Abstract
The Parkinson’s Families Project is a UK-wide study aimed at identifying genetic variation associated with familial and early-onset Parkinson’s disease (PD). We recruited individuals with a clinical diagnosis of PD and age at motor symptom onset ≤45 years and/or a family history of PD in up to third-degree relatives. Where possible, we also recruited affected and unaffected relatives. We analysed DNA samples with a combination of single nucleotide polymorphism (SNP) array genotyping, multiplex ligation-dependent probe amplification (MLPA), and whole-genome sequencing (WGS). We investigated the association between identified pathogenic mutations and demographic and clinical factors such as age at motor symptom onset, family history, motor symptoms (MDS-UPDRS) and cognitive performance (MoCA). We performed baseline genetic analysis in 718 families, of which 205 had sporadic early-onset PD (sEOPD), 113 had familial early-onset PD (fEOPD), and 400 had late-onset familial PD (fLOPD). 69 (9.6%) of these families carried pathogenic variants in known monogenic PD-related genes. The rate of a molecular diagnosis increased to 28.1% in PD with motor onset ≤35 years. We identified pathogenic variants in LRRK2 in 4.2% of families, and biallelic pathogenic variants in PRKN in 3.6% of families. We also identified two families with SNCA duplications and three families with a pathogenic repeat expansion in ATXN2, as well as single families with pathogenic variants in VCP, PINK1, PNPLA6, PLA2G6, SPG7, GCH1, and RAB32. An additional 73 (10.2%) families were carriers of at least one pathogenic or risk GBA1 variant. Most early-onset and familial PD cases do not have a known genetic cause, indicating that there are likely to be further monogenic causes for PD.
Subject terms: Clinical genetics, Parkinson's disease
Introduction
Parkinson’s disease (PD) is the second most common neurodegenerative condition after Alzheimer’s Disease (AD) and its prevalence is rapidly increasing1. PD becomes more common with advancing age, and both common and rare genetic variants can increase the risk of PD. Additionally, rare variants in approximately 20 genes have been reported to cause monogenic PD, although some of these genes have not been widely replicated, and some cause syndromes that are clinically and/or pathologically distinct from sporadic late-onset PD (sLOPD)2,3. First-degree relatives of PD patients have been estimated to have an approximately 2-fold increased risk of developing the condition compared to unrelated individuals4–6. A family history of PD and an early age at onset (AAO) are associated with an increased likelihood of carrying a pathogenic variant7,8. In unselected PD populations, rare causal variants account for around 1–2% of patients, whereas rare causal variants are found in around 5% of patients with familial PD and 20–40% of patients with an age of onset ≤309. Pathogenic variants in LRRK2, SNCA and VPS35 have been consistently identified in autosomal dominant PD, and biallelic pathogenic variants in PRKN, PINK1, DJ-1, and ATP13A2 in autosomal recessive PD. Recently, a single pathogenic variant in RAB32 has been identified in autosomal dominant families10,11. Rare variants in the Gaucher disease-causing GBA1 gene are an important genetic risk factor for PD, with approximately 5–10% of Northern European PD patients carrying single GBA1 variants12. For the vast majority of early-onset and familial PD cases, a known genetic cause has not been identified, suggesting either that there are additional monogenic forms to discover and/or that some PD families have more complex inheritance13,14.
Global efforts are underway to collect clinical and genetic data of diagnosed PD cases to elucidate the multifactorial pathogenesis of this complex disease15–20. However, a major obstacle to identifying and validating candidate monogenic variants is the availability of DNA samples from affected and unaffected family members. Classic linkage analysis and whole-exome/genome sequencing strategies have been used to show a causal relationship between genetic variation and monogenic PD, both of which require access to DNA samples from multiple family members across several generations21.
The Parkinson’s Families Project (PFP) is an ongoing UK-wide study aiming to identify new monogenic forms of PD by recruiting PD patients who are more likely to have a strong genetic contribution to the development of the condition, as well as their affected and unaffected relatives. UK-based studies of PD have previously shown that early-onset PD (EOPD) with age at symptom onset <45 years, as well as PD families with three or more affected members are particularly likely to carry a pathogenic mutation8. Here, we have built on this approach by recruiting early-onset and/or familial PD cases together with their genetically related family members to enable further genetic investigation of PD. The aims of the PFP study are: i) to build a cohort of families in which new monogenic variants may be discovered, and candidate pathogenic variants may be replicated through segregation studies; ii) to define the frequency and clinical features of pathogenic variants in known PD genes in a large-scale multicentre study; iii) to define a cohort of patients eligible for precision drug trials. PFP started recruitment in 2015 and will continue to do so until January 2030, with a target recruitment of over 1500 families, comprising over 3000 participants. Here, we describe the study protocol and the preliminary findings from our genetic screening of the first 718 families.
Results
Cohort description
We recruited 1035 participants from 840 families to the PFP study. Of these, we evaluated 959 individuals from 785 families using at least one of the genetic testing techniques described below. We then excluded 67 index cases from further analysis due to either a diagnosis of secondary parkinsonism (n = 3), atypical parkinsonism (n = 6), non-parkinsonism disorder (n = 2), failure to meet inclusion criteria (n = 30), missing clinical data (n = 14), consent withdrawal (n = 1), duplicated samples (n = 6), or failed genetic testing (n = 5) (Supplementary Fig. 1). Relatives of excluded index cases were also excluded (n = 16 relatives). In total, data were available from 871 eligible participants from 718 families.
Baseline demographics and PD family history for the 718 index cases included are shown in Table 1. 28.6% (205/718) of index cases have sporadic early-onset PD (sEOPD), 15.7% (113/718) have familial early-onset PD (fEOPD), and 55.7% (400/718) have familial late-onset PD (fLOPD). Using genetic principal component analysis (PCA) to define ancestry, 92.8% of all index cases were of European ancestry. In most families only the index case was recruited, but in 16% (n = 117) at least one additional relative was also recruited. Kinship analysis identified four families with cryptic relatedness. In all of these cases, individuals from the same extended family were independently recruited at different study sites. Across all families, we recruited 37 affected and 116 unaffected relatives for segregation studies. Of these multiplex families, 72% consisted of the index case and one single relative, while 20% had two relatives recruited, and 8% had three or more relatives recruited. For 7.9% of early-onset index cases, at least one parent was recruited. In all but one family, we recruited only a single additional affected relative.
Table 1.
Demographic characteristics of index cases by group
| sEOPD N = 205 | fEOPD N = 113 | fLOPD N = 400 | Total N = 718 | Adjusted P-value | |
|---|---|---|---|---|---|
| Sex (% Female) | 40 | 42.5 | 42.8 | 41.9 | 0.803 |
| Age at motor onset (Years, mean ± sd) | 37.7 (6.8) | 36.6 (7.4) | 62.9 (8.89) | 51.6 (15.1) | <2.2e−16a |
| Age at Diagnosis (Years, mean ± sd) | 42.1 (6.1) | 43.8 (8.4) | 65.4 (8.9) | 55.4 (13.9) | <2.2e−16a |
| Age at Assessment (Years, mean ± sd) | 48.8 (8.8) | 51.8 (10.2) | 68.8 (8.6) | 60.4 (13.0) | <2.2e−16b |
| Disease duration at assessment (Years, mean ± sd) | 11.0 (9.4) | 15.0 (12.4) | 5.9 (4.8) | 8.8 (8.6) | <2.2e−16c |
| Family history (%) | 0.751 | ||||
| No family history | 100 | 0 | 0 | 28.6 | – |
| One affected relative | 0 | 67.3 | 63.5 | 46 | – |
| Two affected relatives | 0 | 22.1 | 25.2 | 17.5 | – |
| Three or more affected relatives | 0 | 10.6 | 11.2 | 7.9 | – |
| Genetically determined Ancestry (%) | 0.032d | ||||
| African | 1.5 | 0 | 0.2 | 0.6 | – |
| American | 1 | 0.9 | 0 | 0.4 | – |
| Ashkenazi Jewish | 1.5 | 0.9 | 1.8 | 1.5 | – |
| Central Asian | 1 | 0 | 0.2 | 0.3 | – |
| East Asian | 0 | 0 | 0.2 | 0.1 | – |
| European | 86.8 | 93.8 | 94 | 91.9 | – |
| Finnish | 0.5 | 0 | 0 | 0.1 | – |
| Middle East | 0 | 0 | 0.5 | 0.3 | – |
| South-Asian | 6.3 | 1.8 | 2.8 | 3.6 | – |
| Complex Admixture | 0.5 | 0 | 0 | 0.1 | – |
| Unknown | 1 | 2.7 | 1 | 1 | – |
| Self-reported parental consanguinity (%) | 2.2 | 0.9 | 0.8 | 1.2 | 0.366 |
Categorical variables tested with Chi-square or Fisher’s exact test as appropriate, with post-hoc pairwise comparisons. Continuous variables tested with the Kruskal–Wallis test, followed by pairwise comparisons with the Wilcoxon rank sum test. P-values were FDR-adjusted. Pairwise comparisons: asEOPD vs fLOPD: ****, fEOPD vs fLOPD: ****, sEOPD vs fEOPD: ns. bsEOPD vs fLOPD: ****, fEOPD vs fLOPD: ****, sEOPD vs fEOPD: *. csEOPD vs fLOPD: ****, fEOPD vs fLOPD: ****, sEOPD vs fEOPD: **. dEuropean sEOPD vs fLOPD: *, fEOPD vs fLOPD: *, sEOPD vs fEOPD: ns; all other ancestries: ns.
sEOPD sporadic early-onset PD, fEOPD familial early-onset PD, fLOPD familial late-onset PD, ns not significant.
Bold font indicates statistical significance (P-value <0.05).
Identification of PD-causing variants
Following completion of genetic analysis by a combination of Illumina’s NeuroChip genotyping array (NCA), multiplex ligation-dependent probe amplification (MLPA), whole-genome sequencing (WGS) and/or next-generation targeted sequencing (NGS), we identified known PD-causing variants in 69 families (9.6%, 69/718; Supplementary Table 1). NCA contains probes for hundreds of PD relevant rare variants. We tested the performance of these probes against whole-genome or targeted sequencing (Supplementary Methods and Supplementary Table 2) and found that NCA-derived genotypes showed 95.2% concordance with sequenced-derived genotypes, indicating a high level of accuracy for most probes. Poorly performing probes were excluded from subsequent analyses.
