Abstract
Objective
Investigation of explainable deep learning methods for graph neural networks to predict HIV infections with social network information and performing domain adaptation to evaluate model transferability across different datasets.
Methods
Network data from two cohorts of younger sexual minority men (SMM) from two U.S. cities (Chicago, IL, and Houston, TX) were collected between 2014 and 2016. Feature importance from graph attention network (GAT) models were determined using GNNExplainer. Domain adaptation was performed to examine model transferability from one city dataset to the other dataset, training with 100% of the source dataset with 30% of the target dataset and prediction on the remaining 70% from the target dataset.
Results
Domain adaptation showed the ability of GAT to improve prediction over training with single city datasets. Feature importance analysis with GAT models in single city training indicated similar features across different cities, reinforcing potential application of GAT models in predicting HIV infections through domain adaptation.
Conclusion
GAT models can be used to address the data sparsity issue in HIV study populations. They are powerful tools for predicting individual risk of HIV that can be further explored for better understanding of HIV transmission.
Keywords: Graph machine learning, explainable artificial intelligence, HIV, domain adaptation
KEY MESSAGES
In this study, we conducted domain adaptation between two urban areas to predict HIV status by incorporating social network data.
We employ GNNExplainer to elucidate the model’s predictions on each city dataset, aligning them with knowledge of HIV risk factors.
Domain adaptation resulted in better model performance over individual city training and has great potential for applications in modeling other sexually transmitted infections.
Introduction
In the United States (U.S.), approximately 1.2 million people live with human immunodeficiency virus (HIV), with up to 13% unaware of being infected [1]. Despite advancements in the treatment of HIV through antiretroviral therapy (ART), there is no cure or vaccine for it [2]. Prevention of HIV infections remain a key focus in ending the global HIV epidemic.
Epidemiological network studies have been conducted to examine HIV transmission dynamics and identify network structural features in the spread of HIV by applying various social network methods and stochastic network modeling approaches [3]. A specific type of modeling used are exponential random graph models (ERGMs), which have been used to describe network structures or make statistical inference for possible transmission pathways [4]. ERGMs have been specifically used to explain racial disparities in HIV prevalence as well as understanding the impact of interventions on reducing HIV infections [5–8]. While these studies contribute to better understanding various and complex dynamics of the HIV epidemic, they have limited applications in predicting HIV infections at the individual level.
Abundant health data from various and emerging sources such as electronic health records (EHR), public health surveillance, and research have brought new opportunities to the intersection of predicting HIV infections with machine learning [9–14]. Other studies have also incorporated social media and smartphone survey data to explore better prediction of HIV infections [15,16]. Within these machine learning methods, logistic regression and random forest are the most commonly implemented models. Their strengths lie in high explanatory power in identifying and associating social and clinical factors to the diagnosis of HIV infections within individuals [17].
For predicting HIV infections, it is important to capture complex relationships and interdependencies between individuals in a network, which is best represented in graphs. These networks can contain important contextual information, like individual sexual and social interactions and behaviors to predict HIV infections. Recent developments in deep learning, specifically graph neural networks (GNNs), have incorporated this information and have displayed strong performances. GNN implementations like graph convolutional networks (GCNs) and graph attention networks (GATs) have successfully shown impressive performance in predicting HIV infection status based on social network data [18,19]. Deep learning models like GNNs have high potential for applications but remain limited by the ‘black box’ problem, due to limited understanding of how these models generate their predictions [20]. Efforts have been made to better interpret these models with methods like GNNExplainer to elucidate the underlying mechanisms of GNNs [21].
In this study, we extended upon previously validated GATs to better understand GATs in interpreting their predictions and aligning these predictions to human understanding of HIV transmission mechanisms through applications of domain adaptation and explainable artificial intelligence (AI) [19]. To do this, GNNExplainer was used to explain feature importance in the GAT models. Furthermore, we performed domain adaptation of predicting HIV infections, by adapting the GATs trained on a dataset for one city to another dataset for another city. The two cities, Chicago and Houston, are appropriate for this study because of similar HIV prevalence rates and both cities fall under the jurisdictions of the Ending the HIV Epidemic (EHE) initiative [22,23]. Our objective was to demonstrate whether patterns learned by GATs for one city can be generalized and applied to predicting HIV infections in other cities.
