Abstract
Herpes simplex virus type 1 (HSV-1) immediate-early regulatory protein ICP0 stimulates the initiation of lytic infection and reactivation from quiescence in human fibroblast cells. These functions correlate with its ability to localize to and disrupt centromeres and specific subnuclear structures known as ND10, PML nuclear bodies, or promyelocytic oncogenic domains. Since the natural site of herpesvirus latency is in neurons, we investigated the status of ND10 and centromeres in uninfected and infected human cells with neuronal characteristics. We found that NT2 cells, a neuronally committed human teratocarcinoma cell line, have abnormal ND10 characterized by low expression of the major ND10 component PML and no detectable expression of another major ND10 antigen, Sp100. In addition, PML is less extensively modified by the ubiquitin-like protein SUMO-1 in NT2 cells compared to fibroblasts. After treatment with retinoic acid, NT2 cells differentiate into neuron-like hNT cells which express very high levels of both PML and Sp100. Infection of both NT2 and hNT cells by HSV-1 was poor compared to human fibroblasts, and after low-multiplicity infection yields of virus were reduced by 2 to 3 orders of magnitude. ICP0-deficient mutants were also disabled in the neuron-related cell lines, and cells quiescently infected with an ICP0-null virus could be established. These results correlated with less-efficient disruption of ND10 and centromeres induced by ICP0 in NT2 and hNT cells. Furthermore, the ability of ICP0 to activate gene expression in transfection assays in NT2 cells was poor compared to Vero cells. These results suggest that a contributory factor in the reduced HSV-1 replication in the neuron-related cells is inefficient ICP0 function; it is possible that this is pertinent to the establishment of latent infection in neurons in vivo.
Herpes simplex virus type 1 (HSV-1) is an important human pathogen which causes recurrent infections in epithelia between long periods of latency in neuronal cells (23, 58). The basis of the differences between lytic infection (which involves active and abundant transcription from the whole viral genome) and latency (a state of transcriptional quiescence of the bulk of the genome) has been the subject of intense research. There is now a wealth of information on the lytic transcriptional program of HSV-1 and the viral regulatory proteins which stimulate viral gene expression, but less is known about the difference in host cell functions which might contribute to the differing outcomes of infection in neuronal and non-neuronal cells.
One aspect of host cell biology which has become of interest in recent years and which could conceivably modulate HSV-1 infection is the function and status of small nuclear substructures known as ND10, PML nuclear bodies, or promyelocytic oncogenic domains (11, 37, 38, 53). At the early stages of infection, the parental genomes of HSV-1 and several other DNA viruses preferentially localize in the vicinity of ND10. It has been demonstrated that transcription of viral immediate-early (IE) genes occurs at these sites, from which viral replication centers later originate (26, 36). Many DNA viruses encode regulatory proteins that interact directly with ND10, first colocalizing with the cellular protein constituents and then disrupting these structures. This is especially well understood in the case of HSV-1 regulatory protein ICP0, which disrupts both ND10 and centromeres by inducing the proteasome-dependent degradation of several of their constituent proteins (12, 37, 38). HSV-1 mutants that do not express active ICP0 have a marked defect in the ability to initiate the lytic cycle after low-multiplicity infection and instead are likely to enter a quiescent state in which all viral transcription is silenced. A number of studies have demonstrated that the ability of ICP0 to affect ND10 and centromeres is concordant with its ability to stimulate virus infection, so it has been proposed that degradation of at least one of the known cellular targets of ICP0, or the mechanism by which this occurs, is an important factor in the balance between active and quiescent infection (12). This background led us to investigate the status of ND10 in neuron-like cells.
A number of previous studies have used neuronal cells of various origins. Rat and mouse neuroblastoma cell lines that can be differentiated into a neuron-like morphology have been found to be poorly infected by HSV-1 (56, 57). This defect has been attributed to poor IE gene transcription (29) and can be overcome in part by treatment with butyrate (1, 28) or release of the cells from growth arrest (47). It has been suggested that poor IE transcription in neuronal cells is caused by the repressive effects of Oct-2 transcription factors which bind to the TAATGARAT IE regulatory elements, thereby inhibiting Oct-1- and VP16-mediated transactivation (33, 34), but these studies have remained controversial (24). Whatever the mechanism of repression, the result is that it is possible to establish cultures of rodent neuron-like cells that harbor quiescent HSV-1 genomes for long periods (2, 56). Experiments with neurons explanted from rat embryonic dorsal root ganglia have extended these studies and demonstrated that in these true neuronal cells long-term quiescent infections can be established and used to investigate the processes controlling viral quiescence and reactivation (51, 59).
Rodent cells are not amenable to examination of their ND10 structures since most of the available antibodies recognize only the human forms of the major constituent proteins. Therefore, we sought to use a suitable human neuron-related cell line for our experiments. Human neuroblastoma cell lines have been used in a number of HSV-1 infection studies. Such cells are permissive for viral replication after high-multiplicity infection (4, 5), and the use of infection at supra-optimal temperatures has allowed the establishment of quiescently infected cultures which can later be reactivated (31). We chose to study the well-characterized neuronally committed teratocarcinoma NT2 cells which grow well in culture but can be differentiated into neuron-like hNT cells after treatment with retinoic acid (41, 42). NT2 cells are permissive for high-multiplicity HSV-1 infection (30), but low-multiplicity infections and the influence of ICP0 have not been analyzed in these cells. It had previously been noted that NT2 cells express low amounts of the major ND10 protein PML (32) and that Sp100 (another major ND10 constituent) appeared to be absent (27), but the fate of these proteins during differentiation and HSV-1 infection had not been studied.
The aims of this study were severalfold. We set out to investigate in detail the status of ND10 in both NT2 and hNT cells and to monitor the fate of ND10 and the major constituent proteins during HSV-1 infection. We also studied the ability of HSV-1 to infect both NT2 and hNT cells, and we assessed the role of ICP0 in these infections. Finally, we characterized the ability of ICP0 to affect ND10 proteins, to stimulate virus infection, and to activate gene expression in these cell lines. Our results demonstrate that ND10 are highly aberrant in NT2 cells but become more like those in other cultured cells after differentiation. HSV-1 replicated with reduced efficiency in both cell lines, especially in low-multiplicity infections, and this correlated with a reduced ability of ICP0 to disrupt ND10 and centromeres. Finally, we found that ICP0-deficient viruses can attain a quiescent state in infected NT2 and hNT cells and can be subsequently reactivated by superinfection with viruses which express ICP0. These results suggest that NT2 and hNT cells provide a promising system in which to study HSV-1 infection in neuron-related cultured cells of human origin.
MATERIALS AND METHODS
Cells and cell culture.
The Ntera 2/D1 (NT2) cell line was obtained from Stratagene and was grown in Dulbecco's modified Eagle medium (DMEM) with nutrient mixture F12 (Gibco), supplemented with 4 mM glutamine, 10% fetal calf serum (FCS), and 100 U of penicillin and 100 μg of streptomycin per ml. Differentiation of NT2 cells into the neuronal phenotype hNT cells was done according to the supplier's protocol. Briefly, 106 NT2 cells were seeded into 25-cm2 flasks with 5 ml of medium containing 10 μM all-trans-retinoic acid (ATRA). The culture medium was changed three times per week for 6 weeks, and then the cells were transferred to a 75-cm2 flask in medium without ATRA and cultured for an additional 2 days. The loosely adherent hNT cells were then recovered in the culture medium by striking the flask sharply, resulting in a population of cells with more than 30% having a significant neuronal morphology, and all the recovered hNT cells expressed high levels of Sp100 (see below).
Baby hamster kidney (BHK) cells were used for the propagation and titration of virus stocks, and were grown in Glasgow modified Eagle Medium containing antibiotics as described above and supplemented with 10% newborn calf serum (NBCS) and 10% tryptose phosphate broth. HEp-2 and Vero cells were grown in DMEM supplemented with 10% FCS and antibiotics as described above. Human fetal lung (HFL) diploid fibroblast cells (Imperial Laboratories) and U2OS cells were propagated in DMEM supplemented with 5% FCS, 5% NBCS, 1% nonessential amino acids (Gibco), and antibiotics as described above.
Viruses.
HSV-1 strain 17+ was the wild-type virus used in these experiments, from which the ICP0-null mutant virus dl1403 had been derived (54). Other viruses with lesions in ICP0 were the RING finger deletion mutant FXE (10) and virus M1, which has point mutations in the USP7 binding region (21). Virus tsK contains a temperature-sensitive mutation in ICP4, which results in failure to activate early and late gene expression and high level production of the other IE proteins at the nonpermissive temperature of 38.5°C (44). Virus in1330 carries a human cytomegalovirus (HCMV) lacZ cassette inserted into UL43, the ICP4 tsK mutation, and a deletion in the ICP0 coding region inducing a frameshift in codon 105 (25).
Antibodies.
