Abstract
HeLa cells were transfected with several plasmids that encoded all poliovirus (PV) nonstructural proteins. Viral RNAs were transcribed by T7 RNA polymerase expressed from recombinant vaccinia virus. All plasmids produced similar amounts of viral proteins that were processed identically; however, RNAs were designed either to serve as templates for replication or to contain mutations predicted to prevent RNA replication. The mutations included substitution of the entire PV 5′ noncoding region (NCR) with the encephalomyocarditis virus (EMCV) internal ribosomal entry site, thereby deleting the 5′-terminal cloverleaf-like structure, or insertion of three nucleotides in the 3Dpol coding sequence. Production of viral proteins was sufficient to induce the characteristic reorganization of intracellular membranes into heterogeneous-sized vesicles, independent of RNA replication. The vesicles were stably associated with viral RNA only when RNA replication could occur. Nonreplicating RNAs localized to distinct, nonoverlapping regions in the cell, excluded from the viral protein-membrane complexes. The absence of accumulation of positive-strand RNA from both mutated RNAs in transfected cells was documented. In addition, no minus-strand RNA was produced from the EMCV chimeric template RNA in vitro. These data show that the 5′-terminal sequences of PV RNA are essential for initiation of minus-strand RNA synthesis at its 3′ end.
Recent studies of events associated with poliovirus (PV) RNA replication have contributed to new insights into this complex reaction. In vivo and in vitro studies have implicated interactions between cellular and viral proteins with distinct elements on the viral RNA (vRNA) in various steps leading to production of new vRNA strands. The viral protein 3D catalyzes primer- and template-dependent RNA synthesis and is the only protein required for elongation of RNA chains in vitro (36, 52). It also catalyzes the uridylylation of VPg (viral protein 3B) in vitro, in the presence of poly(A) (42). A short unpaired nucleotide sequence in a highly conserved stem-loop formed by the RNA in the 2C coding region appears to be a component of the natural template for the 3D-catalyzed VPg uridylylation reaction; this reaction is stimulated greatly by uncleaved 3CD (41). Uridylylated VPg is thought to function as the primer for initiation of minus-strand RNA synthesis, and thus uridylylation represents the first step in vRNA replication. In infected cells, however, this reaction appears to require the integrity of a membranous replication complex (RC), which has been demonstrated to serve as the site for vRNA synthesis and is composed of heterogeneous-sized vesicles associated with viral nonstructural proteins and RNA (13). Although all of the viral nonstructural proteins, as well as several of their precursor forms (e.g., 2BC, 3AB, and 3CD), have been implicated in vRNA synthesis, their precise biochemical roles remain uncertain, and detailed analyses of their activities are complicated by the multiple functions manifested by each (reviewed in reference 28).
Association with modified membrane structures appears to be a property of all positive-strand vRNA replication reactions (references 18 and 54 and references therein). Early observations of PV-infected cells showed massive proliferation of intracellular membranes and formation of extensive clusters of membrane vesicles, with which replication proteins and nascent RNAs were associated (9, 11, 12, 15). Electron microscopic (EM) analysis showed that the vesicles are budded from endoplasmic reticulum (9), although markers from several other cellular organelles (e.g., lysosomes and Golgi apparatus) were also found in the RCs, especially at late times postinfection (45).
The formation of membrane-bound RCs requires significant reorganization of cellular membranes. Viral proteins containing 2B, 2C, or 3A sequences can associate with membranes directly (22, 24, 46, 49, 53) and may induce rearrangement of intracellular membrane structures into vesicles, tubules, or other morphological forms (20, 26). Expression of protein 2B, 2BC, or 3A inhibited protein secretory traffic (23, 44), and proteins 2B and 2BC altered plasma membrane permeability (35, 53). Other viral proteins may be recruited to the membrane complex via protein-protein interactions, such as has been shown between 3D and 3AB (32, 55) or by incorporation into the RC in the form of precursors while still attached to membrane-binding carrier sequences. It is not known how RNA is bound to the RC containing the relevant proteins, although several PV nonstructural proteins have been reported to manifest RNA-binding activity (4, 14, 43).
In this study, we show that PV nonstructural proteins, independent of RNA replication, induce morphological changes in the cytoplasm and nucleus indistinguishable from those produced during a PV infection. However, only RNAs capable of replication colocalize with the newly formed vesicles to form an RC. In addition, our data show that synthesis of minus-strand RNA, initiated from the 3′ end of the template, involves simultaneous recognition of the 5′ end of the template.
MATERIALS AND METHODS
Plasmid constructions.
To generate plasmid pPVΔP1, which contains an in-frame deletion of the P1 coding region, overlap extension PCR mutagenesis was used as described previously (17). Two PCRs were performed with pT7-PV1 (30) as a template and the following primers: reaction 1, primers 1 (5′-CGTGGTTGAAAGCGACGG) and 2 (5′-GGTGTCCGAATCCCATTATGATACAATTGTCTGATTG); and reaction 2, primers 3 (5′-GCCATGGTGAAGCATCACAC) and 4 (5′-GACAATTGTATCATAATGGGATTCGGACACCAAAACAAAGCG). A 50-ng portion of the 556-bp product of reaction 1 and a 50-ng portion of the 699-bp product of reaction 2 were mixed and used in a second round of PCR with primers 1 and 3. The resulting 1,224-bp fragment was digested with restriction enzymes AgeI and PstI to produce a 438-bp AgeI-PstI fragment, which was ligated to the 6.8-kb fragment isolated after digestion of the pT7-PV1 vector with the same enzymes. The sequence of the entire region produced by PCR was verified by sequence analysis. Plasmid pPVΔP1-3D∗ was generated by replacement of the BglII-EcoRI fragment of pPVΔP1 by the equivalent fragment from pT7-3D-μ6432 (48). To generate plasmid pE5PVΔP1, the 565-bp PstI-SpeI fragment containing nucleotides (nt) 3417 to 3982 from pT7-PV1 was ligated with plasmid pTM-2BC digested with the same enzymes and double-stranded synthetic oligonucleotide obtained by annealing of the oligonucleotide 2A-NP1 (5′-TATGGATCTGACCACATACGGATTCGGACACCAAAACAAAGCGGTGTACACTGCA) with oligonucleotide 2A-NP2 (5′-GTGTACACCGCTTTGTTTTGGTGTCCGAATCCGTATGTGGTCAGATCCA) to create plasmid pTM-PV-P2. The SpeI-SalI fragment in pTM-PV-P2 was then substituted by the SpeI-SalI fragment from pT7-PV1(Sal) (8). The region introduced by synthetic oligonucleotide was confirmed by sequence analysis.
Plasmid pGEM-PV-NH was generated by conventional subcloning methods using two-fragment ligation reactions. Plasmid pGEM-PV-NH has fragment NheI-HindIII (nt 2470 to 6056 in PV cDNA) inserted between XbaI and HindIII sites of vector pGEM-3Zf(+).
In vitro RNA transcription.
pPVΔP1 and its derivatives were linearized with EcoRI, and pE5PVΔP1 was linearized with SalI. Transcription reactions were performed with T7 RNA polymerase using a MAXIscript in vitro transcription kit (Ambion, Inc.). Transcription reaction mixtures were incubated at 37°C for 90 min, and then reactions were terminated by addition of DNase I. RNA was purified using an RNeasy kit (Qiagen) and analyzed by electrophoresis in a 1% agarose gel stained with ethidium bromide. The RNA concentration was determined by A260 measurement.