Rare pathogenic variants in autosomal dominant genes explained PD occurrence in 38 families (5.3%; Table 2 and Supplementary Table 3). Mutations in LRRK2 were the most commonly identified genetic cause, accounting for PD in 30 families (4.2%). The LRRK2 G2019S variant was identified in all but two of these families. The majority (n = 23; 76.7%) of the LRRK2 mutation-positive families had fLOPD. Interestingly, five LRRK2 G2019S carriers had sEOPD, reflecting incomplete penetrance and the likely presence of disease modifiers. Other pathogenic dominant variants identified include two cases of heterozygous SNCA gene duplication and three cases with expanded trinucleotide repeats in ATXN2. SNCA copy number variants (CNVs) are typically associated with fEOPD22, but both these cases presented as sEOPD. We have also identified pathogenic missense variants in VCP, GCH1 and RAB32. The RAB32 p.Ser71Arg here identified has recently been reported in several autosomal dominant PD families, and has been shown to activate LRRK2 kinase in vitro10,11.
Table 2.
Clinical features of mutation carriers vs. mutation-negative index cases
| LRRK2 | PRKN | GBA1 a | Mutation-negative b | LRRK2 | PRKN | GBA1 | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| N = 30 | N = 26 | N = 70 | N = 431 | Beta (95% CI) | P-value | Beta (95% CI) | P-value | Beta (95% CI) | P-value | |
| Age at onset (mean ± sd) | 57.7 (13.4) | 28.3 (8.7) | 49.1 (14.5) | 52.3 (15.0) | 3.98 (-0.87, 8.84) | 0.108 | −13.3 (-18.8, -7.82) | 2.50E−06 | −2.2 (−5.46, 1.13) | 0.197 |
| Motor features (mean ± sd) | ||||||||||
| MDS-UPDRS Part III | 25.2 (14.7) | 29.7 (16.5) | 27.2 (14.7) | 26.5 (17.4) | −0.85 (−7.67, 5.97) | 0.806 | −4.5 (−13.0, 3.90) | 0.290 | −0.47 (−5.53, 4.58) | 0.855 |
| Motor Severity Score | 6.8 (9.9) | 1.8 (1.7) | 6.2 (5.8) | 6.3 (6.7) | 0.33 (−2.68, 3.35) | 0.829 | −4.34 (−7.51, −1.16) | 0.008 | −0.19 (−2.29, 1.90) | 0.856 |
| Motor subtype (%) | ||||||||||
| Tremor-dominant | 18.2 | 41.2 | 24.4 | 39.1 | ||||||
| PIGD-dominant | 77.3 | 58.8 | 65.8 | 52.1 | 1.14 (0.00, 2.27) | 0.049 | −0.46 (−1.67, 0.75) | 0.457 | 0.68 (−0.12, 1.49) | 0.096 |
| Intermediate | 4.5 | 0 | 9.8 | 8.8 | 0.03 (−2.22, 2.29) | 0.976 | NA | NA | 0.58 (−0.69, 1.84) | 0.371 |
| Hoehn and Yahr stage (%) | ||||||||||
| 0–1.5 | 43.5 | 16.7 | 31.8 | 38.2 | ||||||
| 2 or 2.5 | 30.4 | 38.9 | 40.9 | 37.8 | −0.38 (−1.43, 0.66) | 0.475 | 0.06 (−1.57, 1.70) | 0.939 | −1.79 (−0.62, 0.96) | 0.673 |
| 3+ | 26.1 | 44.4 | 27.3 | 24 | −0.11 (−1.23, 1.00) | 0.843 | 0.71 (−1.12, 2.55) | 0.445 | 0.17 (−0.77, 1.10) | 0.726 |
| Motor complications (%) | ||||||||||
| Dyskinesias | 43.5 | 35.3 | 27.5 | 26 | 1.06 (0.13, 1.96) | 0.022 | −0.94 (−2.38, 0.36) | 0.175 | −0.20 (−1.08, 0.61) | 0.646 |
| Motor fluctuations | 66.7 | 56.2 | 43.6 | 43.4 | 1.22 (0.25, 2.28) | 0.016 | −1.63 (−3.24, −0.15) | 0.037 | −0.29 (−1.12, 0.50) | 0.480 |
| Off dystonia | 27.3 | 35.3 | 20 | 24 | 0.58 (−0.53, 1.57) | 0.274 | −1.15 (−2.58, 0.14) | 0.095 | −0.52 (−1.51, 0.35) | 0.267 |
| Motor aspects of daily living (mean ± sd) | 13.1 (7.8) | 9.8 (7.2) | 13.7 (8.4) | 13.5 (9.2) | −0.12 (−3.24, 3.00) | 0.939 | −9.1 (−12.7, −5.5) | 1.14E−06 | −0.49 (−2.69, 1.70) | 0.658 |
| Autonomic dysfunction (%) | ||||||||||
| Orthostatic hypotension | 42.9 | 38.5 | 46.2 | 48.6 | −0.21 (−1.01, 0.57) | 0.607 | −0.89 (−1.83, 0.00) | 0.056 | −0.13 (−0.67, 0.41) | 0.642 |
| Constipation | 50 | 34.6 | 65.2 | 49.1 | −0.05 (−0.83, 0.74) | 0.906 | −0.82 (−1.83, 0.11) | 0.092 | 0.72 (0.17, 1.29) | 0.012 |
| Urinary dysfunction | 64.3 | 48 | 64.6 | 64.2 | −0.07 (−0.87, 0.78) | 0.865 | −0.82 (−1.80, 0.12) | 0.090 | 0.03 (−0.53, 0.61) | 0.917 |
| REM sleep behaviour disorder (%) | 46.4 | 48 | 54.7 | 39.9 | 0.32 (−0.45, 1.09) | 0.412 | −0.46 (−1.40, 0.45) | 0.323 | 0.58 (0.02, 1.13) | 0.041 |
| Neuropsychiatric symptoms (%) | ||||||||||
| Apathy | 26.1 | 27.8 | 27.7 | 30.7 | −0.12 (−1.17, 0.81) | 0.813 | −0.84 (−2.11, 0.29) | 0.165 | −0.22 (−0.96, 0.45) | 0.528 |
| Depression | 4.3 | 11.1 | 10.4 | 14.8 | −1.19 (−4.09, 0.43) | 0.253 | −0.91 (−2.85, 0.53) | 0.270 | −0.56 (−1.69, 0.37) | 0.281 |
| Anxiety | 17.4 | 27.8 | 16.7 | 20.8 | −0.15 (−1.42, 0.88) | 0.796 | −0.03 (−1.31, 1.12) | 0.958 | −0.38 (−1.27, 0.40) | 0.368 |
| Dopamine dysregulation syndrome | 23.8 | 16.7 | 21.3 | 13.2 | 0.96 (−0.23, 2.01) | 0.088 | −0.58 (−2.18, 0.73) | 0.417 | 0.46 (−0.39, 1.25) | 0.266 |
| Hallucinations | 17.4 | 11.1 | 27.1 | 15.5 | 0.27 (−1.01, 1.33) | 0.637 | −1.36 (−3.36, −0.14) | 0.112 | 0.61 (−0.17, 1.33) | 0.110 |
| MoCA score (mean ± sd) | 26.7 (2.9) | 26.6 (2.6) | 25.5 (3.1) | 26.3 (3.4) | 0.53 (−0.85, 1.92) | 0.451 | 0.02 (−1.57, 1.62) | 0.976 | −0.87 (−1.73, −0.02) | 0.045 |
NA not applicable.
aExcludes cases not investigated with WGS (n = 3) and GBA1 mutations that coexist with pathogenic mutations in LRRK2 (n = 1), PRKN biallelic (n = 2), PRKN monoallelic (n = 3), PINK1 monoallelic (n = 1) and GCH1 (n = 1). Variants classified as ‘severity unknown’ are included.
bExcludes mutation-negative cases not investigated with both WGS and MLPA (n = 122), carriers of GBA1 variants and of monoallelic pathogenic PRKN and PINK1 mutations. Mutation carriers vs. mutation-negative PD were compared with linear, logistic or multinomial regression as appropriate, after adjustment for sex, age and disease duration (except age at onset, which was adjusted only for sex and disease duration, and motor severity, which was adjusted only for sex and age). Significance level set at <0.05. MDS-UPDRS items were used to define the following clinical features: dyskinesias (items 4.1 + 4.2 > 0); motor fluctuations (items 4.3 + 4.4 + 4.5 > 0); off-dystonia (item 4.6 > 0); orthostatic hypotension (item 1.12 > 0); constipation (item 1.11 > 0); urinary dysfunction (item 1.10 > 0); apathy (item 1.5 > 0); depression (item 1.3 > 1); anxiety (item 1.4 > 1), impulse control disorder (item 1.6 > 0); hallucinations (item 1.2 > 0 and <4). Motor severity scores are MDS-UPDRD part III scores divided by disease duration in years. Motor subtypes were defined according to Stebbins et al.82. Motor aspects of daily living scores are the sum of MDS-UPDRS part II items. REM sleep behaviour disorder was defined as RBDSQ score >5.
Bold font indicates statistical significance (P-value <0.05).
Pathogenic biallelic autosomal recessive variants were identified in 31 families (4.3%; Table 2 and Supplementary Table 3). Compound heterozygous or homozygous pathogenic variants in PRKN were the second most common cause of monogenic PD, accounting for PD in 26 families (3.6%). All biallelic PRKN index cases presented with early-onset PD, and 15 (57.7%) cases did not have a family history of PD. Consanguinity was reported in 4.3% of biallelic PRKN mutation carriers compared to 1.2% of early-onset PD cases without mutations (P = 0.298, Fisher’s Exact test). PINK1 homozygous pathogenic variants were identified in two families. The remaining biallelic recessive cases carried homozygous variants in PNPLA6, and compound heterozygous variants in PLA2G6 and SPG7.
Supplementary Table 1 lists genetic findings of all participants with known PD-causing variants and their relatives. We further identified 23 index cases with a single heterozygous pathogenic variant in either PRKN or PINK1, of whom 14 were fully investigated with WGS and MLPA (Supplementary Table 4). A list of all the unique variants identified (n = 72, including GBA1 risk variants) is provided in Supplementary Table 5.
