorhabditis elegans. PLoS ONE7, e40182 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fukuzawa, A. et al. Interactions with titin and myomesin target obscurin and obscurin-like 1 to the M-band: implications for hereditary myopathies. J. Cell Sci.121, 1841–1851 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Hanson, D. et al. The primordial growth disorder 3-M syndrome connects ubiquitination to the cytoskeletal adaptor OBSL1. Am. J. Hum. Genet.84, 801–806 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mayans, O., Benian, G. M., Simkovic, F. & Rigden, D. J. Mechanistic and functional diversity in the mechanosensory kinases of the titin-like family. Biochem. Soc. Trans.41, 1066–1071 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Hu, L. Y. & Kontrogianni-Konstantopoulos, A. The kinse domains of obscurin interact with intercellulrar adhesion proteins. FASEB J.27, 2001–2012 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fleming, J. R. et al. Exploring obscurin and SPEG kinase biology. J. Clin. Med.10, 984 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blondelle, J. et al. Murine obscurin and Obsl1 have functionally redundant roles in sarcolemmal integrity, sarcoplasmic reticulum organization, and muscle metabolism. Commun. Biol.2, 178 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zacharchenko, T. et al. PK1 from Drosophila obscurin is an inactive pseduokinase with scaffolding properties. Open Biol.13, 220350 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Temmerman, K., Simon, B. & Wilmanns, M. Structural and functional diversity in the activity and regulation of DAPK-related protein kinases. FEBS J.280, 5533–5550 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Birukov, K. G. et al. Differential regulation of alternatively spliced endothelial cell myosin light chain kinase isoforms by p60(Src). J. Biol. Chem.276, 8567–8573 (2001). [DOI] [PubMed] [Google Scholar]
- 34.Shohat, G. et al. The pro-apoptotic function of death-associated protein kinase is controlled by a unique inhibitory autophosphorylation-based mechanism. J. Biol. Chem.276, 47460–47470 (2001). [DOI] [PubMed] [Google Scholar]
- 35.von Castelmur, E. et al. Identification of an N-terminal inhibitory extension as the primary mechanosensory regulator of twitchin kinase. Proc. Natl Acad. Sci. USA109, 13608–13613 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taylor, S. S. & Kornev, A. P. Protein kinases: evolution of dynamic regulatory proteins. Trends Biochem. Sci.36, 65–77 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Endicott, J. A., Noble, M. E. & Johnson, L. N. The structural basis for control of eukaryotic protein kinases. Annu. Rev. Biochem.81, 587–613 (2012). [DOI] [PubMed] [Google Scholar]
- 38.Hu, D., Mayeda, A., Trembley, J. H., Lahti, J. M. & Kidd, V. J. CDK11 complexes promote pre-mRNA splicing. J. Biol. Chem.278, 8623–8629 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Abe, Y. et al. Cloning and characterization of a p53-related protein kinase expressed in interleukin-2-activated cytotoxic T-cells, epithelial tumor cell lines, and the testes. J. Biol. Chem.276, 44003–44011 (2001). [DOI] [PubMed] [Google Scholar]
- 40.Iyer, G. H., Moore, M. J. & Taylor, S. S. Consequences of lysine 72 mutation on the phosphorylation and activation state of cAMP-dependent kinase. J. Biol. Chem.280, 8800–8807 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Schymkowitz, J. et al. The FoldX web server: an online force field. Nucleic Acids Res.33, W382–W388 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Randazzo, D. et al. Obscurin is required for ankyrinB-dependent dystrophin localization and sarcolemma integrity. J. Cell Biol.200, 523–536 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rahman, M. et al. NemaFlex: a microfluidics-based technology for standardized measurement of muscular strength of C. elegans. Lab Chip18, 2187–2201 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van der Bliek, A. M., Sedensky, M. M. & Morgan, P. G. Cell biology of the