In vitro translation-replication assays.
In vitro translation or coupled translation-replication assays were performed in micrococcal nuclease-treated HeLa cell extracts essentially as described elsewhere (5, 50), with the following modifications. Reaction mixtures were programmed with in vitro-transcribed RNA at a concentration of 20 nM or with 10 nM vRNA. For translation analysis, 10-μl aliquots of reaction mixtures were supplemented with 15 μCi of [35S]methionine (Amersham). Translation reaction mixtures were incubated at 30°C for 6 h, reactions were terminated by addition of 2× sample buffer, and results were analyzed by electrophoresis on sodium dodecyl sulfate–12.5% polyacrylamide gels. The ability of RNA to replicate in vitro was tested as in method 3 (6). Briefly, preinitiation complexes were formed in 40-μl translation reaction mixtures containing 2 mM guanidine HCl. They were isolated from these reaction mixtures by centrifugation and resuspended in 40 μl of replication mix, containing fresh HeLa S10 extract and 25 μCi of [α-32P]CTP in the absence of guanidine.
RNA stability assay.
Translation reactions were programmed with PVΔP1 and E5PVΔP1 RNA transcripts produced in the presence of 10 μCi of [α-32P]CTP (400 Ci/mmol) in a 40-μl transcription reaction and incubated at 30°C. The RNA transcripts had specific activities of 6 × 105 cpm/μg. RNA was extracted from 10-μl aliquots of translation reaction mixtures at different times of incubation, using an RNeasy kit (Qiagen). After precipitation with ethanol, samples were denatured with glyoxal and analyzed on a 1% agarose gel.
DNA and RNA transfection.
For protein expression, HeLa cells were infected with vaccinia virus vTF7-3 (39) and transfected with plasmid DNA as described previously (46). RNA transfections were performed with DEAE-dextran (molecular weight, 500,000; Sigma Chemical Co.) as described elsewhere (48).
RNA extraction and RNase protection assays.
RNA extractions were performed using an RNeasy kit (Qiagen). To eliminate contamination with plasmid DNA, samples were treated with DNase I (Qiagen) directly on the columns. Total extracted RNA was eluted from columns. RNase protection assays were performed using an RPA III kit (Ambion). For detection of negative-strand RNA, analysis was done using two rounds of RNase protection (40). In the first step, 0.5 μg of PV RNA was added to each sample, and samples were self-annealed in hybridization buffer and treated with a mixture of RNases A and T1. The RNAs were recovered by precipitation, denatured, annealed with 32P-labeled riboprobe, and subjected to a second RNase digestion. Protected RNA fragments were separated on a 6% polyacrylamide–7 M urea gel. Riboprobe for the detection of negative-strand RNA was transcribed by SP6 polymerase from plasmid pGEM-PV-2C∗-3C∗ (19) linearized with NsiI and corresponds to PV nt 4600 to 4830. Riboprobe for the detection of plus-strand RNAs was transcribed by SP6 polymerase from plasmid pPV-NH linearized with KasI and contains antisense RNA corresponding to PV nt 5823 to 6056.
IF, FISH, and EM.
For immunofluorescence (IF) and fluorescent in situ hybridization (FISH), cells were grown and transfected on glass coverslips, fixed with paraformaldehyde, and permeabilized as described elsewhere (15). For IF, the cell preparations were incubated with anti-PV 2B monoclonal antibody 1D3.B1, washed, and incubated with goat anti-mouse antibody coupled to Texas red. Coverslips were mounted in Tris-glycerol (pH 8.5) containing 2.5% 1,4-diazabicyclo(2.2.2)octane (DABCO; Sigma, Buchs, Switzerland) (51). For FISH, a single-stranded RNA probe of minus polarity comprising nt 6012 to 6736 was prepared and labeled with fluorescein isothiocyanate-UTP (Roche Molecular Biochemicals, Mannheim, Germany) during in vitro transcription with T7 polymerase. The probe was hydrolyzed and purified as described elsewhere (25). The FISH protocol to detect RNA of plus polarity has been described in detail previously (15, 25). For double-labeling FISH-IF, IF was performed after completion of FISH. For confocal laser scanning microscopy (CLSM), a Leica TCS4D microscope was used with the photomultiplier settings adjusted to avoid bleeding from one channel into the other. Raw images were adjusted for contrast and background staining with Adobe Photoshop software. For EM, cell cultures were trypsinized, fixed with 2.5% glutaraldehyde–2% OsO4, and embedded in Epon 812 according to standard procedures. Sections were viewed in a Philips CM100 electron microscope.
RESULTS
Construction of plasmids expressing PV nonstructural proteins.
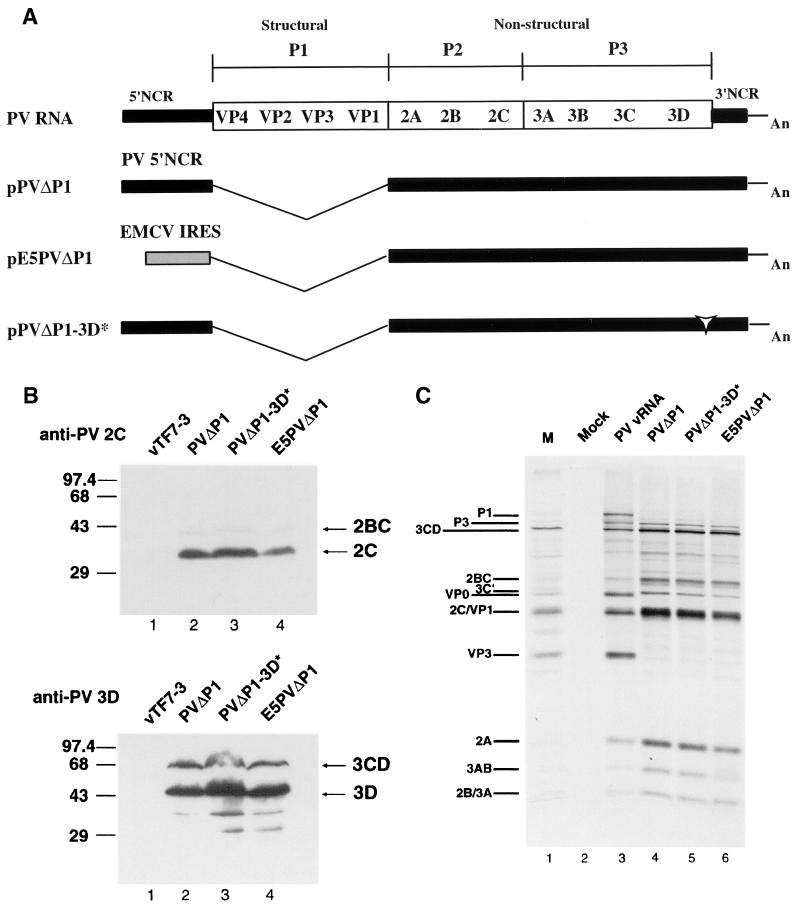
In this study, HeLa cells were transfected with plasmids that expressed proteins required for RNA replication, under conditions in which RNA replication either could or could not occur. We sought to compare the formation of membrane vesicles and the fates of viral proteins and RNAs under these two conditions. In previous reports we described the expression of several individual PV nonstructural proteins, using recombinant vaccinia virus expressing T7 RNA polymerase to transcribe plasmids engineered to contain the appropriate cDNA sequences downstream of a T7 promoter (20, 26, 46). This expression system gives high levels of cytoplasmic expression of heterologous proteins. In this study, we used a similar strategy for the simultaneous expression of all PV nonstructural proteins from replicating or nonreplicating RNAs. The subgenomic plasmid pPVΔP1 is a derivative of the full-length pT7-PV1 from which the entire P1 coding region was deleted (Fig. 1A). In this construct, the first codon of 2A follows the PV initiation AUG. Previous work from several laboratories showed that similar deletions of the P1 coding region could be introduced without affecting replication of viral RNA (21, 29, 33, 34).