Materials and methods
YMAP (Young Men’s Affiliation Project of HIV Risk and Prevention Venue) was a prospective cohort study that investigated the impact of social networks in relation to HIV risk and prevention in younger sexual minority men (SMM), between the ages of 16 and 29, in two United States cities, Houston and Chicago [24]. The study used data collected from SMM participants between 2014 and 2016 through respondent-driven sampling (RDS) method [25]. Respondents were asked to recruit their peers, establishing a network and understanding who recruited whom and the number of social contacts for each respondent. Written informed consent was obtained from all individual participants involved in this study. 378 SMM were recruited from Houston and 377 SMM from Chicago. A participant’s initial HIV infection status was determined based on the ALere Determine TMHIV-1/2 Combo antigen/antibody test. Those with reactive samples received additional HIV-1/HIV-2 multispot differentiation and HIV RNA (viral load) tests during follow up periods (average once a year).
Respondent data were collected in two waves, which identified sociodemographic characteristics, HIV/sexually transmitted infection risk/protective behaviors, social and sexual networks, and venue attendance/affiliation information [26]. A cross-sectional approach was adopted. Features from both waves were aggregated to represent a single point in time, to assess their predictive power and associations with HIV infection. An individual’s HIV infection status was positive if the lab test taken at the first or second wave was positive. There were 130 and 149 HIV positive SMM identified corresponding to HIV prevalence rates of 34.4% and 39.4% in Chicago and Houston, respectively.
The social network for participants in each city was built so that each node represented an individual participant, and edges between nodes represented the type of relationship (social, referral, or sexual) between individuals from RDS. Two nodes were considered to be neighbors if they were connected by an edge. Adjacency matrices were used to represent the edges between nodes of each network, where neighbors of each node are indicated with a value of 1 while other nodes are masked with a value of 0.
Based on these networks, we can define and curate specific network features for the models. The degree centrality of each node reflects the number of edges it has [27]. We also quantify the number of neighbors in the social network each node has, along with the number of social and health venues attended. Important consideration was given to selecting and preprocessing features based on their relationship with HIV infection status. We initially employed logistic regression analysis to quantify the strength and significance of associations between each feature and the outcome. Features that showed strong correlations with HIV infection status were considered highly predictive and excluded from the models.
Graph neural networks
GNNs have emerged in their ability to perform data in non-Euclidean space, in representations like graphs that contain nodes (entities that contain information) and edges (that represent different types of connections between nodes) [28]. Their goal is to learn representations of nodes in a graph and can be used to make predictions on the node-level, edge-level, and graph-level. This is done through message parsing, in which nodes send messages to their neighbors, and this information is aggregated and updated for each node’s representation via GNN. Graph attention networks (GATs) are another type of GNNs that incorporate attention mechanisms [29,30]. The attention mechanism in GATs assigns weights in a manner so that more important neighboring nodes receive higher weights during aggregation.
is a learnable weight matrix applied to every node to transform input feature vector h of dimension f to a hidden vector of dimension d. The attention coefficient between two nodes (i and j) is defined as
The coefficients are normalized with a softmax function
where Ni is a set of all neighbors of node i. Finally, the embeddings from the neighbors are aggregated together to generate a final output feature for each node as
where is a nonlinearity function.
Model construction and analysis
For each city dataset (Houston and Chicago), a GAT model was trained and compared with baseline machine learning models like logistic regression and random forest. We used a 75–25 train-test split. The GAT model architecture contained eight hidden layers with one attention head being used, for consideration of first-order neighbors. Masking vectors were used to separate nodes for training and nodes for GAT model.
The goal of domain adaptation is to improve a model on a ‘target’ domain by using knowledge learned from a ‘source’ domain, due to distribution differences between the two domains [31]. In our study, the two different cities served as the different domains. For training, two sets of training were performed (one where the Houston dataset is the source domain and the other where the Chicago dataset is the source domain). We used 100% of training data from the source domain and combined it with 30% of the target domain with the remaining 70% of the target domain as test data.
Model training was performed through ten-fold cross-validation. In this study, we employed grid search techniques to optimize hyperparameters for logistic regression and random forest models. For logistic regression, we varied the regularization type (L1 and L2) and regularization strength (C values ranging from 0.0001 to 100) to evaluate their impact on model performance. For the random forest models, we varied the maximum tree depths (ranging from 1 to 50). For training the GAT model, training was conducted with an Adam Optimizer for 2400 iterations [32]. Dropout was set at 0.1 to retain more features. To measure model performance, metrics like AUROC (area under the receiver operating characteristics curve), AUPRC (area under the precision-recall curve), and F1 score were used.
