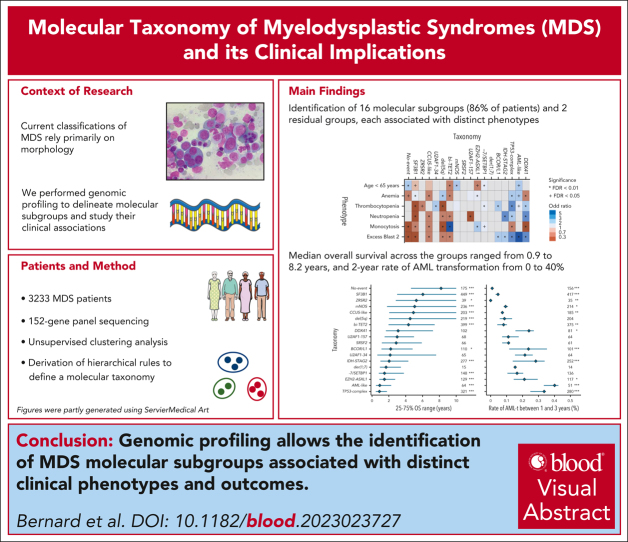
Key Points
-
•
Genomic profiling identifies molecular subgroups of MDS associated with distinct clinical phenotypes and disease courses.
-
•
The molecular taxonomy is a basis for a mechanistic MDS classification that might benefit clinical decision-making and therapeutic research.
Visual Abstract
Abstract
Myelodysplastic syndromes (MDS) are clonal hematologic disorders characterized by morphologic abnormalities of myeloid cells and peripheral cytopenias. Although genetic abnormalities underlie the pathogenesis of these disorders and their heterogeneity, current classifications of MDS rely predominantly on morphology. We performed genomic profiling of 3233 patients with MDS or related disorders to delineate molecular subtypes and define their clinical implications. Gene mutations, copy-number alterations, and copy-neutral loss of heterozygosity were derived from targeted sequencing of a 152-gene panel, with abnormalities identified in 91%, 43%, and 11% of patients, respectively. We characterized 16 molecular groups, encompassing 86% of patients, using information from 21 genes, 6 cytogenetic events, and loss of heterozygosity at the TP53 and TET2 loci. Two residual groups defined by negative findings (molecularly not otherwise specified, absence of recurrent drivers) comprised 14% of patients. The groups varied in size from 0.5% to 14% of patients and were associated with distinct clinical phenotypes and outcomes. The median bone marrow (BM) blast percentage across groups ranged from 1.5% to 10%, and the median overall survival ranged from 0.9 to 8.2 years. We validated 5 well-characterized entities, added further evidence to support 3 previously reported subsets, and described 8 novel groups. The prognostic influence of BM blasts depended on the genetic subtypes. Within genetic subgroups, therapy-related MDS and myelodysplastic/myeloproliferative neoplasms had comparable clinical and outcome profiles to primary MDS. In conclusion, genetically-derived subgroups of MDS are clinically relevant and might inform future classification schemas and translational therapeutic research.
Myelodysplastic syndromes (MDS) are clonal stem cell disorders characterized by abnormal myeloid cell differentiation and cytopenias. The vast majority of MDS have recognizable genetic changes, but current classifications are primarily defined morphologically. Bernard and a group of international contributors performed targeted sequencing of a 152-gene panel on 3233 patients with MDS to assess gene mutations, copy-number changes, and copy-neutral loss of heterozygosity. The authors characterized 16 molecular groups representing 86% of the patients, while 14% of patients lacked identifiable aberrations in driver genes. They confirm previously identified entities but also unveil novel subgroups in work that will likely influence future classification systems.
Introduction
Myelodysplastic syndromes (MDS) comprise a diverse group of myeloid neoplasms characterized by morphologic dysplasia, persistent cytopenias, and a variable risk of evolution to acute myeloid leukemia (AML).1 More than 50 genes are recurrently mutated in MDS implicating the spliceosome complex, epigenetic or transcriptional modifiers, and signaling pathways.2,3 Genomic profiling efforts have demonstrated preferred comutation patterns and robust associations between gene mutations and clinical presentation.2, 3, 4, 5, 6, 7
With current classification schemas relying predominantly on bone marrow (BM) morphology, clinical phenotypes within MDS diagnostic categories exhibit significant heterogeneity.8, 9, 10 This highlights the need for defining genetically-informed disease subtypes to better understand MDS pathogenesis and the variability in responses to treatment and outcomes observed in patients. However, the complexity of the MDS genetic landscape has challenged the delineation of genetic subtypes.6 Formalizing the definition of genetic subtypes would provide a valuable tool for future studies focused on elucidating disease mechanisms and therapeutic targeting.
In this work, we studied how genetic alterations delineate specific disease subtypes using 3233 representative samples from patients with MDS from the cohort built to develop the International Prognostic Scoring System Molecular (IPSS-M).7 We analyzed comutation patterns and genotype-phenotype associations for gene mutations, copy-number alterations (CNAs), and copy-neutral loss of heterozygosity (cnLOH) events. Patients were clustered based on their composite molecular profiles to develop a molecular taxonomy comprising 18 different groups (16 molecular groups and 2 residual groups). The clinical implications of the proposed MDS molecular taxonomy were evaluated.
Methods
Study cohort
The cohort consisted of 3233 treatment-naive diagnostic BM or blood samples from adult patients (median age, 72 years) with MDS or related neoplasms with <30% blasts per the World Health Organization (WHO) 2016 classification (excluding AML with recurrent genetic abnormalities; ie, AML-defining cytogenetic lesions, NPM1, or CEBPA mutations; supplemental Tables 1-3,8 available on the Blood website). To enable the study of molecular subtypes across the borders of diagnostic entities, we included 536 patients with myelodysplastic/myeloproliferative neoplasms (MDS/MPN; 371 and 165 with white blood cell [WBC] count below or above 13 × 109/L, respectively), 267 patients with secondary/therapy-related MDS (s/t-MDS), and 85 patients with AML (61 AML with myelodysplasia-related changes, and 24 AML not otherwise specified). Clinical annotation, cytogenetic, and outcome data were recorded. WHO 2016, WHO 2022, and International Consensus Classification (ICC) classifications were assigned (supplemental Table 4).8, 9, 10 Diagnostic subtypes defined by genetic biomarkers (MDS-5q, MDS-SF3B1, or MDS-biTP53) represented 23% and 22% of cases per WHO 2022 and ICC, respectively.
The study was approved by the institutional review board and Memorial Sloan Kettering Cancer Center after full consideration of the scope and methods of the study.
Genomic analysis
Sequencing was performed with a targeted 152-gene panel enriched with genome-wide CNA probes. Oncogenic mutations with a minimum variant allele frequency (VAF) of 2% were considered. Arm-level CNAs and cnLOH events were called using CNACS,11 complementing cytogenetic. The putative cancer cell fraction per sample was inferred as twice the maximum VAF adjusted for ploidy. Whole-genome sequencing was performed on 30 samples. For further details see the supplemental Methods.
Statistical analysis
Molecular features were constructed from the presence or absence of genetic alterations (gene mutations, CNAs, and cnLOH events). Comutation analysis or prior knowledge further informed the derivation of specific features encoding distinct allelic states or hot spot mutations.12,13 The molecular features were used as input for unsupervised clustering analysis using Bayesian Dirichlet processes to identify genetic groups with high within-group similarity and between-group discrimination.13 The results were then manually curated into a rule-based hierarchical classification tree. The relative chronological order of gene mutations was inferred through Bradley-Terry analysis.2 The IPSS-M score and risk categories were calculated using the open-access R package.7,14
Results
Integrative comutation patterns and implications for genotype-phenotype analysis
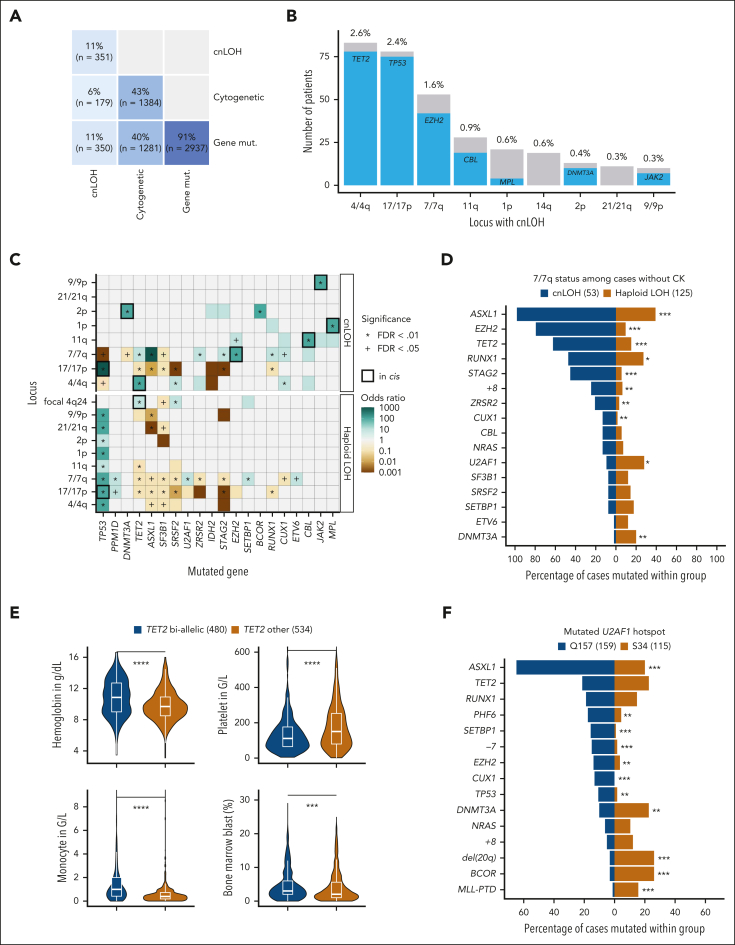
We characterized 10 564 oncogenic mutations across 126 genes in 91% of patients, 3558 chromosomal alterations in 43% of patients, and 375 cnLOH events in 11% of patients (Figure 1A). Gene mutation and CNA comutation patterns were consistent with prior studies (supplemental Figure 1).2, 3, 4,15, 16, 17
Figure 1.
