Abstract
Human immunodeficiency virus type 1 (HIV-1) is frequently attenuated after long-term culture in vitro. The attenuation process probably involves mutations of functions required for replication and pathogenicity in vivo. Analysis of attenuated HIV-1 for replication and pathogenicity in vivo will help to define these functions. In this study, we examined the pathogenicity of an attenuated HIV-1 isolate in a laboratory worker accidentally exposed to a laboratory-adapted HIV-1 isolate. Using heterochimeric SCID-hu Thy/Liv mice as an in vivo model, we previously defined HIV-1 env determinants (HXB/LW) that reverted to replicate in vivo (L. Su, H. Kaneshima, M. L. Bonyhadi, R. Lee, J. Auten, A. Wolf, B. Du, L. Rabin, B. H. Hahn, E. Terwilliger, and J. M. McCune, Virology 227:46–52, 1997). Here we further demonstrate that HIV-1 replication in vivo can be separated from its pathogenic activity, in that the HXB/LW virus replicated to high levels in SCID-hu Thy/Liv mice, with no significant thymocyte depletion. Restoration of the nef gene in the recombinant HXB/LW genome restored its pathogenic activity, with no significant effect on HIV-1 replication in the thymus. Our results suggest that in vitro-attenuated HIV-1 lacks determinants for pathogenicity as well as for replication in vivo. Our data indicate that (i) the replication defect can be recovered in vivo by mutations in the env gene, without an associated pathogenic phenotype, and (ii) nef can function in the HXB/LW clone as a pathogenic factor that does not enhance HIV-1 replication in the thymus. Furthermore, the HXB/LW virus may be used to study mechanisms of HIV-1 nef-mediated pathogenesis in vivo.
Human immunodeficiency virus type 1 (HIV-1) diseases (AIDS) are associated with high levels of HIV-1 viremia and depletion of CD4+ T lymphocytes. In vivo, HIV-1 can infect diverse cell types, including CD4+ T cells, macrophages, dendritic cells, Langerhans cells, and hematopoietic progenitor cells (13, 24, 30, 39, 42). However, the HIV-1 isolates used in many studies have been expanded and maintained in immortalized human T-cell lines. The different selective pressures in vitro have led to the generation of variants with attenuated replication and pathogenicity in vivo (38). Many laboratory-adapted isolates of HIV-1 show defects in the functions of some genes, such as env, vpr, vpu, and nef (38). A good example of such adaptation in vitro is Lai/IIIB (human T-cell lymphotropic virus strain IIIB) (6). Initially derived from a human patient blood sample and cultured in MT2/B cells, Lai/IIIB stock was prepared by infecting the human leukemia T-cell line H9 with infected MT2/B-cell supernatant. Subsequent analyses of the genome from the Lai/IIIB isolate showed that multiple changes accumulated during expansion in vitro (38). For example, the HXB2 genome cloned from Lai/IIIB carries mutations that lead to premature termination of three of the nine open reading frames (ORFs): vpr, vpu, and nef. Many other subtle mutations also may have accumulated. These mutations do not usually affect HIV-1 replication in vitro under specific culture conditions, although some of them may enhance viral replication in certain cell lines. However, it remains unclear which mutations contribute to attenuated HIV-1 replication and pathogenesis in vivo.
It has been difficult to analyze HIV-1 functions specifically involved in pathogenicity. Mutations in putative pathogenic factors such as nef or env in simian immunodeficiency virus (SIV) or HIV-1 have led to reduced viremia in vivo (1, 14, 34, 40). Thus, the reduced pathology may be due to a reduced viral load in vivo. Such pathogenic factors are therefore also replication factors. Transgenic mouse models in which nef is expressed constitutively in thymocytes and CD4+ T and macrophage cells lead to CD4+ T-cell depletion and other AIDS-like symptoms, suggesting that nef may be a principal pathogenic factor (15). However, problems arise with transgenic mouse models during attempts to correlate nef expression with virus-mediated pathogenicity. First, the level and time of transgenic nef expression are very different from those in HIV-1 infection in humans. Second, murine host cells may respond differently to HIV-1 nef proteins. Therefore, whether nef is a factor for replication and/or pathogenicity in vivo is still not clear.
Comparison of HIV-1 isolates attenuated in vitro with pathogenic revertants in vivo will help to identify viral determinants important for replication and pathogenesis in vivo. One example of a factor involved in pathogenicity is found in SHIV (SIV-HIV env chimeric genome) adapted in monkeys. SHIV variants with enhanced replication and pathogenicity have been isolated from monkeys infected with SHIV recombinant viruses (19). Mutations in HIV env genes have been identified which contribute to enhanced replication in monkeys. Interestingly, env determinants have also been defined that specifically contribute to CD4+ T-cell depletion (i.e., pathogenicity) but not replication in monkeys (12). Therefore, unique env determinants have intrinsic pathogenic activity in monkeys.