To evaluate the effect of the features, baseline machine learning models were trained with scikit-learn [33]. Coefficients were extracted from logistic regression models and feature importance from random forest models. For the GAT model, we used the PyTorch library for training on individual city datasets and TensorFlow for the domain adaptation training process [34,35]. To better understand the feature importance for these models, a tool called GNNExplainer was used. GNNExplainer is an optimization task to identify a subgraph of the original graph to maximize the mutual information (node features and edges) important to the prediction task [21]. Specifically, GNNExplainer optimizes the following function to learn an edge mask M and feature mask F
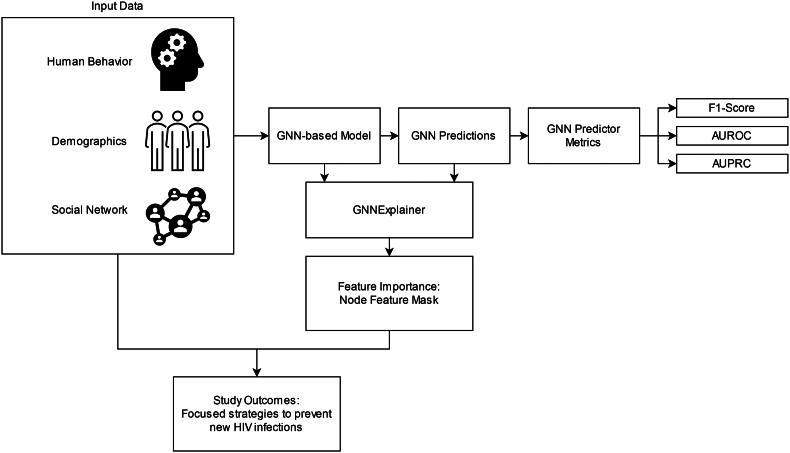
where l is the loss function, y is the original model prediction, is the model prediction with M and F applied, and H is the entropy function. The edge mask and feature mask are learned for each specific node. To quantify feature importance, the feature masks are summed. The workflow of GNNExplainer and the interpretation of its explanations in relation to predicting HIV infection is shown in Figure 1.
Figure 1.
Flow chart explaining incorporation of graph neural networks in this study.
Results
The model performance across three models (logistic regression (LR), random forest (RF), and graph attention network (GAT)) trained on each city dataset individually can be found in Table 1. For domain adaptation, the results can be found in Table 2, where each city serves as a source domain.
Table 1.
Model performance for single city datasets.
| LR | RF | GAT | ||
|---|---|---|---|---|
| Chicago | F1 Score | 0.423 | 0.376 | 0.677 |
| AUROC | 0.611 | 0.643 | 0.737 | |
| AUPRC | 0.419 | 0.443 | 0.777 | |
| Houston | F1 Score | 0.564 | 0.537 | 0.701 |
| AUROC | 0.715 | 0.758 | 0.753 | |
| AUPRC | 0.537 | 0.611 | 0.698 | |
Table 2.
Model performance with domain adaptation.
| LR | RF | GAT | ||
|---|---|---|---|---|
| Chicago → Houston | F1 Score | 0.417 | 0.401 | 0.665 |
| AUROC | 0.643 | 0.687 | 0.743 | |
| AUPRC | 0.436 | 0.477 | 0.608 | |
| Houston → Chicago | F1 Score | 0.483 | 0.348 | 0.639 |
| AUROC | 0.758 | 0.749 | 0.772 | |
| AUPRC | 0.612 | 0.609 | 0.820 |
Training was done with 100% of the source city and 30% of the target city, with the remaining 70% of the target city for predicting HIV infection status.
