Supplemental Digital Content is Available in the Text.
We characterize the expression of microRNA-374 and its targets across pain conditions, species, time, tissue, and sex to identify new pain mechanisms by genetic regulation.
Keywords: Pain, Epigenetics, MicroRNAs, Adipose, Adrenergic receptors
Abstract
Introduction:
Chronic primary pain conditions (CPPCs) are linked to catecholamine activation of peripheral adrenergic receptors. Yet, catecholamine-dependent epigenetic mechanisms, such as microRNA (miRNA) regulation of mRNA transcripts, remain largely unknown.
Objectives:
We sought to identify RNA species correlated with case status in 3 pain cohorts, to validate RNAs found to be dysregulated in a mouse model of CPPC onset, and to directly test the role of adrenergic receptors in miRNA regulation. Furthermore, we tested antinociceptive effects of miR-374 overexpression.
Methods:
We used RNA-seq and quantitative reverse transcription polymerase chain reaction to measure RNA expression in 3 pain cohorts. Next, we validated identified RNAs with quantitative reverse transcription polymerase chain reaction in a mouse model of CPPC onset, measuring expression in plasma, peripheral (adipose, muscle, dorsal root ganglia [DRG]), and central (spinal cord) tissues. Then, we stimulated adrenergic receptors in primary adipocyte and DRG cultures to directly test regulation of microRNAs by adrenergic signaling. Furthermore, we used in vitro calcium imaging to measure the antinociceptive effects of miR-374 overexpression.
Results:
We found that one miRNA family, miR-374, was downregulated in the plasma of individuals with temporomandibular disorder, fibromyalgia syndrome, or widespread pain following a motor vehicle collision. miR-374 was also downregulated in plasma, white adipose tissue, and spinal cord from mice with multisite mechanical sensitivity. miR-374 downregulation in plasma and spinal cord was female specific. Norepinephrine stimulation of primary adipocytes, but not DRG, led to decreased miR-374 expression. Furthermore, we identified tissue-specific and sex-specific changes in the expression of predicted miR-374 mRNA targets, including known (HIF1A, NUMB, TGFBR2) and new (ATXN7, CRK-II) pain targets. Finally, we demonstrated that miR-374 overexpression in DRG neurons reduced capsaicin-induced nociceptor activity.
Conclusions:
Downregulation of miR-374 occurs between adrenergic receptor activation and mechanical hypersensitivity, and its adipocyte source implicates adipose signaling in nociception. Further study of miR-374 may inform therapeutic strategies for the millions worldwide who experience CPPCs.
1. Introduction
Chronic primary pain conditions (CPPCs), such as temporomandibular disorder (TMD) and fibromyalgia syndrome (FMS), are characterized by pain lasting > 3 months in the absence of obvious tissue damage. Chronic primary pain conditions exact a burden on society by lowering life quality and incurring billions annually in healthcare costs.59,61 Despite organic pathology, CPPCs are linked to genetic and environmental factors that increase catecholamine tone. Individuals with CPPCs have loss-of-function mutations in catecholamine metabolism,16,71 resulting in increased catecholamine levels and corresponding increases in pain8,60,68 that are exacerbated by stress.41,44,60 The association between catecholamine metabolism, stress, and pain has also been observed in our rodent model of CPPCs74 and primary pain onset.20,46 This heritable phenotype is driven by catecholamine activation of peripheral adrenergic receptor beta-3 (ADRB3),13 yet epigenetic regulators downstream of ADRB3, such as microRNAs (miRNAs),12 remain underexplored in the context of CPPCs.
miRNAs are small noncoding RNA molecules that inhibit gene expression by blocking protein translation or by degrading downstream messenger RNA (mRNA) transcripts with complementary sequences.4 Evidence implicates miRNAs in the regulation of molecular pathways linked to pain, trauma, inflammation, and immune responses.1,21,26,33,37,51,52,76 As a single miRNA can have many mRNA targets,67 differential expression of a single miRNA or family could trigger a cascade of signaling events leading to chronic pain. MiRNAs are known to be secreted into circulation by white adipose tissue (WAT),3,65 an established endocrine organ14 associated with chronic pain status.50 Furthermore, WAT is a major source of catecholamines,69 proinflammatory cytokines,45 and ADRB3 expression.64 Our laboratory has linked WAT to CPPC development.74 Thus, this article aims to identify miRNAs contributing to high catecholamine pain, their tissue sources, such as WAT, and their mRNA target pathways.
This study evaluated differential expression of miRNAs and corresponding mRNA targets in a discovery TMD cohort and 2 replication cohorts: one with FMS and one with pain following a motor vehicle collision (MVC). Temporomandibular disorder, characterized by orofacial pain, and FMS, characterized by widespread body pain and fatigue, affect 11.5 and 1.1 million Americans, respectively.40,58 MVC frequently causes traumatic injury, with more than 4 million Americans admitted to the emergency department every year, half of whom will go on to suffer chronic pain even after injury healing.9,42 Although these cohorts may differ in clinical presentation, they all have established relationships with stress, mood, and neuroimmune factors,31 which may be driven by shared epigenetic signatures.
Results from our largest TMD RNA sequencing study, to date, identified 16 miRNAs dysregulated in cases relative to controls. Of these, miR-374 family members were downregulated in TMD cases, in replication cohort cases, and in a mouse model of primary pain onset. Furthermore, we identify (1) adrenergic receptor activation in WAT as the likely origin of miR-374 family member downregulation, (2) emerging tissue- and sex-specific regulation of mRNA targets of miR-374, and (3) miR-374's ability to inhibit sensory neuron activity. Our findings begin to elucidate the mechanisms by which the miR-374 family contributes to pain outcomes. This work has the potential to identify druggable targets and advance the treatment for the millions56 of individuals who experience chronic pain.
2. Materials and methods
2.1. Study approval
All participants were enrolled after giving informed consent as approved by the Biomedical Institutional Review Board of the University of North Carolina at Chapel Hill or McGill University. For animal studies, all work followed the National Institutes of Health Guide for the Care and Use of Laboratory Animals,81 the Animal Research: Reporting of In Vivo Experiments guidelines,53 and adhered to the Institutional Animal Care and Use Committee at Duke University.
2.2. Clinical cohorts
Orofacial pain: prospective evaluation and risk assessment (OPPERA) N = 488 (283 control and 205 TMD): As described, the OPPERA observational prospective cohort study assessed individuals with TMD and 4 idiopathic pain conditions (headache, low back pain, irritable bowel syndrome, and widespread body pain) over a median follow-up period of 2.8 years. For study demographics, see Supplemental Table 1, http://links.lww.com/PR9/A254. Here, participants provided blood for RNA-seq analysis and were assigned to either case or pain-free control groups. The 16 differentially expressed miRNAs, their sequence, and genome location can be found in Supplemental Table 2, http://links.lww.com/PR9/A254.
Fibromyalgia syndrome N = 48 (24 control and 24 FMS): As described,70 people with clinically diagnosed FMS or people with no history of a pain or depression diagnosis provided blood for RNA-seq analysis. For study demographics, see Supplemental Table 3, http://links.lww.com/PR9/A254.
African-American CRASH N = 167 (81 low pain and 86 high pain): As described,32 this observational study enrolled Black individuals presenting to emergency department sites within 24 hours of MVC. Participants provided blood for RNA-seq analysis in the emergency department and completed evaluations after MVC. This included an assessment of overall pain severity using a verbal 0 to 10 numeric rating scale. For study demographics, see Supplemental Table 4, http://links.lww.com/PR9/A254. Here, participants were assigned to low pain (<7 of 10) or high pain (≥7) groups based on patient report within 24 hours of MVC.
All blood from clinical cohorts was collected in PAXgene RNA tubes at the time of enrollment, and total RNA was isolated using Qiacube column-based separation, Chemagen magnetic bead separation, or the PAXgene Blood miRNA Kit. Isolation method was controlled for in analyses.
