Abstract
Although the U3 small nucleolar RNA (snoRNA), a member of the box C/D class of snoRNAs, was identified with the spliceosomal small nuclear RNAs (snRNAs) over 30 years ago1,2, its function and its associated protein components have remained more elusive. The U3 snoRNA is ubiquitous in eukaryotes and is required for nucleolar processing of pre-18S ribosomal RNA in all organisms where it has been tested3,4. Biochemical and genetic analyses suggest that U3-pre-rRNA base-pairing interactions mediate endonucleolytic pre-rRNA cleavages3. Here we have purified a large ribonucleoprotein (RNP) complex from Saccharomyces cerevisiae that contains the U3 snoRNA and 28 proteins. Seventeen new proteins (Utp1–17) and Rrp5 were present, as were ten known components. The Utp proteins are nucleolar and specifically associated with the U3 snoRNA. Depletion of the Utp proteins impedes production of the 18S rRNA, indicating that they are part of the active pre-rRNA processing complex. On the basis of its large size (80S; calculated relative molecular mass of at least 2,200,000) and function, this complex may correspond to the terminal knobs present at the 5′ ends of nascent pre-rRNAs. We have termed this large RNP the small subunit (SSU) processome.
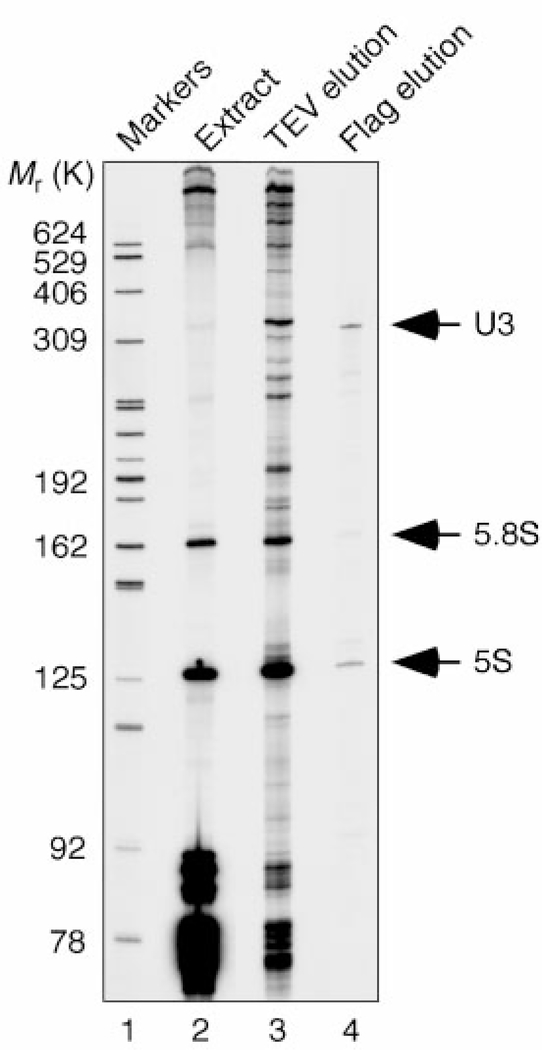
Previous U3 snoRNP purification strategies have used RNA affinity approaches and have resulted in the identification of only a small number of components5,6. We chose a protein-based affinity purification method that relied on simultaneous epitope tagging of two components: Nop5/58, which is common to the box C/D snoRNPs (TAP tag7), and the U3-specific protein Mpp10 (Flag tag). The first step immobilizes the box C/D snoRNPs through the protein A portion of the TAP tag. The snoRNPs are then released by TEV protease cleavage, maintaining native conditions. To isolate the U3 snoRNP, the box C/D snoRNPs are then applied to an anti-Flag antibody column and released with an excess of Flag peptide. The RNA composition of the resulting fractions was analysed (Fig. 1). The predominant enriched small RNA present in the final eluted material is the U3 snoRNA (lane 4), whereas the box C/D snoRNAs are enriched only after the first step (lane 3). The 5S and 5.8S rRNAs are also present in the eluate, although they are not enriched with respect to their relative abundance in the starting extract (lane 2).
Figure 1.
RNA composition of fractions from the purification of the U3 snoRNP. RNA was extracted from the starting material (extract, lane 2), from the first purification step that enriches for the box C/D snoRNAs (TEV elution, lane 3), and from the final eluate (Flag elution, lane 4). The RNA was directly labelled by 32p-labelled pCp and T4 RNA ligase, and analysed on an 8% denaturing polyacrylamide gel.
