Abstract
Background
Chronic bronchopulmonary infection due to MRSA in people with cystic fibrosis (pwCF) has been associated with accelerated decline in lung function, increased hospitalizations and increased mortality.
Material and methods
We studied microbiological and genomic characteristics of MRSA isolates recovered from pwCF in two Spanish multicentre studies (2013, 2021). Antimicrobial susceptibility was performed. WGS was carried out to determine population structure [MLST, spa-typing, staphylococcal cassette chromosome mec (SCCmec)], resistome and virulome. Clinical charts of MRSA-infected and MRSA-non-infected pwCF were also reviewed.
Results
MRSA infection prevalence decreased between 2013 (29/341, 8.5%) and 2021 (21/326, 6.4%) (P = 0.378). Differences in lung function were observed between infected and non-infected patients (P < 0.005). A higher prevalence of hospital-acquired (HA) clones was found compared with community-acquired (CA) clones (2013: 67% versus 33%; and 2021: 71% versus 29%). Overall, we noted clustering of isolates based on year of sampling, type of acquisition and clonal complex (CC). HA-MRSA population was dominated by CC5, with ST125-MRSA-IVc-t067 the most prevalent lineage (37%). A higher clonal diversity was detected among CA-MRSA. One Panton–Valentine leucocidin (PVL)-positive strain (ST8-MRSA-IV) and three strains of porcine origin (two ST398-MRSA-V-t011, one ST398-MRSA-V-t8567) were found. Additionally, acquired resistance genes (n = 24) were detected, including the cfr gene conferring linezolid resistance. A higher gentamicin resistance was found in 2021 (42%) compared with 2013 (7%) (P = 0.046), associated with the aac(6′)-aph(2″) gene.
Conclusions
Despite a decrease in MRSA prevalence, we showed its potential impact on CF severity and progression. Moreover, we observed great genotypic and phenotypic diversity in MRSA isolates from pwCF as well as an MDR trait.
Introduction
Chronic lung infections remain one of the main causes of morbidity and mortality in people with cystic fibrosis (pwCF).1 Among the implicated pathogens, MRSA is particularly significant due to its ability to colonize and cause recurrent and persistent infections. The role of MRSA in exacerbations and its impact on disease progression is attributed to its capacity to induce chronic inflammation, loss of lung function and resistance to multiple antimicrobials.2 A decline in the forced expiratory volume in 1 s (FEV1) is a hallmark of CF progression and is often indicative of worsening lung function.1
Epidemiological studies in Spain have identified significant genetic diversity in MRSA isolates from pwCF, with the ST125/staphylococcal cassette chromosome mec (SCCmec)IV/t067, an allelic variant of ST5, being one of the most prevalent hospital-associated MRSA (HA-MRSA) clones.3–5 Community-associated MRSA (CA-MRSA) clones such as ST30 [clonal complex 30 (CC30)] have also been detected in pwCF. Additionally, a diverse distribution of SCCmec complexes has been observed, with types IV and I generally associated with HA-MRSA, and types IV, VI, VII and IX more related to CA-MRSA.6 Beyond their antibiotic resistance, MRSA also produces virulence factors like Panton–Valentine leucocidin (PVL) (such as lukS-PV and lukF-PV genes) and staphylococcal enterotoxins (SEs), causing tissue damage.7 Additionally, the tst-1 gene encodes toxic shock syndrome toxin-1 protein (TSST-1), leading to toxic shock syndrome, influenced by environmental and genetic factors.8,9
In this study, we present a detailed microbiological characterization of MRSA isolates recovered from pwCF during two multicentre studies conducted in 2013 and 2021 in Spain. Our primary objectives were to determine the prevalence of MRSA pathogenic colonization (hereinafter infection) in pwCF before and after the COVID-19 pandemic, to evaluate the antimicrobial resistance profiles of MRSA isolates and to investigate their genetic diversity in our country.
Material and methods
Study isolates and data collection
In two national multicentre studies conducted in 2013 and 2021, a total of 667 sputum samples (one per patient) were recovered in different CF units from Spanish hospitals (341 in 2013 and 326 in 2021). Overall, samples were collected from 17 centres in 2013 and from 14 centres in 2021. Among these samples, Staphylococcus aureus was detected in 59.9% of pwCF [206/341 (60%) in 2013 and 194/326 (59.5%) in 2021] (Table S1, available as Supplementary data at JAC-AMR Online). Among all patients/samples, 50 isolates were identified as MRSA, 29 in 2013 and 21 in 2021. This subset was selected for antimicrobial susceptibility testing and genomic characterization. All isolates were recovered as part of the routine management of the CF patients and were stored at −80°C for further analysis. Bacterial identification was performed using MALDI-TOF (Bruker, Germany).