2.3. RNA-seq
2.3.1. OPPERA
For small RNA-seq, 75 ng of total RNA was used to generate and sequence libraries on the Illumina HiSeq3000 platform, whereas 250 ng of total RNA was used for mRNA-seq. Deep-sequencing reads were aligned on the human genome version hg38/GRCh38. Quantification of miRNAs were obtained using ShortStack v3.8.52 against miRbase release 22 annotation with hg38 reference genome.
2.3.2. AA CRASH
Template libraries for small RNA sequencing were produced from 1 µg of total RNA, as previously reported.33,49 Reads were aligned on human genome version hg19, and quantification of miRNAs were obtained using RSEM.28
Additional details for both cohorts can be found in the Supplementary Files, http://links.lww.com/PR9/A254.
2.4. RT-qPCR
For miRNA, 10 ng of RNA were transcribed to complementary DNA using the miRCURY LNA RT Kit and Spike In Control Kits. The exogenous controls cel-miR-39-3p and UniSp6 were selected due to a lack of a known control miRNA between tissue types and to control for reverse transcription efficiency. Relative quantification was performed following miRCURY LNA SYBR Green qPCR Kit protocol. For mRNA, 200 ng of RNA were transcribed to complementary DNA using the High-capacity cDNA Reverse Transcription Kit. Relative quantification was performed following the TaqMan Fast Advanced Master Mix protocol. We selected ACTB and 18S as control genes because of their high and stable expression across tissue types. Relative expression of all genes was determined on a QuantStudio 5 Real-Time PCR System. Triplicate CTs were averaged and the log2foldchange of genes calculated using the 2−ΔΔCT method.36 For a complete list of kits and primers, see Supplemental Table 5, http://links.lww.com/PR9/A254.
2.5. Primary pain onset mouse model
Male and female C57BL/6 mice (N = 36) were purchased from The Jackson Laboratory. At 8 to 12 weeks of age, mice were randomly assigned to control or primary pain (PP) onset groups and blinded to the experimenter. The PP onset model incorporated clinically relevant factors (low catechol-O-methyltransferase (COMT) activity and stress). As previously described,79 forced or sham swim stress was conducted on days 1 to 3 (10 minutes on Day 1 and 20 minutes on Days 2 and 3). On Day 3 immediately following swim stress, mice received an intraperitoneal injection of the COMT inhibitor OR486 (30 mg/kg) or vehicle (dimethyl sulfoxide: ethanol: 0.9% sterile saline [5:2:3]). Before model induction, mice were handled and habituated to the testing environment for 3 days. Baseline assessments of plantar mechanical allodynia were determined using the von Frey up–down method (filaments 0.008-4 g).10 Mechanical allodynia was reassessed on Day 3, 3 hours following OR486 injection. The difference in 50% withdrawal threshold between Day 3 and baseline was calculated using established methods.74 Upon completion of behavioral experiments, inguinal subcutaneous WAT, intrascapular brown adipose tissue (BAT), muscle, and spinal cord were collected, flash frozen on dry ice, and kept at −80°C. Whole blood was collected in MiniCollect K3E EDTA Tubes, centrifuged for 15 minutes at 2000g and 4°C to remove blood cells. Total RNA was isolated using TRIzol according to the manufacturer's instructions and stored at −80°C.
2.6. Primary cell culture experiments
Adult mice of both sexes were euthanized and had their inguinal subcutaneous WAT tissue or dorsal root ganglia (DRG) removed aseptically at room temperature. Samples were incubated with collagenase (40 mg/mL) and dispase-II (200 mg/mL) diluted in Hank balanced salt solution at 37°C for 120 minutes. Digestion was halted by DMEM/F12/GlutaMAX-I base medium and homogenate passed through a 70-µm mesh filter. Cells were pelleted at 1000 rpm for 10 minutes at 4°C before resuspension in base medium. Recovered preadipocytes were plated on a T-75 flask and grown to 95% confluency with base media supplemented with 10% fetal bovine serum and 1X penicillin–streptomycin antibiotics. Cells were split following a standard 0.5% trypsin method and plated into 6-well tissue culture–treated plates. After reaching 100% confluency, preadipocytes were differentiated into adipocytes for 4 days with 0.5 mM IBMX, 1 µM rosiglitazone, 5 µM dexamethasone, and 0.5 µg/mL insulin in supplemented medium. Mature adipocytes were maintained for 4 days with only 0.5 µg/mL insulin in supplemented medium. Recovered DRG were plated in FBS and antibiotic supplemented media with 2% B27. No differentiation was necessary, and treatment occurred 2 days following seeding. All cultures were incubated at 37°C and 5% CO2.
For the norepinephrine experiment, each well of mature cells was treated with 1 µM of norepinephrine or vehicle for 3 hours. Medium was removed, and cells were washed with 2 mL of DPBS before collection with 1 mL of TRIzol as described in the TRIzol total RNA isolation protocol.
For the calcium imaging experiment, DRG cultures from Pirt-GCaMP3 mice were grown on a glass coverslip and transfected with a miR-374b mimic or nonsense negative control (scramble) in Polyplus reagents according to manufacturer's protocol; 48 hours following transfection, cells were imaged using a 20X lens at a wavelength of 340 nm in 3-second intervals. As described,57 the imaging buffer had a pH of 7.4 and consisted of 10 mM HEPES, 5 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 10 mM D-(+)-Glucose, and 140 mM NaCl; 200 nM of capsaicin in imaging buffer was used as an inducer. For each neuron, relative calcium influx was determined for each interval using ΔF/F0, which is the ratio of change in fluorescent intensity (Ft-F0) to baseline (F0). Additional measurements included percent responders (ie, % of cells in a trial showing ≥20% increase in fluorescence from baseline) and average max intensity.
2.7. Statistical analysis
Using the G*Power software,22 we calculated the minimum sample size required to detect effect sizes of 0.999, using α = 0.050 and power = 0.800. For RNA-seq experiments, differential expression of RNAs was assessed using moderated statistical tests with DESeq2 after filtering for expressed RNAs at total count of 1 in at least half of the samples.38 Each test was performed with the following covariables: gender, age, self-declared ancestry, and purification kit (MSM1 vs QIACUBE). Correction for multiple testing was performed using Benjamini and Hochberg false discovery rate.5 Heatmaps were created using QLUCore Omics Explorer. Representative volcano plots and bar graphs were created using GraphPad Prism 10. The mRNA target network was created using the STRING Database,63 and the reactome analysis output from the database was used to generate a pathway analysis summary figure.17
For quantitative reverse transcription polymerase chain reaction experiments, fold change values were log-transformed to equally weight under- and overexpression. Within each experiment, initial statistical comparisons were assessed with a single analysis of variance (ANOVA) test to reduce the probability of type 1 errors. Only in instances of interactions P < 0.10 were data subdivided for lower-order ANOVAs, followed by Tukey's Honestly Significant Difference (HSD) test for multiple comparisons between individual effects when appropriate. When no significant interactions were present, only the main treatment effects were assessed. Significance was determined as P < 0.05, 2 tailed. Statistics were calculated using JMP Pro 17. This was the same statistical procedure used to measure 50% paw withdrawal threshold and calcium imaging experiments, sans log transformation of the data. All data are available by reasonable request, and statistical outputs can be accessed in the Supplementary Files, http://links.lww.com/PR9/A254.
