Abstract
Earlier studies have shown that wild-type infected-cell protein 0 (ICP0), a key herpes simplex virus regulatory protein, translocates from the nucleus to the cytoplasm of human embryonic lung (HEL) fibroblasts within several hours after infection (Y. Kawaguchi, R. Bruni, and B. Roizman, J. Virol. 71:1019–1024, 1997). Translocation of ICP0 was also observed in cells infected with the d120 mutant, in which both copies of the gene encoding ICP4, the major regulatory protein, had been deleted (V. Galvan, R. Brandimarti, J. Munger, and B. Roizman, J. Virol. 74:1931–1938, 2000). Furthermore, a mutant (R7914) carrying the D199A substitution in ICP0 does not bind or stabilize cyclin D3 and is retained in the nucleus (C. Van Sant, P. Lopez, S. J. Advani, and B. Roizman, J. Virol. 75:1888–1898, 2001). Studies designed to elucidate the requirements for the translocation of ICP0 between cellular compartments revealed the following. (i) Translocation of ICP0 to the cytoplasm in productive infection maps to the D199 amino acid, inasmuch as wild-type ICP0 delivered in trans to cells infected with an ICP0 null mutant was translocated to the cytoplasm whereas the D199A-substituted mutant ICP0 was not. (ii) Translocation of wild-type ICP0 requires a function expressed late in infection, inasmuch as phosphonoacetate blocked the translocation of ICP0 in wild-type virus-infected cells but not in d120 mutant-infected cells. Moreover, whereas in d120 mutant-infected cells ICP0 was translocated rapidly from the cytoplasm to the nucleus at approximately 5 h after infection, the translocation of ICP0 in wild-type virus-infected cells extended from 5 to at least 9 h after infection. (iii) In wild-type virus-infected cells, the MG132 proteasomal inhibitor blocked the translocation of ICP0 to the cytoplasm early in infection, but when added late in infection, it caused ICP0 to be relocated back to the nucleus from the cytoplasm. (iv) MG132 blocked the translocation of ICP0 in d120 mutant-infected cells early in infection but had no effect on the ICP0 aggregated in vesicle-like structures late in infection. However, in d120 mutant-infected cells treated with MG132 at late times, proteasomes formed a shell-like structure around the aggregated ICP0. These structures were not seen in wild-type virus or R7914 mutant-infected cells. The results indicate the following. (i) In the absence of β or γ protein synthesis, ICP0 dynamically associates with proteasomes and is translocated to the cytoplasm. (ii) In cells productively infected beyond α gene expression, ICP0 is retained in the nucleus until after the onset of viral DNA synthesis and the synthesis of γ2 proteins. (iii) Late in infection, ICP0 is actively sequestered in the cytoplasm by a process mediated by proteasomes, inasmuch as interference with proteasomal function causes rapid relocation of ICP0 to the nucleus.
The infected-cell protein 0 (ICP0), the product of the α0 gene of herpes simplex virus 1 (HSV-1), is a 775-amino-acid protein translated from a spliced mRNA. ICP0 contains a zinc RING finger and is extensively posttranslationally modified by both cellular and viral protein kinases and nucleotidylylated by casein kinase II (1, 12, 20, 22). ICP0 has been previously described to interact with many cellular partners such as the ubiquitin-specific protease 7 (USP-7), the elongation factor 1δ (EF-1δ), cyclin D3, and other proteins (9, 13, 14, 18, 19). ICP0 is essential for viral replication in experimental animals and for efficient replication in cells in culture (2, 6, 14, 24–26). In cells infected with α0 null mutants, viral gene expression is initiated but productive infection ensues in cells infected at high multiplicity or at a phase of the cell cycle that compensates in part for the missing viral gene (2, 6, 26). The mechanisms by which ICP0 enables efficient viral replication are unknown. The available data indicate that during the early phases of the viral replicative cycle, ICP0 colocalizes with PML, a component of the ND10 nuclear structures, and causes the dispersion of PML from these structures (8, 16, 17). In addition, ICP0 has been linked to specific degradation of several cellular proteins including CENP-A, CENP-C, DNA-dependent protein kinase, and Sumo-I-conjugated PML or Sp100 (3, 7, 15, 23). Studies reported from this laboratory have shown that ICP0 binds, stabilizes, and colocalizes with cyclin D3 early in infection (14). The site of interaction with cyclin D3 was mapped to the amino acid D199 (27). ICP0 carrying the substitution D199A cannot be differentiated from wild-type virus with respect to colocalization with PML and the degradation of ND10 structures but does not bind or colocalize with cyclin D3 (28). The recombinant virus R7914 carrying the substitution D199A in ICP0 replicates less well in stationary, serum-deprived, or contact-inhibited human embryonic lung (HEL) fibroblasts and is less virulent than the wild-type virus when administered by a peripheral route (27).
The studies described in this report stemmed from three observations. First, in the course of studies on the interaction of ICP0 with cyclin D3, it was noted that, in productively infected HEL cells, ICP0 could be found in the cytoplasm as early as 3 h after infection and was fully cytoplasmic by late times after infection (14). ICP0 is also translocated into the cytoplasm in SK-N-SH cells but not in Vero cells. Cytoplasmic localization in Vero cells was earlier reported for ICP0 expressed at the nonpermissive temperature by ICP4 ts mutants (e.g., tsK, ts756, tsB21, d2, and n215) or nonsense mutants (n12) but not by ICP0 encoded in cells productively infected by wild-type virus (30, 31). Second, ICP0 encoded by the recombinant virus R7914 was not translocated into the cytoplasm (28). Lastly, ICP0 encoded by an HSV-1 mutant (d120) lacking both copies of the α4 gene encoding the major regulatory protein ICP4 localized in the cytoplasm in structures resembling vesicles (11). The form and distribution of ICP0 in the cytoplasm for d120 mutant-infected cells differed substantially from those of ICP0 accumulating in the cytoplasm of wild-type virus-infected cells.
In this report, we show that, with the possible exception of initial translocation to the nucleus of newly synthesized ICP0, which may be determined by its nuclear localization signal, all subsequent events are governed by proteins which interact with ICP0. ICP0 is translocated to the cytoplasm subsequent to failure of β and γ protein synthesis. In cells undergoing productive infection, ICP0 may be retained in nuclei until after the onset of viral DNA synthesis. A physical interaction of ICP0 with an as yet unknown factor(s) is deduced from the observation that ICP0 carrying the D199A substitution is not transported to the cytoplasm. Finally, of particular interest is the evidence that ICP0 dynamically interacts with proteasomes. This conclusion emerges from two observations. First, in the presence of proteasomal inhibitor MG132, proteasomal proteins formed a shell which surrounded aggregated cytoplasmic ICP0 encoded by the d120 mutant. Second, addition of MG132 to the medium at late times after infection of HEL fibroblasts with wild-type virus resulted in relocation of ICP0 from the cytoplasm to the nucleus.