The following antibodies were used: anti-ICP0 monoclonal antibody (MAB) 11060 (16) and anti-ICP0 polyclonal rabbit serum r190 (19), anti-ICP4 MAB 10176 (15), anti-UL39 rabbit serum r76 (6), anti-UL42 MAB Z1F11 (50), anti-UL29 rabbit serum r515 (35), anti-PML rabbit serum r8 (3), and anti-PML MAB 5E10 (55), anti-Sp100 polyclonal rabbit serum SpGH (52), anti-hDaxx rabbit serum r1866 (43), anti-CENP-C rabbit serum r554 (48), and human anti-centromere autoimmune serum GS (ACA-GS) (7). Anti-β-galactosidase rabbit serum r12741/2 was a gift from Howard Marsden. Secondary antibodies for immunofluorescence were used at the indicated dilutions: fluorescein isothiocyanate-conjugated sheep anti-rabbit immunoglobulin G (IgG; 1/100; Sigma), Cy3-conjugated goat anti-mouse IgG (1/500) and goat anti-rabbit IgG (1/5,000), and Cy5-conjugated goat anti-rabbit IgG (1/500) (Amersham). Horseradish peroxidase-conjugated sheep anti-mouse and goat anti-rabbit secondary antibodies for use in Western blotting were obtained from Sigma.
Immunofluorescence.
Cells were seeded onto coverslips in Linbro wells at a density of 105 cells per well 1 day prior to infection or transfection. After appropriate infection or transfection, cells were fixed with formaldehyde (5% [vol/vol] in phosphate-buffered saline [PBS] containing 2% sucrose) and then permeabilized with 0.5% NP-40 in PBS with 10% sucrose. The primary antibodies were diluted in PBS containing 1% NBCS. Antibodies were used at the following dilutions: 11060, 1/1,000; r8, 1/1,000; r190, 1/200; r515, 1/200; SpGH, 1/1,000; r1866, 1/1,000; ACA-GS, 1/20,000; and r12741/2, 1/1,000. After incubation at room temperature for 1 h, the coverslips were washed at least six times and then treated with secondary antibodies. After a further 60-min incubation, the coverslips were again washed at least six times and mounted using Citifluor AF1.
Confocal microscopy.
Samples were examined using 543-nm and 488-nm excitation lasers of a Zeiss LSM 510 confocal microscope (a Zeiss Axioplan with a ×63 oil immersion objective lens, NA 1.4). The data from the channels were collected sequentially using the appropriate band-pass filters built into the instrument. The scanning conditions were adjusted to ensure that signal overlap between channels was essentially eliminated. Data were collected with fourfold averaging at a resolution of 1,024 × 1,024 pixels using optical slices of between 0.5 and 1 μm. Data sets were processed using the LSM 510 software and then exported for preparation for printing using Photoshop.
Western blotting.
Sodium dodecyl sulfate-polyacrylamide gels were prepared and run in the Bio-Rad MiniProtean II apparatus, and then the proteins were electrophoretically transferred to nitrocellulose membranes according to the manufacturer's recommendations. After a blocking step in PBS containing 0.1% Tween 20 (PBST) and 5% dried milk overnight at 4°C, the membranes were incubated with primary antibody in PBST–5% dried milk at room temperature for 4 h and then washed in PBST at least six times before incubation with horseradish peroxidase-conjugated secondary antibody in PBST–2% dried milk at room temperature for 1 h. After an extensive washing, the filters were soaked in Amersham or NEN Enhanced ECL reagent and exposed to film. Antibodies were stripped from the membranes following the Amersham ECL protocol, and the membranes were reprobed as necessary.
Plasmids, transfection of cultured cells, and CAT assays.
The following plasmids were used for transfection assays: pCI110 (21), pSS80 (18), p175 (9), pCIPIC1 (20), and pgDCAT (8). Plasmid pCI-rtag-cICP0 contains the ICP0 cDNA coding region linked to an N-terminal oligonucleotide encoding an epitope from the C terminus of UL30, inserted into the expression vector pCIneo (Promega). Plasmids were transfected into Vero and NT2 cells using Lipofectamine Plus reagent (Gibco-BRL) using 0.5 μg of total DNA and 1 μl of reagent per 105 cells. For chloramphenicol acetyltransferase (CAT) assay transfections, 10 ng of pSS80 reporter was used, and plasmid pUC9 was used to equalize the total amounts of DNA; the amount of the ICP0 expression plasmid varied between 5 and 500 ng (see Fig. 7). Transfected cells were fixed and stained for immunofluorescence, harvested for Western blotting, or used to prepare CAT assay extracts about 24 h later. The ability of ICP0 to activate gene expression in transfected cells was determined using reporter plasmids pSS80 and pgDCAT, which contain the CAT gene linked to the HSV-1 ICP6 and glycoprotein gD gene promoter regions, respectively. Cells in Linbro wells were transfected as described above, and then whole sonicated cell extracts were prepared and used for estimation of CAT activity using radiolabeled chloramphenicol as substrate, essentially as described elsewhere (8). Initial experiments determined the optimal amounts of reporter and activator plasmids to be used in each cell type. The transfection experiments were repeated on several independent occasions in parallel with positive and negative controls.
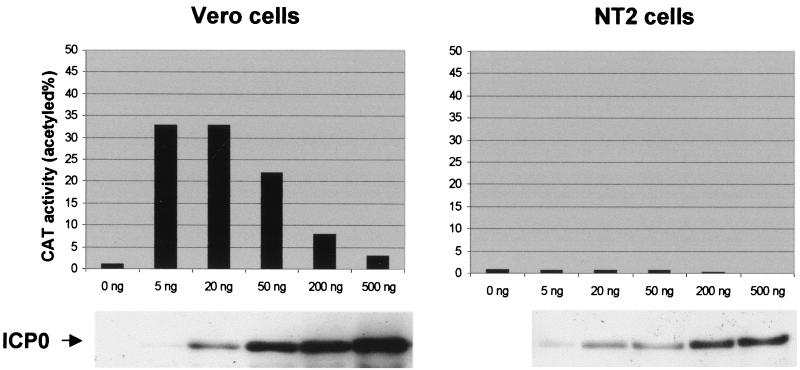
FIG. 7.
Comparison of the transactivation of an ICP6 expression cassette by ICP0 in NT2 and HFL cells. Cells were cotransfected with reporter plasmid pSS80 (ICP6 promoter linked to the CAT gene) and the indicated amounts of the ICP0 expression plasmid pCI-rtag-cICP0 (see Materials and Methods). The Western blot analysis of the total cell proteins of samples transfected in parallel with the same amount of plasmid and probed for ICP0 is shown below. The results of this single experiment were reproduced on several independent occasions, and similar results were obtained with a reporter containing the glycoprotein gD promoter.
RESULTS
Neuronal committed teratocarcinoma NT2 cells have highly abnormal ND10.
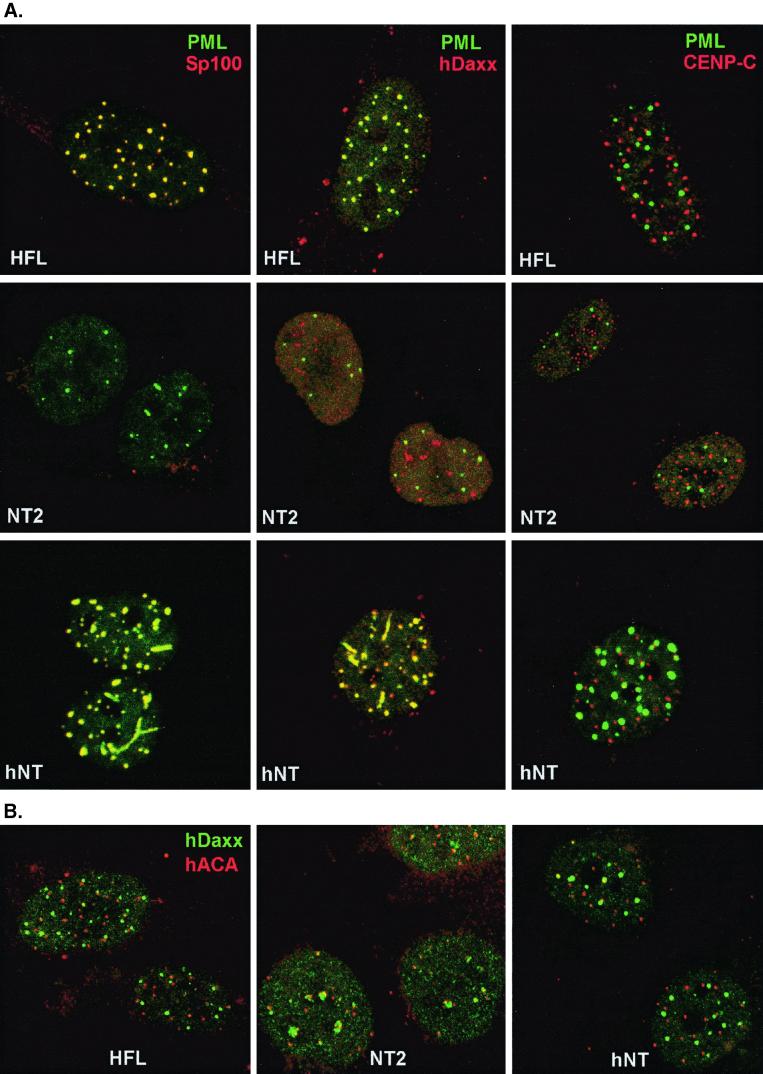
The first stage of this project was to compare the composition and morphology of ND10 in neuronal committed NT2 and fibroblast HFL cells. Double staining for the major ND10 antigens PML and Sp100 showed the normal extensive colocalization of the two proteins in multiple punctate nuclear foci in HFL cells, but the overwhelming majority of NT2 cells had only background levels of Sp100 (27) and fewer PML foci (Fig. 1A). The distribution of PML was unusual in many NT2 cells, commonly with strings, tracks or lines of small foci extending through the nucleoplasm. One of the NT2 cells shown in Fig. 1 has two moderate examples of this phenotype, but many others had PML tracks similar to those in the hNT cells shown in Fig. 1A. In HFL cells, the PML-associated protein hDaxx (27) was present throughout the nucleus and in local accumulations which in many cases strongly colocalized with PML (Fig. 1A). In contrast, in NT2 cells hDaxx was much less well associated with PML (Fig. 1A) and the distinct accumulations of hDaxx were instead preferentially associated with centromeres (Fig. 1B). This is reminiscent of recent data which showed that hDaxx can be present at both ND10 and centromeres, and its relative association with these structures can be influenced by cell status (22). In contrast to the abnormal ND10, the staining pattern of centromere protein CENP-C in NT2 cells was similar to that in HFL cells (Fig. 1A).
