Abstract
A neonatal rat dorsal root ganglion-derived neuronal culture system has been utilized to study herpes simplex virus (HSV) latency establishment, maintenance, and reactivation. We present our initial characterization of viral gene expression in neurons following infection with replication-defective HSV recombinants carrying β-galactosidase and/or green fluorescent protein reporter genes under the control of lytic cycle- or latency-associated promoters. In this system lytic virus reporter promoter activity was detected in up to 58% of neurons 24 h after infection. Lytic cycle reporter promoters were shut down over time, and long-term survival of neurons harboring latent virus genomes was demonstrated. Latency-associated promoter-driven reporter gene expression was detected in neurons from early times postinfection and was stably maintained in up to 83% of neurons for at least 3 weeks. In latently infected cultures, silent lytic cycle promoters could be activated in up to 53% of neurons by nerve growth factor withdrawal or through inhibition of histone deacetylases by trichostatin A. We conclude that the use of recombinant viruses containing reporter genes, under the regulation of lytic and latency promoter control in neuronal cultures in which latency can be established and reactivation can be induced, is a potentially powerful system in which to study the molecular events that occur during HSV infection of neurons.
A defining characteristic of herpesviruses is the ability to establish latent infections in their natural hosts. Herpes simplex virus (HSV) establishes latent infection in neurons of the peripheral nervous system, predominantly in sensory ganglia innervating the site of primary infection (25, 53, 58). Latent virus has the capacity to reactivate, which can give rise to a peripheral lesion in the dermatome relating to the affected ganglia (reviewed in reference 63). During latency the virus genome exists in a form lacking detectable free ends, consistent with the presence of episomal or concatemeric DNA (16, 31, 38). The latency-associated transcripts (LATs) are transcribed from a region within the repeats, mapping antisense to the IE110 gene, giving rise to a family of colinear RNAs (54; reviewed in reference 58). The LAT region is the only region of the genome that is abundantly transcribed during latency, giving rise to RNAs which are predominantly nuclear and consist of two highly abundant, nonpolyadenylated RNAs of 2 and 1.5 kb, termed major LATs. The precise mechanism of synthesis of major LATs is unclear (3), although there is compelling evidence that these transcripts are introns derived from a less abundant 8.5-kb polyadenylated precursor RNA termed a minor LAT (1, 3, 17, 32, 39, 65, 67). The function of the LATs is uncertain; investigations using mouse models have shown that LATs are not essential for the establishment or maintenance of a latent infection or for reactivation (5, 23, 52). There is evidence, however, indicating that LATs can increase the number of neurons in which latency is established. Furthermore, LAT null mutants exhibit a reduced reactivation phenotype in vivo (35, 45, 57), and a 0.38-kb region located downstream of the promoter has been demonstrated to play a significant role in enhancing reactivation (7, 21, 34).
The molecular mechanisms by which HSV establishes and maintains latency and subsequently reactivates are not fully resolved (reviewed in reference 36). A popular hypothesis is that latency is established via a “default” mechanism as a result of failure of the virus to enter the productive cycle. This proposal is consistent with observations that virus mutants which are defective or severely impaired in the ability to initiate a normal lytic cycle of gene expression can establish latency (10, 13, 27, 29). Furthermore, wild-type viruses can establish latency in sensory ganglia that do not innervate the site of peripheral infection and which show no prior evidence of immediate-early (IE) gene expression (26, 50). Consistent with this view, it was demonstrated that the numbers of LAT+ neurons remained constant in the transition from acute to latent infection and that only a minor proportion of these neurons contained virus antigen (Ag) when examined during acute infection (28, 45). A corollary of the “default” hypothesis is that the expression of productive cycle genes and the establishment of latency are mutually exclusive pathways. However, a number of independent calculations imply that latently infected neurons carry multiple copies of the viral genome (16, 42, 44, 46); hence, it has been proposed that latency may be established after an abortive acute infection involving limited replication of the viral genome (46, 50). The biological significance of the “high-copy” latently infected neurons has been demonstrated in two recent reactivation studies. Sawtell (43) showed a direct correlation between latency copy number and reactivation propensity. Studies in our laboratory indicate that reactivation occurs at a higher frequency and with enhanced kinetics in cervical ganglia from levels where there is evidence for virus gene expression prior to the establishment of latency, as opposed to neighboring ganglia where latency is established in the absence of detectable gene expression (26).
Although animal models have proved invaluable in the study of HSV pathogenesis, they are proving less amenable for investigations into the regulatory mechanisms that function during the establishment of and reactivation from latency. The problems are predominantly due to the relatively small proportion of cells within ganglia in which latency is established, the real and perceived asynchronicity of these molecular events in vivo, and the difficulty of addressing latency-dedicated functions of viral gene products that have a role in acute phase replication. To overcome such difficulties, several labs have developed in vitro models of latency using fibroblasts, neuroblastomas, or primary cultures of sensory neurons which, in general, rely on establishing latency through limiting virus replication and spread through the use of antiviral agents or replication-defective virus mutants (6, 18, 19, 60). Such in vitro model systems have several advantages for the study of latency establishment and reactivation. The cells are coordinately and directly infected, negating the need for full replication competency, and high levels of latency may be established. Further, reactivation is inducible in a controlled and uniform manner. Consistent with latency in vivo, no lytic cycle Ags are detectable, and in models utilizing primary neurons, transcription of LATs occurs (14, 18, 20, 48, 56). The best-characterized neuronal model system is that of Wilcox and coworkers (60–62), in which primary cultures of embryonic or neonatal rat sensory neurons are infected with wild-type virus in the presence of acyclovir (ACV). After several days, the cultures are free of infectious virus and LATs are detected in a large proportion of cells (14, 48). In this and other neuron-based models, latency is dependent on the presence of nerve growth factor (NGF) and latent virus can be reactivated by NGF withdrawal, heat shock, or addition of activators of protein kinases (6, 18, 59, 60). The viral genomes harbored in nonneuronal systems are more resistant to reactivation stimuli, and reactivation of gene expression can be induced only by the provision of IE110 (20). It is not yet clear whether this reflects fundamental differences between the neuronal and nonneuronal models with regard to the mechanisms of latency establishment and reactivation employed (reviewed in reference 36).
In this paper we describe the development of a neonatal rat dorsal root ganglion (drg)-derived neuronal culture system for the study of latency establishment, maintenance, and reactivation. We present our initial characterization of virus gene expression in neurons following infection with replication-defective virus recombinants carrying reporter genes under the control of lytic cycle- or latency-associated promoters (LAPs). In this system we are able to show viral lytic cycle gene expression and reporter promoter activation in a large proportion of neurons within 24 h of infection, the shutdown of lytic cycle reporter promoters over time, and the long-term survival of neurons harboring latent viral genomes. LAP-driven reporter gene expression can be detected in neurons at early times postinfection (p.i.) and is stably maintained for the life of the culture. Further, activation of the silent lytic cycle reporter promoters was attained in a significant proportion of neurons by NGF withdrawal or histone deactylase inhibition with trichostatin A (TSA). We conclude that the use of recombinant viruses containing reporter genes under lytic and latent promoter control in conjunction with neuronal cultures in which latency can be established and reactivation can be induced is a potentially powerful system in which to study the molecular events that occur during HSV infection of neurons.
MATERIALS AND METHODS
Cells and virus stocks.
All cell lines were grown in Glasgow modified Eagle medium supplemented with 10% fetal bovine serum. Glycoprotein H-negative (gH−) recombinant viruses were grown and assayed on CR1 helper cells (8), and all other viruses were propagated and assayed on Vero cells.
Cell-released virus stocks for infection of neuronal cultures were prepared from infected tissue culture supernatants purified on a continuous 5 to 25% Ficoll gradient (28,000 × g, 1.5 h) and were resuspended in endotoxin-free phosphate-buffered saline (PBS).
Plasmids.
Plasmid pIMMB34 (8) contains HSV type 1 (HSV-1) nucleotides 41546 to 47856, from which nucleotides 43771 to 47103 are deleted to remove the first 427 bp of UL23 (thymidine kinase [TK]) and all of UL22 (gH). Plasmid pIMMB36 was generated by insertion of a human cytomegalovirus (CMV) IE promoter 1 (CMV IE1) lacZ expression cassette from MV10 (64) into the HpaI site between the gH and TK flanking sequences. Plasmid pIE110LacZ has been described previously (26) and contains the lacZ gene and simian virus 40 polyadenylation signal from pNASSβ (Clontech Laboratories) under the control of the IE110 promoter. The IE110 promoter was derived from KOS (nucleotides 1294 to 2262) and extends from position −818 to position +150 with respect to the transcription start site (33). The plasmid pIMMb34lac1 was made by blunt-end cloning of the HindIII/SalI cassette from pIE110LacZ into HpaI-cut pIMMb34.