FIG. 1.
(A) Schematic representation of full-length and subgenomic PV RNAs used in this study. PVΔP1 and PVΔP1-3D∗ RNAs contain two nonviral guanylate residues at the 5′ ends of the RNAs transcribed by T7 RNA polymerase. Black bars denote PV sequences; the gray box denotes the EMCV IRES sequence in E5PVΔP1 RNA; the open arrowhead indicates insertion of the codon for Ile in PVΔP1-3D∗; An represents the 30-mer poly(A) tail. (B) Immunoblot analysis of HeLa cells expressing PV proteins. HeLa cells were infected with vTF7-3 and simultaneously transfected with pPVΔP1 (lane 2), pPVΔP1-3D∗ (lane 3), or pE5PVΔP1 (lane 4). Cells were harvested 14 hs after transfection and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western immunoblotting with rabbit anti-PV2C serum (top) or rabbit anti-PV3D serum (bottom). Cells infected with vTF7-3 and mock transfected are shown in lane 1. In all lanes, extracts from approximately 5 × 104 cells were loaded. Positions of 2C, 2BC, 3D, and 3CD (right) and protein markers (left, in kilodaltons) are indicated. (C) In vitro translation of subgenomic RNAs. HeLa cell-free translation reactions were programmed with PV RNA (lane 3) or the indicated subgenomic RNAs transcribed in vitro (lanes 4 to 6). The identities of PV proteins are indicated. Lane 1, marker (M) PV proteins produced in infected cells; lane 2, no RNA.
For comparison, we constructed two plasmids that were predicted to produce the same PV nonstructural proteins but whose RNA transcripts were anticipated to be incapable of replication. Plasmid pE5PVΔP1 included cDNA encoding all of the PV nonstructural proteins (P2 and P3 regions), the 3′ noncoding region (NCR), and poly(A) tract, downstream of the encephalomyocarditis virus (EMCV) internal ribosomal entry site (IRES) (Fig. 1A). The RNA transcript obtained with T7 RNA polymerase lacks the PV 5′ NCR, including the 5′-terminal cloverleaf-like structure, a presumed signal for RNA replication. The EMCV IRES sequence was inserted preceding the PV P2-P3 open reading frame to ensure efficient translation to produce all PV nonstructural proteins. The translation product starts with five codons from VP1 to create the natural VP1-2A cleavage site, which forms the authentic N terminus of 2A. A second plasmid, pPVΔP1-3D∗, contains an insertion coding for an Ile residue after position 149 in 3D polymerase. This mutation abolished RNA polymerase activity when assayed in vitro (16) and inhibited vRNA replication when introduced into a full-length PV RNA (48). Thus, RNAs produced from each of the latter two plasmids are predicted not to support their own replication. In the case of RNA E5PVΔP1, the 5′-terminal cloverleaf structure thought to be required for initiation of RNA strand synthesis is absent; for RNA PVΔP1-3D∗, the polymerase protein encoded by the RNA is enzymatically inactive.
Expression and processing of viral proteins.
To examine protein expression from subgenomic plasmids, HeLa cells were transfected with either pPVΔP1, pE5PVΔP1, or pPVΔP1-3D∗ in the presence of recombinant vaccinia virus vTF7-3 as a source of T7 RNA polymerase. IF analysis of cells probed with anti-2C serum showed that the efficiencies of transfection and expression generally varied between 15 and 25% and were similar for each plasmid (data not shown). Immunoblot analysis with antibodies to protein 2C and 3D showed that similar amounts of 2C, 3D, and 3CD proteins (Fig. 1B) were produced in cells transfected with all three plasmids.
Correct translation and processing of other PV nonstructural proteins were demonstrated by translation in vitro. RNA transcripts from each plasmid were translated in extracts derived from uninfected HeLa cells to confirm that protein processing occurred with equal efficiencies and that all three plasmids generated the same protein products (Fig. 1C). The patterns of proteins produced from all three RNAs are virtually identical, and all major bands characteristic of PV nonstructural proteins are present. RNA E5PVΔP1 was translated with a slightly lower efficiency than PVΔP1 or PVΔP1-3D∗, possibly because the translation reaction conditions used were optimized for the PV IRES.
Replication of subgenomic RNA in HeLa cells.
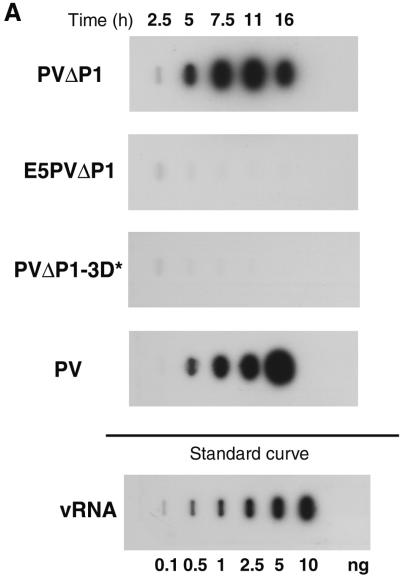
To test for the replicative capacity of the PV subgenomic RNA and the expected loss of replicative ability conferred by substitution of the PV 5′ NCR by the EMCV IRES sequence in the E5PVΔP1 RNA transcript, as well as by the Ile insertion in 3Dpol, we measured the accumulation of virus-specific RNAs after transfection of cells with RNA transcripts produced in vitro from linearized plasmids. Figure 2A shows the analysis of plus-strand RNA by slot blot hybridization. Replication of wild-type, full-length transcripts of pT7-PV1 as well as of the subgenomic transcripts from pPVΔP1 was readily detectable by 5 h posttransfection. The shorter, subgenomic RNA appears to accumulate a bit more rapidly than the full-length RNA, as evidenced by the presence of more RNA at the early time points. RNA PVΔP1 is lacking the capsid protein coding region and thus is unable to spread to untransfected cells. The maximum signal of plus-strand RNA was observed around 11 h after transfection, with the signal diminishing by 16 h after transfection, presumably because of the loss of RNA due to lysis of transfected cells. For the full-length transcript, plus-strand RNA continues to increase up to 16 h due to the spread of virus to additional cells. As expected, no RNA replication was detected in cells transfected with RNA transcripts of pE5PVΔP1 or pPVΔP1-3D∗. A faint band usually observed at 2.5 h after transfection is likely due to the detection of the input RNA transcript (37, 47). A more sensitive RNase protection assay was used to detect potentially very low levels of replication of the mutant transcripts; in agreement with the hybridization data, no accumulation of the E5PVΔP1 or PVΔPV-3D∗ plus-strand RNA was observed (data not shown).
FIG. 2.