FIG. 1.
Distinct expression patterns of ND10 and centromere proteins in HFL, NT2, and hNT cells. Confocal micrographs of the three cell lines are presented, as indicated by white lettering. (A) The labeled proteins are indicated in the top row and are the same in each vertical row of panels. PML was stained with mouse MAb 5E10 (green), and the cells were also costained with SpGH (lefthand panels), rabbit serum r1866 (center panels), or rabbit serum r554 (righthand panels) for Sp100, hDaxx, and CENP-C, respectively. There is no Sp100 expression and less PML expression in NT2 cells, and PML does not colocalize efficiently with hDaxx. In HFL and hNT cells, PML extensively colocalizes (yellow) with Sp100 and hDaxx but not with CENP-C. (B) Localization of hDaxx (green) and centromere proteins (red). Cells were double labeled with r1866 and human serum ACA-GS which contains antibodies against several centromere proteins. In NT2 cells, a significant proportion of centromeres are associated with local accumulations of hDaxx, but this occurs more rarely in HFL and hNT cells.
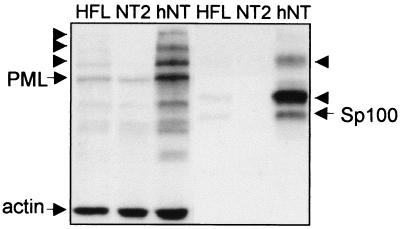
In confirmation of the results of Ishov et al. (27), Western blotting of whole-cell extracts of NT2 compared to HFL cells confirmed that NT2 cells contained negligible amounts of Sp100 (Fig. 2). We also found that NT2 cells contained comparatively low amounts of PML and the proportion of PML that was modified by the small ubiquitin-like protein SUMO-1 was apparently much reduced in NT2 compared to HFL cells (Fig. 2). It is likely that the relatively poor association of hDaxx with PML detected by fluorescence (Fig. 1A) can be explained by the less extensively SUMO-1-modified PML in NT2 cells (Fig. 2) because hDaxx interacts with PML only when the latter is SUMO-1 modified (27).
FIG. 2.
Western blot analysis of the expression patterns of Sp100 and PML in HFL, NT2, and hNT cells. The lefthand three tracks show PML species detected by 5E10, with actin detected simultaneously as the internal control. The righthand three lanes show the same filter after stripping and reprobing for Sp100 with SpGH. The arrowheads mark the presumed SUMO-1-modified forms of PML and Sp100. NT2 cells do not express Sp100, whereas in hNT cells long-term induction of retinoic acid results in a drastic increase of both unmodified and modified forms of Sp100 and PML. The extent of SUMO-1 modification of PML is reduced in NT2 cells.
Differentiation of NT2 cells into neuron-like hNT cells restores ND10 structure.
NT2 cells can be induced to differentiate into nondividing hNT cells by treatment with retinoic acid for several weeks (41). The resultant hNT cells acquire a neuron-like morphology and express a number of neuron-specific markers (42). Staining of hNT cells for PML and Sp100 revealed that, in great contrast to the parental NT2 cells, hNT cells have striking ND10 with prominent foci of both proteins colocalizing precisely (Fig. 1A). Again, tracks or chains of small beads of PML were common. A further difference between NT2 and hNT cells was that in the latter hDaxx was strongly associated with PML (Fig. 1A) and much less well associated with centromeres (Fig. 1B). These data correlate with strikingly increased amounts of both Sp100 and PML, and particularly SUMO-1-modified PML, in hNT cells (Fig. 2). Since hDaxx does not interact with Sp100 (27), this result strengthens the conclusion that the distribution of hDaxx is dependent on PML and its modification. Thus, either ATRA itself or the process of differentiation induces substantial changes in the quantities of PML and Sp100 present in the cells, with consequent alterations in ND10 appearance and composition. Our previous examination of the precursor NT2 cells had found occasional cells with prominent ND10 containing both PML and Sp100 (data not shown), and it is likely that these cells represent the small number of cells which are known to undergo spontaneous differentiation in NT2 cultures. Time course experiments showed that the increased amounts of Sp100 appeared after at least 3 weeks of treatment with retinoic acid, which implies that these changes result from the process of differentiation rather than from a simple induction of PML and Sp100 transcription by retinoic acid.
HSV-1 replicates poorly in NT2 and hNT cells.
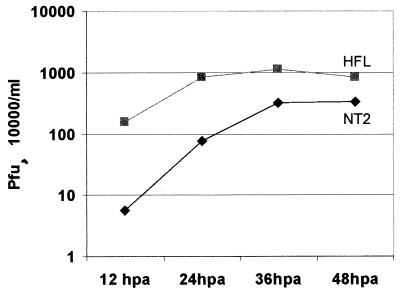
As detailed in the introduction, previous work found that HSV-1 has a growth defect, particularly in low-multiplicity infections, in rodent neuroblastoma cell lines or cells which could be differentiated into a neuron-like state. Equivalent studies on HSV-1 infection of human neuroblastoma cell lines had not investigated low-multiplicity infections in any detail, and there was no information on the requirement for and functionality of ICP0 in such cells. Therefore, we compared the efficiency of HSV-1 replication in HFL and NT2 cells. An initial time course experiment using a multiplicity of 1 PFU per cell showed that the virus replicated slightly more slowly in NT2 cells than in HFL cells, but by 36 h the yield of virus was only moderately reduced. Extending the incubation period to 48 h did not significantly alter the results from those obtained at 36 h (Fig. 3). A series of parallel infections were conducted on NT2 and HFL cells at multiplicities varying from 50 to 0.01 PFU per cell, and the progeny virus was harvested and titrated 36 h later. This length of incubation allows multiple rounds of viral replication in HFL cells, so the final yields were not significantly affected by input multiplicity in this cell type (Table 1). In contrast, at an input multiplicity of 0.01, the yield of virus was reduced 400-fold in NT2 compared to HFL cells (Table 1). Similar results were obtained after extending the incubation period to 48 h, which indicates that the defect cannot be explained simply by delayed kinetics (Table 2). Comparison of NT2 and hNT cells in parallel experiments demonstrated that HSV-1 17+ replicated slightly less well in the differentiated cells at all multiplicities, but the differences between NT2 and hNT cells were small compared to those between NT2 and HFL cells (Table 1). We attempted to compare the plating efficiency of the virus in the three cell lines but, because of the poor growth in NT2 and hNT cells, plaque formation on these cells was too poor to be scored.
FIG. 3.
Growth curves of HSV-1 strain 17+ in HFL and NT2 cells. Cells were infected with the virus at 1 PFU per cell, and both cells and medium were harvested at the indicated times after infection; progeny virus titers were then determined on BHK cells. The data are the average of two independent experiments.
TABLE 1.
Replication of HSV-1 strain 17+ in HFL, NT2, and hNT cellsa
| Input MOIb | Titer (PFU/ml) at 36 h in cell type:
|
Fold reduction
|
|||
|---|---|---|---|---|---|
| HFL | NT2 | hNT | NT2 | hNT | |
| 50 | 9.4 × 107 | 7.4 × 107 | 1.2 × 107 | 1.3 | 7.8 |
| 10 | 6.3 × 107 | 2.7 × 107 | 1.0 × 107 | 2.4 | 6.3 |
| 1 | 6.2 × 107 | 1.9 × 107 | 5.6 × 106 | 3.0 | 11 |
| 0.1 | 7.8 × 107 | 1.1 × 106 | 6.8 × 105 | 70 | 114 |
| 0.01 | 7.3 × 107 | 1.8 × 105 | 5.9 × 104 | 414 | 1,228 |
The three cell types were infected with HSV-1 strain 17+ at the indicated multiplicities, and then the cells and medium were harvested after 36 h and sonicated. The virus progeny was titrated on BHK cells. The fold reduction of the virus titers in NT2 and hNT cells compared to HFL cells is shown. Note that the yields of virus in HFL cells do not decrease with multiplicity because rapid viral replication ensures all the cells become infected during the time course of the experiment.
