Abstract
To better define the effects of sequence variation and tropism on the ability of the simian immunodeficiency virus SIVmac V3 loop to act as a target of antibody-mediated neutralization, a series of experiments were performed. Three SIV strains, SIVmac239, SIVmac316, and SIVmac155/T3, each with defined differences in env sequence and tropism, were used to construct a panel of viruses chimeric for a portion of envelope that includes the V2 and V3 regions. Peptides with sequences corresponding to the V3 loops of the parental viruses were used to immunize rabbits. The polyclonal rabbit antibodies and plasma from SIVmac239-infected animals were then used to assess the neutralization sensitivity of the parental and chimeric viruses. One of the parental viruses, SIVmac316, which is able to replicate to high titer in alveolar macrophages and can infect cells in a CD4-independent fashion, was highly sensitive to neutralization by plasma from SIVmac-infected rhesus macaques, with average 50% neutralization titers of 1:20,480; this same strain was also sensitive to neutralization by the anti-V3 loop peptide sera. Other parental and chimeric viruses were less sensitive to neutralization with this same panel of antibodies, but as seen with SIVmac316, those viruses that were able to productively replicate in alveolar macrophages were more sensitive to antibody-mediated neutralization. To further define the amino acids involved in increased sensitivity to neutralization, a panel of viruses was constructed by changing envelope residues in SIVmac316 to the corresponding SIVmac239 amino acids. The increased neutralization sensitivity observed for SIVmac316 was mapped principally to three amino acid changes spread throughout gp120. In addition, the increased sensitivity to neutralization by V3-directed antibodies correlated with the ability of the various viruses to replicate to high levels in alveolar macrophage cultures and a CD4-negative cell line, BC7/CCR5. These results demonstrate that the V3 loop of SIVmac Env can act as an efficient target of neutralizing antibodies in a fashion that is highly dependent on sequence context. In addition, these studies suggest a correlation between decreased dependence on CD4 and increased sensitivity to antibody-mediated neutralization.
The mechanisms by which different strains of human immunodeficiency virus (HIV) and simian immunodeficiency virus (SIV) display specific cell tropism are slowly becoming elucidated. Members of the G-protein-coupled chemokine receptor family have been shown to be coreceptors, along with CD4, for entry of the immunodeficiency viruses (1, 2, 7, 9–11, 16, 31). Differential expression of these coreceptors among blood-derived monocytes, tissue macrophages, primary T cells, and transformed T-cell lines contributes to selective entry by different viral strains. The exact sequence and structural elements of the viral envelope protein which determine coreceptor usage are becoming clearer, although questions still exist. Several studies have defined the importance of sequences in the V3 loop in coreceptor binding and tropism (6, 20, 27, 44, 54, 56). However, sequences throughout the envelope protein can regulate the ability of virus to productively replicate in tissue macrophages.
Differential coreceptor usage does not completely explain viral tropism. For example, human macrophages express the CXCR4 coreceptor but are not permissive for all strains of CXCR4 utilizing T-cell-tropic viruses (60). Recently, two groups, Mori et al. (36) and Bannert et al. (3), have shown that CD4 expression can dramatically influence the ability of virus to enter into macrophages. The restricted replication of SIVmac239 in macrophages appears to be due principally to the limiting amounts of CD4 on these cells (3, 36). The high replicative capacity of the SIVmac239 variant called SIVmac316 appears to be due to its increased affinity for CD4 and/or its ability to infect cells independent of CD4 (3, 36). Thus, in addition to differential coreceptor usage, sequences throughout the envelope protein that influence CD4 affinity and CD4 dependence also govern the ability of the virus to replicate in different types of cells, particularly macrophages.
Historically, the V3 loop of HIV type 1 (HIV-1) was initially identified as the principal neutralizing determinant of the virus (18, 19, 39, 62). In analogous studies, the V3 loop of SIV was found not to be a target of neutralizing antibodies (23, 24, 51). It is now becoming clear that the V3 loop of HIV-1 is an important epitope for neutralization of T-cell line-adapted strains but not an efficient target of neutralization of primary isolates (47, 55). Passage history and changes in envelope sequence influence the ability of the V3 loop of HIV-1 to act as a target for neutralization. Additionally, Palker and coworkers (40) have shown that the V3 loop of SIV can act as a linear neutralization epitope of SIVmac251, while Javaherian et al. (24) have shown that it contributes to a conformational neutralization epitope. Here we present data that the exposure of neutralization epitopes on SIVmac316 gp120 differs from that on SIVmac239, resulting in increased neutralization sensitivity to both V3 loop-directed antibodies and plasma from SIVmac239-infected macaques. We also present data that the difference in neutralization sensitivity maps principally to three amino acids, all of which are outside the V3 loop and are spread throughout gp120. Furthermore, we present data that this phenomenon of increased neutralization sensitivity extends to other SIVmac variants and correlates, among this small group of closely related SIV strains, with decreased dependence on CD4 for viral entry and the ability to replicate in alveolar macrophages.
MATERIALS AND METHODS
Viruses.
Viruses SIVmac239 (25), SIVmac251 primary stock (8, 30), SIVmac316 (35), and SIVmacMER (35) have been described previously.
Viral genome plasmids.
Plasmids pSIV-239-5′, pSIV-239-3′, pSIV-316-3′, and pSIV-MER-3′ have been described previously (35, 38). Plasmid pSIV-155/T3-3′ was constructed by subcloning an SphI-EcoRI fragment of pSIVmac155/T3 containing the envelope sequences of SIVmac155/T3 into a vector containing the rest of the SIVmac239 3′-half sequences, creating a virus expressing the SIVmac155/T3 envelope in a SIVmac239 backbone.
Production of viral stocks.
Viruses for all experiments were produced by cotransfecting CEMx174 cells by the DEAE-dextran method with the appropriate 3′- and 5′-half viral clones (38). Each half was cut with SphI, column purified, and ligated prior to transfection. Supernatant was collected 3 days after appearance of the first syncytia, around the peak of viral replication, and spun to remove cellular debris. A portion was used to determine p27 antigen levels (described below), and the rest was aliquoted and stored at −80°C.
Sera and sCD4.
All rhesus macaque plasma samples were from monkeys housed at the New England Regional Primate Research Center. All plasma samples were heat inactivated at 56°C for 30 min before use. Soluble CD4 (sCD4) was purchased from NEN Research Products, Boston, Mass. The 2D7 antibody for CCR5 staining was purchased from Pharmingen (San Diego, Calif.), and the OKT4 antibody for CD4 staining was purchased from Becton Dickinson (San Jose, Calif.).
Cells.
Human CD4+ CEMx174 cells (NIH AIDS Research and Reference Reagent Program, Rockville, Md.) were grown in RPMI 1640 medium supplemented with 10% fetal calf serum (FCS). The CEMx174 LTR-SEAP cells have been described previously (34). Alveolar macrophages were obtained by bronchioalveolar lavage performed under sterile conditions on rhesus macaques seronegative for SIV, type D retrovirus, and simian foamy virus. Approximately 150 ml of sterile phosphate-buffered saline (PBS) was introduced into and then removed from the left lung of healthy rhesus macaque, using an endoscope. The aspirate was spun, and the resulting cells were washed twice with PBS containing 5% FCS. The resulting cells were then resuspended in RPMI 1640 containing 10% FCS and 5% human type AB serum at 105 cells/ml and plated into a six-well plate. The cells were allowed to settle for 1 h and were then washed vigorously with PBS–10% FCS to remove any floating or loosely attached cells. Full RPMI 1640–5% human type AB sera was then added, and the cells were maintained in a CO2 incubator.
Construction of the BC7/CCR5 cell line.
Ectopic expression of human CCR5 was achieved using the puromycin resistance-conferring retroviral vector, LP-M/CCR5, and standard retrovirus-mediated gene transfer (17). BC7, a CD4-negative variant of SupT1 (15), was used to derive a human T-cell line that expressed CCR5 in the absence of CD4. Following selection in medium containing puromycin (200 ng/ml), polyclonal populations of cells expressing high levels of surface CCR5 were further selected by cell sorting with a flow cytometer using a CCR5-specific monoclonal antibody, 2D7 (Pharmingen). Cells were then cloned by limiting dilution to derive a line that stably expressed high levels of CCR5, termed BC7/CCR5. In parallel, CD4-positive SupT1 cells were engineered to express high levels of CCR5 by a similar method.
Neutralization assay.