3. Results
3.1. Circulating miRNAs are associated with temporomandibular disorder case status
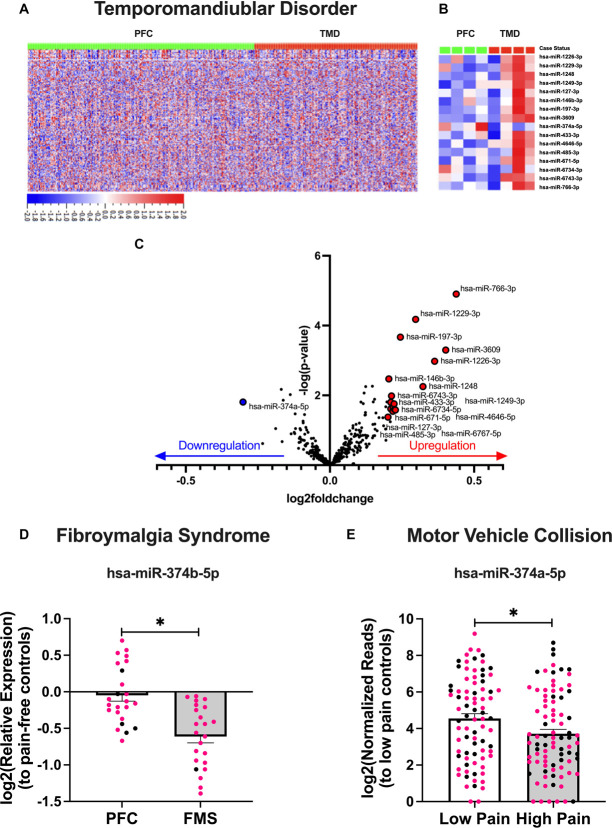
We first sought to define miRNA expression profiles associated with TMD. Using DESeq2 analysis, miRNA expression was determined for TMD cases vs pain-free controls (Fig. 1A). MiRNAs were considered to be significantly associated with TMD case status based on the strength of correlation (adj. P < 0.05) and magnitude of differential expression (log2foldchange > |0.20|) relative to controls. By comparing average expression between the groups, we identified 16 miRNAs associated with TMD case status (Figs. 1B and C; Supplemental Table 2, http://links.lww.com/PR9/A254). Of these, 15 miRNAs were upregulated, and 1 miRNA (miR-374a-5p) was downregulated in TMD cases. Interestingly, we found that miR-374a-5p was downregulated in TMD cases irrespective of race or gender, despite the miRNA originating from the X chromosome (Supplemental Table 2, http://links.lww.com/PR9/A254).
Figure 1.
RNA-seq and RT-qPCR analysis reveals miR-374 family downregulation in multiple chronic pain cohorts. Heatmaps show the relative expression profiles for all miRNAs in each of the 488 study participants (283 control [67% female and 33% male] and 205 TMD [75% female and 25% male]) (A), as well as for the 16 miRNAs associated with TMD case status in a subset of participants (B). A volcano plot illustrates the strength of correlation and log2foldchange of miRNAs upregulated (red) and downregulated (blue) in TMD cases compared with pain-free controls (PFC) (C). MiR-374 is downregulated in FMS patients compared with PFC, shown as a bar graph (N = 48; 24 PFC [79% female and 21% male] and 24 FMS [96% female and 4% male]) (D). MiR-374 is downregulated in those reporting high pain (≥7 of 10) than those reporting low pain (<7) following a motor vehicle collision, shown as a bar graph (N = 167; 81 low pain [63% female and 37% male] and 86 high pain [64% female and 36% male]) (E). For all bar graphs, data from women are shown in pink, while men are shown in black. Data are mean +SEM, and asterisk indicates a Tukey's HSD P < 0.05. FMS, fibromyalgia syndrome; HSD, Honestly Significant Difference; RT-qPCR, quantitative reverse transcription polymerase chain reaction;TMD, temporomandibular disorder.
3.2. miR-374 family downregulation is a signature of multiple pain conditions
We next tested whether any of the 16 miRNAs associated with TMD case status were consistently differentially expressed in plasma from patients with other painful conditions linked to heightened catecholamine tone: FMS and pain following MVC. In the FMS cohort, we determined that miR-374b-5p is significantly downregulated in FMS cases compared with pain-free controls (log2foldchange = −0.61, P < 0.005; Fig. 1D, Supplemental Table 3, http://links.lww.com/PR9/A254).
In the African-American CRASH cohort, we found that levels of miR-374a-5p were significantly lower in individuals with high vs low pain <24 hours following MVC (log2foldchange = −0.93, P < 0.03; Fig. 1E). Together, these data show that downregulation of miR-374 family members is a common signature of multiple pain conditions across time, age, race, and gender. Although miR-374a-5p is not expressed in rodents, miR-374b-5p only differs from miR-374a-5p by 2 base pairs and features 100% sequence homology between humans and mice, so they are predicted to regulate many of the same mRNA targets6 (Supplemental Figure 1, http://links.lww.com/PR9/A254). For these reasons, we selected miR-374b-5p for additional study in our PP onset mouse model.
3.3. White adipose tissue directly contributes to the downregulation of miR-374b-5p
Using our clinically relevant mouse model of PP illustrated in Figure 2A, we found that mechanical sensitivity is established rapidly in both sexes (P < 0.002; Fig. 2B). Plasma levels of miR-374b-5p were downregulated in female mice but not in male mice (sex × group P < 0.07, Tukey's HSD P < 0.005; Fig. 2C). This could be due to the early time point used in this experiment because no sex differences in downregulation were observed in the chronic, clinical studies. In addition, although the TMD and MVC cohorts were powered to detect sex differences, all 3 cohorts show a greater proportion of women enrolled (TMD: 75% women; FMS: 96% women; MVC: 64% women; Supplemental Tables 1, 3 and 4, http://links.lww.com/PR9/A254), whereas equal numbers of male and female mice were used in this experiment. Nevertheless, we continued searching for the source of this miRNA signature in both sexes.
Figure 2.
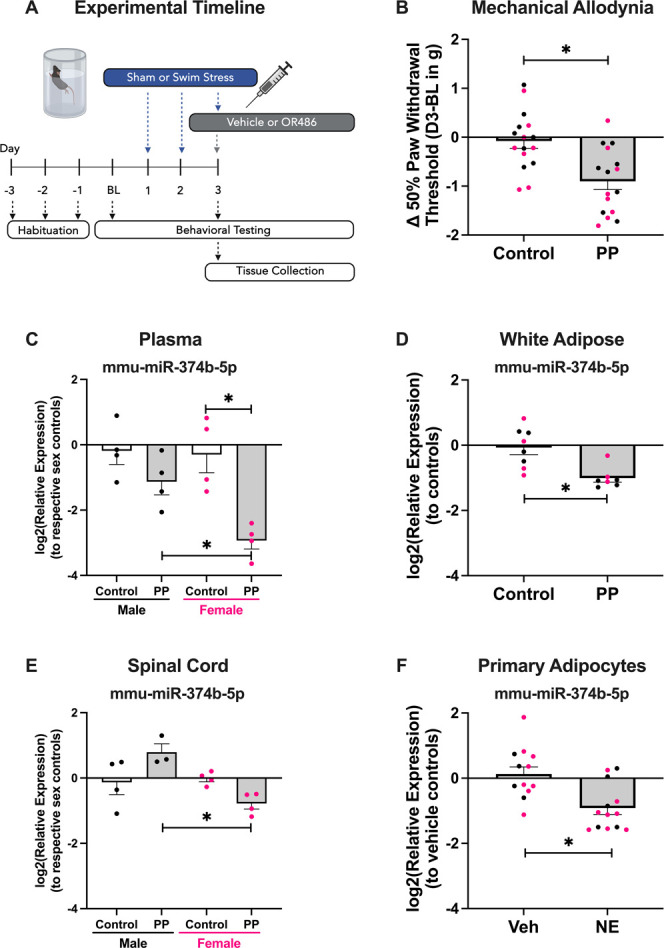
Early time point tissue- and sex-specific miR-374 expression levels in a mouse model of primary pain (PP) onset. The PP mouse model featuring 3 days of forced swim stress followed by delivery of the catechol-o-methyltransferase inhibitor OR486 on Day 3 is illustrated in (A). Evidence of significant reductions in paw withdrawal thresholds of PP mice compared with controls is demonstrated in (B). Compared with control mice of both sexes, only female PP mice had significant reductions in plasma levels of miR-374 (C). miR-374 is significantly downregulated in the white adipose tissue of both sexes compared with control mice (D) but is downregulated only in the spinal cords from female PP mice compared with male PP mice, consistent with the plasma expression data (E). By stimulating primary adipocytes cultured from male and female mice with the pan-adrenergic endogenous agonist norepinephrine (“NE”), the significant downregulation of miR-374 can be observed when compared with the vehicle (“Veh”) treatment control group (F). Data from female mice and their derived cultures are shown in pink, whereas male mice are shown in black. Data are mean +SEM, and asterisk indicates a Tukey's HSD P < 0.05. HSD, Honestly Significant Difference.