Demographic data from patients with MRSA including age, gender or cystic fibrosis transmembrane conductance regulator (CFTR) mutation were recorded. Data regarding presence of pancreatic insufficiency, CF-related diabetes, alterations in carbohydrate metabolism, and the highest percentage predicted FEV1 value, such as exacerbations, days of hospitalization and antibiotic use, were also collected from the year prior to the inclusion in the study. Nevertheless, we used the criteria established in the ‘Spanish Consensus on the Prevention and Treatment of Pseudomonas aeruginosa Bronchial Infections in Cystic Fibrosis Patients’ document that considers pulmonary exacerbation a clinical situation characterized by changes in existing respiratory symptoms and the appearance of new symptoms followed by a decline in respiratory function.10
Antimicrobial susceptibility testing
Methicillin resistance was detected using the 30 μg cefoxitin disc susceptibility. We also determined the antimicrobial susceptibility of gentamicin, levofloxacin, erythromycin, clindamycin, linezolid and rifampicin by agar disc diffusion, and results were interpreted following EUCAST-2024 criteria (https://www.eucast.org/). Inducible resistance to clindamycin was detected by the erythromycin and clindamycin disc approximation test (D-zone test).11
WGS and bioinformatics analysis
Genomic DNA extraction was carried out using the commercial QIAamp DNA Mini Kit QIAgen (Hilden, Germany). WGS was performed using the Illumina NovaSeq 6000 platform (Oxford Genomics Centre, Oxford, UK), with 2 × 150 bp paired-end reads. Sequencing, processing, annotation and bacterial identification were performed as previously described.12
The Bactopia tool was employed to determine MLST, spa and SCCmec type.13 The MRSA strains were defined as HA-MRSA and CA-MRSA based on their belonging to the clonal complex defined by MLST, spa, SCCmec and agr operon types detected.14 Minimum spanning trees based on the MLST were constructed using the algorithm goeBURST (http://www.phyloviz.net/). Assembled genomes were analysed for acquired resistance genes, virulence factors and plasmid replicons using Abricate v.1.0.1 employing the ARG-ANNOT, Resfinder, VFDB and PlasmidFinder databases (threshold: 95% identity/90% coverage). Core genomes were obtained using Snippy (v4.4.3) and S. aureus NCTC 8325 (NC_007795.1) as the reference genome. A core genome maximum-likelihood phylogenetic tree was constructed and visualized using IQtree2 and Microreact (https://microreact.org/showcase), respectively.
The genomes were deposited at DDBJ/ENA/GenBank under the project number PRJNA985233 (JAUBWX000000000-JAUBYU000000000).
Statistical analysis
Differences of clinical characteristics, FEV1 and infection were tested by Fisher’s exact test. A P value of <0.05 was considered significant at a 95% CI. All tests were performed using R software (RStudio, Boston, MA; http://www.rstudio.com/) and GraphPad Prism 8 (San Diego, CA, USA) software.
Results
Demographic data, pathogenic colonization status, pulmonary function and modulator therapy
Comparing both studies, a decrease in MRSA prevalence was observed between 2013 (8.5%, 29/341 patients) and 2021 (6.4%, 21/326 patients), although it was not statistically significant (P = 0.378). Additionally, the percentage of patients infected with MRSA among those infected with S. aureus (MSSA + MRSA) was 14.1% (29/206) and 10.8% (21/194) for the years 2013 and 2021, respectively. Up to 14 (28%) MRSA-infected patients had the F508del mutation in homozygosis, 20 (40%) in heterozygosis, and 16 (32%) had other different mutations.
In both years, statistically significant differences were observed in lung function (FEV1) [%, Q3 (quartil 3)-Q1 (quartil 1)] between MRSA-infected and MSSA-infected patients in both studies [62.0% (Q3-Q1: 75–35) versus 72.9% (Q3-Q1: 86–50) (P < 0.01) in 2013; and 65.3% (Q3-Q1: 84–41) versus 76.3% (Q3-Q1: 98–56) (P < 0.02) in 2021]. A trend towards a higher number of exacerbations in pwCF infected by MRSA versus MSSA was observed in the 2013 study (75.8% versus 70.0%; P = 0.43), which was significant in the 2021 study (57.0% versus 32.0%; P = 0.041). Pathogenic colonization by P. aeruginosa was also more frequent among patients with a positive culture for MRSA (P < 0.001), occurring in 76.0% (38/50) of cases. The colonization status by P. aeruginosa was chronic in 20 patients (40%) and intermittent in 18 (36%) with no differences between years.
The clinical characteristics of the patients infected and non-infected by an MRSA strain are shown in Table 1. The proportion of HA-MRSA and CA-MRSA isolates detected was 68% (65.0% in 2013 and 71.4% in 2021) and 32.0% (35.0% in 2013 and 28.6% in 2021), respectively. The median age [p75 (75th percentil) - p25 (25th percetil)] of patients infected with CA-MRSA strains was similar to that of those infected with HA-MRSA strains [20 (p75-p25: 28–13) versus 21 (p75-p25: 28–16) years]. Clinical and demographic characteristics of both patient populations are shown in Table S2.
Table 1.