U3 snoRNA-associated protein components in the Flag affinity column eluate were digested with trypsin and the resulting peptides were analysed by nanoflow high-performance liquid chromatography/electrospray ionization mass spectrometry. Twenty-eight proteins ranging in relative molecular mass (Mr) from 13,000 to 200,000 were identified (Supplementary Information Table 1). All but one are coded for by essential yeast genes. Ten proteins had been previously described as U3 snoRNA-associated: four proteins common to the box C/D snoRNPs (Nop1, Nop56, Nop5/58 and Snu13) and six proteins specific to the U3 snoRNP (Sof1, Mpp10, Imp3, Imp4, Dhr1, Rrp9). We did not find three proteins previously demonstrated to be U3-associated: Lcp5 (ref. 8), Rcl1 (ref. 9) and Bms1 (ref. 10); however, this does not rule out their presence in the complex as they may be transiently associated, present in substoichiometric amounts or lost during the purification. One protein, Rrp5, had previously been shown to be required for pre-18S processing. Seventeen proteins (U three protein, Utp1–17) were found that had not previously been shown to be associated with U3 or implicated in pre-rRNA processing events. In addition, our analysis yielded five small subunit ribosomal proteins (Rps4, Rps6, Rps7, Rps14 and Rps28). Three proteins (YER087w, YPL110c, YMR029c) subsequently proved to be contaminants, as tagged versions did not co-immunoprecipitate Mpp10 and because the two that could be immunolocalized were found to be cytoplasmic (YER087w and YPL110c).
Bioinformatics analysis of the new protein components revealed motifs characteristic of an RNA–protein machine (Supplementary Information Table 1). Fourteen of the 17 Utp proteins bear protein–protein interaction domains (WD repeats, coiled-coil domains, HEAT repeats and a crooked-neck-like (crn-like) tetratrico peptide repeat (TPR)). The crn-like TPR is found in several proteins involved in other RNA processing events such as pre-mRNA splicing (Prp42, Prp6 and Clf1) and polyadenylation (RNA14)11. Two components have motifs characteristic of catalytically active proteins, an adenylate binding site and an ATP/GTP-binding site (P loop), although none so far identified bears a motif characteristic of an RNA endonuclease. Two of the Utp proteins have been implicated in other aspects of cellular metabolism: Utp17 was co-purified with the RENT complex, which is essential for exit from mitosis12, and Utp3 was discovered because it disrupts transcriptional silencing on overexpression13. For 11 of the 17 Utp proteins, probable human homologues exist; two of the remaining six Utp proteins have invertebrate homologues. For one of these, a probable homologue exists in Schizosaccharomyces pombe. This degree of evolutionary conservation suggests preservation of the fundamental mechanism of pre-rRNA processing from unicellular organisms to humans.
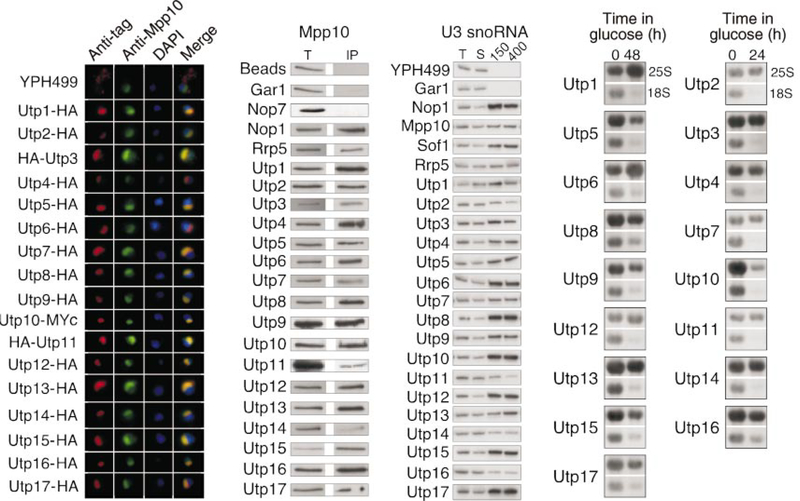
The previously identified U3 snoRNP-specific proteins are nucleolar and co-immunoprecipitate the U3 snoRNA. To verify that the new Utp proteins meet these criteria, we carried out the following experiments. The 17 Utp proteins were tagged with the haemagglutinin (HA)-epitope by chromosomal integration. Their subcellular localization was evaluated by indirect immunofluorescence microscopy. All 17 Utp proteins co-localized with the Mpp10 protein, indicating their nucleolar localization (Fig. 2a). Consistent with the co-localization studies, all 17 Utp proteins, as well as Rrp5, co-immunoprecipitated the Mpp10 protein (Fig. 2b). Other non-U3-associated nucleolar proteins, such as Gar1 and Nop7, did not co-immunoprecipitate Mpp10, suggesting that the Utp proteins and Rrp5 belong to a distinct nucleolar complex. Finally, all 17 Utp proteins and Rrp5 co-immunoprecipitated the U3 snoRNA (Fig. 2c). Furthermore, the protein–RNA association was stable after washes in 400 mM NaCl. In contrast, there was essentially no co-immunoprecipitation of the U14, snR10 or snR30 snoRNAs with any of the Utp proteins or with Rrp5 at either of the salt concentrations used (data not shown). The Utp proteins and Rrp5 are therefore part of a nucleolar complex that contains both Mpp10 and the U3 snoRNA but excludes many other nucleolar factors.
