Abstract
Background
Dalbavancin is a lipoglycopeptide antibiotic with a wide spectrum of activity against Gram-positive bacteria, including MDR isolates. Its pharmacokinetic properties and administration patterns could be useful for the treatment of bone and joint infections, especially prosthetic joint infections (PJIs).
Introduction
We report the case of an 80-year-old man who experienced an acute periprosthetic joint infection of his right total knee arthroplasty (TKA). A DAIR procedure was done with tissue sampling, which allowed identification of a linezolid-resistant MDR S. epidermidis (LR-MDRSE) strain. The patient was then treated with dalbavancin (four injections).
Methods
We studied the phenotypic and genomic evolution of the strains and plasma through concentrations of dalbavancin at different points in time.
Results
After four injections (1500 mg IV) of dalbavancin over a 6 month period, the dalbavancin MIC increased 4-fold. Calculated fAUC0–24/MIC ratios were 945, 1239 and 766.5, respectively, at Days 49, 71 and 106, assuming an MIC of 0.032 mg/L. The PFGE dendrogram revealed 97% similarity among all the isolates. These results suggest acquisition by the S. epidermidis strain of dalbavancin resistance when the patient underwent dalbavancin treatment. A 4-amino-acid deletion in the walK gene coinciding with the emergence of phenotypic resistance was revealed by WGS without any other relevant indels.
Conclusions
Despite dalbavancin treatment with pharmacokinetic management, emerging dalbavancin resistance in S. epidermidis was observed, resulting in treatment failure. This outcome led to a prosthesis revision and long-term suppressive antibiotic therapy, with no recurrence of PJI after an 18 month follow-up.
Introduction
Dalbavancin is a long-acting lipoglycopeptide active against Gram-positive bacteria, including MRSA and VRE harbouring vanB genes.1–3 Regardless of authorization for the treatment of acute bacterial skin and skin structure infections (ABSSSIs), dalbavancin is mainly used in off-label indications such as bone and joint infections (BJIs), including periprosthetic infections (PJIs), endocarditis or vascular graft infections.4
Its pharmacokinetic profile with a long half-life makes it particularly suitable for the treatment of BJI. In addition, dalbavancin allows spaced administrations along with good bone penetration5 and an excellent tolerability profile.6 Moreover, in vitro models have shown dalbavancin is also active against staphylococcal biofilm in association with rifampicin.7–9
Dalbavancin has a similar mechanism of action and spectrum as vancomycin but with increased potency, higher protein binding and an elimination half-life up to 60 times longer.10 Due to their pharmacodynamical similarities, cross-resistance among vancomycin, daptomycin and dalbavancin was observed in vitro after exposure to dalbavancin alone had selected for daptomycin- and vancomycin-non-susceptible strains of Staphylococcus aureus.11,12 Based on these results, the authors suggested that dalbavancin exposure may also increase the collateral risk of vancomycin and daptomycin resistance. Neutropenic murine thigh and lung infection models have shown that the 24 h drug-free period under the concentration-versus-time curve/MIC ratio (fAUC0–24/MIC) best correlated with treatment efficacy and supports the use of high doses spaced as far apart as possible.13,14
Dalbavancin primary resistance remains infrequent; the emergence of reduced susceptibility during dalbavancin exposure was described in vitro for S. aureus.15 Recently, Zhang et al.16 reported the occurrence of dalbavancin resistance associated with walK and scrA mutations during dalbavancin treatment. These mutations induced a reduction in long-chain lipid content causing a decrease in membrane fluidity.
Methicillin resistance in CoNS is frequently associated with resistance to other antibiotics, except for glycopeptides, which for many years were the drugs of choice in the treatment of staphylococcal infections. Over recent decades, treatment options for Gram-positive infections have expanded significantly with new glycopeptides, β-lactams, lipopeptides, glycylcyclines and oxazolidinones (linezolid and tedizolid). However, high rates of oxazolidinone consumption or the use of long courses of therapy promote resistance.17 Côrtes et al. and Coustillères et al. described MDR Staphylococcus epidermidis (MDRSE) ST2 clones known to be involved in hospital-acquired infections and capable of persisting and spreading for several years in the same hospital.18,19 These methicillin-resistant S. epidermidis (MRSE) clones belonged to ST2 and harboured both cfr and mutation in genes encoding 23S rRNA and substitution in ribosomal protein L3. This combination conferred high-level resistance to linezolid, but also to tedizolid, further limiting antimicrobial treatment options.