Demographic characteristics of pathogenic variant carriers
As expected, pathogenic variants in PD-related genes were more common in participants with an early AAO, defined by symptom onset before age 45 years (Supplementary Table 6). We identified a monogenic cause in 12.9% (41/318) of patients with EOPD (≤45 years) compared to 7% (28/400) of patients with LOPD (χ2 = 7.1, df = 1, 95% confidence interval [CI] = 0.01–0.10, P = 0.008, Chi-squared test). Moreover, when looking into juvenile and young onset PD (≤35 years), a monogenic cause was present in 28.1% (27/96) of patients with symptom onset ≤35, compared to 6.7% (42/622) of patients with AAO > 35 (χ2 = 43.7, df = 1, 95% CI = 0.12–0.31, P = 3.76e−11, Chi-squared test). In particular, 26% (25/96) of patients with symptom onset ≤35 carried homozygous or compound heterozygous mutations in recessive genes, compared to only 0.96% in patients with onset >35 (6/622; P = 2.2e−16, Fisher’s exact test). Among patients with a family history of PD, dominant mutations were more frequent than biallelic recessive mutations (6.0% vs 2.5%; χ2 = 6.9, df = 1, 95% CI = 0.01–0.06, P = 0.009, Chi-squared test). Furthermore, each additional affected family member increased the odds of having a dominant mutation by a factor of 1.6, after adjusting the logistic regression for sex and age at symptom onset (95% CI = 1.21–2.02, P = 5.34e−04). The majority of pathogenic mutation carriers were of European ancestry, except for one participant of South East Asian ancestry with homozygous pathogenic mutations in PINK1 (Y258*), and four participants of Ashkenazi Jewish ancestry (three heterozygous LRRK2 G2019S carriers and one homozygous PNPLA6 P1297S carrier).
Clinical features of LRRK2 mutation carriers
Among LRRK2 mutation carriers, 83.3% (25/30) had a positive family history of PD, and the majority experienced symptom onset >45 years (76.7%, 23/30). Demographic characteristics of LRRK2 mutation carriers are described in Supplementary Table 7. Clinical features of PD-LRRK2 mutation carriers compared to mutation-negative index cases (i.e., no identified dominant or biallelic/monoallelic recessive variants in PD-related genes or GBA1) are presented in Table 2. Age at onset was similar in PD-LRRK2 and mutation-negative PD (57.7 ± 13.4 vs 52.3 ± 15.0 years; r = 0.09, P = 0.063, Mann–Whitney U test). While the majority of LRRK2 mutation carriers were European, 10% were of Ashkenazi Jewish ancestry compared to 0.91% of mutation-negative PD (P = 0.006, Fisher’s exact test). We compared the PD motor subtype in PD-LRRK2 and mutation-negative PD using multinomial logistic regression, adjusted for sex, age, and disease duration. PD-LRRK2 cases had an increased odds ratio (OR) of having a postural instability and gait difficulty (PIGD)-dominant compared to a tremor-dominant motor subtype (OR = 3.1, 95% CI = 1.00–9.71, P = 0.049). There was no difference in motor severity, as measured by MDS-UPDRS part III, between PD-LRRK2 and mutation-negative PD (25.2 ± 14.7 vs 26.5 vs 17.4, respectively; r = 0.01, P = 0.821, Mann–Whitney U test). Regarding motor complications, motor fluctuations were more common in PD-LRRK2 (Chi-squared test: χ2 = 4.2, df = 1, 95% CI = 0.02–0.44, P = 0.039) and there was also a tendency towards a higher rate of dyskinesia in PD-LRRK2 (Chi-squared test: χ2 = 3.2, df = 1, 95% CI = −0.03–0.38, P = 0.071). We then adjusted for sex, age, and disease duration in a logistic regression model, which confirmed the association between LRRK2 mutations and dyskinesia and motor fluctuations (dyskinesia: OR = 2.9, 95% CI = 1.14–7.12, P = 0.022; motor fluctuations: OR = 3.4, 95% CI = 1.29–9.75, P = 0.016). No other comparisons of clinical features between PD-LRRK2 and mutation-negative PD cases reached significance.
Clinical features of biallelic PRKN mutation carriers
The demographic and clinical features of biallelic PRKN mutation carriers are summarised in Supplementary Table 7 and Table 2, respectively. 42.3% (11/26) of biallelic PRKN mutation carriers had a positive family history of PD. The majority had symptom onset ≤35 years (80.8%, 21/26), while 23.1% (6/26) had juvenile PD (i.e., symptom onset ≤21). Accordingly, biallelic PRKN mutation carriers had a significantly earlier age of symptom onset compared to mutation-negative PD (28.3 ± 8.7 vs 52.3 ± 15.0 years; r = 0.34, P = 5.49e−13, Mann–Whitney U test). Disease duration was also significantly longer at study assessment (21.7 ± 14.0 vs 8.37 ± 8.31; r = 0.25, P = 8.34e−08, Mann–Whitney U test). All biallelic PRKN mutation carriers were of European ancestry. There were no differences in motor scores or motor subtypes between groups. However, given that biallelic PRKN mutation carriers had significantly longer disease duration, we adjusted motor severity to disease duration by dividing MDS-UPDRS part III scores at assessment by disease duration. Biallelic PRKN mutation carriers had significantly lower adjusted motor severity scores compared to PD without a monogenic cause (1.8 ± 1.7 vs 6.3 ± 6.7; r = 0.26, P = 1.76e−05, Mann–Whitney U test), indicating a slower rate of motor symptom progression. Concordantly, individuals with biallelic PRKN mutations performed better in motor aspects of activities of daily living, as measured by MDS-UPDRS part II, after adjusting for confounding variables including disease duration (linear regression: beta = −9.1, standard error (sd) = 1.8, P = 1.14e−06). Biallelic PRKN carriers had increased rate of motor fluctuations at baseline (56.2% vs 30.9%; χ2 = 4.5, df = 1, 95% CI = 0.00–0.50, P = 0.0339, Chi-squared test). However, biallelic PRKN mutations were associated with a reduced likelihood of experiencing motor fluctuations compared to mutation-negative PD after adjusting for disease duration (OR = 0.20, 95% CI = 0.04–0.91, P = 0.0369). No other clinical features differentiated biallelic PRKN mutation carriers from mutation-negative PD cases. In addition to biallelic PRKN mutation carriers, there were 12 index cases fully investigated with WGS and MLPA for whom only a single pathogenic mutation could be found. Interestingly, monoallelic PRKN pathogenic variant carriers were more similar to mutation-negative PD than to biallelic PRKN pathogenic variant carriers (Supplementary Table 8).
Demographic and clinical features of pathogenic and risk GBA1 variant carriers
We screened GBA1 for rare pathogenic Gaucher disease (GD)-causing variants and common PD risk variants. We identified 73 carriers of GBA1 variants and an additional eight index cases with concomitant pathogenic mutations in LRRK2, PRKN, PINK1 and GCH1 (Supplementary Table 9). 3.7% (3/81) of GBA1 carriers were of Ashkenazi Jewish ancestry. After excluding GBA1 carriers with coexistent pathogenic mutations in other PD-related genes (n = 8) and those who did not complete WGS (n = 3), 70 individuals were available for subsequent analysis (Table 2). A family history of PD was present in 71.4% (50/70) of GBA1 mutation carriers, and in 93.9% of these families affected individuals were present in at least two generations. 50% (35/70) of GBA1 mutation carriers had motor symptom onset ≤45 years, in line with previous studies suggesting earlier symptom onset in GBA1 mutation carriers23,24. Compared to mutation-negative PD, GBA1 mutation carriers had decreased MoCA scores after adjusting for age at assessment and disease duration (beta = −0.87, sd = 0.43, P = 0.045). In addition, the odds of REM sleep behaviour disorder, which is often a precursor of cognitive decline and dementia in PD25–27, were significantly increased in GBA1 carriers (OR = 1.79, 95% CI = 1.02–3.11, P = 0.041). We also found an association between constipation and GBA1 status (OR = 2.05, 95% CI = 1.18–3.64, P = 0.012), which is interesting as constipation has also been found to be predictive of cognitive decline in PD28,29. Finally, the frequency of hallucinations, which again have been shown to be a risk factor for dementia in PD30, was increased in GBA1 mutation carriers (27.1% vs 15.5%; χ2 = 3.9, df = 1, 95% CI = −0.02–0.25, P = 0.049), albeit the association of GBA1 mutations with hallucinations was not significant after correcting for confounders. Interestingly, when analysing the effect of GBA1 variants by their severity31 (Supplementary Table 10), the association with decreased MoCA scores was only observed in mutations classified as “severe” (beta = −1.49, sd = 0.72, P = 0.039).
Polygenic risk score analysis
Despite the significant enrichment of cases with early onset and/or family history of PD, who carry an increased a priori probability of a positive genetic finding, a monogenic cause for PD was not identified in 90.4% of families, of which 66.4% completed WGS and MLPA. A further 11.9% of cases carried a GBA1 variant that significantly increases the risk of PD. We therefore wondered if other seemingly monogenic cases could be the result of increased risk of PD due to the cumulative effect of several risk variants, each contributing only a small fraction to the overall PD risk32. To answer this question, we calculated the PD polygenic risk score (PD-PRS) for each individual, but found that unit changes in the z-transformed PD-PRS were not positively or negatively associated with PD mutation status (Supplementary Fig. 2a; OR = 1.07, 95% CI = 0.82–1.41, P = 0.624). Looking in more detail at the mutation-negative group, we found an association between the PD-PRS and a family history of PD specifically in cases with early onset (Supplementary Fig. 2b; OR = 1.41, 95% CI = 1.02–1.94, P = 0.036), which suggests that a subset of mutation-negative early onset PD families might have pseudo-autosomal inheritance due to a shared increased load of common risk variants.
Discussion
The UK-based PFP study consists of early-onset and familial PD cases and their relatives, with a collection of detailed demographic, clinical, lifestyle, and environmental data, as well as biological samples for genetic testing. It aims to provide support for monogenic PD gene discovery while contributing to the characterisation of genotype-phenotype relationships of known monogenic forms of PD. The first phase of genetic screening for mutations in genes known to cause PD has been successfully completed for 718 families. Pathogenic causal mutations have been identified in 69 families, providing an overall diagnostic yield of 9.6% (13.8% in EOPD and 6% in fLOPD). This is in line with previous studies that found pathogenic mutations in known PD-related genes account for 5–10% of familial PD cases33.
Unsurprisingly, mutations in LRRK2 were the most common cause of monogenic PD and were more frequent in the fLOPD group, although 16.7% of cases did not report a family history of PD and age of motor symptom onset ranged between 34 and 80 years. Age of symptom onset for LRRK2 is reported to average 58–61 years, yet it frequently varies even within the same family34, probably reflecting the presence of disease-modifying genetic factors35,36. In addition, the seemingly sporadic nature of LRRK2-associated PD in many individuals is also likely due to its incomplete penetrance, which has been extensively described elsewhere34,37,38. While clinical characteristics are largely indistinguishable from idiopathic PD34, it has been suggested that LRRK2-associated PD has a milder phenotype and slower disease progression39. We found that LRRK2 mutations were associated with an increased risk of dyskinesia and motor fluctuations compared to mutation-negative PD. This is in line with a large meta-analysis reporting an increased likelihood of motor complications in LRRK2 G2019S carriers40. However, other studies comparing LRRK2-PD with idiopathic PD did not find an association between LRRK2 status and incidence of dyskinesias41,42.