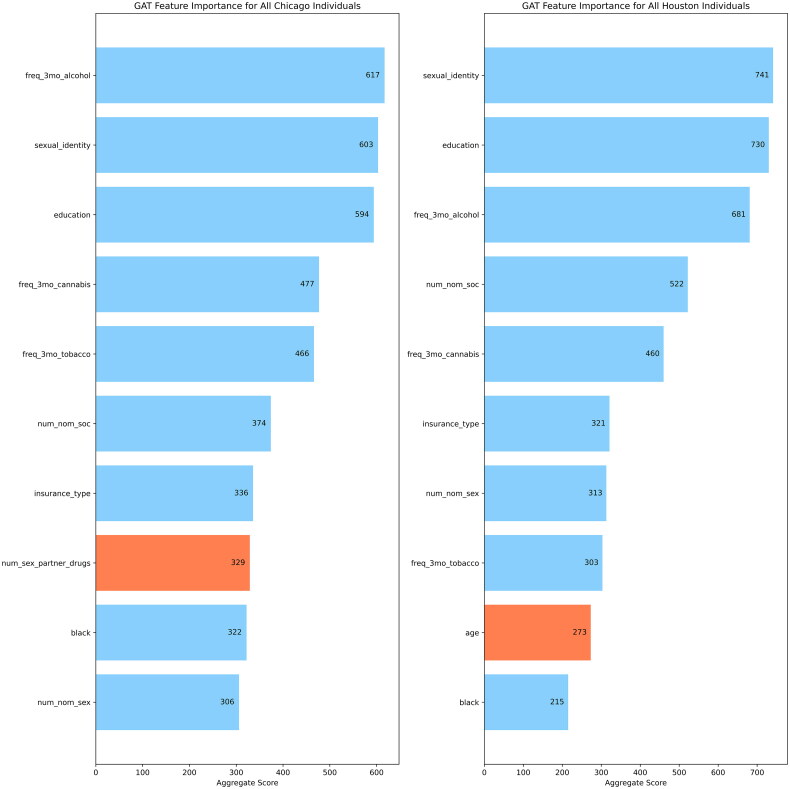
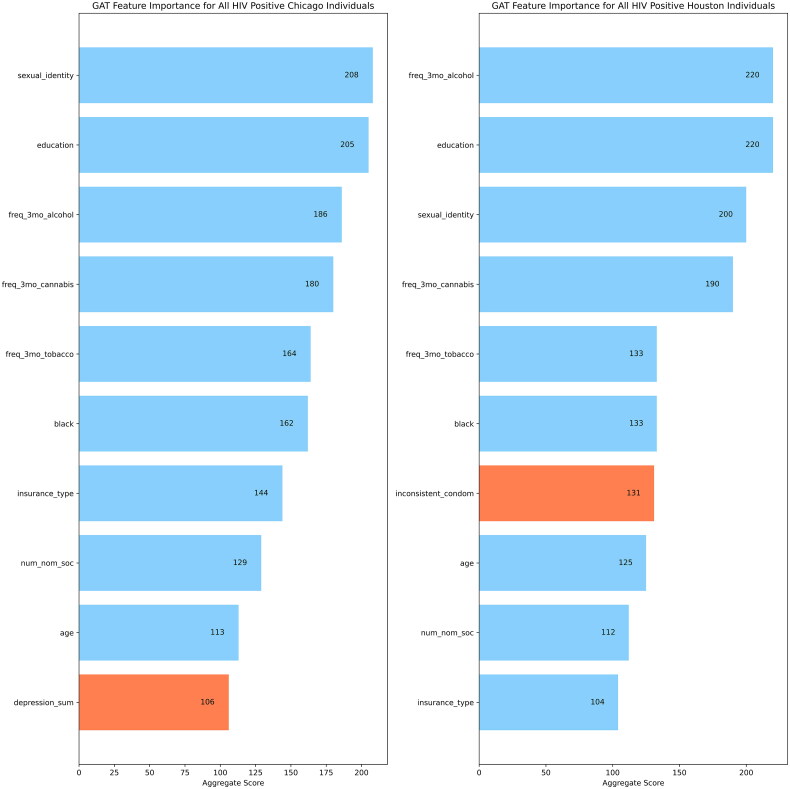
GNNExplainer was used to quantify feature importance for the GAT models performed on the Chicago and Houston datasets. The top ten important features were determined across two sets of populations of each city: all individuals (Figure 2(a)) and all HIV positive individuals (Figure 2(b)).
Figure 2.
(a) Top 10 features identified by GNNExplainer for the Chicago and Houston datasets for all individuals in each city. (b) Top 10 features identified by GNNExplainer for the Chicago and Houston datasets for all HIV positive individuals in each city.
Across all the individuals for each city dataset, GNNExplainer identified 9 similar features for the GAT model: education, insurance type, sexual identity, number of venue neighbors, frequency of alcohol, cannabis, and tobacco use in the last 3 months, number of nominated sexual partners, and number of nominated sexual partners. When we look at the top 10 features for only HIV positive individuals in each city dataset, GNNExplainer identified these 9 similar features: education, black racial identity, age, sexual identity, frequency of alcohol, cannabis, and tobacco use in the last 3 months, insurance type, and number of nominated social partners. The features with blue bars in the plots of Figure 2(a and b) are used to show similar important features across the two cities while the features with orange bars are used to show features with no overlap between the two cities.