Clinical characteristics and demographic data of patients
| MRSA | MSSA | P valuea | |||||
|---|---|---|---|---|---|---|---|
| 2013 | 2021 | Total | 2013 | 2021 | Total | ||
| No. of patients | 29 | 21 | 50 | 177 | 173 | 350 | — |
| No. females (%) | 13 (45) | 11 (52) | 25 (49) | 76 (42.9) | 80 (46.2) | 156 (44.5) | — |
| Age, y | |||||||
| Mean (SD) | 21 (10) | 25 (11) | 22 (11) | 18.2 (10) | 20.2 (10) | 19.1 (10) | — |
| Range (Q3–Q1)b | 27–13 | 30–17 | 27–15 | 24–8 | 25–9 | 25–9 | — |
| Mean FEV1% (Q3–Q1)b | 63.15 [84–41] | 67.3 [85–41] | 65.3 [84–41] | 79 [89–50] | 92.8 [99–57] | 87.3 [98–56] | <0.02 |
| 0–10 y: No. of pwCF (FEV1 %) | 2 (89.5) | 3 (85.3) | 5 (86.9) | 47 (82.5) | 41 (98.6) | 88 (90) | — |
| 11–20 y: No. of pwCF (FEV1 %) | 5 (83.4) | 14 (76.7) | 19 (77) | 58 (84.5) | 71 (84.1) | 129 (84.2) | — |
| 21–30 y: No. of pwCF (FEV1 %) | 9 (48.11) | 7 (70.1) | 16 (57.7) | 45 (60) | 32 (96) | 77 (75) | — |
| 31–40 y: No. of pwCF (FEV1 %) | 2 (68.5) | 4 (65) | 6 (66.1) | 21 (59) | 19 (78.8) | 40 (68) | — |
| >40 y: No. of pwCF (FEV1 %) | 3 (68.7) | 1 (68.5) | 4 (68.7) | 6 (55.8) | 10 (49.8) | 16 (52) | — |
| P. aeruginosa co-infection | 22 (76) | 16 (76) | 38 (76) | 123 (70) | 63 (36) | 186 (53) | <0.01 |
| Intermittent | 9 (31) | 9 (43) | 18 (36) | 74 (41.8) | 25 (14) | 99 (28) | 0.32 |
| Chronic | 13 (45) | 7 (33) | 20 (40) | 57 (32.2) | 30 (22) | 87 (25) | <0.02 |
| Pulmonary exacerbation, n (%) | 22 (76) | 12 (57) | 34 (68) | 135 (76.2) | 79 (45) | 214 (61.3) | 0.58 |
| Pancreatic insufficiency, n (%) | 28 (99) | 19 (90.4) | 47 (94.2) | 154 (87) | 139 (80.1) | 293 (83.7) | 0.09 |
| Hydrocarbon intolerance, n (%) | 6 (21) | 11 (52.3) | 16 (31.2) | 141 (79.6) | 70 (40) | 211 (60.3) | 0.39 |
| Insulin therapy, n (%) | 2 (7) | 3 (14.2) | 5 (9.8) | 132 (74.5) | 56 (32.3) | 188 (53.8) | <0.01 |
aThe statistical differences observed are between MRSA and MSSA, taking into account the total number of isolates and not study years. Bold values indicate statistical significance.
bQ3-Q1, quartil 3-quartil 1.
In the 2013 study, therapy with CFTR modulators was not recorded due to their very limited use at the time. In the 2021 study, nine patients (43%) received a CFTR modulator. The most commonly used combination was ivacaftor + tezacaftor (n = 6), followed by ivacaftor alone (n = 2), and only one patient received elexacaftor + tezacaftor + ivacaftor. The median FEV1 among these patients was 48%, with four patients having less than 40% lung function. Only three patients had chronic colonization by P. aeruginosa and MRSA, whereas 66% (n = 6) experienced an exacerbation.
Antimicrobial susceptibility
The antimicrobial susceptibility of the isolates is shown in Figure S1. A total of 96.0% of the isolates were MDR and 4.0% were XDR. Generally, high resistance rates were observed in quinolones (78.0%) and aminoglycosides (24.0%) in both study years. Among the isolates resistant to erythromycin, the macrolides, lincosamides and streptogramins B (MLSB) constitutive phenotype was detected in 56.0% of the isolates, and the MLSB inducible phenotype in 22%. When comparing the two study years, we observed very similar resistance rates for all tested antimicrobials except for gentamicin. The percentage of gentamicin-resistant isolates (42.0%) was higher in 2021 than in 2013 (7.0%) (P = 0.047). It is worth noting that we found three isolates resistant to linezolid using diffusion discs (one in 2013 and two in 2021).
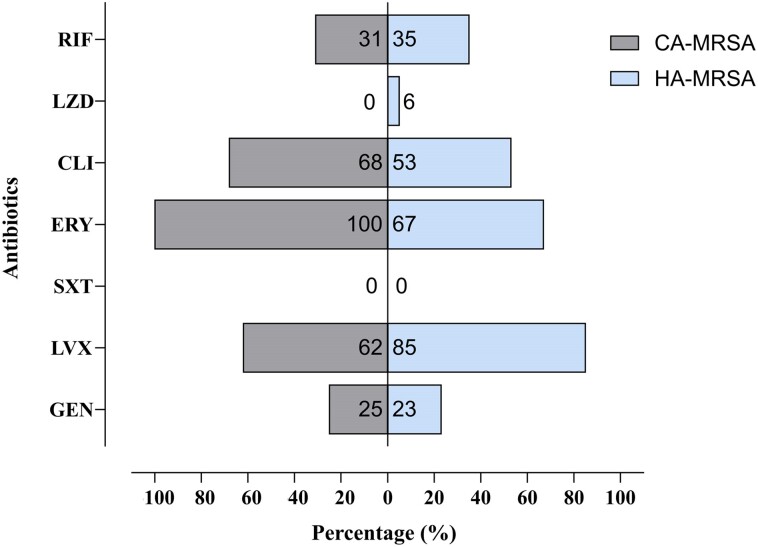
Considering the distinction between HA-MRSA and CA-MRSA strains, higher resistance rates were found to quinolones in HA-MRSA strains (85.0%) than in CA-MRSA (62.5%), whereas CA-MRSA strains showed higher MIC values of macrolides (100% versus 67.0%) and clindamycin (69.0% versus 56.0%) (Figure 1).
Figure 1.
Percentage of resistant strains in HA-MRSA and CA-MRSA. CLI, clindamycin; ERY, erythromycin; GEN, gentamicin; LVX, levofloxacin; LZD, linezolid; RIF, rifampicin; SXT, trimethoprim/sulfamethoxazole.
Bacterial typing and WGS
Genome characteristics
The genome size of our isolates was 2.69–5.51 Mb, with a G + C content of 33.0% and an average of 2845.32 protein-coding sequences. Genomic information based on contig size 500 bp is detailed in Table S3. All isolates were identified as S. aureus.
