Abstract
Peptide substrate reporters are short chains of amino acids designed to act as substrates for enzymes of interest. Combined with capillary electrophoresis and laser-induced fluorescence detection (CE-LIF), they are powerful molecular tools for quantitative measurements of enzyme activity even at the level of single cells. Although most peptide substrate reporters have been optimized for human or murine cells in health-related applications, their performance in nonmammalian organisms remains largely unexplored. In this study, we evaluated three peptide substrate reporters for protein kinase B (PKB) in two eukaryotic microbes, Dictyostelium discoideum and Tetrahymena thermophila, which are evolutionarily distant from mammals and from each other yet express PKB homologues. All three peptide substrate reporters were phosphorylated in lysates from both organisms but with varying phosphorylation kinetics and stability. To demonstrate reporter utility, we used one to screen for and identify the previously unknown stimulus needed to activate PHK5, the PKB homologue in T. thermophila. In D. discoideum, we employed the highly quantitative nature of these assays using CE-LIF to make precise measurements of PKB activity in response to transient stimulation, drug treatment, and genetic mutation. These results underscore the broad applicability of peptide substrate reporters across diverse species while highlighting the need for further research to determine effective peptide stabilization strategies across different biological contexts.
Keywords: capillary electrophoresis, enzyme assay, peptide substrate reporter, kinase, phosphorylation
Peptide substrate reporters are molecular tools used to measure the activity of an enzyme of interest. Each reporter is a short (3–20 amino acid) peptide, often tagged with a fluorescent label, that is incubated in intact cells or lysates.1,2 For highly quantitative assays, electrophoretic separation is used to resolve the unmodified reporter from any enzymatic products, such as peptide fragments or phosphorylated peptide, and products are sensitively detected by laser-induced fluorescence (LIF). These experiments provide direct measures of enzyme activity and complement studies of enzymes based on mRNA transcript levels or antibody-based methods. Even when full-length, native substrates are known, peptide substrates have several advantages. Full-length protein substrates are typically quantified by Western blotting, but traditional implementations of this technique are labor-intensive, semiquantitative, and require relatively large quantities of samples and reagents, including validated antibodies against the protein of interest. Additionally, for enzymes such as kinases, total enzyme concentration is sometimes poorly correlated with enzyme activity, which is further regulated by post-translational modifications, localization, and other mechanisms. Peptide substrate reporters address these factors because modifications directly reflect net enzyme activity, and reporters and enzymatic products are readily separated by capillary electrophoresis (CE), a highly efficient, quantitative separation technique that is compatible with antibody-free analysis and small sample volumes, including single-cell analysis.3
Despite these advantages, peptide substrate reporters have not been as widely adopted for enzyme assays as more traditional methods. The development of peptide reporters is challenging because the amino acid sequence must be tailored to obtain stable substrates that have rapid reaction kinetics and are specific to an enzyme of interest. Peptide substrate reporters are often based on the sequence of a native substrate or the consensus sequence from several native substrates. However, because peptides are shorter than protein substrates, they are more susceptible to degradation by peptidases. Short peptides also lack secondary and tertiary structure, so they may have fewer favorable interactions with the enzyme of interest or undergo off-target interactions with related enzymes. For these reasons, peptide substrate reporter design is typically an iterative process involving both library screening and rational design or machine learning.4−9 Peptide substrate reporters have been developed for a range of enzymes, including kinases,2,4,7,10−16 phosphatases,17 deacylases,18 transferases,9,19,20 and many proteases. These substrates have typically been developed and validated using purified enzymes, lysates, or intact cells from humans or mice, and their applicability to other species is unclear. The advantages of peptide substrate reporters and the high investment in their development make it appealing to validate their application beyond mammalian cells for research in other organisms.
In this work, we evaluate three peptide substrate reporters for protein kinase B (PKB, also known as Akt) for measurements in two unicellular eukaryotes, the social amoeba Dictyostelium discoideum and the ciliate Tetrahymena thermophila. PKB is one of 39 primordial kinase families conserved throughout eukaryotic evolution.21,22 Thus, it is recognizably present across great evolutionary distances while maintaining high levels of sequence similarity (Figure 1). In human cells, the PI3K-PKB signaling pathway is associated with neutrophil chemotaxis, metabolism, and stress response and is commonly implicated in cancers.23,24 Because of its implications for human health, PKB has been a popular target for peptide substrate reporter research. All three peptide substrate reporters tested were initially developed and validated in human cell lines.12,23,25−27 Applications of these peptide substrate reporters have involved patient cells and mammalian cell lines for pancreatic cancer28 and rheumatoid arthritis.29 While previous work has focused on the role of PKB in human health, the PI3K-PKB pathway is integral to cell stress response in many organisms. In this work, we assess the suitability of three PKB reporters for enzyme assays in two organisms, D. discoideum and T. thermophila, demonstrating the applicability of peptide substrate reporters across evolutionary distance to biological investigations beyond human health.
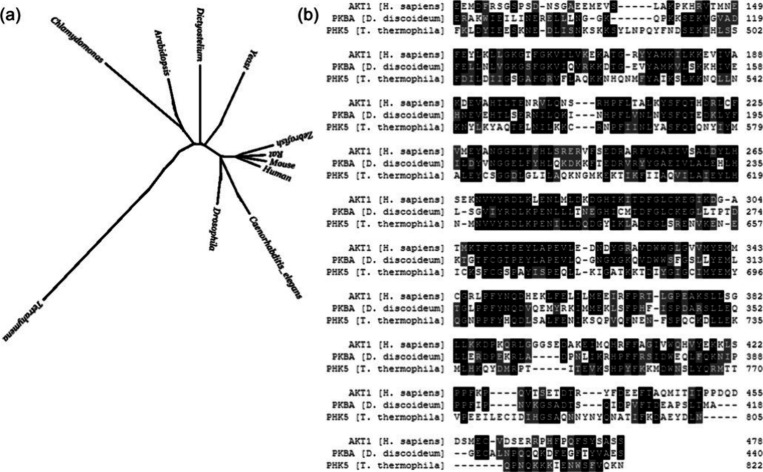
Figure 1.
(a) Phylogenetic tree based on conserved proteins showing the evolutionary distance between human cells, the unicellular eukaryotes used in this study, and other common model organisms. Adapted with permission from ref (30). Copyright 2010 IEEE. (b) Sequence alignment of PKB homologues from human cells (AKT1, H. sapiens), D. discoideum (PKBA), and T. thermophila (PHK5).
Methods
Sequence Alignment
Protein sequences were obtained in the FASTA format from the NCBI database, aligned using T-Coffee,31 and formatted using Multiple Align Show from Bioinformatics.org.32
Cell Culture
Axenic K-AX3 D. discoideum (DBS0236487) and mutant strain cARA- (DBS0236898) were obtained from the Dicty Stock Center.33,34 Cells were grown in shaking suspension at 180 rpm at 22 °C in HL-5 media supplemented with 13.5 g/L glucose, 300 μg/mL streptomycin and 100 μg/mL ampicillin and maintained at a cell density below 5 × 106 cells/mL.35 HL-5 media was composed of 14 g/L proteose peptone no. 3 (BD), 7 g/L yeast extract, 3.5 mM dibasic sodium phosphate, and 11 mM monobasic potassium phosphate, pH 6.4–6.7. New cultures were started from frozen stocks every 2–3 weeks.
T. thermophila B2086.2 (SD00709) and CU428.2 (TSC_SD00178) strains were obtained from the Tetrahymena Stock Center,36 and cultured according to established protocols.37 Stock tubes were maintained in room temperature modified Neff media and replaced every 2–3 weeks. Modified Neff media was composed of 2.5 g/L proteose peptide, 2.5 g/L yeast extract, 5 g/L glucose, and 33.3 μM FeCl3. For experiments, cultures were expanded to 100,000–300,000 cells/mL in proteose peptone (PP) media in unshaken cultures at 30 °C.