Integrative comutation patterns in MDS and implications for genotype-phenotype analysis. (A) Proportion and number of patients with or without gene mutations (mut.), cytogenetic alterations, and cnLOH events. (B) Number of patients with cnLOH at the most recurrent loci and co-occurrence of gene mutations in cis. The most recurrent cnLOH loci were 4/4q (2.6%), 17/17p (2.3%), and 7/7q (1.6%). Mutations in TET2, TP53, and EZH2 occurred in 94% (78/83), 96% (75/78), and 79% (42/53) of cases with cnLOH at the 4/4q, 17/17p, and 7/7q loci, respectively. (C) Heat map representing mutual exclusivity (brown) or co-occurrence (green) between gene mutations and loci with haploid LOH or cnLOH. Apart from the strong cnLOH–gene mutation interaction in cis (black thick line), focal deletions at the TET2 locus (4q24) also co-occurred with TET2 mutations (OR, 2.4; P = .0007). P values are from Fisher exact test with Benjamini-Hochberg (BH) multiple testing correction. (D) Comparison of comutation frequencies between cases with cnLOH (blue) or haploid LOH (gold) at 7/7q in the absence of CK. For example, mutations in EZH2 were enriched in the 53 cases with cnLOH at 7/7q (79%) compared with the 125 cases with isolated haploid LOH at 7/7q (10%). Conversely, haploid LOH at 7/7q was significantly enriched for U2AF1 and DNMT3A mutations (28% and 20%) compared with cnLOH at 7/7q (9% and 2%). P values are from Fisher exact test with BH multiple testing correction. ∗∗∗P < .001; ∗∗P < .01; ∗P < .05. (E) Comparison of the distributions of blood counts and of the percentage of BM blasts between cases classified as TET2 biallelic (blue) or TET2 other (ie, likely monoallelic, gold). P values are from the Wilcoxon rank-sum test. ∗∗∗∗P < .0001; ∗∗∗P < .001. (F) Comparison of comutation frequencies between cases with U2AF1 Q157 or S34 hot spot mutations. For example, ASXL1 mutations were present in 65% and 20% of patients with U2AF1 Q157 and S34 mutations, respectively (OR, 7.3; P < .0001). Mutations in CUX1, EZH2, PHF6, SETBP1, TP53, and monosomy 7 were also significantly more frequent in the Q157 subset. Conversely, BCOR mutations and del(20q) were more common in patients with S34 (26%) compared with Q157 (3%) mutations (P < .0001). P values are from Fisher exact test with BH multiple testing correction. ∗∗∗P < .001; ∗∗P < .01. FDR, false discovery rate; PTD, partial tandem duplication.
Interactions in cis between gene mutation and cnLOH
Of the patients with cytogenetic or cnLOH alterations, 93% (1281/1384) and 99% (350/351) had co-occurring gene mutations, respectively (Figure 1A). The interactions between cnLOH events and gene mutations in cis were highly specific, most frequently 4q-TET2, 17p-TP53, and 7q-EZH2 (odds ratio [OR], 37, 247, and 68, respectively; P < .0001), but also 11q-CBL, 2p-DNMT3A, 9p-JAK2, and 1p-MPL (Figures 1B-C). Large chromosomal losses (haploid LOH) but not cnLOH events were systematically linked to TP53 mutations (Figure 1C). Haploid LOH and cnLOH events had distinct genome-wide comutation patterns (Figure 1D). As observed in clonal hematopoiesis,18,19 cnLOH events likely contribute to biallelic inactivation and/or allele dosage adjustment of specific mutated genes in MDS.20
TET2 allelic states are associated with distinct comutation patterns and clinical correlates
Consistent with published methodology,12,21 we integrated the number of TET2 mutations, their cumulative VAFs, and 4q24 LOH status to stratify 1014 patients with mutated TET2 into 2 groups: (1) “TET2 biallelic” (47%), and (2) “TET2 other” (53%) representing likely monoallelic mutation (supplemental Figure 2). SRSF2 mutations were more frequent in the TET2 biallelic group, whereas SF3B1 mutations prevailed in TET2 other (OR, 2.7 and 0.45, respectively; P < .0001; supplemental Figure 3). Chronic myelomonocytic leukemia (CMML) was enriched in the biallelic group (38% vs 9%; OR, 6.4; P < .0001). Blood counts and BM blast percentages differed between the 2 TET2 subsets (Figure 1E), but not overall survival (OS; supplemental Figure 3).
Distinct comutation patterns between U2AF1 hot spot mutations
We systematically assessed comutation patterns across hot spot mutations within genes. Among 278 patients with mutated U2AF1, 58% had a Q157 mutation, 41% had a S34 mutation, and 1% had both. Patients with Q157 or S34 mutations had distinct comutation patterns (Figure 1F), in addition to previously reported different RNA splicing events.22,23 However, their clinical profiles were similar (supplemental Figure 4). For SF3B1 hot spot mutations, the only significant difference was an overrepresentation of STAG2 mutations in patients with K666 mutation, with no other clear clinical associations (supplemental Figure 4).24,25
Together, these results emphasize the importance of incorporating cnLOH events, allelic states, and hot spot mutations to study genetic subtypes. Such discrimination lacked in recent prior MDS molecular classification studies.5,26
Derivation of MDS molecular subgroups and their clinical relevance
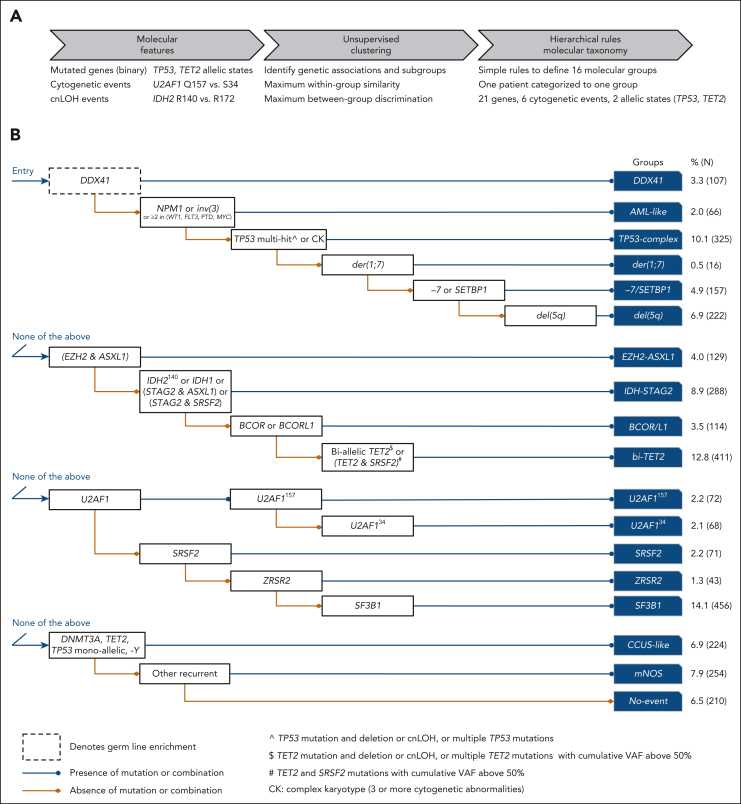
We characterized 18 molecular subgroups using unequivocal and hierarchical rules (Figure 2; supplemental Figure 5). The first 16 groups (86%, n = 2769) were based on the presence of mutations in 21 genes (ASXL1, BCOR, BCORL1, DDX41, DNMT3A, EZH2, FLT3, IDH1, IDH2, MLL [ie, KMT2A], MYC, NPM1, SETBP1, SF3B1, SRSF2, STAG2, TET2, TP53, U2AF1, WT1, and ZRSR2), 6 cytogenetic events (inv(3), complex karyotype [CK], der(1;7), −7, del(5q), −Y), and LOH at the TP53 and TET2 loci. The molecularly not otherwise specified (mNOS) group (8%, n = 254) corresponded to the presence of other cytogenetic abnormalities and/or mutations in 51 other recurrently mutated genes (>0.5%). The No-event category (6%, n = 210) was characterized by the absence of any recurrent drivers evaluated in this study.
Figure 2.
Derivation of 16 MDS molecular groups. (A) Schematic of the analytical workflow to identify molecular groups. Features were based on the presence or absence of genetic alterations (gene mutations, cytogenetic, and cnLOH events). Distinct allelic states or hot spots were also included based on prior knowledge (TP53 allelic state, IDH2 hot spots) or univariate comutation analysis (TET2 allelic state, U2AF1 hot spots). (B) Rules of co-occurrence and mutual exclusivity of genetic alterations organized in a hierarchical classification tree.