Here we report a case of linezolid-resistant MDRSE (LR-MDRSE) PJI with the selection of a dalbavancin-resistant clone with a resistance mechanism, albeit not yet described. We investigated the phenotypic resistance evolution of the different isolates. A comparative genomic analysis by WGS was conducted to investigate the genomic alterations that could be responsible for reduced susceptibility to dalbavancin.
Materials and methods
Case presentation
An 80-year-old man underwent a revision of a medial unicompartmental knee arthroplasty to total knee arthroplasty (TKA) for progressive arthritis of the lateral joint. Three weeks after surgery, the patient had a scar disunion, with large exposure of the prosthesis, suggesting an early surgical site infection.
It was subsequently decided to perform debridement, antibiotics and implant retention (DAIR) with a change of modular implants. Three bacteriological samples were collected, ultimately revealing a positive LR-MDRSE strain resistant to methicillin, rifampicin and linezolid but still susceptible to dalbavancin and ceftaroline with MICs of 0.032 and 1 mg/L, respectively [STA1, 24 days post arthroplasty (Day 24)]. A treatment shift to dalbavancin 1500 mg IV on Days 35, 49, 71 and 106, combined with doxycycline 200 mg/day for 3 months was initiated.
Seven months after the surgery, the patient was seen for his follow-up consultation. The evolution was considered unfavourable due to a loco-regional inflammatory syndrome with persistent chronic pain. A joint aspirate revealed a dalbavancin-resistant LR-MDRSE strain that was still susceptible to ceftaroline (STA3, Day 240, dalbavancin and ceftaroline MICs of 0.5/0.75 mg/L, respectively).
A revision of the knee was decided upon, with five bacteriological samples collected. As previously, they tested positive with the same LR-MDRSE strain (Day 371). Ceftaroline (1800 mg/day) and doxycycline (200 mg/day) combination treatment was administered for the next 12 weeks. The treatment turned out to be well tolerated and the evolution was favourable (with a clean, closed and non-inflammatory scar without discharge). Eighteen months after the revision TKA, the patient did not report any pain and was in good overall condition. His C-reactive protein level was <4 mg/L (reference range: <4 mg/L). However, suppressive doxycycline therapy was maintained (200 mg/day).
Bacterial isolates
Isolates were identified by MALDI-TOF mass spectrometry (VITEK MS, bioMérieux, Marcy-l’Étoile, France). Antibiotic susceptibility testing was performed using the AST-P668 bioMérieux card using a VITEK XL automated system (bioMérieux). Dalbavancin, ceftaroline, delafloxacin, tigecycline and daptomycin MICs were determined using a 0.5 McFarland bacterial inoculum on Mueller–Hinton agar plates with gradient strips (Liofilchem, Roseto degli Abruzzi, Italy) incubated for 24 h at 35°C. Interpretation of the susceptibility was made according to the EUCAST 2022 criteria.
PFGE
The PFGE procedure was performed as previously described.20 Briefly, a washed bacterial suspension was mixed with 2% SeaPlaque agarose (FMC, Rockland, ME, USA) at 55°C and allowed to solidify into plug moulds at room temperature. Chromosomal DNA was prepared in several incubation and washing steps by using EC buffer (Tris-HCl, NaCl, EDTA, deoxycholate, Sarkosyl), lysostaphin and proteinase K. After digestion of the genomic DNA by SmaI, the restriction fragments were separated by PFGE using a temperature-controlled CHEF DRIII system (Bio-Rad, Hertfordshire, UK). After staining with 0.5 μg/mL ethidium bromide, the fragments were visualized by using a UV transilluminator and then documented using a video gel documentation system. For PFGE pattern analysis, GelCompar software (Applied Maths) was applied. The dendrograms were calculated, and isolates were considered, respectively, identical or closely related when there were either 0 or ≤2 banding differences.