MATERIALS AND METHODS
Cells and viruses.
Human embryonic lung (HEL) fibroblasts were obtained from Aviron (Mountain View, Calif.) and grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum. HSV-1 strain F [HSV-1(F)], a limited-passage isolate, is the prototype strain used in this laboratory (5). The construction and phenotypic properties of the recombinant viruses R7914 and R7915 have been previously described (27). The HSV-1 mutant d120 (a gift of N. DeLuca) carries a deletion in both copies of the α4 gene and was grown in a Vero cell line (E5) expressing α4 (4). The construction and phenotypic properties of the recombinant virus R7910 lacking both copies of the α0 gene were described elsewhere (14).
Phosphonoacetic acid (PAA) and MG132 were obtained from Sigma (St. Louis, Mo.) and Biomol Research Laboratories, Inc. (Plymouth Meeting, Pa.), respectively.
Baculovirus transfer vector.
pAcSG2-ICP0 was constructed by cloning the ICP0 cDNA (a gift of S. Silverstein) (29) into the NcoI-BglII site of the pSAcSG2 baculovirus transfer vector (PharMingen, San Diego, Calif.). The XhoI-EcoRI fragment containing the human cytomegalovirus (CMV) immediate-early 1 promoter/enhancer sequences was inserted in the XhoI-EcoRI site of pAcSG2-ICP0 to generate pAcCMV-ICP0, in which the ICP0 coding sequence was controlled by the human CMV immediate-early promoter. pAcCMV-D199A was constructed by cloning a KpnI-SalI fragment from pRB4986 containing the D199A substitution into the KpnI-SalI site of pAcCMV-ICP0. The plasmids were sequenced, amplified, and purified with the aid of the Qiagen plasmid purification kit (Chatsworth, Calif.).
Construction of recombinant baculovirus.
Baculoviruses encoding ICP0 (Bac-ICP0) or D199A ICP0 (Bac-D199A) were generated by cotransfecting sf9 cells with BaculoGold linearized baculovirus DNA (PharMingen) with pAcCMV-ICP0 or pAcCMV-D199A transfer vectors, respectively, according to the manufacturer's instructions. The viruses were propagated in sf9 cells grown in 150-cm3 flasks in TNM-FH insect medium (PharMingen). Virus stocks were prepared as described by the manufacturer.
Preparation of HEL fibroblast cultures for immunofluorescence analyses.
HEL fibroblasts were seeded onto glass slides (Cell-Line, Newfield, N.J.) at a density of 104 cells per well 1 day prior to infection. The slide cultures were exposed for 2 h at 37°C in mixture 199 supplemented with 1% calf serum (199V) to 20 PFU of HSV-1(F), R7914, R7915, or d120 virus per cell. The inoculum was replaced with fresh DMEM containing 10% serum and reincubated at 37°C for time intervals stated in Results.
Exposure of mammalian cells to recombinant baculovirus expressing ICP0 and recombinant R7910.
Cells seeded onto glass slides were exposed to approximately 20 PFU of baculovirus per cell and incubated at 37°C for 2 h in 199V. The culture medium was replaced with fresh DMEM containing 10% fetal bovine serum and 5 mM sodium butyrate. Cells were fixed at 15 h after infection in ice-cold methanol. In coinfection experiments, after exposure to baculovirus for 2 h and incubation for an additional 1 h in DMEM containing 10% fetal bovine serum and 5 mM sodium butyrate, the cells were exposed to 0.5 PFU of R7910 recombinant virus per cell for 2 h in 199V supplemented with 5 mM sodium butyrate. The inoculum was then replaced with DMEM containing 10% fetal bovine serum and 5 mM sodium butyrate. Cells were fixed 12 h after infection with the R7910 mutant.
Immunofluorescence analyses.
At times indicated in the text, the slide cultures were fixed with cold methanol for 2 h and then blocked for 1 h in phosphate-buffered saline (PBS) containing 1% bovine serum albumin (BSA) and 20% normal human serum. The primary antibodies were diluted in PBS containing 1% BSA and 10% normal human serum. The mouse monoclonal antibody to ICP0 (H1083) and mouse monoclonal antibody to gB (H12817) were obtained from the Goodwin Cancer Research Institute (Plantation, Fla.) and used at 1:1,000 and 1:500 dilutions, respectively. The rabbit polyclonal antibody to ICP0 (14), the mouse monoclonal antibody to PML (Santa Cruz Biotechnology, Santa Cruz, Calif.), and the rabbit polyclonal antibody to proteasomal proteins (Affiniti Research Products Ltd., Mamhead, Exeter, Devon, United Kingdom; catalogue no. PW 8155) were used at 1:1,000, 1:200, and 1:2,500 dilutions, respectively. The anti-proteasomal protein antibody designated core antibody was raised by immunization of rabbits with a proteasomal preparation purified from human red blood cells. After reaction overnight at 4°C, the slide cultures were rinsed at least five times with PBS and exposed to the secondary antibodies in PBS containing 1% BSA and 10% normal human serum. Goat anti-rabbit and goat anti-mouse secondary antibodies coupled with Texas Red (Molecular Probes, Eugene, Oreg.) were used at a 1:400 dilution, and goat anti-rabbit and goat anti-mouse secondary antibodies coupled with fluorescein isothiocyanate (FITC; Sigma) were used at 1:160 and 1:64 dilutions, respectively. After 1 h of reaction with secondary antibody, the slide cultures were again rinsed at least five times with PBS and mounted in 90% glycerol containing 1 mg of 1-4-phenylenediamine per ml (Aldrich Chemical Co., Milwaukee, Wis.). The slides were examined in a Zeiss confocal microscope. Digitized images of the fluorescent antibody-stained cells were acquired with software provided by Zeiss. Cell counts were done by two different examiners.
RESULTS
Wild-type ICP0 but not the ICP0 carrying the D199A substitution is translocated into the cytoplasm by a process mediated by a viral function.