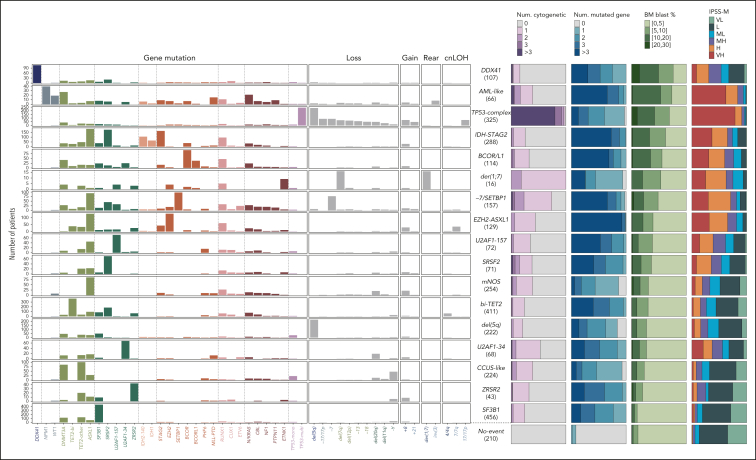
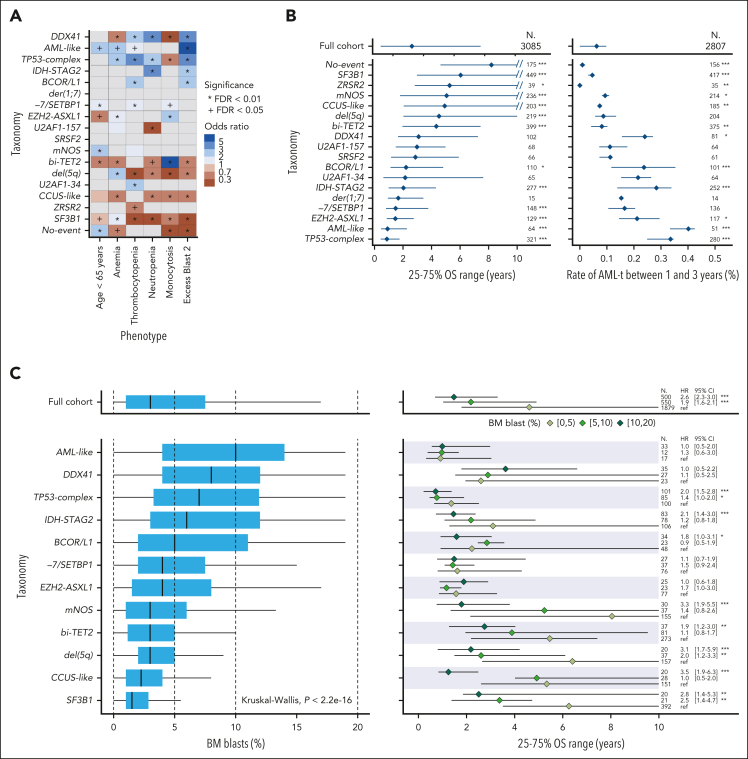
The groups varied in size (0.5%-14%) and in molecular complexity (median, 0-5 mutated genes per patient; Figure 3; supplemental Figures 6-7; supplemental Table 5). They were associated with distinct demographics, clinical presentations, and patient outcomes (Figure 3; Figure 4A-B; supplemental Figures 8-11). Median age of diagnosis across groups ranged from 64 to 75 years, median BM blast percentage ranged from 1.5% to 10%, median OS ranged from 0.9 to 8.2 years, and 2-year rate of leukemic transformation ranged from 0% to 40%. The IPSS-M captured genetic heterogeneity in relation to outcomes within molecular groups (Figure 3) and retained superior predictive performances compared with a model based on molecular groups, consistent with prior studies (supplemental Figure 12).5
Figure 3.
Molecular composition and main characteristics of MDS molecular groups. Each row represents a molecular subgroup, and molecular features are ordered by blocks of gene mutations, chromosomal losses, gains, rearrangements, and cnLOH events. For each subgroup, the relative proportions of (1) the number of mutated genes per patient, (2) the number of cytogenetic alterations per patients, (3) the proportion of BM blasts, and (4) IPSS-M risk category are indicated as stacked bar plots. H, high; L, low; MH, moderate high; ML, moderate low; VH, very high; VL, very low.
Figure 4.
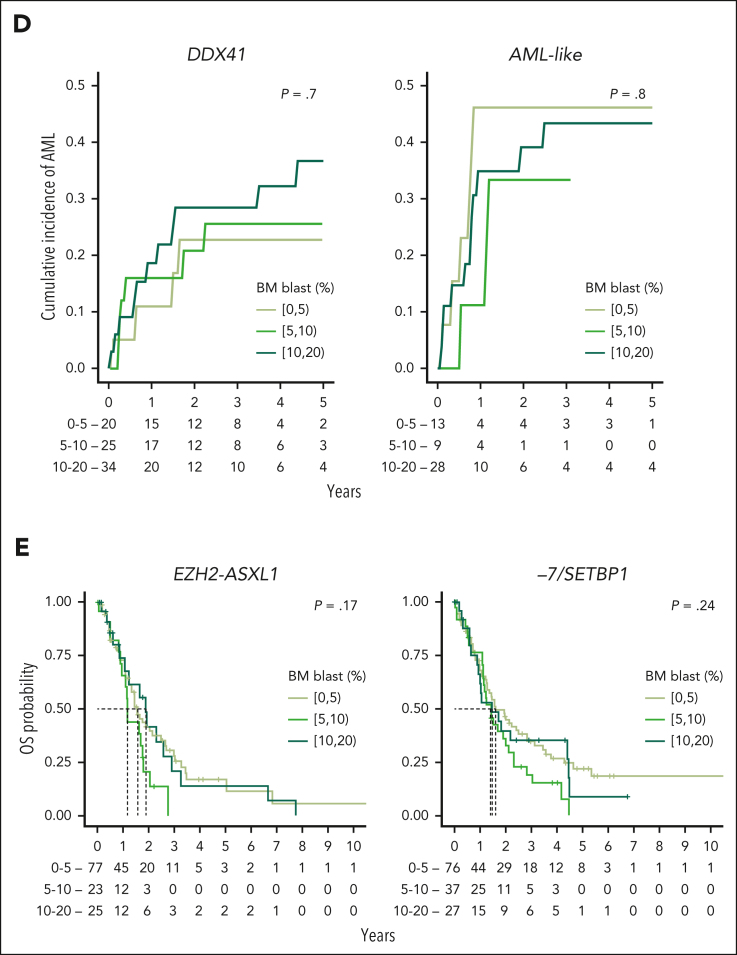
Associations between MDS molecular groups, clinical phenotypes, and outcomes. (A) Association between molecular groups and clinical phenotypes. Darker blue indicates co-occurrence, and darker red indicates mutual exclusivity. (B) Association between molecular groups and outcomes, for OS (left) and AML transformation (AML-t, right). Dots indicate median OS and lines extend to the interquartile range (left). Dots indicate the 2-year incidence of AML-t and lines extend to the 1-year and 3-year incidences (right). The outcome metrics on the full cohort are provided on top for comparison. P values are from the log-rank test (OS) and the Gray test (AML-t). ∗∗∗P < .001; ∗∗P < .01; ∗P < .05. (C) Distribution of the percentage of BM blast for 12 molecular groups with at least 10 patients within each subset of BM blast, that is, 0% to 5%, 5% to 10%, and 10% to 20% (left). Median survival (dots) and interquartile range (lines) for each blast subset within each molecular group (right). P values are from a univariate Cox model with the 0% to 5% blast subset used as the reference level. The distributions of the full cohort are provided on top for comparison. (D) Cumulative incidence curves of AML-t stratified with the range of percentage of BM blast within the DDX41 and AML-like subgroups. P values are from the Gray test. (E) Kaplan-Meier probability estimates of OS stratified with the range of percentage of BM blast within the EZH2-ASXL1 and −7/SETBP1 subgroups. P values are from the log-rank test. CI, confidence interval; HR, hazard ratio; ref, reference level.
From the 16 groups excluding mNOS and No-event groups (accounting together for 14% of patients), 5 subgroups (36%, n = 1176) validated established (TP53-complex, del(5q), and SF3B1) or well-characterized entities (DDX41 and AML-like; supplemental Figures 13-19), 3 subgroups (20%, n = 651) supported previously reported subsets (bi-TET2, der(1;7), and CCUS-like; supplemental Figures 20-22), and 8 subgroups were novel (30%, n = 942; supplemental Figures 23-28).
Established or well-characterized entities: DDX41, AML-like, TP53-complex, del(5q), and SF3B1
The DDX41 group (3.3%, n = 107; supplemental Figure 13) aligned with the results of Makishima et al.27 In our study, 56% of patients with mutated DDX41 had both a putative germ line DDX41 variant (defined here as >30% VAF) and a somatic DDX41 mutation, 37% only a putative germ line DDX41 variant, and 7% only somatic DDX41 mutations (≥1; supplemental Table 6). DDX41 mutations were the sole alteration in 23% of cases, more frequently biallelic than monoallelic (16 and 9 cases, respectively). CUX1 was the only gene significantly comutated with DDX41 (OR, 3.4; P < .001). TET2 mutations were mutually exclusive with biallelic but not monoallelic DDX41 mutations (P = .0003). The DDX41 group had a unique clinical profile including elevated blasts and increased risk of leukemic transformation yet no excess risk of death.27
The AML-like group (2%, n = 66; supplemental Figure 14) was defined by NPM1 mutations (n = 39), inv(3)/t(3;3) (n = 8), or at least 2 events from WT1, FLT3, MLLPTD, or MYC mutations (n = 22). Of the 8 cases with inv(3)/t(3;3), 5 had a SF3B1 mutation (VAF, 5%-36%). The AML-like group showed the highest percentage of BM blast (median, 10% vs 3%; P < .0001; supplemental Figure 9). It was also associated with a younger age (median, 65 vs 72 years; P < .0001), a female predominance (53% vs 39%; OR, 1.7; P = .03), short OS (median, 0.9 years [interquartile range [IQR]; 0.5-2.3]), and had the highest rate of leukemic transformation (40% vs 14% for other MDS at 2 years; Figure 4B). Those associations were preserved when separating patients with established AML-defining NPM1 mutations or inv(3) vs other AML-like mutations (supplemental Figure 14).