Peptide Substrate Reporters
Three peptide substrate reporters were obtained from Anaspec and stored at −80 °C as 500 μM aqueous stocks.
Degradation Assays
Degradation assays were conducted as described previously.38 Briefly, aliquots of vegetative cells were washed twice, resuspended in ice-cold phosphate buffered saline at a density of 2 × 108 cells/mL and lysed by three freeze–thaw cycles using liquid nitrogen. The resulting lysate was clarified by 5 min centrifugation at 15,000 × g, and the supernatant was stored at −80 °C. The total protein concentration was determined using a fluorescamine assay with a bovine serum albumin (BSA) calibration curve.39 Briefly, three parts diluted lysate (or BSA standard) in 30 mM borate buffer (pH 9) and one part 3 mg/mL fluorescamine in acetone were incubated for 5 min in the dark at room temperature. The fluorescence at 475 nm was then measured using 390 nm excitation. Based on this assay, the lysate was diluted to a concentration of 3 mg/mL in phosphate buffered saline. The degradation reaction was run at 25 °C and initiated by addition of 10 μM reporter to the diluted lysate. At fixed time points, aliquots were removed, and the reaction was stopped by incubation for 4 min at 95 °C.
Phosphorylation in Lysates
For D. discoideum lysates, chemotaxis-competent cells were prepared as described previously.40 Briefly, cells were washed three times in ice-cold development buffer (DB; 10 mM phosphate buffer pH 6.5, 1 mM CaCl2, 2 mM MgCl2), resuspended at a density of 2 × 107 cells/mL in room temperature DB, and shaken at 100 rpm. Prior to lysis, cells were basalated by addition of 4.3 mM caffeine for 20 min with 200 rpm shaking. Cells were then washed with ice-cold development buffer, stimulated with 1 μM cAMP, and lysed with an equal volume of 2× lysis buffer (50 mM MOPS, pH 7.6, 200 mM sodium chloride, 2 mM pervanadate, 50 mM glycerophosphate, 6 mM sodium pyrophosphate, 20 mM sodium fluoride, 2 mM EDTA, 2 mM EGTA, 4 μg/mL aprotinin, 4 μg/mL leupeptin, 2 mM DTT, 1% Triton X-100, and 20% glycerol).41 For assays with LY294002, cells were incubated with 15 μM LY294002 or an equivalent volume of DMSO as a vehicle control for 1 min immediately before cAMP stimulation and lysis.
For T. thermophila lysates, mating was induced by mixing equal cell numbers of B2086.2 and CU428.2 strains that had been starved for 16 h at 30 °C in unshaken Dryl’s buffer (2 mM phosphate buffer, 2 mM citrate, 1.5 mM CaCl2). Two hours after cells were mixed, microscopy was used to confirm that >75% of cells were paired. Samples were then lysed with an equal volume of ice-cold 2× lysis buffer, before and 30 s after addition of 20% proteose peptone stock to a final concentration of 2% or other stimuli as noted.
The protein concentrations of lysates were determined using the fluorescamine assay described above, and lysates were diluted to the same final concentration (0.4–0.6 mg/mL) in 1× lysis buffer. For T. thermophila stimulated with proteose peptone, it was not possible to determine protein concentrations in the cell lysate without interference from peptone, so these samples were diluted proportionately to unstimulated controls. Phosphorylation reactions were conducted in kinase buffer (25 mM MOPS, pH 7.4, 20 mM magnesium chloride, 25 mM glycerophosphate, 1 mM DTT).41 The peptide substrate reporter was added to a final concentration of 5 μM, and the reaction was started by addition of a final concentration of 2 mM ATP. The reaction was run at room temperature and stopped by the addition of CE run buffer. Negative control experiments were performed by substituting deionized water for ATP or denaturing the lysate with heat before the addition of peptide.
Capillary Electrophoresis
Samples were analyzed using micellar electrokinetic chromatography (MEKC) in a PA-800 Plus capillary electrophoresis instrument (Beckman Coulter). For all assays of B-5/I, the CE run buffer was 100 mM borate, 100 mM SDS, pH 7.7, which was shown previously to resolve the reporter from all enzymatic products (i.e., phosphorylated peptide and fragments formed by cleavage of peptide bonds).13 The same buffer was used for phosphorylation assays of Crosstide. For degradation assays of Crosstide, the run buffer was 100 mM HEPES, 10 mM SDS, pH 8.0 since this buffer resolved all fragment peaks. For all VI–B assays, the run buffer was 100 mM borate, 15 mM SDS, pH 11.4. The capillary was 50 μm diameter bare silica, 21 cm effective length with an applied potential of 400 V/cm. Peaks for the unmodified reporters, the phosphorylated product, and some peptide fragments were identified by their migration times and confirmed by addition of purified standards. Peak integration was performed using 32 Karat (v. 10.1) software.
Results and Discussion
In this work, we evaluated three peptide substrate reporters (Table 1) designed for human Akt for assays of PKB homologues in the eukaryotic microbes D. discoideum and T. thermophila. Crosstide is derived from the phosphorylation site of GSK3, a native PKB substrate,26 but is generically phosphorylated by other AGC kinases, including MAPKAP kinase-1, p70 S6 kinase, and PKA.25,42 B-5/I was developed using a screen to identify peptide sequences that would be specific for human PKB over MAPKAP kinase-1 and p70 S6 kinase.25 Then, the sequence for B-5/I was iteratively modified to make it resistant to peptidases by incorporating non-native amino acids while retaining a low KM for human PKB.13D. discoideum and T. thermophila express the PKB homologues, PKBA and PHK5, respectively, during nutrient deprivation.41,43 Despite their sequence similarity to human PKB/AKT1 (Figure 1b), variations in sequence and structure were expected to change their phosphorylation activity toward the peptide substrate reporters. Promisingly, Crosstide has been used previously for phosphorylation assays of related AGC kinases, including an Akt homologue, in S. cerevisiae.44−47 However, the kinome of S. cerevisiae is closer to that of human cells than those of D. discoideum or T. thermophila.21
Table 1. Peptide Substrate Reporter Sequencesa.
| name | sequence |
|---|---|
| Crosstide | 5FAM-GRPRTSSFAEG |
| B-5/I | 6FAM-GRPRAATFAEG-NH2 |
| VI–B | 6FAM-GRPRAFTFA-NH2 |
Boldface underlined amino acids are N-methylated.
Thus, an initial step was to demonstrate phosphorylation of the reporters by enzymes from each organism. Purified, active enzymes from these organisms are not commercially available, and multiple attempts at custom protein expression of D. discoideum PKBA in various vectors were unsuccessful in yielding active enzyme. Consequently, we confirmed phosphorylation of the peptide substrate reporters in cell lysates from both unstimulated and stimulated cells that had been nutrient deprived (Figure 2). In D. discoideum, transient activation of PKBA coordinates chemotaxis and gene expression during social development, when individual cells form a differentiated, multicellular structure in response to nutrient deprivation.48 Much less is known about the kinome of T. thermophila. However, genome analysis identified 12 kinases containing a Pleckstrin homology; of these, PHK5 is most like mammalian Akt. Preliminary mRNA expression data show spikes in PHK5 levels 2 and 6 h after initiation of starvation-induced sexual reproduction (also called conjugation or mating).43
Figure 2.