Discussion
Individual city model interpretation
First, we performed experiments to establish that the GAT model outperforms logistic regression and random forest for both the Chicago and Houston datasets across AUROC, AUPRC, and F1 score metrics. The only exception was for the Houston dataset, where the AUROC score of random forest outperformed that of the GAT model (0.758 vs 0.753). We also observe that the Houston-trained models tended to outperform the Chicago-trained models, but the Chicago-trained GAT model performed similarly to the Houston-trained GAT model. This could be explained by the importance of similar features for the GAT models across both cities, as identified by GNNExplainer.
Domain adaptation model interpretation
The results for model performances after domain adaptation across the two cities indicated general improvements across all metrics (AUC, AUPRC, F1) over single city dataset training. This shows that the domain adaptation strategy does allow GAT models to improve prediction of HIV infection status across different datasets. For the GAT model with the Chicago dataset as the source domain, it can be inferred that the improved performance can be attributed to their processing of network contextual information from the graph data rather than defined features that traditional machine learning methods rely on.
GNNExplainer interpretation
We used GNNExplainer to interpret the feature importance for the GAT model trained on each individual city and align it to our findings in the models used for domain adaptation. We specifically wanted to analyze the feature importance for the HIV positive individuals in both cities.
In terms of sociodemographic data, individuals who identify as black are disproportionately affected by HIV [36]. The GAT model was able to associate the importance of individuals who identified as black to their HIV status. Age was also an important factor identified by GNNExplainer. This study is focused on the young SMM population, because this age gap reflects the largest number of new HIV infections annually [37].
Factors like substance abuse (cannabis, alcohol, and tobacco) align with previously described risk factors that can result in HIV infections [38]. Similarly, education showed the highest importance across both cities from the GAT models, which the CDC has linked those with lower education levels as having higher HIV prevalence rates [39]. This also continues to support the importance social determinants of health play in HIV infection [40].
Network features like the number of nominated social partners showed high importance. The nominated social partners are those identified as individuals that participants in this study shared personal information with. This continues to show the importance of incorporating network information into studies in understanding patterns related to HIV transmission [41,42].
For the Houston dataset, inconsistent use of condom was an important feature not shared with the Chicago dataset. This reflects findings from previous studies that emphasize the importance of prioritizing HIV prevention strategies that promote consistent condom use [43,44]. For the Chicago dataset, the depression sum score was a differing important feature from the Houston dataset. This score is based upon the Brief Symptom Inventory-18 (BSI-18) self-report questionnaire [45]. The depression sum score used in our study was generated from questions specifically related to the depression subscale of BSI-18. This finding reinforces that individuals who are depressed may engage in substance abuse and sexual risk behaviors that may lead to HIV infection [46,47].
Strengths and limitations
We first note that the domain adaptation approach presented in this study of model training from one city dataset to another city dataset can be achieved in the task of HIV infection prediction. We also provide a better understanding in explaining how GATs perform with real world data and in the challenging task of predicting HIV infection status. This gives further opportunity to evaluate GATs in similar prediction tasks for other sexually transmitted infections.
There are some limitations to our study. The current implementation of GNNExplainer cannot be used to quantify feature importance across different datasets for the domain adaptation process. We hope to contribute to that in the future and improve better understanding and explanation of GNN models. Furthermore, we have no validation datasets to further enhance our results.
Conclusion
We showed that the proposed framework of domain adaptation for predicting HIV status in younger SMM from one city to another led to better performance in GAT models than training on individual datasets. Such a framework is especially valuable in HIV studies where social network data among this population is limited and sparse. We were also able to determine the feature importance of the GAT models with GNNExplainer to align current knowledge of HIV transmission factors to the populations used in this study. Our findings continue to support the need for interdisciplinary work between public health experts, computer scientists, and clinicians in ending the HIV epidemic. There is viability in this strategy in better understanding specific factors in HIV transmission for local populations.
There are several directions for future work. We plan to externally validate our domain adaptation models on a third dataset. In addition, we would like to encode more advanced features by defining edge types between nodes and explore adapting these models to multi-relational graph data. This would allow training and tuning of models for HIV prediction and using these models for status prediction in other sexually transmitted infections.
Funding Statement
This research was supported by a fellowship from the Gulf Coast Consortia, on the NLM Training Program in Biomedical Informatics and Data Science T15 LM007093. This research was also supported by the National Institutes of Health under awards R56AI150272, R01MH100021 and U24AI171008.