Figure 2.
Function of the new components of the SSU processome. a, The Utp proteins are nucleolar. Anti-HA or anti-Myc antibodies were used to detect the tagged Utp proteins (red), whereas anti-Mpp10 antibodies were used to decorate the nucleolus (green). DAPI was used to stain the nucleus (blue). Utp proteins were triply tagged by HA or Myc. b, The Utp proteins and Rrp5 are complexed with Mpp10. Anti-HA immunoprecipitations performed on cell extracts were analysed for the presence of Mpp10 by western blotting with anti-Mpp10 antibodies. T, total (5% of the input for immunoprecipitation); IP, immunoprecipitate. c, The Utp proteins and Rrp5 are associated with U3 snoRNA. Anti-HA immunoprecipitations were performed on cell extracts and were washed at 150 mM (150) or 400 mM (400) NaCl. RNA was extracted and analysed for the presence of the U3 snoRNA by northern blotting. T, total (20% of the input); S, supernatant (20%). d, The Utp proteins are required for 18S rRNA biogenesis. Yeast strains conditionally expressing Utp proteins were grown in medium containing galactose/raffinose (0 h), but switched to glucose for protein depletion (24 or 48 h). RNA was extracted at the indicated time points and analysed for the presence of 25S and 18S rRNAs by northern blot.
To define the function of the Utp proteins we constructed yeast strains in which each of the essential proteins could be conditionally depleted by growth in glucose. After depletion, we assessed the levels of 25S and 18S rRNAs by northern blotting. Depletion of each of the essential Utp proteins resulted in reduction of 18S rRNA levels with respect to the levels of 25S rRNA (Fig. 2d). Depletion of each Utp protein did not, however, affect the levels of the U3 snoRNA (Supplementary Information Fig. 1). These results suggest that the reduction in 18S rRNA levels upon Utp protein depletion is not due to a general defect in U3 snoRNP biogenesis but instead is due to a specific protein deficiency. The Utp proteins are therefore required for biogenesis of the small ribosomal subunit RNA, as are all of the other known U3 snoRNP-specific components.
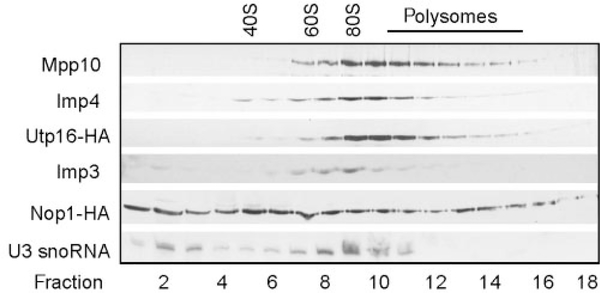
The finding of 28 U3-associated proteins prompted us to examine the size of the U3 snoRNA-associated protein complex on sucrose gradients (Fig. 3). The U3-specific proteins Mpp10, Imp4, Imp3, Utp16 (as an example of a new protein) and the U3 snoRNA co-migrate at a size that approximately corresponds to that of 80S ribosomes. In contrast, Nop1 (fibrillarin), which is a protein common to all box C/D snoRNPs, does not share the same sedimentation profile. Therefore the large U3–protein complex is approximately the same size as 80S ribosomes.