Multihit TP53 alterations have been incorporated in the WHO 2022/ICC MDS-biTP53 entity.9,10 In the TP53-complex group (10%, n = 325; supplemental Figure 15), multihit TP53 mutations were present in 241 (74%) cases, of which 219 (91%) had CK. Only 13 cases (4%) had monoallelic TP53 mutations and CK. Among the 303 (93%) cases with CK, 71 (23%) had no detected TP53 mutation. This allowed us to compare CK cases with (n = 232) or without (n = 71) TP53 mutation (supplemental Figure 18). As expected,28 CK cases with TP53 mutations (CK:TP53) had more cytogenetic abnormalities compared with those with CK only (73% vs 20% with ≥6 events; OR, 10.8; P < .0001); deletion of 5q occurred in 87% and 32%, respectively. Only trisomy 8 and deletion of 11q were more frequent in the CK-only group. The CK-only subset had more mutated genes than the CK:TP53 group (46% vs 24% with ≥3 mutated genes; OR, 2.8; P = .0005), with enrichment for RUNX1, SRSF2, and SF3B1 mutations. OS was poor in both CK subsets, albeit shorter in the presence of TP53 mutation (median, 0.7 vs 1.5 years; P < .0001; supplemental Figure 18). Therefore, although our categorization did not formally distinguish between CK with or without TP53 mutation, differences in molecular profiles and outcomes support a subcategorization, as suggested by Huber et al.26
The del(5q) group (6.9%, n = 222; supplemental Figure 16) was defined by the presence of del(5q) as the sole cytogenetic abnormality or with 1 additional abnormality excluding −7/7q.9,10,29 In this group, 84% of patients had at least 1 gene mutation, most commonly SF3B1 (22%), DTA (DNMT3A, TET2, ASXL1: 21%, 17%, 14%), and monoallelic TP53 (13%) mutations. Monoallelic TP53 mutations were significantly enriched in this group (OR, 4.8; P < .0001), along with mutations in CSNK1A1 (10%; OR, 16.4; P < .0001), IRF1 (5.4%; OR, 19.0; P < .0001), and RAD50 (4.1%; OR, 14.1; P < .0001), all located on 5q; and NFE2 (3.6%; OR, 5.1; P = .0006). Gene mutations occurred secondary to del(5q) (supplemental Figure 19).30 Del(5q) was associated with a female bias (75% vs 37%; P < .0001), low hemoglobin values, high platelet counts, and favorable OS (Figure 4B). Notably, 22% (49/222) of cases from our del(5q) group would be excluded from the WHO 2022/ICC MDS with isolated del(5q) category because of excess blasts. Only 1% of cases were CMML per WHO 2016 compared with 8% per the new WHO 2022 definition of CMML.
The SF3B1 group (14%, n = 456; supplemental Figure 17) was the most prevalent. Comutations were relatively infrequent, with 23% of cases having isolated SF3B1 mutations, and 27% of cases having only SF3B1 and DTA mutations.31,32 SF3B1 mutations were frequently secondary to DNMT3A mutations and dominant to TET2 mutations. The group had low BM blasts (median, 1.5% [IQR, 1-3]) and favorable outcomes (median OS, 6 years [IQR, 3-12]). Per WHO 2016, 80% of cases had low-blast MDS, 8% had excess blasts, and 12% had MDS/MPN. Per WHO 2022 and ICC, 60% and 54% of cases were MDS-SF3B1, respectively (25 discordant cases classified as MDS-NOS per ICC, with SF3B1 VAF of <10% or RUNX1 mutations). Of 355 cases with low-blast MDS, 85 (24%) met monocyte criteria for CMML per WHO 2022.
Previously reported subsets: bi-TET2, der(1;7), and CCUS-like
The second largest group was defined by biallelic TET2 mutations (13%, n = 411; supplemental Figure 20), which has been recognized in few studies despite its prevalence.21,33 Patients in this group had distinct clinicohematological features, including older age, milder anemia, increased monocytes, and a CMML enrichment (42%; Figure 4A).
The bi-TET2 group was defined by early biallelic TET2 mutations with splicing factor mutations in 80% of patients, most commonly affecting SRSF2, SF3B1, or ZRSR2 (185/411, 86/411, and 65/411, respectively; supplemental Figures 20-21). The association with elevated monocytes within bi-TET2 was preserved across all splicing factor mutation subsets. However, comutations further modulated the phenotypes: in multivariable analysis, SF3B1 and JAK2 mutations were both predictors of higher platelet count, whereas SF3B1 mutations were predictive of lower hemoglobin level. RUNX1 mutations were predictive of higher blasts, and ASXL1 and RAS pathway mutations of higher WBC and monocyte counts (supplemental Figure 21). Although secondary events influenced the overall disease phenotype, all molecular trajectories converged on a bias toward monocytic differentiation.
The der(1;7) group (0.5%, n = 16) was a rare subset characterized by der(1;7)(q10;p10), previously reported to be enriched in Japanese populations.34,35 There was a male predominance (14/16) and a relatively young age (median, 64 years [IQR, 55-70]). ETNK1 mutations were enriched in this group (56%; OR, 61; P < .0001) as secondary events (median VAF, 25%).
In the CCUS-like group (6.9%, n = 224; supplemental Figure 22), 104 (46%) patients had a single mutated gene (45 TET2, and 37 DNMT3A), 17 (8%) had loss of Y without gene mutations, and 14 (6%) only had ≥2 DTA mutations. CCUS-like had low cancer cell fractions compared with other groups (median, 42% vs 84%; P < .0001), reflecting smaller clonal cytopenia of unknown significance (CCUS) clone sizes.36 The group was associated with lower risk (68% IPSS-M very low/low) and favorable outcomes (median OS, 4.9 years [IQR, 2 to not reached]).
Novel molecular groups: −7/SETBP1, EZH2-ASXL1, IDH-STAG2, BCOR/L1, U2AF1157, U2AF134, SRSF2, and ZRSR2
The −7/SETBP1 group (4.9%, n = 157; supplemental Figure 23) was defined by SETBP1 mutations and/or −7 in the absence of CK (OR, 9; P < .0001).37,38 Within this group, 11% of patients had both SETBP1 mutation and −7, 51% only SETBP1 mutation, and 38% only −7. SETBP1 mutations were secondary events (median VAF, 24%). Clonally dominant events included ASXL1, SRSF2 (enriched in the SETBP1-only subset), or U2AF1 Q157 mutations. GATA2 mutations were also overrepresented (OR, 6.8; P < .0001), with VAFs suggestive of germ line variants in 6 (4%) patients (median age, 59 years; range, 18-80).39 The −7/SETBP1 group was associated with a younger age (median, 67 vs 72 years; P < .0001), particularly for the subset with both SETBP1 mutation and −7 (median, 61 years [IQR, 56-69]). The sequencing panel excluded SAMD9/L genes linked to germ line driven −7 reversion in MDS.40 Further research is needed to investigate SAMD9/L or other germ line variants in the −7/SETBP1 group. Last, −7/SETBP1 was characterized by higher risk (71% IPSS-M very high/high) and poor outcomes (median OS, 1.5 years [IQR, 0.8-3.8]).
The EZH2-ASXL1 group (4%, n = 129; supplemental Figure 24) was defined by ASXL1 and EZH2 mutation co-occurrence. It was characterized by a high molecular complexity (75% of patients with ≥5 mutated genes). Half of cases had splicing factor mutations in ZRSR2 (20% [26/129]), U2AF1 Q157 (12%), SRSF2 (11%), and SF3B1 (8%). EZH2 mutations were frequently subclonal to TET2, ASXL1, and splicing factor mutations. This group had the highest proportion (47%) of secondary RUNX1 mutations. Although 60% of cases had BM blasts of <5%, this group was associated with poor OS (median, 1.5 years [IQR, 0.9-2.8]). Splicing factor mutations in this context were not associated with differing survival or comutation patterns (supplemental Figure 25).
The IDH-STAG2 group (8.9%, n = 288; supplemental Figure 26) was defined by mutations at the IDH2 R140 hot spot, IDH1, and/or STAG2 co-occurring with either SRSF2 or ASXL1 mutations. Mutations in IDH2, SRSF2, and ASXL1 were generally acquired early whereas STAG2 mutations were late. The composite genotypes of this group mirrored secondary AML,41,42 and the myelodysplasia-related AML (WHO2022) or AML with MDS-related gene mutations (ICC) subtypes.43,44 Accordingly, this group had elevated BM blasts (median, 6% [IQR, 3-12]; supplemental Figure 9). Median BM blasts of cases with both IDH1/2 and STAG2 mutations, STAG2 only, and IDH1/2 only were 12% (IQR, 7-15), 8% (IQR, 4-13), and 4% (IQR, 2-8), respectively. This group had the third highest 2-year incidence rate of AML transformation (28%) after AML-like and TP53-complex (Figure 4B).
In the BCOR/L1 group (3.5%, n = 114; supplemental Figure 27), 83% of patients had mutations in BCOR, 33% in BCORL1, and 17% in both genes. BCOR/L1 mutations were secondary to DTA and splicing factor mutations. The latter occurred in 61% of patients: U2AF1 S34 mutations were most frequent (22%; OR, 9.4; P < .0001), followed by SF3B1 (21%) and SRSF2 (11%) mutations. Subclonal RUNX1 mutations were frequent (41%). BCOR/L1 was characterized by pronounced thrombocytopenia, elevated BM blasts, short OS (median, 2.2 years [IQR, 1.1-4.8]), and a 24% 2-year incidence of AML transformation. These phenotypic associations were irrespective of splicing factor mutation status (supplemental Figure 28).
The last 4 groups, U2AF1157 (2.2%, n = 72), U2AF134 (2.1%, n = 68), SRSF2 (2.2%, n = 71), and ZRSR2 (1.3%, n = 43), were defined based on the exclusion of the aforementioned group-defining events and the residual presence of distinct splicing factor mutations other than SF3B1. Patients from these groups had fewer mutations, with 38% (96/254) having only 1 or 2 mutated genes (Figure 3). The ZRSR2 group, specific to males, correlated with indolent clinical phenotypes and favorable OS (median, 5.3 years [IQR, 3.5 to not reached]; Figure 4A-B).
Residual groups: mNOS and No-event
The last 2 groups were defined by negative findings: mNOS (absence of group-defining patterns) and No-event (absence of recurrent mutations or cytogenetic). Both groups were associated with mild phenotypes and favorable outcomes (Figure 4A-B). Compared with cases with oncogenic events, patients without identified drivers were younger (median age, 65 vs 72 years; P < .001), with a female bias (61% vs 38%; OR, 2.5; P < .001), frequent classification as MDS with single-/multilineage dysplasia (73% vs 27%; OR, 7.1; P < .001), lower risk IPSS-M (75% vs 36% as very low/low; OR, 5.5; P < .001), and favorable OS (median, 7.6 vs 3.0 years; P < .001; supplemental Figure 29). A concomitant study demonstrated that the male subset of the No-event group was enriched for UBA1 mutations (7%) and VEXAS-like clinical presentation.45
Nonetheless, 11% (21/193) of cases without identified drivers had excess blasts (median BM blasts, 7% [range, 5%-15%]). We therefore performed whole-genome sequencing from 30 patients with excess blasts that did not have an identified driver (n = 20) or only had −Y or DTA mutation (n = 10; −Y in 1, DNMT3A in 4, TET2 in 4, and ASXL1 in 1). We identified additional putative oncogenic events in 27% (8/30) of cases. This included 1 NUP98-MLL fusion, chromosome 20 and 22 abnormalities, and 1 UBA1 M41T mutation in a male patient (5% BM blasts), and possibly germ line frameshift mutations in ATR, FANCC, INPPL1, and RTEL1 (supplemental Table 7).