Based on the unique RNA expression patterns in target tissues,23,24,78 one miRNA can cause diverse regulation across tissue type. To determine possible sources and targets of miR-374b-5p downregulation during pain, we assessed the expression of miR-374b-5p in WAT, BAT, muscle, DRG, and spinal cord tissue in our model. We found that miR-374b-5p in WAT was downregulated in both sexes (P < 0.006; Fig. 2D). Consistent with the plasma findings, miR-374b-5p in spinal cord was downregulated in female mice only (sex × group P < 0.006, Tukey's HSD P < 0.007; Fig. 2E). miR-374b-5p was not differentially expressed in BAT, DRG, or muscle, despite miR-374's established role as a myomir66,73 (Supplemental Figure 1A-C, http://links.lww.com/PR9/A254). Therefore, we hypothesized a circuit whereby WAT packages miR-374b-5p to send to nervous system sites, such as the spinal cord, but this signal is lost during pain acquisition, particularly in female mice.
To test the potential adipose vs neuronal origin of miR-374b-5p, we stimulated primary adipocytes or DRG neurons cultured from male or female mice with the endogenous catecholamine norepinephrine. Stimulation of adipocytes with norepinephrine directly reduced the expression of miR-374b-5p compared with vehicle-treated cultures that came from male or female mice (Student's t P < 0.02; Fig. 2F). However, when this experiment was repeated in primary DRG cultures, norepinephrine had no effect (Supplemental Figure 1D, http://links.lww.com/PR9/A254). Together, these data show that adrenergic receptors in WAT are a key contributor to the common miR-374b-5p downregulation signature observed during pain in both sexes. We next aimed to investigate the mechanism behind miR-374 downregulation and pain development.
3.4. Circulating mRNA targets of miR-374a-5p
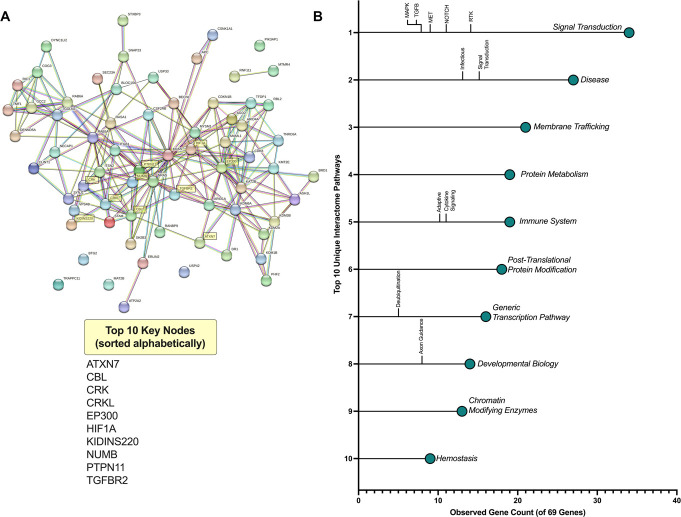
Using the TMD cohort samples, we performed mRNA-seq to analyze the mRNA targets with significant relationships to miR-374a-5p. We identified 69 predicted targets of miR-374a-5p that met the following criteria: (1) significant negative correlation with miR-374a-5p expression, (2) upregulation above 0.25 log2foldchange, and (3) Bonferroni corrected P < 3.6E-6. These targets were used as input in the STRING query, which builds protein networks from gene data sets63 (Fig. 3A). The top 10 of 171 unique pathways highlighted by our query reveal significant changes in signal transduction, immune regulation, neurodevelopment, and pain and inflammation pathways (Fig. 3B). The strength and false discovery rate of these enriched pathways can be found in Supplemental Table 6, http://links.lww.com/PR9/A254. Based on these results, we selected 10 mRNA targets (ATXN7, CBL, CRK, CRKL, EP300, HIF1A, KIDINS220, NUMB, PTPN11, and TGFBR2) for tissue-specific analysis in our mouse model by measuring the strength of their association, appearance in multiple pathways, and outputs from in silico predictive modeling tools such as STarMIR.55
Figure 3.
STRING predicts functional interactions among circulating mRNA targets of miR-374. STRING predicted functional interactions of the 69 predicted mRNA binding partners of miR-374 are visualized as a gene network, with the top 10 mRNA nodes highlighted (A). Colors correspond with mRNA hubs organized by cluster of function. Of the 171 gene pathways that feature 2 or more of the mRNA targets, the top 10 sorted by the number of genes in a pathway are shown, along with their significant subfamilies of gene pathways (B).
3.5. Tissue- and sex-specific expression of predicted miR-374 family targets
Using the same tissue samples that were used to measure miR-374b-5p expression, we performed an mRNA screen in both WAT and spinal cord because these were sites of miR-374b-5p dysregulation. MiRNAs canonically inhibit target mRNA expression, so we predicted that expression of miR-374b-5p and its mRNA targets would be negatively correlated. In WAT, where miR-374b-5p was downregulated in both sexes, PP females had significant upregulation of a gene cluster, including HIF1A, NUMB, and TGFBR2, compared with control females and downregulation of CRK-II compared with PP males (sex × group ×tissue ×gene P < 0.001, Tukey's HSD P < 0.02, Fig. 4A). In the spinal cord, where miR-374b-5p was upregulated in males only, PP males had significant downregulation of the genes ATXN7 and TGFBR2 compared with control males and females of either condition (Tukey's HSD P < 0.001, Fig. 4B). Sexually dimorphic RNA pathways have been reported previously in the context of stress in humans and rodents.23,27,29 These RNA changes may impact cell physiology.
Figure 4.
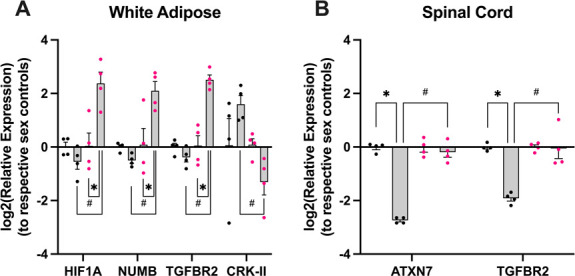
Tissue- and sex-specific mRNA targets negatively corelated with miR-374 expression. In the same mouse samples used to quantify tissue-specific expression of miR-374, we found a female-specific cluster of mRNA target overexpression in the white adipose tissue of female PP mice compared with female controls and male PP mice (HIF1A, NUMB, TGFBR2), whereas male PP mice showed overexpression of the mRNA target CRK-II compared with female PP mice (A). In the spinal cord, where male PP mice show significantly elevated levels of miR-374 compared with female PP mice, male mice show marked downregulation of the predicted mRNA binding partners ATXN7 and TGFBR2 (B). Data from female mice and their derived cultures are shown in pink, whereas male mice are shown in black. Data are mean +SEM, and asterisk is P < 0.05 by Tukey's HSD compared with same sex control, while # is P < 0.05 by Tukey's HSD compared with PP group of the opposite sex. PP, primary pain; HSD, Honestly Significant Difference.