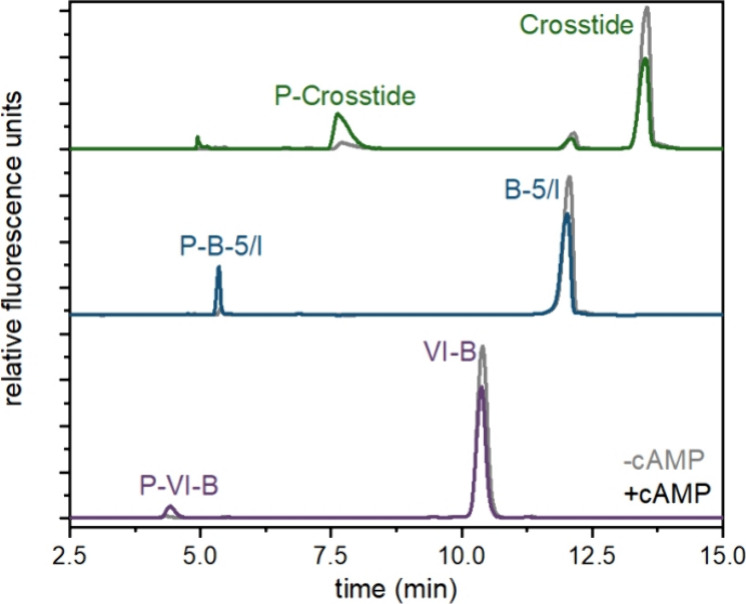
Phosphorylation of peptide substrate reporters in D. discoideum lysates. Electropherograms of the three peptide substrate reporters (Crosstide, green; B-5/I, blue; VI–B, purple) incubated with D. discoideum lysates from chemotaxis-competent cells unstimulated (gray) and stimulated (color) cells with cAMP. Cells were lysed just before or 20 s after stimulation. For all traces, x-axes have been normalized to align peaks for display only.
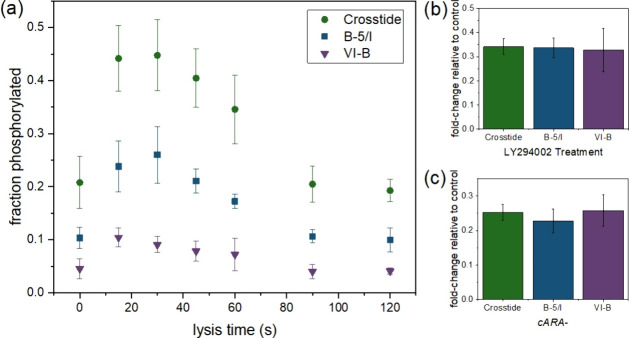
Lysates from D. discoideum demonstrated low levels of basal phosphorylation activity in unstimulated cells and increased phosphorylation activity toward all three reporters upon stimulation with cAMP (Figure 2). In T. thermophila, all three reporters were phosphorylated in lysates from unstimulated cells, but the phosphorylation activity was low and did not increase after initiation of mating as expected. PHK5 expression increases alongside the expression of the opposing phosphatase PTEN, suggesting that activation may be transient, as in D. discoideum.43 Thus, we hypothesized that an as-yet unidentified stimulus may be necessary to fully activate PHK5 and that activation may be transient due to concurrently high levels of PTEN.
Using the peptide substrate reporter Crosstide, we screened several known hormones in T. thermophila, including cGMP, serotonin, histamine, insulin, and concavalin A.49 Cells were treated with a final concentration of 1 μM stimulus for 20 s prior to lysis. None of these stimuli resulted in increased phosphorylation activity (Figure 3a). (It is worth noting that mechanical agitation caused a slight increase in activity, and we modified our procedure to eliminate this interference.) Since conjugation is induced by nutrient deprivation, we then screened for the effect of nutrients on phosphorylation by adding proteose peptone to a final concentration of 2%. This treatment resulted in a dramatic increase in phosphorylation (Figure 3b). To our knowledge, this is the first assay of purported PHK5 activity, which has previously only been studied using mRNA expression,43 and the first indication that it may respond to nutrient levels. Because peptide substrate reporters are less specific than full-length native protein substrates, additional biochemical experiments are needed to confirm these results. However, these data demonstrate the utility of peptide substrate reporters for hypothesis testing and discovery. In particular, these experiments demonstrate the importance of analytical methods that do not rely on validated antibodies, which may not be available or which may not accurately reflect enzyme activity due to post-translational modifications and upstream signaling.
Figure 3.
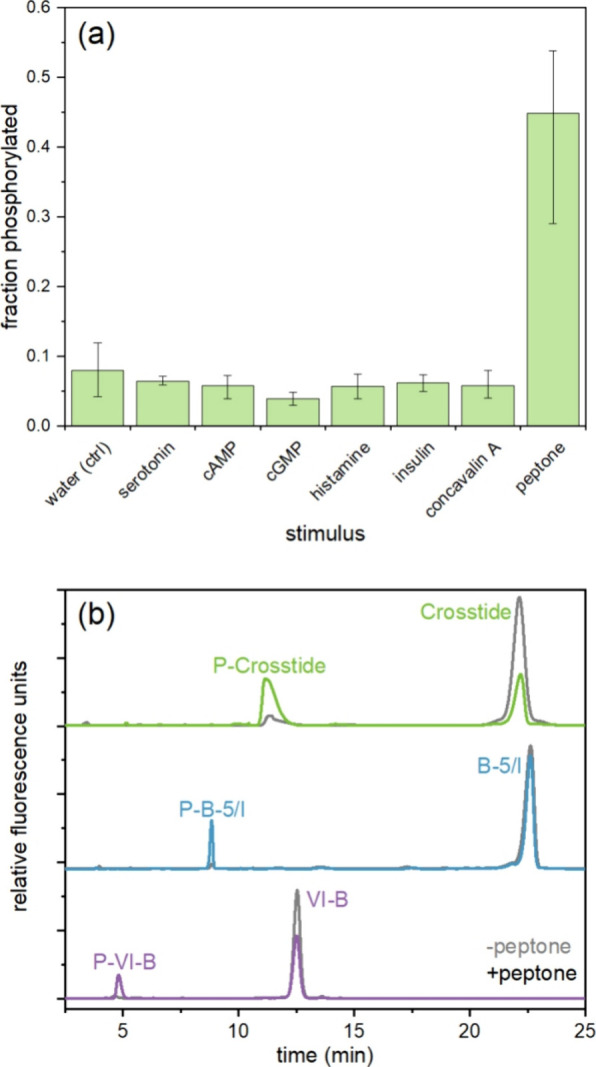
Phosphorylation of peptide substrate reporters in T. thermophila lysates. (a) Average fraction of Crosstide that was phosphorylated after 15 min incubation in lysates prepared 20 s after stimulation with 1 μM stimulus (or 2% for proteose peptone). Error bars represent the maximum and minimum values for n = 2–3 biological replicates on different days. (b) Electropherograms of the three peptide substrate reporters (Crosstide, green; B-5/I, blue; VI–B, purple) incubated with T. thermophila lysates from cells 2 h into conjugation for unstimulated (gray) and stimulated (color) cells with 2% proteose peptone. Cells were lysed just before or 30 s after stimulation. For all traces, x-axes have been normalized to align peaks for display only.
In addition to confirming that all three reporters were phosphorylated in lysates from both cell types, we also characterized the kinetics of the phosphorylation, which varied substantially between organisms. Using reactions with ≤10% total phosphorylation, we determined the phosphorylation rate for lysates from stimulated cells in units of zeptomoles of phosphorylated product per picogram of total protein per second (zmol pg–1 s–1; Table 2). Crosstide, which is based on the native substrate GSK-3,26 showed the fastest phosphorylation kinetics of the three peptides in both organisms. For D. discoideum, B-5/I was the next most preferred substrate. This kinetic trend is similar to that observed in human cells. Crosstide and a truncated version of B-5/I have similar values for Vmax with human Akt1, but the KM value is about 5-fold lower for Crosstide.25 The value of Vmax has not been determined for VI–B with Akt1, but its phosphorylation rate is known to be slower than that of B-5/I.13 In contrast, T. thermophila lysates showed a preference for VI–B over B-5/I.