Earlier, this laboratory reported that, in HEL fibroblasts infected with wild-type virus HSV-1(F), ICP0 is translocated at late times after infection from the nucleus to the cytoplasm (13). In contrast to the ICP0 encoded by wild-type parent virus, the ICP0 carrying the D199A substitution is restricted to the nucleus throughout the replicative cycle of the virus (28). To ascertain whether the D199A substitution is solely responsible for the nuclear retention of ICP0 and to determine whether the translocation requires a viral function, two series of experiments were done.
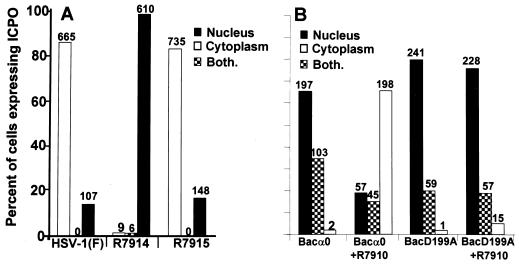
In the first, replicate slide cultures of HEL fibroblasts were exposed (20 PFU/cell) to HSV-1(F), R7914 carrying the D199A substitution, or R7915, in which the mutated amino acid was replaced with the native one (27). The cells were fixed at 12 h after infection and reacted with rabbit polyclonal antibody to exon II of ICP0 and then with FITC-conjugated antibody to rabbit immunoglobulin G (IgG). The cells expressing ICP0 were counted as described in Materials and Methods. The results (Fig. 1A) show that at 12 h after infection ICP0 encoded by wild-type virus or by R7915 (A199D-repaired virus) accumulated in the cytoplasm. ICP0 carrying the D199A mutation accumulated in the nucleus. These results support the hypothesis that the D199A substitution caused ICP0 to be retained in the nucleus.
FIG. 1.
The requirements for the translocation of ICP0 from the nucleus to the cytoplasm. (A) ICP0 carrying the D199A substitution encoded by R7914 is not translocated to the cytoplasm late in infection of HEL fibroblasts. Replicate slide cultures of HEL fibroblasts were infected with 20 PFU of HSV-1(F), the recombinant virus R7914, or the repaired virus R7915 per cell and maintained at 37°C. At 12 h after infection, the cells were fixed and reacted first with polyclonal rabbit serum directed against exon II of ICP0 and second with FITC-conjugated goat anti-rabbit immunoglobulin antibodies. Sequential fields were examined in a Zeiss confocal microscope, and the numbers of cells exhibiting nuclear, cytoplasmic, or both nuclear and cytoplasmic localization of ICP0 were tabulated as shown in the histogram. The numbers above the bars indicate the numbers of cells showing a specific distribution of ICP0. (B) A viral gene function mediates the translocation of ICP0 from the nucleus to the cytoplasm during productive infection. Replicate slide cultures of HEL fibroblasts were exposed first to recombinant baculoviruses encoding wild-type or mutant ICP0 (D199A). After 2 h, cells were treated with 5 mM Na-butyrate. At 3 h after exposure to baculoviruses, the cells were exposed to the recombinant virus R7910, from which both copies of the α0 gene had been deleted. After 15 h of exposure to baculoviruses alone or 12 h after infection with R7910, the cells were fixed and reacted with rabbit polyclonal antibody against ICP0 and mouse monoclonal antibody against gD and then reacted with FITC-conjugated goat anti-rabbit IgG and Texas Red-conjugated goat anti-mouse IgG antibodies. Cell counts were done as described above, except that only cells exhibiting gD encoded by R7910 and ICP0 encoded by baculoviruses were counted.
In the second series of experiments, we cloned both the wild-type and the mutant ICP0 carrying the D199A substitution in baculoviruses under the CMV immediate-early promoter. In these experiments, replicate slide cultures of HEL fibroblasts were exposed to baculoviruses encoding wild-type or D199A-substituted ICP0. At 3 h after exposure to the recombinant baculoviruses, the slide cultures were exposed to 0.5 PFU of α0 null mutant R7910 per cell in the presence of 5 mM sodium butyrate. At 15 h after exposure of HEL fibroblasts to baculoviruses alone or 12 h after exposure of cells to both R7910 and baculoviruses, the cell cultures were fixed, blocked, and reacted with the rabbit polyclonal antibody to exon II of ICP0 and with the mouse monoclonal antibody to gD. Among singly infected cells, all ICP0-positive cells were counted. Among doubly infected cells, only cells exhibiting both ICP0 and gD were counted. The results, shown in Fig. 1B, were as follows.
(i) In contrast to the exclusive nuclear localization of ICP0 in cells transfected with plasmids encoding ICP0 (data not shown), in cells singly infected with baculoviruses expressing the wild-type or mutant α0 gene, ICP0 localized predominantly but not exclusively in the nucleus. In approximately 20 to 35% of baculovirus-infected HEL cells, ICP0 accumulated in both the cytoplasm and the nucleus.
(ii) In cells infected with R7910 and baculoviruses encoding wild-type α0, ICP0 was localized in the cytoplasm of more than 80% of the HEL fibroblasts expressing gD (Fig. 1). In contrast, in cells doubly infected with both R7910 and baculovirus carrying the D199A mutation of the α0 gene, the distribution of ICP0 was similar to that of cells infected with the baculovirus only.
We conclude from these complementation experiments that one or more viral functions expressed during lytic infection are required to translocate wild-type ICP0 from the nucleus to the cytoplasm. ICP0 carrying the D199A mutation is not translocated to the cytoplasm by the functions expressed by the ICP0 null mutant.
Viral DNA synthesis and/or γ2 gene expression is required for the translocation of ICP0 from the cytoplasm to the nucleus.
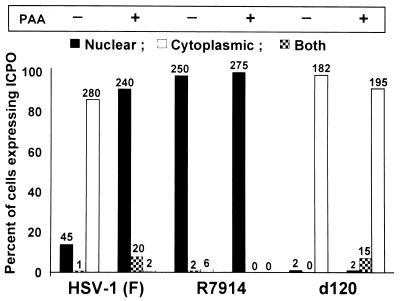
Studies described elsewhere have established that translocation of wild-type ICP0 from the nucleus to the cytoplasm of HEL fibroblasts takes place between 5 and 9 h after infection with wild-type virus, that is, at the time of or after the onset of viral DNA synthesis (28). However, in cells infected with the d120 mutant, ICP0 is translocated even in the apparent absence of viral DNA synthesis. To determine the role of viral DNA synthesis and/or the requirement for the expression of γ2 proteins, slide cultures of HEL cells were exposed to 20 PFU of HSV-1(F), R7914, or the d120 mutant per cell in the presence or absence of 300 μg of PAA per ml of medium. The cultures were fixed 12 h after infection and then reacted with antibody to ICP0 or to both PML and ICP0. The results, shown in Fig. 2 and 3, were as follows.