Construction of recombinant viruses.
All virus mutants were constructed on an HSV-1 strain SC16 background. Unless stated otherwise, all recombinant viruses were generated by cotransfection into CR1 cells of 10 μg of parent-virus-infected-cell DNA with 3 to 6 μg of linearized plasmid DNA by a CaCl2-dimethyl sulfoxide boost method (55). gH−TK− recombinant transfection progeny were isolated by two passages at low multiplicity (0.001 PFU/cell) in 5 μg of ACV/ml and were plaque purified two or three times, and the phenotype of the isolated viruses was checked biologically for resistance to ACV, lack of growth in a noncomplementing cell line (Vero), and, where appropriate, by staining for β-galactosidase (β-gal) activity.
Virus SC16LβA (described previously in reference 25) contains the lacZ gene linked to the encephalomyocarditis virus (EMCV) internal ribosome entry site (IRES) inserted approximately 1.5 kb downstream of the LAP.
Recombinant virus CS1 (Fig. 1) was generated by coinfection of CR1 cells with 5 PFU of viruses SC16LβA and SC16gH−TK− per cell (8).
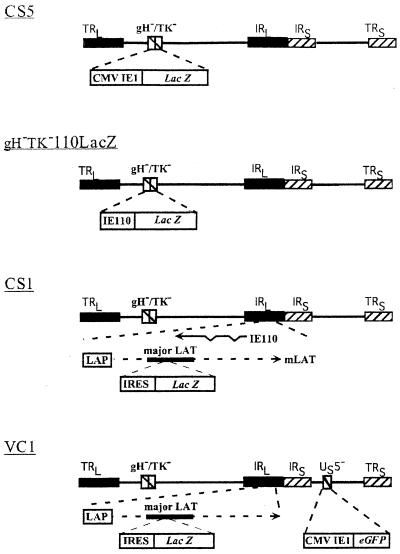
FIG. 1.
Schematic of gH−TK− recombinant viruses. All viruses were made gH−TK− through the deletion of nucleotides 43771 to 47103 spanning the gH and TK coding sequences. Virus CS5 contains a CMV IE1 promoter-driven lacZ expression cassette replacing gH and TK sequences. Virus gH−TK−110LacZ contains an IE110 promoter-driven lacZ expression cassette inserted at the same locus. Virus CS1 contains an EMCV IRES-linked lacZ gene inserted between the HpaI sites within major LATs and was generated by the deletion of gH and TK from SC16 LβA. Virus VC1 is derived from coinfection of CS1 and C12 and consists of the CMV IE1 promoter-driven EGFP reporter gene cassette in the Us5 locus from C12 on the CS1 backbone. In VC1 and CS1, β-gal expression is driven by the endogenous LAP.
Virus CS5 contains the CMV IE1 promoter-driven lacZ cassette inserted into the gH-TK locus by recombination with pIMMb36.
Virus gH−TK−110LacZ contains the IE110 promoter-driven lacZ expression cassette from pIMMB34lac1 replacing gH and TK coding sequences (Fig. 1).
Virus C12 (a gift from N. Babic) contains the CMV IE1 promoter-driven enhanced green fluorescent protein (EGFP) cassette from pEGFP-C1 (Clontech) inserted in the nonessential Us5 locus.
Double-tagged recombinant virus VC1 (Fig. 1) was generated by coinfection of CR1 cells with 5 PFU each of viruses CS1 and C12 per cell. VC1 was isolated as described above, except that GFP+ plaques were identified under UV illumination prior to picking.
Further confirmation of all viral structures was obtained by restriction endonuclease digestion and Southern blot analysis.
drg-derived primary neuronal cultures.
drgs from spinal levels C1 to L6 were removed from newborn (days 1 to 3) Sprague-Dawley rats to F14+ (Ham's F14 medium supplemented with 0.5 μg of amphotericin B/ml, 100 U of penicillin/ml, 100 μg of streptomycin/ml, and 1% Ultroser G [USG; GibcoBRL]). The drgs were incubated in trypsin-EDTA (GibcoBRL) for 15 min at 37°C and were mechanically dissociated by pipetting (approximately 50 passes), and any ganglionic debris remaining was removed to fresh trypsin, reincubated, and dissociated further. The cell suspensions were diluted in 5 volumes of F14+ with 5% USG, pelleted (200 × g, 10 min), and then resuspended in growth medium (F14+ containing 40 ng of NGF 2.5S/ml, 0.1× B27 supplement [USG; GibcoBRL], and 40 μM 5-fluoro-5′-deoxyuridine [FdU]). Nonneuronal cells were removed by preplating on culture dishes coated with 0.5 mg of rat tail collagen type VII/ml (3 h, 37°C, 5% CO2). The neuron-enriched cell suspension was pelleted (200 × g, 10 min), resuspended in growth medium containing 5% USG, and plated onto 13-mm coverslips coated with MATRIGEL (BD Biosciences) in 24-well culture dishes. Neurons were plated at an approximate density of 200 to 400/well or 800 to 1,000/well. Neuronal cultures were transferred to growth medium lacking FdU (FdU−) 48 h prior to infection (F14+ containing 40 ng of NGF/ml and 0.1× B27), and virus infections were routinely performed at 5 × 105 or 1 × 106 PFU per well in 300 μl of FdU− growth medium. To control for variation in neuronal preparations, experiments were conducted using coverslip cultures from a single drg preparation and generally two to three coverslips were examined per data point.
Detection and quantitation of reporter gene expression.
Neuronal cultures were fixed in 2% paraformaldehyde–PBS for 20 min, permeabilized with 0.2% Triton–PBS (15 min), and incubated overnight at 4°C in 2% normal horse serum–PBS prior to the addition of specific antisera. β-Gal was detected using rabbit polyclonal antisera (5 Prime → 3 Prime, Inc.) and an Alexa 488-conjugated goat anti-rabbit immoglobulin G (IgG) antibody (Molecular Probes). Virus Ags were detected with rabbit polyclonal antisera to HSV-1 (MacIntyre strain; DAKO) and were visualized using the Alexa 488-conjugated goat anti-rabbit IgG. Neuron-specific β-tubulin III (β-tub) was detected using mouse monoclonal antibodies (Sigma) and Cy3-conjugated donkey anti-mouse IgG (Amersham). IE110 expression was detected using monoclonal antibody 17060 (mouse ascites fluid; binds N-terminal epitope) and rabbit polyclonal antiserum 191 (binds C-terminal epitope). Both antisera were kindly donated by R. Everett (Glasgow). Cell nuclei were counterstained with 100 ng of 4′,6′-diamidino-2-phenylindole (DAPI)/ml. Fluorescence was visualized using an Olympus I×70 inverted fluorescence microscope and either a fluorescein isothiocyanate (FITC) filter (excitation, 455 to 495 nm; emission, 510 to 555 nm; Chroma) for Alexa and GFP only or a DAPI-FITC-tetramethyl rhodamine isocyanate triple band-pass filter (emission ranges, 425 to 465 nm, 500 to 540 nm, and 582 to 650 nm; Chroma) allowing covisualization of Alexa-GFP (green), Cy3 (red), and DAPI (blue) signals. Photographs were generated as digital images using a color-chilled 3CCD camera (Hamamatsu) and Image-Pro Plus 4.0 software (Media Cybernetics). Photographic montages were prepared using Adobe Illustrator 7.0.
For the purpose of enumeration, the total number of neurons present on each coverslip was counted unless stated otherwise. The data in Table 2 were generated by making a series of photographs spanning the entire coverslip and then counting neurons in alternate photographs. Data were pooled from three coverslips per time point and represent counts from 136 photos (day 4 p.i.), 210 photos (day 7 p.i.), 159 photos (day 20 p.i.), or 147 photos (uninfected, day 7). We have determined that this method of counting gives an accurate assessment of the percentage of neurons expressing β-gal. For example, following infection with 106 PFU of CS1, the total number of neurons on one coverslip from day 7 p.i. was 766, 674 of which showed LAP-driven β-gal expression (88%). Counting every photograph from this sample (79 in all) gave 160 neurons (21% of the total neurons), 139 of which were β-gal+ (91%). Counting alternate photographs (40 in all) gave 80 neurons (10% of total), 69 of which were β-gal+ (86%). Thus, the maximum variation between methods was 5%. A second coverslip from this infection was photographed, yielding 71 photographs containing 159 neurons, 137 of which were β-gal+ (86%). Counting alternate photographs (36 in all) gave 82 neurons, 71 of which were β-gal+ (87%), indicating that the variation between wells was minor.
TABLE 2.