The neutralization sensitivity of each virus was tested using the secreted, engineered alkaline phosphatase (SEAP) reporter cell assay previously described by our laboratory (34). Briefly, 96-well plates were set up as follows. To the first three columns, 25 μl of RPMI 1640–10% FCS containing a 1:20 dilution of mock plasma was added. To each of the other columns, 4 through 12, 25-μl aliquots of successive twofold dilutions of test sera or plasma in full RPMI 1640 were added. Next, 75 μl of test virus at the indicated concentration was added to each well in columns 3 through 12. Virus-free complete RPMI 1640 was added to columns 1 and 2. The plate was incubated for 1 h at room temperature with occasional mixing. After incubation, 40,000 cells in a volume of 100 μl were added to each well. The first column received the parental cell line without the SEAP reporter construct. All of the other columns received the appropriate pLNLTR-SEAP cell line. The plate was then placed into a humidified chamber within a CO2 incubator at 37°C for 52 to 72 h. SEAP activity was measured according to the manufacturer's recommendations, with slight modifications as described previously (34). Neutralization by sCD4 was performed in a similar manner, substituting various concentrations of sCD4 in place of the potentially neutralizing sera or plasma.
Production of anti-V3 antibodies.
Peptides containing the sequence of the parental viruses, SIVmac239, -316, and -155/T3, as well as a peptide containing the sequence of a related virus, SIVmac239/321,325, were produced and chemically circularized. These peptides were then resuspended at 0.5 mg/ml in complete Freund's adjuvant (Sigma) and injected subcutaneously into New Zealand White rabbits. After 3 weeks, the rabbits were boosted by subcutaneous injections of peptide in incomplete Freund's adjuvant. One additional boost was performed 3 weeks later. Two weeks after the second boost, blood was collected by cardiac puncture and the animals were sacrificed.
Antibody quantitation.
The levels of specific antibodies produced in peptide-inoculated rabbits were measured in two ways: against peptide and against lysed virions. Reactivity against peptide was measured by coating the wells of a 96-well plate with the appropriate peptide. Each peptide was resuspended in PBS at a concentration of 1 μg/ml, and 100 μl of this suspension was put into each well and allowed to air dry. The wells were washed three times with distilled H2O, and then 100 μl of 10% bovine serum albumin was added. After a 1-h incubation the wells were washed three times with distilled H2O. The plates were tapped dry and used for antibody quantitation. Serial twofold dilutions of each antibody were added to the wells and allowed to react for 1 h at 37°C. Wells were then washed three times with PBS containing 0.05% Tween 20, and an appropriate dilution of alkaline peroxidase-conjugated goat anti-rabbit antibody (Kirkegaard & Perry, Gaithersburg, Md.) was added. Next, each well received 200 μl of p-nitrophenyl phosphate (Kirkegaard & Perry) substrate solution. After 30 min, 50 μl of 3 N NaOH was added to each well, and the absorbance at 410 nm was read.
To measure reactivity against virus, the same protocol as described above was used except that 1 μg of p27 antigen/ml of virus in PBS was added to the wells instead of peptide. Blocking and quantitation of antibody binding was performed as above.
Peptide competition.
The ability of peptide to compete away neutralizing activity of serum was measured using a modification of the standard neutralization assay. Rows of a 96-well plate received either no sera or peptide, a 25-μl aliquot of a 1:40 dilution of serum 321,325-2, which had been previously shown to give 90% neutralization of SIVmac316, a 25-μl aliquot of serial twofold dilutions of peptide 321,325 or 25 μl containing both serum 321,325-1 at a 1:40 dilution, and serial dilutions of peptide 321,325. Virus was added and allowed to incubate for 1 h before the addition of cells. The plate was then put into a humidified chamber within a CO2 incubator at 37°C for 52 to 72 h. After this incubation, SEAP activity was measured as described above.
Chimeric envelope construction.
Chimeric envelope protein-containing viruses were constructed through use of two unique restriction sites, SpeI (nucleotide 7045) and XcmI (nucleotide 7625), in the 3′-half plasmids, pSIV-239-3′, pSIV-316-3′, and pSIV-155/T3-3′. These sites flank the upstream side of the V2 loop after V1 and the downstream side of the V3 loop, respectively. The V2V3 chimeric plasmids were constructed by cutting each of the parental plasmids with both SpeI and XcmI, and the resulting fragments were purified by gel extraction. The appropriate fragments were then ligated to give each of the combinations desired.
For construction of the 239SX316 and 316SX239 plasmids, plasmids pSIV-239-3′ and pSIV-316-3′ were cut with SphI, which cuts at the 5′ end of the viral DNA contained in the parental plasmids, along with XcmI. The resulting fragments were purified by gel extraction and ligated to give the desired clones. The 239SS316 and 316SS239 plasmids were made in a similar way except that the parental 239-3′ and 316-3′ plasmids were first cut with SpeI and then EcoRI, which cuts at the 3′ end of the viral DNA contained in the parental plasmids. Each ligation mix was transformed into XL2-Blue Supercompetent Escherichia coli (Stratagene) according to the manufacturer's recommendations. Plasmid DNA from individual colonies was screened using the appropriate enzymes, and those plasmids with the correct pattern were further screened by sequencing.
The 239SS316R/G and 239SS316Y/H mutations were introduced into the 239SS316 plasmid by splice overlap PCR mutagenesis. Two overlapping, mutagenic primers, one positive sense and one negative sense, were used along with nonmutagenic primers of the opposite orientation to amplify overlapping PCR fragments for each mutation. These fragments were then spliced together by additional rounds of PCR using the two outer primers, producing a single PCR fragment for each of the two mutations. This product was cleaved with XcmI and SacI and cloned into 239SS316 with the XcmI-SacI fragment removed. The resulting plasmids were sequenced across the entire PCR insert to ensure that there were no additional mutations.
The 239SS316R-G/D-N, 239SS316R-G/E-K, and 239SS316R-G/I-M mutants were also created by splice overlap PCR mutagenesis. The mutagenized PCR products were cut with SpeI and XcmI and cloned into 239SS316R/G with the XcmI-SpeI fragment removed. The resulting plasmids were sequenced across the entire PCR insert to ensure that there were no additional mutations.
Splice overlap PCR mutagenesis primers.
The same outer primers were used for each mutation. Primer env F3 5′-AACAGCATCAACAACATCAACGAC-3′ was used as a positive-sense primer, and primer env R13 5′-GGCCTCACTGATACCCCTACCA-3′ was used as a negative-sense primer. For the Arg-to-Gly mutation of 239SS316 to create 239SS316R-G, primer R-G(+) 5′-ACGGCTCCTGGAGGAGGAGATCCGGAAG-3′ and primer R-G(−) 5′-TCCTCCAGGAGCCGTCAAATTGATTTTATC-3′ were used. To introduce the Tyr-to-His mutation of 239SS316 to create 239SS316Y-H, primer T-H(+) 5′-ACTTGGCATAAAGTAGGCAAAAATGTTTATTTG-3′ and primer T-H(−) 5′-TTATGCCAAGTGTTGATTATTTGTCTAATATG-3′ were used. For the Asp-to-Asn mutation of 239SS316R-G to create 239SS316R-G/D-N, primer D-N(+) 5′-GGTTAACACTGGTAATGAAAGTAGATGTTAC-3′ and primer D-N(−) 5′-CAGTGTTATTCCCTTGTTCACATAC-3′ were used. For the Glu-to-Lys mutation of 239SS316R-G to create 239SS316R-G/E-K, primers E-K(+) 5′-GTTAAAAAGAGACAAGAAAAAAGAGTAC-3′ and E-K(−) 5′-GTCCTCTTTTTAACCCTGTCATGTTGAAT were used. For the Ile-to-Met mutation of 239SS316R-G to create 239SS316R-G/I-M, primers I-M(+) 5′-GCAAATGATAAGCTGTAAATTCAACAACATGACAG-3′ and I-M(−) 5′-GCTTATCATTTGCTCTTGTTCCAAG-3′ were used. The primers were purchased from Gene Link, Inc. (Thornwood, N.Y.).
Viral growth curves.
For growth curves in CEMx174 cells, 2.5 × 106 cells were infected with 10 ng of p27 of each virus. One day postinfection, the cells were pelleted and resuspended in virus-free RPMI 1640–10% FCS. Cell-free supernatant was harvested on the indicated days, and the amount of p27 antigen was determined with a commercial antigen capture kit (Coulter Co., Hialeah, Fla.). Rhesus macrophage culture growth curves were performed as described above but in Dulbecco modified Eagle medium containing 10% human type AB sera and 5% FCS. Growth curves in CD4+ SupT1/CCR5 cells and CD4 BC7/CCR5 cells were performed by infecting 2 × 106 cells with 20 ng of p27 of each virus in six-well plates. One day postinfection, the cells were thoroughly washed with RPMI 1640–10% FCS and resuspended in 2 ml of fresh virus free RPMI 1640–10% FCS. Virus production was quantitated at various times postinfection as described above.