MOI, multiplicity of infection.
TABLE 2.
Efficiency of replication of HSV-1 strain 17+ and ICP0 mutants dl1403 and M1 in HFL, NT2, and hNT cellsa
| Virus and input MOIb | Titer (PFU/ml) at 36 h in cell type:
|
Fold reduction
|
|||
|---|---|---|---|---|---|
| HFL | NT2 | HFL vs. NT2 at 36 h | HFL vs. NT2 at 48 h | NT2 vs. hNT at 36 h | |
| 17+ | |||||
| 0.01 | 2.0 × 108 | 4.3 × 105 | 465 | 244 | 3.0 |
| 0.1 | 8.7 × 107 | 8.7 × 106 | 10 | 12 | 1.6 |
| 1 | 7.1 × 107 | 2.8 × 107 | 3 | 3 | 3.7 |
| dl1403 | |||||
| 5 | 3.8 × 106 | 5.5 × 104 | 69 | 77 | 4.7 |
| 50 | 8.2 × 106 | 5.1 × 105 | 16 | 10 | 1.2 |
| 500 | 7.6 × 106 | 5.1 × 105 | 15 | 10 | 2.5 |
| M1 | |||||
| 0.01 | 1.5 × 107 | 5.7 × 103 | 2,667 | 26,000 | 0.3 |
| 0.1 | 7.1 × 107 | 6.1 × 104 | 1,164 | 562 | 0.46 |
| 1 | 7.3 × 107 | 1.4 × 105 | 514 | 17 | 0.57 |
Viruses 17+, dl1403, and M1 were used to infect the three cell types at the indicated multiplicities (calculated on the basis of titration in U2OS cells). Cells and medium were harvested 36 and 48 h later and sonicated, and the virus progeny was titrated on U2OS cells. Actual titration figures are given for the 36-h samples. The fold reduction of the virus titers in NT2 compared to HFL cells are shown at the two time points. Note that the yields of wild-type virus in HFL cells do not decrease with multiplicity because rapid viral replication ensures all the cells become infected during the time course of the experiment. The high figure for the reduction of M1 replication at the lowest multiplicity in NT2 cells was due to further replication of the virus between 36 and 48 h in HFL cells but not in NT2 cells. Comparative yields of the viruses in NT2 and hNT cells from a separate experiment are given in the last column. vs., versus.
MOI, multiplicity of infection.
ICP0-deficient mutants also replicate poorly in NT2 and hNT cells.
Next, we compared the growth efficiencies in HFL and NT2 cells of ICP0-null mutant virus dl1403 and the USP7-binding negative mutant M1. Deletion dl1403 has been characterized as having a severe multiplicity-dependent growth defect in HFL cells (54), while the interaction with the ubiquitin-specific protease USP7 was found to contribute to ICP0 function in BHK cells and to a lesser extent in HFL cells (13, 21). To compare the yields of these viruses in the various cell types, we first determined the titers of the viral stocks on U2OS cells in which ICP0 function is not required, thus allowing efficient replication of wild-type and all ICP0 mutant viruses (60). We then infected HFL and NT2 cells with each virus at multiplicities based on the titers in U2OS cells and titrated the progeny virus in U2OS cells. Increased input multiplicities of dl1403 were used in order to initiate a productive infection. As expected, at 36 h after infection, dl1403 replicated poorly in HFL cells compared to the wild-type virus; even with the input virus increased 500-fold, yields were reduced by an order of magnitude (Table 2). At the highest multiplicity used, the yield of dl1403 was reduced by a further order of magnitude in NT2 cells compared to HFL cells, and extending the incubation period to 48 h did not significantly affect the results (Table 2). Virus M1 also replicated much less efficiently in NT2 compared to HFL cells at all multiplicities tested (Table 2), indicating that the ability of ICP0 to bind to USP7 is apparently very important for virus replication in NT2 cells. These data provide the most striking indication obtained so far that the ICP0-USP7 interaction is important for HSV-1 replication in certain situations. As with wild-type virus (Table 1), the replication efficiencies of the two mutant viruses in hNT cells were similar to those in NT2 cells, with dl1403 replicating slightly less well and M1 replicating slightly better in the differentiated cells (Table 2).
Viral gene expression is abnormal in NT2 and hNT cells.
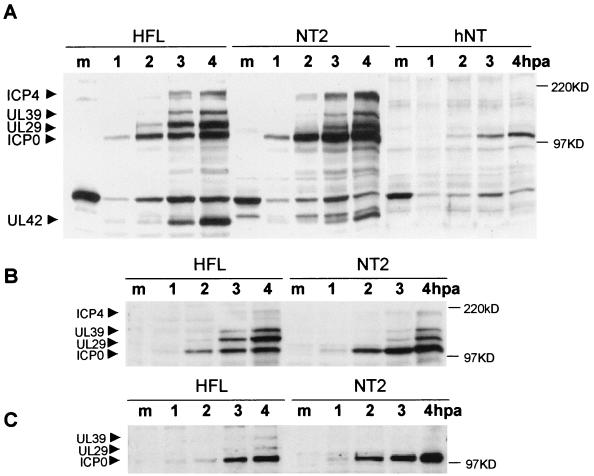
To investigate the basis of the poor growth of HSV-1 in NT2 and hNT cells, we compared viral gene expression in the two cell types with that in the fully permissive HFL cells. Whole-cell extracts were prepared at various times after infection and probed with a mixture of antibodies recognizing representative IE and early gene products; we probed for ICP0, ICP4, the large subunit of ribonucleotide reductase UL39 (ICP6), the major DNA-binding protein UL29 (ICP8), and the DNA polymerase accessory factor UL42. It should be noted that, because of the varied antibody sensitivities, this Western blot approach compares the relative expression of this group of proteins in the different samples rather than measuring absolute protein levels. At a multiplicity of infection of 5 there was, as might be expected on the basis of the titration data, little difference in the patterns and time course of viral gene expression in HFL and NT2 cells (Fig. 4A). However, reduction of the multiplicity to 1 PFU per cell revealed an intriguing phenotype. At 2 h postinfection, some UL29 expression was detectable in HFL cells, but viral gene expression in NT2 cells was restricted to ICP0 (Fig. 4B). At 3 h UL29 and UL39 expression was well marked in HFL cells but, compared to the expression of ICP0, was much reduced in NT2 cells. By 4 h the early viral gene products accumulated in NT2 cells, but their expression levels compared to ICP0 were still reduced from those seen in HFL cells. These effects were more pronounced at a multiplicity of infection of 0.2 (Fig. 4C). In hNT cells, even at 4 h after infection with a multiplicity of 5, the expression of UL29 and UL39 was only just detectable (Fig. 4A). Since ICP0 is expressed with greater or equivalent efficiency in NT2 and hNT compared to HFL cells, these data suggest that the virus is entering NT2 and hNT cells but that the infection progresses from the IE to later stages inefficiently.
FIG. 4.
Comparison of the rates of viral gene expression in HFL, NT2, and differentiated hNT cells. Cells were infected with 5 (A), 1 (B), or 0.2 (C) PFU of wild-type HSV-1 strain 17+ per cell and harvested before infection (lanes m) and at 1, 2, 3, and 4 h after absorption (hpa) and then processed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blot analysis. A mixture of antibodies recognizing several viral proteins was employed to detect IE proteins ICP0 and ICP4 and also the early genes UL39, UL29, and UL42 (as indicated on the left).
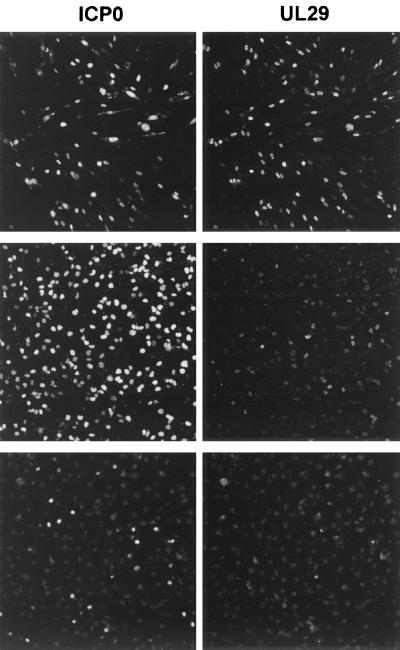
The results were supported by immunofluorescence analysis of infected HFL, NT2, and hNT cells. At 8 h postinfection, most infected HFL cells express abundant UL29, whereas in NT2 cells (seeded at a higher density in this example) few of the cells had progressed to this stage of infection (Fig. 5). At this time point, none of the infected hNT cells expressing ICP0 were expressing detectable levels of UL29 (Fig. 5).
FIG. 5.