WGS and genomic analysis
Briefly, WGS was performed on the four isolates using the MiSeq platform. The raw reads were trimmed, then assembled into contigs and scaffolded using, respectively, Trim Reads 2.5 and De Novo Assembly 1.5 tools from the CLC Genomic Workbench version 22.0 (QIAGEN). Contigs with length below 500 bp were discarded. Genome assemblies were submitted to PlasmidFinder for plasmid contigs identification. This prediction was expanded using an extra step. Briefly, trimmed reads were assembled by plasmidSPAdes to distinguish plasmids and chromosomes by coverage. Plasmids contigs were then mapped back to the CLC assemblies for identification using BLAST and pulled apart. The resulting four strain sets of chromosome contigs were ordered by alignment against the genome of the S. epidermidis RP62A reference strain (GenBank accession NC_002976.3) using the Whole Genome Alignment tool from the CLC Genomic Workbench version 22.0. Annotation of protein coding genes within ordered chromosome contigs and plasmid contigs were performed using the MicroScope platform. Prediction of resistomes were performed using the Comprehensive Antibiotic Resistance Database (CARD, version 3.0.2).
Dalbavancin therapeutic drug monitoring
Because of the long-term duration, dalbavancin therapeutic drug monitoring was performed. Plasma trough concentrations were measured using a validated LC coupled with MS method (Acquity UPLC® BEH C18 and Acquity QDA, Waters, France). The lower limit of quantification for dalbavancin was 1 mg/L and the limit of linearity was 200 mg/L. Knowing that dalbavancin is 93% bound to plasma proteins and taking into account the long terminal half-life, some authors consider a single plasma concentration measurement, at least 1 week after the injection, enough to extrapolate fAUC0–24.21,22 We therefore used this methodology for fAUC0–24 determination.
Results
Antibiotic susceptibility testing
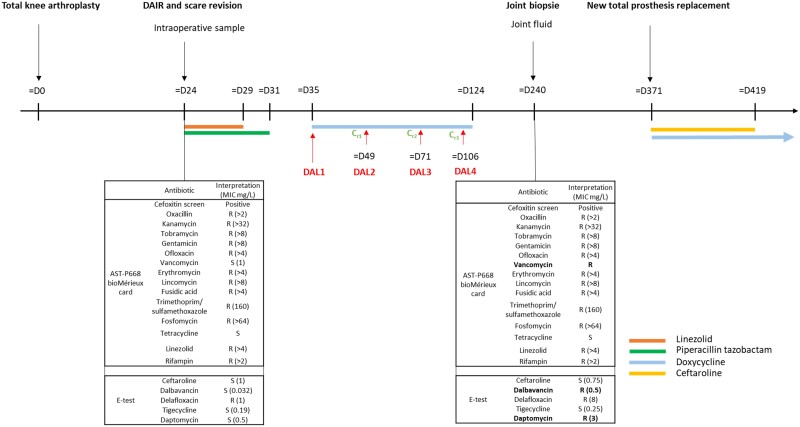
Susceptibilities to 21 antibiotics for two types of isolates (STA1 at Day24 and STA3 at Day240) are listed in Figure 1. Dalbavancin MIC increased 4-fold between the two instances (isolate 1 MIC = 0.032 mg/L; isolate 3 MIC = 0.5). It is interesting to note that, beside the increase in dalbavancin MIC, we also observed an increase of the MICs of vancomycin (>5-fold) and daptomycin (6-fold). Finally, ceftaroline, delafloxacin, tigecycline MICs were unchanged.
Figure 1.
Timeline of diagnostic and therapeutic management. D, Day; DAL, dalbavancin; Cr, residual concentration (mg/L); R, resistant; S, susceptible.