LAP activity in CS1-infected neuronal culturesa
| Day p.i.b | No. of neurons countedc | % β-Gal+d | % Intense β-gal staininge |
|---|---|---|---|
| 4 | 185 | 55 | 9 |
| 7f | 454 | 71 | 10 |
| 20 | 320 | 83 | 11 |
Nine wells were infected with 106 PFU of CS1/well.
At various days p.i., wells were fixed and dually immunostained for β-gal and β-tub.
Total neurons (β-tub+) counted in photographic maps (see Materials and Methods) of three wells per data point.
Percentage of neurons showing β-gal expression.
Percentage of β-gal+ neurons showing intense staining.
A total of 475 neurons were counted in the photographs from three uninfected control wells fixed on day 7.
Reactivation.
To withdraw NGF, the growth medium was replaced with NGF− medium (F14+ with 0.1× B27 and 0.1 μl of rabbit anti-NGF antiserum [Sigma]/ml). To induce reactivation with TSA, the growth medium was replaced with fresh growth medium containing 660 nM TSA. Activation of silenced viral promoters was assessed by immunohistochemical detection of β-gal (CS5 and gH−TK−110LacZ) or by direct visualization of GFP (VC1).
RESULTS
Establishment of primary neuronal cultures.
Primary dissociated drg cultures were prepared from neonatal rats as described in Materials and Methods. Variation in neuron numbers per well within a drg preparation was tested in five separate preparations (A to E), where two to four coverslips were fixed either 3 (A), 7 (B to D), or 14 (E) days after plating and were immunostained for the neuron-specific marker β-tub. The total number of neurons per well was assayed, and the maximum variation between wells was as follows: A, 1.2-fold; B, 1.1-fold; C, 1.3-fold; D, 1.4-fold; and E, 1.6-fold.
All recombinant viruses used (Fig. 1) were based on a strain SC16 gH−TK− backbone to limit virus replication in neurons and eliminate secondary spread resulting from productive infection of the nonneuronal population. Thus, antiviral agents such as ACV are not required to control virus infection in this model. Initial dose dilution experiments using gH−TK− recombinant viruses indicated that 5 × 105 to 1 × 106 PFU per well was optimal for establishing viral gene expression in 40 to 70% of neurons by 24 h p.i. (data not shown). This dose range corresponds to a theoretical multiplicity of infection of approximately 50 to 100 PFU/cell; however, as has been noted previously for neuronal culture systems, the majority of the coverslip surface area is available for nonspecific binding of virions, and thus, the actual multiplicity of infection is likely to be significantly lower (4).
Characterization of primary infection.
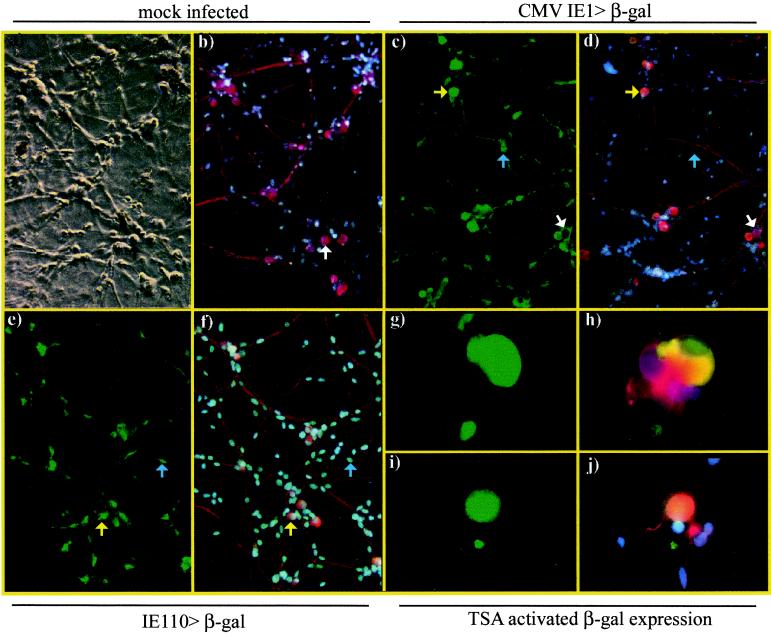
To assess IE viral gene expression, neuronal cultures were infected with 106 PFU of gH−TK− recombinant viruses/well carrying the lacZ reporter gene under the control of the human CMV IE1 promoter or the HSV-1 IE110 promoter (Fig. 1). A single coverslip per virus was fixed at 1, 3, 4, and 6 days p.i. and was immunostained for β-gal expression and β-tub as described (Fig. 2). The β-gal+ neurons and total neurons in the cultures were counted (Table 1). Activation of the CMV IE1 and IE110 reporter promoters was observed within 24 h of infection (Fig. 2c to f), and β-gal expression peaked at day 1 p.i. with the CMV IE1 promoter (CS5; 58%) and at day 3 p.i. with the IE110 promoter (gH−TK−110LacZ; 67%). The intensity of β-gal staining was generally greater for the CMV promoter than for the IE110 promoter. Following the initial burst of reporter promoter activity, the number of β-gal+ neurons and the intensity of signal declined with time p.i. and by day 6 p.i. fell to 1% (gH−TK−110LacZ) or 10% (CS5) of neurons expressing β-gal (Table 1). The range in neuron numbers in this experiment makes it difficult to conclude whether the decline in β-gal+ neurons from day 3 to day 6 p.i. (by 48 or 99%) is due to neuronal death or promoter shutdown. However, the data indicate that it is unlikely that all neurons in which the IE110 promoter was active died. Should the detection of IE110 and CMV IE1 reporter promoter activity accurately reflect expression of IE110 protein following neuronal infection, our data would suggest that neurons can survive at least limited IE gene expression. This raises the possibility that viral gene expression may occur prior to the establishment of latency.
FIG. 2.
Representative examples of mock-infected neuronal cultures or cultures infected with 106 PFU of CS5 (CMV IE1>β-gal) or gH−TK−110LacZ (IE110>β-gal) per well. Cultures were fixed and dually immunostained for expression of β-gal (FITC, green) and neuron-specific β-tub (Cy3, red) and were counterstained with DAPI to show all cell nuclei (blue). An example is given of mock-infected cultures showing identification of neurons by β-tub staining (a and b, arrows). Examples are given of β-gal expression at day 1 p.i. in neurons (i.e., yellow arrows) and nonneuronal cells (i.e., blue arrows) in wells infected with CS5 (c, d) and gH−TK−110LacZ (e, f). An example of β-gal-negative neurons is indicated (white arrows). Examples are given of β-gal expression in CS5-infected (g, h) and gH−TK−110LacZ-infected (i, j) cultures at 15 days p.i., 24 h after the addition of 660 nM TSA. The small areas of β-gal fluorescence do not have nuclei and likely indicate antibody binding to cellular debris. Digital photomicrographs were taken as phase-contrast (a) or fluorescence images using either the FITC filter for β-gal (c, e, g, i) or the triple band-pass filter (b, d, f, h, j) to allow covisualization of FITC, Cy3, and DAPI fluorescence in which colocalization of β-gal and β-tub gives a yellow-orange signal.
TABLE 1.
Activation of CMV and IE110 promoters on initial infection of neuronal culturesa
| Reporter cassette | No. of neurons countedc (% β-gal-expressing neurons)d on day p.i.
|
|||
|---|---|---|---|---|
| 1 | 3 | 4 | 6 | |
| CMV > lacZb | 127 (58) | 375 (46) | ND (ND)f | 246 (10) |
| IE110 > lacZe | 341 (54) | 281 (67) | 285 (19) | 287 (1) |
Neuronal cultures infected with 106 PFU of either CS5 or gH−TK−IE110LacZ per well.
CMV IE1 reporter promoter driving lacZ expression in CS5-infected wells.
One well per data point was fixed and dually immunostained for β-gal and β-tub, and the number of neurons (β-tub+) was counted.
Percentage of neurons showing β-gal expression.
IE110 reporter promoter driving lacZ expression in gH−TK−IE110LacZ-infected wells.
ND, not determined.
Assessment of LAP activity during latent infection.