FIG. 2.
Translocation of ICP0 from the nucleus to the cytoplasm in HEL fibroblasts infected with wild-type virus is blocked by PAA. Replicate slide cultures of HEL fibroblasts were infected with 20 PFU of HSV-1(F), R7914, or d120 mutant virus per cell in the presence or absence of 300 μg of PAA per ml of medium. The cells were fixed at 12 h after infection and reacted with polyclonal rabbit antibody against ICP0 and then with FITC-conjugated goat anti-rabbit IgG. The tabulation of cells exhibiting ICP0 in the nucleus, the cytoplasm, or both was done as described in the legend to Fig. 1.
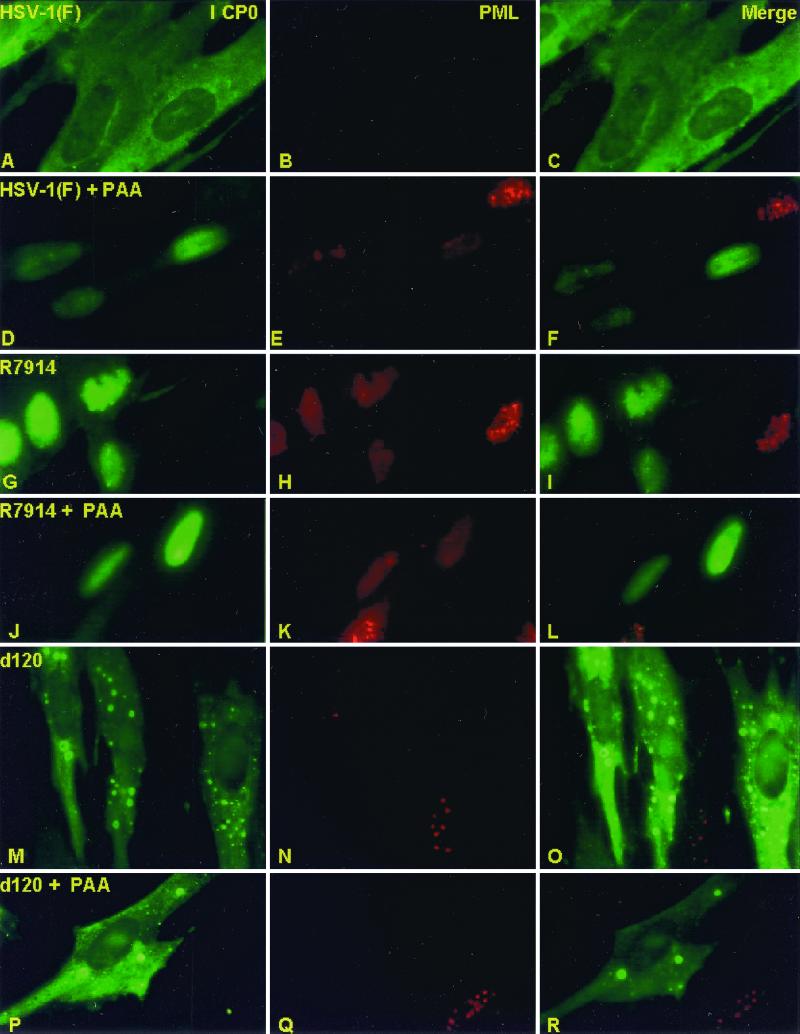
FIG. 3.
Immunofluorescent images of HEL fibroblasts infected with HSV-1(F), R7914, or d120 mutant and either treated or not treated with PAA. The images shown are representative of infected cells treated, generated, and counted as described in the legend to Fig. 2. The cells were reacted with rabbit polyclonal antibody to ICP0 and mouse monoclonal antibody to PML and then reacted with goat anti-rabbit IgG conjugated to FITC and goat anti-mouse IgG conjugated to Texas Red. The left, middle, and right columns show the cell localization of ICP0 and PML and merged images, respectively. The images were captured with a Zeiss confocal microscope with the aid of software provided by the manufacturer.
(i) In the presence of PAA, wild-type ICP0 remained in the nucleus in more than 90% of the infected cells (Fig. 2). PAA had no effect on the disaggregation of ND10 structures in these cells (Fig. 3A to F). PAA also had no effect on the nuclear localization of ICP0 encoded by the R7914 mutant (Fig. 2 and 3G to L).
(ii) As could be expected, PAA had no effect on the cytoplasmic localization of ICP0 encoded by the d120 mutant (Fig. 2 and Fig. 3M to R).
We conclude from these studies the following.
(i) Translocation of ICP0 from the nucleus to the cytoplasm required a function expressed after the onset of viral DNA synthesis. This function may be mediated by a viral protein (γ2) requiring viral DNA synthesis for its expression.
(ii) The retention of ICP0 in the nucleus during the early stages of infection also requires a viral function. This function may be expressed by ICP0, by α4, or by a gene product expressed by a post-α gene. We are led to this conclusion by the observation that, in wild-type-infected cells, ICP0 is retained in the nucleus and requires a late function to be translocated from that site. In d120 mutant-infected cells, ICP0 is initially contained in the nucleus and is translocated to the cytoplasm in the absence of the late viral function required in the wild-type-infected cells. A necessary conclusion is that the retention of ICP0 in the nucleus in wild-type-infected cells is mediated by a viral function that is not expressed in d120 mutant-infected cells.
The translocation of ICP0 in cells infected with the d120 mutant takes place earlier than that of ICP0 in wild-type virus-infected cells.