Table 2. Phosphorylation Rates of Each Reporter in Lysates from Each Cell Type 20–30 s after Stimulation with cAMP (D. discoideum) or Proteose Peptone (T. thermophila).
| phosphorylation
rate (zmol pg–1 s–1) |
||
|---|---|---|
| peptide | D. discoideum | T. thermophila |
| Crosstide | 5.8 ± 0.8 | 9.1 ± 2.4 |
| B-5/I | 3.1 ± 0.6 | 1.8 ± 1.1 |
| VI–B | 1.4 ± 0.2 | 4.1 ± 0.9 |
Notably, in both microbes, initial phosphorylation rates for VI–B ranged from 1 to 4 zmol pg–1 s–1 after stimulation. These rates are much higher than rates obtained in intact human prostate cancer cells (LNCaP, 0.04 zmol pg–1 s–1)6 and pancreatic cancer cell lines (0.02–0.1 zmol pg–1 s–1)28,50 with the same reporter. Our assays were conducted in lysates containing phosphatase inhibitors, which will increase the apparent activity of PKB by removing the effects of any opposing phosphatases. Additionally, higher phosphorylation rates may arise from differing expression levels of PKB between cell types. For both D. discoideum and T. thermophila, PKB expression is linked to specific life cycle stages, and high levels of expression may be necessary to transduce short, transient signals.
Application to Biochemical Assays in D. discoideum
To further validate the use of these reporters in an evolutionarily distant organism, we conducted several quantitative biochemical assays of known physiological responses in D. discoideum. D. discoideum activates the PI3K-PKBA signaling pathway during chemotaxis in the early stages of its social life cycle.51,52 Under nutrient deprivation, D. discoideum undergoes a 24 h long social development process and transitions from free-living single cells into a differentiated, multicellular aggregate. Early in this process, cAMP secreted in pulses binds to a G-protein coupled receptor, cAR1, resulting in transient activation of the PI3K-PKBA pathway, which initiates actin polymerization and chemotaxis.41 We examined phosphorylation activity in relation to three aspects of this signaling pathway: the time scale of the transient activation after cAMP stimulus (Figure 4a); the pharmacological effect of the PI3K inhibitor LY294002 (Figure 4b); and genetic mutation to abolish the main isoform of the G-protein coupled receptor cAR1 (Figure 4c). In all three cases, results from peptide substrate reporters were in agreement with Western blotting results from our lab using an antibody against phospho-Akt substrates (Figures S1–S3) and from past research.41 However, the results from peptide substrate reporters highlight the highly quantitative nature of these assays using CE-LIF compared to semiquantitative Western blots.
Figure 4.
Confirmation of known changes to PKBA activity in D. discoideum using peptide substrate reporters. (a) Transient activation of kinase activity by cAMP stimulation. (b) Decreased kinase activity after treatment with LY294002. (c) Decreased kinase activity in cARA– mutant cell lysates. For all experiments, social development proceeded for 1 h prior to stimulation and lysis. For (b) and (c) cells were lysed 15 s after stimulation with cAMP. Error bars represent the standard deviation of n = 3 biological replicates.
For the first assay, we confirmed the rapid, transient increase in PKBA activity in response to cAMP stimulation. In D. discoideum, PKBA expression levels are high throughout the early stages of social development; however PKBA activity remains low until cAMP stimulation, which activates PI3K, resulting in a rapid, but transient, increase in PKB activity within 10–20 s that subsides 1–3 min poststimulus.53,54 Phosphorylation data for all three reporters reflected this transient activation. Reporters were maximally phosphorylated when cells were lysed 15–30 s after cAMP stimulation (Figure 4a), and the time course of activation was remarkably similar for all three peptides (Figure S4), even though the kinetics of phosphorylation differed, reflecting trends observed in Table 2.
Next, we tested pharmacological treatment of D. discoideum. We treated cells for 1 min with 15 μM LY294002, a PI3K inhibitor. LY294002 has been shown previously to reduce PKBA activity in D. discoideum.53,55−57 As expected, LY294002 treatment decreased phosphorylation activity toward all three reporters (Figure 4b). The magnitude of this effect was quite similar for all three peptides; percent phosphorylation of cAMP-stimulated cells decreased to 33–34% of the untreated phosphorylation levels upon LY294002 treatment. The LY294002 dose (15 μM) was previously identified as the EC50 for PKBA in D. discoideum,53 making this reduction in activity reasonable if PKBA is the enzyme primarily responsible for substrate phosphorylation.
Finally, we evaluated the ability of the peptide substrate reporters to reflect differences in signaling between “wild type” cells and a genetic mutant. The major G-protein coupled receptor for cAMP, cAR1, is deleted in the cARA– strain, preventing cAMP stimulus from initiating PKBA activation. As expected, the cARA– cell lysates showed minimal increase in phosphorylation activity toward the reporters in response to cAMP stimulation, resulting in phosphorylation levels roughly one-fourth of that observed for K-AX3 “wild type” controls (Figure 4c). As in past studies,41 phosphorylation activity was not completely abolished due to the presence of another cAR isoform, cAR3.
It is somewhat surprising that the results of these assays were so similar for the three peptides. If the peptides showed different levels of off-target phosphorylation by enzymes other than PKBA, then differences might be expected between peptides in their phosphorylation as a function of time, drug treatment, or mutation. Past research suggested that B-5/I was more specific for human Akt than Crosstide,12,25 but specificity may be one aspect of peptide substrate reporters that does not readily transfer between species. PKB is a member of the AGC group of small, cytoplasmic kinases, and the Dictyostelium kinome includes 21 kinases from this group, including the two PKB isoforms (PKBA and PKBR1) and an SGK ortholog.58 In D. discoideum, the PKBR1 isoform is highly homologous with PKBA,55 making it likely that any reporter peptide could be phosphorylated by both isoforms. Similarly, the D. discoideum SGK ortholog is likely to show activity toward the substrates since a previous screen of ∼50 kinases found that Crosstide and a truncated version of B-5/I were phosphorylated by SGK.5 However, the timing of PKBA expression during development differs from that of PKBR1 and SGK. Transcriptomic experiments show pkbA transcript levels peak during the first hour of development, then decline to basal levels by 4–6 h. In contrast, levels of pkgB transcripts (which code for PKBR1) and DDB_G0277449 transcripts (which code for the SGK ortholog) only rise after 5 h of development and remain high at least through 12 h.59 Our experiments, conducted 1 h into social development, likely reflect PKBA activity over the activity of PKBR1 and SGK. Other AGC kinases may also contribute to phosphorylation of these reporters; however, evolutionary analysis of phospho-recognition sites shows that the R-x-x-T/S–F motif, associated with PKB substrates and present in the three peptides used here, is enriched in the human and yeast phosphoproteomes but not those of D. discoideum or T. thermophila.60 This may suggest that fewer kinases in these organisms recognize the motif. Nevertheless, further work with purified enzymes is required to evaluate specificity.