3.6. miR-374 inhibits neuron activity
To test miR-374's effects on neuron function, DRG cultures from mice with a peripheral sensory neuron–specific fluorescent reporter of calcium (Pirt-GCaMP3) were transfected with a miR-374b-5p mimic or scramble control sequence 48 hours before a capsaicin challenge. Compared with naive or scramble transfected cultures, miR-374b-5p transfection resulted in a lower percentage of neuron activation (Group P < 0.01, Tukey's HSD P < 0.03, Fig. 5A) and average maximum fluorescent intensity (group P < 0.06, Tukey's HSD P ≤ 0.05, Fig. 5B). In addition, when relative calcium influx is plotted over time, the miR-374b-5p–treated group had a significantly lower area under the curve compared with controls (group P < 0.02, Tukey's HSD P < 0.05, Figs. 5C and D). Effects were greater in cultures from female mice than from male mice, but these comparisons were not significant (Supplementary Figure 3, http://links.lww.com/PR9/A254). These data indicate that miR-374b-5p overexpression inhibits neuron activity, supporting a purported role for miR-374 downregulation in primary pain onset.
Figure 5.
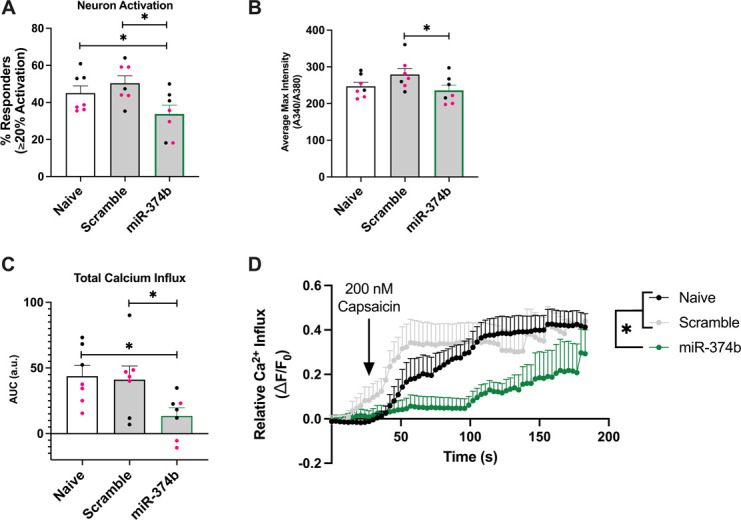
miR-374 inhibits neuron activity. Using primary DRG cultures from Pirt-GCaMP3 mice, measurements of calcium signaling following a capsaicin challenge, a proxy for neuron activity, were collected. Wells were either not transfected (naive) or transfected with a nonsense sequence negative control (scramble) or an mmu-miR-374b-5p mimic (miR-374b). miR-374b treatment significantly lowered the percentage of neurons that reached an activation threshold of ≥20% compared with baseline recordings (A) and reduced average maximum fluorescent intensity (B). miR-374b treatment group has significantly lower calcium signaling compared with the other treatments (C), illustrated in a tracing plot (D). Data from female-derived cultures are shown in pink, whereas male-derived cultures are shown in black. Data are mean +SEM, and asterisk indicates a Tukey's HSD P < 0.05. DRG, dorsal root ganglia; HSD, Honestly Significant Difference.
4. Discussion
Chronic primary pain conditions represent a heterogenous group of conditions that share genetic and environmental dispositions associated with heightened catecholamine tone. Although genetic risk is well-documented, the epigenetic regulation of genes in pain pathways remains understudied in these disorders. Here, we sought to identify miRNA determinants of pain. We conducted the largest RNA sequencing study to date in individuals with TMD and identified 16 miRNAs associated with case status, including miR-374. By analyzing that circulating mRNAs negatively correlated with miR-374 expression in TMD, we identified a diverse interactome, which includes pronociceptive pathways. Furthermore, we replicated or validated the downregulation of the miR-374 family (1) in a cohort of individuals with FMS, (2) those with pain following MVC, and (3) in a mouse model of primary pain onset. Although miR-374 downregulation has been linked to FMS previously,7,47 we are the first to implicate miR-374 in TMD and show replication across cohorts and species. We next identified WAT as a key source of miR-374 family downregulation, showed tissue- and sex-specific regulation of predicted mRNA target binding partners, and demonstrated neuron silencing by this miRNA. These data highlight the potential role of miRNAs in the pathophysiology of pain.
The miR-374 family is encoded on the X-inactivating site of the X chromosome in a region known to escape X inactivation,6 which can partially explain some of the observed sex differences reported here. In women, differential expression of genes that escape X chromosome inactivation predicts pain acquisition following trauma exposure.77 Furthermore, miR-374 downregulation has been previously linked to nervous system39,72,75 and immune cell dysfunction.15,54
We present evidence that WAT contributes to miR-374 downregulation in both sexes by observing the downregulation of miR-374 after adrenergic receptor activation in primary adipocytes but not in primary DRG cultures. Our in vitro calcium imaging experiment shows a novel, direct role for miR-374 in modulating neuron activity. In addition, we screened for predicted miR-374 targets in WAT and spinal cord because this was where altered miR-374 expression occurred. This revealed a female-specific cluster of genes in WAT (HIF1A, NUMB, TGFBR2), and a male-specific gene in WAT (CRK-II) and cluster in the spinal cord (ATXN7, TGFBR2). Hypoxia inducible factor 1 alpha (HIF1A) and NUMB endocytic adaptor protein (NUMB) expression is controlled by a positive-feedback loop through transforming growth factor-β (TGFB) signaling.43,80 This cluster, which has been previously reported as female specific in the context of neuropathic pain,19 is known to regulate proinflammatory signaling in immune cells.11,18,25 TGFB type II receptor (TGFBR2) is a key mediator of TGFB signaling, which has been shown to reduce nociception in the central nervous system.35 Thus, TGFBR2 downregulation in the spinal cord of males following overexpression of miR-374 at this site may be pronociceptive. However, TGFBR2 downregulation by miR-20a-5p promotes inflammation18 that is essential to the acute healing process.62
Of note, this work reveals 2 genes that, to our knowledge, have not previously been linked to pain: ATXN7 is a transcription factor related to neurodegeneration48 and CRK-II plays diverse roles in immune cell function,34 including promoting proinflammatory cytokine IL-2 secretion in T cells.30 These gene functions can partially explain the sexual dimorphism in mechanical hypersensitivity exhibited by our primary pain mice at early time points. However, women are more likely to present with CPPCs,40 highlighting the need for additional study in the sex differences underlying the chronification of pain.
The study does have some limitations and unanswered questions. First, the clinical cohorts were predominately female. Second, future work should investigate adipocyte-neuron crosstalk to determine how miR-374 might be trafficked between the sites, such as exosome packaging. Finally, interactome pathway analysis and literature search on our mRNA targets heavily implicate the immune system during pain acquisition, but these neuroimmune contributions need to be directly tested. Despite these limitations, this study remains the largest RNA-seq study across multiple pain cohorts and identifies a single miRNA family whose expression is consistent across a range of ages, races, or genders in humans, providing a foundation for future basic and translational studies into this miRNA and its mRNA targets.
5. Conclusion
In summary, downregulation of the miR-374 family is associated with multiple pain conditions linked to heightened catecholamine tone and regulates a diverse array of tissue- and sex-specific gene pathways related to pain and inflammation, likely through secretion by adipocytes. Future work will directly test these relationships and determine the ability of miR-374 overexpression to resolve CPPCs that lack etiology-based relief.
Disclosures
The authors have no conflict of interest to declare.
Appendix A. Supplemental digital content
Supplemental digital content associated with this article can be found online at http://links.lww.com/PR9/A254.