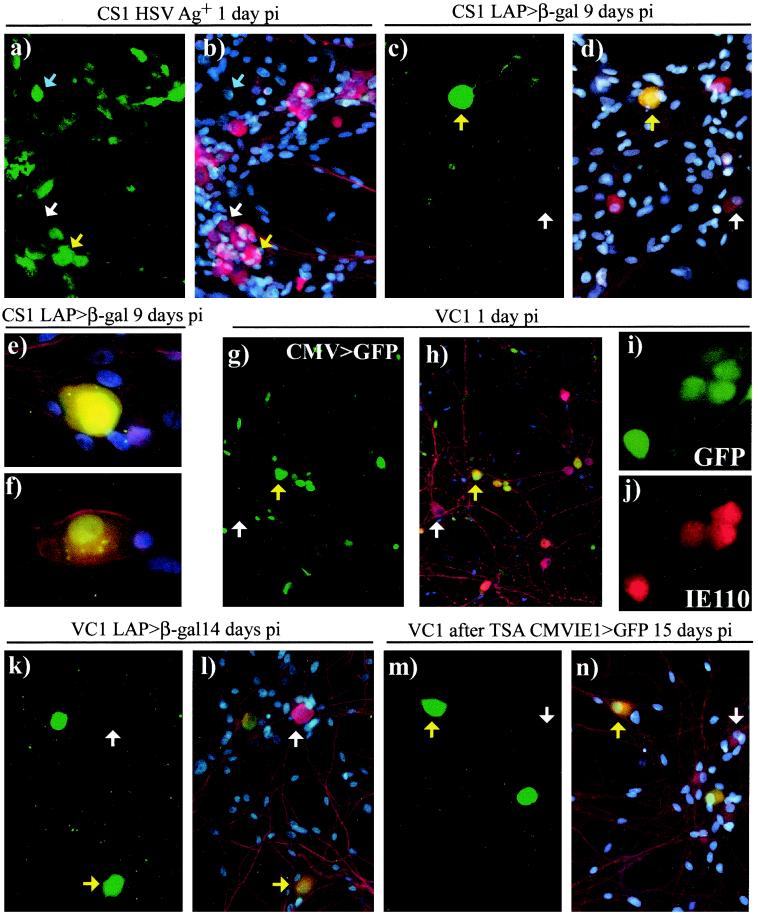
One of the key characteristics of latently infected neurons is the detection of nuclear LATs. To address whether the LAP is active in our neuronal model, we have used the recombinant virus CS1, which contains an EMCV IRES-linked lacZ reporter gene inserted approximately 1.5 kb downstream of the LAP (Fig. 1). It has previously been demonstrated that this expression cassette is not active during acute viral infection of fibroblasts but is active from day 7 p.i. in drg neurons in vivo (47). Following infection with 5 × 105 PFU of CS1/well, 484 out of 893 neurons (54%) from a single well were positive for HSV Ags on day 1 p.i. (Fig. 3a and b), consistent with our previous observations for lacZ reporter gene expression. At 9 days p.i. β-gal expression in 3 separate wells was detected in 334 of 697 neurons (48%), 366 of 763 neurons (48%), and 321 of 721 neurons (46%). That is, a total of 1,021 β-gal+ neurons in the 2,181 neurons counted (47%) (Fig. 3c to f). Between days 1 and 9 p.i. there was an average neuron loss of 19% (ranging from 15 to 22% of neurons/well). Thus, even if all neurons lost from day 1 p.i. were from the HSV Ag+ pool, 40 to 72% of the HSV Ag+ neurons had survived to day 9 p.i. Hence, we conclude that at minimum, 40% of neurons present at day 9 p.i. had survived some degree of HSV-1 protein expression. However, we are not able to determine if any of the β-gal+ neurons (47% of the total neurons) at day 9 p.i. had undergone prior viral protein expression.
FIG. 3.
Representative examples of neuronal cultures infected with 5 × 105 PFU of either CS1 (a to f) or VC1 (g, h, j to m) per well or 3 × 105 PFU of VC1 (i, j) per well. Fixed cultures were stained by dual (a to f, k, l) or single (g to k, m, n) immunofluorescence to detect expression of viral gene products (green; also see panel h) and/or β-tub (red). Cultures were counterstained with DAPI to show cell nuclei (blue) and were visualized via fluorescence by using either the FITC filter to show FITC (a, c, k) and GFP (g, i, m) or the triple band-pass filter to allow covisualization of DAPI, Cy3 with FITC (b, d to f, k), or GFP signals (h, m). Examples indicated are HSV Ag+, GFP+, or β-gal+ neurons (yellow arrows); HSV Ag+ nonneuronal cells (blue arrows); or neurons in which viral gene products are not detected (white arrows). At day 1 p.i., HSV Ag expression from CS1 (a, b) and CMV IE1 promoter-driven GFP expression from VC1 (k, l) or IE110 (i, j) were detected in neurons and nonneuronal cells. LAP-driven β-gal expression was detected in a proportion of neurons 9 days p.i. with CS1 (c to f) or 14 days p.i. with VC1 (k, l), and higher magnifications show examples of the intense (e) or average (f) β-gal staining obtained. Addition of 660 nM TSA to VC1-infected cultures 14 days p.i. resulted in detection of GFP expression in some neurons 24 h later (m, n).
Variation in β-gal expression levels was noted (compare Fig. 3e and f) with intense β-gal staining observed in 8% of β-gal+ neurons (83 of 1,021 β-gal+ neurons). This variation in expression levels is consistent with previous observations in vivo for LATs and LAP-driven β-gal expression and may reflect differences in LAP activity in neuronal subtypes (26, 28, 42, 66).
In order to follow LAP activity over time p.i. and to assess stability of latency in this system, cultures were infected with 106 PFU of CS1/well, and on days 4, 7, and 20 p.i., cells were fixed and immunostained (Table 2). Photographic maps (see Materials and Methods) of three coverslips per time point were assessed, and on day 4 p.i., 55% of neurons counted (31 of 54, 38 of 67, and 33 of 64 neurons) showed β-gal expression, indicating that in a proportion of neurons the LAT region is transcribed at early times p.i., consistent with in vivo observations of LAP activity (28, 51). The number of β-gal+ neurons increased from day 4 to 71% on day 7 p.i. (106 of 116, 100 of 147, and 118 of 191 neurons counted) and to 83% on day 20 p.i. (97 of 126, 77 of 89, and 93 of 105 neurons counted). The number of neurons (475) enumerated from three uninfected control wells fixed on day 7 was comparable to that observed for the CS1-infected wells at this time. Thus, we conclude that stable LAP+ latency was established in the majority of neurons in these cultures. Again, the level of reporter gene expression varied between neurons, with about 10% displaying high levels of β-gal. The survival in the previous experiment of the majority of CS1-infected neurons that expressed HSV Ags at day 1 p.i. suggests that a proportion of the LAP+ latently infected neurons had undergone some viral gene expression prior to the establishment of latency.
Assessment of latency establishment and reactivation using a “double-tagged” recombinant virus.
A “double-tagged” recombinant virus designated VC1 was constructed by incorporating a CMV IE1 promoter-driven EGFP cassette into the nonessential Us5 locus (gJ) on the CS1 backbone (Fig. 1). Following infection with 5 × 105 PFU of VC1/well, EGFP expression was monitored in the living cultures daily and all wells showed comparable high levels of neuronal infection on days 1 to 3 p.i. (not shown). In order to generate a more accurate assessment of the numbers of neurons infected, coverslips were fixed and immunostained for β-tub on days 3 and 14 p.i. (Fig. 3g and h). CMV IE1-driven EGFP expression was detected in 55% of neurons on day 3 p.i. and in 2% of neurons on day 14 p.i. (Table 3). Over this 11-day period the number of GFP+ neurons decreased by an average of 404 neurons per well, which was greater than any discernible loss of neurons (an average of 40 neurons/well). We estimate that 50 to 58% of the neurons surviving on day 14 p.i. had undergone prior CMV IE1 promoter-driven expression, conclusively demonstrating that neurons can survive at least limited activation of lytic viral promoters. To determine the degree of LAP+ latency, further coverslips fixed on day 14 p.i. were immunostained for β-gal (Fig. 3a and k) and 38% of neurons harbored LAP+ latent genomes, 10% of which (76 of 749 β-gal+ neurons) displayed the intense β-gal staining observed previously with CS1. In this experiment we were therefore unable to determine if the LAP+ latent population had undergone prior reporter gene expression. Thus, either neurons which survived initial GFP expression went on to establish LAP+ latency, and our β-gal staining failed to detect 12 to 20% of this population, or the LAP+ population did not undergo prior GFP expression. In both scenarios the potential exists for a population of neurons in the culture to harbor latent genomes in which the LAP is inactive.
TABLE 3.
VC1-infected neuronal culturesa
| Reporter cassette | Days p.i.b | Total no. of neurons/wellc | No. of positive neurons/welld | % Positive neuronse |
|---|---|---|---|---|
| CMV>GFPf | 3 | 897 | 440 | 49 |
| 574 | 367 | 64 | ||
| CMV>GFP | 14 | 789 | 13 | 2 |
| LAP>β-galg | 14 | 693 | 238 | 34 |
| 506 | 231 | 47 | ||
| 795 | 280 | 35 |
Neuronal cultures infected with 106 PFU of VC1 per well.
For enumeration, wells were fixed at the stated times p.i. and were immunostained for β-tub alone or β-tub and β-gal.