Stability of the Reporter in Cell Lysates
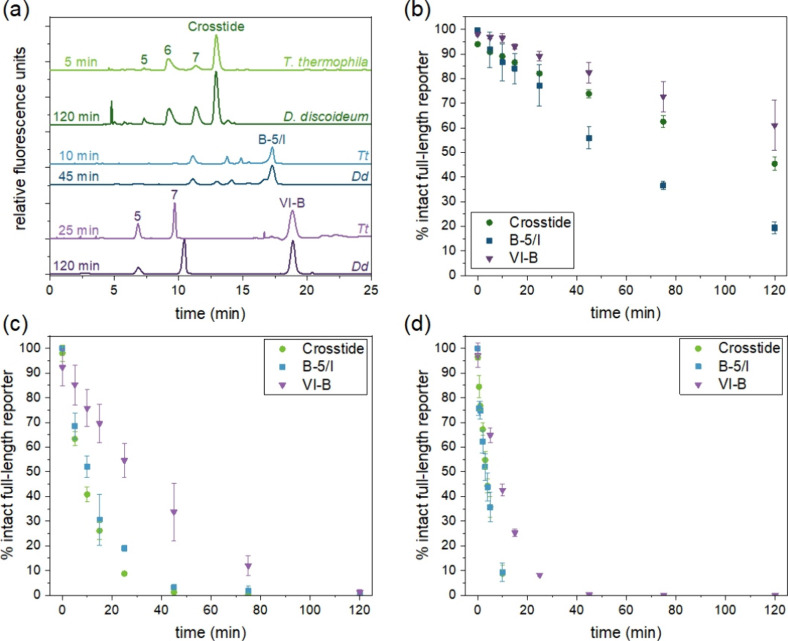
In lysates, peptide substrate reporters were used in combination with a cocktail of protease inhibitors to prevent degradation. However, application of peptide substrate reporters to advanced applications like single-cell analysis requires that they be resistant to degradation in intact cells. To check for stability, we incubated the reporters in freeze–thaw lysates from both organisms in the absence of protease inhibitors. We have previously shown that the stability of VI–B and its major degradation products varies widely across species.38 In the current work, we expanded our previous studies to include T. thermophila lysates and to look at degradation of B-5/I and Crosstide in addition to VI–B. We have previously shown that organisms that express different classes of proteolytic enzymes produce different fragmentation patterns.38 Genomic analysis of peptidase expression across organisms shows that D. discoideum and T. thermophila share the same core of eukaryotic peptidases despite their evolutionary distance.61 While fragmentation rates varied widely between D. discoideum and T. thermophila, the fragments observed were the same (Figure 5a). For Crosstide, the major fragments detected were those with 5, 6, or 7 amino acids remaining on the fluorescently labeled N-terminus, corresponding to 5FAM-GRPRT, 5FAM-GRPRTS, and 5FAM-GRPRTSS. For VI–B, the major fragments were 6FAM-GRPRA and 6FAM-GRPRAFT, where R is N-methylated arginine. Individual standards were not used to identify the fragments of B-5/I, but the peak patterns in electropherograms suggest that this peptide also formed the same fragments in lysates from both organisms. The observation of the same qualitative fragmentation patterns for all three peptides in both organisms supports the expectation based on genomic data that D. discoideum and T. thermophila express similar proteolytic enzymes.
Figure 5.
Peptide degradation data in freeze–thaw lysates. (a) Representative electropherograms of peptides incubated in vegetative cell lysates from T. thermophila B2086.2 (lighter) and D. discoideum (darker). Time points were selected to represent roughly one-half-life for each peptide (i.e., when the full-length reporter was approximately 50% degraded). Numbered peaks indicate peptide fragments identified by spiking with known standards and represented by the number of N-terminal amino acids remaining from the original unmodified reporter. (b-d) Degradation of the reporters over time in lysates from (b) D. discoideum, (c) T. thermophila B2086.2, and (d) T. thermophila CU428.2. Error bars represent the standard deviation for n = 3 biological replicates.
To study degradation kinetics, we fit the data for each full-length peptide to a first-order reaction model and determined the half-life for each peptide for three biological replicates (Figure 5b-d, Table 3). Degradation rates and half-lives varied widely between organisms and peptides; however, some general trends were apparent. For all peptides, degradation was much faster in T. thermophila than in D. discoideum. T. thermophila is known to produce and secrete cysteine proteases and is a large (∼50 μm wide and ∼20 μm long), fast-growing cell. Its doubling time is nearly as rapid as that of S. cerevisiae, despite a cell volume that is ∼170-fold larger;62,63 however, our past research did not identify any correlation between VI–B degradation rate and the proliferation rate of the cells.38 Interestingly, the two strains used to produce mating pairs for phosphorylation experiments had different degradation kinetics; degradation was consistently 2–4 times faster in lysates from CU428.2 cells than from the B2086.2 strain. Further characterization of protease expression in these strains may be helpful identifying the enzyme(s) primarily responsible for degradation of these reporters.
Table 3. Half-Life of Each Reporter in Freeze-Thaw Lysatesa.
| half-life
(min) |
|||
|---|---|---|---|
| Crosstide | B-5/I | VI–B | |
| D. discoideum | 117 ± 10 | 51 ± 4 | 184 ± 65 |
| T. thermophila B2086.2 | 8 ± 1 | 12 ± 4 | 22 ± 4 |
| T. thermophila C428.2 | 2.9 ± 0.5 | 3.1 ± 0.5 | 6.7 ± 0.2 |
Uncertainty values represent the standard deviation for 3 biological replicates.
Of the three peptides, VI–B had the longest half-life in both organisms. VI–B was optimized to be degradation-resistant in HeLa lysates through substitution of non-native N-methylated amino acids.6 This change appeared to improve its stability in the organisms studied here as well. That said, degradation of VI–B in T. thermophila was still very fast. In D. discoideum lysates, the initial rate of VI–B of fragmentation in lysates (based on the first 15 min of each reaction) was 0.3 ± 0.1 pmol of peptide per mg of total protein per second (pmol mg–1 s–1), In contrast, the initial degradation rate of VI–B in T. thermophila lysates (based on the first 5 min of each reaction) was 1.1 ± 0.5 pmol mg–1 s–1 for strain B2048.2 and 5.1 ± 0.3 pmol mg–1 s–1 for strain CU428.2. These rates are comparable to previous observations for peptide substrate reporter degradation (0.02–3 pmol mg–1 s–1),28,29,38,64 but the rate in T. thermophila is roughly an order of magnitude faster than in D. discoideum. Given the half-life of the peptides in T. thermophila, kinase assays in intact cells would likely require treatment with protease inhibitors to maintain intact reporter on the time scale of the experiment. Alternatively, a peptide substrate reporter could be designed for stability in eukaryotic microbes. As noted above, the 5-mer and 7-mer were prominent fragments of Crosstide and VI–B in both D. discoideum and T. thermophila. In human cells, non-native amino acids at position 5 yielded poor substrates for PKB, so this bond may be challenging to stabilize with compromising kinase activity toward the substrate. However, the bond between amino acids 7 and 8 has not previously been targeted for modifications because the 7-mer is not a major fragment in human cells.13 Our past research in D. discoideum has shown that formation of the 7-mer from the parent peptide has the highest rate constant of all possible fragmentation reactions and that formation of the 5-mer preferentially occurs via the 7-mer rather than from direct fragmentation of the parent peptide.38 Thus, stabilization of the bond between amino acids 7 and 8 represents a promising avenue for improved stability. Bulky N-terminal modifications and peptide cyclization or “stapling” are additional methods that may be employed.65
Conclusions
Peptide substrate reporters are often developed and validated in a single cell type or a few cell types derived from the same species. However, these reporters could have broad applications across different organisms if their behavior in evolutionarily distant cell types were known or predictable. Here, we have demonstrated that three different peptide substrate reporters for the primordial kinase PKB are all well-suited to applications in evolutionarily distant organisms, suggesting that transfer of reporters to new species may be readily achievable. Peptide substrate reporters may be especially useful for cross-species comparisons because they are small and interact primarily with the enzyme’s active site. While antibodies may bind to epitopes that vary between organisms, the active site of most enzymes is highly conserved. As a result, reporter phosphorylation may be the most predictable and successful for peptide substrate reporters such as Crosstide that are based closely on native protein substrates.