Dalbavancin therapeutic drug monitoring
Three dalbavancin plasma trough concentrations (before administration) were measured. Concentrations were 18 mg/L at Day 49 (measured 14 days after the last dose), 23.6 mg/L at Day 71 (measured 22 days after the last dose) and 14.6 mg/L at Day 106 (measured 35 days after the last dose). Calculated fAUC0–24/MIC ratios were 945, 1239 and 766.5, respectively, at Days 49, 71 and 106, assuming an MIC of 0.032 mg/L. It is interesting to note that the calculated fAUC0–24/MIC ratios would be 60, 79 and 49, respectively, at Days 49, 71 and 106 assuming an MIC of 0.5 mg/L.
PFGE
PFGE was performed after restriction by the SmaI enzyme. The macrorestriction profiles of the isolates were perfectly identical for three of them [100% similarity; two strains from episode 1 (STA1 and STA2) and one strain from episode 2 (STA4)], suggesting a strong relatedness between these isolates according to Tenover’s criteria.23 The last isolate [second strain from episode 2 (STA3)] had only one band difference and thus had a very similar macrorestriction profile (97% similarity), suggesting tight ties with the clone. Thus, despite the evolution towards resistance, it turns out to be clearly the same strain present in both episode 1 and episode 2, despite happening 216 days apart.
WGS and genomic analysis
Genomic comparison analysis of the four isolates (STA1 to STA4) revealed that they all belonged to ST2 with several indels. The genomes were identical between STA1 and STA2 (strains from episode 1) and STA3 and STA4 (strains from episode 2), except for a loss of about 50 genes (mainly plasmid genes, which were not found in STA3 and STA4) and a ‘gain’ of 8 genes (mainly genes encoding ‘proteins of unknown function’ and a merT gene corresponding to a mercury transporter or a close homologue). All strains contained blaZ, mecA, fusB, dfrC, norA, aph (2″)-Ia, ant (4′)-Ib, qacA and mupA genes.
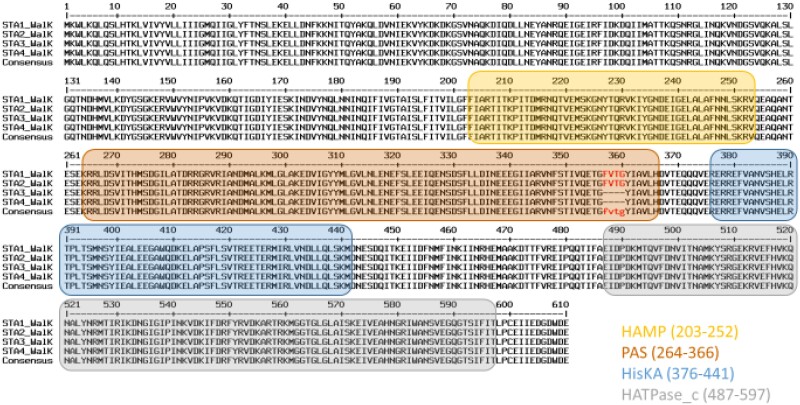
Moreover, it is worth noting that a 12 bp deletion in the walK gene, was observed in STA3 and STA4 but not in STA1 and STA2 (Figure 2). Mutations in this gene have been previously described and linked to reduced dalbavancin susceptibility.15
Figure 2.
Amino acid sequences of walK gene alignment of the four S. epidermidis study isolates against NCBI Reference Sequence WP_002437327.1. Cytoplasmic domains are highlighted in colours according to InterPro analysis and classification. HAMP (InterPro entry: IPR003660): histidine kinases, adenylyl cyclases, methyl binding proteins, phosphatases domain; PAS (IPR000014): signal sensor domain; his_kinase_dom (IPR005467): histidine kinase domain. Amino acids highlighted in red were found to be deleted within the genomic sequences of STA3 and STA4 strains. STA1 and STA2 correspond to episode 1 (24 days after arthroplasty) and STA3 and STA4 correspond to episode 2 (240 days after arthroplasty).