RESULTS
Sensitivity of SIVmac strains to neutralization.
The neutralization sensitivities of three SIVmac virus isolates were tested using the SEAP induction assay. The strain of SIVmac316 used in these experiments differs by six amino acids in gp120 and two amino acids in gp41 from SIVmac239 (Fig. 1A). SIVmac155/T3 was isolated from the lymph node of an SIVmac239-infected macaque, and its envelope protein has 22 amino acid differences compared with SIVmac239 gp120, including five substitutions and one insertion in the V3 loop (Fig. 1A and 2). These changes from SIVmac239 in SIVmac316 and SIVmac155/T3 have been shown to influence the ability of these viruses to replicate in various cell types. SIVmac239 and SIVmac155/T3 replicate poorly in alveolar macrophage cultures, while SIVmac316 replicates to high levels. Plasma samples from naive or SIVmac239-infected rhesus macaques were pooled and tested for the ability to neutralize the three viruses. The same amount of input p27 protein (1 ng per well) was used for each of the virus stocks, and the induced SEAP activity per nanogram of p27 differed by less than 20% among the different viruses. The neutralization curves were normalized to a mock-neutralized control where the same concentration of serum from an uninfected rhesus macaque was added in place of the test serum. The plasma from the SIV-positive macaques gave 50% neutralization titers of >1:5,120 against SIVmac316 (Fig. 3). Further titration showed that the 50% neutralization titer against SIVmac316 was 1:20,480 (data not shown). Plasma titers against SIVmac239 were much lower, with an average 50% neutralization titer of 1:80. Neutralization titers against SIVmac155/T3, which differs from SIVmac239 at 23 amino acids in gp120, had an average 50% neutralization titer of only 1:20. Similar results were obtained when each of the plasma in the pool were tested individually (data not shown).
FIG. 1.
Schematic overview of the chimeras and mutants used. The gp120 open reading frame of each virus is designated by a box; red lollipops above the boxes indicate locations of amino acid differences versus SIVmac239. The three parental viruses are indicated by boxes of different colors: SIVmac239, white; SIVmac316, black; SIVmac155/T3, gray. Also indicated are a scale of amino acid numbers, locations of the variable regions, and locations in the SIVmac239 plasmid of SpeI and XcmI restriction sites used in this study. A green arrow shows the location of an amino acid insertion in SIVmac155/T3 with respect to SIVmac239. SIVmac316 gp41 also contains two amino acid changes versus SIVmac239 gp41 that are not shown.
FIG. 2.
Schematic representation of peptides used to inoculate rabbits. Peptides containing the amino acid sequences of the V3 loops of SIVmac239, SIVmac316, SIVmac321, 325 and SIVmac155/T3 were produced and chemically circularized. Each peptide was used to subcutaneously inoculate two rabbits, for a total of six rabbits. The rabbits received two additional booster inoculations with the appropriate peptide. Sera were harvested 3 weeks after the second boost. Amino acid differences from SIVmac239 are shown in black.
FIG. 3.
Neutralization of SIVmac239, SIVmac316, and SIVmac155/T3 by rhesus macaque plasma. Plasma samples were taken from naive or SIVmac239-infected rhesus macaques and pooled. Twofold dilutions of plasma were added to wells containing 2 ng of p27 of the indicated virus. After 1 h of incubation, 3 × 104 CEMx174 SIV-SEAP cells were added to each well, and the plate was transferred to a humidified chamber within a 37°C CO2 incubator. Approximately 60 h later, SEAP activity in the supernatant was measured using a Phospha-light kit according to the manufacturer's recommendations, with slight modifications as described previously (34). Activity is expressed as a percentage of the amount of SEAP activity secreted from cells infected with mock-neutralized virus.
Characterization of anti-V3 loop peptide antibodies.
Peptides were synthesized that corresponded to the V3 loop sequences of each of the three viruses plus one additional virus, SIVmac321,325, a variant containing two amino acid changes in the V3 loop which abrogates the ability of virus to replicate in CEMx174 and macrophages (26). SIVmac316 and SIVmac239 have identical V3 loop sequences; thus, three peptides, corresponding to SIVmac239 and 316, SIVmac321,325, and the 155/T3 V3 loop sequences, were produced (Fig. 2). These peptides were used to induce antibodies in New Zealand White rabbits. An initial inoculation and two boosts were given subcutaneously. Six rabbits, two per peptide, were used; 3 weeks after the final boost, sera were collected and tested for reactivity with the immunizing peptide by enzyme-linked immunosorbent assay. Each of the sera, in general, bound the homologous peptide better than the heterologous peptide (data not shown). However, there was not a strict correlation, likely due to animal-to-animal variation in the immune response and the high degree of sequence homology between the three peptides. The sera were next tested for the ability to bind to detergent-lysed virion proteins by enzyme-linked immunosorbent assay. The sera generally bound better to wells containing protein from virus containing the homologous V3 loop sequence than to those containing proteins from virus with heterologous V3 loop sequences (data not shown). Two of the sera, 239/316-1 and 321,325-2, had consistently high titers against all of the viruses.
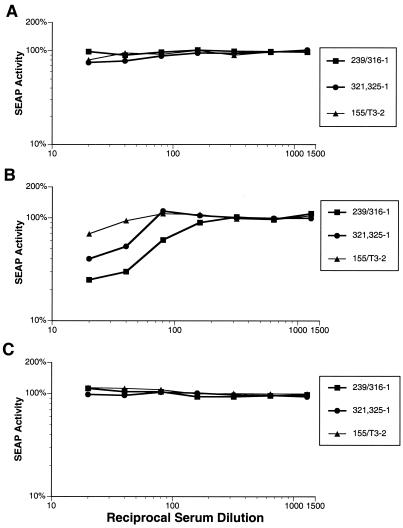
Differences in the ability of SIV strains to be neutralized by anti-V3 loop peptide antibodies.
Each of the rabbit sera was examined for its ability to neutralize parental virus infectivity in the SEAP neutralization assay. SIVmac239 and SIVmac155/T3 were essentially insensitive to neutralization by the anti-V3 loop peptide sera (Fig. 4). SIVmac239 was detectably neutralized at a 1:10 dilution of serum 321,325-2, but the 50% neutralization titer was <1:10 (data not shown). SIVmac316, however, was quite sensitive to neutralization by several of the sera (Fig. 4B). Sera 239/316-1, 321,325-1, and 321,325-2 gave 50% neutralization titers of 1:60, 1:25, and 1:80, respectively. Serum 155/T3-2 was able to significantly neutralize SIVmac316 virus infectivity, but not by 50% at the dilutions that were tested. The other sera were not able to significantly reduce SIVmac316 infectivity (data not shown). SIVmac155/T3 was not detectably neutralized by any of the antipeptide sera (Fig. 4C and data not shown).
FIG. 4.
Neutralization of SIVmac239 (A), SIVmac316 (B), and SIVmac155/T3 (C) by peptide-inoculated rabbit sera. Each of the three viruses was tested for neutralization sensitivity to sera from the peptide-inoculated rabbits as described for Fig. 3.
Neutralization of SIVmac316 by serum 321,325-2 can be competed by peptide 321,325.
To verify that the neutralizing activities of the rabbit sera were directed against sequences contained in the inoculated peptide, we performed a modified neutralization assay in which the amount of serum and virus was kept constant but an increasing amount of the immunizing peptide was added. An assay plate was set up with twofold serial dilutions of peptide 321,325 from 0.004 up to 1.0 μg per well. To these wells, SIVmac316 containing 2 ng of p27 and a 1:40 dilution of serum 321,325-2 were added. The presence of peptide 321,325 at high concentrations was able to completely block the neutralization of SIVmac316 by serum 321,325-2 (Fig. 5). The presence of peptide without serum had no effect on viral infectivity, while serum controls without peptide neutralized viral infectivity by 84%.
FIG. 5.
Peptide competition assay. The ability of peptide 321,325 V3 loop to compete away neutralization of serum 321,325-2 was tested in a modified SEAP neutralization assay as described in Materials and Methods. SIVmac316 was incubated in the presence of twofold dilutions of peptide along with a 1:40 dilution of serum 321,325-2. As controls, part of the wells received peptide only, part received serum only, and part received neither. CEMx174 LTR-SEAP cells were added to each of the wells; approximately 60 h postinfection, SEAP activity was measured and normalized as described in the legend to Fig. 3. The solid line shows the amount of SEAP activity in the medium of cells infected with virus in the presence of a 1:40 dilution of serum 321,325-2; the dotted line represents the amount of SEAP activity in the medium of cells infected with virus in the absence of both serum and peptide.