Accumulation of PV-specific RNAs in transfected cells. HeLa cell monolayers were transfected with RNA transcripts in the presence of DEAE-dextran. Cells were harvested at the indicated times after transfection. (A) Accumulation of plus-strand RNA. Total cytoplasmic RNA isolated from approximately 104 cells was bound to a nylon membrane and hybridized to a 32P-labeled riboprobe complementary to nt 5823 to 6056 of the vRNA. The standard curve shows increasing amounts of purified PV RNA (0.1 to 10 ng) hybridized in parallel. (B) Accumulation of negative-strand RNA. Total cytoplasmic RNA isolated from approximately 105 cells was subjected to two-step RNase protection (40). Positions of unprotected (258-nt) and protected (230-nt) probes are indicated. Top, mock-transfected cells (lanes 1 to 5) and cells transfected with PV full-length RNA transcript (lanes 6 to 10); bottom, cells transfected with PVΔP1 (lanes 1 to 5), E5PVΔP1 (lanes 6 to 10), and PVΔP1-3D∗ (lanes 11 to 15) RNA transcripts.
PV 5′-terminal sequences are required for synthesis of negative-strand RNA.
The failure to replicate the chimeric RNA lacking the PV 5′-terminal sequences was anticipated, since the cloverleaf-like structure formed by the first ∼90 nt of PV RNA has been shown to be an essential determinant for assembly of the complex of proteins required for initiation of RNA synthesis (2, 3). It has been suggested that assembly of this complex is essential for minus-strand synthesis (27) initiated at the 3′ end of the template. We wished to determine directly whether the absence of the PV 5′-terminal sequences in E5PVΔP1 RNA prevents a single round of negative-strand synthesis. The presence of negative-strand RNA in cells after transfection with RNA transcripts was first analyzed using a two-step RNase protection assay, described previously by Novak and Kirkegaard (40). Negative-strand RNA was detected in cells 2.5 h after transfection with PVΔP1 RNA or at 5 h after transfection with full-length PV RNA (Fig. 2B). Although this method may be only semiquantitative, the detection of negative-strand subgenomic PVΔP1 RNA at earlier times than full-length negative-strand RNA is consistent with the relative kinetics observed for positive-strand RNA accumulation (Fig. 2A). No signal was detected for the negative strand of E5PVΔP1 or PVΔP1-3D∗ after transfection of these RNAs. However, comparison of the kinetics of accumulation of plus-and minus-sense RNAs shown in Fig. 2 indicates that negative-strand RNA was detected only after some amplification of input RNA had occurred. Furthermore, the presence of small amounts (<0.1%) of negative-strand RNA in the input transcripts produced by T7 RNA polymerase in vitro limited our attempts to increase the sensitivity of the detection method. We also have observed production of small amounts of negative-strand RNA by T7 RNA polymerase, presumably from a cryptic promoter, in cells transfected with pE5PVΔP1 or pPVΔP1-3D∗ and infected with vTF7-3, raising some doubts as to the origin of very small amounts of negative-strand RNA that might be detected.
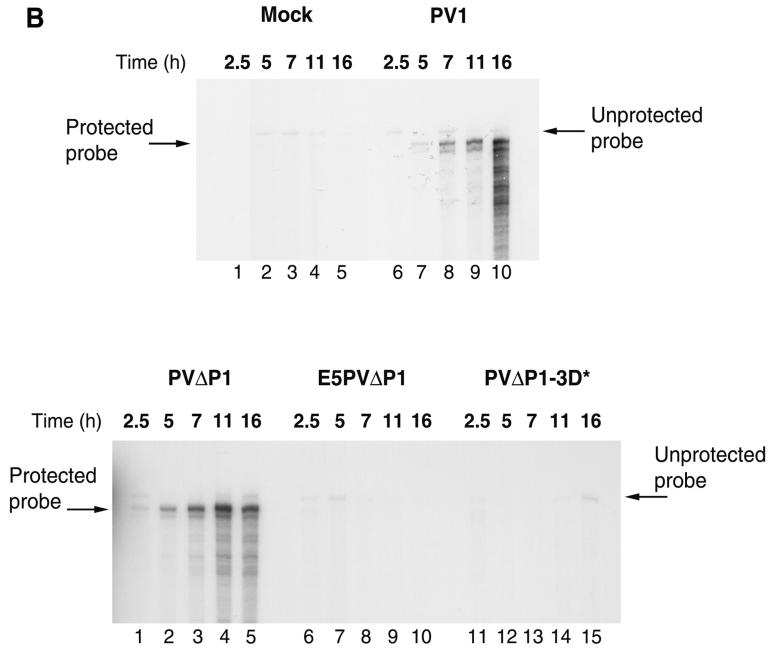
To circumvent difficulties with detection of minus strands produced in vivo, we utilized the translation and replication properties manifested by uninfected HeLa cell extracts. Synthesis of minus-strand RNA can be detected in extracts that efficiently produce all viral replication proteins by translation of added template RNAs that contain the necessary replication signals (6). When the 5′ ends of such template RNAs contain the two non-PV guanylate residues generated by transcription from the T7 promoter, minus strands are produced but no amplification of plus strands is detected (7, 31). Indeed, virus production in extracts programmed with T7 RNA transcripts is reduced 100- to 1,000-fold compared with extracts programmed with virion RNA containing authentic 5′ termini. We therefore analyzed RNA synthesis in HeLa cell extracts programmed with PVΔP1 or E5PVΔP1 RNA. Figure 3A shows 32P-labeled RNA synthesis by isolated preinitiation RNA RCs formed during translation of several templates in the presence of 2 mM guanidine HCl.
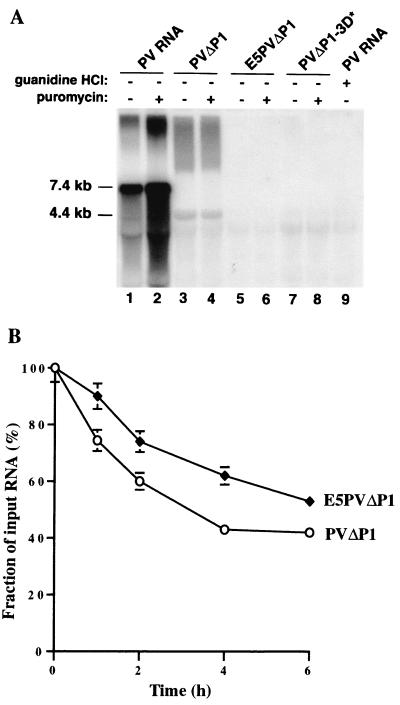
FIG. 3.
(A) Synthesis of RNA in vitro. Preinitiation RNA replication complexes were isolated from 40-μl HeLa translation-replication reaction mixtures containing 2 mM guanidine HCl and the indicated RNAs after incubation at 30°C for 6 h. The complexes were resuspended in 40-μl reaction mixtures containing fresh HeLa S10 extract, translation initiation factors, and 25 μCi of [α-32P]CTP. Reaction mixtures 2, 4, 6, and 8 also contained 100 μg of puromycin/ml; reaction mixture 9 contained 2 mM guanidine HCl. The reaction mixtures were incubated at 34°C for 2 h; the labeled RNA products were denatured with glyoxal and characterized by electrophoresis in a 1.1% agarose gel followed by autoradiography (38). The positions of migration of RNA markers run on the same gel and visualized by staining with ethidium bromide are indicated. (B) Stability of RNA templates. 32P-labeled RNAs were synthesized and incubated for various times in HeLa translation-replication reactions, and samples were analyzed on agarose gels as described in Materials and Methods. Intact RNA migrating to the correct position on the gel was quantitated by phosphorimaging. The amounts of radioactivity at the start of the incubation were the same for both samples.