Discussion
Infections caused by ST2 S. epidermidis are a major problem due to their increasing resistance to antibiotics such as rifampicin and linezolid, which drastically limits antimicrobial treatment options.
Moreover, the clinical infections caused by biofilms poses a serious threat to public health. Staphylococcus spp. biofilms stand out as one of the most prevalent, especially in osteoarticular infection and PJI. The biofilm lifecycle represents a significant hurdle for treatment because bacterial cells become highly tolerant to a wide range of antimicrobial compounds, which are usually effective against the planktonic forms. Thus, new therapeutic strategies targeting biofilms and MDR bacteria are urgently needed. Recent studies have shown dalbavancin could successfully reduce MRSA, methicillin-resistant S. epidermidis and vancomycin-susceptible enterococci in biofilms.7–9,24 The particular characteristics of this antibiotic are its prolonged half-life (about 14 days), its good bone penetration and excellent tolerability profile.5,6 Thus, dalbavancin is a promising option for the treatment of infections associated with biofilms.
Here, we present an LR-MDRSE PJI case with emerging dalbavancin resistance under treatment, resulting in treatment failure and, consequently, the need for long-term suppressive antibiotic therapy.
fAUC0–24/MIC best correlates with in vivo efficacy of dalbavancin.13 Indeed, Lepak et al.14 showed that for seven isolates of S. aureus (including four vancomycin-intermediate S. aureus strains) that fAUC0–24/MIC values were, respectively, 27.1, 53.3 and 111.1 for net stasis, 1-log kill and 2-log kill. For our patient, calculated fAUC0–24/MIC ratios were 945, 1239 and 766.5, respectively, at Days 49, 71 and 106, assuming an MIC of 0.032 mg/L, but only 60, 79 and 49, respectively, assuming an MIC of 0.5 mg/L. Indeed, it is possible that between the first MIC measurement (Day 24) and the days of dalbavancin injections (Days 35, 49, 71 and 106), the MIC could have increased due to insufficient antibiotic pressure within the biofilm.
Analyses of the genomic data for the patient isolates revealed disappearance of the walK gene. The WalKR system, a two-component system (TCS) specific to low G + C Gram-positive bacteria, is the only essential TCS in S. aureus viability.20,21 The WalKR plays a role in cell wall biosynthesis, virulence and antibiotic resistance. Thus, the disappearance of the walK gene observed in the MDR strains could lead to an increase in wall thickness, explaining the observed resistance of this bacterium to dalbavancin, daptomycin and vancomycin.
Previous in vitro studies of S. aureus have demonstrated cross-resistance among dalbavancin, vancomycin and daptomycin.11,12 Thus, we included determination of daptomycin and vancomycin MICs for the four S. epidermidis isolates studied in this work. We observed a significant change in MICs of these different molecules, with an increase of 6-fold and >5 fold, respectively, for daptomycin and vancomycin, validating the cross-resistance.
Using WGS, we were able to establish that this S. epidermidis strain belongs to ST2. Recent studies have shown that this ST2 lineage is particularly enriched in S. epidermidis strains isolated from patients with PJI.18,19 In addition, Trobos et al.25 have shown that this ST2 lineage is responsible for the majority of relapses and is associated with multidrug resistance and high biofilm production, which may also explain the treatment failure.
For these particular strains, source control (especially bacterial inoculum) combined with adequate antibiotic concentrations is important to prevent reduced susceptibility from emerging during dalbavancin treatment.
This case highlighted that good pharmacokinetic follow-up should probably be established to monitor prolonged treatment during PJI. Antibiotic bi-monthly monitoring ought to be mandatory in order to properly manage this off-label cure.
This case raises some questions about the management of these ST2 S. epidermidis LR-MDRSE infections. Given the WalKR deletions, the thickening of the wall, and the capacity to overproduce biofilm, should we continue to use lipoglycopeptides on these LR-MDRSE strains or should we rather prescribe a β-lactam such as ceftaroline, which is still very active, even if the diffusion is not as good?