The prognostic influence of BM blasts within genetic subtypes
Blast count thresholds (<5% for low blast, 5%-9% for excess blasts 1, and 10%-19% for excess blasts 2) are defining features in MDS classifications.8, 9, 10 We evaluated how genetic subtypes relate to BM blast strata. In our data set, 61% (1974) of patients had <5% blasts, 18% (566) had 5% to 9%, and 16% (507) had 10% to 19%.
Seven to 9 different molecular groups accounted for >5% of patients within each blast stratum (supplemental Figure 30). This molecular heterogeneity translated into variability in outcomes, whereby the OS of patients within each blast stratum stratified with the molecular groups. However, contrary to prior findings,26 both molecular groups and blast percentages were independently predictive of OS (supplemental Figure 30). We reasoned that the impact of BM blast counts on outcomes might depend on the specific molecular subtype, and we therefore investigated the effect of blast counts on the OS of patients within each molecular group.
We focused on the 12 groups with at least 10 patients with outcome data per blast stratum (Figure 4C; supplemental Figure 31). In 8 groups (SF3B1, CCUS-like, del(5q), bi-TET2, mNOS, BCOR/L1, IDH-STAG2, and TP53-complex), blast count differentiated OS. Of note, blast stratification was significant in the TP53-complex group but outcomes were all very poor (median OS: 1.4, 0.8, and 0.7 years across the 3 strata; supplemental Figure 32). For 4 groups (EZH2-ASXL1, −7/SETBP1, DDX41, and AML-like), OS was not stratified by blast count. DDX41 and AML-like had the highest blasts of all groups: 49% (51/104) and 53% (34/64) of patients had ≥10% blasts, respectively. Yet, within the DDX41 and AML-like groups, both the OS and the rate of leukemic transformation were comparable across the 3 blast strata (Figure 4D; supplemental Figure 31). This effect in AML-like was independent of NPM1 mutations (supplemental Figure 32). Conversely, the EZH2-ASXL1 and −7/SETBP1 groups were enriched for low blasts, with 61% (77/126) and 52% (79/152) of patients having <5% blasts, respectively. However, the OS of patients in those 2 groups was poor independently of blasts (median OS, <2 years in each stratum; Figure 4E). This demonstrates that specific genetic subtype outweighs blast count as a predictor of outcomes, which should be considered in a future MDS classification.
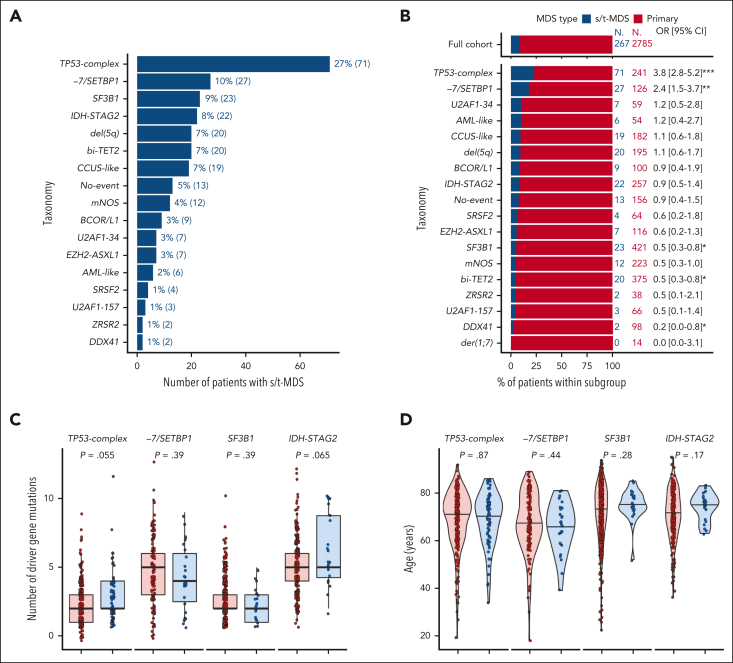
Genetic heterogeneity of s/t-MDS
We used the molecular groups to investigate genetic heterogeneity in s/t-MDS. We first evaluated the repartition of 267 patients with s/t-MDS across molecular groups (Figures 5A-B). As expected, the TP53-complex group was the most frequent within s/t-MDS (27%) and enriched in this subset compared with primary MDS (OR, 3.8; P < .001).46, 47, 48 The remaining 73% of patients with s/t-MDS spread across 16 other groups, predominantly −7/SETBP1 (10%, 27/267), SF3B1 (9%), and IDH-STAG2 (8%). The −7/SETBP1 group was enriched in s/t-MDS (OR, 2.4; P < .01), whereas the SF3B1, bi-TET2, and DDX41 groups were underrepresented (OR: 0.5, 0.5, and 0.2, respectively; P < .05).
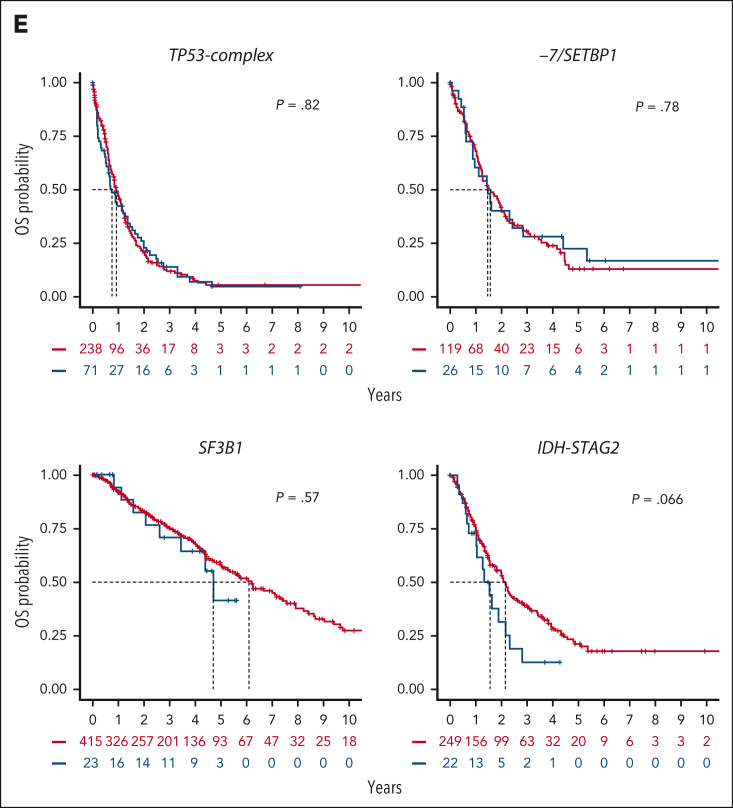
Figure 5.
Genetic heterogeneity of s/t-MDS. (A) Prevalence of molecular groups in 267 patients with s/t-MDS. (B) Proportion of primary (red) and s/t-MDS (blue) within each molecular group and comparison with the full cohort (top). (C) Distribution of the number of mutated genes per patient in primary or s/t-MDS within each molecular group (TP53-complex, −7/SETBP1, SF3B1, and IDH-STAG2), showing similar molecular complexity of the 2 disease subsets. For example, the median number of mutated genes per patient was equal to 2 for both primary and s/t-MDS within TP53-complex, whereas it was equal to 5 for both subsets within IDH-STAG2. (D) Age distribution in primary or s/t-MDS within each molecular group. (E) Kaplan-Meier probability estimates of OS for primary or s/t-MDS within each molecular group. P values are from the log-rank test.
When selecting groups with >20 patients with s/t-MDS (TP53-complex, −7/SETBP1, SF3B1, and IDH-STAG2), and comparing s/t-MDS with primary MDS, we observed similar molecular (Figure 5C; supplemental Figure 33), clinical (Figure 5D; supplemental Figure 34), and outcome (Figure 5E) profiles. The younger age characterizing −7/SETBP1 was conserved in s/t-MDS (median age, 67 years; range, 39-81). We, and others, have shown that the IPSS-M efficiently risk stratifies patients with s/t-MDS.3,49 Here, we demonstrated that the heterogeneity in outcomes within s/t-MDS can be partially explained by the underlying molecular subtypes. Indeed, the median OS of patients with s/t-MDS across TP53-complex, −7/SETBP1, IDH-STAG2, and SF3B1 groups were 0.8, 1.5, 1.6, and 4.7 years, respectively, which was similar to the OS of the respective primary MDS subsets (Figure 5E).