After translation of PVΔP1 RNA, production of minus-strand RNA was detected when RNA replication was allowed to occur (Fig. 3A, lane 3). In agreement with previous reports, significantly greater RNA synthesis was observed from vRNA, when both negative- and positive-strand synthesis occur in this reaction (compare lanes 1 and 2 with lanes 3 and 4). vRNA synthesis in vitro was reported recently to be stimulated by the presence of puromycin (7), presumably by the drug's clearance of ribosomes from the positive-strand RNA, facilitating its utilization as a template for RNA synthesis. This stimulation by puromycin is seen clearly in Fig. 3A, lane 2. No such stimulation was observed for RNA synthesis from PVΔP1 RNA (compare lanes 3 and 4). Since the PVΔP1 RNA transcript does not support synthesis of new plus-strand RNA (7), the reaction generating minus strand is complete by 2 h of incubation, when natural clearance of ribosomes has had time to occur. This effect was demonstrated previously by Barton et al. (7), who showed stimulation of negative-strand production by puromycin only during the first 60 min of incubation. As expected, a mutation in 3Dpol prevented any RNA synthesis on PVΔP1-3D∗ RNA (lanes 7 and 8). Similarly, no RNA was detected in the E5PVΔP1 RNA reaction (lanes 5 and 6). Some unresolved labeled material is present at the top of the gel in lanes 1 to 4 in addition to labeled RNA bands of the expected size. This material presumably represents incompletely denaturated RNA products and is present only in those lanes where production of RNA was observed. Some concern remained, however, that the slightly less efficient production of proteins from E5PVΔP1 RNA under our standard reaction conditions (Fig. 1C) might cause our failure to detect RNA synthesis in these reactions. We therefore developed conditions to equalize translation levels from PVΔP1 RNA and E5PVΔP1 RNA. This was accomplished in two ways: (i) increasing the concentration of added magnesium acetate from 0.3 to 0.6 mM (which decreased translation from the PV IRES and increased translation from the EMCV IRES) or (ii) reducing the concentration of PVΔP1 RNA from 20 to 1.4 nM. Both of these altered reaction conditions generated viral protein in equal amounts from the two RNAs (data not shown). Under both of these conditions, synthesis of negative-strand RNA from preinitiation complexes formed with PVΔP1 RNA was observed, whereas none was detected from E5PVΔP1 (data not shown). To determine whether the absence of detectable synthesis of negative-strand RNA from E5PVΔP1 RNA might be due to increased degradation of E5PVΔP1 RNA compared to PVΔP1 RNA in the HeLa cell extract, we incubated 32P-labeled PVΔP1 and E5PVΔP1 RNAs under conditions used for translation and production of preinitiation complexes and analyzed the kinetics of RNA decay on agarose gels. Surprisingly, PVΔP1 RNA was degraded more rapidly, with a half-life in the range of 3 h, while E5PVΔP1 RNA was more stable, with half-life of approximately 6 to 6.5 h (Fig. 3B). Thus, the inability of E5PVΔP1 RNA to produce negative-strand RNA could not be attributed to relative instability of this RNA. Taken together, these results demonstrate that the 5′ end of PV RNA contains signals essential for initiation of minus-strand synthesis at the 3′ end.
Formation of vesicles in cells expressing PV nonstructural proteins.
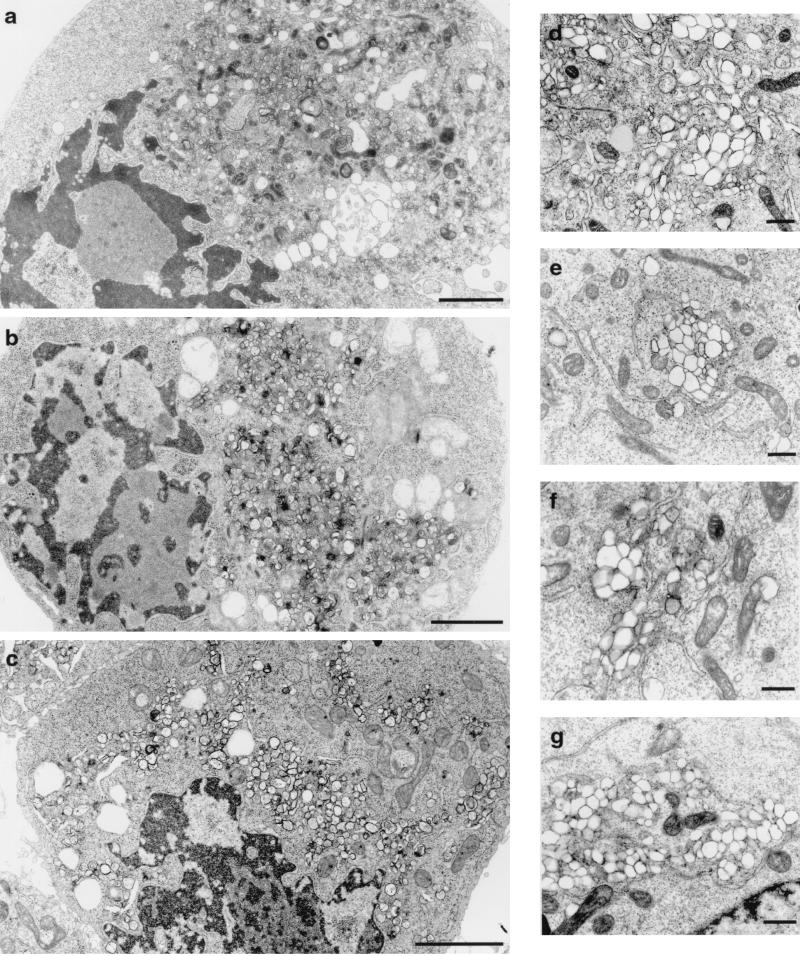
A hallmark of PV infection is the reorganization of endoplasmic reticulum and other intracellular membranes to generate clusters of vesicles that serve as the sites for RNA replication. Several laboratories have observed that expression of individual viral proteins can induce a variety of morphological rearrangements of intracellular membranes, but the pathway of formation of functional RCs is not known. We therefore compared the patterns of membrane rearrangement in cells transfected with plasmids expressing replicating PVΔP1 RNA or the nonreplicating E5PVΔP1 or PVΔP1-3D∗ RNA. Figure 4 shows that vesicles very similar in appearance are found in cells producing all of the PV nonstructural proteins, independent of vRNA replication. The overall cell morphology (Fig. 4a to c), as well as the appearance of the vesicles (Fig. 4d to f), is indistinguishable from that seen in PV-infected cells (Fig. 4g) (9), manifesting characteristic clusters of smooth membrane vesicles of heterogeneous sizes. Interestingly, in addition to cytoplasmic vesicle formation, even in the absence of RNA replication, nuclei show peripheral positioning, indentation, and chromatin condensation similar to those in PV-infected cells. Expression of individual protein 2BC, 2C, or 3AB did not induce these nuclear morphological changes (D. Eggar and K. Bienz, unpublished observations).
FIG. 4.
Electron micrographs of HeLa cells expressing PV nonstructural proteins. Cell cultures were infected with vTF7-3 and transfected with plasmid pPVΔP1 (a and d), pE5PVΔP1 (b and e), or pPVΔP1-3D∗(c and f). (g) PV-infected cell. The morphologies of cells (a to c) and of individual vesicles (d to g) appeared similar in all panels. Bars: 200 (a to c) and 500 (d to g) nm.
Active RNA replication is necessary for the association of RNA with virus-induced vesicles.