Supplementary Material
Acknowledgements
This work was supported by NINDS/NIH R01NS109541, NINDS R61/R33NS123753, and NIDCR/NIH R56DE025298 to Dr. Nackley, by NIAMS/NIH K01AR071504 and NIAMS/NIH R01AR081454 to Dr. Linnstaedt, by NIDCR/NIH U01DE017018 and CIHR SCA-145102 to Dr. Diatchenko, and by NINDS/NIH F31 NS130861 and NIGMS/NIH T32 GM133352 to Mr. Hernandez. The authors acknowledge the University of North Carolina BioSpecimen Facility Lab for blood/sample processing, storage, and disbursements.
Footnotes
Sponsorships or competing interests that may be relevant to content are disclosed at the end of this article.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's Web site (www.painrpts.com).
Contributor Information
Nathaniel P. Hernandez, Email: nh124@duke.edu.
Ashleigh Rawls, Email: ashleigh.rawls@duke.edu.
Jiegen Chen, Email: jiegen.chen@duke.edu.
Xin Zhang, Email: xin.zhang3@duke.edu.
Yaomin Wang, Email: yaomin.wang@duke.edu.
Xianglong Gao, Email: xianglong.gao@duke.edu.
Marc Parisien, Email: marc.parisien@mcgill.ca.
Mohamad Karaky, Email: mohamad.karaky@mcgill.ca.
Carolina Beraldo Meloto, Email: carol.meloto@mcgill.ca.
Francesca Montagna, Email: francesca.montagna@mcgill.ca.
Hong Dang, Email: dangh@email.unc.edu.
Yue Pan, Email: yue221@live.unc.edu.
Ying Zhao, Email: ying_zhao@med.unc.edu.
Samuel McLean, Email: smclean@aims.unc.edu.
Sarah Linnstaedt, Email: sarah_linnstaedt@med.unc.edu.
Luda Diatchenko, Email: luda.diatchenko@mcgill.ca.
References
- [1].Ajit SK. Circulating microRNAs as biomarkers, therapeutic targets, and signaling molecules. Sensors (Basel) 2012;12:3359–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Axtell MJ. ShortStack: comprehensive annotation and quantification of small RNA genes. RNA 2013;19:740–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Bak M, Silahtaroglu A, Moller M, Christensen M, Rath MF, Skryabin B, Tommerup N, Kauppinen S. MicroRNA expression in the adult mouse central nervous system. RNA 2008;14:432–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009;136:215–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Stat Methodol 1995;57:289–300. [Google Scholar]
- [6].Bian H, Zhou Y, Zhou D, Zhang Y, Shang D, Qi J. The latest progress on miR-374 and its functional implications in physiological and pathological processes. J Cell Mol Med 2019;23:3063–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Bjersing JL, Bokarewa MI, Mannerkorpi K. Profile of circulating microRNAs in fibromyalgia and their relation to symptom severity: an exploratory study. Rheumatol Int 2015;35:635–42. [DOI] [PubMed] [Google Scholar]
- [8].Bote ME, Garcia JJ, Hinchado MD, Ortega E. An exploratory study of the effect of regular aquatic exercise on the function of neutrophils from women with fibromyalgia: role of IL-8 and noradrenaline. Brain Behav Immun 2014;39:107–12. [DOI] [PubMed] [Google Scholar]
- [9].Carroll LJ, Holm LW, Hogg-Johnson S, Cote P, Cassidy JD, Haldeman S, Nordin M, Hurwitz EL, Carragee EJ, van der Velde G, Peloso PM, Guzman J, Bone and Joint Decade 2000-2010 Task Force on Neck Pain and Its Associated Disorders. Course and prognostic factors for neck pain in whiplash-associated disorders (WAD): results of the bone and joint decade 2000-2010 task force on neck pain and its associated disorders. Spine (Phila Pa 1976) 2008;33:S83–92. [DOI] [PubMed] [Google Scholar]
- [10].Chaplan SR, Bach FW, Pogrel JW, Chung JM, Yaksh TL. Quantitative assessment of tactile allodynia in the rat paw. J Neurosci Methods 1994;53:55–63. [DOI] [PubMed] [Google Scholar]
- [11].Chen Y, Gaber T. Hypoxia/HIF modulates immune responses. Biomedicines 2021;9:260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Cho YK, Son Y, Kim SN, Song HD, Kim M, Park JH, Jung YS, Ahn SY, Saha A, Granneman JG, Lee YH. MicroRNA-10a-5p regulates macrophage polarization and promotes therapeutic adipose tissue remodeling. Mol Metab 2019;29:86–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ciszek BP, O'Buckley SC, Nackley AG. Persistent catechol-O-methyltransferase-dependent pain is initiated by peripheral beta-adrenergic receptors. Anesthesiology 2016;124:1122–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Coelho M, Oliveira T, Fernandes R. Biochemistry of adipose tissue: an endocrine organ. Arch Med Sci 2013;9:191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Crossland RE, Norden J, Juric MK, Green K, Pearce KF, Lendrem C, Greinix HT, Dickinson AM. Expression of serum microRNAs is altered during acute graft-versus-host disease. Front Immunol 2017;8:308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Diatchenko L, Slade GD, Nackley AG, Bhalang K, Sigurdsson A, Belfer I, Goldman D, Xu K, Shabalina SA, Shagin D, Max MB, Makarov SS, Maixner W. Genetic basis for individual variations in pain perception and the development of a chronic pain condition. Hum Mol Genet 2005;14:135–43. [DOI] [PubMed] [Google Scholar]
- [17].Fabregat A, Sidiropoulos K, Viteri G, Forner O, Marin-Garcia P, Arnau V, D'Eustachio P, Stein L, Hermjakob H. Reactome pathway analysis: a high-performance in-memory approach. BMC Bioinformatics 2017;18:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Fu X, Qie J, Fu Q, Chen J, Jin Y, Ding Z. miR-20a-5p/TGFBR2 Axis affects pro-inflammatory macrophages and aggravates liver fibrosis. Front Oncol 2020;10:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Gomez K, Ran D, Madura CL, Moutal A, Khanna R. Non-SUMOylated CRMP2 decreases Na(V)1.7 currents via the endocytic proteins Numb, Nedd4-2 and Eps15. Mol Brain 2021;14:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hartung JE, Ciszek BP, Nackley AG. β2- and β3-adrenergic receptors drive COMT-dependent pain by increasing production of nitric oxide and cytokines. PAIN 2014;155:1346–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Jean-Toussaint R, Lin Z, Tian Y, Gupta R, Pande R, Luo X, Hu H, Sacan A, Ajit SK. Therapeutic and prophylactic effects of macrophage-derived small extracellular vesicles in the attenuation of inflammatory pain. Brain Behav Immun 2021;94:210–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kang H. Sample size determination and power analysis using the G*Power software. J Educ Eval Health Prof 2021;18:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kassam I, Wu Y, Yang J, Visscher PM, McRae AF. Tissue-specific sex differences in human gene expression. Hum Mol Genet 2019;28:2976–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kim-Hellmuth S, Aguet F, Oliva M, Munoz-Aguirre M, Kasela S, Wucher V, Castel SE, Hamel AR, Vinuela A, Roberts AL, Mangul S, Wen X, Wang G, Barbeira AN, Garrido-Martin D, Nadel BB, Zou Y, Bonazzola R, Quan J, Brown A, Martinez-Perez A, Soria JM, GTEx Consortium, Getz G, Dermitzakis ET, Small KS, Stephens M, Xi HS, Im HK, Guigo R, Segre AV, Stranger BE, Ardlie KG, Lappalainen T. Cell type-specific genetic regulation of gene expression across human tissues. Science 2020;369:eaaz8528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kueanjinda P, Roytrakul S, Palaga T. A novel role of numb as A regulator of pro-inflammatory cytokine production in macrophages in response to toll-like receptor 4. Sci Rep 