Inefficient progression of viral infection in NT2 and hNT cells. HFL (top panels), NT2 (middle panels), and hNT (bottom panels) cells were infected with wild-type HSV stain 17+ at 1 PFU per cell. Infected cells were fixed 8 h after absorption and prepared for immunofluorescence and simultaneous staining for ICP0 (lefthand panels) and UL29 (righthand panels), using MAb 11060 and serum r515 as markers of viral IE and E gene expression. In hNT cells, the number of infected cells expressing ICP0 and UL29 is much less than in HFL and NT2 cells. The number of NT2 cells expressing ICP0 is equivalent to the HFL infections, but relatively few NT2 cells are expressing large amounts of UL29.
The efficient expression of ICP0 but not of other viral polypeptides in NT2 and hNT cells suggests that the activation of gene expression induced by ICP0 is compromised. It is conceivable that one factor in the poor growth of HSV-1 in these cells is that ICP0 is unable to function normally. Therefore, we designed further experiments to test this possibility.
ICP0 affects ND10 and centromeres more slowly in NT2 cells than in HFL cells.
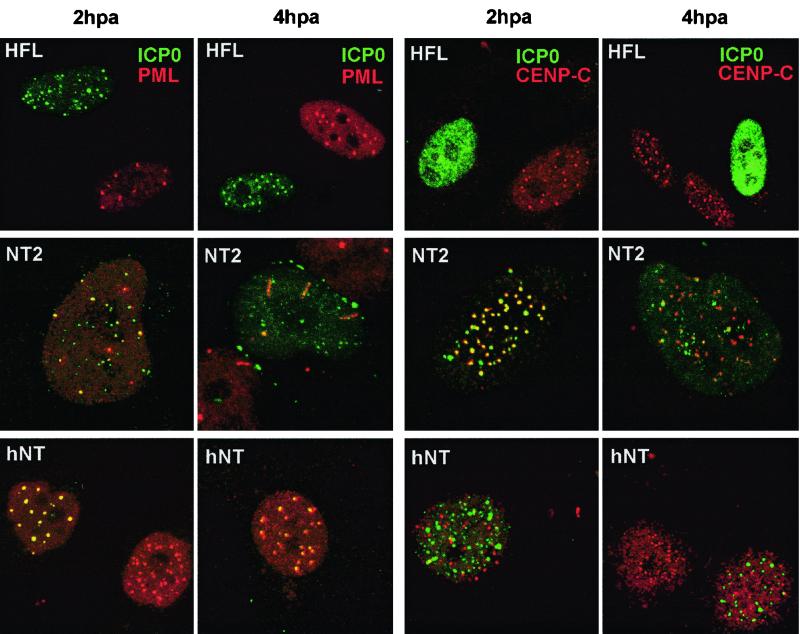
ICP0 causes major effects on the cell by inducing the degradation of PML, Sp100, and CENP-C, thereby disrupting both ND10 and centromeres (12). We therefore compared the efficiency by which ICP0 induced these effects in HFL, NT2, and hNT cells. Cells were infected with HSV-1 17+ at a multiplicity of 1 and then stained for ICP0 and the cellular proteins PML, CENP-C, and hDaxx at various times postinfection. In the early stages of infection, ICP0 colocalized with PML in all cell types, but as time progressed the rate at which the PML foci were disrupted in NT2 and hNT cells was noticeably slower than in HFL cells (Fig. 6 and Table 3). In this experiment, only 4% of infected HFL cells retained PML foci by 2 h postinfection, whereas 56% of NT2 cells did so. Even by 4 h postinfection, 54% of infected NT2 cells retained punctate PML staining (Table 3). Similar results were obtained in infected cells stained for CENP-C (Fig. 6 and Table 3) and hDaxx (Table 3). Loss of CENP-C from centromeres in infected HFL cells was extremely rapid, but in NT2 cells ICP0 markedly colocalized with CENP-C at early times of infection to a greater degree than in any other cell type we have studied (Fig. 6). It is intriguing that ICP0 associates so well with centromeres in this cell type, in which PML is poorly expressed and less well modified by SUMO-1 (Fig. 2) and in which hDaxx becomes more highly associated with centromeres than with ND10 (Fig. 1B). This suggests that ICP0 associates with a factor which can be present in both structures but is predominantly associated with centromeres in NT2 cells. At later times of infection, although about two-thirds of infected NT2 cells had lost CENP-C from centromeres (Table 3), in the remaining one-third ICP0 either remained at centromeres and CENP-C was much less completely degraded or ICP0 became more diffuse yet CENP-C remained in characteristic centromeric foci (Fig. 6). The results in hNT cells were similar to those in NT2 cells, except that with the increased expression of PML and its modification by SUMO-1 in hNT cells (Fig. 2), ICP0 strongly colocalized with PML yet inefficiently disrupted ND10 (Fig. 6). Correspondingly, ICP0 colocalized with centromeres less efficiently in hNT cells and again caused their disruption inefficiently (Fig. 6).
FIG. 6.
Comparison of the stability of ND10 and centromeres in HFL, NT2, and hNT cells at 2 and 4 h after the start of infection. HFL (top panels), NT2 (middle panels), and hNT (bottom panels) cells were infected with wild-type HSV stain 17+ at 1 PFU per cell. After various times of infection, cells were fixed and stained for ICP0 (MAb 11060) and PML (r8) (lefthand pairs of panels) or ICP0 and CENP-C (r554) (righthand pairs of panels). Both ND10 and centromeres, as judged by PML and CENP-C staining, are more stable in infected NT2 and hNT than in HFL cells.
TABLE 3.
Proportion of infected cells retaining ND10 proteins and CENP-C in infected HFL and NT2 cellsa
| Cell type | % of infected cells retaining:
|
|||||
|---|---|---|---|---|---|---|
| PML at:
|
CENP-C at:
|
hDaxx at:
|
||||
| 2 h | 4 h | 2 h | 4 h | 2 h | 4 h | |
| HFL | 4 | 0 | 0 | 0 | 0 | 0 |
| NT2 | 56 | 54 | 55 | 35 | 49 | 32 |
Cells were infected with wild-type HSV-1 stain 17+ at 1 PFU per cell and then fixed and double stained for ICP0 and PML, CENP-C, or hDaxx separately. The proportion of infected cells retaining punctate staining for PML, CENP-C, or hDaxx was calculated after counting 300 infected cells per coverslip at the indicated times after virus adsorption.
These results are consistent with either of the following hypotheses: either ICP0 activity is compromised in NT2 and hNT cells, so that it affects ND10 proteins more slowly and this correlates with less efficient infection or ICP0 stimulates viral gene expression and infection less efficiently, causing reduced effects on the ND10 proteins. Either way, it appears that ICP0 functions less efficiently in NT2 and hNT cells. Since it has been shown that ICP0 alone can disrupt ND10 and induce the degradation of PML (17, 39, 40), the former explanation seems the more likely.
Evidence that ICP0 activates gene expression poorly in NT2 cells.
To obtain more direct evidence that ICP0 activates gene expression less efficiently in NT2 cells, we assessed the activation of gene expression induced by HSV-1 IE proteins in transfected cells. Because HFL cells transfect very poorly, we compared the results using Vero and NT2 cells. Initially, we optimized the amount of pSS80 (a reporter plasmid containing the ICP6 promoter linked to the CAT gene) required to give low levels of basal activity, and then we cotransfected the cells with increasing amounts of the ICP0 expression vector pCI-rtag-cICP0. In Vero cells, maximal activation was obtained using 5 to 50 ng of pCI-rtag-cICP0, while higher amounts had less effect (Fig. 7). However, in NT2 cells pCI-rtag-ICP0 did not give any significant activation of gene expression (Fig. 7). Although Western blotting of the extracts showed that ICP0 was slightly less well expressed in NT2 cells, 20 ng of pCI-rtag-ICP0 in NT2 cells gave greater expression of ICP0 than 5 ng in Vero cells, yet it failed to activate gene expression (Fig. 7).
Since ICP0 and ICP4 have been shown to activate gene expression synergistically in some situations (14), we compared the activation of gene expression induced by ICP4 and the combination of ICP4 and ICP0 in transfected cells. Again, we found that both ICP4 and, particularly, the ICP4-ICP0 combination were poor activators of gene expression in transfected NT2 cells compared to Vero cells (data not shown). Western blotting showed that ICP4 was less well expressed in NT2 cells, but this could not by itself explain the poor activation of gene expression observed. Thus, the reduced activation of viral gene expression seen in NT2 cells (Fig. 4) could be in part a consequence of reduced activity of both ICP0 and ICP4.
These transfection experiments suggest that ICP0 and ICP4 are poor activators of gene expression in NT2 cells, but a number of factors need to be considered. Previous studies have suggested that HSV-1 IE proteins are poorly expressed in neuroblastoma cells because Oct-1, a factor required for basal and VP16-activated IE promoter activation, is less abundantly expressed than the related repressive Oct-2 factors (33, 34). This cannot explain our current results since ICP0 and ICP4 are expressed from the HCMV or simian virus 40 SV40 promoter-enhancer regions in our plasmids. In addition, the results cannot be explained by intrinsic poor transfection of NT2 cells since similar HCMV enhancer plasmids expressing control proteins PML and SUMO-1 gave at least 30% of positive transfected NT2 cells as estimated by immunofluorescence, and similar amounts of these proteins were expressed in transfected NT2 compared to Vero cells as detected by Western blotting (data not shown).