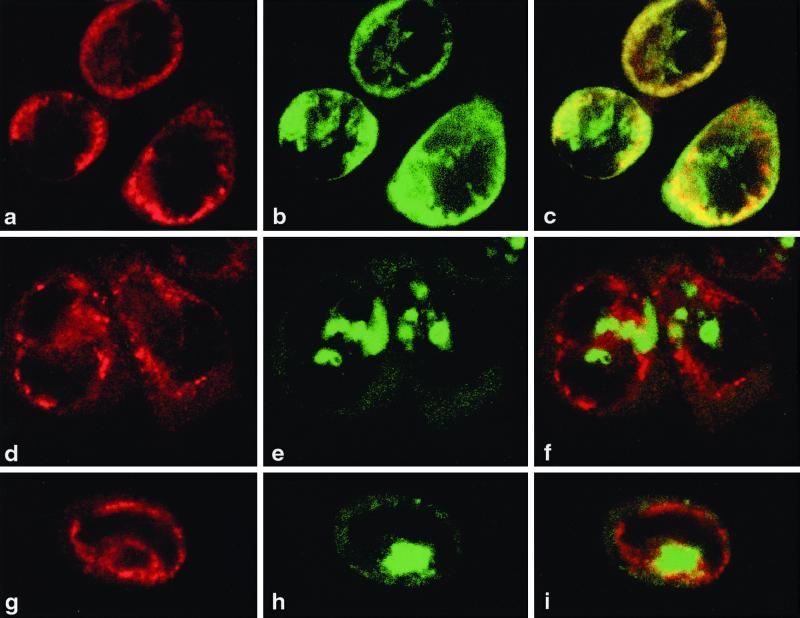
The above experiments demonstrate that vRNA replication is not required for vesicle formation. We therefore tested the abilities of replicating and nonreplicating RNAs to associate with the induced protein-membrane complexes. Cells expressing PVΔP1 RNA, E5PVΔP1 RNA, or PVΔP1-3D∗ RNA in the presence of vTF7-3 were analyzed simultaneously in situ for intracellular localization of the RNA and of viral protein 2B-containing sequences, which have been shown previously to be exclusively associated with induced membranous vesicles (10). RNA was identified by FISH analysis, and 2B-containing proteins were demonstrated by IF and CLSM at 7 to 11 h posttransfection. Figure 5 shows that only replicating RNA colocalized with the viral protein-induced vesicles (26). pPVΔP1 induced the formation of structures in which viral protein and RNA colocalize and which resemble virus-induced replication complexes early in infection (15) (Fig. 5a to c). On the other hand, cells transfected with pE5PVΔP1 (Fig. 5d to f) or pPVΔP1-3D∗ (Fig. 5g to i) accumulate viral protein and plus-strand RNA in distinct, nonoverlapping regions. The same pattern of aggregates of vRNA separated from viral protein-induced vesicles was seen in cells transfected with PVΔP1 RNA in the presence of 2 mM guanidine HCl (data not shown), which also prevents the first step in RNA replication, that of synthesis of minus-strand RNA (6). The exclusion of the nonreplicating RNAs from the protein-membranous vesicle complex suggests that active RNA replication is required for the stable association of viral RNA with such complexes.
FIG. 5.
Localization of viral proteins and RNA in transfected HeLa cells. Cells were infected with vTF7-3 and transfected with pPVΔP1 (a to c), pE5PVΔP1 (d to f), or pPVΔP1-3D∗ (g to i). 2B and 2BC were visualized by IF and CLSM with anti-2B monoclonal antibody and Texas red-labeled secondary antibody (a, d, and g). RNA was localized by FISH with fluorescein isothiocyanate-labeled riboprobe (b, e, and h). Merged images (c, f, and i) show replicating RNA associated with membranes carrying viral nonstructural proteins (c), whereas nonreplicating RNA remains separate (f and i). The horizontal axis of each picture corresponds to 38 μm.
DISCUSSION
In this study, we constructed several plasmids that direct expression of all PV nonstructural proteins from RNAs that either could or could not serve as templates for replication. Nonreplicating RNAs were RNAs lacking the 5′ NCR of PV sequence, for which the IRES of EMCV was substituted, RNAs containing a mutation that abolished vRNA polymerase activity, or wild-type RNAs in the presence of 2 mM guanidine HCl, which prevents vRNA synthesis. In all cases, viral proteins were synthesized and processed in similar amounts, since all of the mRNAs were actively transcribed from a T7 promoter by RNA polymerase expressed from a recombinant vaccinia virus. When the PV RNAs were capable of replication, they were found localized with viral proteins associated with vesicles that formed a characteristic RC. In all cases when they could not be replicated, they were found in separate aggregates, independent of the protein-induced vesicles, which nevertheless developed an ultrastructural morphology apparently identical to that of authentic replication complexes. As some of these nonreplicating RNAs contain a full complement of PV coding and noncoding sequences, the data indicate that the presence of specific sequences or structures in the plus-strand RNA are not sufficient to signal incorporation or association of the RNA into a replication complex. Rather, only those RNAs actively undergoing replication remain stably associated with the viral protein-induced vesicles. It remains uncertain whether sequences present on the minus-strand RNA contain signals to link the RNA into a replication complex.
Individual PV proteins containing 2B, 2C, or 3A sequences all manifest properties that reflect their inherent affinities for membranes, and each can independently produce alterations in intracellular membrane morphology and function. Indeed, characteristic cytoplasmic and nuclear changes of a PV-infected cell require only the combined protein-protein and protein-lipid interactions of all of the involved viral proteins together with preexisting cellular organelles, independent of whether the RNA from which they were translated was replicated. The nuclear changes observed during a PV infection or after expression of all PV nonstructural proteins did not occur following expression of 2B, 2BC, or 3AB individually; thus, these changes may be triggered by proteins outside these regions or may emerge by the combined action of these proteins with other viral proteins or with each other.
It is not known whether individual viral proteins are synthesized and subsequently associate with endoplasmic reticulum, or whether the nascent polyprotein associates with endoplasmic reticulum during translation, perhaps carrying protein, RNA, and ribosomes to the membranes en bloc. The latter model is attractive and is supported by observations that specific protein defects in viral RNA synthesis in a coupled translation-replication system in vitro are more efficiently complemented by coexpression of large precursor proteins than by production of the defective protein itself (50). If the model is correct, however, the nonreplicating RNAs, along with the ribosome, must be released from the membrane site when translation is completed, if RNA synthesis does not start.
Substitution of the PV 5′ NCR sequences in PVΔP1 by the EMCV IRES in E5PVΔP1 completely abolished RNA synthesis and in particular prevented synthesis of complementary negative strands initiated from the 3′ end of the template. Previously, Alexander et al. (1) reported construction of a chimeric PV-EMCV cDNA (pP108ENPO) in which the PV IRES was substituted by the EMCV IRES but which retained the 5′-terminal PV cloverleaf sequence. RNAs transcribed from the chimeric sequence were able to replicate and produce viable virus. Taken together, these data prove that the cloverleaf sequence element is essential for negative-strand synthesis. Work by Gamarnik and Andino (27) has suggested previously that assembly of a ribonucleoprotein complex between viral protein 3CD, cellular poly(C)-binding protein, and the 5′ cloverleaf-like structure is required to stop translation before negative-strand synthesis can start. RNAs containing mutations in the cloverleaf region that were defective in binding 3CD translated more efficiently than the wild type but did not accumulate negative-strand RNA. Their data did not distinguish between the direct participation of the cloverleaf structure in minus-strand RNA synthesis or failure to initiate RNA synthesis due to an inability to shut down translation. Our studies show that E5PVΔP1 RNA did not support negative-strand RNA synthesis even in the presence of puromycin, when the template was not being translated. Thus, our data provide evidence that there is cross talk between the two ends of PV RNA in order to initiate RNA replication.