Total number of neurons (β-tub+ cells) in the wells examined.
Total number of neurons per well showing staining for reporter gene expression.
Percentage of neurons in the well showing reporter gene expression.
CMV IE1 promoter-driven expression of GFP.
LAP-driven expression of β-gal.
To assess the number of neurons which harbored latent viral genomes that were responsive to reactivation stimuli, further wells were examined for GFP expression, initially on day 14 p.i. to estimate the background number of GFP+ neurons in each well and then again on day 15 p.i., 24 h after the application of reactivation stimuli (NGF withdrawal or TSA). The number of neurons exhibiting CMV IE1 promoter activity (i.e., GFP+) in the wells given normal medium remained low at 2 to 6% (Table 4), indicating that neither the change of medium nor the illumination for fluorescence on day 14 p.i. had resulted in activation of GFP expression. In contrast, GFP was observed in 31 to 68% of neurons following the withdrawal of NGF or 26% of neurons given TSA (Fig. 3l and m), an average increase of 6.3- or 4.3-fold above the pretreatment estimates (Table 4). Activation of GFP expression was not observed in nonneuronal cells following either treatment. To test the stability of this latent population with the potential for activation, further wells were left until day 27 p.i. and were then given the same reactivation stimuli (Table 4). In the normal medium control, the background level of GFP+ neurons on day 28 p.i. was 1 to 10% (Table 4). However, 31 to 39% of neurons were GFP+ after NGF withdrawal (6.4-fold greater on average than for medium containing NGF), and 33 to 39% of neurons were GFP+ after addition of TSA (an average 6.5-fold greater than for normal growth medium). On both days 15 and 28 p.i., the level of CMV IE1 activation observed with either NGF withdrawal or TSA induction was similar to the level of LAP+ latency observed on day 14 p.i.. We noted that NGF withdrawal affected neuronal survival or β-tub expression, with an average loss of 40 to 55% of neurons, compared to the normally fed control, in the 24 h following withdrawal (Table 4). Further, we observed that activation of the CMV IE1 promoter by NGF withdrawal (but not by TSA) decreased with extended time in culture and speculate that this is a consequence of loss of an NGF-dependent neuronal subpopulation from the cultures over time either through death or maturation to an NGF-independent state. TSA treatment at the dose used (660 nM) did not result in adverse effects on either neuronal numbers or morphology, and we conclude that this is the more reproducible method of inducing CMV IE1 promoter activity in our model system.
TABLE 4.
VC1-infected cultures on different days p.i.
| Day p.i. | Stimulus | Total no. of neurons/well | GFP+/well prereactivationa | GFP+/well after reactivationb | % Positive neurons |
|---|---|---|---|---|---|
| 15 | Normal mediumc | 845 | 18 | 19 | 2 |
| 758 | 55 | 42 | 6 | ||
| NGF−d | 389 | 38 | 264 | 68 | |
| 327 | 18 | 103 | 31 | ||
| TSA+e | 941 | 50 | 213 | 26 | |
| 28 | Normal medium | 655 | NAf | 9 | 1 |
| 657 | NA | 69 | 10 | ||
| NGF− | 334 | NA | 106 | 31 | |
| 262 | NA | 101 | 39 | ||
| TSA+ | 653 | NA | 213 | 33 | |
| 1,012 | NA | 418 | 39 |
Numbers of GPP+ neuron-like cells (identified by morphology) in living cultures prior to replacement of culture medium on day 14 p.i.
Values in top half of table reflect number of GFP+ neurons per well 24 h after replacement of culture medium (i.e., day 15), which was enumerated from fixed cultures immunostained for β-tub. Values in lower half reflect number of GFP+ neurons per well on day 28 p.i. 24 h after replacement of culture medium.
Culture medium was replaced with fresh normal growth medium.
Culture medium was replaced with fresh growth medium lacking NGF and containing 0.1 pl of rabbit anti-NGF antiserum/ml.
Culture medium was replaced with fresh growth medium supplemented with 660 nM TSA.
NA, not applicable.
To confirm that activation of reporter promoters such as CMV IE1 reflected expression of native viral proteins in neurons, cultures infected with 3 × 105 PFU/well were fixed 24 h p.i. and were immunostained for IE110 expression using a mouse monoclonal antibody (Fig. 3i and j). Of 1,007 neurons (identified by morphology) in which IE110 or GFP was detected, 972 neurons (97%) showed dual expression of GFP and IE110, 27 (3%) stained for IE110 alone, and 8 (1%) were only GFP+ (example shown in Fig. 3i). This indicates that activation of lytic cycle reporter promoters accurately reflects the expression of native viral proteins. As the antibody used was directed against an N-terminal epitope, synthesis of full-length IE110 was confirmed using a polyclonal antiserum raised to C-terminal epitopes (data not shown). Consistent with the recent observations of Chen and coworkers (9), we observe that IE110 staining in neurons is predominantly cytoplasmic, although some neurons display both nuclear and cytoplasmic staining. In nonneuronal cells IE110 staining was generally nuclear.
To address the relevance of activation of the CMV IE1 promoter in latently infected neurons, we compared the levels of TSA-induced activation with that of the HSV IE110 reporter promoter. Neuronal cultures were infected with CS5 or gH−TK−110LacZ at 106 PFU/well and were left to establish latency. On day 21 p.i. culture medium was replaced with either normal growth medium or growth medium containing 660 nM TSA. The wells were fixed 24 h later and were dually immunostained for β-gal and β-tub (Fig. 2g to j). The number of neurons expressing β-gal was assessed (Table 5). Addition of normal growth medium resulted in low levels (2%) of β-gal+ neurons for both viruses. Application of medium containing TSA, however, resulted in 53% of neurons in CS5-infected wells, or 18% of neurons in gH−TK−110LacZ-infected wells, showing expression of β-gal. Thus, activation of the CMV IE1 promoter following the addition of TSA reflects the response of a legitimate HSV IE promoter. The difference in the levels of activation observed may reflect the previously noted differences in promoter strength or may indicate that the CMV IE1 promoter is more sensitive than IE110 to TSA induction.
TABLE 5.
Activation of CMV and IE110 promoters in latently infected culturesa
| Reporter cassette | Stimulusef | No. of neurons countedb (% β-gal+)c |
|---|---|---|
| CMV>lacZd | Normal feed | 187 (2) |
| TSA+ | 163 (53) | |
| IE110>lacZg | Normal feed | 189 (2) |
| TSA+ | 397 (18) |
Neuronal cultures 22 days after infection with 106 PFU of either CS5 or gH−TK−IE110LacZ per well. They were fixed and dually immunostained for β-gal and β-tub.
Total neurons (β-tub+) examined (two wells per data point).
Percentage of neurons showing β-gal expression.
CMV IE1 promoter-driven lacZ expression in CS5-infected wells.
Culture medium for normal feed was replaced with fresh growth medium on day 21 p.i.
Culture medium for TSA+ was replaced with fresh growth medium supplemented with 660 nM TSA on day 21 p.i.
IE110 promoter-driven lacZ expression in gH−TK−IE110LacZ-infected wells.
In conclusion, our data indicate that in this model system, following infection with gH−TK− recombinant viruses, latent HSV genomes are harbored for extended periods in a state that remains responsive to reactivation stimuli.
DISCUSSION
In order to address fundamental questions regarding the molecular mechanisms of latency establishment, maintenance, and reactivation, we have established an in vitro model of neuronal latency using primary cultures of dissociated drg and gH−TK− recombinant viruses bearing reporter genes under the control of IE gene promoters (CMV IE1 and HSV-1 IE110) and the LAP. Following infection, HSV Ag- or IE promoter-driven reporter gene expression is detectable in neurons within 24 h. Reporter gene expression from either the CMV IE1 or IE110 promoter peaked within the first 3 days p.i., with 58 to 67% of neurons positive, depending on the infection dose used. This activation of lytic viral reporter promoters reflected the initiation of viral protein synthesis in neurons, as observed with the CS1 mutant (in 54% of neurons) and in the dual detection of IE110- and CMV IE1-driven GFP expression in 97% of neurons infected with VC1. Lytic phase reporter promoter expression decreased from day 3 p.i., in terms of both overall numbers of positive neurons and the intensity of signal, with loss of β-gal essentially complete by 6 to 8 days p.i.. Throughout the initial infection, CMV IE1 promoter activity in neurons is greater and more prolonged than that observed for the IE110 promoter and may reflect relative promoter strengths or differences in promoter regulation in neurons. We have demonstrated that the loss of β-gal+ neurons from day 3 p.i. is not accounted for by neuronal death, indicating reporter promoter shutdown and suggesting that neurons can tolerate some degree of lytic phase reporter gene expression. It will be of interest to examine expression of the individual viral protein classes during neuronal infection in this model to determine the extent of progression through the viral gene cascade. The low frequency of IE promoter activity observed at later times (in 2 to 5% of neurons) most likely represents spontaneous abortive reactivation events; however, it is possible that in a subpopulation of neurons, complete lytic cycle promoter shutdown does not occur. More extensive analysis will be required to accurately determine the degree of neuronal death in this system and, in particular, to ascertain whether the expression of viral proteins such as IE110 affects neuronal survival and latency establishment.