mitochondrion. Genetics207, 843–871 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matsunaga, Y. et al. Twitchin kinase inhibits muscle activity. Mol. Biol. Cell28, 1591–1600 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qadota, H. et al. The SH3 domain of UNC-89 (obscurin) interacts with paramyosin, a coiled-coil protein, in Caenorhabditis elegans muscle. Mol. Biol. Cell27, 1606–1620 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scholtes, C. et al. DRP-1-mediated apoptosis induces muscle degeneration in dystrophin mutants. Sci. Rep.8, 7354 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu, C., Huang, Y. & Li, L. Drp1-dependent mitochondrial fission plays critical roles in physiological and pathological processes in mammals. Int. J. Mol. Sci.18, 144 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen, H., Chomyn, A. & Chan, D. C. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J. Biol. Chem.280, 26185–26192 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Jheng, H.-F. et al. Mitochondrial fission contributes to mitochondrial dysfunction and insulin resistance in skeletal muscle. Mol. Cell Biol.32, 309–319 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Metallo, C. M. & Vander Heiden, M. G. Understanding metabolic regulation and its influence on cellular physiology. Mol. Cell49, 388–398 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berg, J., Hung, Y. P. & Yellen, G. A genetically encoded fluorescent reporter of ATP:ADP ratio. Nat. Methods6, 161–166 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tantama, M., Martinez-Francois, J. R., Mongeon, R. & Yellen, G. Imaging energy status in live cells with a fluorescent biosensor of the intracellular ATP-to-ADP ratio. Nat. Commun.4, 2550l (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zanotelli, M. R. et al. Regulation of ATP utilization during metastatic cell migration by collagen architecture. Mol. Biol. Cell29, 1–9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bess, A. S. et al. Effects of mutations in mitochondrial dynamics-related genes on the mitochondrial response to ultraviolet C radiation in developing Caenorhabditis elegans. Worm2, e23763 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ahier, A. et al. Affinity purification of cell-specific mitochondria from whole animals resolves patterns of genetic mosaicism. Nat. Cell Biol.20, 352–360 (2018). [DOI] [PubMed] [Google Scholar]
- 57.Liemburg-Apers, D. C., Schirris, T. J., Russel, F. G., Willems, P. H. & Koopman, W. J. Mitoenergetic dysfunction triggers a rapid compensatory increase in steady-state glucose flux. Biophys. J.109, 1372–1386 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Slocinska, M., Barylski, J. & Jarmuszkiewicz, W. Uncoupling proteins of invertebrates: a review. IUBMB Life68, 691–699 (2016). [DOI] [PubMed] [Google Scholar]
- 59.Iser, W. B., Kim, D., Bachman, E. & Wolkow, C. Examination of the requirement for ucp-4, a putative homolog of mammalian uncoupling proteins, for stress tolerance and longevity in C. elegans. Mech. Ageing Dev.126, 1090–1096 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tilokani, L., Nagashima, S., Paupe, V. & Prudent, J. Mitochondrial dynamics: overview of molecular mechanisms. Essay Biochem.62, 341–360 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Glancy, B. et al. Mitochondrial reticulum for cellular energy distribution in muscle. Nature523, 617–620 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pasqualini, F. S., Nesmith, A. P., Horton, R. E., Sheehy, S. P. & Parker, K. K. Mechanotransduction and metabolism in cardiomyocyte microdomains. BioMed. Res. Int.2016, 4081638 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Avellaneda, J. et al. Myofibril and mitochondria morphogenesis are coordinated by a mechanical feedback mechanism in muscle. Nat. Commun.12, 2091 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bach, D. et al. Mitofusin-2 determines mitochondrial network architecture and mitochondrial metabolism. A novel regulatory mechanism altered in obesity. J. Biol. Chem.278, 17190–17197 (2003). [DOI] [PubMed] [Google Scholar]