Population structure
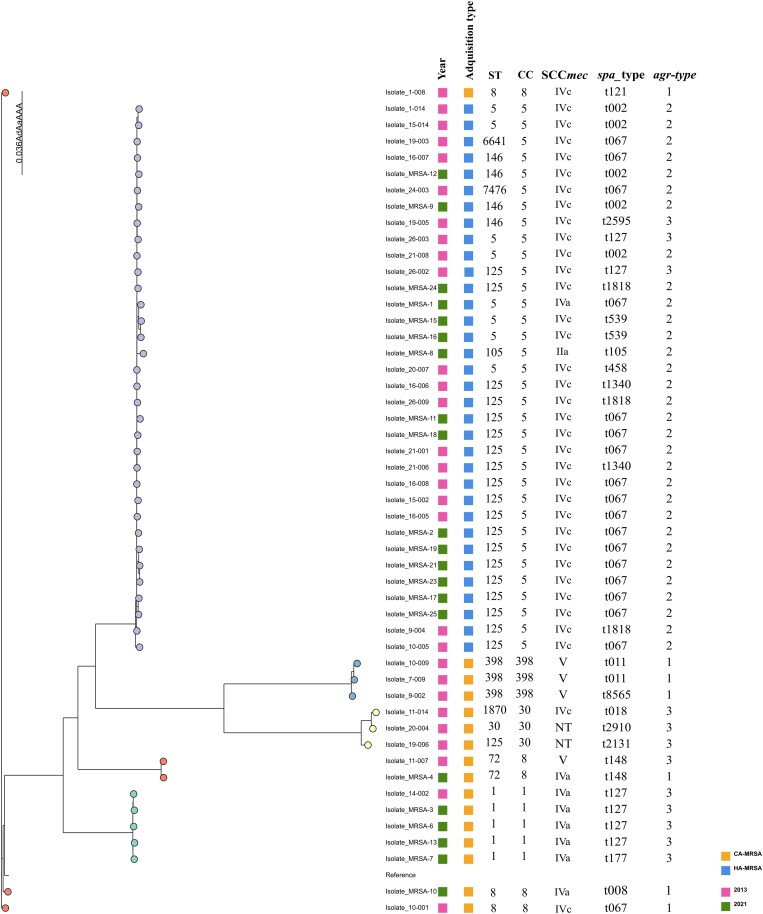
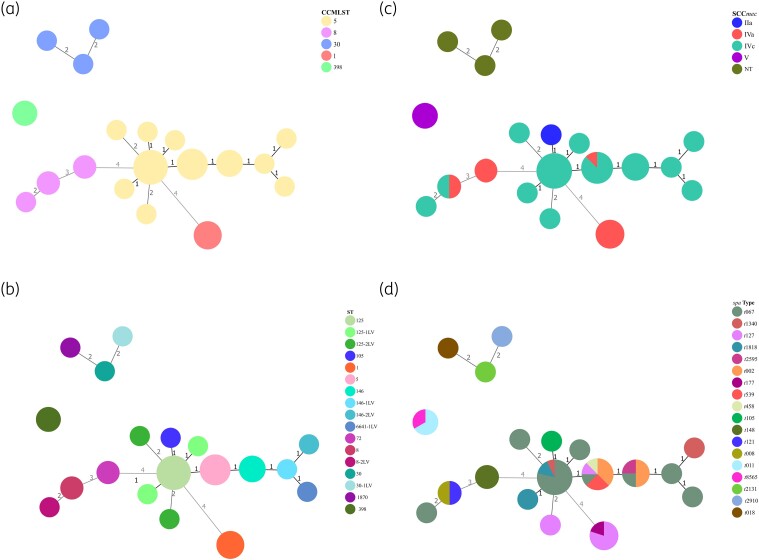
A total of 18 different spa types were obtained. Overall, t067 was the most frequent (18/50, 36%), followed by t127 (6/50, 12%) and t002 (5/50, 10%). The most prevalent operon agr types obtained were type II (33/50, 66%) and type III (11/50, 22%), with type I being the least prevalent (6/50, 12%). MLST showed a lower degree of diversity, revealing the existence of 12 different STs, with ST125 (14/50, 30%) being predominant, followed by ST5 (8/50, 16%). Regarding the obtained clonal complexes, CC5 was the most frequent [68% (32/50)] and was distributed across various hospitals without predominance in any of them. To a lesser extent, we also detected CC1 (10%), CC8 (6%), CC30 (6%), CC398 (6%) and CC72 (4%). The predominant SCCmec type was IVc (34/50, 70%), followed by type IVa (8/50, 16%) and type V (4/50, 8%). One strain presented an SCCmec type IIa, and three others were non-typable (ST30, spa t2131, agr III; ST30-1LV, spa t2910, agr III; and ST1870, spa t018, agr III) (Figure 2). Among the CA-MRSA strains, three isolates (two in 2013 and one in 2021) belonged to the porcine clone ST398-MRSA-V, two of which had the spa type t011 and another one the spa type t8565 (Figure 3).
Figure 2.
Core genome maximum-likelihood phylogenetic tree of all MRSA isolates and S. aureus reference genome (NC_007795.1) analysed by WGS.
Figure 3.
Minimum spanning trees constructed using the algorithm goeBURST (http://www.phyloviz.net/) and annotated by: (a) clonal complex multilocus sequence type (CCMLST); (b) sequence type (ST); (c) staphylococcal cassette chromosome mec (SCCmec); and (d) spa type.
The core genome phylogenetic analysis revealed that our MRSA isolates clustered by acquisition type and MLST results. Moreover, the isolates were also grouped according to the year of collection.