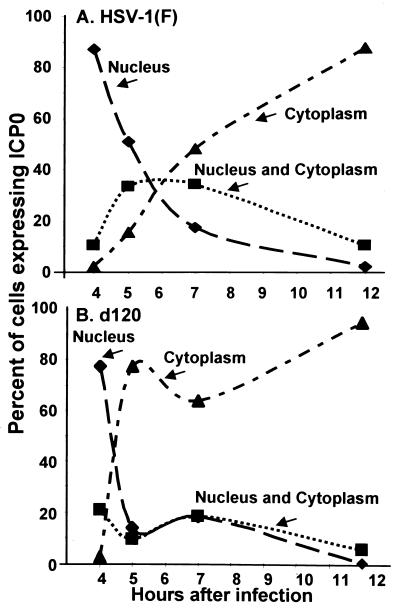
Because the requirements for the translocation of ICP0 encoded by the d120 mutant and wild-type virus were different, it was of interest to assess the rate of translocation of these proteins in infected HEL fibroblasts. In this series of experiments, replicate slide cultures of HEL fibroblasts were exposed to 20 PFU of HSV-1(F) or d120 mutant per cell. The cells were fixed at 4, 5, 7, or 12 h after infection and reacted with rabbit polyclonal antibody to ICP0. The distribution of cells expressing nuclear, cytoplasmic, or both nuclear and cytoplasmic ICP0 is shown in Fig. 4. In essence, by 5 h after infection with the d120 mutant, more than 80% of infected HEL cells contained only cytoplasmic ICP0. Cells exhibiting both nuclear and cytoplasmic ICP0 constituted no more than 20% of the total. In contrast, by 5 h after infection with wild-type virus, less than 20% of HEL cells exhibited strictly cytoplasmic localization of ICP0. In these cells, ICP0 transited slowly from the nucleus to the cytoplasm, and full cytoplasmic localization required at least 12 h from the time of infection.
FIG. 4.
Temporal pattern of translocation of ICP0 encoded by wild-type and d120 mutant viruses from the nucleus to the cytoplasm. Replicate slide cultures of HEL fibroblasts were exposed to 20 PFU of HSV-1(F) or d120 mutant virus per cell. At 4, 5, 7, or 12 h after infection and incubation at 37°C, replicate cultures were fixed and reacted with rabbit polyclonal antibody against ICP0 and then with goat anti-rabbit IgG conjugated to FITC. The cells were examined and tabulated as described in the legend to Fig. 1.
We conclude that, in cells infected with the d120 mutant, ICP0 transits from the nucleus to the cytoplasm relatively rapidly at approximately 4 to 5 h after infection. In contrast, the translocation of ICP0 from cells infected with the wild-type virus takes place at a lower rate during a much longer interval. This interval includes an extensive phase in which ICP0 is found in both the nucleus and the cytoplasm. The results suggest that, in wild-type virus-infected cells expressing the full gamut of viral genes, ICP0 is retained in the nucleus until after the onset of viral DNA synthesis and concomitant expression of γ2 genes.
The effect of proteasomal inhibitor MG132 on the localization of ICP0 early and late in infection of HEL fibroblasts with wild-type and mutant viruses.
Everett et al. (10) reported that the proteasomal inhibitor MG132, added to the medium 30 min before HSV-1 infection, blocked the translocation of wild-type ICP0 from the nucleus to the cytoplasm. We employed a different experimental design to examine the effect of MG132 inhibitor on the localization of ICP0 in cells infected with the d120 mutant. These studies led us to reexamine the effect of MG132 on the localization of ICP0 in cells infected with wild-type virus with startling results.
The experimental design of these studies was as follows. Replicate slide cultures of HEL fibroblasts were exposed to 20 PFU of HSV-1(F) or the d120 mutant per cell. One set of replicate slide cultures was exposed to MG132 (5 μM) at 2 or 9 h after infection. All cultures were fixed at 12 h after infection and reacted with antibody to ICP0. The results were as follows (Fig. 5).
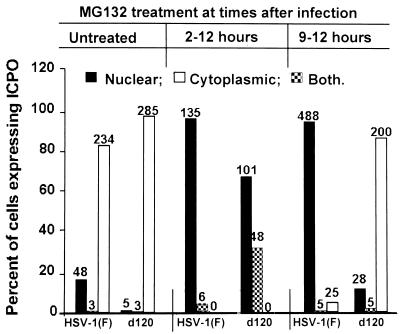
FIG. 5.
Effect of MG132 proteasomal inhibitor on the translocation of ICP0 from the nucleus to the cytoplasm. Replicate slide cultures of HEL fibroblasts were exposed to 20 PFU of HSV-1(F) or d120 mutant virus per cell. At 2 h after infection (middle bars) or 9 h after infection (right bars), the cells were replenished with medium containing MG132 (5 μM). At 12 h after infection, the cells were fixed and reacted with polyclonal rabbit antibody against ICP0 and then with goat anti-rabbit IgG conjugated to FITC. The cells were examined and quantified as described in the legend to Fig. 1.
(i) As described earlier in the text, in untreated control cultures, ICP0 accumulated predominantly in the cytoplasm of wild-type virus-infected cells by 12 h after infection with wild-type virus. The addition of the drug at 2 h after infection completely blocked the translocation of ICP0 from the nucleus to the cytoplasm. In this respect, the results were similar to those of Everett et al. (10).
(ii) The effect of MG132 added at 2 h after infection on the localization of ICP0 encoded by the d120 mutant was only slightly less inhibitory. Thus, 67% of the infected cells contained ICP0 localized only in the nucleus. In the remaining infected cells, ICP0 was present in both the nucleus and the cytoplasm.
(iii) The localization of ICP0 was not affected in d120 mutant-infected cells exposed to MG132 at 9 h after infection.
(iv) In HEL cultures exposed to MG132 between 9 and 12 h after infection, ICP0 encoded by wild-type virus was found exclusively in the nucleus whereas in untreated cells ICP0 accumulated in the cytoplasm.
We conclude the following.
(i) When added at 2 h after infection, MG132 arrested the translocation of ICP0 in cells infected with the d120 mutant. When added late (9 h) after infection, the distribution of ICP0 at the end of the treatment interval resembled that at the time of addition of the drug. As noted in the Discussion, this observation does not exclude an interaction between ICP0 and proteasomes.
(ii) The situation appears to be different in cells exposed to MG132 at 9 h after infection with wild-type virus, at the time when ICP0 was mostly cytoplasmic. In these cells, ICP0 was translocated to the nucleus. These results suggest that translocation of ICP0 from the nucleus to the cytoplasm requires a function mediated by or affected by proteasomes and that MG132 abrogates this function, thereby releasing ICP0 to return to the nucleus.
Proteasomal proteins aggregate near or around ICP0 accumulating in cells infected with the d120 mutant and exposed to MG132.