2015;5:12784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kusuda R, Cadetti F, Ravanelli MI, Sousa TA, Zanon S, De Lucca FL, Lucas G. Differential expression of microRNAs in mouse pain models. Mol Pain 2011;7:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lee AG, Hagenauer M, Absher D, Morrison KE, Bale TL, Myers RM, Watson SJ, Akil H, Schatzberg AF, Lyons DM. Stress amplifies sex differences in primate prefrontal profiles of gene expression. Biol Sex Differ 2017;8:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 2011;12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Liao YY, Tsai HC, Chou PY, Wang SW, Chen HT, Lin YM, Chiang IP, Chang TM, Hsu SK, Chou MC, Tang CH, Fong YC. CCL3 promotes angiogenesis by dysregulation of miR-374b/VEGF-A axis in human osteosarcoma cells. Oncotarget 2016;7:4310–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Ling P, Yao Z, Meyer CF, Wang XS, Oehrl W, Feller SM, Tan TH. Interaction of hematopoietic progenitor kinase 1 with adapter proteins Crk and CrkL leads to synergistic activation of c-Jun N-terminal kinase. Mol Cell Biol 1999;19:1359–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Linnstaedt SD, Bortsov AV, Soward AC, Swor R, Peak DA, Jones J, Rathlev N, Lee DC, Domeier R, Hendry PL, McLean SA. CRHBP polymorphisms predict chronic pain development following motor vehicle collision. PAIN 2016;157:273–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Linnstaedt SD, Hu J, Liu AY, Soward AC, Bollen KA, Wang HE, Hendry PL, Zimny E, Lewandowski C, Velilla MA, Damiron K, Pearson C, Domeier R, Kaushik S, Feldman J, Rosenberg M, Jones J, Swor R, Rathlev N, McLean SA. Methodology of AA CRASH: a prospective observational study evaluating the incidence and pathogenesis of adverse post-traumatic sequelae in African-Americans experiencing motor vehicle collision. BMJ Open 2016;6:e012222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Linnstaedt SD, Rueckeis CA, Riker KD, Pan Y, Wu A, Yu S, Wanstrath B, Gonzalez M, Harmon E, Green P, Chen CV, King T, Lewandowski C, Hendry PL, Pearson C, Kurz MC, Datner E, Velilla MA, Domeier R, Liberzon I, Mogil JS, Levine J, McLean SA. MicroRNA-19b predicts widespread pain and posttraumatic stress symptom risk in a sex-dependent manner following trauma exposure. PAIN 2020;161:47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Liu D. The adaptor protein Crk in immune response. Immunol Cell Biol 2014;92:80–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Liu L, Zhu Y, Noë M, Li Q, Pasricha PJ. Neuronal transforming growth factor beta signaling via SMAD3 contributes to pain in animal models of chronic pancreatitis. Gastroenterology 2018;154:2252–65.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402–8. [DOI] [PubMed] [Google Scholar]
- [37].Lopez-Gonzalez MJ, Landry M, Favereaux A. MicroRNA and chronic pain: from mechanisms to therapeutic potential. Pharmacol Ther 2017;180:1–15. [DOI] [PubMed] [Google Scholar]
- [38].Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Manzine PR, Pelucchi S, Horst MA, Vale FAC, Pavarini SCI, Audano M, Mitro N, Di Luca M, Marcello E, Cominetti MR. microRNA 221 targets ADAM10 mRNA and is downregulated in Alzheimer's disease. J Alzheimers Dis 2018;61:113–23. [DOI] [PubMed] [Google Scholar]
- [40].Marques AP, Santo A, Berssaneti AA, Matsutani LA, Yuan SLK. Prevalence of fibromyalgia: literature review update. Rev Bras Reumatol Engl Ed 2017;57:356–63. [DOI] [PubMed] [Google Scholar]
- [41].McLean SA, Diatchenko L, Lee YM, Swor RA, Domeier RM, Jones JS, Jones CW, Reed C, Harris RE, Maixner W, Clauw DJ, Liberzon I. Catechol O-methyltransferase haplotype predicts immediate musculoskeletal neck pain and psychological symptoms after motor vehicle collision. J Pain 2011;12:101–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].McLean SA, Ulirsch JC, Slade GD, Soward AC, Swor RA, Peak DA, Jones JS, Rathlev NK, Lee DC, Domeier RM, Hendry PL, Bortsov AV, Bair E. Incidence and predictors of neck and widespread pain after motor vehicle collision among US litigants and nonlitigants. PAIN 2014;155:309–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].McMahon S, Charbonneau M, Grandmont S, Richard DE, Dubois CM. Transforming growth factor beta1 induces hypoxia-inducible factor-1 stabilization through selective inhibition of PHD2 expression. J Biol Chem 2006;281:24171–81. [DOI] [PubMed] [Google Scholar]
- [44].Mladenovic I, Supic G, Kozomara R, Dodic S, Ivkovic N, Milicevic B, Simic I, Magic Z. Genetic polymorphisms of catechol-O-methyltransferase: association with temporomandibular disorders and postoperative pain. J Oral Facial Pain Headache 2016;30:302–10. [DOI] [PubMed] [Google Scholar]
- [45].Mohamed-Ali V, Flower L, Sethi J, Hotamisligil G, Gray R, Humphries SE, York DA, Pinkney J. beta-Adrenergic regulation of IL-6 release from adipose tissue: in vivo and in vitro studies. J Clin Endocrinol Metab 2001;86:5864–9. [DOI] [PubMed] [Google Scholar]
- [46].Nackley AG, Tan KS, Fecho K, Flood P, Diatchenko L, Maixner W. Catechol-O-methyltransferase inhibition increases pain sensitivity through activation of both beta2- and beta3-adrenergic receptors. PAIN 2007;128:199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Nepotchatykh E, Caraus I, Elremaly W, Leveau C, Elbakry M, Godbout C, Rostami-Afshari B, Petre D, Khatami N, Franco A, Moreau A. Circulating microRNA expression signatures accurately discriminate myalgic encephalomyelitis from fibromyalgia and comorbid conditions. Scientific Rep 2023;13:1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Niewiadomska-Cimicka A, Hache A, Trottier Y. Gene deregulation and underlying mechanisms in spinocerebellar ataxias with polyglutamine expansion. Front Neurosci 2020;14:571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Nolet PS, Emary PC, Kristman VL, Murnaghan K, Zeegers MP, Freeman MD. Exposure to a motor vehicle collision and the risk of future back pain: a systematic review and meta-analysis. Accid Anal Prev 2020;142:105546. [DOI] [PubMed] [Google Scholar]
- [50].Okifuji A, Hare BD. The association between chronic pain and obesity. J Pain Res 2015;8:399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Orlova IA, Alexander GM, Qureshi RA, Sacan A, Graziano A, Barrett JE, Schwartzman RJ, Ajit SK. MicroRNA modulation in complex regional pain syndrome. J Transl Med 2011;9:195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Pauley KM, Cha S, Chan EK. MicroRNA in autoimmunity and autoimmune diseases. J Autoimmun 2009;32:189–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Percie du Sert N, Hurst V, Ahluwalia A, Alam S, Avey MT, Baker M, Browne WJ, Clark A, Cuthill IC, Dirnagl U, Emerson M, Garner P, Holgate ST, Howells DW, Karp NA, Lazic SE, Lidster K, MacCallum CJ, Macleod M, Pearl EJ, Petersen OH, Rawle F, Reynolds P, Rooney K, Sena ES, Silberberg SD, Steckler T, Wurbel H. The ARRIVE guidelines 2.0: updated guidelines for reporting animal research. BMJ Open Sci 2020;4:e100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Qian D, Chen K, Deng H, Rao H, Huang H, Liao Y, Sun X, Lu S, Yuan Z, Xie D, Cai Q. MicroRNA-374b suppresses proliferation and promotes apoptosis in T-cell lymphoblastic lymphoma by repressing AKT1 and wnt-16. Clin Cancer Res 2015;21:4881–91. [DOI] [PubMed] [Google Scholar]