Antibiotic resistance genes
The WGS analysis confirmed the presence of genes conferring resistance to aminoglycosides [aac(6′)-aph(2″), aadD, ant(6)-Ia, ant(9)-Ia, aph(3′)-IIb, aph(3′)-III], penicillins (blaZ, mecA), chloramphenicol (catB7), trimethoprim/sulfamethoxazole (dfrK), tetracyclines [tet(K), tet(L), tet(M)] and macrolides [erm(A), erm(C), mph(C), msr(A)]. We detected the aph(2″)-aac(6′) gene related to gentamicin resistance in eight (72%) MRSA isolates, most of them from the 2021 study (6/8).
We found the cfr gene in three isolates, but resistance to linezolid by disc diffusion was detected in only one of them (ST125 CC5 IVc, t1340). The other MRSA carrying the cfr gene belonged to the same clonal complex (CC5) but varied in the ST (5 and 125) and the spa type (t458 and t002). Both isolates also exhibited resistance to aminoglycosides, tetracyclines and macrolides.
Regarding the two XDR isolates, both carried aph(2″)-aac(6′), mecA, erm(C) and mup(A) genes. Additionally, one of them presented resistance genes to erythromycin and azithromycin [mph(C) and msr(A), respectively].
Only 6 of the 21 isolates (28.6%) resistant to tetracycline carried tet(K), tet(L) or/and tet(M) genes. The gene related to resistance to trimethoprim/sulfamethoxazole, dfrK, was found in five isolates that were susceptible to this antibiotic. However, dfrK was not detected in the only isolate that was resistant to trimethoprim/sulfamethoxazole. The resistome of the remaining isolates is shown in Figure S2.
Virulome
Overall, the virulence genes detected in our isolates were related to adherence, biofilm formation, capsular polysaccharide expression, toxin and cytotoxin production, immune modulation, iron scavenging, protease production and type VII/ESS secretion system (Table S4, Figure S3).
The lukS-PV and lukF-PV virulence factors, encoding the PVL toxin, were detected in one isolate (ST8, CC8, IVa, t008, gp1), which also carried other virulence genes such as selq and selk, precursors of staphylococcal enterotoxins. Regarding the tst-1 gene, it was detected in four CA-MRSA isolates from the 2013 study, with different STs (72, 1870, 125, 30) and belonging to CC30, except for one grouped into CC8.
Discussion
Among the bacteria that cause lung infections in pwCF, MRSA has increasingly been detected, affecting up to 25% of individuals with this disease. Additionally, MRSA is resistant to multiple antibiotics, and bronchopulmonary infections caused by this bacterium often become chronic.1 According to recent European reports, the overall prevalence of MRSA pathogenic colonization is decreasing.15 Coincidentally, we found a downward trend of MRSA prevalence between 2013 (8.5%) and 2021 (6.4%), although this was higher than that in a previous study performed also in our institution in 2008 (4.4%).16 Furthermore, the observed higher number of exacerbations among MRSA-infected patients compared with MSSA underscores the potential impact of MRSA infection on disease severity and progression. Overall, the demographic data revealed a balanced gender distribution among the patients included in the study.
In the study, the community or hospital origin of the strains was determined based on the CC to which they belonged, defined by MLST, spa type and agr operon, as previously described.4,17 The SCCmec type was not useful for this distinction, as types I and II, traditionally associated with hospital strains, have been largely replaced by type IV, which was initially described in community strains.17 Considering all these factors, the proportion of HA-MRSA and CA-MRSA isolates obtained was 68% (35/50) and 32% (15/50), respectively. In both years, the predominant hospital clone found was the ST125-SCCmec-IV-t067, belonging to CC5. This clone is also the most common in the nosocomial context of our country, accounting for more than 50% of hospital-acquired colonizations, with rare presence in other European countries.17,18 In contrast, the most prevalent SCCmec found in HA-MRSA strains was IVc (94%) in both 2013 and 2021, contradicting published studies in which IVa (2%) was identified as the most prevalent in the Spanish hospital setting.4,19 In the context of CF, an explanation could be that type IVc carries a Tn4001 transposon for resistance to aminoglycosides, antibiotics commonly administered to pwCF, and hence may have a selective advantage.20
Consistent with previous studies, we found higher clonal diversity in CA-MRSA strains than in HA-MRSA strains.4,21–23 In Spain, the incidence of CA-MRSA is low, usually associated with sporadic cases and outbreaks; the most frequently described clone in Spain is ST8-SCCmec-IVc, which aligns with our study findings. Additionally, we detected three strains belonging to a lineage of MRSA of porcine origin (ST398-SCCmec-V spa t011 and another to spa t8565) that is rarely described in pwCF24–26 The CA-MRSA strains often carry virulence factors like lukF-PV and lukS-PV, which encode the PVL toxin, potentially contributing to more severe infections and rapid spread.4 In our study, these genes were observed only in a community isolate, ST8-CC8-IVa-t008, consistent with previous reports in hospitalized patients.26–28 Although MDR is not typically associated with CA-MRSA, it is increasingly reported among PVL-positive MRSA populations.28 Our PVL-MRSA isolates showed resistance to gentamicin, levofloxacin and erythromycin. In the case of our patients with CA-MRSA, they had slightly lower lung function than patients with hospital clones but, in contrast, they had a lower frequency of exacerbations and hospitalization episodes. The worse prognosis of patients with HA-MRSA may be due to their slightly older age and more advanced stage of the disease.