Author contributions
Study conception/design: Cui Tao, Jingcheng Du, Kayo Fujimoto; data analysis and performing experiments: Evan Yu, Jingna Feng, Xinyue Hu, Yang Xiang; drafting of manuscript: Evan Yu; Interpretation of results: Evan Yu, Kayo Fujimoto, Cui Tao, John A. Schneider; critical revision of manuscript: Cui Tao, Kayo Fujimoto, Xi Luo, Degui Zhi. All authors have read and agreed to the published version of the manuscript.
Consent form
Informed consent was obtained from all individual participants involved in this study. All participating institutions in YMAP (The University of Chicago, Ann & Robert H. Lurie Children’s Hospital of Chicago, and The University of Texas Health Science Center at Houston School of Public Health) received approval from the institutional review boards (IRB #HSCSPH120830).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are available on request from the corresponding author.
The source code is available at: https://github.com/Tao-AI-group/Domain_Adaptation_GNN_HIV.
References
- 1.CDC . Basic Statistics | HIV Basics | HIV/AIDS |; 2021; [cited 2022 Mar 9]. Available from:https://www.cdc.gov/hiv/basics/statistics.html.
- 2.Hargrave A, Mustafa AS, Hanif A, et al. Current status of HIV-1 vaccines. Vaccines. 2021;9(9):1026. doi: 10.3390/vaccines9091026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amirkhanian YA. Social networks, sexual networks and HIV risk in men who have sex with men. Curr HIV/AIDS Rep. 2014;11(1):81–92. doi: 10.1007/s11904-013-0194-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jenness SM, Goodreau SM, Morris M.. EpiModel: an R package for mathematical modeling of infectious disease over networks. J Stat Softw. 2018;84(8):8. doi: 10.18637/jss.v084.i08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krivitsky PN, Morris M.. Inference for social network models from egocentrically sampled data, with application to understanding persistent racial disparities in HIV prevalence in the US. Ann Appl Stat. 2017;11(1):427–455. doi: 10.1214/16-AOAS1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goodreau SM, Rosenberg ES, Jenness SM, et al. Sources of racial disparities in HIV prevalence in men who have sex with men in Atlanta, GA, USA: a modelling study. Lancet HIV. 2017;4(7):e311–e320. doi: 10.1016/S2352-3018(17)30067-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cassels S, Clark SJ, Morris M.. Mathematical models for HIV transmission dynamics. J. Acquir. Immune Defic Syndr. 2008;47(Supplement 1):S34–S39. doi: 10.1097/QAI.0b013e3181605da3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jenness SM, Johnson JA, Hoover KW, et al. Modeling an integrated HIV prevention and care continuum to achieve the ending the HIV epidemic goals. AIDS. 2020;34(14):2103–2113. doi: 10.1097/QAD.0000000000002681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marcus JL, Sewell WC, Balzer LB, et al. Artificial intelligence and machine learning for HIV prevention: emerging approaches to ending the epidemic. Curr HIV/AIDS Rep. 2020;17(3):171–179. doi: 10.1007/s11904-020-00490-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krakower DS, Gruber S, Hsu K, et al. Development and validation of an automated HIV prediction algorithm to identify candidates for pre-exposure prophylaxis: a modelling study. Lancet HIV. 2019;6(10):e696–e704. doi: 10.1016/S2352-3018(19)30139-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haas O, Maier A, Rothgang E.. Machine learning-based HIV risk estimation using incidence rate ratios. Front Reprod Health. 2021;3:756405. https://www.frontiersin.org/article/10.3389/frph.2021.756405. doi: 10.3389/frph.2021.756405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mutai CK, McSharry PE, Ngaruye I, et al. Use of machine learning techniques to identify HIV predictors for screening in sub-Saharan Africa. BMC Med Res Methodol. 2021;21(1):159. doi: 10.1186/s12874-021-01346-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marcus JL, Hurley LB, Krakower DS, et al. Use of electronic health record data and machine learning to identify potential candidates for HIV preexposure prophylaxis: a modelling study. Lancet HIV. 2019;6(10):e688–e695. doi: 10.1016/S2352-3018(19)30137-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nisa SU, Mahmood A, Ujager FS, et al. HIV/AIDS predictive model using random forest based on socio-demographical, biological and behavioral data. Egypt. Inform. J. 2023;24(1):107–115. doi: 10.1016/j.eij.2022.12.005. [DOI] [Google Scholar]