For these cases where strains overproduce biofilm and where surgery is not optimal (DAIR), should we continue to use dalbavancin-based monotherapy for the treatment of BJI or should we always use it in combination with rifampicin?
In our opinion, several factors contributed to this patient’s treatment failure: unsuitable surgery (DAIR); a particular bacterial strain (LR-MDRSE) that was multi-resistant and overproduced biofilm; and the lack of combination of dalbavancin with rifampicin.
Finally, further clinical research studies should be performed to optimize this treatment, especially for LR-MDRSE strains, which may have derepressed metabolic pathways leading to biofilm overproduction.
Acknowledgements
We acknowledge the other members of the Centre de Référence des Infections Ostéo-articulaires du Grand Ouest (CRIOGO).
Contributor Information
L Ruffier d’Epenoux, Institut de Biologie des Hôpitaux de Nantes, Service de Bactériologie et des Contrôles Microbiologiques, CHU de Nantes, 9 quai Moncousu, 44093 Nantes Cedex 01, France; INSERM, Immunology and New Concepts in ImmunoTherapy, INCIT, UMR 1302, Nantes Université, Nantes, France; Membre du CRIOGO (Centre de Référence des Infections Ostéo-articulaires du Grand Ouest), Nantes, France.
P Barbier, Institut de Biologie des Hôpitaux de Nantes, Service de Bactériologie et des Contrôles Microbiologiques, CHU de Nantes, 9 quai Moncousu, 44093 Nantes Cedex 01, France.
E Fayoux, Institut de Biologie des Hôpitaux de Nantes, Service de Bactériologie et des Contrôles Microbiologiques, CHU de Nantes, 9 quai Moncousu, 44093 Nantes Cedex 01, France.
A Guillouzouic, Institut de Biologie des Hôpitaux de Nantes, Service de Bactériologie et des Contrôles Microbiologiques, CHU de Nantes, 9 quai Moncousu, 44093 Nantes Cedex 01, France.
R Lecomte, Membre du CRIOGO (Centre de Référence des Infections Ostéo-articulaires du Grand Ouest), Nantes, France; Service des Maladies Infectieuses, Hôtel-Dieu, Centre Hospitalier Universitaire, Nantes, France; Centre d’Investigation Clinique Unité d’Investigation Clinique, Centre Hospitalier Universitaire, Nantes, France.
C Deschanvres, Membre du CRIOGO (Centre de Référence des Infections Ostéo-articulaires du Grand Ouest), Nantes, France; Service des Maladies Infectieuses, Hôtel-Dieu, Centre Hospitalier Universitaire, Nantes, France; Centre d’Investigation Clinique Unité d’Investigation Clinique, Centre Hospitalier Universitaire, Nantes, France.
C Nich, Membre du CRIOGO (Centre de Référence des Infections Ostéo-articulaires du Grand Ouest), Nantes, France; Nantes Université, CHU Nantes, Clinique Chirurgicale Orthopédique et Traumatologique, F-44000 Nantes, France; Nantes Université, INSERM, UMRS 1229, Regeneration Medicine and Skeleton (RMeS), ONIRIS, F-44042 Nantes, France.
P Bémer, Institut de Biologie des Hôpitaux de Nantes, Service de Bactériologie et des Contrôles Microbiologiques, CHU de Nantes, 9 quai Moncousu, 44093 Nantes Cedex 01, France; Membre du CRIOGO (Centre de Référence des Infections Ostéo-articulaires du Grand Ouest), Nantes, France.
M Grégoire, Service de Pharmacologie, CHU Nantes, Nantes, France; UMR Inserm 1235, The Enteric Nervous System in Gut and Brain Disorders, Nantes Université, Nantes, France.
S Corvec, Institut de Biologie des Hôpitaux de Nantes, Service de Bactériologie et des Contrôles Microbiologiques, CHU de Nantes, 9 quai Moncousu, 44093 Nantes Cedex 01, France; INSERM, Immunology and New Concepts in ImmunoTherapy, INCIT, UMR 1302, Nantes Université, Nantes, France; Membre du CRIOGO (Centre de Référence des Infections Ostéo-articulaires du Grand Ouest), Nantes, France; ESGIAI (ESCMID Study Group for Implant-Associated Infections) Member.