Figure 3.
The SSU processome sediments at 80S on sucrose gradients. Yeast extracts were analysed on 10–47% sucrose gradients. Anti-Mpp10, anti-Imp3, anti-Imp4 and anti-HA antibodies were used to detect the indicated proteins in each fraction by western blotting. Northern blotting was used to detect the U3 snoRNA. The migration of the 40S, 60S and 80S ribosomal subunits is specified. Utp16 and Nop1 proteins were triply tagged by HA.
Our results, combined with those of others, demonstrate that the 80S U3–protein complex has at least 28 physically and functionally associated proteins. Thus, the active U3 particle is not a small nucleolar RNP but rather a large nucleolar RNP. We have termed this complex the small subunit (SSU) processome because its components are required for pre-18S rRNA processing14,15. By analogy with the spliceosome, the SSU processome may be present in forms that differ slightly in composition, and it may be a dynamic entity. Similarly, there may be other proteins and snoRNPs transiently associated with it or present in the SSU processome in substoichiometric amounts.
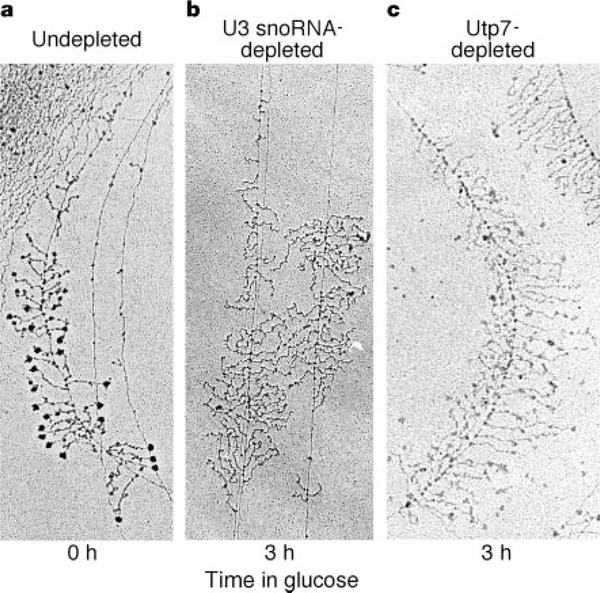
The presence of terminal knobs on amphibian pre-rRNA transcripts was first noted in 1969 (ref. 16), and the structures were termed ‘Christmas trees’ for their characteristic appearance. More than 20 years later, it was proposed that the terminal knobs included some of the pre-rRNA processing machinery17. Electron micrographs of chromatin spreads from yeast indicate that their nascent rRNA transcripts, like those in amphibians, also possess 5′ terminal knobs (Fig. 4a). In a strain where either the U3 snoRNA or Utp7 can be conditionally depleted by growth in glucose, we found that their depletion leads to loss of the terminal knobs on nascent pre-rRNAs (Fig. 4b, c). Depletion of Imp3 and Imp4 also leads to loss of the terminal knobs (data not shown). Switch of a parent strain from galactose to glucose, however, has no effect on the terminal knobs (data not shown); neither does a mutation in Mpp10 that does not lower protein levels (data not shown)18. Therefore the presence both of the U3 snoRNA and of SSU processome proteins is required for terminal knob formation.
Figure 4.
The U3 snoRNA and Utp7 are required for terminal knob formation on nascent pre-rRNAs. Yeast strains conditionally expressing either the U3 snoRNA or Utp7 from a galactose promoter were used to make the chromatin spreads. Strains were undepleted (a) or depleted for U3 snoRNA (b) or Utp7 (c). For depletion, the strains were switched from growth in galactose to growth in glucose. Chromatin spreads were made before (0 h) and after (3 h) the switch to glucose and analysed by electron microscopy. Scale, the width of panel a is 0.85 μm.
The size of the yeast terminal knobs measured using electron micrographs is variable, with an average size (42 × 34 nm) similar to that of ribosomes measured in the same way (28 × 30 nm). The size of the SSU processome estimated by sedimentation analysis (80S) and its calculated minimal mass (Mr 2,200,000, assuming each component is present once) are also similar to that of ribosomes. Therefore the SSU processome is roughly the same size as the terminal knobs. Owing to the approximate size correlation between the SSU processome and the terminal knobs, the requirement for the U3 snoRNA, Utp7, Imp3 and Imp4 in terminal knob formation, and the requirement for the SSU processome and the terminal knob in rRNA biogenesis17, we propose that the terminal knobs on nascent pre-rRNAs may represent the SSU processome. Further support comes from the observation that one of the SSU processome components, Imp4, co-immunoprecipitates the nascent 35S rRNA near the 5′ end19, as might be expected if it were a component of the terminal knob.