We have shown that following infection with 106 PFU of CS1/well, LAP activity is detected in 55% of neurons at early times p.i. (day 4), as is observed for LAT expression in vivo (28, 51). The signal intensity and numbers of β-gal+ neurons continue to increase with time p.i. (up to 83% by day 21 p.i.), as has been noted for other latency systems (29, 48). As total neuron numbers were not determined in this experiment, we cannot rule out the possibility that the increase in LAP+ neurons over time results from selective loss of infected neurons in which the LAP is not active. We observe a wide variation in LAP activity between infected neurons, consistent with our previous observations of LATs and LAP activity in vivo (2, 25) and with the findings that certain neuronal subtypes are more permissive for LAT expression during latency in vivo (28, 40, 66). Further studies are required to determine if there is population restriction on latency establishment and LAP activity in our model.
Following shutdown of lytic phase promoter activity, viral genomes are harbored in neurons and can be induced to activate lytic phase promoters using NGF withdrawal (49) or addition of TSA. In latently infected ganglia, TSA has been reported to increase the number of latently infected neurons undergoing reactivation during the first 22 h postexplant by 10-fold, with a commensurate increase in infectious virus production. In this same study, the ICP0 promoter was shown to be upregulated by TSA in the setting of the latent viral genome in explanted ganglia (N. M. Sawtell and S. K. Burridge, Abstr. 24th Int. Herpesvirus Workshop, abstr. 2.0016, 1999). It will now be of interest to determine the pattern of legitimate viral gene expression following reactivation stimuli by using antisera to specific protein classes. The ability of TSA to induce IE promoter activation is noteworthy; Deshmane and Fraser (12) determined that latent HSV DNA is associated with nucleosomes in vivo, raising the possibility that chromatin modification may have an important regulatory role in governing viral gene activity during latency. It is conceivable that the acetylation of histones associated with one or more IE gene promoters may be a critical step in facilitating viral reactivation. Such a hypothesis is not without precedent in herpesvirus latency. In the case of Epstein-Barr virus, latent DNA is organized in nucleosomes (15) and a key step in the switch from latency to the productive cycle is the expression of the IE gene BZLF1 (11). Recently Jenkins and coworkers (24) have determined that the acetylation of histones in chromatin around the BZLF1 promoter region is involved in activation of its expression. Alternatively, derepression of viral gene expression through histone modification may be controlled by a viral gene product. Hobbs and DeLuca (22) have recently demonstrated that IE110 and TSA have similar effects on gene expression from cellular and quiescent HSV genomes. They propose that like TSA, IE110 derepresses gene expression by altering chromatin structure, possibly by reducing the stability and abundance of proteins involved in histone metabolism or by IE110 induction of proteins which bind and alter nucleosome structure, such as Gadd45. Lastly, our TSA data may reflect a role for a host cell product(s) controlled by chromatin condensation in the initiation of reactivation. Such TSA-induced changes in neural gene expression pathways have been demonstrated, and in PC12 cells TSA can induce de novo synthesis of cellular factors that specifically block an NGF-induced signal transduction pathway required for neurite outgrowth (41). It is feasible that such blockage of NGF signal transduction pathways by TSA may simulate the aspects of NGF withdrawal that induce virus reactivation.
We have demonstrated that neurons can survive activation of IE reporter promoters and also limited viral gene expression following infection with TK− viruses; however, we are at present unable to determine whether any of the LAP+ cells present at late times p.i. had undergone prior viral gene expression. Attempts to increase the proportion of neurons showing lytic cycle gene expression by increasing the viral load resulted in significant neuronal loss, indicating that there is a threshold for the control of virus infection in this system. However, by using the “double-tagged” virus VC1, it should be possible to follow the infection of individual neurons in this system to determine if neurons in which the CMV IE1 promoter is activated subsequently establish latency. Although our focus was neuronal infection, it is clear that nonneuronal cells were infected, and although no accurate assessment of nonneuronal cell loss was taken, our general impression was that the viruses used were cytotoxic to these cells. Furthermore, LAP-driven β-gal expression was not detected in nonneuronal cells and, following TSA treatment, induction of reporter promoters was restricted to neurons.
If the default model of latency is absolute, then neurons that initiated the viral gene cascade would not establish latency. Therefore, in our VC1 infection, for example, we could infer that the majority of neurons were infected, that 38% entered LAP+ latency directly, and that 55% initiated viral gene expression which, but for the absence of TK, would result in productive infection and presumably death of the cell. However, it is known that latently infected neurons in vivo can harbor high-copy latency (44, 46), suggesting that neurons can survive limited viral gene expression, including DNA replication. Further, there is evidence to suggest that although neurons may enter latency in the absence of lytic cycle gene expression, efficient establishment requires some IE110 expression (61). Therefore, there may be multiple routes into latency, and the activation of viral gene expression and subsequent survival of neurons observed in our model system may well reflect what occurs in vivo. Our observation that neurons survive the initiation of reporter promoter activity and limited viral gene expression (at least IE110) is consistent with the theory that there are two routes of latency establishment. That is, in one population of neurons, virus directly enters into latency and establishes low-copy infection, and in a second population, there is an abortive infection where limited viral replication could potentially give rise to high-genome-copy latency. LATs may be expressed in all the latently infected cells, and the variation in signal intensity could be directly related to virus copy number. Alternatively, it is conceivable that LAT expression may be restricted to one of the two latent populations, and in vivo it has been shown that more neurons harbor viral DNA than are detected by in situ hybridization for LATs (30, 37). Transcriptionally silent or “quiescent” viral genomes harbored long-term in nonneuronal cells in vitro may reactivate but are resistant to many stimuli that reactivate latent virus in LAT+ neuronal models in vitro and in vivo (20, 56). This has led to the proposal that there are different degrees of viral genome repression which may be influenced by the host cell or route of entry into latency (36). Thus, it will be of interest to determine whether reactivation is restricted to LAP+ neurons and, if not, whether the genomes harbored in neurons which do not synthesize LATs are, like quiescent genomes in nonneuronal cells, more resistant to reactivation stimuli. Studies are currently under way to assess whether activation of CMV IE1-driven GFP expression following TSA induction is restricted to β-gal+ neurons. Finally, although this model uses replication-defective virus mutants to infect neurons in the absence of the normal immunological controls present in vivo, this system offers a potentially powerful tool for the analysis of the primary molecular events that occur during latency establishment and reactivation.
ACKNOWLEDGMENTS
We thank Janet May for technical assistance and Nathalie Babic for the provision of C12 virus and help with the establishment of the neuronal culture system. We also thank Roger Everett for the provision of IE110 antisera and Chris Preston for valuable comments on the manuscript.
This work was supported by the United Kingdom Medical Research Council.