- [55].Rennie W, Liu C, Carmack CS, Wolenc A, Kanoria S, Lu J, Long D, Ding Y. STarMir: a web server for prediction of microRNA binding sites. Nucleic Acids Res 2014;42:W114–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Rikard SM, Strahan AE, Schmit KM, Guy GP, Jr. Prevalence of pharmacologic and nonpharmacologic pain management therapies among adults with chronic pain-United States, 2020. Ann Intern Med 2023;176:1571–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Scarneo S, Zhang X, Wang Y, Camacho-Domenech J, Ricano J, Hughes P, Haystead T, Nackley AG. Transforming growth factor-β-activated kinase 1 (TAK1) mediates chronic pain and cytokine production in mouse models of inflammatory, neuropathic, and primary pain. J Pain 2023;24:1633–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Slade GD, Bair E, Greenspan JD, Dubner R, Fillingim RB, Diatchenko L, Maixner W, Knott C, Ohrbach R. Signs and symptoms of first-onset TMD and sociodemographic predictors of its development: the OPPERA prospective cohort study. J Pain 2013;14:T20–32.e323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Slade GD, Ohrbach R, Greenspan JD, Fillingim RB, Bair E, Sanders AE, Dubner R, Diatchenko L, Meloto CB, Smith S, Maixner W. Painful temporomandibular disorder: decade of discovery from OPPERA studies. J Dent Res 2016;95:1084–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Slade GD, Sanders AE, Ohrbach R, Bair E, Maixner W, Greenspan JD, Fillingim RB, Smith S, Diatchenko L. COMT diplotype amplifies effect of stress on risk of temporomandibular pain. J Dent Res 2015;94:1187–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Smith TJ, Hillner BE. The cost of pain. JAMA Netw Open 2019;2:e191532. [DOI] [PubMed] [Google Scholar]
- [62].Soliman AM, Barreda DR. Acute inflammation in tissue healing. Int J Mol Sci 2022;24:641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, Mering CV. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47:D607–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Thomas RF, Holt BD, Schwinn DA, Liggett SB. Long-term agonist exposure induces upregulation of beta 3-adrenergic receptor expression via multiple cAMP response elements. Proc Natl Acad Sci U S A 1992;89:4490–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Thomou T, Mori MA, Dreyfuss JM, Konishi M, Sakaguchi M, Wolfrum C, Rao TN, Winnay JN, Garcia-Martin R, Grinspoon SK, Gorden P, Kahn CR. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature 2017;542:450–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Umehara T, Kagawa S, Tomida A, Murase T, Abe Y, Shingu K, Ikematsu K. Body temperature-dependent microRNA expression analysis in rats: rno-miR-374-5p regulates apoptosis in skeletal muscle cells via Mex3B under hypothermia. Sci Rep 2020;10:15432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Valinezhad Orang A, Safaralizadeh R, Kazemzadeh-Bavili M. Mechanisms of miRNA-mediated gene regulation from common downregulation to mRNA-specific upregulation. Int J Genomics 2014;2014:970607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Vargas-Alarcon G, Fragoso JM, Cruz-Robles D, Vargas A, Vargas A, Lao-Villadoniga JI, Garcia-Fructuoso F, Ramos-Kuri M, Hernandez F, Springall R, Bojalil R, Vallejo M, Martinez-Lavin M. Catechol-O-methyltransferase gene haplotypes in Mexican and Spanish patients with fibromyalgia. Arthritis Res Ther 2007;9:R110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Vargovic P, Ukropec J, Laukova M, Cleary S, Manz B, Pacak K, Kvetnansky R. Adipocytes as a new source of catecholamine production. FEBS Lett 2011;585:2279–84. [DOI] [PubMed] [Google Scholar]
- [70].Verma V, Drury GL, Parisien M, Ozdag Acarli AN, Al-Aubodah TA, Nijnik A, Wen X, Tugarinov N, Verner M, Klares R, III, Linton A, Krock E, Morado Urbina CE, Winsvold B, Fritsche LG, Fors EA, Piccirillo C, Khoutorsky A, Svensson CI, Fitzcharles MA, Ingelmo PM, Bernard NF, Dupuy FP, Uceyler N, Sommer C, King IL, Meloto CB, Diatchenko L, HUNT-All In Pain. Unbiased immune profiling reveals a natural killer cell-peripheral nerve axis in fibromyalgia. PAIN 2022;163:e821–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Vetterlein A, Monzel M, Reuter M. Are catechol-O-methyltransferase gene polymorphisms genetic markers for pain sensitivity after all? - a review and meta-analysis. Neurosci Biobehav Rev 2023;148:105112. [DOI] [PubMed] [Google Scholar]
- [72].Waller R, Goodall EF, Milo M, Cooper-Knock J, Da Costa M, Hobson E, Kazoka M, Wollff H, Heath PR, Shaw PJ, Kirby J. Serum miRNAs miR-206, 143-3p and 374b-5p as potential biomarkers for amyotrophic lateral sclerosis (ALS). Neurobiol Aging 2017;55:123–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Wang W, Ma F, Zhang H. MicroRNA-374 is a potential diagnostic biomarker for atherosclerosis and regulates the proliferation and migration of vascular smooth muscle cells. Cardiovasc Diagn Ther 2020;10:687–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Wang Y, Kim SH, Klein ME, Chen J, Gu E, Smith S, Bortsov A, Slade GD, Zhang X, Nackley AG. A mouse model of chronic primary pain that integrates clinically relevant genetic vulnerability, stress, and minor injury. Sci Transl Med 2024;16:eadj0395. [DOI] [PubMed] [Google Scholar]
- [75].Wang Z, Liu Y, Shao M, Wang D, Zhang Y. Combined prediction of miR-210 and miR-374a for severity and prognosis of hypoxic-ischemic encephalopathy. Brain Behav 2018;8:e00835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Willemen HL, Huo XJ, Mao-Ying QL, Zijlstra J, Heijnen CJ, Kavelaars A. MicroRNA-124 as a novel treatment for persistent hyperalgesia. J Neuroinflammation 2012;9:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Yu S, Chen C, Pan Y, Kurz MC, Datner E, Hendry PL, Velilla MA, Lewandowski C, Pearson C, Domeier R, McLean SA, Linnstaedt SD. Genes known to escape X chromosome inactivation predict co-morbid chronic musculoskeletal pain and posttraumatic stress symptom development in women following trauma exposure. Am J Med Genet B Neuropsychiatr Genet 2019;180:415–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Yuan F, Pan X, Zeng T, Zhang YH, Chen L, Gan Z, Huang T, Cai YD. Identifying cell-type specific genes and expression rules based on single-cell transcriptomic atlas data. Front Bioeng Biotechnol 2020;8:350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Zhang X, Kanter K, Chen J, Kim S, Wang Y, Adeyemi C, O'Buckley SC, Nackley AG. Low catechol-O-methyltransferase and stress potentiate functional pain and depressive behavior, especially in female mice. PAIN 2020;161:446–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Zhu F, Li H, Long T, Zhou M, Wan J, Tian J, Zhou Z, Hu Z, Nie J. Tubular Numb promotes renal interstitial fibrosis via modulating HIF-1α protein stability. Biochim Biophys Acta Mol Basis Dis 2021;1867:166081. [DOI] [PubMed] [Google Scholar]
- [81].Zimmermann M. Ethical guidelines for investigations of experimental pain in conscious animals. PAIN 1983;16:109–10. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental digital content associated with this article can be found online at http://links.lww.com/PR9/A254.