In addition to enabling assays for enzymes that lack validated antibodies, peptide substrate reporters offer several additional advantages. CE-LIF detection is highly quantitative and facilitates precise measures of even small changes in enzyme activity relative to controls. Electrophoretic separation and LIF detection of peptide substrate reporters are also compatible with single-cell analysis.8 Even when antibodies are available, traditional Western blots require large numbers of cells and report on population-level changes in signaling, obscuring variation at the single-cell level. However, cell-to-cell variation in enzyme activity is critical in many biological processes.3 For example, during D. discoideum social development, genetically identical cells differentiate into stalk cells, which vacuolate and die, and spore cells, which enter a resting state until their dispersal to a more favorable environment.66 This profound difference in cell fate is based on differences in enzyme activity between cells, and validated tools to measure enzyme activity at the single-cell level are necessary to study the role of cellular heterogeneity in this process. Although the work presented here was conducted in cell lysates, peptide substrate reporters combined with capillary or microchip electrophoresis are a well-established method for single-cell enzyme assays.3,65 One challenge to single-cell assays with peptide substrate reporters is stabilizing reporters in intact cells, where degradation rates are an important consideration. As evidenced by the short half-life of VI–B in T. thermophila lysates, reporters that have been iteratively optimized in one species may not retain the same level of optimization in new species. Thus, further research is needed to determine which peptide stabilization strategies are effective across species. A deeper understanding of how peptide substrates behave in evolutionarily diverse cells will facilitate the full breadth of applications of these tools from basic biology to human health.
Acknowledgments
The authors thank Eric Cole for advice on T. thermophila conjugation; Hebe Guardiola-Diaz for assistance with Western blotting; Qunzhao Wang of the Lawrence laboratory at the University of North Carolina for synthesis of the VI–B peptide fragment standards and the Allbritton laboratory, currently at the University of Washington, for providing them; and Emilie Mainz, Jeremiah Marden, Angela Proctor, and Abigail Turner for helpful discussions. TOC graphic made using biorender.com. This material is based upon work supported by the National Science Foundation under RUI grants MCB-1615482 and MCB-2126177 and by Trinity College.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsmeasuresciau.4c00030.
Author Present Address
# M.J.F.: Department of Chemistry, Weinberg College of Arts and Science, Northwestern University, Evanston, Illinois 60208, USA
Author Present Address
§ K.Y.: Department of Chemistry, Michigan State University, East Lansing, Michigan 48824, USA.
Author Present Address
$ R.S.C.: Division of Chemistry and Chemical Engineering, California Institute of Technology, Pasadena, California 91125, USA. ORCID: https://orcid.org/0000-0002-3805-6144
Author Contributions
† M.J.F. and M.M. contributed equally to this paper. CRediT: Mengqi Jonathan Fan investigation, methodology, visualization, writing-original draft, writing-review & editing; Misha Mehra investigation, methodology, writing-original draft, writing-review & editing; Kunwei Yang methodology, writing-original draft, writing-review & editing; Rahuljeet S. Chadha investigation, methodology, visualization; Sababa Anber investigation, methodology; Michelle L. Kovarik conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, project administration, resources, supervision, visualization, writing-original draft, writing-review & editing.
The authors declare no competing financial interest.
Supplementary Material
References
- Hardie D. G.Peptide Assay of Protein Kinases and Use of Variant Peptides to Determine Recognition Motifs. In Stress Response; Humana Press: NJ, 2000; Vol. 99, pp 191–201. 10.1385/1-59259-054-3:191 [DOI] [PubMed] [Google Scholar]
- Wu D.; Sylvester J. E.; Parker L. L.; Zhou G.; Kron S. J. Peptide Reporters of Kinase Activity in Whole Cell Lysates. Biopolymers 2010, 94 (4), 475–486. 10.1002/bip.21401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovarik M. L.; Allbritton N. L. Measuring Enzyme Activity in Single Cells. Trends Biotechnol. 2011, 29 (5), 222–230. 10.1016/j.tibtech.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shults M. D.; Imperiali B. Versatile Fluorescence Probes of Protein Kinase Activity. J. Am. Chem. Soc. 2003, 125 (47), 14248–14249. 10.1021/ja0380502. [DOI] [PubMed] [Google Scholar]
- Shults M. D.; Kozlov I. A.; Nelson N.; Kermani B. G.; Melnyk P. C.; Shevchenko V.; Srinivasan A.; Musmacker J.; Hachmann J. P.; Barker D. L.; Lebl M.; Zhao C. A Multiplexed Protein Kinase Assay. ChemBioChem. 2007, 8 (8), 933–942. 10.1002/cbic.200600522. [DOI] [PubMed] [Google Scholar]
- Proctor A.; Wang Q.; Lawrence D. S.; Allbritton N. L. Development of a Peptidase-Resistant Substrate for Single-Cell Measurement of Protein Kinase B Activation. Anal. Chem. 2012, 84 (16), 7195–7202. 10.1021/ac301489d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner A. H.; Lebhar M. S.; Proctor A.; Wang Q.; Lawrence D. S.; Allbritton N. L. Rational Design of a Dephosphorylation-Resistant Reporter Enables Single-Cell Measurement of Tyrosine Kinase Activity. ACS Chem. Biol. 2016, 11 (2), 355–362. 10.1021/acschembio.5b00667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor A.; Wang Q.; Lawrence D. S.; Allbritton N. L. Selection and Optimization of Enzyme Reporters for Chemical Cytometry. Methods Enzymol 2019, 622, 221–248. 10.1016/bs.mie.2019.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallorin L.; Wang J.; Kim W. E.; Sahu S.; Kosa N. M.; Yang P.; Thompson M.; Gilson M. K.; Frazier P. I.; Burkart M. D.; Gianneschi N. C. Discovering de Novo Peptide Substrates for Enzymes Using Machine Learning. Nat. Commun. 2018, 9 (1), 5253. 10.1038/s41467-018-07717-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.-A.; Yeh R.-H.; Lawrence D. S. Design and Synthesis of a Fluorescent Reporter of Protein Kinase Activity. J. Am. Chem. Soc. 2002, 124 (15), 3840–3841. 10.1021/ja017530v. [DOI] [PubMed] [Google Scholar]
- Wang Q.; Cahill S. M.; Blumenstein M.; Lawrence D. S. Self-Reporting Fluorescent Substrates of Protein Tyrosine Kinases. J. Am. Chem. Soc. 2006, 128 (6), 1808–1809. 10.1021/ja0577692. [DOI] [PubMed] [Google Scholar]
- Bozinovski S.; Cristiano B. E.; Marmy-Conus N.; Pearson R. B. The Synthetic Peptide RPRAATF Allows Specific Assay of Akt Activity in Cell Lysates. Anal. Biochem. 2002, 305 (1), 32–39. 10.1006/abio.2002.5659. [DOI] [PubMed] [Google Scholar]
- Proctor A.; Wang Q.; Lawrence D. S.; Allbritton N. L. Development of a Peptidase-Resistant Substrate for Single-Cell Measurement of Protein Kinase B Activation. Anal. Chem. 2012, 84 (16), 7195–7202. 10.1021/ac301489d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anttila M. M.; Vickerman B. M.; Wang Q.; Lawrence D. S.; Allbritton N. L. A Novel Class of Photoactivatable Reporter to Perform Multiplexed and Temporally Controlled Measurements of Kinase and Protease Activity in Single Cells. Anal. Chem. 2021, 93 (49), 16664–16672. 10.1021/acs.analchem.1c04225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Banion C. P.; Priestman M. A.; Hughes R. M.; Herring L. E.; Capuzzi S. J.; Lawrence D. S. Design and Profiling of a Subcellular Targeted Optogenetic cAMP-Dependent Protein Kinase. Cell Chem. Biol. 2018, 25 (1), 100–109.e8. 10.1016/j.chembiol.2017.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soamalala J.; Diot S.; Pellerano M.; Blanquart C.; Galibert M.; Jullian M.; Puget K.; Morris M. C. Fluorescent Peptide Biosensor for Probing CDK6 Kinase Activity in Lung Cancer Cell Extracts. ChemBioChem. 2021, 22 (6), 1065–1071. 10.1002/cbic.202000677. [DOI] [PubMed] [Google Scholar]
- Phillips R. M.; Bair E.; Lawrence D. S.; Sims C. E.; Allbritton N. L. Measurement of Protein Tyrosine Phosphatase Activity in Single Cells by Capillary Electrophoresis. Anal. Chem. 2013, 85 (12), 6136–6142. 10.1021/ac401106e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guetschow E. D.; Kumar S.; Lombard D. B.; Kennedy R. T. Identification of Sirtuin 5 Inhibitors by Ultrafast Microchip Electrophoresis Using Nanoliter Volume Samples. Anal Bioanal Chem. 2016, 408 (3), 721–731. 10.1007/s00216-015-9206-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezovski M.; Li W.-P.; Poulter C. D.; Krylov S. N. Measuring the Activity of Farnesyltransferase by Capillary Electrophoresis with Laser-Induced Fluorescence Detection. Electrophoresis 2002, 23 (19), 3398–3403. . [DOI] [PubMed] [Google Scholar]
- Arkhipov S. N.; Berezovski M.; Jitkova J.; Krylov S. N. Chemical Cytometry for Monitoring Metabolism of a Ras-Mimicking Substrate in Single Cells. Cytometry, Part A 2005, 63A (1), 41–47. 10.1002/cyto.a.20100. [DOI] [PubMed] [Google Scholar]
- Manning G.; Hunter T.. Chapter 56 - Eukaryotic Kinomes: Genomics and Evolution of Protein Kinases. In Handbook of Cell Signaling (Second ed.); Bradshaw R. A., Dennis E. A., Eds.; Elsevier: San Diego, 2010; pp 393–397. 10.1016/B978-0-12-374145-5.00056-5 [DOI] [Google Scholar]
- Protein Kinase Evolution, http://kinase.com/web/current/evolution/.