Funding
This research project was carried out as part of our routine work and received no external funding.
Transparency declarations
The authors declare no conflicts of interest.
References
- 1. Soriano A, Rossolini GM, Pea F. The role of dalbavancin in the treatment of acute bacterial skin and skin structure infections (ABSSSIs). Expert Rev Anti Infect Ther 2020; 18: 415–22. 10.1080/14787210.2020.1746643 [DOI] [PubMed] [Google Scholar]
- 2. David MZ, Dryden M, Gottlieb T et al. Recently approved antibacterials for methicillin-resistant Staphylococcus aureus (MRSA) and other Gram-positive pathogens: the shock of the new. Int J Antimicrob Agents 2017; 50: 303–7. 10.1016/j.ijantimicag.2017.05.006 [DOI] [PubMed] [Google Scholar]
- 3. Simonetti O, Lucarini G, Morroni G et al. New evidence and insights on dalbavancin and wound healing in a mouse model of skin infection. Antimicrob Agents Chemother 2020; 64: e02062-19. 10.1128/AAC.02062-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dinh A, Duran C, Pavese P et al. French national cohort of first use of dalbavancin: a high proportion of off-label use. Int J Antimicrob Agents 2019; 54: 668–72. 10.1016/j.ijantimicag.2019.08.006 [DOI] [PubMed] [Google Scholar]
- 5. Dunne MW, Puttagunta S, Sprenger CR et al. Extended-duration dosing and distribution of dalbavancin into bone and articular tissue. Antimicrob Agents Chemother 2015; 59: 1849–55. 10.1128/AAC.04550-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Leighton A, Gottlieb AB, Dorr MB et al. Tolerability, pharmacokinetics, and serum bactericidal activity of intravenous dalbavancin in healthy volunteers. Antimicrob Agents Chemother 2004; 48: 940–5. 10.1128/AAC.48.3.940-945.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Knafl D, Tobudic S, Cheng SC et al. Dalbavancin reduces biofilms of methicillin-resistant Staphylococcus aureus (MRSA) and methicillin-resistant Staphylococcus epidermidis (MRSE). Eur J Clin Microbiol Infect Dis 2017; 36: 677–80. 10.1007/s10096-016-2845-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. El Haj C, Benavent E, Sierra Y et al. Comparative efficacy of dalbavancin alone and with rifampicin against in vitro biofilms in a pharmacodynamic model with methicillin-resistant Staphylococcus aureus. Int J Antimicrob Agents 2022; 60: 106664. 10.1016/j.ijantimicag.2022.106664 [DOI] [PubMed] [Google Scholar]
- 9. Jacob B, Makarewicz O, Hartung A et al. In vitro additive effects of dalbavancin and rifampicin against biofilm of Staphylococcus aureus. Sci Rep 2021; 11: 23425. 10.1038/s41598-021-02709-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. FDA . Dalbavancin for Injection, NDA 021-883. 2014. https://www.accessdata.fda.gov/drugsatfda_docs/nda/2014/021883Orig1s000ClinPharmR.pdf.