Low-multiplicity infection of NT2 and hNT cells produces quiescently infected cells which can be reactivated by ICP0 expressed by a superinfecting virus.
Previous studies have shown that infection by ICP0 mutant viruses can result in the maintenance of quiescent genomes in nonproductively infected cells and that these genomes can be reactivated by later expression of exogenous ICP0 (46). The poor infection of NT2 cells by both wild-type and ICP0 mutant viruses and the apparently poor function of ICP0 in NT2 cells suggests that a similar situation could be occurring in these cells. Recent studies of this type have used heavily defective viruses to reduce the initial viral infectivity as much as possible, but the very poor growth of dl1403 on NT2 cells allowed the examination of quiescent infection with dl1403/lacZ, a relatively viable virus which carries a lacZ gene driven by the HCMV promoter and the dl1403 ICP0 deletion.
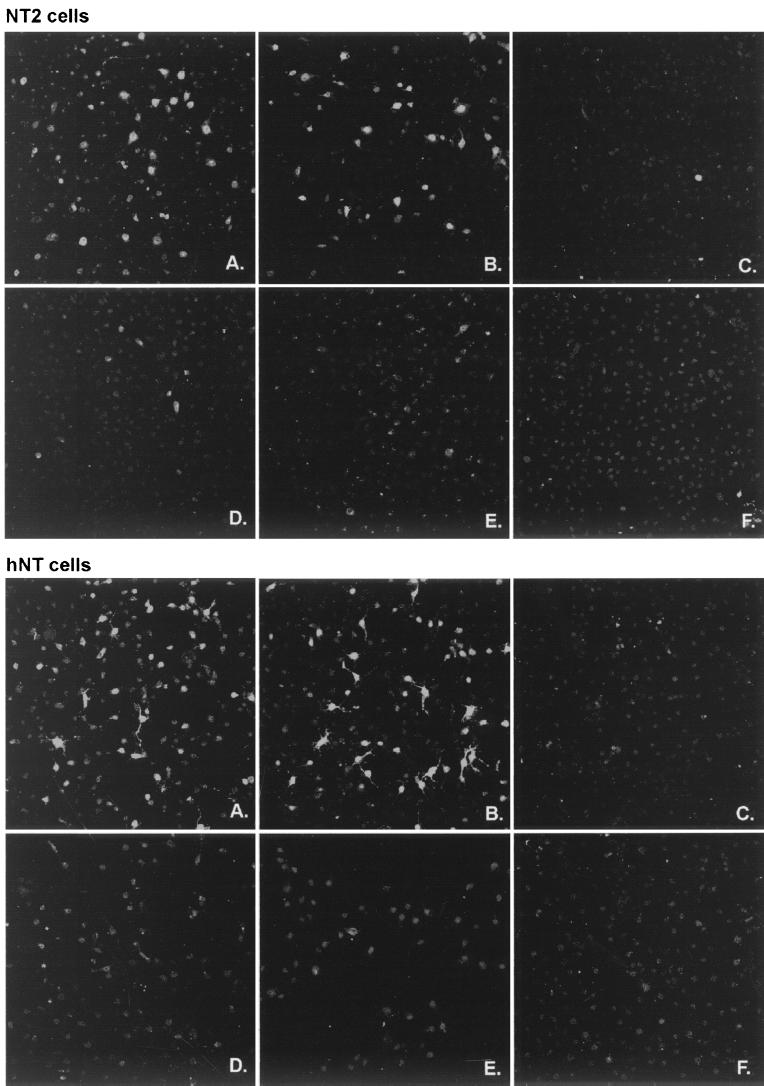
NT2 cells were infected with dl1403/lacZ at a multiplicity of 0.02 PFU per cell (BHK cell titer) and then maintained for 2 days at 37°C. Except for the occasional plaque of replicating virus, indirect immunofluorescence staining showed that the cells in the monolayer did not express β-galactosidase: the HCMV promoter was inactive. The cultures were then superinfected at 38.5°C with 3 PFU per cell of wild-type HSV-1 strain 17+, ICP0 RING finger deletion mutant FXE, ICP4 mutant tsK, or the ICP4 tsK-ICP0 double mutant in1330 and then stained for β-galactosidase expression 8 h later. A similar experiment was conducted with hNT cells, except that they were stained 16 h after superinfection because of the relatively poor growth of strain 17+ in these cells (Table 1). The results (Fig. 8) showed that superinfection with both 17+ and tsK caused the expression of β-galactosidase as a result of reactivation of the quiescent HCMV promoter in the initially infecting virus. The equivalent ICP0-deficient viruses FXE and in1330 were unable to reactivate the quiescent virus efficiently, thus demonstrating that ICP0 is required for this effect. The addition of the proteasome inhibitor MG132 to the reactivation experiment demonstrated that, as in a previous study (19), this activity of ICP0 requires an active proteasome degradation pathway (Fig. 8).
FIG. 8.
Establishment of quiescently infected NT2 and hNT cells using an ICP0-deficient virus and reactivation by superinfecting ICP0-positive virus. Cells were infected as described in Materials and Methods. NT2 (upper field) and hNT (lower field) cells were initially infected with dl1403/lacZ virus and then superinfected with HSV-1 strain 17+ (A), tsK (B), FXE (C), in1330 (D), 17+ in the presence of MG132 (E), or else were mock infected (F). NT2 and hNT cells were fixed 8 and 16 h postsuperinfection, respectively, and then stained for β-galactosidase expression by immunofluorescence. Quiescent virus can be reactivated by viruses expressing functional ICP0 viruses (A and B) but not by ICP0-deficient viruses (C and D) or if the ICP0 activity is inhibited by MG132 (E).
DISCUSSION
In this study we have compared the expression and distribution of selected ND10 proteins in human fibroblast HFL, neuronal precursor NT2, and differentiated neuron-like hNT cells and then compared their infection by HSV-1 and the functionality of ICP0 in the three situations. We found striking differences in ND10 structure between HFL and NT2 cells and further substantial changes in the differentiated hNT cells. Both NT2 and hNT cells were poor hosts for HSV-1 replication, particularly in low-multiplicity infections, and the data suggest that a contributing factor to this poor growth could be the apparently poor functionality of ICP0 in both cell types.
Our results demonstrating negligible expression of Sp100 and low-level expression of PML in NT2 cells correlate well with previous studies (27, 32), but we have extended the previous data by demonstrating highly aberrant PML track structures in many NT2 and hNT cells and a relatively low proportion of SUMO-1-modified PML in NT2 compared to HFL cells. Differentiation of NT2 into nondividing hNT cells resulted in massive increases in the expression of both PML and Sp100, which correlated with the establishment of prominent ND10 foci in which the two proteins strongly colocalized. Since this effect occurred only after several weeks of ATRA treatment, it is likely that it is in some way associated with the process of differentiation rather than with the ATRA treatment itself. This notion is consistent with the presence of low numbers of cells in NT2 populations which express high levels of Sp100, which are probably in the process of spontaneously differentiating.
Previous studies using cultured cells with neuronal features for HSV-1 infection have concentrated on rodent cell lines such as mouse C1300 neuroblastoma cells and derivatives (28, 29) or cells derived from rat PC12 cells (47, 56). Similar to our present results, these rodent cell lines were found to be poor hosts for HSV-1 infection in low-multiplicity infections. The advantage of the human NT2 and hNT cells is that it is possible to correlate the efficiency of viral infection with the effects of ICP0 on ND10 and centromeres since most available antibodies against components of these structures recognize the human but not the equivalent rodent proteins. We found that despite early expression of ICP0 in NT2 and hNT cells, the progression of viral gene expression into the pattern characteristic of lytic infection was delayed. This correlated with relatively slow disruption of ND10, prolonged and striking association of ICP0 with centromeres (in NT2 cells), and inefficient induced loss of centromere protein CENP-C. It is unlikely that these effects are a nonspecific consequence of the generally poor infection because ICP0 can disrupt ND10 and centromeres in the absence of other viral proteins. Indeed, the observations that ICP0 is relatively abundantly expressed in NT2 cells, as detected by Western blotting, and the slow disruption of ND10 and centromeres in cells that are clearly expressing ICP0 suggests that ICP0 function itself may be compromised in these cells. This contention is supported by the transfection experiments which suggest that ICP0 is a poor activator of gene expression in NT2 cells.
Our results are also relevant to the debate on whether the degradation of ND10 proteins PML and Sp100 specifically contributes to ICP0 function or whether it is the process by which this degradation occurs which is important. Although NT2 cells express relatively low levels of PML and no detectable Sp100, while hNT cells express hugely increased amounts of both proteins, hNT cells are only slightly less permissive for HSV-1 infection than are NT2 cells. While many other factors may be affecting the efficiency of viral replication in these two cell types, it is clear that the endogenous expression levels of PML and Sp100 are not dominant factors.