Construction of viruses with envelopes chimeric for V2 and V3.
Plasmids containing the envelope sequences of each of the three parental viruses were cut with the restriction enzymes SpeI and XcmI, each recognizing a single site in the plasmid. The SpeI recognition site is located just 5′ of the V2 loop sequences, and the XcmI site is located just 5′ of the sequences for the downstream cysteine of the V3 loop region (Fig. 1A). SpeI-XcmI fragments from each of the parental viruses were then inserted into SpeI-XcmI-digested vectors that contained SIV 3′ sequences, thus creating six SIVs containing envelope proteins chimeric for their V2 and V3 loop sequences (Fig. 1B). The resulting plasmids were named according to the source of their backbone and V2-V3 loop region. Each plasmid was used to create a virus by cotransfection into CEMx174 cells with a plasmid encoding the 5′-half SIVmac239 sequences.
Growth of V2V3 chimeric viruses in macrophage and T cells.
Since SIVmac239, SIVmac316, and SIVmac155/T3 differ in tropism, we examined the ability of each of the chimeric viruses to grow in both CEMx174 T cells and rhesus alveolar macrophages. The parental viruses grew with nearly identical kinetics in CEMx174 cells (Fig. 6A, top). Four of the six chimeras also grew with wild-type kinetics. Two of the chimeras, 239 V2V3 155/T3 and 316 V2V3 155/T3, grew with delayed kinetics and to a lower peak of viral production than the parental viruses. SIVmac316 replicated well in macrophage cultures, giving a peak p27 antigen production of almost 100 ng/ml, while the other two parental viruses produced levels of p27 that were undetectable (Fig. 6A, bottom). Four of the chimeric viruses were able to grow to detectable levels in macrophage cultures. One, 316 V2V3 239, grew as well as SIVmac316, while the other three, 239 V2V3 316, 155/T3 V2V3 316, and 155/T3 V2V3 239, were able to replicate in macrophage cultures, but not to as high a level as SIVmac316. The ability of the 155/T3 V2V3 239 and 155/T3 V2V3 316 viruses to replicate in macrophage cultures is interesting since growth of the parental virus SIVmac155/T3 is undetectable in macrophages. Thus, the restriction of SIVmac155/T3 for replication in macrophages is determined principally by sequences in the V2-V3 region. Compared with SIVmac239, SIVmac155/T3 has 10 amino acid differences in this region, including 7 within the V3 loop.
FIG. 6.
Characterization of the V2V3 chimeric viruses. Stocks of the chimeric viruses were produced by transfection into CEMx174 cells and then tested for growth kinetics (A) and neutralization sensitivity (B). (A) Growth in CEMx174 cells (top) or rhesus alveolar macrophages (bottom) was tested by infection of cultures with equal amounts of virus and then measurement of viral p27 antigen levels at various days postinfection (P.I.). The p27 antigen levels were determined with a Coulter SIV antigen capture kit. (B) Dilutions of serum 239/316-1 (top), serum 321,325-2 (middle), and pooled rhesus macaque plasma (bottom) required to neutralize 50% of viral infectivity of each of the parental and V2V3 chimeric viruses under study. These values were derived from curves of neutralization sensitivity measured using the SEAP neutralization assay described in the legend to Fig. 3.
V2V3 chimeric viruses are mostly insensitive to neutralization.
Each of the chimeric viruses was tested for neutralization sensitivity using the SEAP neutralization assay. Plasma from SIVmac239-infected rhesus macaques and sera from the V3 loop peptide-immunized rabbits were tested for their ability to neutralize viral infectivity. None of the chimeras was as sensitive to neutralization as SIVmac316 (Fig. 6B and data not shown). One chimeric virus, 239 V2V3 155/T3, displayed increased sensitivity to serum 239/316-1 but no increase in sensitivity to the pooled rhesus macaque plasma or the other anti-V3 loop peptide antibodies. 155/T3 V2V3 316 was neutralized to comparable levels as SIVmac316 by two of the rabbit sera, but it was not neutralized by the rhesus macaque plasma to the same extent as was SIVmac316 (Fig. 6B and data not shown). This may be due at least in part to the divergence of the envelope sequence in this virus compared with SIVmac239, the virus used to infect the rhesus macaques.
Construction and characterization of additional SIVmac239 and SIVmac316 chimeras.
To better define the amino acids responsible for the increased neutralization sensitivity of SIVmac316 versus SIVmac239, four additional chimeras were constructed. Restriction enzyme recognition sites at the extreme 3′ and 5′ ends of the envelope sequence were used to digest the viral gp120 sequence. Using the two additional restriction sites, the 5′ or 3′ third of the one parental virus envelope was exchanged with the corresponding fragment of the other parental virus. The resulting viruses were named 316SS239, 316SX239, 239SS316, and 239SX316, based on the fragments that were exchanged (Fig. 1C). The middle-third exchanges, 316 V2V3 239 and 239 V2V3 316, had already been constructed in the original set of chimeras (Fig. 1B).
After production of viral stocks, each of the viruses was tested for the ability to grow in CEMx174 and macrophage cultures. All of the viruses grew with kinetics and peak p27 antigen production similar to those for the parental viruses in CEMx174 cells (Table 1 and data not shown). While none of the chimeras replicated as well as SIVmac316 in macrophages, all but one replicated better than SIVmac239 in macrophage cultures (Fig. 7 and Table 1). Chimera 239SS316 grew to the highest titer of all the chimeras, while 316SS239, with only a single amino acid change from SIVmac239, had replication kinetics only slightly higher than that of SIVmac239 (Fig. 7 and data not shown).
TABLE 1.
Properties of chimeric viruses
| Virusa | Growthb (peak viral p27 antigen [ng/ml])
|
Neutralizationc
|
|||
|---|---|---|---|---|---|
| CEMx174 cells | Macrophages | Pooled macaque | 239/316-1 | 321/325-2 | |
| 239 | 600 | — | 25 | <20 | <20 |
| 316 | 750 | 105 | >20,000 | 40 | 80 |
| MER | 750 | 80 | 6,500 | <20 | <20 |
| 316SS239 | 810 | 5 | 20 | <20 | 20 |
| 316SX239 | 750 | 27 | 5,000 | 20 | 20 |
| 239SS316 | 700 | 55 | >20,000 | 20 | 45 |
| 239SX316 | 570 | 40 | >20,000 | <20 | 20 |
| 239SS316 R-G | 620 | 50 | >20,000 | 20 | 80 |
239, SIVmac239; 316, SIVmac316; MER, SIVmacMER. Other viruses are as named in the text.
Data from one representative experiment are shown. The kinetics of viral growth in CEMx174 cells were essentially identical, while differences in the kinetics of growth in macrophages are shown in Fig. 7. The dash indicates a level of p27 antigen below the limit of detection. Growth curves were performed in triplicate.
50% reciprocal neutralization titer as determined in the SEAP neutralization assay (see Materials and Methods).
FIG. 7.
Growth of the SS and SX viruses in macrophage cultures. Stocks of the chimeric viruses were produced by transfection of CEMx174 cells and then tested for growth kinetics in rhesus alveolar macrophages. Cultures were infected with equal amounts of each virus, and p27 antigen levels were determined with a Coulter SIV antigen capture kit at the indicated days postinfection (P.I.).
All of the SS and SX series chimeras were tested for neutralization sensitivity. Two of the chimeras, 239SS316 and 239SX316, demonstrated neutralization sensitivity to pooled plasma from infected macaques that was equal to the sensitivity of SIVmac316 (Table 1 and data not shown). In addition, 239SS316 was sensitive to neutralization by the V3 loop-directed rabbit sera to almost the same extent as SIVmac316 (Table 1 and data not shown). Chimera 316SX239 also demonstrated increased neutralization sensitivity to all of the antibodies, but not to the extent seen with 239SS316 (Table 1 and data not shown). Chimera 239SX316, although highly sensitive to neutralization by rhesus macaque plasma, was only weakly sensitive to neutralization by one of the rabbit sera (Table 1 and data not shown).
239SS316R-G is sensitive to neutralization and grows in macrophages.