Why are there so many proteins required for the pre-rRNA cleavage reactions? The results presented here indicate that there are at least 28 proteins in the SSU processome. All of the proteins essential for growth are required for 18S rRNA biogenesis. We suggest that the complexity of the SSU processome reflects multiple functions. For example, the SSU processome may also have a critical role in folding the pre-18S rRNA. Consistent with this is the proposal that specific U3 snoRNA sequences are required for formation of the central, conserved pseudoknot in 18S rRNA3. The SSU processome may therefore function both in pre-rRNA cleavage and as a pre-rRNA chaperone.
Other pre-ribosomal complexes have recently been isolated using protein affinity techniques coupled with mass spectrometry20. In contrast to the function of these complexes in later steps in pre-rRNA processing or in transport, the SSU processome functions in the earliest steps in ribosome biogenesis. Of note, none of the non-ribosomal protein components of the SSU processome were found in the other identified complexes. These distinctions suggest that the different pre-ribosomal particles are each tailored for a specific function.
This work identifies the SSU processome as the third large RNP in eukaryotic cells, the two others being the ribosome and the spliceosome. Our results show that the SSU processome is an RNP larger and more complex than originally thought, with a definitive role in pre-rRNA processing and a putative role in pre-rRNA folding. Thus, it takes a large RNP (the SSU processome) to make a large RNP (the ribosome).
Methods
For purification of the U3 snoRNP, mass spectrometry and bioinformatics analysis, see Supplementary Information.
Validation and functional analysis
Saccharomyces cerevisiae strains expressing carboxy-terminal triple-HA-tagged versions of the Utp proteins, Gar1, Nop7, Nop1, Sof1 and Rrp5 were created by homologous recombination of polymerase chain reaction products at chromosomal loci in strain YPH499 (ref. 21). Strains expressing amino-terminal triple-HA-tagged versions of the Utp proteins were similarly created22. The genes N-terminally tagged were expressed under the control of galactose promoters to generate strains conditionally expressing the Utp proteins. In both cases, kanMX6 was used as the selection marker. Utp10 in Fig. 2a was tagged at its C terminus by chromosomal integration of triple-Myc using URA3 as a marker23. Successful tagging was verified by western blot analysis. The HA-tagged Mpp10 was obtained from the laboratory of M. Snyder24.
Yeast expressing tagged proteins were processed for indirect immunofluorescence microscopy as described25. Mouse anti-HA monoclonal antibody HA.11 (diluted 1:1,000; Covance) and rabbit anti-Mpp10 polyclonal antibodies26 (diluted 1:2,000) were detected with tetramethyl rhodamine isothiocyanate-conjugated goat anti-mouse immunoglobulin-γ (IgG) (diluted 1:100) and fluorescein isothiocyanate-conjugated donkey anti-rabbit IgG (diluted 1:200) secondary antibodies (Jackson ImmunoResearch). For localization of tagged Utp10, mouse anti-Myc monoclonal antibody 9E10 was used in place of the anti-HA antibody. Nuclear DNA was stained with 4,6-diamidino-2-phenylindole (DAPI, 1 μgml−1; Sigma).
For analysis of Mpp10 co-immunoprecipitation, anti-HA (12CA5) immunoprecipitations were carried out on extracts from strains bearing tagged proteins and western blotted with anti-Mpp10 antibodies as described27. Except for Utp11, each immunoprecipitation was performed with strains expressing C-terminal triple-HA-tagged proteins. Utp11 was triply HA-tagged at the N terminus.
For analysis of snoRNA co-immunoprecipitation, extracts were made and immunoprecipitations performed as described27, except that HA beads were prepared by incubating 200 μl anti-HA (12CA5) with 3 mg protein A Sepharose beads (Amersham Pharmacia Biotech) for 1–2 h at room temperature. The HA-tagged strains are described above.