Earlier, this laboratory reported that ICP0 encoded by d120 accumulates in the cytoplasm in vesicle-like structures (11). In the course of the studies presented above, we noted that the morphology of the structures containing ICP0 in HEL fibroblasts infected with d120 and treated with MG132 could not be differentiated from that of untreated cells infected with this mutant. Analyses of the distribution of proteasomes in infected cells were, however, more rewarding. Thus, MG132 was added at 9 h after infection of a series of replicate slide cultures with either the wild type, the R7914 mutant, or the d120 mutant. In the experiment illustrated in Fig. 6, the mock-treated and MG132-treated cultures were fixed at 9 and 12 h after infection and reacted with antibody to ICP0 and antibody to proteasomes designated as core antibody (Materials and Methods). The results were as follows.
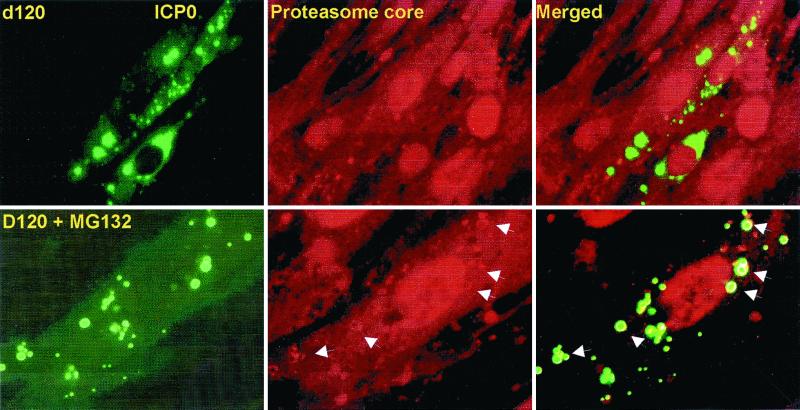
FIG. 6.
Colocalization of proteasome and ICP0 in cells infected with the d120 mutant and exposed to MG132. The experimental design of this series of experiments was identical to that described in the legend to Fig. 3, except that the cells were reacted with mouse monoclonal antibody against ICP0 and rabbit polyclonal antibody to the whole proteasome designated as core antibody (Affiniti Research Products Ltd.). The secondary antibodies were goat anti-mouse IgG conjugated to FITC and goat anti-rabbit IgG conjugated to Texas Red. Shown are images of cells infected with d120 mutant virus and either left untreated (upper panels) or exposed at 9 h after infection to 5 μM MG132 (lower panels). The images were captured with a Zeiss confocal microscope with the aid of software provided by the manufacturer. The arrows point to proteasomal core proteins aggregated in the form of a shell surrounding ICP0 in the cytoplasm of MG132-treated cells. These structures were seen only in d120 mutant-infected cells treated with MG132.
Proteasomes did not colocalize with ICP0 in an unambiguous fashion in either treated or untreated cells infected with wild-type virus or the R7914 (D199A) mutant. The overlap in distribution (e.g., in nuclei of infected cells) was not credible evidence of colocalization (data not shown). In contrast, in HEL cells infected with d120 mutant and treated with MG132, proteasomal proteins formed a shell surrounding the structures containing ICP0. The shell formed by the proteasomal proteins was uniform in appearance (Fig. 6, arrows). Moreover, these structures were not detected in untreated, d120 mutant-infected cells. The results were reproduced with antibody to individual α-proteasomal subunits such as HC3, HC8, and XAPC7 (Affiniti Research Products Ltd.) and also with MG262 (Biomol), a different proteasomal inhibitor (data not shown). We have also found the same structures in ICP0 localizing in similar vesicle-like structures in the cytoplasm of HEp-2 cells overexpressing Bcl-2 (VAX-3 cells, data not shown).
DISCUSSION
The studies described in this report were done in HEL fibroblasts. The salient features of this report are as follows.
(i) Immediately after its synthesis, ICP0 is transported to the nucleus of the infected cell. ICP0 was translocated to the nucleus irrespective of whether it was synthesized from a transfected plasmid, a baculovirus under a CMV immediate promoter, a viral genome lacking the α4 genes, or a wild-type HSV-1 genome.
(ii) Translocation of ICP0 from the nucleus to the cytoplasm appears to take place under three entirely different conditions. The first was observed in HEL fibroblasts infected with baculoviruses carrying the α0 gene regulated by the CMV immediate-early promoter. Unlike ICP0 encoded by recombinant plasmids, which is retained in the nucleus, ICP0 encoded by baculoviruses was retained in the nucleus but in approximately 35% of the cells was also present in the cytoplasm. The basis for this difference in the site of accumulation of ICP0 is unknown. Baculoviruses are not known to express their genes in mammalian cells at 37°C. We noted, however, that baculoviruses induce the activation of cdc2 at relatively low levels in mammalian cells. ICP0 appears to be a substrate of cdc2, and this posttranslational modification may potentially account for the partial displacement of ICP0 (S. J. Advani and B. Roizman, unpublished studies).
The second type of translocation of ICP0 from the nucleus to the cytoplasm was noted in cells infected with the d120 mutant. The translocation of ICP0 from cells infected with d120 mutant virus differs in several respects from that of ICP0 encoded by wild-type virus. Thus, the d120 mutant-encoded ICP0 was translocated earlier and more efficiently, aggregated into vesicle-like structures, and was unaffected by MG132.
The third type of translocation was exhibited by ICP0 encoded by wild-type virus. In this instance, ICP0 was largely translocated and dispersed in the cytoplasm between 5 and 9 h after infection. Translocation required post-DNA synthesis events, since PAA in concentrations sufficient to inhibit DNA synthesis blocked the translocation of ICP0. In contrast to the d120 mutant-infected cells, in cells infected with wild-type virus and exposed to MG132 late in infection, ICP0 was relocated back to the nucleus. The relocation of ICP0 to the nucleus in the presence of MG132 is of considerable interest since it points to a role of proteasomes in the cytoplasmic retention of ICP0. While Everett et al. (10) noted that MG132 blocked the translocation of ICP0 from the nucleus to the cytoplasm when added before infection, they missed the relocation of ICP0 back to the nucleus, since they examined the effects of MG132 added only from the start of infection.
These studies indicate the following.
(i) Initial localization of ICP0 in the nucleus may reflect the nuclear localization signal encoded in the protein and mapped to exon III (21).