ACKNOWLEDGMENTS
This work was supported by the National Institute of Allergy and Infectious Diseases, NIH, and by grant 31-055397.98 from the Swiss National Science Foundation (K.B.) and by Public Health Service grant AI 22693 from the National Institutes of Health (B.L.S.). D.M.B. was a predoctoral trainee funded by Public Health Service training grant AI07319.
REFERENCES
- 1.Alexander L, Lu H-H, Wimmer E. Poliovirus containing picornavirus type 1 and/or type 2 internal ribosomal entry site elements: genetic hybrids and the expression of a foreign gene. Proc Natl Acad Sci USA. 1994;91:1406–1410. doi: 10.1073/pnas.91.4.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andino R, Rieckhof G E, Achacoso P L, Baltimore D. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J. 1993;12:3587–3598. doi: 10.1002/j.1460-2075.1993.tb06032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andino R, Rieckhof G E, Baltimore D. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell. 1990;63:369–380. doi: 10.1016/0092-8674(90)90170-j. [DOI] [PubMed] [Google Scholar]
- 4.Banerjee R, Echeverri A, Dasgupta A. Poliovirus-encoded 2C polypeptide specifically binds to the 3′-terminal sequences of viral negative-strand RNA. J Virol. 1997;71:9570–9578. doi: 10.1128/jvi.71.12.9570-9578.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barton D J, Black E P, Flanegan J B. Complete replication of poliovirus in vitro: preinitiation RNA replication complexes require soluble cellular factors for the synthesis of VPg-linked RNA. J Virol. 1995;69:5516–5527. doi: 10.1128/jvi.69.9.5516-5527.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barton D J, Flanegan J B. Synchronous replication of poliovirus RNA: initiation of negative-strand RNA synthesis requires the guanidine-inhibited activity of protein 2C. J Virol. 1997;71:8482–8489. doi: 10.1128/jvi.71.11.8482-8489.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barton D J, Morasco B J, Flanegan J B. Translating ribosomes inhibit poliovirus negative-strand RNA synthesis. J Virol. 1999;73:10104–10112. doi: 10.1128/jvi.73.12.10104-10112.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bell Y C, Semler B L, Ehrenfeld E. Requirements for RNA replication of a poliovirus replicon by coxsackievirus B3 RNA polymerase. J Virol. 1999;73:9413–9421. doi: 10.1128/jvi.73.11.9413-9421.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bienz K, Egger D, Pasamontes L. Association of polioviral proteins of the P2 genomic region with the viral replication complex and virus-induced membrane synthesis as visualized by electron microscopic immunocytochemistry and autoradiography. Virology. 1987;160:220–226. doi: 10.1016/0042-6822(87)90063-8. [DOI] [PubMed] [Google Scholar]
- 10.Bienz K, Egger D, Pfister T. Characteristics of the poliovirus replication complex. Arch Virol Suppl. 1994;9:147–157. doi: 10.1007/978-3-7091-9326-6_15. [DOI] [PubMed] [Google Scholar]
- 11.Bienz K, Egger D, Pfister T, Troxler M. Structural and functional characterization of the poliovirus replication complex. J Virol. 1992;66:2740–2747. doi: 10.1128/jvi.66.5.2740-2747.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bienz K, Egger D, Rasser Y, Bossart W. Intracellular distribution of poliovirus proteins and the induction of virus-specific cytoplasmic structures. Virology. 1983;131:39–48. doi: 10.1016/0042-6822(83)90531-7. [DOI] [PubMed] [Google Scholar]
- 13.Bienz K, Egger D, Troxler M, Pasamontes L. Structural organization of poliovirus RNA replication is mediated by viral proteins of P2 genomic region. J Virol. 1990;64:1156–1163. doi: 10.1128/jvi.64.3.1156-1163.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blair W S, Parsley T B, Bogerd H P, Towner J S, Semler B L, Cullen B R. Utilization of a mammalian cell-based RNA binding assay to characterize the RNA binding properties of picornavirus 3C proteinases. RNA. 1998;4:215–225. [PMC free article] [PubMed] [Google Scholar]
- 15.Bolten R, Egger D, Gosert R, Schaub G, Landmann L, Bienz K. Intracellular localization of poliovirus plus- and minus-strand RNA visualized by strand-specific fluorescent in situ hybridization. J Virol. 1998;72:8578–8585. doi: 10.1128/jvi.72.11.8578-8585.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burns C C, Lawson M A, Semler B L, Ehrenfeld E. Effects of mutations in poliovirus 3Dpol on polymerase activity and on polyprotein cleavage. J Virol. 1989;63:4866–4874. doi: 10.1128/jvi.63.11.4866-4874.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burns C C, Richards O C, Ehrenfeld E. Temperature-sensitive polioviruses containing mutations in RNA polymerase. Virology. 1992;189:568–582. doi: 10.1016/0042-6822(92)90580-i. [DOI] [PubMed] [Google Scholar]
- 18.Chen J, Ahlquist P. Brome mosaic virus polymerase-like protein 2a is directed to the endoplasmic reticulum by helicase-like viral protein 1a. J Virol. 2000;74:4310–4318. doi: 10.1128/jvi.74.9.4310-4318.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cho M W, Richards O C, Dmitrieva T M, Agol V, Ehrenfeld E. RNA duplex unwinding activity of poliovirus RNA-dependent RNA polymerase 3Dpol. J Virol. 1993;67:3010–3018. doi: 10.1128/jvi.67.6.3010-3018.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho M W, Teterina N, Egger D, Bienz K, Ehrenfeld E. Membrane rearrangement and vesicle induction by recombinant poliovirus 2C and 2BC in human cells. Virology. 1994;202:129–145. doi: 10.1006/viro.1994.1329. [DOI] [PubMed] [Google Scholar]
- 21.Collis P S, O'Donnell B J, Barton D J, Rogers J A, Flanegan J B. Replication of poliovirus RNA and subgenomic RNA transcripts in transfected cells. J Virol. 1992;66:6480–6488. doi: 10.1128/jvi.66.11.6480-6488.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Datta U, Dasgupta A. Expression and subcellular localization of poliovirus VPg-precursor protein 3AB in eukaryotic cells: evidence for glycosylation in vitro. J Virol. 1994;68:4468–4477. doi: 10.1128/jvi.68.7.4468-4477.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doedens J R, Kirkegaard K. Inhibition of cellular protein secretion by poliovirus proteins 2B and 3A. EMBO J. 1995;14:894–907. doi: 10.1002/j.1460-2075.1995.tb07071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Echeverri A C, Dasgupta A. Amino terminal regions of poliovirus 2C protein mediate membrane binding. Virology. 1995;208:540–553. doi: 10.1006/viro.1995.1185. [DOI] [PubMed] [Google Scholar]
- 25.Egger D, Bolten R, Rahner C, Bienz K. Fluorochrome-labeled RNA as a sensitive, strand-specific probe for direct fluorescence in situ hybridization. Histochem Cell Biol. 1999;111:319–324. doi: 10.1007/s004180050363. [DOI] [PubMed] [Google Scholar]