- Alessi D. R.; Cohen P. Mechanism of Activation and Function of Protein Kinase B. Current Opinion in Genetics & Development 1998, 8 (1), 55–62. 10.1016/S0959-437X(98)80062-2. [DOI] [PubMed] [Google Scholar]
- Carnero A. The PKB/AKT Pathway in Cancer. Curr. Pharm. Des. 2010, 16 (1), 34–44. 10.2174/138161210789941865. [DOI] [PubMed] [Google Scholar]
- Alessi D. R.; Barry Caudwell F.; Andjelkovic M.; Hemmings B. A.; Cohen P. Molecular Basis for the Substrate Specificity of Protein Kinase B; Comparison with MAPKAP Kinase-1 and P70 S6 Kinase. FEBS Lett. 1996, 399 (3), 333–338. 10.1016/S0014-5793(96)01370-1. [DOI] [PubMed] [Google Scholar]
- Cross D. A. E.; Alessi D. R.; Cohen P.; Andjelkovich M.; Hemmings B. A. Inhibition of Glycogen Synthase Kinase-3 by Insulin Mediated by Protein Kinase B. Nature 1995, 378 (6559), 785–789. 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- Li Y.; Park J.; Piao L.; Kong G.; Kim Y.; Park K. A.; Zhang T.; Hong J.; Hur G. M.; Seok J. H.; Choi S.-W.; Yoo B. C.; Hemmings B. A.; Brazil D. P.; Kim S.-H.; Park J. PKB-Mediated PHF20 Phosphorylation on Ser291 Is Required for P53 Function in DNA Damage. Cell. Signal. 2013, 25 (1), 74–84. 10.1016/j.cellsig.2012.09.009. [DOI] [PubMed] [Google Scholar]
- Proctor A.; Herrera-Loeza S. G.; Wang Q.; Lawrence D. S.; Yeh J. J.; Allbritton N. L. Measurement of Protein Kinase B Activity in Single Primary Human Pancreatic Cancer Cells. Anal. Chem. 2014, 86 (9), 4573–4580. 10.1021/ac500616q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainz E. R.; Serafin D. S.; Nguyen T. T.; Tarrant T. K.; Sims C. E.; Allbritton N. L. Single Cell Chemical Cytometry of Akt Activity in Rheumatoid Arthritis and Normal Fibroblast-like Synoviocytes in Response to Tumor Necrosis Factor α. Anal. Chem. 2016, 88 (15), 7786–7792. 10.1021/acs.analchem.6b01801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan A. L.; Dutcher S. K.; Stormo G. D.. Detecting Coevolution of Functionally Related Proteins for Automated Protein Annotation. In 2010 IEEE International Conference on BioInformatics and BioEngineering; IEEE; 2010; pp 99–105. 10.1109/BIBE.2010.24 [DOI] [PMC free article] [PubMed]
- Di Tommaso P.; Moretti S.; Xenarios I.; Orobitg M.; Montanyola A.; Chang J.-M.; Taly J.-F.; Notredame C. T-Coffee: A Web Server for the Multiple Sequence Alignment of Protein and RNA Sequences Using Structural Information and Homology Extension. Nucleic Acids Res. 2011, 39, W13–17. 10.1093/nar/gkr245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Multiple Align Show, https://www.bioinformatics.org/sms/multi_align.html.
- Nellen W.; Silan C.; Firtel R. A. DNA-Mediated Transformation in Dictyostelium Discoideum: Regulated Expression of an Actin Gene Fusion. Mol. Cell. Biol. 1984, 4 (12), 2890–2898. 10.1128/MCB.4.12.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insall R. H.; Soede R. D.; Schaap P.; Devreotes P. N. Two cAMP Receptors Activate Common Signaling Pathways in Dictyostelium. Mol. Biol. Cell 1994, 5 (6), 703–711. 10.1091/mbc.5.6.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fey P.; Kowal A. S.; Gaudet P.; Pilcher K. E.; Chisholm R. L. Protocols for Growth and Development of Dictyostelium Discoideum. Nat. Protoc 2007, 2 (6), 1307–1316. 10.1038/nprot.2007.178. [DOI] [PubMed] [Google Scholar]
- Tetrahymena Stock Center. https://tetrahymena.vet.cornell.edu/.