Biallelic mutations in PRKN were the second most frequently identified cause of monogenic PD and were present in 3.6% of families, all with EOPD. These individuals had an earlier age at symptom onset compared to mutation-negative PD cases, consistent with findings reported elsewhere7,8,43. We also observed lower MDS-UPDRS motor severity scores after adjusting for disease duration, indicating slower progression of motor symptoms compared to mutation-negative PD cases. In line with slower disease progression, there was significant association between biallelic PRKN carrier status and a decrease in the MDS-UPDRS part II scores, indicating reduced impact of motor symptoms on experiences of daily living. These findings are consistent with other studies, which have shown slower progression in biallelic PRKN carriers7. Previous studies have reported that postural symptoms8, dystonia, and psychiatric symptoms may be more common in PRKN carriers7,44, but we did not find evidence of this in our cohort.
In addition to monogenic PD-related genes, GBA1 mutations were present in 10.2% of families, thus confirming GBA1 as the most important genetic risk factor for PD. Family history of PD was present in most GBA1 mutation carriers, often in a pattern akin to autosomal dominant inheritance. REM sleep behaviour disorder, a precursor of dementia in PD25–27, was more frequent in GBA1 mutation carriers, as previously reported by others45,46. As expected, GBA1 mutation carriers also performed worse in cognitive testing, in line with several studies showing worse cognitive outcomes in PD GBA1 mutation carriers47–52. The detrimental effect of GBA1 mutations on cognition was observed only in cases harbouring severe mutations (i.e., pathogenic mutations associated with neuronopathic forms of Gaucher disease), again corroborating previous studies53. However, it should be noted that other studies have found an association between the common risk variant E365K and cognitive decline in PD46,51,54.
In 90.4% of cases, no pathogenic mutations could be identified, which suggests that additional causative or contributing genetic factors are yet to be uncovered. It is possible that not all cases with early onset and/or familial PD have a monogenic form of the disease. We have found GBA1 risk variants in 10.2% of our mutation-negative cohort, which increase the risk of PD in families that share GBA1 risk variants. The incidence of GBA1 mutations is significantly higher among PD patients, but the degree of pathogenicity and penetrance of different mutations is still debated55. Likewise, we have found a single heterozygous mutation in a recessive PD-related gene in another 1.9% of all index cases fully investigated with WGS and MLPA. These could represent truly monogenic PD, where the second mutation has yet to be identified due to technical constraints. Recently, long-read sequencing has identified complex structural variants in PRKN not detected by MLPA, including large inversions56,57. Conversely, there have been reports that heterozygous PRKN and PINK1 carriers may have increased risk of developing PD symptoms with highly reduced penetrance58–60. However, other studies did not find an association between single heterozygous mutations in recessive PD-related genes and the risk of PD61,62. Interestingly, a recent study found that symptomatic heterozygous PRKN carriers had significantly reduced PRKN expression in peripheral blood mononuclear cells63. Furthermore, PRKN expression levels were decreased in symptomatic relative to asymptomatic family members carrying the same variants, suggesting the existence of additional genetic or epigenic mechanisms that regulate PRKN expression and could contribute to the risk of PD in monoallelic PRKN carriers63. Another possibility is that familial PD can be polygenic in nature, with relatives sharing multiple risk variants, each with a small risk effect, that increase the overall risk of PD among family members that share the same genetic background32. We did not find an association between the PD polygenic risk score and mutation status. However, in early-onset mutation-negative PD cases, an increasing PD-PRS was associated with familial status, which suggests that, at least in some families, a polygenic PD risk, compounded by the cumulative effect of many common risk variants, might contribute to a familial risk of PD, giving the appearance of pseudo-autosomal inheritance.
In addition to pathogenic mutations in well-established PD-related genes (LRRK2, PRKN, PINK1, SNCA, PLA2G6 and GBA1), we identified pathogenic mutations in genes that have been reported to present as levodopa-responsive parkinsonism but typically present with alternative or atypical phenotypes. Mutations in PNPLA6 cause Hereditary Spastic Paraplegia 39 (OMIM #612020), but levodopa-responsive parkinsonism has been reported in association with biallelic mutations, generally with additional clinical features64,65. Mutations in SPG7 cause Hereditary Spastic Paraplegia 7 (OMIM #607259), which typically presents as pure spastic paraplegia but is often associated with complex phenotypes. Cases presenting with levodopa-responsive parkinsonism in association with biallelic SPG7 mutations have been previously reported66–68. The VCP gene is typically associated with autosomal dominant Charcot-Marie Tooth type 2Y (OMIM #616687), frontotemporal dementia and/or amyotrophic lateral sclerosis 6 (OMIM #613954), or inclusion body myopathy with early-onset Paget disease and frontotemporal dementia (OMIM #167320). There are several reports of levodopa-responsive parkinsonism in association with pathogenic mutations in VCP69–71. Likewise, mutations in GCH1 typically manifest as dopa-responsive dystonia (OMIM #128230), but several cases manifesting with autosomal dominant PD have been reported72–74. Finally, we found three individuals with fLOPD due to a pathogenic repeat expansion in ATXN2. Although typically manifesting as spinocerebellar ataxia 2 (OMIM #183090), ATNX2 expanded CAG trinucleotide repeats have been identified in PD cases across multiple ancestries, most often in association with a family history of autosomal dominance75–79. Even though ATXN2 repeat expansions have generally been considered a rather rare cause of PD80, they were the third most common cause of familial PD (and the second most common in late-onset disease) in this cohort.
Our study as some limitations. Over 90% of all recruited participants are of European ancestry, meaning that mutation rates cannot be generalised across populations. Further efforts are needed to recruit individuals from other ancestry groups. Despite our efforts to recruit family members, the number of recruited relatives is still relatively small. Several reasons account for this, namely, the fact that in adult-onset disorders such as PD, family members from older generations might no longer be available for study participation. In addition, the fact that this is a cross-sectional study without longitudinal follow-up might hamper recruitment of newly affected relatives at a future date. We cannot rule out a recruitment bias inherent to the study design, given the inability to recruit all eligible PD cases in a clinic-based study as compared to a community-based study.
In summary, we have identified a monogenic form of PD in 9.6% of recruited families. An additional 10.2% of families carried a GBA1 variant. We have succeeded in building a cohort enriched for known pathogenic variants in PD-related genes, which will aid further characterization of genotype-phenotype associations, important for accurate diagnosis and prognosis prediction. The large number of families with a seemingly strong genetic component that remain without a molecular diagnosis presents an opportunity to uncover novel causative or high-risk conferring genetic variants and will be the focus of the next phase of the analysis. Currently, efforts are being made to recruit additional relatives from these unexplained families, in particular targeting families with a very early age at symptom onset or with multiple affected family members. As more samples are whole-genome sequenced from both affected and unaffected family members, segregation studies will be possible for demonstrating gene-disease associations, thereby facilitating new genetic discoveries. In addition, unaffected mutation carriers will allow for the examination of penetrance modifiers, thus providing insights into disease mechanisms and potential drug targets. PFP will continue to recruit from currently participating and new families until 2030.
Methods
Subjects and clinical data collection
The PFP study has been reviewed and approved by the London Camden and King’s Cross Research Ethics Committee (REC – 15/LO/0097; IRAS ID – 162268) and is sponsored by the University College London Joint Research Office. The study is conducted in compliance with UK General Data Protection Regulation (GDPR) and the principles expressed in the Helsinki Declaration. PFP is registered with www. clinicaltrials.gov (NCT02760108). All participants provided written informed consent to study participation and data sharing. Participants could also opt to consent to confirmatory diagnostic genetic testing in case of a positive genetic finding, and to being re-contacted for further research studies, including therapeutic drug trials.
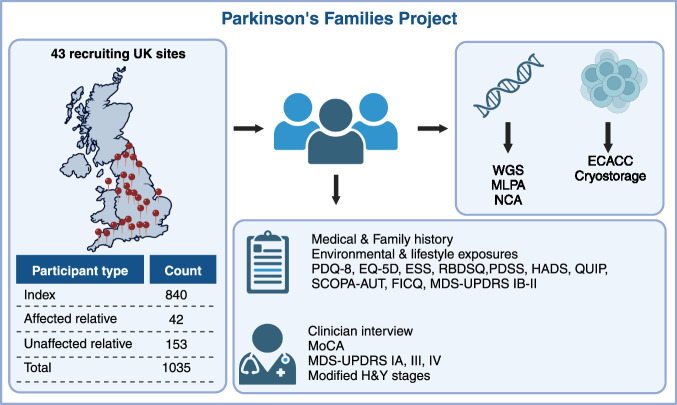
For this analysis, we included families recruited to PFP between 01/01/2015 and 24/02/2020, at 43 study sites across the UK (Fig. 1). Eligible index cases had a clinical diagnosis of PD and met at least one of the following criteria: i) Motor symptom onset at or before the age of 45 (early onset PD); ii) At least one relative up to 3rd degree affected by PD (familial PD). We set the cut-off for early-onset disease at 45 years to specifically target individuals with higher a priori probability of recessive PD, given previous studies showing that the cumulative rate of pathogenic recessive mutations is considerably higher in younger age groups8. Whenever possible we also recruited affected and unaffected relatives of index cases. Participating individuals were at least 16 years old and had capacity to consent to participation. Participants were assessed only once during the study. For all participants, we collected demographic, environmental, medical, and family history data through questionnaires and a peripheral blood or saliva sample for DNA extraction. We also facilitated remote participation of participants who did not live near a study site. These participants completed shortened and simplified assessment booklets from home and donated samples through their local doctor. Patient questionnaires included: Parkinson’s Disease Quality of Life Questionnaire (PDQ-8), EQ-5D, Epworth Sleepiness Scale (ESS), REM Sleep Behavior Disorder Screening Questionnaire (RBDSQ), Hospital Anxiety and Depression Scale (HADS), Questionnaire for Impulsive-Compulsive Disorders in Parkinson’s Disease (QUIP), Fecal Incontinence and Constipation Questionnaire, Scales for Outcomes in Parkinson’s Disease - Autonomic (SCOPA-AUT), Parkinson’s Disease Sleep Scale (PDSS). Affected participants recruited on-site were also subject to a standardised structured interview and completed validated scales and questionnaires by experience raters to assess motor and non-motor symptoms, including: Montreal Cognitive Assessment (MoCA), Movement Disorder Society Unified Parkinson’s Disease Rating Scale (MDS-UPDRS), and the Modified Hoehn and Yahr Stages. Figure 1 shows an overview of the study protocol.