Genetic heterogeneity of MDS/MPN with ubiquitous RAS pathway mutations enrichment
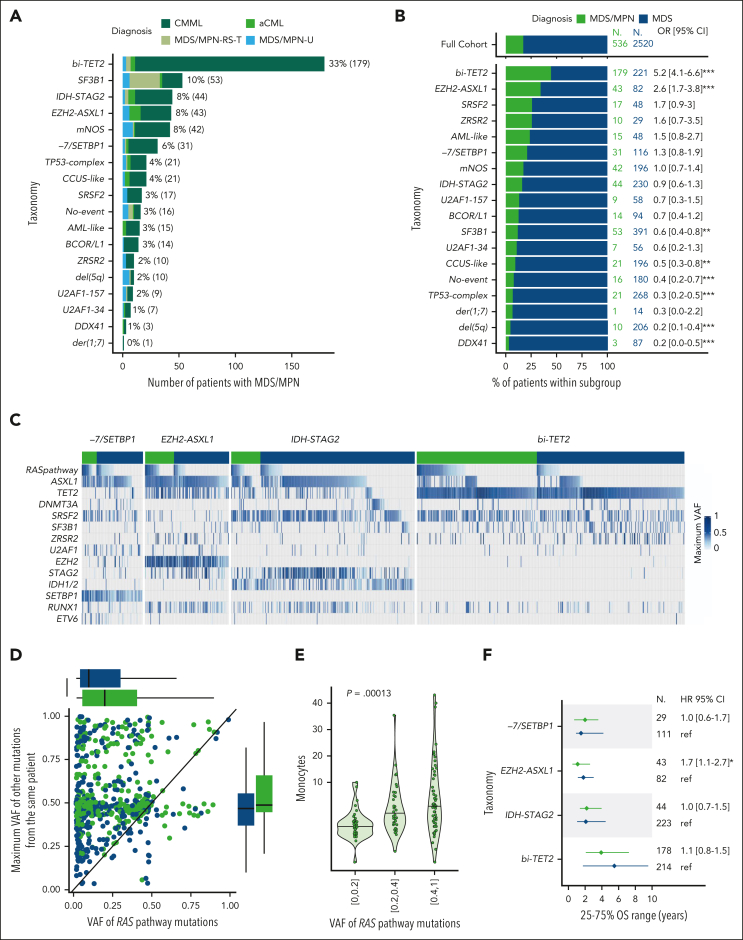
The distribution of molecular subgroups in 536 patients with MDS/MPN (WHO 2016) was heterogeneous and consistent with prior studies (Figure 6A).50 Fifteen groups were observed in >1% of patients, and 6 groups (bi-TET2, SF3B1, IDH-STAG2, EZH2-ASXL1, mNOS, and −7/SETBP1) in >5% of patients. Among those, bi-TET2 and EZH2-ASXL1 were enriched in MDS/MPN compared with MDS (OR, 5.2 and 2.6, respectively; P < .0001; Figure 6B). Beyond the genes defining the enriched groups (TET2, EZH2, ASXL1, and SRSF2), RAS signaling pathway mutations (OR, 4.8; P < .0001) and, to a lesser extent, RUNX1 mutations (OR, 1.4; P < .01) were enriched in MDS/MPN. RAS pathway mutations were consistently overrepresented in MDS/MPN across the −7/SETBP1, EZH2-ASXL1, IDH-STAG2, and bi-TET2 groups (Figure 6C; supplemental Figure 35). Moreover, the VAF of RAS pathway mutations was higher in MDS/MPN (median, 20% [IQR, 6-42]) than in MDS (median, 10% [IQR, 4-31]; P < .0001) across molecular subgroups (Figure 6C-D; supplemental Figure 35).
Figure 6.
Genetic heterogeneity of MDS/MPN with ubiquitous RAS pathway mutations enrichment. (A) Prevalence of molecular groups in 536 patients with MDS/MPN color-coded by specific subtypes. The bi-TET2 group accounted for 33% of MDS/MPN cases and 42% (168/399) of CMML cases. The EZH2-ASXL1 group comprised 8% of MDS/MPN cases and 26% (10/38) of atypical chronic myeloid leukemia (aCML) cases. The majority of MDS/MPN with ring sideroblasts and thrombocytosis (MDS/MPN-RS-T) cases (63%, 27/43) were part of the SF3B1 group. (B) Proportion of MDS/MPN (green) and MDS (blue) within each molecular group and comparison with the full cohort (top). (C) Oncoplots for each molecular group (−7/SETBP1, EZH2-ASXL1, IDH-STAG2, and bi-TET2) separating MDS/MPN (green) and MDS (blue). Each column corresponds to a patient. The presence of a mutation in each gene for each patient is color-coded by the VAF (maximum VAF if several mutations within the same gene). RAS pathway mutations include mutations in N/KRAS, CBL, NF1, and PTPN11. They were observed in 68%, 51%, 45%, and 40% of patients with MDS/MPN within groups −7/SETBP1, EZH2-ASXL1, IDH-STAG2, and bi-TET2, respectively, compared with 33%, 21%, 14%, and 15% of patients with MDS within the same groups. (D) Scatterplot representing 1 dot per patients with RAS pathway mutation, with the VAF of RAS pathway mutations on the x-axis and the maximum VAF of all other mutations in the same patient on the y-axis. (E) Monocyte level in 109/L as a function of the VAF of RAS pathway mutations in the subset of 297 patients with MDS/MPN from the same molecular groups as in panel C. (F) Median survival (dots) and interquartile range (lines) for MDS/MPN (green) and MDS (blue) within each molecular group. Patients from the bi-TET2 group had favorable OS (median, 5.5 and 3.9 years for MDS and MDS/MPN, respectively). Conversely, patients from the IDH-STAG2, EZH2-ASXL1, and −7/SETBP1 groups had dismal OS in both subsets (median, 2.1, 1.8, and 1.5 years, respectively, in MDS and 2.2, 1.1, and 2.0 years, respectively, in MDS/MPN).
We explored molecular predictors of WBC and monocyte levels. In multivariable regressions for leukocytosis and monocytosis, the presence of RAS pathway, SRSF2, biallelic TET2 (but not monoallelic), and ASXL1 mutations were positive predictors for both parameters (supplemental Figure 36). EZH2 mutations predicted for leukocytosis but not monocytosis, consistent with the atypical CML enrichment in the EZH2-ASXL1 group (OR, 8.8; P < .001). The 185 patients with MDS from the bi-TET2 group had higher monocyte counts than the 1675 patients with MDS from other groups (48% vs 24% with ≥0.5 × 109/L; supplemental Figure 37). In addition, the VAF of RAS pathway mutations strongly predicted increased monocyte counts in multivariable analysis (Figure 6E; supplemental Figure 36).51,52 This effect was stronger in MDS/MPN than MDS (supplemental Figures 36-37), suggesting that considerable expansion of RAS pathway mutated clones is necessary to drive elevated monocyte counts at diagnosis.
Last, we compared the clinical phenotypes and outcomes of patients categorized as MDS or MDS/MPN within molecular groups. Despite expected differences in WBC and monocyte counts, age, blast, hemoglobin, and platelet distributions were comparable within each group (supplemental Figure 38), as well as OS (Figure 6F; supplemental Figure 39). Thus, similar to our observations in s/t-MDS, the genetic subtype plays a dominant role in the overall clinical phenotype (aside from the MDS/MPN-defining features of WBC and monocyte counts) and outcomes irrespective of morphologic classification as MDS or MDS/MPN.
Discussion
Diagnostic classification of hematologic neoplasms rely predominantly on morphological features, which do not adequately account for heterogeneous clinical phenotypes and outcomes. Deletion 5q, SF3B1, and biallelic TP53 mutations are established subtype-defining MDS biomarkers.9,10 However, the majority of genetic lesions are not yet considered. A broader integration of genetic alterations will improve our understanding of disease ontogeny; the design of clinical trials; and, ultimately, the diagnosis and treatment of MDS.
We characterized 18 MDS subtypes (16 genetic groups and 2 residual groups) each associated with specific clinical phenotypes and disease courses. The subtypes were defined by combinations of chromosomal aneuploidies, cnLOH events (TP53 and TET2 loci), gene mutations, and gene–gene interactions. We validated established (TP53-complex, del(5q), and SF3B1) or well-characterized entities (DDX41 and AML-like), substantiated support for emerging subtypes (bi-TET2, der(1;7), and CCUS-like), and unveiled novel groups (−7/SETBP1, EZH2-ASXL1, IDH-STAG2, BCOR/L1, U2AF157, U2AF134, SRSF2, and ZRSR2). Unlike published MDS genetic hierarchies,5 splicing factor mutations were not at the top of the classification tree. For several subgroups, the defining events were secondary to DTA and/or splicing factor mutations (SETBP1, EZH2, STAG2, or BCOR/L1 mutations), reminiscent to NPM1 mutations defining a unique AML subtype while often being secondary to DNMT3A mutations.53 Study limitations included the use of targeted sequencing, which might have omitted pertinent biomarkers (eg, SAMD9/L variants and UBTF tandem duplication), the absence of germ line control tissues, and the lack of correlative analysis with some relevant morphological features, such as cellularity and fibrosis.
We evaluated the relationship between genetic subtypes and blasts. In several groups (AML-like, DDX41, −7/SETBP1, and EZH2-ASXL1), blast percentages did not stratify patient outcomes. However, for groups enriched for lower-risk disease (for example del(5q), bi-TET2, SF3B1, CCUS-like, and mNOS) blasts percentages differentiated outcomes. This suggests a novel paradigm in which the genetic subtypes should be the overarching basis of disease classification, whereby blasts designate disease stage within certain genetic contexts, for example del(5q) or IDH-STAG2 with increased blasts, as Kewan et al also have suggested.6
These molecular subgroups facilitated the evaluation of genetic heterogeneity in s/t-MDS and MDS/MPN. Although enriched in specific subtypes (TP53-complex and −7/SETBP1), s/t-MDS were represented across most genetic groups, including those associated with favorable outcomes (SF3B1). This suggests that although some chemotherapy-resistant molecular subtypes are promoted by prior therapy,54 others may arise from other mechanisms. Patients with s/t-MDS and primary MDS had comparable clinical profiles and outcomes within genetic subtypes. MDS/MPN were also represented across diverse genetic subgroups (SF3B1, IDH-STAG2, and EZH2-ASXL1) but were strongly enriched in the bi-TET2 group, which was associated with elevated monocytes across disease categories. A higher frequency and clonal expansion of RAS pathway mutations in MDS/MPN over MDS was ubiquitous across genetic subtypes. This argues for the recognition of bi-TET2 as a unique entity encompassing both MDS and MDS/MPN, and for the prospective monitoring of RAS pathway mutations as potential biomarkers predictive of monocytic proliferation.