REFERENCES
- 1.Alvira R M, Goins W F, Cohen J B, Glorioso J G. Genetic studies exposing the splicing events involved in herpes simplex virus type 1 latency-associated transcript production during lytic and latent infection. J Virol. 1999;73:3866–3878. doi: 10.1128/jvi.73.5.3866-3876.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arthur J, Efstathiou S, Simmons A. Intranuclear foci containing low abundance herpes simplex virus latency-associated transcripts visualized by non-isotopic in situ hybridization. J Gen Virol. 1993;74:1363–1370. doi: 10.1099/0022-1317-74-7-1363. [DOI] [PubMed] [Google Scholar]
- 3.Arthur J L, Everett R, Brierley I, Efstathiou S. Disruption of the 5′ and 3′ splice sites flanking the 2-kilobase latency-associated transcripts: evidence for alternate splicing in lytic and latent infections. J Gen Virol. 1998;79:107–116. doi: 10.1099/0022-1317-79-1-107. [DOI] [PubMed] [Google Scholar]
- 4.Babic N, Rodger G, Arthur J, Minson A C. A study of primary neuronal infection by mutants of herpes simplex virus type 1 lacking dispensable and non-dispensable glycoproteins. J Gen Virol. 1999;80:2403–2409. doi: 10.1099/0022-1317-80-9-2403. [DOI] [PubMed] [Google Scholar]
- 5.Block T M, Spivack J G, Steiner I, Deshmane S, McIntosh M T, Lirette R P, Fraser N W. A herpes simplex virus type 1 latency-associated transcript mutant reactivates with normal kinetics from latent infection. J Virol. 1990;64:3417–3426. doi: 10.1128/jvi.64.7.3417-3426.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Block T M, Barney S, Masonis J, Maggioncalda J, Rattan S, Valyi-Nagy T, Fraser N W. Long-term herpes simplex virus infections of nerve growth factor-differentiated PC12 cells. J Gen Virol. 1994;75:2481–2487. doi: 10.1099/0022-1317-75-9-2481. [DOI] [PubMed] [Google Scholar]
- 7.Bloom D C, Hill J M, Devi-Rao G, Wagner E K, Feldman L T, Stevens J G. A 348-base-pair region in the latency-associated transcript facilitates herpes simplex virus type 1 reactivation. J Virol. 1996;70:2449–2459. doi: 10.1128/jvi.70.4.2449-2459.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boursnell E G M, Entwistle C, Blakeley D, Roberts C, Duncan I A, Chisholm S E, Martin G M, Jennings R, Ni Challanain D, Sobek I, Inglis S C, McLean C S. A genetically inactivated herpes simplex virus type 2 (HSV-2) vaccine provides effective protection against primary and recurrent HSV-2 disease. J Infect Dis. 1997;175:16–25. doi: 10.1093/infdis/175.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen X, Li J, Mata M, Goss J, Wolfe D, Glorioso J C, Fink D J. Herpes simplex virus type 1 ICP0 protein does not accumulate in the nucleus of primary neurons in culture. J Virol. 2000;74:10132–10141. doi: 10.1128/jvi.74.21.10132-10141.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clements G B, Jamieson F E. Reactivation of latent herpes simplex virus-1 (HSV) from mouse footpad cells demonstrated by in situ hybridization. Arch Virol. 1989;104:95–106. doi: 10.1007/BF01313811. [DOI] [PubMed] [Google Scholar]
- 11.Countryman J, Miller G. Activation of expression of latent Epstein-Barr herpesvirus after gene transfer with a small cloned subfragment of heterogeneous DNA. Proc Natl Acad Sci USA. 1985;82:4085–4089. doi: 10.1073/pnas.82.12.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deshmane S L, Fraser N W. During latency, herpes simplex virus type 1 DNA is associated with nucleosomes in a chromosome structure. J Virol. 1989;63:943–947. doi: 10.1128/jvi.63.2.943-947.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dobson A T, Margolis T P, Sederati F, Stevens J G, Feldman L T. A latent, nonpathogenic HSV-1-derived vector stably expresses β-galactosidase in mouse neurons. Neuron. 1990;5:353–360. doi: 10.1016/0896-6273(90)90171-b. [DOI] [PubMed] [Google Scholar]
- 14.Doerig C, Pizer L I, Wilcox C L. Detection of the latency-associated transcript in neuronal cultures during the latent infection with herpes simplex virus type 1. Virology. 1991;183:423–426. doi: 10.1016/0042-6822(91)90159-9. [DOI] [PubMed] [Google Scholar]
- 15.Dyson P J, Farrell P J. Chromatin structure of Epstein-Barr virus. J Gen Virol. 1985;66:1931–1940. doi: 10.1099/0022-1317-66-9-1931. [DOI] [PubMed] [Google Scholar]
- 16.Efstathiou S, Minson A C, Field H J, Anderson J R, Wildy P. Detection of herpes simplex virus-specific DNA sequences in latently infected mice and in humans. J Virol. 1986;57:446–455. doi: 10.1128/jvi.57.2.446-455.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farrell J M, Dobson A T, Feldman L T. Herpes simplex virus latency-associated transcript is a stable intron. Proc Natl Acad Sci USA. 1991;88:790–794. doi: 10.1073/pnas.88.3.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Halford W P, Gebhardt B M, Carr D J. Mechanisms of herpes simplex virus type 1 reactivation. J Virol. 1996;70:5051–5060. doi: 10.1128/jvi.70.8.5051-5060.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harris R A, Everett R D, Zhu X, Silverstein S, Preston C M. Herpes simplex virus type 1 immediate-early protein Vmw110 reactivates latent herpes simplex virus type 2 in an in vitro system. J Virol. 1989;63:3513–3515. doi: 10.1128/jvi.63.8.3513-3515.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harris R A, Preston C M. 1814. J. Gen. Virol. 72:907–913. 1991. Establishment of latency in vitro by the herpes virus type 1 mutant. [DOI] [PubMed] [Google Scholar]
- 21.Hill J M, Garza H H, Jr, Su Y-H, Meegalla R, Hanna L A, Loutsch J M, Thompson H W, Varnell E D, Bloom D C, Block T M. A 437-base-pair deletion at the beginning of the latency-associated transcript promoter significantly reduced adrenergically induced herpes simplex virus type 1 ocular reactivation in latently infected rabbits. J Virol. 1997;71:6555–6559. doi: 10.1128/jvi.71.9.6555-6559.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hobbs W E I, DeLuca N A. Perturbation of cell cycle progression and cellular gene expression as a function of herpes simplex virus ICP0. J Virol. 1999;73:8245–8255. doi: 10.1128/jvi.73.10.8245-8255.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Javier R T, Stevens J G, Dissette V B, Wagner E K. A herpes simplex virus transcript abundant in latently infected neurons is dispensable for establishment of the latent state. Virology. 1988;166:254–257. doi: 10.1016/0042-6822(88)90169-9. [DOI] [PubMed] [Google Scholar]
- 24.Jenkins P J, Binne U K, Farrell P J. Histone acetylation and reactivation of Epstein-Barr virus from latency. J Virol. 2000;74:710–720. doi: 10.1128/jvi.74.2.710-720.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lachmann R H, Efstathiou S. Utilization of the herpes simplex virus type 1 latency-associated regulatory region to drive stable reporter gene expression in the nervous system. J Virol. 1997;71:3197–3207. doi: 10.1128/jvi.71.4.3197-3207.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lachmann R H, Sadarangani M, Atkinson H R, Efstathiou S. An analysis of herpes simplex virus gene expression during latency establishment and reactivation. J Gen Virol. 1999;80:1271–1282. doi: 10.1099/0022-1317-80-5-1271. [DOI] [PubMed] [Google Scholar]
- 27.Leib D A, Coen D M, Bogard C L, Hicks K A, Yager D R, Knipe D M, Tyler K L, Schaffer P A. Immediate-early regulatory gene mutants define different stages in the establishment and reactivation of herpes simplex virus latency. J Virol. 1989;63:759–768. doi: 10.1128/jvi.63.2.759-768.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Margolis T P, Sedarati F, Dobson A T, Feldman L T, Stevens J G. Pathways of viral gene expression during acute neuronal infection with HSV-1. Virology. 1992;189:150–160. doi: 10.1016/0042-6822(92)90690-q. [DOI] [PubMed] [Google Scholar]
- 29.Marshall K R, Lachmann R H, Efstathiou E, Rinaldi A, Preston C M. Long-term transgene expression in mice infected with a herpes simplex virus type 1 mutant severely impaired for immediate-early gene expression. J Virol. 2000;74:956–964. doi: 10.1128/jvi.74.2.956-964.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mehta A, Maggioncalda J, Bagasra O, Thikkavarapu S, Saikumari P, Valyi N T, Fraser N W, Block T M. In situ DNA PCR and RNA hybridization detection of herpes simplex virus sequences in trigeminal ganglia of latently infected mice. Virology. 1995;206:633–640. doi: 10.1016/s0042-6822(95)80080-8. [DOI] [PubMed] [Google Scholar]
- 31.Mellerick D M, Fraser N W. Physical state of the latent herpes simplex virus genome in a mouse model system: evidence suggesting an episomal state. Virology. 1987;158:265–275. doi: 10.1016/0042-6822(87)90198-x. [DOI] [PubMed] [Google Scholar]
- 32.Mitchell W J, Lirette R P, Fraser N W. Mapping of low abundance latency-associated RNA in the trigeminal ganglia of mice latently infected with herpes simplex virus type 1. J Gen Virol. 1990;71:125–132. doi: 10.1099/0022-1317-71-1-125. [DOI] [PubMed] [Google Scholar]