Our study showed diverse and complex resistance profiles among our MRSA isolates, with 96% of them being MDR. Resistance rates were very similar in both studies, showing high resistance to levofloxacin (78%) and erythromycin (78%). This aligns with several studies on MRSA antimicrobial susceptibility, which have also revealed high rates of resistance to erythromycin, clindamycin, levofloxacin and ciprofloxacin.29 In 2021, the aminoglycoside that showed the highest resistance rate was gentamicin, probably due to the presence of the aph2-aac6 gene (72%). The administration of inhaled tobramycin for the treatment of patients colonized by P. aeruginosa can also generate isolates resistant to it over time.
The presence of genes conferring resistance to different antibiotic families underscores the multifaceted nature of antibiotic resistance in the studied MRSA strains. In fact, the linezolid-resistant isolate in which we found the cfr gene (ST125 CC5 IVc, t1340) additionally harboured other resistance genes: blaZ, dfrK, erm(C), mecA and tet(L). Despite being susceptible to erythromycin, this strain demonstrated resistance to clindamycin, with additional resistance to chloramphenicol and linezolid (PhLOPSa phenotype).30 The existence of MRSA isolates carrying the cfr gene within the same clonal complex (CC5) but exhibiting variations in STs and spa typing raises questions about the dynamics of resistance gene dissemination within this clonal complex.
Interestingly, all isolates that carried a resistance gene related to trimethoprim (dfrK) were susceptible to trimethoprim/sulfamethoxazole combination. Conversely, only one isolate from the 2021 survey was resistant to trimethoprim/sulfamethoxazole, but no other known resistance-related genes were detected. Furthermore, regarding tetracyclines, we found a low correlation between the presence of the tetK, tetM and tetL genes and a resistant phenotype (29%).
Few studies have analysed the clinical impact of community clones in pwCF, limiting our understanding of their implications and prevalence. The small sample size and geographical limitations of our study reduce the generalizability of our findings. We also consider the absence of the analysis of data for FEV1 over time as a limitation, along with the lack of information regarding the duration of colonization by various bacterial isolates. These factors introduce uncertainties that may affect the interpretation of our findings. On the other hand, only one patient received the triple therapy (elexacaftor + tezacaftor + ivacaftor), so we could not estimate the effect of this treatment on lung function and its impact on the evolution of MRSA infection. It should be noted that this treatment was approved in Spain at the end of 2021, and patients receive this treatment generally on compassionate grounds.
In conclusion, our study revealed a reduction in the prevalence of MRSA bronchopulmonary infection among pwCF in Spain between 2013 and 2021, although this decrease was not statistically significant. Despite this trend, MRSA infection continues to pose a significant risk to lung function and exacerbations in pwCF. The high genetic and phenotypic diversity observed in MRSA isolates highlights the complex epidemiology of this pathogen in CF. HA-MRSA strains, particularly those of the ST125 lineage, remain predominant and exhibited resistance to multiple antibiotics. In contrast, CA-MRSA strains showed greater genetic diversity and carried different virulence factors. Our findings underscore the need for continued surveillance to control MRSA infection in pwCF, addressing both the persistent challenges posed by hospital-associated strains and the emerging threat from community-associated lineages.
Supplementary Material
Acknowledgements
We thank the members of the GEIFQ group (Grupos Español para el Estudio de la Colonización/Infección Broncopulmonar en Fibrosis Quística). GEIFQ members include the following: Ainhize Maruri-Aransolo, Juan de Dios-Caballero, Rafael Cantón, Malkoa Michelena-González, Luis Máiz, Saioa Vicente (Hospital Ramón y Cajal, Madrid); Esther Quintana (Hospital Virgen del Rocío, Sevilla); María Dolores Pastor-Vivero (Hospital de Cruces, Barakaldo); Antonio Álvarez (Hospital Vall d'Hebrón, Barcelona); Rosa Girón, Teresa Alarcón (Hospital de la Princesa, Madrid); Carmen Luna-Paredes (Hospital 12 de Octubre, Madrid); Marta Ruiz de Valbuena, María Concepción Prados (Hospital de la Paz, Madrid); Silvia Castillo-Corullón (Hospital Clinic de Valencia, Valencia); María José Selma, Amparo Solé (Hospital la Fe, Valencia); Maria Cols-Roig (Hospital Sant Joan de Deu, Barcelona); Pedro Mondéjar-López (Hospital Virgen de la Arrixaca, Murcia); Estela Pérez Ruiz, Casilda Olveira, Pilar Caro Aguilera, Pilar Bermúdez Ruiz (Hospital General de Málaga, Málaga); Carla López Causapé, Joan Figuerola, Antonio Oliver (Hospital Son Espases, Palma de Mallorca); Oscar Asensio (Consorci Corporació Sanitària Parc Taulí, Sabadell).
Contributor Information
Ainhize Maruri-Aransolo, Servicio de Microbiología, Hospital Universitario Ramón y Cajal and Instituto Ramón y Cajal de Investigación Sanitaria (IRYCIS), Madrid, España.
Marta Hernandez-García, Servicio de Microbiología, Hospital Universitario Ramón y Cajal and Instituto Ramón y Cajal de Investigación Sanitaria (IRYCIS), Madrid, España; CIBER de Enfermedades Infecciosas (CIBERINFEC), Instituto de Salud Carlos III, Madrid, España.
Raquel Barbero, Servicio de Microbiología, Hospital Universitario Ramón y Cajal and Instituto Ramón y Cajal de Investigación Sanitaria (IRYCIS), Madrid, España.
Malkoa Michelena, Servicio de Microbiología, Hospital Universitario Ramón y Cajal and Instituto Ramón y Cajal de Investigación Sanitaria (IRYCIS), Madrid, España.
María Dolores Pastor-Vivero, Servicio de neumología pediátrica, Hospital Universitario Cruces and IIS Biobizkaia, Bizkaia, Spain.