Fig. 1. Parkinson’s families project overview.
Participants are recruited across 43 sites in the UK. Index cases must be ≥18 years, have capacity to consent, have a diagnosis of PD with symptom onset ≤45 and/or family history of PD. All participants donate a blood sample for DNA extraction. Affected participants additionally donate blood for peripheral blood lymphocyte extraction, which are sent to the European Collection of Authenticated Cell Cultures (ECACC) for transformation into lymphoblastoid cell lines and storage. All affected participants fill out a questionnaire with detailed medical and family history, environmental, drug and lifestyle exposures, as well as the following questionnaires: Parkinson’s Disease Questionnaire (PDQ-8), EQ-5D, Epworth Sleepiness Scale (ESS), REM Sleep Behavior Disorder Screening Questionnaire (RBDSQ), Panic Disorder Severity Scale (PDSS), Hospital Anxiety and Depression Scale (HADS), Questionnaire for Impulsive-Compulsive Disorders in Parkinson’s Disease (QUIP), Scales for Outcomes in Parkinson’s disease - Autonomic Dysfunction (SCOPA-AUT), Fecal Incontinence and Constipation Questionnaire (FICQ), MDS-UPDRS parts IB and II. Affected participants recruited on-site are also assessed by an experienced investigator, who rates the MDS-UPDRS parts IA, III and IV, MoCA and Hoehn & Yahr scales. WGS whole-genome sequencing, MLPA multiplex ligation-dependent probe amplification assay, NCA NeuroChip genotyping array.
Participants with partially completed MDS-UPDRS ratings that fell below the threshold defined by Goetz and colleagues were excluded from downstream analyses81. Subjects were classified into motor subtypes (tremor dominant [TD], postural instability and gait difficulty [PIGD] or intermediate) based on the methodology defined by Stebbins and colleagues82. If items required for classification were missing, individuals were labelled as “unclassifiable”. To account for differences in disease duration at assessment, we computed a motor severity score that consists of the ratio between the total MDS-UPDRS part III score and disease duration from reported symptom onset. Based on the MDS-UPDRS part IV, we also computed composite scores for dyskinesia (sum of items 4.1 and 4.2) and motor fluctuations (sum of items 4.3–4.5). Items of the MDS-UPDRS were categorised as present if the composite score was ≥1, except depression (item 1.3) and apathy (item 1.4), which were considered present only if sustained over more than one day at a time (score ≥2). REM sleep behaviour disorder was considered present if the RBDSQ was >5.
Clinical data storage and management
Data collected is held on REDCap® (Research Electronic Data Capture), a secure web-based Hypertext Preprocessor (PHP) software with a MySQL database back-end (https://www.project-redcap.org). It is tried and tested for use in managing clinical studies and trials, longitudinal studies and surveys83. The web host, network connection and storage is Information Governance Toolkit (IGT)-compliant and ISO27001-certified, according to data security best practices. Personally identifiable information is held in a database that is separated from the main study database. Members of the study team at each site only have access to records for participants recruited at their site. The databases will be maintained until 2034 for genetic/epidemiological research, under the custodianship of Prof. Huw Morris to enable the long-term follow-up of patients recruited to this study. All clinical data were processed, stored, and disposed in accordance with all applicable legal and regulatory requirements, including the Data Protection Act 1998 and any amendments thereto.
Sample collection and storage
DNA was extracted from EDTA blood or saliva samples (saliva collection kit: Oragene® OG-500, DNA Genotek Inc.) by LGC Biosearch Technologies™. DNA is stored in secure freezers at University College London. Affected participants additionally donated ACD blood that was sent to the European Collection of Authenticated Cell Cultures (ECCAC, https://www.culturecollections.org.uk/collections/ecacc.aspx), in Wiltshire, UK, for peripheral blood lymphocytes (PBLs) extraction and transformation into lymphoblastoid cell lines. These cell lines provide an ongoing source of DNA for future studies, and may be used for disease models or the generation of induced pluripotent cell lines. Cell lines are stored at the ECACC encoded by the unique PFP study identifier.
Genetic analysis
Whole-genome sequencing (WGS)
DNA samples from 585 participants were sequenced within the Global Parkinson’s Genetics Program (GP2) Monogenic Network84,85. Briefly, samples were sequenced with Illumina short-read WGS at Psomagen, with a mean coverage of 30x. 150 bp paired-end reads were aligned to the human reference genome (GRCh38 build) using the functional equivalence pipeline86. Sample processing and variant calling were performed using DeepVariant v.1.6.187. Joint-genotyping was performed using GLnexus v1.4.3 with the preset DeepVariant WGS configuration88. Samples were retained for downstream analyses after passing the quality control with the quality metrics defined by the Accelerating Medicines Partnership Parkinson’s Disease program (AMP-PD; https://amp-pd.org)89. Variant annotation was performed with Ensembl Variant Effect Predictor90. A target list of GBA1 variants were called using the Gauchian v.1.0.2 tool (https://github.com/Illumina/Gauchian)91. The length of STRs in ATXN2 and ATXN3 was estimated in whole-genome sequence data using the ExpansionHunter v.5.0.0 software92. All the pipelines used are available on GitHub (https://github.com/GP2code/GP2-WorkingGroups/tree/main/MN-DAWG-Monogenic-Data-Analysis). Additional details on variant interpretation are available in Supplementary Materials. A further 39 participants were analysed with WGS as part of the 100,000 Genomes Project93.
Next-generation targeted sequencing (NGS)
DNA samples of an additional three participants underwent diagnostic genetic screening using next-generation sequencing (Illumina MiSeq or HiSeq) of a panel of seven genes (FBXO7, LRRK2, PRKN, PARK7, PINK1, SNCA, VPS35) and MLPA gene dosage analysis of three genes (PRKN, PINK1, SNCA), as described in the next section. Pathogenic or likely pathogenic variants were confirmed with bi-directional Sanger sequencing.
Multiplex ligation-dependent probe amplification (MLPA)
Samples from 827 participants were screened for copy number variants (CNVs) using the SALSA MLPA EK5-FAM reagent kit and the SALSA MLPA Probemix P051, according to the manufacturer’s instructions (MRC-Holland, Amsterdam, The Netherlands). Where DNA was available, we additionally screened relatives of index cases with a CNV. PCR fragments were analysed by capillary electrophoresis using an ABI 3730XL genetic analyzer (Applied Biosystems). Data was analysed using the Coffalyser.Net™ (MRC-Holland) or GeneMarker® (SoftGenetics®, PA, USA) software packages, according to the supplied protocols.
SNP array genotyping
Quantity and purity of DNA were determined with a Qubit fluorometric assay (Invitrogen) and a NanoDrop spectrophotometer (Thermo Fisher Scientific, UK), respectively. Samples were diluted to a standard concentration in molecular grade nuclease-free water (Thermo Fisher Scientific, UK). We genotyped 849 DNA samples from 698 families using the Illumina NeuroChip array (NCA), which consists of a 306,670 SNP backbone (Infinium HumanCore-24 v1.0) with added custom content covering 179,467 neurodegenerative disease-related variants94. We manually clustered the genotypes using Illumina GenomeStudio v2.0 (Illumina Inc., San Diego, CA, USA), based on the protocol by Guo and colleagues95. We curated a list of GBA1 PD risk variants and GD-causing mutations, as well as pathogenic and likely pathogenic SNVs and indels from 10 PD causing genes (PRKN, DJ-1, PINK1, ATP13A2, FBXO7, SCNA, LRRK2, VCP, VPS35, DCTN1), from ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/, accessed on the 18/01/2023)96. We added any additional variants from PD-related genes classified as definitely pathogenic in the MDSGene database (https://www.mdsgene.org/, accessed on the 21/02/2023)97. Probes for 131 of these variants were present in the Neurochip array and were systematically screened for in all index cases using a custom R script (Supplementary Table 2). We evaluated the accuracy of the NCA probes of interest by comparing their performance against other methods, as described in Supplementary Materials.
For additional downstream analyses, we performed standard quality control in PLINK v1.998. Briefly, we excluded samples with genotype missingness >5% (which can indicate poor quality of DNA sample), mismatch between clinical and genetically determined sex (which could be due to a sample mix-up), and excess heterozygosity defined as individuals who deviate >3 SD from the mean heterozygosity rate (which can indicate sample contamination)99. We excluded variants if the call rate was <95%. Pairwise identity-by-descent (IBD) analysis was performed to infer relatedness across all samples and identify cryptic familial relationships using the KING tool (https://www.kingrelatedness.com/)100. Ancestry was genetically determined using GenoTools (https://github.com/dvitale199/GenoTools)101,102. To perform polygenic risk score analysis, genotypes were imputed against the TOPMed reference panel (version R2; https://www.nhlbiwgs.org/) using the TOPMed Imputation Server (https://imputation.biodatacatalyst.nhlbi.nih.gov) using Minimac4 (version 1.7.3)103. Imputed variants were excluded if the imputation info R2 score was ≤0.3. Following imputation, variants with missingness >5% and minor allele frequencies <1% were also excluded. Polygenic risk scores were computed in PRSice-2 (https://choishingwan.github.io/PRSice/)104 based on summary statistics from the largest Parkinson’s disease case-control genome-wide association study (GWAS) to date105.
Statistical analyses
For statistical analysis, we classified PD cases into the following categories: i) Sporadic early-onset PD (sEOPD): motor symptom onset ≤45 years, no family history of PD; ii) Familial early-onset PD (fEOPD): motor symptom onset ≤45 years, positive family history of PD; iii) Familial late-onset PD (fLOPD): motor symptom onset >45 years, positive family history of PD. GBA1 variants were classified by severity according to the GBA1-PD browser (https://pdgenetics.shinyapps.io/gba1browser/, accessed on the 25th May 2024)31. For statistical purposes, the mutation-negative and monoallelic PRKN mutation groups comprise only individuals fully investigated with WGS and MLPS, to ensure that no undetected mutations are present. Likewise, the GBA1 mutation group excludes individuals not investigated with WGS. We compared demographic and clinical features using Mann–Whitney U-test for continuous variables and Fisher’s exact tests or Chi-squared tests for proportions. We investigated the effect of the LRRK2, PRKN and GBA1 genetic status on clinical features using linear regression for continuous scores or logistic regression for categorical scores, adjusting for sex, age at assessment, and disease duration at assessment, where appropriate. We used multinomial logistic regression to analyse motor subtype, using the tremor dominant group as the reference. For analysis of the modified Hoehn & Yahr stages, we used the 0–1.5 group as the reference. For the polygenic risk score analysis, scores were z-transformed and used in logistic regression models to predict the dependent variables. All p-values are two-tailed. We used R version 4.2.1 to perform statistical analyses106.