(ii) In cells infected with mutants in which progression of infection from α to genes expressed later in infection is blocked, ICP0 is actively translocated into the cytoplasm. The function of viral or cellular gene products in this process is inferred from the observation that, since ICP0 has a nuclear localization signal, its transport from the nucleus to the cytoplasm must overcome the directionality imposed by this signal through the interaction of one or more proteins. This is the case for ICP0 encoded by the d120 mutant. The focus on proteasomal subunits as possible candidates in this process is based on the observation that these coalesce with and form a shell around ICP0 in cells treated with MG132. Since the proteasomes are rarely and only faintly observed around the ICP0 aggregates in the absence of MG132 (data not shown), the conclusion is that the process is dynamic in the sense that proteasomes shuttle to and from the aggregated cytoplasmic ICP0 and remain aggregated with it only in the presence of the drug.
There is additional circumstantial evidence that implicates proteasomes in this process. Thus, ICP0 causes degradation of nuclear structures known as ND10. Concurrently with the degradation of these structures, ICP0 causes the degradation of several proteins including CENP-A, CENP-C, DNA-dependent protein kinase, and Sumo-I-conjugated Sp100 and PML (3, 7, 15, 23). The available data suggest a causal relationship between the degradation of these proteins, ICP0, and association with proteasomes. The association of ICP0 with proteasomal functions prior to the translocation to the cytoplasm adds weight to a dynamic association of ICP0 with proteasomal subunits in the cytoplasm.
(iii) A necessary corollary to the conclusion listed above is that, in cells in which HSV-1 progresses beyond the expression of α genes, ICP0 is actively retained in the nucleus. Since progression to β and γ gene expression requires functional ICP4 and since ICP0 has been reported to physically interact with this protein, it is conceivable that ICP4 directly causes the retention of ICP0 in the nucleus. An alternative scenario is that ICP0 is posttranslationally modified or interacts with proteins expressed later in infection.
(iv) As shown in this report, the translocation of ICP0 from the nucleus into the cytoplasm of wild-type virus-infected cells requires the synthesis of viral DNA and, more likely, the expression of γ2 proteins whose synthesis requires the synthesis of viral DNA. There is considerable information but no firm conclusion regarding this translocation. First, the rate of translocation is cell type specific. Translocation is relatively early in primary HEL fibroblasts and in SK-N-SH cells but not in Vero or HEp-2 cells, suggesting that a cellular factor also plays a role in the translocation. Second, this translocation requires a function mapping at or near amino acid D199 in exon II of ICP0. The ICP0 of the mutant virus carrying the D199A substitution is retained in the nucleus and is not translocated into the cytoplasm. The translocation is independent of the destruction of ND10 since the wild-type virus and the mutant carrying the D199A substitution cannot be differentiated with respect to the degradation of ND10 structures (28). In addition, dispersion of ND10 and translocation of ICP0 to the cytoplasm are sequential events since the exposure of infected cells to PAA blocks the latter but not the former. Third, the functions mapped to the D199A locus are the stabilization of cyclin D3 and activation of cdk4. In cells infected with a wild-type virus expressing cyclin D3, ICP0 was translocated more rapidly than in cells infected with the wild-type parent virus. Insertion of the cyclin D3 gene into the same location in the mutant carrying the D199A substitution did not alter the retention of ICP0 in the nucleus. The data suggest that cyclin D3 mediates the translocation of ICP0.
(v) The observation that MG132 causes the relocation of ICP0 to the nucleus suggests that proteasomal components arrested in their activity by MG132 release ICP0 from its cytoplasmic shuttle vector. The simplest explanation for the relocation is that the nuclear localization signal becomes once again functional. What is less clear is the nature of the shuttle vector from the nucleus to the cytoplasm. Above in the text, we have linked proteasomes to the translocation of the d120-mutant encoded ICP0. The evidence linking proteasomal components as the shuttle vectors for the translocation of ICP0 late in infection is less compelling but not refutable. The observation that MG132 blocks translocation from the nucleus to the cytoplasm when cells are exposed to it early in infection could imply an inhibition of a vital step in viral replication, a block in the degradation of a specific protein necessary to effect the translocation of ICP0, or interference with the function of proteasomes as a shuttle vector.
A puzzling issue is the function of ICP0 in the cytoplasm. The only known interaction of ICP0 with cytoplasmic proteins is that between wild-type ICP0 and elongation factor 1δ (13). Is ICP0 an impediment to the infectious process which must be removed forthwith from the nucleus late in infection? Is ICP0 a shuttle for removal of proteasomal subunits from the nucleus to the cytoplasm? These and other possible functions of ICP0 remain to be investigated.
ACKNOWLEDGMENTS
These studies were aided by grants from the National Cancer Institute (CA47451, CA71933, and CA78766), United States Public Health Service. P.L. is a postdoctoral fellow of L'Association pour la Recherche sur le Cancer (A.R.C. France).