Pedro Mondejar-Lopez, Servicio de neumología pediátrica, Hospital Clínico Universitario Virgen de la Arrixaca, Murcia, España.
Amparo Solé, Servicio de neumología, Hospital Universitario y Politécnico La Fe and Universitat de Valencia, Valencia, España.
Rafael Cantón, Servicio de Microbiología, Hospital Universitario Ramón y Cajal and Instituto Ramón y Cajal de Investigación Sanitaria (IRYCIS), Madrid, España; CIBER de Enfermedades Infecciosas (CIBERINFEC), Instituto de Salud Carlos III, Madrid, España.
Juan de Dios Caballero-Pérez, Servicio de Microbiología, Hospital Universitario Ramón y Cajal and Instituto Ramón y Cajal de Investigación Sanitaria (IRYCIS), Madrid, España; CIBER de Enfermedades Infecciosas (CIBERINFEC), Instituto de Salud Carlos III, Madrid, España.
GEIFQ study group:
Ainhize Maruri-Aransolo, Juan de Dios-Caballero, Rafael Cantón, Malkoa Michelena-González, Luis Máiz, Saioa Vicente, Esther Quintana, María Dolores Pastor-Vivero, Antonio Álvarez, Rosa Girón, Teresa Alarcón, Carmen Luna-Paredes, Marta Ruiz de Valbuena, María Concepción Prados, Silvia Castillo-Corullón, María José Selma, Amparo Solé, Maria Cols-Roig, Pedro Mondéjar-López, Estela Pérez Ruiz, Casilda Olveira, Pilar Caro Aguilera, Pilar Bermúdez Ruiz, Carla López Causapé, Joan Figuerola, Antonio Oliver, and Oscar Asensio
Funding
This study was funded by Plan Estatal de Investigacion Cientifica y técnica y de Innovación (I+D+I) 2017–2020 (PI19/01043 and PI22/1045) and CIBER de Enfermedades Infecciosas (CIBERINFEC) (CB21/13/00084), Instituto de Salud Carlos III, Madrid, Spain. A.M.-A. is supported by a pre-doctoral contract associated with PI19/01043 project, M.H.-G. by a postdoctoral contract by CIBERINFEC (CB21/13/00084) and J.dD.C.-P. by Juan Rodes contract (Ref. JR18/00034).
Transparency declarations
R.C. has participated in educational programmes organized by MSD, Pfizer and Shionogi. Other authors declare no conflicts of interest regarding the content of this article.
Supplementary data
Figures S1 to S3 and Tables S1 to S4 are available as Supplementary data at JAC-AMR Online.
References
- 1. Akil N, Muhlebach MS. Biology and management of methicillin resistant Staphylococcus aureus in cystic fibrosis. Pediatr Pulmonol 2018: 53 Suppl 3: S64–74. 10.1002/ppul.24139 [DOI] [PubMed] [Google Scholar]
- 2. Fusco NM, Toussaint KA, Prescott WA. Antibiotic management of methicillin-resistant Staphylococcus aureus–associated acute pulmonary exacerbations in cystic fibrosis. Ann Pharmacother 2015; 49: 458–68. 10.1177/1060028014567526 [DOI] [PubMed] [Google Scholar]
- 3. Cercenado E, de Gopegui ER. Staphylococcus aureus resistente a la meticilina de origen comunitario. Enferm Infecc Microbiol Clin 2008; 26: 19–24. 10.1157/13128776 [DOI] [PubMed] [Google Scholar]
- 4. Vindel A, Trincado P, Cuevas O et al. Molecular epidemiology of community-associated methicillin-resistant Staphylococcus aureus in Spain: 2004–12. J Antimicrob Chemother 2014; 69: 2913–9. 10.1093/jac/dku232 [DOI] [PubMed] [Google Scholar]
- 5. Rodríguez-Baño J, Domínguez MA, Millán AB et al. Clinical and molecular epidemiology of community-acquired, healthcare-associated and nosocomial methicillin-resistant Staphylococcus aureus in Spain. Clin Microbiol Infect 2009; 15: 1111–8. 10.1111/j.1469-0691.2009.02717.x [DOI] [PubMed] [Google Scholar]
- 6. Mama OM, Aspiroz C, Ruiz-Ripa L et al. Prevalence and genetic characteristics of Staphylococcus aureus CC398 isolates from invasive infections in Spanish hospitals, focusing on the livestock-independent CC398-MSSA clade. Front Microbiol 2021; 12: 623108. 10.3389/fmicb.2021.623108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ayepola OO, Olasupo NA, Egwari LO et al. Characterization of Panton–Valentine leukocidin-positive Staphylococcus aureus from skin and soft tissue infections and wounds in Nigeria: a cross-sectional study. F1000Res 2018; 7: 1155. 10.12688/f1000research.15484.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Spaulding AR, Salgado-Pabón W, Kohler PL et al. Staphylococcal and streptococcal superantigen exotoxins. Clin Microbiol Rev 2013; 26: 422–47. 10.1128/CMR.00104-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Goudarzi M, Fazeli M, Goudarzi H et al. Spa typing of Staphylococcus aureus strains isolated from clinical specimens of patients with nosocomial infections in Tehran, Iran. Jundishapur J Microbiol 2016; 9: e35685. 10.5812/jjm.35685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cantón R, Máiz L, Escribano A et al. Spanish consensus on the prevention and treatment of Pseudomonas aeruginosa bronchial infections in cystic fibrosis patients. Arch Bronconeumol 2015; 51: 140–50. 10.1016/j.arbres.2014.09.021 [DOI] [PubMed] [Google Scholar]
- 11. Swenson JM, Brasso WB, Ferraro MJ et al. Detection of inducible clindamycin resistance in staphylococci by broth microdilution using erythromycin-clindamycin combination wells. J Clin Microbiol 2007; 45: 3954–7. 10.1128/JCM.01501-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hernández-García M, García-Castillo M, García-Fernández S et al. Distinct epidemiology and resistance mechanisms affecting ceftolozane/tazobactam in Pseudomonas aeruginosa isolates recovered from ICU patients in Spain and Portugal depicted by WGS. J Antimicrob Chemother 2021; 76: 370–9. 10.1093/jac/dkaa430 [DOI] [PubMed] [Google Scholar]
- 13. Petit RA, Read TD. Bactopia—a flexible pipeline for complete analysis of bacterial genomes. mSystems 2020; 5: 10–128. 10.1128/mSystems.00190-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sanchez-Herrero JF, Sullivan M. spaTyper: staphylococcal protein A (spa) characterization pipeline. 2020.