Due to its high sensitivity to neutralization, 239SS316 was used to further define the sequences necessary for increased V3 loop-directed neutralization sensitivity. It contains five amino acid differences compared to SIVmac239, including the 176Glu and 382Arg changes found in SIVmacMER (35). The other amino acid differences, 165Ile, 199Asp, and 442Tyr, are present only in SIVmac316. We constructed mutants of 239SS316 by changing the two C-terminal envelope differences of 239SS316 versus SIVmac239, 382Arg and 442Tyr, individually to the residues found in SIVmac239 to create the viruses 239SS316R-G and 239SS316Y-H (Fig. 1D). Plasmids for the 3′ sequences of these two viruses were cotransfected with plasmids containing the 5′-half SIVmac239 sequences into CEMx174 cells. 239SS316R-G grew in these cells, but 239SS316Y-H did not produce detectable p27 antigen or infectious virus in three separate transfections. The kinetics of growth and peak p27 antigen production of 239SS316R-G was essentially identical to those for SIVmac316 in CEMx174 cells (Table 1 and data not shown). Stocks of the viruses were tested for growth in macrophages and for neutralization sensitivity. 239SS316R-G grew to almost the same level as SIVmac316 in macrophage cultures (Fig. 7 and Table 1), and its neutralization sensitivity was almost equal to the sensitivity of 239SS316 (Table 1). Sensitivity to serum 239/316-1 was slightly less than the sensitivity of SIVmac316; however, neutralization sensitivity to pooled 239-infected rhesus macaque plasma and to serum 321,325-2 was equal to that of SIVmac316. Having observed a correlation between growth in macrophages and increased neutralization sensitivity of SIVmac, we also tested SIVmacMER for neutralization sensitivity. SIVmacMER contains three amino acid changes with respect to SIVmac239, 67Met, 176Glu, and 382Arg, that are also present in SIVmac316 (Fig. 1D). These three changes allow for replication in macrophage cultures, although not to as high a level as SIVmac316 (Table 1). SIVmacMER was more sensitive than SIVmac239 to neutralization by plasma from SIVmac239-infected rhesus macaques (Table 1). However, it was insensitive to neutralization by the V3 loop-directed polyclonal rabbit antibodies (Table 1).
Construction and testing of D-N, E-K, and I-M mutants.
The neutralization sensitive 239SS316R-G was next used as the starting point for construction of one final set of viruses. The 239SS316R-G virus gp120 contains four amino acid differences versus SIVmac239 (Fig. 1D). Since the 442Tyr-to-His mutation in the context of 239SS316 abrogated viral growth, we focused on mutation of each of the upstream amino acids from the 239SS316R-G residue to the amino acid found in SIVmac239. This created the viruses 239SS316R-G/D-N, 239SS316R-G/E-K, and 239SS316R-G/I-M (Fig. 1E). One other virus, 239SS316R-G/9 was created by swapping the XcmI-EcoRI fragment of 239SS316R-G into the SIVmac239 3′-half plasmid, effectively eliminating all three of the upstream residues from 239SS316R-G (Fig. 1E). Each of these constructs was transfected into CEMx174 cells and grew with the same kinetics and peak p27 antigen level as the 239SS316R-G parental virus (Table 2 and data not shown). The viruses were next tested for the ability to replicate in alveolar macrophages. SIVmac239 and 239SS316R-G/9 were essentially incompetent for growth in macrophages, while each of the other viruses grew to significant titer (Fig. 8 and Table 2). None of the viruses grew to as great a peak titer as SIVmac316, with 239SS316R-G/I-M having the most significantly reduced growth kinetics.
TABLE 2.
Properties of mutant viruses
| Virusa | Growthb (peak viral p27 antigen [ng/ml])
|
Neutralization
|
|||||
|---|---|---|---|---|---|---|---|
| CEMx174 cells | Macrophages | BC7/CCR5 cells | Pooled macaquec | 239/316-1c | 321/325-2c | sCD4d (ng/ml) | |
| 239 | 540 | 5 | 0.5 | 20 | <20 | <20 | 280 |
| 316 | 600 | 120 | 5 | 15,000 | 20 | 90 | 20 |
| R-G | 580 | 85 | ND | 15,000 | 20 | 90 | 25 |
| R-G/D-N | 520 | 65 | 2.0 | 8,000 | 20 | 40 | 28 |
| R-G/E-K | 575 | 65 | 7.5 | 500 | <20e | 20 | 40 |
| R-G/I-M | 520 | 25 | 1.8 | 250 | <20e | <20e | 50 |
| R-G/9 | 530 | 5 | 0.5 | 20 | <20 | <20 | 230 |
239, SIVmac239; 316, SIVmac316; R-G, 239SS316R-G; R-G/D-N, 239SS316R-G/D-N; R-G/D-N, 239SS316R-G/E-K; R-G/I-M, 239SS316R-G/I-M; R-G/9, 239SS316R-G/9.
Data from one representative experiment are shown. The kinetics of viral growth in CEMx174 cells were essentially identical, while differences in the kinetics of growth in macrophages are shown in Fig. 8. Growth curves were performed in triplicate. ND, not determined.
50% reciprocal neutralization titer as determined in the SEAP neutralization assay (see Materials and Methods).
Concentration required to neutralize 50% of viral infectivity as determined in the SEAP neutralization assay.
While the 50% reciprocal neutralization titer was not achieved, significant viral neutralization occurred.
FIG. 8.
Growth of the 239SS316R-G point mutants in macrophage cultures. Stocks of the chimeric viruses were produced by transfection of CEMx174 cells and then tested for growth kinetics in rhesus alveolar macrophages. Cultures were infected with equal amounts of each virus, and p27 antigen levels were determined with a Coulter SIV antigen capture kit at the indicated days postinfection (P.I.).
Each of the newly created viruses were tested for sensitivity to neutralization by plasma from SIVmac239-infected rhesus macaques and by anti-V3 peptide antibodies. The 239SS316R-G/9 virus demonstrated the same sensitivity to each of these agents as SIVmac239 (Table 2). The 239SS316R-G/D-N virus was almost as sensitive to neutralization by pooled plasma and V3-directed sera as SIVmac316 and 239SS316R-G. The 239SS316R-G/E-K and 239SS316R-G/I-M viruses were only slightly more sensitive than SIVmac239 to neutralization.
Finally, the viruses were tested for sensitivity to neutralization by sCD4. As we have previously reported (33), SIVmac316 is much more sensitive than SIVmac239 to neutralization by sCD4. Each of the viruses in this panel capable of significant growth in macrophage also displayed increased sensitivity to sCD4 neutralization (Table 2). 239SS316R-G/9, which did not grow significantly in macrophages, was neutralized to the same extent as SIVmac239 (Fig. 8 and Table 2).
Increased neutralization sensitivity and ability to grow in macrophages correlate with ability of virus to grow in a CD4− cell line.
It was previously observed that SIVmac316 is able to enter into cells in a CD4-independent manner (3). To investigate whether any of the newly constructed SIVmac316 derivatives also had this trait, we first constructed a CD4-negative, CCR5-positive cell line called BC7/CCR5. BC7, a CD4-negative variant of SupT1 (15), was used to derive a human T-cell line that expressed CCR5 in the absence of CD4. Cells were selected in puromycin, sorted by flow cytometry for high CCR5 expression, and then cloned by limiting dilution. In parallel, CD4-positive SupT1 cells were also engineered to express high CCR5 levels. Both cell lines stably express high levels of CCR5 as detected by flow cytometry (Fig. 9, left and middle), but unlike the SupT1/CCR5 cells, the BC7/CCR5 cells do not express CD4 (Fig. 9, right).
FIG. 9.
Receptor expression on SupT1/CCR5 and BC7/CCR5 cells. Flow cytometry was used to examine CCR5 and CD4 expression on the SupT1/CCR5, BC7/CCR5, and parental cell lines, using 2D7, an anti-CCR5 antibody, and OKT4, an anti-CD4 antibody. Levels of CCR5 expression on BC7/CCR5 (left) and SupT1/CCR5 (middle) cells are shown as filled curves, while levels on the parental cells are shown as open histograms, on the right, cell surface expression of CD4 for the SupT1/CCR5 cells is shown as a bold line and levels on the BC7/CCR5 cells is shown as a thin line.
Using the two new cell lines, we next performed growth curves. All of the newly constructed SIVmac316 derivatives grew with similar kinetics in the SupT1/CCR5 cells (Fig. 10A). However, SIVmac239 and 239SS316R-G/9 replicated poorly if at all in the BC7/CCR5 cells (Fig. 10B). SIVmac316 and 239SS316R-G/E-K replicated the best of this group of viruses, while the peak titers of 239SS316R-G/D-N and 239SS316R-G/I-M were slightly lower.
FIG. 10.