Depletion was carried out on strains expressing Utp proteins under the control of galactose promoters. The strains were grown in medium containing galactose and raffinose, and then switched to glucose for depletion. For some strains, growth was severely impeded after 24 h in glucose, and RNA was extracted. For others, no change in growth was observed after 24 h in glucose, and the yeast were re-diluted and grown for another 24 h (48 h total) before RNA extraction. For these strains, growth was severely impeded after dilution at 24 h of growth in glucose. All strains were grown at 30 °C except for the strain expressing Utp16 from a galactose promoter. This strain is cold-sensitive when grown in glucose and was grown at room temperature. We carried out RNA extraction and northern blots as described27.
Sucrose gradients
Extracts were prepared27 and analysed on 10–7% sucrose gradients in 25 mM Tris pH 7.6, 150 mM KCl, 10 mM MgCl2, 0.1% NP-40. The gradients were spun in a SW41 rotor at 39,000 r.p.m. (260,000g) for 2 h. We collected 18 570-μl fractions. The migration of the 40S, 60S and 80S ribosomes was determined from the ultraviolet profile provided by the ISCO Model 185 density gradient fractionator. Fractions were analysed for the indicated proteins by western blotting with rabbit anti-Mpp10 (ref. 26), guinea-pig anti-Imp3, guinea-pig anti-Imp4 or anti-HA (12CA5). The guinea-pig anti-Imp3 and Imp4 antibodies were raised to recombinant proteins expressed and purified from Escherichia coli as described26. The U3 snoRNA was detected by northern blotting27.
Chromatin spreads and electron microscopy
We used strain JH84 for U3 snoRNA depletion28,29. In this strain the U3A snoRNA gene is under the control of a galactose promoter and the U3B gene is disrupted. The strain for depletion of Utp7 was constructed as described above. Depletion of the U3 snoRNA and Utp7 was performed by growth in glucose for 3 h. For chromatin spreads, yeast cultures were grown in YP medium plus galactose or glucose plus 1 M sorbitol to an absorbance at 600 nm of about 0.4. One millilitre of culture was digested with 5 mg zymolyase for 4 min at 30 °C. The yeast were then pelleted and 1 ml of 0.025% TritonX-100 at pH 9 was added to the pellet. After resuspension the yeast solution was mixed with 3 ml of 0.025% Triton and allowed to disperse for 20 min with swirling. One-tenth volume of 0.1 M sucrose-10% formalin, pH 8.5, was then added and grids were made in the usual manner30. Measurements of the size of the terminal balls were made on chromatin spreads of the YPH499 strain.
Supplementary Material
Acknowledgements
We thank A. Djikeng, G. Dreyfuss, P. Gordon, J. Laney, T. Serio and T. Stone for assistance and advice. We also thank P. Glazer, M. Snyder and J. Steitz for critical reading of the manuscript. F.D. was supported by the Anna Fuller Fund for Molecular Oncology. This work was supported by federal grants from the NIH to D.F.H and S.J.B. and the NSF to A.L.B., and by The Patterson Trust to S.J.B.
Footnotes
Competing interests statement
The authors declare that they have no competing financial interests.
Supplementary Information accompanies the paper on Nature’s website (http://www.nature.com/nature).
References
- 1.Weinberg RA & Penman S Small molecular weight monodisperse nuclear RNA. J. Mol. Biol 38, 289–304 (1968). [DOI] [PubMed] [Google Scholar]
- 2.Nakamura T, Prestayko AW & Busch H Studies on nucleolar 4 to 6S ribonucleic acid of Novikoff hepatoma cells. J. Biol. Chem 243, 1368–1375 (1968). [PubMed] [Google Scholar]
- 3.Venema J & Tollervey D Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet 33, 261–311 (1999). [DOI] [PubMed] [Google Scholar]
- 4.Maxwell ES & Fournier MJ The small nucleolar RNAs. Ann. Rev. Biochem 64, 897–934 (1995). [DOI] [PubMed] [Google Scholar]
- 5.Watkins NJ et al. A common core RNP structure shared between the small nucleolar box C/D RNPs and the spliceosomal U4 snRNP. Cell 103, 457–466 (2000). [DOI] [PubMed] [Google Scholar]
- 6.Lubben B,Marshallsay C, Rottman N & Luhrmann R Isolation of U3 snoRNP from CHO cells: a novel 55 kDa protein binds to the central part of U3 snoRNA. Nucleic Acids Res. 21, 5377–5385 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rigaut G, Shevchenko A, Rutz B, Wilm M & Seraphin B A generic protein purification method for protein complex characterization and proteome exploration. Nature Biotechnol. 17, 1030–1032 (1999). [DOI] [PubMed] [Google Scholar]