- 15.Wray TB, Luo X, Ke J, et al. Using smartphone survey data and machine learning to identify situational and contextual risk factors for HIV risk behavior among men who have sex with men who are not on PrEP. Prev Sci. 2019;20(6):904–913. doi: 10.1007/s11121-019-01019-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Young SD, Yu W, Wang W.. Toward automating HIV identification: machine learning for rapid identification of HIV-related social media data. J Acquir Immune Defic Syndr. 2017;74(Suppl 2):S128–S131. doi: 10.1097/QAI.0000000000001240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiang Y, Du J, Fujimoto K, et al. Review of application of artificial intelligence and machine learning for HIV prevention interventions to eliminate HIV. Lancet HIV. 2022;9(1):e54–e62. doi: 10.1016/S2352-3018(21)00247-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiang Y, Fujimoto K, Schneider J, et al. Network context matters: graph convolutional network model over social networks improves the detection of unknown HIV infections among young men who have sex with men. J Am Med Inform Assoc. 2019;26(11):1263–1271. doi: 10.1093/jamia/ocz070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiang Y, Fujimoto K, Li F, et al. Identifying influential neighbors in social networks and venue affiliations among young MSM: a data science approach to predict HIV infection. AIDS. 2021;35(Suppl 1):S65–S73. doi: 10.1097/QAD.0000000000002784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang G, Ye Q, Xia J.. Unbox the black-box for the medical explainable AI via multi-modal and multi-centre data fusion: a mini-review, two showcases and beyond. Inf Fusion. 2022;77:29–52. doi: 10.1016/j.inffus.2021.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ying Z, Bourgeois D, You J, et al. GNNExplainer: generating explanations for graph neural networks. Adv Neural Inf Process Syst. Curran Associates, Inc.; 2019; [cited 2023 Sep 16]. Available from: https://papers.nips.cc/paper_files/paper/2019/hash/d80b7040b773199015de6d3b4293c8ff-Abstract.html. [PMC free article] [PubMed]
- 22.Hall HI, Espinoza L, Benbow N, et al. Epidemiology of HIV infection in large urban areas in the United States. PLoS One. 2010;5(9):e12756. doi: 10.1371/journal.pone.0012756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ending the HIV Epidemic, HIV.Gov .; n.d.; [cited 2023 Dec 7]. Available from: https://www.hiv.gov/federal-response/ending-the-hiv-epidemic/overview.
- 24.Fujimoto K, Turner R, Kuhns LM, et al. Network centrality and geographical concentration of social and service venues that serve young men who have sex with men. AIDS Behav. 2017;21(12):3578–3589. doi: 10.1007/s10461-017-1711-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schonlau M, Liebau E.. Respondent-driven sampling. The Stata Journal. 2012;12(1):72–93. doi: 10.1177/1536867X1201200106. [DOI] [Google Scholar]
- 26.Fujimoto K, Cao M, Kuhns LM, et al. Statistical adjustment of network degree in respondent-driven sampling estimators: venue attendance as a proxy for network size among young MSM. Soc Networks. 2018;54:118–131. doi: 10.1016/j.socnet.2018.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Freeman LC, Borgatti SP, White DR.. Centrality in valued graphs: a measure of betweenness based on network flow. Soc. Netw. 1991;13(2):141–154. doi: 10.1016/0378-8733(91)90017-N. [DOI] [Google Scholar]
- 28.Wu Z, Pan S, Chen F, et al. A comprehensive survey on graph neural networks. IEEE Trans Neural Netw Learn Syst. 2021;32(1):4–24. doi: 10.1109/TNNLS.2020.2978386. [DOI] [PubMed] [Google Scholar]
- 29.Veličković P, Cucurull G, Casanova A, et al. Graph attention networks. 2018. doi: 10.48550/arXiv.1710.10903. [DOI]
- 30.Vaswani A, Shazeer N, Parmar N, et al. Attention is all you need. Adv. Neural Inf. Process. Syst. Curran Associates, Inc.; 2017. ; [cited 2023 Sep 16]. Available from: https://papers.nips.cc/paper_files/paper/2017/hash/3f5ee243547dee91fbd053c1c4a845aa-Abstract.html.
- 31.Farahani A, Voghoei S, Rasheed K, et al. A brief review of domain adaptation. In Stahlbock R, Weiss GM, Abou-Nasr M, Yang C-Y, Arabnia HR, Deligiannidis L, editors. Advances in Data Science and Information Engineering. Cham: Springer International Publishing; 2021. p. 877–894. doi: 10.1007/978-3-030-71704-9_65. [DOI] [Google Scholar]
- 32.Kingma DP, Ba J.. Adam: a method for stochastic optimization; 2017; [cited 2023 May 12]. Available from: http://arxiv.org/abs/1412.6980.