- 11. Werth BJ, Jain R, Hahn A et al. Emergence of dalbavancin non-susceptible, vancomycin-intermediate Staphylococcus aureus (VISA) after treatment of MRSA central line-associated bloodstream infection with a dalbavancin- and vancomycin-containing regimen. Clin Microbiol Infect 2018; 24: 429.e1–e5. 10.1016/j.cmi.2017.07.028 [DOI] [PubMed] [Google Scholar]
- 12. Hines KM, Shen T, Ashford NK et al. Occurrence of cross-resistance and β-lactam seesaw effect in glycopeptide-, lipopeptide- and lipoglycopeptide-resistant MRSA correlates with membrane phosphatidylglycerol levels. J Antimicrob Chemother 2020; 75: 1182–6. 10.1093/jac/dkz562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Andes D, Craig WA. In vivo pharmacodynamic activity of the glycopeptide dalbavancin. Antimicrob Agents Chemother 2007; 51: 1633–42. 10.1128/AAC.01264-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lepak A, Marchillo K, VanHecker J et al. Impact of glycopeptide resistance in Staphylococcus aureus on the dalbavancin in vivo pharmacodynamic target. Antimicrob Agents Chemother 2015; 59: 7833–6. 10.1128/AAC.01717-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Werth BJ, Ashford N, Penewit K et al. Dalbavancin exposure in vitro selects for dalbavancin-non-susceptible and vancomycin-intermediate strains of methicillin-resistant Staphylococcus aureus. Clin Microbiol Infect 2021; 27: 910.e1–e8. 10.1016/j.cmi.2020.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang R, Polenakovik H, Barreras Beltran IA et al. Emergence of dalbavancin, vancomycin, and daptomycin nonsusceptible Staphylococcus aureus in a patient treated with dalbavancin: case report and isolate characterization. Clin Infect Dis 2022; 75: 1641–4. 10.1093/cid/ciac341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dortet L, Glaser P, Kassis-Chikhani N et al. Long-lasting successful dissemination of resistance to oxazolidinones in MDR Staphylococcus epidermidis clinical isolates in a tertiary care hospital in France. J Antimicrob Chemother 2018; 73: 41–51. 10.1093/jac/dkx370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Côrtes MF, André C, Martins Simões P et al. Persistence of a multidrug-resistant worldwide-disseminated methicillin-resistant Staphylococcus epidermidis clone harbouring the cfr linezolid resistance gene in a French hospital with evidence of interspecies transfer to several Staphylococcus aureus lineages. J Antimicrob Chemother 2022; 77: 1838–46. 10.1093/jac/dkac119 [DOI] [PubMed] [Google Scholar]
- 19. Coustillères F, Renault V, Corvec S et al. Clinical, bacteriological, and genetic characterization of bone and joint infections involving linezolid-resistant Staphylococcus epidermidis: a retrospective multicenter study in French reference centers. Microbiol Spectr 2023; 11: e0419022. 10.1128/spectrum.04190-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. d’Ersu J, Aubin GG, Mercier P et al. Characterization of Staphylococcus caprae clinical isolates involved in human bone and joint infections, compared with goat mastitis isolates. J Clin Microbiol 2016; 54: 106–13. 10.1128/JCM.01696-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cojutti PG, Tedeschi S, Gatti M et al. Population pharmacokinetic and pharmacodynamic analysis of dalbavancin for long-term treatment of subacute and/or chronic infectious diseases: the major role of therapeutic drug monitoring. Antibiotics (Basel) 2022; 11: 996. 10.3390/antibiotics11080996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cojutti PG, Rinaldi M, Gatti M et al. Usefulness of therapeutic drug monitoring in estimating the duration of dalbavancin optimal target attainment in staphylococcal osteoarticular infections: a proof-of-concept. Int J Antimicrob Agents 2021; 58: 106445. 10.1016/j.ijantimicag.2021.106445 [DOI] [PubMed] [Google Scholar]
- 23. Tenover FC, Arbeit RD, Goering RV et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol 1995; 33: 2233–9. 10.1128/jcm.33.9.2233-2239.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Oliva A, Stefani S, Venditti M et al. Biofilm-related infections in Gram-positive bacteria and the potential role of the long-acting agent dalbavancin. Front Microbiol 2021; 12: 749685. 10.3389/fmicb.2021.749685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Trobos M, Firdaus R, Svensson Malchau K et al. Genomics of Staphylococcus aureus and Staphylococcus epidermidis from periprosthetic joint infections and correlation to clinical outcome. Microbiol Spectr 2022; 10: e0218121. 10.1128/spectrum.02181-21 [DOI] [PMC free article] [PubMed] [Google Scholar]