The replication efficiency of ICP0-deficient viruses is known to be very dependent on cell type, such that the probability of an ICP0 mutant establishing a lytic infection in HFL cells is reduced about 2,000-fold in HFL cells compared to U2OS cells (13, 60). NT2 and hNT cells are even more resistant than HFL cells to productive infection by dl1403 (Table 2). It is noteworthy that the poor growth of dl1403 in NT2 and hNT cells is not a trivial consequence of failure of the virus to enter the cells since, in a manner analogous to the establishment of quiescent infection in cultured cells using multiply defective viral genomes (45, 50), at least a proportion of the input dl1403 genomes can be reactivated by superinfection with viruses expressing ICP0. A potential application of these findings could be to investigate the control of gene expression from quiescent viral genomes in cultured human cells with neuronal characteristics.
We have shown that cultured human cells with neuronal characteristics are poor hosts for productive HSV-1 infection in a way that correlates at least in part with poor ICP0 functionality. Cells infected at a low multiplicity progress inefficiently into the lytic cycle, and in the absence of ICP0 many cells become quiescently infected. It is an interesting speculation that, if these observations hold true in human neuronal cells in vivo, a controlling factor in the establishment of latent infections in neurons in natural infections could be the relative activity of ICP0 in these cells.
ACKNOWLEDGMENTS
We thank Duncan McGeoch and Chris Preston for helpful comments on the manuscript. We also thank Chris Preston for providing the multiply defective HSV-1 strains and for advice on the establishment of quiescently infected cultures. Paul Freemont, Roel van Driel, Thomas Sternsdorf, Ann Pluta, Bill Earnshaw, and Howard Marsden kindly provided antibodies for use in these studies.
Work in the laboratory of R.D.E. is funded by the Medical Research Council; W.-L.H. was supported by a Glasgow University Overseas Research Student Scholarship.
REFERENCES
- 1.Ash R J. Butyrate-induced reversal of herpes simplex virus restriction in neuroblastoma cells. Virology. 1986;155:584–592. doi: 10.1016/0042-6822(86)90218-7. [DOI] [PubMed] [Google Scholar]
- 2.Block T, Barney S, Masonis J, Maggioncalda J, Valyi-Nagy T, Fraser N W. Long term herpes simplex virus type 1 infection of nerve growth factor-treated PC12 cells. J Gen Virol. 1994;75:2481–2487. doi: 10.1099/0022-1317-75-9-2481. [DOI] [PubMed] [Google Scholar]
- 3.Boddy M N, Howe K, Etkin L D, Solomon E, Freemont P S. PIC 1, a novel ubiquitin-like protein which interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukaemia. Oncogene. 1996;13:971–982. [PubMed] [Google Scholar]
- 4.Chatterjee S, Burns P, Koga J. Effect of protein kinase C inhibitors on the antiviral activity of human alpha interferon in herpes simplex virus-infected human neuroblastoma cells. J Virol. 1995;69:1315–1318. doi: 10.1128/jvi.69.2.1315-1318.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chou J, Roizman B. The gamma 1(34.5) gene of herpes simplex virus 1 precludes neuroblastoma cells from triggering total shutoff of protein synthesis characteristic of programed cell death in neuronal cells. Proc Natl Acad Sci USA. 1992;89:3266–3270. doi: 10.1073/pnas.89.8.3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conner J, Murray J, Cross A, Clements J B, Marsden H S. Intracellular localisation of herpes simplex virus type 1 ribonucleotide reductase subunits during infection of cultured cells. Virology. 1995;213:615–623. doi: 10.1006/viro.1995.0033. [DOI] [PubMed] [Google Scholar]
- 7.Earnshaw W C, Rothfield N. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma. 1985;91:313–321. doi: 10.1007/BF00328227. [DOI] [PubMed] [Google Scholar]
- 8.Everett R D. The products of herpes simplex virus type 1 (HSV-1) immediate early genes 1, 2 and 3 can activate HSV-1 gene expression in trans. J Gen Virol. 1986;67:2507–2513. doi: 10.1099/0022-1317-67-11-2507. [DOI] [PubMed] [Google Scholar]
- 9.Everett R D. A detailed mutational analysis of Vmw110, a trans-acting transcriptional activator encoded by herpes simplex virus type 1. EMBO J. 1987;6:2069–2076. doi: 10.1002/j.1460-2075.1987.tb02472.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Everett R D. Construction and characterization of herpes simplex virus type 1 mutants with defined lesions in immediate early gene 1. J Gen Virol. 1989;70:1185–1202. doi: 10.1099/0022-1317-70-5-1185. [DOI] [PubMed] [Google Scholar]
- 11.Everett R D. A surprising role for the proteasome in the regulation of herpesvirus infection. Trends Biochem Sci. 1999;24:293–295. doi: 10.1016/s0968-0004(99)01433-4. [DOI] [PubMed] [Google Scholar]
- 12.Everett R D. ICP0—a regulator of herpes simplex virus lytic and latent infection. Bioessays. 2000;22:761–770. doi: 10.1002/1521-1878(200008)22:8<761::AID-BIES10>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 13.Everett R D. ICP0 induces the accumulation of co-localizing conjugated ubiquitin. J Virol. 2000;74:9994–10005. doi: 10.1128/jvi.74.21.9994-10005.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Everett R D, Preston C M, Stow N D. Functional and genetic analysis of the role of Vmw110 in herpes simplex virus replication. In: Wagner E K, editor. The control of herpes virus gene expression. Boca Raton, Fla: CRC Press, Inc.; 1991. p. 50-76. [Google Scholar]
- 15.Everett R, Cross A, Tyler J, Orr A. An epitope within the DNA-binding domain of the herpes simplex virus immediate early protein Vmw175 is conserved in the varicella-zoster virus gene 62 protein. J Gen Virol. 1993;74:1955–1958. doi: 10.1099/0022-1317-74-9-1955. [DOI] [PubMed] [Google Scholar]
- 16.Everett R D, Cross A, Orr A. A truncated form of herpes simplex virus type 1 immediate-early protein Vmw110 is expressed in a cell type dependent manner. Virology. 1993;197:751–756. doi: 10.1006/viro.1993.1651. [DOI] [PubMed] [Google Scholar]
- 17.Everett R D, Maul G G. HSV-1 immediate-early protein Vmw110 causes redistribution of PML. EMBO J. 1994;13:5062–5069. doi: 10.1002/j.1460-2075.1994.tb06835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Everett R, O'Hare P, O'Rourke D, Barlow P, Orr A. Point mutations in the herpes simplex virus type 1 Vmw110 RING finger helix affect activation of gene expression, viral growth, and interaction with PML-containing nuclear structures. J Virol. 1995;69:7339–7344. doi: 10.1128/jvi.69.11.7339-7344.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Everett R D, Orr A, Preston C M. A viral activator of gene expression functions via the ubiquitin-proteasome pathway. EMBO J. 1998;17:7161–7169. doi: 10.1093/emboj/17.24.7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Everett R D, Freemont P, Saitoh H, Dasso M, Orr A, Kathoria M, Parkinson J. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J Virol. 1998;72:6581–6591. doi: 10.1128/jvi.72.8.6581-6591.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Everett R D, Meredith M, Orr A. The ability of herpes simplex virus type 1 immediate-early protein Vmw110 to bind to a ubiquitin-specific protease contributes to its roles in the activation of gene expression and stimulation of virus replication. J Virol. 1999;73:417–426. doi: 10.1128/jvi.73.1.417-426.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Everett R D, Earnshaw W C, Pluta A F, Sternsdorf T, Ainsztein A M, Carmena M, Ruchaud S, Hsu W L, Orr A. A dynamic connection between centromeres and ND10 proteins. J Cell Sci. 1999;112:3443–3454. doi: 10.1242/jcs.112.20.3443. [DOI] [PubMed] [Google Scholar]
- 23.Fields B N, Knipe D M, Howley P M, editors. Fields virology, 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. [Google Scholar]
- 24.Hagmann M, Georgiev O, Schaffner W, Douville P. Transcription factors interacting with herpes simplex virus alpha gene promoters in sensory neurons. Nucleic Acids Res. 1995;23:4978–4985. doi: 10.1093/nar/23.24.4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Homer E G, Rinaldi A, Nicholl M J, Preston C M. Activation of herpesvirus gene expression by the human cytomegalovirus protein pp71. J Virol. 1999;73:8512–8518. doi: 10.1128/jvi.73.10.8512-8518.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ishov A M, Maul G G. The periphery of nuclear domain 10 (ND10) as sites of DNA virus deposition. J Cell Biol. 1996;134:815–826. doi: 10.1083/jcb.134.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishov A M, Sotnikov A G, Negorev D, Vladimirova O V, Neff N, Kamitani T, Yeh E T, Strauss III J F, Maul G G. PML is critical for ND10 formation and recruits the PML-interacting protein Daxx to this nuclear structure when modified by SUMO-1. J Cell Biol. 1999;147:221–234. doi: 10.1083/jcb.147.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kemp L M, Latchman D S. Regulated transcription of herpes simplex virus immediate-early genes in neuroblastoma cells. Virology. 1989;171:607–610. doi: 10.1016/0042-6822(89)90631-4. [DOI] [PubMed] [Google Scholar]