- 33.Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: machine learning in Python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- 34.Paszke A, Gross S, Chintala S, et al. Automatic differentiation in PyTorch; 2017; [cited 2023 Sep 23]. Available from: https://openreview.net/forum?id=BJJsrmfCZ.
- 35.Abadi M, Agarwal A, Barham P, et al. TensorFlow: a system for large-scale machine learning. In Proceedings of the 12th USENIX conference on Operating Systems Design and Implementation (OSDI’16). USENIX Association; 2016; 265–283. 10.48550/arXiv.1603.04467. [DOI] [Google Scholar]
- 36.CDC . HIV in the U.S. by the Numbers – 2021 | Fact Sheets | Newsroom | NCHHSTP | 2023; [cited 2021 Nov 17]. Available from: https://www.cdc.gov/nchhstp/newsroom/fact-sheets/hiv/hiv-in-the-us-by-the-numbers.html.
- 37.HIV.Gov . HIV & AIDS Trends and U.S. Statistics Overview; n.d.; [cited 2023 Apr 25]. Available from:https://www.hiv.gov/hiv-basics/overview/data-and-trends/statistics.
- 38.Koblin BA, Husnik MJ, Colfax G, et al. Risk factors for HIV infection among men who have sex with men. AIDS. 2006;20(5):731–739. doi: 10.1097/01.aids.0000216374.61442.55. [DOI] [PubMed] [Google Scholar]
- 39.Centers for Disease Control and Prevention (CDC) . Characteristics associated with HIV infection among heterosexuals in urban areas with high AIDS prevalence – 24 cities, United States, 2006-2007. MMWR Morb Mortal Wkly Rep. 2011;60(31):1045–1049. [PubMed] [Google Scholar]
- 40.Dean HD, Fenton KA.. Addressing social determinants of health in the prevention and control of HIV/AIDS, viral hepatitis, sexually transmitted infections, and tuberculosis. Public Health Rep. 2010;125(Suppl 4):1–5. doi: 10.1177/00333549101250S401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Friedman SR, Kippax SC, Phaswana-Mafuya N, et al. Emerging future issues in HIV/AIDS social research. AIDS. 2006;20(7):959–965. doi: 10.1097/01.aids.0000222066.30125.b9. [DOI] [PubMed] [Google Scholar]
- 42.Adimora AA, Schoenbach VJ, Doherty IA.. HIV and African Americans in the Southern United States: sexual networks and social context. Sex Transm Dis. 2006;33(7 Suppl):S39–S45. doi: 10.1097/01.olq.0000228298.07826.68. [DOI] [PubMed] [Google Scholar]
- 43.Pinkerton SD, Abramson PR.. Effectiveness of condoms in preventing HIV transmission. Soc Sci Med. 1997;44(9):1303–1312. doi: 10.1016/S0277-9536(96)00258-4. [DOI] [PubMed] [Google Scholar]
- 44.Effectiveness of condoms in preventing sexually transmitted infections. Database Abstr. Rev. Eff. DARE Qual.-Assess. Rev. Internet. UK Centre for Reviews and Dissemination; 2004; [cited 2021 Oct 12]. Available from:https://www.ncbi.nlm.nih.gov/books/NBK70881/ (accessed December 13, 2023). [Google Scholar]
- 45.Rath JF, Fox LM.. Brief symptom inventory. In Kreutzer JS, DeLuca J, Caplan B, editors. Encyclopedia of Clinical Neuropsychology. Cham: Springer International Publishing; 2018. p. 633–636. doi: 10.1007/978-3-319-57111-9_1977. [DOI] [Google Scholar]
- 46.Hutton HE, Lyketsos CG, Zenilman JM, et al. Depression and HIV risk behaviors among patients in a sexually transmitted disease clinic. Am J Psychiatry. 2004;161(5):912–914. doi: 10.1176/appi.ajp.161.5.912. [DOI] [PubMed] [Google Scholar]
- 47.Taniguchi T, Shacham E, Onen NF, et al. Depression severity is associated with increased risk behaviors and decreased CD4 cell counts. AIDS Care. 2014;26(8):1004–1012. doi: 10.1080/09540121.2014.880399. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Veličković P, Cucurull G, Casanova A, et al. Graph attention networks. 2018. doi: 10.48550/arXiv.1710.10903. [DOI]
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author.
The source code is available at: https://github.com/Tao-AI-group/Domain_Adaptation_GNN_HIV.