The molecular taxonomy outlined in this study identifies groups of patients with MDS with shared molecular pathogenesis, providing the foundation for a broader genetically informed classification of MDS. Its purpose is different from that of prognostic assessments, such as IPSS-M, whereas both inform clinical decision-making. Prognostic schemas consider clinical and molecular features to estimate patient outcomes, thus guiding risk-adapted therapies. However, there is no equivalence between risk and molecular groups: 2 patients from the TP53-complex or EZH2-ASXL1 molecular groups may both be categorized as high risk although distinct. The definition of molecular groups is key to identifying determinants of disease progression and treatment response. This molecular taxonomy should be a useful tool for future correlative studies and exploratory analyses within clinical trials evaluating the efficacy of therapeutic compounds and for the development of targeted therapies.
Conflict-of-interest disclosure: A.A.v.d.L. reports research funding from Roche, Bristol Myers Squibb (BMS), and Celgene; and reports membership on the board of directors or advisory committees of BMS and Celgene. F.T. reports membership on the board of directors of Novartis and AbbVie. M.G.D.P. reports honoraria from, and membership on the board of directors or advisory committees of BMS. P.F. reports consultancy with, and honoraria from and research funding from Novartis, AbbVie, Janssen, Jazz Pharmaceuticals, and BMS; and reports honoraria from French MDS Group. M.R.S. reports membership on the board of directors or advisory committees of Savona, AbbVie Inc, BMS, Geron Corporation, Forma Therapeutics, CTI BioPharma Corp, Karyopharm Therapeutics Inc, Novartis, Sierra Oncology, Inc, Taiho, Takeda Pharmaceutical, and TG Therapeutics Inc; reports consultancy with Forma Therapeutics Inc, Karyopharm Therapeutics Inc, Ryvu Therapeutics; is a current equity holder in publicly traded companies, Karyopharm Therapeutics Inc and Ryvu Therapeutics; received research funding from Takeda Pharmaceutical, TG Therapeutics Inc, ALX Oncology, Astex Pharmaceuticals, Incyte Corporation; and reports patents with, and royalties from, Boehringer Ingelheim. P.V. reports honoraria from BMS, Novartis, Pfizer, Incyte, Blueprint, and Stemline. I.K. reports consultancy with Novartis, BMS, Genesis, and AbbVie; received research funding from Novartis; and received honoraria from Genesis and AbbVie. V.S. reports membership on the board of directors or advisory committees of Santini, BMS, AbbVie, Geron, Gilead, CTI BioPharma, Otsuka, Servier, Janssen, and Syros. U.P. received research funding from Janssen Biotech, Geron, Fibrogen, Silence Therapeutics, Takeda, Curis, Merck, Servier, Syros, Novartis, Celgene, BMS, Amgen, Roche, Jazz Pharmaceuticals, and BeiGene; serves in a consulting role for Janssen Biotech, Geron, Silence Therapeutics, Takeda, Curis, Servier, Jazz Pharmaceuticals, Syros, AbbVie, Novartis, BMS, and Amgen; received honoraria from Silence Therapeutics, Celgene, Takeda, Servier, Jazz Pharmaceuticals, Syros, Novartis, and BMS; reports membership on the board of directors or advisory committees of MDS Foundation and BMS; and received other support (travel support and medical writing) from BMS. M.H. reports consultancy with Pfizer, PinotBio, AbbVie, Servier, BMS, Glycostem, Jazz Pharmaceuticals, Amgen, and Delbert Lab; received honoraria from Pfizer, Novartis, Sobi, Certara, Janssen, and Jazz Pharmaceuticals; and received research funding from PinotBio, BerGenBio, Astellas, Agios, AbbVie, Loxo Oncology, BMS, Glycostem, Karyopharm, and Jazz Pharmaceuticals. M.T.V. serves on the advisory board of Jazz Pharmaceuticals, Celgene/BMS, and Syros; received research funding from Novartis and Celgene/BMS; and is a member of the speakers bureau of AstraZeneca, Novartis, AbbVie, Jazz Pharmaceuticals, Astellas, and Celgene/BMS. B.L.E. is a current equity holder in private company, Skyhawk Therapeutics, Exo Therapeutics, TenSixteen Bio, and Neomorph Inc; reports membership on the board of directors or advisory committees of Skyhawk Therapeutics, Exo Therapeutics, TenSixteen Bio, and Neomorph Inc; serves in a consulting role for AbbVie; and received research funding from Calico and Novartis. P.L.G. reports consultancy with, and research funding from BMS, Novartis, and Gilead. N.G. received research funding from Takeda; and honoraria from Novartis and BMS. E.P. is cofounder of, and holds a fiduciary role in a private company, Isabl Inc; and holds stock options in a private company, TenSixteen Bio. The remaining authors declare no competing financial interests.
Acknowledgments
This work was supported in part by grants from Celgene Corporation through the MDS Foundation. E.B. was supported by the Edward P. Evans Foundation and the INSERM ATIP-Avenir Program. L.M. was supported by the Associazione Italiana per la Ricerca sul Cancro (AIRC), Milan, Italy (investigator grant 20125; AIRC 5×1000 project 21267); Cancer Research UK, Fundación Científica de la Asociacion Española Contra el Cáncer's (FC AECC) and AIRC-International Accelerator Award Program (project C355/A26819 and 22796). V.S. was supported by AIRC project IG-26537. L.-Y.S. was supported by MOHW111-TDU-B-221-01400. U.G. and N.G. were supported by the Deutsche Krebshilfe grant 70113711. M.G.D.P. was supported by AIRC project 22053 and AIRC 5×1000 project 21267. M.Y.F. was supported by Italian MIUR-PRIN (grant 2017RKWNJT) and the Fondazione Carisbo. E.P. was supported by the Josie Robertson Investigators Program.
Authorship
Contribution: E.B. and E.P. designed the study, analyzed the data, and wrote the paper; E.B. created the figures; E.B., J.E.A.O., J.G.-A., D.D., J.S.M-M., M.L., K.L., and N.F. executed bioinformatics pipelines; R.P.H, P.L.G., H.T., B.L.E, R.B., L.M., M. Cazzola., M. Creignou., S.O., and E.H.-L critically reviewed the data; R.P.H reviewed established classification of all included cases; L.M., F.P.S.S., M. Creignou, U.G., A.A.v.d.L., M.J., M.T., O.K., M.Y.F., F.T., R.F.P., V.S., I.K., J.B., F.P.S.S., V.M.K., M.R.S., M.B., C.G., L.P., L.A., M.G.D.P., P.F., A.P., U.P., M.H., P.V., C.F., M.T.V., L.-Y.S., M.F., J.H.J., J.C., N.G., M. Cazzola, and E.H.-L. provided clinical data and DNA specimens; and all authors provided feedback on the manuscript.
Footnotes
Deidentified patient data including clinical annotations and molecular data have been deposited to cBioPortal under the study name Myelodysplastic Syndrome International Working Group.
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Supplementary Material
References
- 1.Cazzola M. Myelodysplastic syndromes. N Engl J Med. 2020;383(14):1358–1374. doi: 10.1056/NEJMra1904794. [DOI] [PubMed] [Google Scholar]
- 2.Papaemmanuil E, Gerstung M, Malcovati L, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122(22):3616–3699. doi: 10.1182/blood-2013-08-518886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haferlach T, Nagata Y, Grossmann V, et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2014;28(2):241–247. doi: 10.1038/leu.2013.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nazha A, Komrokji R, Meggendorfer M, et al. Personalized prediction model to risk stratify patients with myelodysplastic syndromes. J Clin Oncol. 2021;39(33):3737–3746. doi: 10.1200/JCO.20.02810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bersanelli M, Travaglino E, Meggendorfer M, et al. Classification and personalized prognostic assessment on the basis of clinical and genomic features in myelodysplastic syndromes. J Clin Oncol. 2021;39(11):1223–1233. doi: 10.1200/JCO.20.01659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kewan T, Durmaz A, Bahaj W, et al. Molecular patterns identify distinct subclasses of myeloid neoplasia. Nat Commun. 2023;14(1):3136. doi: 10.1038/s41467-023-38515-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernard E, Tuechler H, Greenberg PL, et al. Molecular international prognostic scoring system for myelodysplastic syndromes. NEJM Evid. 2022;1(7) doi: 10.1056/EVIDoa2200008. [DOI] [PubMed] [Google Scholar]
- 8.Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391–2405. doi: 10.1182/blood-2016-03-643544. [DOI] [PubMed] [Google Scholar]
- 9.Arber DA, Orazi A, Hasserjian RP, et al. International consensus classification of myeloid neoplasms and acute leukemias: integrating morphologic, clinical, and genomic data. Blood. 2022;140(11):1200–1228. doi: 10.1182/blood.2022015850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khoury JD, Solary E, Abla O, et al. The 5th edition of the World Health Organization classification of haematolymphoid tumours: myeloid and histiocytic/dendritic neoplasms. Leukemia. 2022;36(7):1703–1719. doi: 10.1038/s41375-022-01613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoshizato T, Nannya Y, Atsuta Y, et al. Genetic abnormalities in myelodysplasia and secondary acute myeloid leukemia: impact on outcome of stem cell transplantation. Blood. 2017;129(17):2347–2358. doi: 10.1182/blood-2016-12-754796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernard E, Nannya Y, Hasserjian RP, et al. Implications of TP53 allelic state for genome stability, clinical presentation and outcomes in myelodysplastic syndromes. Nat Med. 2020;26(10):1549–1556. doi: 10.1038/s41591-020-1008-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Papaemmanuil E, Gerstung M, Bullinger L, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374(23):2209–2221. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernard E. R package for the IPSS-M. https://github.com/papaemmelab/ipssm
- 15.Smith AE, Kulasekararaj AG, Jiang J, et al. CSNK1A1 mutations and isolated del(5q) abnormality in myelodysplastic syndrome: a retrospective mutational analysis. Lancet Haematol. 2015;2(5):e212–e221. doi: 10.1016/S2352-3026(15)00050-2. [DOI] [PubMed] [Google Scholar]