Supplementary information
Acknowledgements
P.F.P. has received support from the Janet Owen’s bequest fund, the Walker-Peltz charitable fund, the Medical Research Council (MRC-G1100643), Cure Parkinson’s Trust, Parkinson’s UK (K-1501), and the National Institute for Health Research (NIHR) Clinical Research Network (CRN) North Thames. The funders played no role in study design, data collection, analysis and interpretation of data, or the writing of this manuscript. A full list of PFP Study Group members is available in Supplementary Materials. This research was funded in part by Aligning Science Across Parkinson’s [Grant number: ASAP-000478] through the Michael J. Fox Foundation for Parkinson’s Research (MJFF). For the purpose of open access, the author has applied a CC BY public copyright licence to all Author Accepted Manuscripts arising from this submission. Part of the whole-genome sequencing data used in the preparation of this article were obtained from Global Parkinson’s Genetics Program (GP2). GP2 is funded by the Aligning Science Across Parkinson’s (ASAP) initiative and implemented by The Michael J. Fox Foundation for Parkinson’s Research (https://gp2.org). For a complete list of GP2 members see https://gp2.org. This research was in part made possible through access to data in the National Genomic Research Library, which is managed by Genomics England Limited (a wholly owned company of the Department of Health and Social Care). The National Genomic Research Library holds data provided by patients and collected by the NHS as part of their care and data collected as part of their participation in research. The National Genomic Research Library is funded by the National Institute for Health Research and NHS England. The Wellcome Trust, Cancer Research UK and the Medical Research Council have also funded research infrastructure. The authors would like to thank study participants and referring clinicians, without whom this study would not be possible. Figures created with BioRender.com.
Author contributions
R.R., C.T., M.M.X.T. and H.R.M. designed the study. M.M.X.T., L.W. and R.R. prepared samples for SNP array genotyping. R.R. performed clustering, quality control and analysis of SNP array data. E.J.S. prepared samples for WGS. M.M.X.T., C.T. and R.R. performed and analysed MLPA experiments. R.R. performed and analysed ATXN2 and ATXN3 fragment analysis experiments. Z.H.F. performed WGS variant calling and annotation of GP2-generated WGS data, and developed the pipeline for STR expansion calling. R.R. performed variant interpretation of GP2-generated WGS data. Z.H.F. and R.R. performed STR expansion analysis on GP2 and Genomics England WGS data, respectively. J.H., R.L. and J.P. performed and interpreted diagnostic targeted sequencing data. M.H., M.M.X.T., M.P., R.R., R.T., S.J. and T.M.S. collected and processed clinical data. R.R. performed statistical analysis and interpreted the data. A.B.S. and C.B. are the leads of the Global Parkinson’s Genetics Program. C.K., L.M.L. and Z.H.F. are members of the GP2 Monogenic Network, of which CK is the lead. R.R., H.R.M., P.R.J., H.H. and N.W.W. are the leads of the Parkinson’s Families Project. KPB and AHVS advised on the project. C.T., R.R. and T.M.S. wrote the initial draft of the manuscript. All authors read and approved the final manuscript.
Data availability
A pseudo-anonymised cleaned dataset is available from 10.5281/zenodo.12549399. The data, code, protocols, and key lab materials used and generated in this study are listed in a Key Resource Table alongside their persistent identifiers at 10.5281/zenodo.12549398. Array data has been deposited at the European Genome-phenome Archive (EGA), which is hosted by the EBI and the CRG, under accession number EGAS00001007906. Further information about EGA can be found on https://ega-archive.org and “The European Genome-phenome Archive in 2021” (10.1093/nar/gkab1059). For whole-genome sequence data obtained from the 100,000 Genomes Project, research on the de-identified patient data used in this publication can be carried out in the Genomics England Research Environment subject to a collaborative agreement that adheres to patient led governance. All interested readers will be able to access the data in the same manner that the authors accessed the data. For more information about accessing the data, interested readers may contact research-network@genomicsengland.co.uk or access the relevant information on the Genomics England website: https://www.genomicsengland.co.uk/research. Data (10.5281/zenodo.10962119, release 7) used in the preparation of this article were partially obtained from the Global Parkinson’s Genetics Program (GP2). To obtain access to de-identified individual level data, interested readers must register to access the AMP PD Knowledge Platform: https://amp-pd.org/researchers/data-use-agreement.
Code availability
Raw SNP array data was clustered in GenomeStudio v2.0 (RRID:SCR_010973) according to the protocol described by Guo et al. (ref. 95) and quality control performed in Plink v1.9 (RRID:SCR_001757). Genetic ancestry was determined using Genotools (10.5281/zenodo.10443258). Sample relatedness was inferred using KING (RRID:SCR_009251). Polygenic risk scores were computed in PRSice-2 (RRID:SCR_017057). WGS processing, quality control, joint genotyping and variant calling of data generated in Genomic England in the 100,000 Genomes Project was done according to the protocol defined in ref. 93. WGS processing, quality control, joint genotyping and variant calling of data generated in GP2 was performed using DeepVariant v.1.6.1 (https://github.com/google/deepvariant) and GLnexus v1.4.3 (https://github.com/dnanexus-rnd/GLnexus) according to pipelines available at https://github.com/GP2code. Variants were annotated with Ensembl Variant Effect Predictor (RRID:SCR_007931). GBA1 variants were called using Gauchian v.1.0.2 (https://github.com/Illumina/Gauchian). Short tandem repeat sizing was performed using ExpansionHunter v.5.0.0 (https://github.com/Illumina/ExpansionHunter). For data generated by fragment analysis, GeneMapper® v5.0 (RRID:SCR_014290) was used. MLPA data was analysed using GeneMarker® (RRID:SCR_015661) or Coffalyser.Net (freely available from https://www.mrcholland.com/technology/software). Statistical analyses were performed in R v4.2.1 (RRID:SCR_001905) using basic statistical packages (stats v4.2.1, nnet v7.3.19). Other packages used include dplyr (v1.1.4), tidyr (v1.3.0), ggplot2 (v3.4.4), data.table (v1.14.8), broom (v1.0.5), purrr (v1.0.2), knitr (v1.45), forcats (v1.0.0) and plinkQC (v0.3.4).
Competing interests
H.R.M. reports paid consultancy from Biogen, Biohaven, Lundbeck and lecture fees/honoraria from the Wellcome Trust and Movement Disorders Society; H.R.M. is also a co-applicant on a patent application related to C9ORF72 - Method for diagnosing a neurodegenerative disease (PCT/GB2012/052140). A.B.S. has received royalty payments related to a diagnostic for stroke. A.B.S. is an editor for npj Parkinson’s Disease. A.B.S. was not involved in the journal’s review of, or decisions related to, this manuscript. C.K. serves as a medical advisor to Centogene, Retromer Therapeutics, and Takeda, and she received Speakers’ honoraria from Desitin and Bial. All other authors declare no financial or non-financial competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Lists of authors and their affiliations appear at the end of the paper.
A full list of members and their affiliations appears in the Supplementary Information.
Contributor Information
Huw R. Morris, Email: h.morris@ucl.ac.uk
Raquel Real, Email: r.real@ucl.ac.uk.
Parkinson’s Families Project (PFP) Study Group:
Manuela M. X. Tan, Russel Tilney, Huw R. Morris, Raquel Real, Paul R. Jarman, Nicholas W. Wood, Simona Jasaityte, Megan Hodgson, Clodagh Towns, Miriam Pollard, Elizabeth Wakeman, Tabish Saifee, Sam Arianayagam, Saifuddin Shaik, Sophie Molloy, Ralph Gregory, Mirdhu Wickremaratchi, Rosaria Buccoliero, Oliver Bandmann, Dominic Paviour, Diran Padiachy, Anjum Misbahuddin, Jeremy Cosgrove, Sunku Guptha, Ray Chaudhuri, Yen Tai, Sukaina Asad, Ayano Funaki, Marek Kunc, Charlotte Brierley, Ray Sheridan, Rena Truscott, Suzanne Dean, Carinna Vickers, Rani Sophia, Sion Jones, Erica Capps, Neil Archibald, Louise Wiblin, Sean J. Slaght, Edward Jones, Colin Barnes, Dominick D’Costa, Carl Mann, Uma Nath, Anette Schrag, Sarah Williams, Gillian Webster, Sigurlaug Sveinbjornsdottir, Lucy Strens, Annette Hand, Richard Walker, Rosemary Crouch, Jason Raw, Stephanie Tuck, Khaled Amar, Emma Wales, Irene Gentilini, Aileen Nacorda, Louise Hartley, and Henry Houlden
Global Parkinson’s Genetics Program (GP2):
Zih-Hua Fang, Manuela M. X. Tan, Simona Jasaityte, Eleanor J. Stafford, Lara M. Lange, Anthony H. V. Schapira, Kailash P. Bhatia, Andrew B. Singleton, Cornelis Blauwendraat, Christine Klein, Henry Houlden, Nicholas W. Wood, Huw R. Morris, and Raquel Real
Supplementary information
The online version contains supplementary material available at 10.1038/s41531-024-00778-z.