- Cassidy-Hanley D. M. Tetrahymena in the Laboratory: Strain Resources, Methods for Culture, Maintenance, and Storage. Methods Cell Biol. 2012, 109, 237–276. 10.1016/B978-0-12-385967-9.00008-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tierney A. J.; Pham N.; Yang K.; Emerick B. K.; Kovarik M. L. Interspecies Comparison of Peptide Substrate Reporter Metabolism Using Compartment-Based Modeling. Anal Bioanal Chem. 2017, 409 (5), 1173–1183. 10.1007/s00216-016-0085-9. [DOI] [PubMed] [Google Scholar]
- Nakamura H.; Tamura Z. Fluorometric Determination of Secondary Amines Based on Their Reaction with Fluorescamine. Anal. Chem. 1980, 52 (13), 2087–2092. 10.1021/ac50063a023. [DOI] [Google Scholar]
- Cai H.; Huang C.-H.; Devreotes P. N.; Iijima M. Analysis of Chemotaxis in Dictyostelium. Methods Mol. Biol. 2011, 757, 451–468. 10.1007/978-1-61779-166-6_26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meili R.; Ellsworth C.; Lee S.; Reddy T. B.; Ma H.; Firtel R. A. Chemoattractant-Mediated Transient Activation and Membrane Localization of Akt/PKB Is Required for Efficient Chemotaxis to cAMP in Dictyostelium. EMBO J. 1999, 18 (8), 2092–2105. 10.1093/emboj/18.8.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheow L. F.; Sarkar A.; Kolitz S.; Lauffenburger D.; Han J. Detecting Kinase Activities from Single Cell Lysate Using Concentration-Enhanced Mobility Shift Assay. Anal. Chem. 2014, 86 (15), 7455–7462. 10.1021/ac502185v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leondaritis G.; Siokos J.; Skaripa I.; Galanopoulou D. Genome-Wide Analysis of the Phosphoinositide Kinome from Two Ciliates Reveals Novel Evolutionary Links for Phosphoinositide Kinases in Eukaryotic Cells. PLoS One 2013, 8 (11), e78848 10.1371/journal.pone.0078848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban J.; Soulard A.; Huber A.; Lippman S.; Mukhopadhyay D.; Deloche O.; Wanke V.; Anrather D.; Ammerer G.; Riezman H.; Broach J. R.; De Virgilio C.; Hall M. N.; Loewith R. Sch9 Is a Major Target of TORC1 in Saccharomyces Cerevisiae. Mol. Cell 2007, 26 (5), 663–674. 10.1016/j.molcel.2007.04.020. [DOI] [PubMed] [Google Scholar]
- Casamayor A.; Torrance P. D.; Kobayashi T.; Thorner J.; Alessi D. R. Functional Counterparts of Mammalian Protein Kinases PDK1 and SGK in Budding Yeast. Curr. Biol. 1999, 9 (4), 186–S4. 10.1016/S0960-9822(99)80088-8. [DOI] [PubMed] [Google Scholar]
- Roelants F. M.; Torrance P. D.; Bezman N.; Thorner J. Pkh1 and Pkh2 Differentially Phosphorylate and Activate Ypk1 and Ykr2 and Define Protein Kinase Modules Required for Maintenance of Cell Wall Integrity. Mol. Biol. Cell 2002, 13 (9), 3005–3028. 10.1091/mbc.e02-04-0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y.; Taniguchi R.; Tanoue D.; Yamaji T.; Takematsu H.; Mori K.; Fujita T.; Kawasaki T.; Kozutsumi Y. Sli2 (Ypk1), a Homologue of Mammalian Protein Kinase SGK, Is a Downstream Kinase in the Sphingolipid-Mediated Signaling Pathway of Yeast. Mol. Cell. Biol. 2000, 20 (12), 4411–4419. 10.1128/MCB.20.12.4411-4419.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimura Y.; Tang M.; Devreotes P. Assays for Chemotaxis and Chemoattractant-Stimulated TorC2 Activation and PKB Substrate Phosphorylation in Dictyostelium. Methods Mol. Biol. 2009, 571, 255–270. 10.1007/978-1-60761-198-1_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csaba G. The Hormonal System of the Unicellular Tetrahymena: A Review with Evolutionary Aspects. Acta Microbiol Immunol Hung 2012, 59 (2), 131–156. 10.1556/amicr.59.2012.2.1. [DOI] [PubMed] [Google Scholar]
- Mainz E. R.; Wang Q.; Lawrence D. S.; Allbritton N. L. An Integrated Chemical Cytometry Method: Shining a Light on Akt Activity in Single Cells. Angew. Chem., Int. Ed. 2016, 55 (42), 13095–13098. 10.1002/anie.201606914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm R. L.; Firtel R. A. Insights into Morphogenesis from a Simple Developmental System. Nat. Rev. Mol. Cell Biol. 2004, 5 (7), 531–541. 10.1038/nrm1427. [DOI] [PubMed] [Google Scholar]
- Loomis W. F.Dictyostelium Discoideum: A Developmental System; Elsevier: New York, 1975. [Google Scholar]
- Liao X.-H.; Buggey J.; Kimmel A. R. Chemotactic Activation of Dictyostelium AGC-Family Kinases AKT and PKBR1 Requires Separate but Coordinated Functions of PDK1 and TORC2. J. Cell. Sci. 2010, 123 (Pt 6), 983–992. 10.1242/jcs.064022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimura Y.; Xiong Y.; Iglesias P. A.; Hoeller O.; Bolourani P.; Devreotes P. N. PIP3-Independent Activation of TorC2 and PKB at the Cell’s Leading Edge Mediates Chemotaxis. Curr. Biol. 2008, 18 (14), 1034–1043. 10.1016/j.cub.2008.06.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meili R.; Ellsworth C.; Firtel R. A. A Novel Akt/PKB-Related Kinase Is Essential for Morphogenesis in Dictyostelium. Curr. Biol. 2000, 10 (12), 708–717. 10.1016/S0960-9822(00)00536-4. [DOI] [PubMed] [Google Scholar]
- Rupper A.; Lee K.; Knecht D.; Cardelli J. Sequential Activities of Phosphoinositide 3-Kinase, PKB/Akt, and Rab7 during Macropinosome Formation in Dictyostelium. Mol. Biol. Cell 2001, 12 (9), 2813–2824. 10.1091/mbc.12.9.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier B.; Zielinski A.; Weber C.; Arcizet D.; Youssef S.; Franosch T.; Radler J. O.; Heinrich D. Chemotactic Cell Trapping in Controlled Alternating Gradient Fields. Proc. Natl. Acad. Sci. U. S. A. 2011, 108 (28), 11417–11422. 10.1073/pnas.1014853108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg J. M.; Manning G.; Liu A.; Fey P.; Pilcher K. E.; Xu Y.; Smith J. L. The Dictyostelium Kinome—Analysis of the Protein Kinases from a Simple Model Organism. PLoS Genet 2006, 2 (3), e38 10.1371/journal.pgen.0020038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King J. S.; Insall R. H. Chemotaxis: TorC before You Akt···. Curr. Biol. 2008, 18 (18), R864–R866. 10.1016/j.cub.2008.07.051. [DOI] [PubMed] [Google Scholar]
- Bradley D.; Beltrao P. Evolution of Protein Kinase Substrate Recognition at the Active Site. PLOS Biology 2019, 17 (6), e3000341 10.1371/journal.pbio.3000341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page M. J.; Di Cera E. Evolution of Peptidase Diversity. J. Biol. Chem. 2008, 283 (44), 30010–30014. 10.1074/jbc.M804650200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyfert H. M.; Hipke H.; Schmidt W. Isolation and Phenotypic Characterization of Tetrahymena Thermophila Size Mutants: The Relationship between Cell Size and Regulation of DNA Content. J. Cell Sci. 1984, 67, 203–215. 10.1242/jcs.67.1.203. [DOI] [PubMed] [Google Scholar]
- Chan Y.-H. M.; Marshall W. F. Organelle Size Scaling of the Budding Yeast Vacuole Is Tuned by Membrane Trafficking Rates. Biophys. J. 2014, 106 (9), 1986–1996. 10.1016/j.bpj.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.; Proctor A.; Cline L. L.; Houston K. M.; Waters M. L.; Allbritton N. L. β-Turn Sequences Promote Stability of Peptide Substrates for Kinases within the Cytosolic Environment. Analyst 2013, 138 (15), 4305. 10.1039/c3an00874f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickerman B. M.; Anttila M. M.; Petersen B. V.; Allbritton N. L.; Lawrence D. S. Design and Application of Sensors for Chemical Cytometry. ACS Chem. Biol. 2018, 13 (7), 1741–1751. 10.1021/acschembio.7b01009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annesley S. J.; Fisher P. R. Dictyostelium Discoideum--a Model for Many Reasons. Mol. Cell. Biochem. 2009, 329 (1–2), 73–91. 10.1007/s11010-009-0111-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.