- 8.Wiederkehr T, Pretot RF & Minvielle-Sebastia L Synthetic lethal interactions with conditional poly(A) polymerase alleles identify LCP5, a gene involved in 18S rRNA maturation. RNA 4, 1357–1372 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Billy E, Wegierski T, Nasr F & Filipowicz W Rcl1p, the yeast protein similar to the RNA 3′ -phosphate cyclase, associates with U3 snoRNP and is required for 18S rRNA biogenesis. EMBO J. 19, 2115–2126 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wegierski T, Billy E, Nasr F & Filipowicz W Bms1p, a G-domain-containing protein, associates with Rcl1p and is required for 18S rRNA biogenesis in yeast. RNA 7, 1254–1267 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chung S, McLean MR & Rymond BC Yeast ortholog of the Drosophila crooked neck protein promotes spliceosome assembly through stable U4/U6.U5 snRNP addition. RNA 5, 1042–1054 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shou W et al. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell 97, 233–244 (1999). [DOI] [PubMed] [Google Scholar]
- 13.Kamakaka R & Rine J Sir- and silencer-independent disruption of silencing in Saccharomyces by Sas10p. Genetics 149, 903–914 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fournier MJ & Maxwell ES The nucleolar snRNAs: catching up with the spliceosomal snRNAs. Trends Biochem. Sci 18, 131–135 (1993). [DOI] [PubMed] [Google Scholar]
- 15.Venema J & Tollervey D Processing of pre-ribosomal RNA in Saccharomyces cerevisiae. Yeast 11, 1629–1650 (1995). [DOI] [PubMed] [Google Scholar]
- 16.Miller OL Jr & Beatty BR Visualization of nucleolar genes. Science 164, 955–957 (1969). [DOI] [PubMed] [Google Scholar]
- 17.Mougey EB et al. The terminal balls characteristic of eucaryotic rRNA transcription units in chromatin spread are rRNA processing complexes. Genes Dev. 7, 1609–1619 (1993). [DOI] [PubMed] [Google Scholar]
- 18.Lee SJ & Baserga SJ Functional separation of pre-rRNA processing steps revealed by truncation of the U3 small nucleolar ribonucleoprotein component, Mpp10. Proc. Natl Acad. Sci. USA 94, 13536–13541 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wehner KA & Baserga SJ The σ70-like motif: a eukaryotic RNA binding domain unique to a superfamily of proteins required for ribosome biogenesis. Mol. Cell 9, 329–339 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Warner JR Nascent ribosomes. Cell 107, 133–136 (2001). [DOI] [PubMed] [Google Scholar]
- 21.Knop M et al. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15, 963–972 (1999). [DOI] [PubMed] [Google Scholar]
- 22.Longtine MS et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 (1998). [DOI] [PubMed] [Google Scholar]
- 23.Schneider BL, Seufert W, Steiner B, Yang QH & Futcher AB Use of polymerase chain reaction epitope tagging for protein tagging in Saccharomyces cerevisiae. Yeast 11, 1265–1274 (1995). [DOI] [PubMed] [Google Scholar]
- 24.Ross-MacDonald P et al. Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature 402, 413–418 (1999). [DOI] [PubMed] [Google Scholar]
- 25.Dunbar DA, Dragon F, Lee SJ & Baserga SJ A nucleolar protein related to ribosomal protein L7 is required for an early step in large ribosomal subunit biogenesis. Proc. Natl Acad. Sci. USA 97, 13027–13032 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dunbar DA, Wormsley S, Agentis TM & Baserga SJ Mpp10p, a U3 small nucleolar ribonucleoprotein component required for pre-18S rRNA processing in yeast. Mol. Cell. Biol 17, 5803–5812 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee SJ & Baserga SJ Imp3p and Imp4p: two specific components of the U3 small nucleolar ribonucleoprotein that are essential for pre-18S rRNA processing. Mol. Cell. Biol 19, 5441–5452 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hughes JMX & Ares M Jr Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and prevents formation of 18S ribosomal RNA.EMBO J. 10, 4231–4239 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Samarsky DA & Fournier MJ Functional mapping of the U3 small nucleolar RNA from the yeast Saccharomyces cerevisiae. Mol. Cell. Biol 18, 3431–3444 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Osheim YN & Beyer AL Electron microscopy of RNP complexes on nascent RNA using the Miller chromatin spreading method. Methods Enzymol. 180, 481–509 (1989). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.