Growth of various viruses in CD4-negative cells. Growth curves in CD4+ SupT1/CCR5 cells (A) and CD4− BC7/CCR5 cells (B) were performed by infecting 2 × 106 cells with 20 ng of p27 of each virus in six-well plates. One day postinfection (P.I.), the cells were thoroughly washed with RPMI 1640–10% FCS and resuspended in 2 ml of fresh virus-free RPMI 1640–10% FCS. Virus p27 antigen production was quantitated at various times postinfection using a Coulter SIV antigen capture kit.
DISCUSSION
Using a series of chimeras and point mutants, we have mapped amino acid differences between SIVmac316 and SIVmac239 gp120 that allow for increased replication in CD4-negative cells and confer increased sensitivity to V3-directed neutralizing antibodies (Fig. 8 and Table 2). The 239SS316R-G/D-N virus contains two amino acid changes in the C2 region between the V1 and V2 loops and a third amino acid difference in the C5 region, amino terminal of V4 (Fig. 1E). It is known that the V1/V2 loop region of HIV-1 plays a critical role in governing second receptor interactions and growth in a variety of cell types (4, 28, 37, 46). In addition, antibody binding experiments predict that the V1 and V2 loops shield the coreceptor binding site and change in conformation after CD4 binding (29, 42, 58). Thus, it is reasonable that the two changes in this region might contribute to an altered envelope structure capable of interacting with CCR5 in the absence of CD4.
Several studies have demonstrated that while SIVmac239 and SIVmac316 both use CCR5 as their second receptor, their specific interactions with CCR5 differ (12, 13, 57). SIVmac316 is able to utilize CCR5 either in the absence or at very low concentrations of CD4 for entry into cells, while SIVmac239 requires CD4 to enter cells. Two groups, Mori et al. (36) and Bannert et al. (3), have recently shown that this differential usage in combination with variably low or absent CD4 expression on macrophages is largely responsible for the block to SIVmac239 replication in these cells. These observations provide a rational framework for the linkage of decreased dependence on CD4 with ability to replicate in tissue macrophages that exhibit low or no CD4 expression. In our studies described here, the neutralization sensitivity of various viruses correlated with high-level growth in alveolar macrophage cultures and with CD4-independent replication in the BC7/CCR5 cell line. The linkage of these three properties is not expected to be absolute; certainly there are neutralization-sensitive strains of SIV and HIV that are not CD4 independent and not highly competent for replication in tissue macrophages. However, CD4 independence could conceivably be inexorably linked with increased sensitivity to antibody-mediated neutralization. Hoffman et al. (21) made the observation that adaptation of HIV-1IIIB to CD4-independent growth led to increased sensitivity to neutralization by HIV-positive human sera and anti-gp120 polyclonal antibodies. Similarly, the neurovirulent SIV varient 17E is capable of replicating in macrophage cultures, utilizes CCR5 for entry, and is CD4 independent (14). Compared with SIVmac239, this virus displays increased sensitivity to neutralization by SIV-infected rhesus macaque plasma on a variety of target cells (61). Nonetheless, examination of SIV and HIV strains in other contexts will be needed to determine whether CD4 independence is uniformly linked to increased neutralization sensitivity.
In studies of SIVMne, Rudensey et al. (43) found that the parental SIVMneCL8 cloned virus accumulated sequence changes in the V1 loop region of envelope during infection of pig-tailed macaques. These changes were shown to allow the virus to escape from neutralizing antibodies and to decrease the ability of the virus to replicate in macrophages. Separate studies with viruses containing these same envelope sequences indicated that changes in the pattern of N-linked glycosylation were responsible for the decreased neutralization sensitivity (5). While the sequence changes mapped in the present study do not alter any potential N-linked glycosylation sites, the overall conclusions of these studies are similar; increased ability to grow in macrophages correlated with increased susceptibility to neutralization. Other groups have also reported that changes in the V1 and V2 loop regions of both HIV-1 and SIV affect the ability of the viruses to replicate in macrophages and alter sensitivity to neutralization by sCD4 and antibodies (28, 32, 37, 41).
What potential models exist to explain the linkage between increased neutralization sensitivity and decreased reliance on CD4 for cell entry? A number of groups (48–50, 52, 53, 59) have demonstrated that binding of virus to sCD4 induces a number of conformational changes important for viral entry that also increase viral neutralization sensitivity and/or specific epitope exposure. Edinger et al. (13) have shown that several strains of SIV with decreased dependence on CD4 for cell entry also display increased CCR5 affinity. Taken together, this suggests that amino acid changes mapped in these experiments could alter protein structure such that it more closely resembles the post-CD4 binding conformation. This potential structure, like the sCD4-bound envelope, would have greater exposure of a number of neutralizing epitopes that are normally protected by the relatively closed conformation of circulating virus or by close proximity to the cell surface after CD4 binding. In addition, this altered structure would likely have a greater affinity than wild-type envelope for binding to second receptor, allowing for entry into cells expressing normal levels of second receptor but low or nonexistent levels of CD4.
Another possible mechanism is that the amino acid changes mapped in this work could be altering how the viral envelope interacts with second receptor. Edinger et al. (12) have shown that macrophagetropic and T-cell-tropic SIVs both utilize CCR5 for entry in to cells, but in functionally different ways. In experiments with chimeric receptors, it was found that the macrophagetropic SIVmac316 interacts largely with the amino-terminal region of CCR5, while SIVmac239 has stronger interactions with the second extracellular loop (13). Changes in gp120 which alter viral envelope interactions with CCR5 might also change which regions of envelope constitute neutralizing epitopes. These two models are not mutually exclusive and both could contribute.
Our studies demonstrate that the V3 loop of SIVmac is a target of neutralization. However, its ability to act as a target of neutralizing antibodies is linked to the sequence context in which it is found. Increased sensitivity to neutralization was mapped to three of the six amino acids of SIVmac316 gp120 that differ from SIVmac239 gp120 (Fig. 1E and Table 2); these same amino acid changes allow for high-level replication in macrophage cultures and decreased dependence on CD4 (Fig. 8 and Table 2). The ability of V3 to serve as a target for antibody-mediated neutralization is likely linked to its degree of exposure on virions (22, 45). Our experiments suggest that increased exposure of V3 can result from amino acid changes that confer CD4 independence in addition to those that confer adaptation in the laboratory to growth in lymphoid cell cultures (34).
Early studies of the immunological structures of HIV-1 and SIV envelope proteins suggested differences between the two viruses with respect to the antigenicity and immunogenicity of the V3 loop (18, 19, 24, 39, 62). V3 region-directed antibodies from HIV-infected individuals were able to efficiently neutralize laboratory-adapted strains of HIV, leading researchers to call the V3 loop the principal neutralizing determinant (62). Early attempts to raise neutralizing antibodies to the V3 loop of SIV were unsuccessful (24). The results presented here suggest that the earlier findings may have been dependent on the strains of virus that were used. Our data indicate that envelope structure strongly influences neutralization by V3 loop-directed antibodies for SIV similar to what has been described for HIV-1 (47, 55). Thus, SIVmac appears similar to HIV-1 in that V3 can be a target for neutralization by antibodies but its ability to do so is greatly influenced by the envelope sequence context.
ACKNOWLEDGMENTS
We thank Susan Czajak and Lou Alexander for technical assistance and advice, Patrick Lyden and Jennifer Morgan for critical reading, and members of the Desrosiers laboratory for helpful discussions.
This work was supported by Public Health Service grants AI25328, AI35365, and RR00168 from the National Institutes of Health.