- 16.Beauchamp EM, Leventhal M, Bernard E, et al. is mutated in clonal hematopoiesis and myelodysplastic syndromes and impacts RNA splicing. Blood Cancer Discov. 2021;2(5):500–517. doi: 10.1158/2643-3230.BCD-20-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okuda R, Nannya Y, Ochi Y, et al. Der(1;7)(q10;p10) presents with a unique genetic profile and frequent ETNK1 mutations in myeloid neoplasms [abstract] Blood. 2021;138(suppl 1):1513. [Google Scholar]
- 18.Gao T, Ptashkin R, Bolton KL, et al. Interplay between chromosomal alterations and gene mutations shapes the evolutionary trajectory of clonal hematopoiesis. Nat Commun. 2021;12(1):338. doi: 10.1038/s41467-020-20565-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saiki R, Momozawa Y, Nannya Y, et al. Combined landscape of single-nucleotide variants and copy number alterations in clonal hematopoiesis. Nat Med. 2021;27(7):1239–1249. doi: 10.1038/s41591-021-01411-9. [DOI] [PubMed] [Google Scholar]
- 20.Sanada M, Suzuki T, Shih L-Y, et al. Gain-of-function of mutated C-CBL tumour suppressor in myeloid neoplasms. Nature. 2009;460(7257):904–908. doi: 10.1038/nature08240. [DOI] [PubMed] [Google Scholar]
- 21.Awada H, Nagata Y, Goyal A, et al. Invariant phenotype and molecular association of biallelic mutant myeloid neoplasia. Blood Adv. 2019;3(3):339–349. doi: 10.1182/bloodadvances.2018024216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ilagan JO, Ramakrishnan A, Hayes B, et al. U2AF1 mutations alter splice site recognition in hematological malignancies. Genome Res. 2015;25(1):14–26. doi: 10.1101/gr.181016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shiozawa Y, Malcovati L, Gallì A, et al. Aberrant splicing and defective mRNA production induced by somatic spliceosome mutations in myelodysplasia. Nat Commun. 2018;9(1):3649. doi: 10.1038/s41467-018-06063-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dalton WB, Helmenstine E, Pieterse L, et al. The K666N mutation in SF3B1 is associated with increased progression of MDS and distinct RNA splicing. Blood Adv. 2020;4(7):1192–1196. doi: 10.1182/bloodadvances.2019001127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adema V, Khouri J, Ni Y, et al. Analysis of distinct hotspot mutations in relation to clinical phenotypes and response to therapy in myeloid neoplasia. Leuk Lymphoma. 2021;62(3):735–738. doi: 10.1080/10428194.2020.1839647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huber S, Haferlach T, Müller H, et al. MDS subclassification-do we still have to count blasts? Leukemia. 2023;37(4):942–945. doi: 10.1038/s41375-023-01855-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Makishima H, Saiki R, Nannya Y, et al. Germ line DDX41 mutations define a unique subtype of myeloid neoplasms. Blood. 2023;141(5):534–549. doi: 10.1182/blood.2022018221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haase D, Stevenson KE, Neuberg D, et al. TP53 mutation status divides myelodysplastic syndromes with complex karyotypes into distinct prognostic subgroups. Leukemia. 2019;33(7):1747–1758. doi: 10.1038/s41375-018-0351-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Acha P, Mallo M, Solé F. Myelodysplastic syndromes with Isolated del(5q): Value of molecular alterations for diagnostic and prognostic assessment. Cancers. 2022;14(22) doi: 10.3390/cancers14225531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Woll PS, Kjällquist U, Chowdhury O, et al. Myelodysplastic syndromes are propagated by rare and distinct human cancer stem cells in vivo. Cancer Cell. 2014;25(6):794–808. doi: 10.1016/j.ccr.2014.03.036. [DOI] [PubMed] [Google Scholar]
- 31.Malcovati L, Papaemmanuil E, Bowen DT, et al. Clinical significance of SF3B1 mutations in myelodysplastic syndromes and myelodysplastic/myeloproliferative neoplasms. Blood. 2011;118(24):6239–6246. doi: 10.1182/blood-2011-09-377275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malcovati L, Karimi M, Papaemmanuil E, et al. SF3B1 mutation identifies a distinct subset of myelodysplastic syndrome with ring sideroblasts. Blood. 2015;126(2):233–241. doi: 10.1182/blood-2015-03-633537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garcia-Gisbert N, Arenillas L, Roman-Bravo D, et al. Multi-hit TET2 mutations as a differential molecular signature of oligomonocytic and overt chronic myelomonocytic leukemia. Leukemia. 2022;36(12):2922–2926. doi: 10.1038/s41375-022-01733-8. [DOI] [PubMed] [Google Scholar]
- 34.Sanada M, Uike N, Ohyashiki K, et al. Unbalanced translocation der(1;7)(q10;p10) defines a unique clinicopathological subgroup of myeloid neoplasms. Leukemia. 2007;21(5):992–997. doi: 10.1038/sj.leu.2404619. [DOI] [PubMed] [Google Scholar]
- 35.Ganster C, Müller-Thomas C, Haferlach C, et al. Comprehensive analysis of isolated der(1;7)(q10;p10) in a large international homogenous cohort of patients with myelodysplastic syndromes. Genes Chromosomes Cancer. 2019;58(10):689–697. doi: 10.1002/gcc.22760. [DOI] [PubMed] [Google Scholar]
- 36.Gallì A, Todisco G, Catamo E, et al. Relationship between clone metrics and clinical outcome in clonal cytopenia. Blood. 2021;138(11):965–976. doi: 10.1182/blood.2021011323. [DOI] [PubMed] [Google Scholar]
- 37.Meggendorfer M, Bacher U, Alpermann T, et al. SETBP1 mutations occur in 9% of MDS/MPN and in 4% of MPN cases and are strongly associated with atypical CML, monosomy 7, isochromosome i(17)(q10), ASXL1 and CBL mutations. Leukemia. 2013;27(9):1852–1860. doi: 10.1038/leu.2013.133. [DOI] [PubMed] [Google Scholar]
- 38.Gurnari C, Pagliuca S, Prata PH, et al. Clinical and molecular determinants of clonal evolution in aplastic anemia and paroxysmal nocturnal hemoglobinuria. J Clin Oncol. 2023;41(1):132–142. doi: 10.1200/JCO.22.00710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pastor V, Hirabayashi S, Karow A, et al. Mutational landscape in children with myelodysplastic syndromes is distinct from adults: specific somatic drivers and novel germline variants. Leukemia. 2017;31(3):759–762. doi: 10.1038/leu.2016.342. [DOI] [PubMed] [Google Scholar]
- 40.Sahoo SS, Pastor VB, Goodings C, et al. Clinical evolution, genetic landscape and trajectories of clonal hematopoiesis in SAMD9/SAMD9L syndromes. Nat Med. 2021;27(10):1806–1817. doi: 10.1038/s41591-021-01511-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lindsley RC, Mar BG, Mazzola E, et al. Acute myeloid leukemia ontogeny is defined by distinct somatic mutations. Blood. 2015;125(9):1367–1376. doi: 10.1182/blood-2014-11-610543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tazi Y, Arango-Ossa JE, Zhou Y, et al. Unified classification and risk-stratification in acute myeloid leukemia. Nat Commun. 2022;13(1):4622. doi: 10.1038/s41467-022-32103-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cazzola M, Sehn LH. Developing a classification of hematologic neoplasms in the era of precision medicine. Blood. 2022;140(11):1193–1199. doi: 10.1182/blood.2022015849. [DOI] [PubMed] [Google Scholar]
- 44.Duncavage EJ, Bagg A, Hasserjian RP, et al. Genomic profiling for clinical decision making in myeloid neoplasms and acute leukemia. Blood. 2022;140(21):2228–2247. doi: 10.1182/blood.2022015853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sirenko M, Bernard E, Creignou M, et al. UBA1 mutations identify a rare but distinct subtype of myelodysplastic syndromes [abstract] Blood. 2023;142(suppl 1):1862. [Google Scholar]
- 46.Lindsley RC, Saber W, Mar BG, et al. Prognostic mutations in myelodysplastic syndrome after stem-cell transplantation. N Engl J Med. 2017;376(6):536–547. doi: 10.1056/NEJMoa1611604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weinberg OK, Siddon A, Madanat YF, et al. TP53 mutation defines a unique subgroup within complex karyotype de novo and therapy-related MDS/AML. Blood Adv. 2022;6(9):2847–2853. doi: 10.1182/bloodadvances.2021006239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuendgen A, Nomdedeu M, Tuechler H, et al. Therapy-related myelodysplastic syndromes deserve specific diagnostic sub-classification and risk-stratification-an approach to classification of patients with t-MDS. Leukemia. 2021;35(3):835–849. doi: 10.1038/s41375-020-0917-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aguirre LE, Al Ali N, Sallman DA, et al. Assessment and validation of the molecular international prognostic scoring system for myelodysplastic syndromes. Leukemia. 2023;37(7):1530–1539. doi: 10.1038/s41375-023-01910-3. [DOI] [PubMed] [Google Scholar]
- 50.Palomo L, Meggendorfer M, Hutter S, et al. Molecular landscape and clonal architecture of adult myelodysplastic/myeloproliferative neoplasms. Blood. 2020;136(16):1851–1862. doi: 10.1182/blood.2019004229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Geissler K, Jäger E, Barna A, et al. Correlation of RAS-pathway mutations and spontaneous myeloid colony growth with progression and transformation in chronic myelomonocytic leukemia-a retrospective analysis in 337 patients. Int J Mol Sci. 2020;21(8) doi: 10.3390/ijms21083025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carr RM, Vorobyev D, Lasho T, et al. RAS mutations drive proliferative chronic myelomonocytic leukemia via a KMT2A-PLK1 axis. Nat Commun. 2021;12(1):2901. doi: 10.1038/s41467-021-23186-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Papaemmanuil E, Döhner H, Campbell PJ. Genomic classification in acute myeloid leukemia. N Engl J Med. 2016;375(9):900–901. doi: 10.1056/NEJMc1608739. [DOI] [PubMed] [Google Scholar]
- 54.Bolton KL, Ptashkin RN, Gao T, et al. Cancer therapy shapes the fitness landscape of clonal hematopoiesis. Nat Genet. 2020;52(11):1219–1226. doi: 10.1038/s41588-020-00710-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.