REFERENCES
- 1.Blaho J A, Mitchell C, Roizman B. An amino acid sequence shared by the herpes simplex virus 1 alpha regulatory proteins 0, 4, 22, and 27 predicts the nucleotidylylation of the UL21, UL31, UL47, and UL49 gene products. J Biol Chem. 1994;269:17401–17410. [PubMed] [Google Scholar]
- 2.Cai W Z, Schaffer P A. Herpes simplex virus type 1 ICP0 plays a critical role in the de novo synthesis of infectious virus following transfection of viral DNA. J Virol. 1989;63:4579–4589. doi: 10.1128/jvi.63.11.4579-4589.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chelbi-Alix K M, de The H. Herpes virus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene. 1999;18:935–941. doi: 10.1038/sj.onc.1202366. [DOI] [PubMed] [Google Scholar]
- 4.DeLuca N A, McCarthy A M, Schaffer P A. Isolation and characterization of deletion mutants of herpes simplex virus type 1 in the gene encoding immediate-early regulatory protein ICP4. J Virol. 1985;56:558–570. doi: 10.1128/jvi.56.2.558-570.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ejercito P M, Kieff E D, Roizman B. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J Gen Virol. 1968;2:357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- 6.Everett R D. Construction and characterization of herpes simplex virus type 1 mutants with defined lesions in immediate early gene 1. J Gen Virol. 1989;70:1185–1202. doi: 10.1099/0022-1317-70-5-1185. [DOI] [PubMed] [Google Scholar]
- 7.Everett R D, Earnshaw W C, Findlay J, Lomonte P. Specific destruction of kinetochore protein CENP-C and disruption of cell division by herpes simplex virus immediate-early protein Vmw110. EMBO J. 1999;18:1526–1538. doi: 10.1093/emboj/18.6.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Everett R D, Maul G G. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 1994;13:5062–5069. doi: 10.1002/j.1460-2075.1994.tb06835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Everett R D, Meredith M, Orr A, Cross A, Kathoria M, Parkinson J. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 1997;16:566–577. doi: 10.1093/emboj/16.3.566. . (Corrected and republished, 16:1519–1530.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Everett R D, Orr A, Preston C M. A viral activator of gene expression functions via the ubiquitin-proteasome pathway. EMBO J. 1998;17:7161–7169. doi: 10.1093/emboj/17.24.7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galvan V, Brandimarti R, Munger J, Roizman B. Bcl-2 blocks a caspase-dependent pathway of apoptosis activated by herpes simplex virus 1 infection in HEp-2 cells. J Virol. 2000;74:1931–1938. doi: 10.1128/jvi.74.4.1931-1938.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Honess R W, Roizman B. Proteins specified by herpes simplex virus. XIII. Glycosylation of viral polypeptides. J Virol. 1975;16:1308–1326. doi: 10.1128/jvi.16.5.1308-1326.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawaguchi Y, Bruni R, Roizman B. Interaction of herpes simplex virus 1 α regulatory protein ICP0 with elongation factor 1δ: ICP0 affects translational machinery. J Virol. 1997;71:1019–1024. doi: 10.1128/jvi.71.2.1019-1024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawaguchi Y, Van Sant C, Roizman B. Herpes simplex virus 1 α regulatory protein ICP0 interacts with and stabilizes the cell cycle regulator cyclin D3. J Virol. 1997;71:7328–7336. doi: 10.1128/jvi.71.10.7328-7336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lomonte, P., K. F. Sullivan, and R. D. Everett. Degradation of nucleosome-associated centromeric histone H3-like protein CENP-A induced by herpes simplex virus type 1 protein ICP0. J. Biol. Chem., in press. [DOI] [PubMed]
- 16.Maul G G, Everett R D. The nuclear location of PML, a cellular member of the C3.HC4 zinc-binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J Gen Virol. 1994;75:1223–1233. doi: 10.1099/0022-1317-75-6-1223. [DOI] [PubMed] [Google Scholar]
- 17.Maul G G, Guldner H H, Spivack J G. Modification of discrete nuclear domains induced by herpes simplex virus type 1 immediate early gene 1 product (ICP0) J Gen Virol. 1993;74:2679–2690. doi: 10.1099/0022-1317-74-12-2679. [DOI] [PubMed] [Google Scholar]
- 18.Meredith M, Orr A, Elliott M, Everett R. Separation of sequence requirements for HSV-1 Vmw110 multimerisation and interaction with a 135-kDa cellular protein. Virology. 1995;209:174–187. doi: 10.1006/viro.1995.1241. [DOI] [PubMed] [Google Scholar]
- 19.Meredith M, Orr A, Everett R. Herpes simplex virus type 1 immediate-early protein Vmw110 binds strongly and specifically to a 135-kDa cellular protein. Virology. 1994;200:457–469. doi: 10.1006/viro.1994.1209. [DOI] [PubMed] [Google Scholar]
- 20.Mitchell C, Blaho J A, Roizman B. Casein kinase II specifically nucleotidylylates in vitro the amino acid sequence of the protein encoded by the alpha 22 gene of herpes simplex virus 1. Proc Natl Acad Sci USA. 1994;91:11864–11868. doi: 10.1073/pnas.91.25.11864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mullen M A, Ciufo D M, Hayward G S. Mapping of intracellular localization domains and evidence for colocalization interactions between the IE110 and IE175 nuclear transactivator proteins of herpes simplex virus. J Virol. 1994;68:3250–3266. doi: 10.1128/jvi.68.5.3250-3266.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogle W O, Ng T I, Carter K L, Roizman B. The UL13 protein kinase and the infected cell type are determinants of posttranslational modification of ICP0. Virology. 1997;235:406–413. doi: 10.1006/viro.1997.8710. [DOI] [PubMed] [Google Scholar]
- 23.Parkinson J, Everett R D. Alphaherpesvirus proteins related to herpes simplex virus type 1 ICP0 affect cellular structures and proteins. J Virol. 2000;74:10006–10017. doi: 10.1128/jvi.74.21.10006-10017.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sacks W R, Schaffer P A. Deletion mutants in the gene encoding the herpes simplex virus type 1 immediate-early protein ICP0 exhibit impaired growth in cell culture. J Virol. 1987;61:829–839. doi: 10.1128/jvi.61.3.829-839.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stow E C, Stow N D. Complementation of a herpes simplex virus type 1 Vmw110 deletion mutant by human cytomegalovirus. J Gen Virol. 1989;70:695–704. doi: 10.1099/0022-1317-70-3-695. [DOI] [PubMed] [Google Scholar]
- 26.Stow N D, Stow E C. Isolation and characterization of a herpes simplex virus type 1 mutant containing a deletion within the gene encoding the immediate early polypeptide Vmw110. J Gen Virol. 1986;67:2571–2585. doi: 10.1099/0022-1317-67-12-2571. [DOI] [PubMed] [Google Scholar]
- 27.Van Sant C, Kawaguchi Y, Roizman B. A single amino acid substitution in the cyclin D binding domain of the infected cell protein no. 0 abrogates the neuroinvasiveness of herpes simplex virus without affecting its ability to replicate. Proc Natl Acad Sci USA. 1999;96:8184–8189. doi: 10.1073/pnas.96.14.8184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Sant C, Lopez P, Advani S J, Roizman B. Role of cyclin D3 in the biology of herpes simplex virus 1 ICP0. J Virol. 2001;75:1888–1898. doi: 10.1128/JVI.75.4.1888-1898.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu X X, Chen J X, Silverstein S. Isolation and characterization of a functional cDNA encoding ICP0 from herpes simplex virus type 1. J Virol. 1991;65:957–960. doi: 10.1128/jvi.65.2.957-960.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu Z, Cai W, Schaffer P A. Cooperativity among herpes simplex virus type 1 immediate-early regulatory proteins: ICP4 and ICP27 affect the intracellular localization of ICP0. J Virol. 1994;68:3027–3040. doi: 10.1128/jvi.68.5.3027-3040.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu Z, DeLuca N A, Schaffer P A. Overexpression of the herpes simplex virus type 1 immediate-early regulatory protein, ICP27, is responsible for the aberrant localization of ICP0 and mutant forms of ICP4 in ICP4 mutant virus-infected cells. J Virol. 1996;70:5346–5356. doi: 10.1128/jvi.70.8.5346-5356.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]