REFERENCES
- 1.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 2.Alkhatib G, Liao F, Berger E A, Farber J M, Peden K W. A new SIV co-receptor, STRL33. Nature. 1997;388:238. doi: 10.1038/40789. [DOI] [PubMed] [Google Scholar]
- 3.Bannert N, Schenten D, Craig S, Sodroski J. The level of CD4 expression limits infection of primary rhesus monkey macrophages by a T-tropic simian immunodeficiency virus and macrophagetropic human immunodeficiency viruses. J Virol. 2000;74:10984–10993. doi: 10.1128/jvi.74.23.10984-10993.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyd M T, Simpson G R, Cann A J, Johnson M A, Weiss R A. A single amino acid substitution in the V1 loop of human immunodeficiency virus type 1 gp120 alters cellular tropism. J Virol. 1993;67:3649–3652. doi: 10.1128/jvi.67.6.3649-3652.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chackerian B, Rudensey L M, Overbaugh J. Specific N-linked and O-linked glycosylation modifications in the envelope V1 domain of simian immunodeficiency virus variants that evolve in the host alter recognition by neutralizing antibodies. J Virol. 1997;71:7719–7727. doi: 10.1128/jvi.71.10.7719-7727.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan S Y, Speck R F, Power C, Gaffen S L, Chesebro B, Goldsmith M A. V3 recombinants indicate a central role for CCR5 as a coreceptor in tissue infection by human immunodeficiency virus type 1. J Virol. 1999;73:2350–2358. doi: 10.1128/jvi.73.3.2350-2358.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 8.Daniel M D, Letvin N L, King N W, Kannagi M, Sehgal P K, Hunt R D, Kanki P J, Essex M, Desrosiers R C. Isolation of T-cell tropic HTLV-III-like retrovirus from macaques. Science. 1985;228:1201–1204. doi: 10.1126/science.3159089. [DOI] [PubMed] [Google Scholar]
- 9.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 10.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic primary HIV-1 isolate that uses fusion and the b-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 11.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC- CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 12.Edinger A L, Amedee A, Miller K, Doranz B J, Endres M, Sharron M, Samson M, Lu Z H, Clements J E, Murphey-Corb M, Peiper S C, Parmentier M, Broder C C, Doms R W. Differential utilization of CCR5 by macrophage and T cell tropic simian immunodeficiency virus strains. Proc Natl Acad Sci USA. 1997;94:4005–4010. doi: 10.1073/pnas.94.8.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edinger A L, Blanpain C, Kunstman K J, Wolinsky S M, Parmentier M, Doms R W. Functional dissection of CCR5 coreceptor function through the use of CD4-independent simian immunodeficiency virus strains. J Virol. 1999;73:4062–4073. doi: 10.1128/jvi.73.5.4062-4073.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edinger A L, Mankowski J L, Doranz B J, Margulies B J, Lee B, Rucker J, Sharron M, Hoffman T L, Berson J F, Zink M C, Hirsch V M, Clements J E, Doms R W. CD4-independent, CCR5-dependent infection of brain capillary endothelial cells by a neurovirulent simian immunodeficiency virus strain. Proc Natl Acad Sci USA. 1997;94:14742–14747. doi: 10.1073/pnas.94.26.14742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Endres M J, Clapham P R, Marsh M, Ahuja M, Turner J D, McKnight A, Thomas J F, Stoebenau-Haggarty B, Choe S, Vance P J, Wells T N, Power C A, Sutterwala S S, Doms R W, Landau N R, Hoxie J A. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell. 1996;87:745–756. doi: 10.1016/s0092-8674(00)81393-8. [DOI] [PubMed] [Google Scholar]
- 16.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 17.Fouchier R A, Meyer B E, Simon J H, Fischer U, Malim M H. HIV-1 infection of non-dividing cells: evidence that the amino-terminal basic region of the viral matrix protein is important for Gag processing but not for post-entry nuclear import. EMBO J. 1997;16:4531–4539. doi: 10.1093/emboj/16.15.4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goudsmit J, Bakker M, Smit L. Immunodominant B-cell clones responsive to an HIV-1 neutralization and cell fusion inhibition epitope in chimpanzee-to-chimpanzee passages of HTLV-IIIB and LAV-1. Res Virol. 1989;140:405–418. doi: 10.1016/s0923-2516(89)80119-0. [DOI] [PubMed] [Google Scholar]
- 19.Goudsmit J, Geelen J, Keulen W, Notermans D, Kuiken C, Ramautarsing C, Smit L, Koole L, van Genderen M, Buck H, et al. Characterization of the African HIV-1 isolate CBL-4 (RUT) by partial sequence analysis and virus neutralization with peptide antibody and antisense phosphate-methylated DNA. AIDS. 1990;4:559–564. doi: 10.1097/00002030-199006000-00010. . (Errata, 4(7): following A44 and 4(12):A72.) [DOI] [PubMed] [Google Scholar]
- 20.Hirsch V M, Martin J E, Dapolito G, Elkins W R, London W T, Goldstein S, Johnson P R. Spontaneous substitutions in the vicinity of the V3 analog affect cell tropism and pathogenicity of simian immunodeficiency virus. J Virol. 1994;68:2649–2661. doi: 10.1128/jvi.68.4.2649-2661.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoffman T L, LaBranche C C, Zhang W, Canziani G, Robinson J, Chaiken I, Hoxie J A, Doms R W. Stable exposure of the coreceptor-binding site in a CD4-independent HIV-1 envelope protein. Proc Natl Acad Sci USA. 1999;96:6359–6364. doi: 10.1073/pnas.96.11.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hogervorst E, de Jong J, van Wijk A, Bakker M, Valk M, Nara P, Goudsmit J. Insertion of primary syncytium-inducing (SI) and non-SI envelope V3 loops in human immunodeficiency virus type 1 (HIV-1) LAI reduces neutralization sensitivity to autologous, but not heterologous, HIV-1 antibodies. J Virol. 1995;69:6342–6351. doi: 10.1128/jvi.69.10.6342-6351.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Javaherian K, Langlois A, McDanal C, Ross K, Eckler L, Jellis C, Profy A, Rusche J, Bolognesi D, Putney S, Matthews T. Principle neutralizing domain of the human immunodeficiency virus type 1 envelope. Proc Natl Acad Sci USA. 1989;86:6768–6772. doi: 10.1073/pnas.86.17.6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Javaherian K, Langlois A J, Schmidt S, Kaufmann M, Cates N, Langedijk J P, Meloen R H, Desrosiers R C, Burns D P, Bolognesi D P, et al. The principal neutralization determinant of simian immunodeficiency virus differs from that of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1992;89:1418–1422. doi: 10.1073/pnas.89.4.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kestler H, Kodama T, Ringler D, Marthas M, Pedersen N, Lackner A, Regier D, Sehgal P, Daniel M, King N, Desrosiers R. Induction of AIDS in rhesus monkeys by molecularly cloned simian immunodeficiency virus. Science. 1990;248:1109–1112. doi: 10.1126/science.2160735. [DOI] [PubMed] [Google Scholar]
- 26.Kirchhoff F, Desrosiers R C. A PCR-derived library of random point mutations within the V3 region of simian immunodeficiency virus. PCR Methods Appl. 1993;2:301–304. doi: 10.1101/gr.2.4.301. [DOI] [PubMed] [Google Scholar]
- 27.Kirchhoff F, Mori K, Desrosiers R C. The “V3” domain is a determinant of simian immunodeficiency virus cell tropism. J Virol. 1994;68:3682–3692. doi: 10.1128/jvi.68.6.3682-3692.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koito A, Harrowe G, Levy J A, Cheng-Mayer C. Functional role of the V1/V2 region of human immunodeficiency virus type 1 envelope glycoprotein gp 120 in infection of primary macrophages and soluble CD4 neutralization. J Virol. 1994;68:2253–2259. doi: 10.1128/jvi.68.4.2253-2259.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kwong P D, Wyatt R, Robinson J, Sweet R W, Sodroski J, Hendrickson W A. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewis M G, Bellah S, McKinnon K, Yalley-Ogunro J, Zack P M, Elkins W R, Desrosiers R C, Eddy G A. Titration and characterization of two rhesus-derived SIVmac challenge stocks. AIDS Res Hum Retroviruses. 1994;10:213–220. doi: 10.1089/aid.1994.10.213. [DOI] [PubMed] [Google Scholar]