- 33.O'Hare, P., and C. R. Goding. 1988. Herpes simplex virus regulatory elements and the immunoglobulin octamer domain bind a common factor and are both targets for virion transactivation. Cell 52:435–445. [DOI] [PubMed]
- 34.Perng G C, Slanina S M, Yukht A, Drolet B S, Keleher W J, Ghiasi H, Nesburn A B, Wechsler S L. A herpes virus simplex virus type 1 latency-associated transcript mutant with increased virulence and reduced spontaneous reactivation. J Virol. 1999;73:920–929. doi: 10.1128/jvi.73.2.920-929.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perng G C, Slanina S M, Yukht A, Ghiasi H, Nesburn A B, Wechsler S L. The latency-associated transcript gene enhances establishment of herpes simplex virus type 1 latency in rabbits. J Virol. 2000;74:1885–1891. doi: 10.1128/jvi.74.4.1885-1891.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Preston C M. Repression of viral transcription during herpes simplex virus latency. J Gen Virol. 2000;81:1–19. doi: 10.1099/0022-1317-81-1-1. [DOI] [PubMed] [Google Scholar]
- 37.Ramakrishnan R, Levine M, Fink D J. PCR-based analysis of herpes simplex virus type 1 latency in the rat trigeminal ganglion established with a ribonucleotide reductase-deficient mutant. J Virol. 1994;68:7083–7091. doi: 10.1128/jvi.68.11.7083-7091.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rock D L, Fraser N W. Detection of HSV-1 genome in central nervous system of latently infected mice. Nature (London) 1983;302:523–525. doi: 10.1038/302523a0. [DOI] [PubMed] [Google Scholar]
- 39.Rødahl E, Haarr L. Analysis of the 2-kilobase latency-associated transcript expressed in PC12 cells productively infected with herpes simplex virus type 1: evidence for a stable, nonlinear structure. J Virol. 1997;71:1703–1707. doi: 10.1128/jvi.71.2.1703-1707.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rodahl E, Stevens J G. Differential accumulation of herpes simplex type 1 latency-associated transcripts in sensory automatic ganglia. Virology. 1992;189:385–388. doi: 10.1016/0042-6822(92)90721-z. [DOI] [PubMed] [Google Scholar]
- 41.Sano M, Kitajima S. Inhibition of nerve growth factor-induced outgrowth of neurites by trichostatin A requires de novo protein synthesis in PC12D cells. Brain Res. 1996;742:195–202. doi: 10.1016/s0006-8993(96)01007-4. [DOI] [PubMed] [Google Scholar]
- 42.Sawtell N M. Comprehensive quantification of herpes simplex virus latency at the single-cell level. J Virol. 1997;71:5423–5431. doi: 10.1128/jvi.71.7.5423-5431.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sawtell N M. The probability of in vivo reactivation of herpes simplex virus type 1 increases with the number of latently infected neurons in the ganglia. J Virol. 1998;72:6888–6892. doi: 10.1128/jvi.72.8.6888-6892.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sawtell N M, Poon D K, Tansky C S, Thompson R L. The latent herpes simplex virus type 1 genome copy number in individual neurons is virus strain specific and correlates with reactivation. J Virol. 1998;72:5343–5350. doi: 10.1128/jvi.72.7.5343-5350.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sawtell N M, Thompson R L. Herpes simplex virus type 1 latency-associated transcription unit promotes anatomical site-dependent establishment and reactivation from latency. J Virol. 1992;66:2157–2169. doi: 10.1128/jvi.66.4.2157-2169.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Slobedman B, Efstathiou S, Simmons A. Quantitative analysis of herpes simplex virus DNA and transcriptional activity in ganglia of mice latently infected with wild-type and thymidine kinase-deficient viral strains. J Gen Virol. 1994;75:2469–2474. doi: 10.1099/0022-1317-75-9-2469. [DOI] [PubMed] [Google Scholar]
- 47.Smith, C., R. H. Lachmann, and S. Efstathiou. 2000. Expression from the herpes simplex virus type 1 latency-associated promoter in the murine central nervous system. J. Gen. Virol. 81:649–662. [DOI] [PubMed]
- 48.Smith R L, Escudero J M, Wilcox C L. Regulation of the herpes simplex virus latency-associated transcripts during establishment of latency in sensory neurons in vitro. Virology. 1994;202:49–60. doi: 10.1006/viro.1994.1321. [DOI] [PubMed] [Google Scholar]
- 49.Smith R L, Pizer L I, Johnson E M, Wilcox C L. Activation of second messenger pathways reactivates latent herpes simplex virus in neuronal cultures. Virology. 1992;188:311–318. doi: 10.1016/0042-6822(92)90760-m. [DOI] [PubMed] [Google Scholar]
- 50.Speck P G, Simmons A. Divergent molecular pathways of productive and latent infection with a virulent strain of herpes simplex virus type 1. J Virol. 1991;65:4001–4005. doi: 10.1128/jvi.65.8.4001-4005.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Speck P G, Simmons A. Synchronous appearance of antigen-positive and latently infected neurons in spinal ganglia of mice infected with a virulent strain of herpes simplex virus. J Gen Virol. 1992;73:1281–1285. doi: 10.1099/0022-1317-73-5-1281. [DOI] [PubMed] [Google Scholar]
- 52.Steiner I, Spivack J G, Lirette R P, Brown S M, MacLean A R, Subak-Sharpe J, Fraser N W. Herpes simplex virus type 1 latency-associated transcripts are evidently not essential for latent infection. EMBO J. 1989;8:505–511. doi: 10.1002/j.1460-2075.1989.tb03404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stevens J G, Cook M L. Latent herpes simplex virus in spinal ganglia of mice. Science. 1971;173:843–845. doi: 10.1126/science.173.3999.843. [DOI] [PubMed] [Google Scholar]
- 54.Stevens J G, Wagner E K, Devi-Rao G B, Cook M L, Feldman L T. RNA complementary to a herpesvirus α gene mRNA is prominent in latently infected neurons. Science. 1987;235:1056–1059. doi: 10.1126/science.2434993. [DOI] [PubMed] [Google Scholar]
- 55.Stow N D, Wilkie N M. An improved technique for obtaining enhanced infectivity with herpes simplex virus type 1 DNA. J Gen Virol. 1976;33:447–458. doi: 10.1099/0022-1317-33-3-447. [DOI] [PubMed] [Google Scholar]
- 56.Su Y-H, Meegalla R L, Chowhan R, Cubitt C, Oakes J E, Lausch R N, Fraser N W, Block T M. Human corneal cells and other fibroblasts can stimulate the appearance of herpes simplex virus from quiescently infected PC12 cells. J Virol. 1999;73:4171–4180. doi: 10.1128/jvi.73.5.4171-4180.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thompson R L, Sawtell N M. The herpes simplex virus type 1 latency-associated transcript gene regulates the establishment of latency. J Virol. 1997;71:5432–5440. doi: 10.1128/jvi.71.7.5432-5440.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wagner E K, Bloom D C. Experimental investigation of herpes simplex virus latency. Clin Microbiol Rev. 1997;10:419–443. doi: 10.1128/cmr.10.3.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilcox C L, Johnson E M., Jr Characterization of nerve growth factor-dependent herpes simplex virus latency in neurons in vitro. J Virol. 1988;62:393–399. doi: 10.1128/jvi.62.2.393-399.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wilcox C L, Johnson E M., Jr Nerve growth factor deprivation results in the reactivation of latent herpes simplex virus in vitro. J Virol. 1987;61:2311–2315. doi: 10.1128/jvi.61.7.2311-2315.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wilcox C L, Smith R L, Everett R D, Mysofski D. The herpes simplex virus type 1 immediate-early protein ICP0 is necessary for the efficient establishment of latent infection. J Virol. 1997;71:6777–6785. doi: 10.1128/jvi.71.9.6777-6785.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wilcox C L, Smith R L, Freed C R, Johnson E M. Nerve growth factor-dependent herpes simplex virus latency in neurons in vitro. J Neurosci. 1990;104:1268–1275. doi: 10.1523/JNEUROSCI.10-04-01268.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wildy P, Field H J, Nash A A. Classical herpes latency revisited. In: Mahy B W J, Minson A C, Darby G K, editors. Virus persistence symposium 33. Cambridge, United Kingdom: Cambridge University Press; 1982. pp. 133–167. [Google Scholar]
- 64.Wilkinson G W, Akrigg A. Constitutive and enhanced expression from the CMV major IE promoter in a defective adenovirus vector. Nucleic Acids Res. 1992;20:2233–2239. doi: 10.1093/nar/20.9.2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu T T, Su Y H, Block T M, Taylor J M. Atypical splicing of the latency-associated transcripts of herpes simplex type 1. Virology. 1998;243:140–149. doi: 10.1006/viro.1998.9036. [DOI] [PubMed] [Google Scholar]
- 66.Yang L, Voytek C C, Margolis T P. Immunohistochemical analysis of primary sensory neurons latently infected with herpes simplex virus type 1. J Virol. 2000;74:209–217. doi: 10.1128/jvi.74.1.209-217.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zabolotny J M, Krummenacher C, Fraser N W. The herpes simplex virus type 1 2.0-kilobase latency-associated transcript is a stable intron which branches at a guanosine. J Virol. 1997;71:4199–4208. doi: 10.1128/jvi.71.6.4199-4208.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]