- 26.Egger D, Teterina N, Ehrenfeld E, Bienz K. Formation of the poliovirus replication complex requires coupled viral translation, vesicle production, and viral RNA synthesis. J Virol. 2000;74:6570–6580. doi: 10.1128/jvi.74.14.6570-6580.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gamarnik A V, Andino R. Switch from translation to RNA replication in positive-stranded RNA virus. Genes Dev. 1998;12:2293–2304. doi: 10.1101/gad.12.15.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gromeier M, Wimmer E, Gorbalenya A E. Genetics, pathogenesis and evolution of picornaviruses. In: Domingo E, Webster R G, Holland J, editors. Origin and evolution of viruses. San Diego, Calif: Academic Press; 1999. pp. 287–343. [Google Scholar]
- 29.Hagino-Yamagishi K, Nomoto A. In vitro construction of poliovirus defective-interfering particles. J Virol. 1990;63:5386–5392. doi: 10.1128/jvi.63.12.5386-5392.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haller A A, Semler B L. Linker scanning mutagenesis of the internal ribosome entry site of poliovirus RNA. J Virol. 1992;66:5075–5086. doi: 10.1128/jvi.66.8.5075-5086.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herold J, Andino R. Poliovirus requires a precise 5′ end for efficient positive-strand RNA synthesis. J Virol. 2000;74:6394–6400. doi: 10.1128/jvi.74.14.6394-6400.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hope D A, Diamond S E, Kirkegaard K. Genetic dissection of interaction between poliovirus 3D polymerase and viral protein 3AB. J Virol. 1997;71:9490–9498. doi: 10.1128/jvi.71.12.9490-9498.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaplan G, Racaniello V R. Construction and characterization of poliovirus subgenomic replicons. J Virol. 1988;62:1678–1696. doi: 10.1128/jvi.62.5.1687-1696.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuge S, Nomoto A. Construction of viable deletion and insertion mutants of the Sabin strain of type 1 poliovirus: function of the 5′ noncoding sequence in viral replication. J Virol. 1987;61:1478–1487. doi: 10.1128/jvi.61.5.1478-1487.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lama J, Carrasco L. Expression of poliovirus nonstructural proteins in Escherichia coli cells. Modification of membrane permeability induced by 2B and 3A. J Biol Chem. 1992;267:15932–15937. [PubMed] [Google Scholar]
- 36.Lundquist R E, Ehrenfeld E, Maizel J V. Isolation of a viral polypeptide associated with poliovirus RNA polymerase. Proc Natl Acad Sci USA. 1974;71:773–777. doi: 10.1073/pnas.71.12.4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marc D, Drugeon G, Haenni A-L, Girard M, van der Werf S. Role of myristoylation of poliovirus capsid protein VP4 as determined by site-directed mutagenesis of its N-terminal sequence. EMBO J. 1989;8:2661–2668. doi: 10.1002/j.1460-2075.1989.tb08406.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McMasters G K, Carmichael G G. Analysis of single- and double-stranded nucleic acids on polyacrylamide and agarose gels by using glyoxal and acridine orange. Proc Natl Acad Sci USA. 1977;74:4835–4838. doi: 10.1073/pnas.74.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moss B, Elroy-Stein O, Mizukami T, Alexander W A, Fuerst T R. New mammalian expression vectors. Nature. 1990;348:91–92. doi: 10.1038/348091a0. [DOI] [PubMed] [Google Scholar]
- 40.Novak J E, Kirkegaard K. Improved method for detecting poliovirus negative strands used to demonstrate specificity of positive-strand encapsidation and the ratio of positive to negative strands in infected cells. J Virol. 1991;65:3384–3387. doi: 10.1128/jvi.65.6.3384-3387.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paul A V, Reider E, Kim D W, Van Boom J H, Wimmer E. Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg. J Virol. 2000;74:10359–10370. doi: 10.1128/jvi.74.22.10359-10370.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paul A V, van Boom J H, Fillipov D, Wimmer E. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature. 1998;393:280–284. doi: 10.1038/30529. [DOI] [PubMed] [Google Scholar]
- 43.Rodriguez P L, Carrasco L. Poliovirus protein 2C contains two regions involved in RNA binding activity. J Biol Chem. 1995;270:10105–10112. doi: 10.1074/jbc.270.17.10105. [DOI] [PubMed] [Google Scholar]
- 44.Sandoval I V, Carrasco L. Poliovirus infection and expression of the poliovirus protein 2B provoke the disassembly of the Golgi complex, the organelle target for the antipoliovirus drug Ro-090179. J Virol. 1997;71:4679–4693. doi: 10.1128/jvi.71.6.4679-4693.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schlegel A, Giddings T H, Ladinsky M S, Kirkegaard K. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J Virol. 1996;70:6576–6588. doi: 10.1128/jvi.70.10.6576-6588.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Teterina N L, Gorbalenya A E, Egger D, Bienz K, Ehrenfeld E. Poliovirus 2C protein determinants of membrane binding and rearrangements in mammalian cells. J Virol. 1997;71:8962–8972. doi: 10.1128/jvi.71.12.8962-8972.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Teterina N L, Kean K M, Gorbalenya E, Agol V I, Girard M. Analysis of the functional significance of amino acid residues in the putative NTP-binding pattern of the poliovirus 2C protein. J Gen Virol. 1992;73:1977–1986. doi: 10.1099/0022-1317-73-8-1977. [DOI] [PubMed] [Google Scholar]
- 48.Teterina N L, Zhou W D, Cho M W, Ehrenfeld E. Inefficient complementation activity of poliovirus 2C and 3D proteins for rescue of lethal mutations. J Virol. 1995;69:4245–4254. doi: 10.1128/jvi.69.7.4245-4254.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Towner J S, Ho T V, Semler B L. Determinants of membrane association for poliovirus protein 3AB. J Biol Chem. 1996;271:26810–26818. doi: 10.1074/jbc.271.43.26810. [DOI] [PubMed] [Google Scholar]
- 50.Towner J S, Mazanet M M, Semler B L. Rescue of defective poliovirus RNA replication by 3AB-containing precursor polyproteins. J Virol. 1998;72:7191–7200. doi: 10.1128/jvi.72.9.7191-7200.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Valnes K, Brandtzaeg P. Retardation of immunofluorescence fading during microscopy. J Histochem Cytochem. 1985;33:755–761. doi: 10.1177/33.8.3926864. [DOI] [PubMed] [Google Scholar]
- 52.Van Dyke T A, Flanegan J. Identification of poliovirus polypeptide P63 as a soluble RNA-dependent RNA polymerase. J Virol. 1980;35:732–740. doi: 10.1128/jvi.35.3.732-740.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Kuppeveld F G M, Hoenderop J G J, Smeets R L L, Willems P H G M, Dijkman H B P M, Galama J M D, Melchers W J G. Coxsackie-virus protein 2B modifies the ER membrane and plasma membrane permeability and facilitates virus release. EMBO J. 1997;16:3519–3532. doi: 10.1093/emboj/16.12.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wimmer E, Hellen C U T, Cao X M. Genetics of poliovirus. Annu Rev Genet. 1993;27:353–436. doi: 10.1146/annurev.ge.27.120193.002033. [DOI] [PubMed] [Google Scholar]
- 55.Xiang W, Cuconati A, Hope D, Kirkegaard K, Wimmer E. Complete protein linkage map of poliovirus P3 proteins: interaction of polymerase 3Dpol with VPg and with genetic variants of 3AB. J Virol. 1998;72:6732–6741. doi: 10.1128/jvi.72.8.6732-6741.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]