- 31.Liao F, Alkhatib G, Peden K W, Sharma G, Berger E A, Farber J M. STRL33, a novel chemokine receptor-like protein, functions as a fusion cofactor for both macrophage-tropic and T cell line-tropic HIV-1. J Exp Med. 1997;185:2015–2023. doi: 10.1084/jem.185.11.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ly A, Stamatatos L. V2 loop glycosylation of the human immunodeficiency virus type 1 SF162 envelope facilitates interaction of this protein with CD4 and CCR5 receptors and protects the virus from neutralization by anti-V3 loop and anti-CD4 binding site antibodies. J Virol. 2000;74:6769–6776. doi: 10.1128/jvi.74.15.6769-6776.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Means R E, Desrosiers R C. Resistance of native, oligomeric envelope on simian immunodeficiency virus to digestion by glycosidases. J Virol. 2000;74:11181–11190. doi: 10.1128/jvi.74.23.11181-11190.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Means R E, Greenough T, Desrosiers R C. Neutralization sensitivity of cell culture-passaged simian immunodeficiency virus. J Virol. 1997;71:7895–7902. doi: 10.1128/jvi.71.10.7895-7902.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mori K, Ringler D, Kodama T, Desrosiers R. Complex determinants of macrophage tropism in Env of simian immunodeficiency virus. J Virol. 1992;66:2067–2075. doi: 10.1128/jvi.66.4.2067-2075.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mori K, Rosenzweig M, Desrosiers R C. Mechanisms for adaptation of simian immunodeficiency virus to replication in alveolar macrophages. J Virol. 2000;74:10852–10859. doi: 10.1128/jvi.74.22.10852-10859.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morikita T, Maeda Y, Fujii S, Matsushita S, Obaru K, Takatsuki K. The V1/V2 region of human immunodeficiency virus type 1 modulates the sensitivity to neutralization by soluble CD4 and cellular tropism. AIDS Res Hum Retroviruses. 1997;13:1291–1299. doi: 10.1089/aid.1997.13.1291. [DOI] [PubMed] [Google Scholar]
- 38.Naidu Y M, Kestler H W, Li Y, Butler C V, Silva D P, Schmidt D K, Troupp C D, Seghal P K, Sonigo P, Daniel M D, Desrosiers R C. Characterization of infectious molecular clones of simian immunodeficiency virus (SIVmac) and human immunodeficiency virus type 2: persistent infection of rhesus monkeys with molecularly cloned SIVmac. J Virol. 1988;62:4691–4696. doi: 10.1128/jvi.62.12.4691-4696.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nara P, Smit L, Dunlop N, Hatch W, Merges M, Waters D, Kelliher J, Krone W, Goudsmit J. Evidence for rapid selection and deletion of HIV-1 subpopulations in vivo by V3-specific neutralizing antibody: a model of humoral-associated selection. Dev Biol Stand. 1990;72:315–341. [PubMed] [Google Scholar]
- 40.Palker T J, Muir A J, Spragion D E, Staats H F, Langlois A, Montefiori D C. The V3 domain of SIVmac251 gp120 contains a linear neutralizing epitope. Virology. 1996;224:415–426. doi: 10.1006/viro.1996.0548. [DOI] [PubMed] [Google Scholar]
- 41.Reitter J N, Means R E, Desrosiers R C. A role for carbohydrates in immune evasion in AIDS. Nat Med. 1998;4:679–684. doi: 10.1038/nm0698-679. [DOI] [PubMed] [Google Scholar]
- 42.Rizzuto C D, Wyatt R, Hernandez-Ramos N, Sun Y, Kwong P D, Hendrickson W A, Sodroski J. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science. 1998;280:1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 43.Rudensey L M, Kimata J T, Long E M, Chackerian B, Overbaugh J. Changes in the extracellular envelope glycoprotein of variants that evolve during the course of simian immunodeficiency virus SIVMne infection affect neutralizing antibody recognition, syncytium formation, and macrophage tropism but not replication, cytopathicity, or CCR-5 coreceptor recognition. J Virol. 1998;72:209–217. doi: 10.1128/jvi.72.1.209-217.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakaida H, Hori T, Yonezawa A, Sato A, Isaka Y, Yoshie O, Hattori T, Uchiyama T. T-tropic human immunodeficiency virus type 1 (HIV-1)-derived V3 loop peptides directly bind to CXCR-4 and inhibit T-tropic HIV-1 infection. J Virol. 1998;72:9763–9770. doi: 10.1128/jvi.72.12.9763-9770.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schonning K, Jansson B, Olofsson S, Nielsen J O, Hansen J S. Resistance to V3-directed neutralization caused by an N-linked oligosaccharide depends on the quaternary structure of the HIV-1 envelope oligomer. Virology. 1996;218:134–140. doi: 10.1006/viro.1996.0173. [DOI] [PubMed] [Google Scholar]
- 46.Shieh J T, Martin J, Baltuch G, Malim M H, Gonzalez-Scarano F. Determinants of syncytium formation in microglia by human immunodeficiency virus type 1: role of the V1/V2 domains. J Virol. 2000;74:693–701. doi: 10.1128/jvi.74.2.693-701.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spenlehauer C, Saragosti S, Fleury H J, Kirn A, Aubertin A M, Moog C. Study of the V3 loop as a target epitope for antibodies involved in the neutralization of primary isolates versus T-cell-line-adapted strains of human immunodeficiency virus type 1. J Virol. 1998;72:9855–9864. doi: 10.1128/jvi.72.12.9855-9864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sullivan N, Sun Y, Binley J, Lee J, Barbas III C F, Parren P W, Burton D R, Sodroski J. Determinants of human immunodeficiency virus type 1 envelope glycoprotein activation by soluble CD4 and monoclonal antibodies. J Virol. 1998;72:6332–6338. doi: 10.1128/jvi.72.8.6332-6338.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sullivan N, Sun Y, Sattentau Q, Thali M, Wu D, Denisova G, Gershoni J, Robinson J, Moore J, Sodroski J. CD4-induced conformational changes in the human immunodeficiency virus type 1 gp120 glycoprotein: consequences for virus entry and neutralization. J Virol. 1998;72:4694–4703. doi: 10.1128/jvi.72.6.4694-4703.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sullivan N, Thali M, Furman C, Ho D D, Sodroski J. Effect of amino acid changes in the V1/V2 region of the human immunodeficiency virus type 1 gp120 glycoprotein on subunit association, syncytium formation, and recognition by a neutralizing antibody. J Virol. 1993;67:3674–3679. doi: 10.1128/jvi.67.6.3674-3679.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thali M, Furman C, Wahren B, Posner M, Ho D D, Robinson J, Sodroski J. Cooperativity of neutralizing antibodies directed against the V3 and CD4 binding regions of the human immunodeficiency virus gp120 envelope glycoprotein. J Acquir Immune Defic Syndr. 1992;5:591–599. [PubMed] [Google Scholar]
- 52.Thali M, Moore J P, Furman C, Charles M, Ho D D, Robinson J, Sodroski J. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J Virol. 1993;67:3978–3988. doi: 10.1128/jvi.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trkola A, Dragic T, Arthos J, Binley J M, Olson W C, Allaway G P, Cheng-Mayer C, Robinson J, Maddon P J, Moore J P. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature. 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 54.Vallejo A, Heredia A, Mas A, Lee S F, Epstein J S, Soriano V, Hewlett I K. Tropism, coreceptor use, and phylogenetic analysis of both the V3 loop and the protease gene of three novel HIV-1 group O isolates. J Acquir Immune Defic Syndr Hum Retrovirol. 1998;18:417–425. doi: 10.1097/00042560-199808150-00002. [DOI] [PubMed] [Google Scholar]
- 55.Vancott T C, Polonis V R, Loomis L D, Michael N L, Nara P L, Birx D L. Differential role of V3-specific antibodies in neutralization assays involving primary and laboratory-adapted isolates of HIV type 1. AIDS Res Hum Retroviruses. 1995;11:1379–1391. doi: 10.1089/aid.1995.11.1379. [DOI] [PubMed] [Google Scholar]
- 56.Wang W K, Dudek T, Essex M, Lee T H. Hypervariable region 3 residues of HIV type 1 gp120 involved in CCR5 coreceptor utilization: therapeutic and prophylactic implications. Proc Natl Acad Sci USA. 1999;96:4558–4562. doi: 10.1073/pnas.96.8.4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weissman D, Rabin R L, Arthos J, Rubbert A, Dybul M, Swofford R, Venkatesan S, Farber J M, Fauci A S. Macrophage-tropic HIV and SIV envelope proteins induce a signal through the CCR5 chemokine receptor. Nature. 1997;389:981–985. doi: 10.1038/40173. [DOI] [PubMed] [Google Scholar]
- 58.Wyatt R, Kwong P D, Desjardins E, Sweet R W, Robinson J, Hendrickson W A, Sodroski J G. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393:705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 59.Wyatt R, Moore J, Accola M, Desjardin E, Robinson J, Sodroski J. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J Virol. 1995;69:5723–5733. doi: 10.1128/jvi.69.9.5723-5733.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yi Y, Rana S, Turner J D, Gaddis N, Collman R G. CXCR-4 is expressed by primary macrophages and supports CCR5-independent infection by dual-tropic but not T-tropic isolates of human immunodeficiency virus type 1. J Virol. 1998;72:772–777. doi: 10.1128/jvi.72.1.772-777.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhuge W, Jia F, Adany I, Narayan O, Stephens E B. Plasmas from lymphocyte- and macrophage-tropic SIVmac-infected macaques have antibodies with a broader spectrum of virus neutralization activity in macrophage versus lymphocyte cultures. Virology. 1997;227:24–33. doi: 10.1006/viro.1996.8300. [DOI] [PubMed] [Google Scholar]
- 62.Zwart G, Langedijk H, van der Hoek L, de Jong J J, Wolfs T F, Ramautarsing C, Bakker M, de Ronde A, Goudsmit J. Immunodominance and antigenic variation of the principal neutralization domain of HIV-1. Virology. 1991;181:481–489. doi: 10.1016/0042-6822(91)90880-k. [DOI] [PubMed] [Google Scholar]