References
- 1.Dorsey, E. R., Sherer, T., Okun, M. S. & Bloem, B. R. The emerging evidence of the Parkinson pandemic. J. Parkinsons. Dis.8, S3–S8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blauwendraat, C., Nalls, M. A. & Singleton, A. B. The genetic architecture of Parkinson’s disease. Lancet Neurol.19, 170–178 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lange, L. M. et al. Nomenclature of Genetic Movement Disorders: Recommendations of the International Parkinson and Movement Disorder Society Task Force - an update. Mov. Disord.37, 905–935 (2022). [DOI] [PubMed] [Google Scholar]
- 4.Marder, K. et al. Risk of Parkinson’s disease among first-degree relatives: A community-based study. Neurology47, 155–160 (1996). [DOI] [PubMed] [Google Scholar]
- 5.Torti, M. et al. Effect of family history, occupation and diet on the risk of Parkinson disease: A case-control study. PLoS One15, e0243612 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu, F.-C. et al. Familial aggregation of Parkinson’s disease and coaggregation with neuropsychiatric diseases: a population-based cohort study. Clin. Epidemiol.10, 631–641 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kasten, M. et al. Genotype-phenotype relations for the Parkinson’s disease genes Parkin, PINK1, DJ1: MDSGene systematic review. Mov. Disord.33, 730–741 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Tan, M. M. X. et al. Genetic analysis of Mendelian mutations in a large UK population-based Parkinson’s disease study. Brain142, 2828–2844 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alcalay, R. N. et al. Frequency of known mutations in early-onset Parkinson disease: implication for genetic counseling: the consortium on risk for early onset Parkinson disease study. Arch. Neurol.67, 1116–1122 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gustavsson, E. K. et al. RAB32 Ser71Arg in autosomal dominant Parkinson’s disease: linkage, association, and functional analyses. Lancet Neurol23, 603–614 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hop, P. J. et al. Systematic rare variant analyses identify RAB32 as a susceptibility gene for familial Parkinson’s disease. Nat. Genet. 56, 1371–1376 (2024). [DOI] [PMC free article] [PubMed]
- 12.Smith, L. & Schapira, A. H. V. GBA variants and Parkinson disease: Mechanisms and treatments. Cells11, 1261 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bandres-Ciga, S., Diez-Fairen, M., Kim, J. J. & Singleton, A. B. Genetics of Parkinson’s disease: An introspection of its journey towards precision medicine. Neurobiol. Dis.137, 104782 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skrahina, V. et al. The Rostock International Parkinson’s Disease (ROPAD) study: Protocol and initial findings. Mov. Disord.36, 1005–1010 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malek, N. et al. Tracking Parkinson’s: Study Design and Baseline Patient Data. J. Parkinsons. Dis.5, 947–959 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao, Y. et al. The role of genetics in Parkinson’s disease: a large cohort study in Chinese mainland population. Brain143, 2220–2234 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Kovanda, A. et al. A multicenter study of genetic testing for Parkinson’s disease in the clinical setting. NPJ Parkinsons Dis8, 149 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cristina, T.-P. et al. A genetic analysis of a Spanish population with early onset Parkinson’s disease. PLoS One15, e0238098 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Towns, C. et al. Defining the causes of sporadic Parkinson’s disease in the global Parkinson’s genetics program (GP2). NPJ Parkinsons Dis9, 131 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun, Y.-M. et al. The genetic spectrum of a cohort of patients clinically diagnosed as Parkinson’s disease in mainland China. NPJ Parkinsons Dis. 9, 76 (2023). [DOI] [PMC free article] [PubMed]
- 21.Klein, C. & Westenberger, A. Genetics of Parkinson’s disease. Cold Spring Harb. Perspect. Med.2, a008888–a008888 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siddiqui, I. J., Pervaiz, N. & Abbasi, A. A. The Parkinson Disease gene SNCA: Evolutionary and structural insights with pathological implication. Sci. Rep.6, 24475 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sidransky, E. & Lopez, G. The link between the GBA gene and parkinsonism. Lancet Neurol11, 986–998 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malek, N. et al. Features of GBA -associated Parkinson’s disease at presentation in the UK Tracking Parkinson’s study. J. Neurol. Neurosurg. Psychiatry89, 702–709 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marion, M.-H., Qurashi, M., Marshall, G. & Foster, O. Is REM sleep Behaviour Disorder (RBD) a risk factor of dementia in idiopathic Parkinson’s disease? J. Neurol.255, 192–196 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Postuma, R. B. et al. Rapid eye movement sleep behavior disorder and risk of dementia in Parkinson’s disease: A prospective study. Mov. Disord.27, 720–726 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Nagy, A. V. et al. Cognitive impairment in REM-sleep behaviour disorder and individuals at risk of Parkinson’s disease. Parkinsonism Relat. Disord.109, 105312 (2023). [DOI] [PubMed] [Google Scholar]
- 28.Camacho, M. et al. Early constipation predicts faster dementia onset in Parkinson’s disease. NPJ Parkinsons Dis. 7, 45 (2021). [DOI] [PMC free article] [PubMed]
- 29.Jones, J. D., Rahmani, E., Garcia, E. & Jacobs, J. P. Gastrointestinal symptoms are predictive of trajectories of cognitive functioning in de novo Parkinson’s disease. Parkinsonism Relat. Disord.72, 7–12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gryc, W. et al. Hallucinations and development of dementia in Parkinson’s disease. J. Parkinsons. Dis.10, 1643–1648 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parlar, S. C., Grenn, F. P., Kim, J. J., Baluwendraat, C. & Gan-Or, Z. Classification of GBA1 variants in Parkinson’s disease: The GBA1‐PD browser. Mov. Disord.38, 489–495 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khera, A. V. et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet.50, 1219–1224 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deng, H., Wang, P. & Jankovic, J. The genetics of Parkinson disease. Ageing Res. Rev.42, 72–85 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Marder, K. et al. Age-specific penetrance of LRRK2 G2019S in the Michael J. fox Ashkenazi Jewish LRRK2 Consortium. Neurology85, 89–95 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hamza, T. H. & Payami, H. The heritability of risk and age at onset of Parkinson’s disease after accounting for known genetic risk factors. J. Hum. Genet.55, 241–243 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trinh, J. et al. DNM3 and genetic modifiers of age of onset in LRRK2 Gly2019Ser parkinsonism: a genome-wide linkage and association study. Lancet Neurol15, 1248–1256 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Lee, A. J. et al. Penetrance estimate of LRRK2 p.G2019S mutation in individuals of non-Ashkenazi Jewish ancestry. Mov. Disord.32, 1432–1438 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goldwurm, S. et al. Evaluation of LRRK2 G2019S penetrance: relevance for genetic counseling in Parkinson disease. Neurology68, 1141–1143 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Saunders-Pullman, R. et al. Progression in the LRRK2-asssociated Parkinson disease population. JAMA Neurol.75, 312–319 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shu, L. et al. Clinical Heterogeneity Among LRRK2 Variants in Parkinson’s Disease: A Meta-Analysis. Front. Aging Neurosci.10, 283 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yahalom, G. et al. Dyskinesias in patients with Parkinson’s disease: effect of the leucine-rich repeat kinase 2 (LRRK2) G2019S mutation. Parkinsonism Relat. Disord.18, 1039–1041 (2012). [DOI] [PubMed] [Google Scholar]
- 42.Healy, D. G. et al. Phenotype, genotype, and worldwide genetic penetrance of LRRK2-associated Parkinson’s disease: a case-control study. Lancet Neurol.7, 583–590 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Menon, P. J. et al. Genotype-phenotype correlation in PRKN-associated Parkinson’s disease. NPJ Parkinsons Dis.10, 72 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koros, C., Simitsi, A. & Stefanis, L. Genetics of Parkinson’s disease: Genotype-phenotype correlations. Int. Rev. Neurobiol.132, 197–231 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Zhou, Y. et al. Mutational spectrum and clinical features of GBA1 variants in a Chinese cohort with Parkinson’s disease. NPJ Parkinsons Dis. 9, 129 (2023). [DOI] [PMC free article] [PubMed]
- 46.Iwaki, H. et al. Genetic risk of Parkinson disease and progression:: An analysis of 13 longitudinal cohorts. Neurol Genet5, e348 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alcalay, R. N. et al. Cognitive performance of GBA mutation carriers with early-onset PD: the CORE-PD study. Neurology78, 1434–1440 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Setó-Salvia, N. et al. Glucocerebrosidase mutations confer a greater risk of dementia during Parkinson’s disease course. Mov. Disord.27, 393–399 (2012). [DOI] [PubMed] [Google Scholar]
- 49.Mata, I. F. et al. GBA Variants are associated with a distinct pattern of cognitive deficits in Parkinson’s disease. Mov. Disord.31, 95–102 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stoker, T. B. et al. Impact of GBA1 variants on long-term clinical progression and mortality in incident Parkinson’s disease. J. Neurol. Neurosurg. Psychiatry91, 695–702 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Davis, M. Y. et al. Association of GBA Mutations and the E326K Polymorphism With Motor and Cognitive Progression in Parkinson Disease. JAMA Neurol.73, 1217–1217 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Szwedo, A. A. et al. GBA and APOE Impact Cognitive Decline in Parkinson’s Disease: A 10-Year Population-Based Study. Mov. Disord.37, 1016–1027 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu, G. et al. Specifically neuropathic Gaucher’s mutations accelerate cognitive decline in Parkinson’s. Ann. Neurol.80, 674–685 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Straniero, L. et al. The SPID-GBA study: Sex distribution, Penetrance, Incidence, and Dementia in GBA-PD. Neurol. Genet.6, e523 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Riboldi, G. M., Di Fonzo, A. B. & GBA gaucher disease, and Parkinson’s disease: From genetic to clinic to new therapeutic approaches. Cells8, 364 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Daida, K. et al. Long‐read sequencing resolves a complex structural variant in PRKN Parkinson’s disease. Mov. Disord.38, 2249–2257 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cogan, G. et al. Long-read sequencing unravels the complexity of structural variants inPRKNin two individuals with early-onset Parkinson’s disease. bioRxiv10.1101/2024.05.02.24306523 (2024) [DOI] [PMC free article] [PubMed]
- 58.Castelo Rueda, M. P. et al. Frequency of heterozygous Parkin (PRKN) variants and penetrance of Parkinson’s disease risk markers in the population-based CHRIS cohort. Front. Neurol.12, 706145 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weissbach, A. et al. Influence of L-dopa on subtle motor signs in heterozygous Parkin- and PINK1 mutation carriers. Parkinsonism Relat. Disord.42, 95–99 (2017). [DOI] [PubMed] [Google Scholar]