- 29.Kemp L M, Gelman I H, Silverstein S J, Latchman D S. Regulation of herpes simplex virus immediate-early gene promoters in mouse neuroblastoma cells. Neurosci Lett. 1990;118:185–188. doi: 10.1016/0304-3940(90)90622-g. [DOI] [PubMed] [Google Scholar]
- 30.Kesari S, Randazzo B P, Valyi-Nagy T, Huang Q S, Brown S M, MacLean A R, Lee V M, Trojanowski J Q, Fraser N W. Therapy of experimental human brain tumors using a neuroattenuated herpes simplex virus mutant. Lab Investig. 1995;73:636–648. [PubMed] [Google Scholar]
- 31.Kondo Y, Yura Y, Iga H, Yanagawa T, Yoshida H, Furumoto N, Sato M. Effect of hexamethylene bisacetamide and cyclosporin A on recovery of herpes simplex virus type 2 from the in vitro model of latency in a human neuroblastoma cell line. Cancer Res. 1990;50:7852–7857. [PubMed] [Google Scholar]
- 32.Li H, Leo C, Zhu J, Wu X, O'Neil J, Park E J, Chen J D. Sequestration and inhibition of Daxx-mediated transcriptional repression by PML. Mol Cell Biol. 2000;20:1784–1796. doi: 10.1128/mcb.20.5.1784-1796.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lillycrop K A, Dent C L, Wheatley S C, Beech M N, Ninkina N N, Wood J N, Latchman D S. The octamer-binding protein Oct-2 represses HSV immediate-early genes in cell lines derived from latently infectable sensory neurons. Neuron. 1991;7:381–390. doi: 10.1016/0896-6273(91)90290-g. [DOI] [PubMed] [Google Scholar]
- 34.Lillycrop K A, Estridge J K, Latchman D S. The octamer binding protein Oct-2 inhibits transactivation of the herpes simplex virus immediate-early genes by the virion protein Vmw65. Virology. 1993;196:888–891. doi: 10.1006/viro.1993.1552. [DOI] [PubMed] [Google Scholar]
- 35.Marsden H S, Cross A M, Francis G J, Patel A H, MacEachran K, Murphy M, McVey G, Haydon D, Abbotts A, Stow N D. The herpes simplex virus type 1 UL8 protein influences the intracellular localization of the UL52 but not the ICP8 or POL replication proteins in virus-infected cells. J Gen Virol. 1996;77:2241–2249. doi: 10.1099/0022-1317-77-9-2241. [DOI] [PubMed] [Google Scholar]
- 36.Maul G G, Ishov A M, Everett R D. Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type-1. Virology. 1996;217:67–75. doi: 10.1006/viro.1996.0094. [DOI] [PubMed] [Google Scholar]
- 37.Maul G G. Nuclear domain 10, the site of DNA virus transcription and replication. Bioessays. 1998;20:660–667. doi: 10.1002/(SICI)1521-1878(199808)20:8<660::AID-BIES9>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 38.Maul G G, Negorev D, Bell P, Ishov A M. Review: properties and assembly mechanisms of ND10, PML bodies, or PODs. J Struct Biol. 2000;129:278–287. doi: 10.1006/jsbi.2000.4239. [DOI] [PubMed] [Google Scholar]
- 39.Muller S, Dejean A. Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins, correlating with nuclear body disruption. J Virol. 1999;73:5137–5143. doi: 10.1128/jvi.73.6.5137-5143.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parkinson J, Everett R. Alphaherpesvirus proteins related to herpes simplex virus type 1 ICP0 affect cellular structures and proteins. J Virol. 2000;74:10006–10017. doi: 10.1128/jvi.74.21.10006-10017.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pleasure S J, Page C, Lee V M. Pure, postmitotic, polarized human neurons derived from NTera 2 cells provide a system for expressing exogenous proteins in terminally differentiated neurons. J Neurosci. 1992;12:1802–1815. doi: 10.1523/JNEUROSCI.12-05-01802.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pleasure S J, Lee V M. NTera 2 cells: a human cell line which displays characteristics expected of a human committed neuronal progenitor cell. J Neurosci Res. 1993;35:585–602. doi: 10.1002/jnr.490350603. [DOI] [PubMed] [Google Scholar]
- 43.Pluta A F, Earnshaw W C, Goldberg I G. Interphase-specific association of intrinsic centromere protein CENP-C with HDaxx, a death domain-binding protein implicated in Fas-mediated cell death. J Cell Sci. 1998;111:2029–2041. doi: 10.1242/jcs.111.14.2029. [DOI] [PubMed] [Google Scholar]
- 44.Preston C M. Control of herpes simplex virus type 1 mRNA synthesis in cells infected with wild-type virus or the temperature-sensitive mutant tsK. J Virol. 1979;29:275–284. doi: 10.1128/jvi.29.1.275-284.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Preston C M, Nicholl M J. Repression of gene expression upon infection of cells with herpes simplex virus type 1 mutants impaired for immediate-early protein synthesis. J Virol. 1997;71:7807–7813. doi: 10.1128/jvi.71.10.7807-7813.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Preston C M. Repression of viral transcription during herpes simplex virus latency. J Gen Virol. 2000;81:1–19. doi: 10.1099/0022-1317-81-1-1. [DOI] [PubMed] [Google Scholar]
- 47.Ralph W M, Jr, Cabatingan M S, Schaffer P A. Induction of herpes simplex virus type 1 immediate-early gene expression by a cellular activity expressed in Vero and NB41A3 cells after growth arrest-release. J Virol. 1994;68:6871–6882. doi: 10.1128/jvi.68.11.6871-6882.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saitoh H, Tomkiel J, Cooke C A, Ratrie H D, Maurer M, Rothfield N F, Earnshaw W C. CENP-C, an autoantigen in scleroderma, is a component of the human inner kinetochore plate. Cell. 1992;70:115–125. doi: 10.1016/0092-8674(92)90538-n. [DOI] [PubMed] [Google Scholar]
- 49.Samaniego L A, Neiderhiser L, DeLuca N A. Persistence and expression of the herpes simplex virus genome in the absence of immediate-early proteins. J Virol. 1998;72:3307–3320. doi: 10.1128/jvi.72.4.3307-3320.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schenk P, Ludwig H. The 65 K DNA binding protein appears early in HSV-1 replication. Arch Virol. 1988;102:119–123. doi: 10.1007/BF01315568. [DOI] [PubMed] [Google Scholar]
- 51.Smith R L, Escudero J M, Wilcox C L. Regulation of the herpes simplex virus latency-associated transcripts during establishment of latency in sensory neurons in vitro. Virology. 1994;202:49–60. doi: 10.1006/viro.1994.1321. [DOI] [PubMed] [Google Scholar]
- 52.Sternsdorf T, Guldner H H, Szostecki C, Grotzinger T, Will H. Two nuclear dot-associated proteins, PML and Sp100, are often co-autoimmunogenic in patients with primary biliary cirrhosis. Scand J Immunol. 1995;42:257–268. doi: 10.1111/j.1365-3083.1995.tb03652.x. [DOI] [PubMed] [Google Scholar]
- 53.Sternsdorf T, Grotzinger T, Jensen K, Will H. Nuclear dots: actors on many stages. Immunobiology. 1997;198:307–331. doi: 10.1016/S0171-2985(97)80051-4. [DOI] [PubMed] [Google Scholar]
- 54.Stow N D, Stow E C. Isolation and characterization of a herpes simplex virus type 1 mutant containing a deletion within the gene encoding the immediate early polypeptide Vmw110. J Gen Virol. 1986;67:2571–2585. doi: 10.1099/0022-1317-67-12-2571. [DOI] [PubMed] [Google Scholar]
- 55.Stuurman N, de Graaf A, Floore A, Josso A, Humbel B, de Jong L, van Driel R. A monoclonal antibody recognizing nuclear matrix-associated nuclear bodies. J Cell Sci. 1992;101:773–784. doi: 10.1242/jcs.101.4.773. [DOI] [PubMed] [Google Scholar]
- 56.Su Y H, Meegalla R L, Chowhan R, Cubitt C, Oakes J E, Lausch R N, Fraser N W, Block T M. Human corneal cells and other fibroblasts can stimulate the appearance of herpes simplex virus from quiescently infected PC12 cells. J Virol. 1999;73:4171–4180. doi: 10.1128/jvi.73.5.4171-4180.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vahlne A, Lycke E. Herpes simplex virus infection of in vitro cultured neuronal cells (mouse neuroblastoma C 1300 cells) J Gen Virol. 1978;39:321–332. doi: 10.1099/0022-1317-39-2-321. [DOI] [PubMed] [Google Scholar]
- 58.Wagner E K, Bloom D C. Experimental investigation of herpes simplex virus latency. Clin Microbiol Rev. 1997;10:419–443. doi: 10.1128/cmr.10.3.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilcox C L, Smith R L, Everett R D, Mysofski D. The herpes simplex virus type 1 immediate-early protein ICP0 is necessary for the efficient establishment of latent infection. J Virol. 1997;71:6777–6785. doi: 10.1128/jvi.71.9.6777-6785.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yao F, Schaffer P A. An activity specified by the osteosarcoma line U2OS can substitute functionally for ICP0, a major regulatory protein of herpes simplex virus type 1. J Virol. 1995;69:6249–6258. doi: 10.1128/jvi.69.10.6249-6258.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]