- 65.Kitaoka, Y., Ogasawara, R., Tamura, Y., Fujita, S. & Hatta, H. Effect of electrical stimulation-induced resistance exercise on mitochondrial fission and fusion proteins in rat skeletal muscle. Appl. Physiol. Nutr. Metab.40, 1137–1142 (2015). [DOI] [PubMed] [Google Scholar]
- 66.Feng, Z. et al. Mitochondrial dynamic remodeling in strenuous exercise-induced muscle and mitochondrial dysfunction: regulatory effects of hydroxytyrosol. Free Rad. Biol. Med.50, 1437–1446 (2011). [DOI] [PubMed] [Google Scholar]
- 67.Coronado, M. et al. Physiological mitochondrial fragmentation is a normal cardiac adaptation to increased energy demand. Circ. Res.122, 282–295 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bogomolovas, J., Gravenhorst, P. & Mayans, O. Production and analysis of titin kinase: exploiting active/inactive kinase homologs in pseudokinase validation. Methods Enzymol.667, 147–181 (2022). [DOI] [PubMed] [Google Scholar]
- 70.Hannak, E. et al. The kinetically dominant assembly pathway for centrosomal asters in Caenorhabditis elegans is gamma-tubulin dependent. J. Cell Biol.157, 591–602 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nonet, M. L., Grundahl, K., Meyer, B. J. & Rand, J. B. Synaptic function is impaired but not eliminated in C. elegans mutants lacking synaptotagmin. Cell73, 1291–1305 (1993). [DOI] [PubMed] [Google Scholar]
- 72.Qadota, H., Mercer, K. B., Miller, R. K., Kaibuchi, K. & Benian, G. M. Two LIM domain proteins and UNC-96 link UNC-97/pinch to myosin thick filaments in Caenorhabditis elegans muscle. Mol. Biol. Cell18, 4317–4326 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Miller, D. M., Ortiz, I., Berliner, G. C. & Epstein, H. F. Differential localization of two myosins within nematode thick filaments. Cell34, 477–490 (1983). [DOI] [PubMed] [Google Scholar]
- 74.Waterston, R. H., Hirsh, D. & Lane, T. R. Dominant mutations affecting muscle structure in C. elegans that map near the actin gene cluster. J. Mol. Biol.180, 473–496 (1984). [DOI] [PubMed] [Google Scholar]
- 75.Miller, D. M., Stockdale, F. E. & Karn, J. Immunological identification of the genes encoding the four myosin heavy chain isoforms of C. elegans. Proc. Natl Acad. Sci. USA83, 2305–2309 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hewitt, J. E. et al. Muscle strength deficiency and mitochondrial dysfunction in a muscular dystrophy model of Caenorhabditis elegans and its functional response to drugs. Dis. Model Mech.11, 12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mercer, K. B. et al. Caenorhabditis elegans UNC-96 is a new component of M-lines that interacts with UNC-98 and paramyosin and is required in adult muscle for assembly and/or maintenance of thick filaments. Mol. Biol. Cell17, 3832–3847 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mercer, K. B. et al. C. elegans UNC-98, a C2H2 Zn finger protein, is a novel partner of UNC-97/PINCH in muscle adhesion complexes. Mol. Biol. Cell14, 2492–2507 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mitani, S. Genetic regulation of mec-3 gene expression implicated in the specification of the mechanosensory neuron cell types in Caenorhabditis elegans. Dev. Growth Diff.37, 551–557 (1995). [DOI] [PubMed] [Google Scholar]
- 80.Hwang, H., Krajniak, J., Matsunaga, Y., Benian, G. M. & Lu, H. On-demand optical immobilization of C. elegans for high-resolution imaging and microinjection. Lab Chip14, 3498–3501 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ghazal, N., Peoples, J. N., Mohiuddin, T. A. & Kwong, J. Q. Mitochondrial functional resilience after TFAM ablation in the adult heart. Am. J. Physiol. Cell Physiol.320, C929–C942 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary File
Data Availability Statement
The authors declare that the data supporting the results of this study are displayed in the Figures and the Supplementary Figs. The numerical source data for the graphs presented in the figures are available in the Supplementary Data.