- 15. European Cystic Fibrosis Society . ECFSPR 2021 Annual Data Report. 2023. https://www.ecfs.eu/sites/default/files/Annual%20Report_2021_09Jun2023.pdf
- 16. Molina A, Del Campo R, Máiz L et al. High prevalence in cystic fibrosis patients of multiresistant hospital-acquired methicillin-resistant Staphylococcus aureus ST228-SCCmecI capable of biofilm formation. J Antimicrob Chemother 2008; 62: 961–7. 10.1093/jac/dkn302 [DOI] [PubMed] [Google Scholar]
- 17. Vindel A, Cuevas O, Cercenado E et al. Methicillin-resistant Staphylococcus aureus in Spain: molecular epidemiology and utility of different typing methods. J Clin Microbiol 2009; 47: 1620–7. 10.1128/JCM.01579-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Goudarzi M, Eslami G, Rezaee R et al. Clonal dissemination of Staphylococcus aureus isolates causing nosocomial infections, Tehran, Iran. Iran J Basic Med Sci 2019; 22: 238–45. 10.22038/ijbms.2018.30067.7245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sánchez-Serrano A, García-González N, Bonillo D et al. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in a tertiary hospital from the Comunidad Valenciana (Spain). Microb Drug Resist 2022; 28: 1071–8. 10.1089/mdr.2022.0027 [DOI] [PubMed] [Google Scholar]
- 20. Depardieu F, Podglajen I, Leclercq R et al. Modes and modulations of antibiotic resistance gene expression. Clin Microbiol Rev 2007; 20: 79–114. 10.1128/CMR.00015-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rolo J, Miragaia M, Turlej-Rogacka A et al. High genetic diversity among community-associated Staphylococcus aureus in Europe: results from a multicenter study. PLoS One 2012; 7: e34768. 10.1371/journal.pone.0034768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Otter JA, French GL. Molecular epidemiology of community-associated meticillin-resistant Staphylococcus aureus in Europe. Lancet Infect Dis 2010; 10: 227–39. 10.1016/S1473-3099(10)70053-0 [DOI] [PubMed] [Google Scholar]
- 23. Junie LM, Jeican II, Matroș L et al. Molecular epidemiology of the community-associated methicillin-resistant Staphylococcus aureus clones: a synthetic review. Med Pharm Rep 2018; 91: 7–11. 10.15386/cjmed-807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rodríguez-Lázaro D, Oniciuc E-A, García PG et al. Detection and characterization of Staphylococcus aureus and methicillin-resistant S. aureus in foods confiscated in EU borders. Front Microbiol 2017: 8: 1344. 10.3389/fmicb.2017.01344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lima DF, Cohen RW, Rocha GA et al. Genomic information on multidrug-resistant livestock-associated methicillin-resistant Staphylococcus aureus ST398 isolated from a Brazilian patient with cystic fibrosis. Mem Inst Oswaldo Cruz 2017; 112: 79–80. 10.1590/0074-02760160342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Aschbacher R, Pichon B, Spoladore G et al. High clonal heterogeneity of Panton–Valentine leukocidin-positive meticillin-resistant Staphylococcus aureus strains from skin and soft-tissue infections in the province of Bolzano, Northern Italy. Int J Antimicrob Agents 2012; 39: 522–5. 10.1016/j.ijantimicag.2012.02.004 [DOI] [PubMed] [Google Scholar]
- 27. Kudo K, Noda T, Sasaoka Y et al. Severe cervical abscess due to PVL-positive ST6562 MRSA-IVa, a presumptive variant of ST8-IVa USA300 clone in northern Japan. New Microbes New Infect 2024; 58: 101230. 10.1016/j.nmni.2024.101230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McManus BA, Aloba BK, Earls MR et al. Multiple distinct outbreaks of Panton–Valentine leucocidin-positive community-associated meticillin-resistant Staphylococcus aureus in Ireland investigated by whole-genome sequencing. J Hosp Infect 2021; 108: 72–80. 10.1016/j.jhin.2020.11.021 [DOI] [PubMed] [Google Scholar]
- 29. Liang Y, Tu C, Tan C et al. Antimicrobial resistance, virulence genes profiling and molecular relatedness of methicillin-resistant Staphylococcus aureus strains isolated from hospitalized patients in Guangdong Province, China. Infect Drug Resist 2019; 12: 447–59. 10.2147/IDR.S192611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. de Dios Caballero J, Pastor MD, Vindel A et al. Emergence of cfr-mediated linezolid resistance in a methicillin-resistant Staphylococcus aureus epidemic clone isolated from patients with cystic fibrosis. Antimicrob Agents Chemother 2016; 60: 1878–82. 10.1128/AAC.02067-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.