- 60.Lubbe, S. J. et al. Assessing the relationship between monoallelic PRKN mutations and Parkinson’s risk. Hum. Mol. Genet.30, 78–86 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu, W. et al. Heterozygous PRKN mutations are common but do not increase the risk of Parkinson’s disease. Brain145, 2077–2091 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Krohn, L. et al. Comprehensive assessment of PINK1 variants in Parkinson’s disease. Neurobiol. Aging91, 168.e1–168.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Papagiannakis, N. et al. Parkin mRNA expression levels in peripheral blood mononuclear cells in Parkin‐related Parkinson’s disease. Mov. Disord.39, 715–722 (2024). [DOI] [PubMed] [Google Scholar]
- 64.Sen, K., Finau, M. & Ghosh, P. Bi-allelic variants in PNPLA6 possibly associated with Parkinsonian features in addition to spastic paraplegia phenotype. J. Neurol.267, 2749–2753 (2020). [DOI] [PubMed] [Google Scholar]
- 65.Kazanci, S. et al. PNPLA6-related disorder with levodopa-responsive parkinsonism. Mov. Disord. Clin. Pract.10, 338–340 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sáenz-Farret, M. et al. Spastic paraplegia type 7 and movement disorders: Beyond the spastic paraplegia. Mov. Disord. Clin. Pract.9, 522–529 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pedroso, J. L. et al. SPG7 with parkinsonism responsive to levodopa and dopaminergic deficit. Parkinsonism Relat. Disord.47, 88–90 (2018). [DOI] [PubMed] [Google Scholar]
- 68.Phillips, O., Amato, A. M. & Fernandez, H. H. Early-onset parkinsonism and hereditary spastic paraplegia type 7: pearls and pitfalls. Parkinsonism Relat. Disord.110, 105315 (2023). [DOI] [PubMed] [Google Scholar]
- 69.Regensburger, M., Türk, M., Pagenstecher, A., Schröder, R. & Winkler, J. VCP-related multisystem proteinopathy presenting as early-onset Parkinson disease. Neurology89, 746–748 (2017). [DOI] [PubMed] [Google Scholar]
- 70.Alshaikh, J. T., Paul, A., Moukheiber, E., Scholz, S. W. & Pantelyat, A. VCP mutations and parkinsonism: An emerging link. Clin. Park. Relat. Disord.10, 100230 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Al-Obeidi, E. et al. Genotype‐phenotype study in patients with valosin‐containing protein mutations associated with multisystem proteinopathy. Clin. Genet.93, 119–125 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yoshino, H. et al. GCH1 mutations in dopa-responsive dystonia and Parkinson’s disease. J. Neurol.265, 1860–1870 (2018). [DOI] [PubMed] [Google Scholar]
- 73.Rudakou, U. et al. Common and rare GCH1 variants are associated with Parkinson’s disease. Neurobiol. Aging73, 231.e1–231.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mencacci, N. E. et al. Parkinson’s disease in GTP cyclohydrolase 1 mutation carriers. Brain137, 2480–2492 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Casse, F. et al. Detection of ATXN2 expansions in an exome dataset: An underdiagnosed cause of parkinsonism. Mov. Disord. Clin. Pract.10, 664–669 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Payami, H. et al. SCA2 may present as levodopa‐responsive parkinsonism. Mov. Disord.18, 425–429 (2003). [DOI] [PubMed] [Google Scholar]
- 77.Charles, P. et al. Are interrupted SCA2 CAG repeat expansions responsible for parkinsonism? Neurology69, 1970–1975 (2007). [DOI] [PubMed] [Google Scholar]
- 78.Lu, C.-S., Wu Chou, Y.-H., Kuo, P.-C., Chang, H.-C. & Weng, Y.-H. The parkinsonian phenotype of spinocerebellar ataxia type 2. Arch. Neurol.61, 35 (2004). [DOI] [PubMed] [Google Scholar]
- 79.Shan, D.-E. et al. Spinocerebellar ataxia type 2 presenting as familial levodopa‐responsive parkinsonism. Ann. Neurol.50, 812–815 (2001). [DOI] [PubMed] [Google Scholar]
- 80.Kock, N. et al. Role of SCA2 mutations in early‐ and late‐onset dopa‐responsive parkinsonism. Ann. Neurol.52, 257–258 (2002). [DOI] [PubMed] [Google Scholar]
- 81.Goetz, C. G. et al. Handling missing values in the MDS-UPDRS. Mov. Disord.30, 1632–1638 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stebbins, G. T. et al. How to identify tremor dominant and postural instability/gait difficulty groups with the movement disorder society unified Parkinson’s disease rating scale: comparison with the unified Parkinson’s disease rating scale. Mov. Disord.28, 668–670 (2013). [DOI] [PubMed] [Google Scholar]
- 83.Harris, P. A. et al. Research electronic data capture (REDCap)–a metadata-driven methodology and workflow process for providing translational research informatics support. J. Biomed. Inform.42, 377–381 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lange, L. M. et al. Elucidating causative gene variants in hereditary Parkinson’s disease in the Global Parkinson’s Genetics Program (GP2). NPJ Parkinsons Dis. 9, 100 (2023). [DOI] [PMC free article] [PubMed]
- 85.Leonard, H. et al. Global Parkinson’s Genetics Program data release 7. Zenodo,10.5281/ZENODO.10962119 (2024).
- 86.Regier, A. A. et al. Functional equivalence of genome sequencing analysis pipelines enables harmonized variant calling across human genetics projects. Nat. Commun. 9, 4038 (2018). [DOI] [PMC free article] [PubMed]
- 87.Poplin, R. et al. A universal SNP and small-indel variant caller using deep neural networks. Nat. Biotechnol.36, 983–987 (2018). [DOI] [PubMed] [Google Scholar]
- 88.Yun, T. et al. Accurate, scalable cohort variant calls using DeepVariant and GLnexus. Bioinformatics36, 5582–5589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Iwaki, H. et al. Accelerating Medicines Partnership: Parkinson’s Disease. Genetic Resource. Mov. Disord.36, 1795–1804 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McLaren, W. et al. The ensembl variant effect predictor. Genome Biol. 17, 122 (2016). [DOI] [PMC free article] [PubMed]
- 91.Toffoli, M. et al. Comprehensive short and long read sequencing analysis for the Gaucher and Parkinson’s disease-associated GBA gene. Commun. Biol. 5, 670 (2022). [DOI] [PMC free article] [PubMed]
- 92.Halman, A., Dolzhenko, E. & Oshlack, A. STRipy: A graphical application for enhanced genotyping of pathogenic short tandem repeats in sequencing data. Hum. Mutat.43, 859–868 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Caulfield, M. et al. National Genomic Research Library. figshare 10.6084/M9.FIGSHARE.4530893.V7 (2020).
- 94.Blauwendraat, C. et al. NeuroChip, an updated version of the NeuroX genotyping platform to rapidly screen for variants associated with neurological diseases. Neurobiol. Aging57, 247.e9–247.e13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Guo, Y. et al. Illumina human exome genotyping array clustering and quality control. Nat. Protoc.9, 2643–2662 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Landrum, M. J. et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res.46, D1062–D1067 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Klein, C., Hattori, N. & Marras, C. MDSGene: Closing data gaps in genotype-phenotype correlations of monogenic Parkinson’s disease. J. Parkinsons. Dis.8, S25–S30 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chang, C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience4, 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Marees, A. T. et al. A tutorial on conducting genome-wide association studies: Quality control and statistical analysis. Int. J. Methods Psychiatr. Res.27, e1608 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Manichaikul, A. et al. Robust relationship inference in genome-wide association studies. Bioinformatics26, 2867–2873 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Vitale, D. et al. GenoTools: An open-source Python package for efficient genotype data quality control and analysis. bioRxiv10.1101/2024.03.26.586362 (2024).
- 102.Vitale, D. et al. Dvitale199/GenoTools: Zenodo Release. Zenodo, 10.5281/ZENODO.10443258 (2023).
- 103.Das, S. et al. Next-generation genotype imputation service and methods. Nat. Genet.48, 1284–1287 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Choi, S. W. & O’Reilly, P. F. PRSice-2: Polygenic Risk Score software for biobank-scale data. Gigascience8, giz082 (2019). [DOI] [PMC free article] [PubMed]
- 105.Nalls, M. A. et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol.18, 1091–1102 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ripley, B. D. The R project in statistical computing. MSOR Connect.1, 23–25 (2001). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
A pseudo-anonymised cleaned dataset is available from 10.5281/zenodo.12549399. The data, code, protocols, and key lab materials used and generated in this study are listed in a Key Resource Table alongside their persistent identifiers at 10.5281/zenodo.12549398. Array data has been deposited at the European Genome-phenome Archive (EGA), which is hosted by the EBI and the CRG, under accession number EGAS00001007906. Further information about EGA can be found on https://ega-archive.org and “The European Genome-phenome Archive in 2021” (10.1093/nar/gkab1059). For whole-genome sequence data obtained from the 100,000 Genomes Project, research on the de-identified patient data used in this publication can be carried out in the Genomics England Research Environment subject to a collaborative agreement that adheres to patient led governance. All interested readers will be able to access the data in the same manner that the authors accessed the data. For more information about accessing the data, interested readers may contact research-network@genomicsengland.co.uk or access the relevant information on the Genomics England website: https://www.genomicsengland.co.uk/research. Data (10.5281/zenodo.10962119, release 7) used in the preparation of this article were partially obtained from the Global Parkinson’s Genetics Program (GP2). To obtain access to de-identified individual level data, interested readers must register to access the AMP PD Knowledge Platform: https://amp-pd.org/researchers/data-use-agreement.
Raw SNP array data was clustered in GenomeStudio v2.0 (RRID:SCR_010973) according to the protocol described by Guo et al. (ref. 95) and quality control performed in Plink v1.9 (RRID:SCR_001757). Genetic ancestry was determined using Genotools (10.5281/zenodo.10443258). Sample relatedness was inferred using KING (RRID:SCR_009251). Polygenic risk scores were computed in PRSice-2 (RRID:SCR_017057). WGS processing, quality control, joint genotyping and variant calling of data generated in Genomic England in the 100,000 Genomes Project was done according to the protocol defined in ref. 93. WGS processing, quality control, joint genotyping and variant calling of data generated in GP2 was performed using DeepVariant v.1.6.1 (https://github.com/google/deepvariant) and GLnexus v1.4.3 (https://github.com/dnanexus-rnd/GLnexus) according to pipelines available at https://github.com/GP2code. Variants were annotated with Ensembl Variant Effect Predictor (RRID:SCR_007931). GBA1 variants were called using Gauchian v.1.0.2 (https://github.com/Illumina/Gauchian). Short tandem repeat sizing was performed using ExpansionHunter v.5.0.0 (https://github.com/Illumina/ExpansionHunter). For data generated by fragment analysis, GeneMapper® v5.0 (RRID:SCR_014290) was used. MLPA data was analysed using GeneMarker® (RRID:SCR_015661) or Coffalyser.Net (freely available from https://www.mrcholland.com/technology/software). Statistical analyses were performed in R v4.2.1 (RRID:SCR_001905) using basic statistical packages (stats v4.2.1, nnet v7.3.19). Other packages used include dplyr (v1.1.4), tidyr (v1.3.0), ggplot2 (v3.4.4), data.table (v1.14.8), broom (v1.0.5), purrr (v1.0.2), knitr (v1.45), forcats (v1.0.0) and plinkQC (v0.3.4).