The Lai/IIIB isolate and its associated infectious molecular clones (e.g., HXB2) were found to infect T-cell lines such as H9 as well as peripheral blood mononuclear cells (PBMCs) in vitro but to be replication defective in vivo (14, 34, 40). When a laboratory worker was accidentally infected by Lai/IIIB, infectious virus was isolated from plasma by infection of primary PBMCs but not by infection of T-cell lines (22, 43). We have previously used SCID-hu Thy/Liv mice as an in vivo model (29, 31) to study the replication of HXB2 and of HXB2 recombinant viruses containing HIV-1 fragments isolated from the infected laboratory worker (40). Like Lai/IIIB, HXB2 failed to replicate in the Thy/Liv organ (4, 40). Replacement of an HXB2 subgenomic fragment containing the env ORF with the corresponding fragment from the laboratory worker isolate (LW12.3) generated a recombinant virus (HXB/LW) which replicated in SCID-hu Thy/Liv mice and in the human fetal thymus organ culture (HF-TOC) model (23, 40). The specific in vivo replication determinants were mapped to the region from V1 to V3 of the HXB/LW env gene (40). Therefore, the attenuated Lai/IIIB isolate acquired in vivo replication activity by mutational reversion of the env gene in the infected laboratory worker.
We used the SCID-hu Thy/Liv model to study the pathogenic activity of HXB/LW. HXB/LW replicated to high levels in the SCID-hu Thy/Liv mouse and HF-TOC models. However, the pathogenic activity of HXB/LW was significantly reduced. Restoration of the defective nef ORF in HXB/LW resulted in enhanced viral pathogenicity, with no significant effect on viral replication, in both the SCID-hu Thy/Liv mouse and the HF-TOC models. Thus, we demonstrated in this study that it was possible to separate the replication activity of a recombinant HIV-1 clone from its pathogenicity in vivo and that an intact nef ORF was able to serve exclusively as an HIV-1 pathogenic factor in the replication-competent recombinant virus.
MATERIALS AND METHODS
Construction of recombinant HIV-1 genomes.
HXB2 genomic DNA was digested with SalI and BamHI to remove a 2.7-kb fragment, and the corresponding fragment from LW12.3 was ligated into the provirus DNA to generate the HXB/LW provirus. The mutations in vpr and nef are still present in the HXB/LW genome (22, 40).
Construction of HXB/LW-nef+ (HXB/LWn+) was carried out by first subcloning the 3′ half of the HXB/LW provirus into pBluescript (pBS) SK(+) (Invitrogen). The HXB/LW nef gene was then repaired by a recombination PCR (RPCR) site-directed mutagenesis method described previously (11). First, the 3′ half of the HXB/LW proviral genome was amplified by PCR with primer HC5 (GGGTGTCGACATAGCAGAATAGGC), which includes the SalI site in the vpr gene, and primer HIV3′-X (CGGCTCTAGAGATTTTCCACACTGACTAAAAGG), which anneals to the 3′ terminus of the genome and contains a 3′-terminal XbaI site. This genomic fragment was subcloned into pBS. The resulting pBS.HIV-LW.Sal/Xba plasmid (pBS-3′HX/LW) was then used as template DNA for RPCR mutagenesis as previously described in detail (11). Briefly, two pairs of overlapping primers were used in RPCR mutagenesis. The mutagenizing primer pair bound to overlapping regions of the HXB/LW nef gene. The primers (forward, 5′-CCCTGATTGGCAGAACTACACACC-3′; reverse, 5′-GTAGTTCTGCCAATCAGGGAAGTAGCC-3′) incorporated an AT-GC base change (underlined residues) which repaired the premature stop codon in the HXB/LW nef gene. The nonmutagenizing primer pair bound to overlapping regions of the pBS Ampr gene (5′-GATGTAACCCACTCGTGCACCCAACTGAT-3′; 5′-GGGTGCAGCAGTGGGTTACATC-3′). One primer from each pair was used in two separate PCRs, each of which amplified about half of the plasmid template and incorporated the base change from the mutagenizing primers. The two sets of reaction products were pooled, cleaned, and transformed directly into library-competent Escherichia coli DH5α cells (Gibco-BRL). The mutagenized plasmid was then generated by in vivo recombination between the overlapping ends of the PCR products (44).
Two clones were sequenced; both carried the repaired nef gene. One of these was used to generate a recombinant proviral genome by subcloning the 3′ fragments back into the HXB/LW genome. A low-mutation-rate polymerase with proofreading capability (Boehringer Mannheim Biochemicals) was used in all PCRs. However, as additional confirmation that the DNA amplification did not interfere with either the replication or the pathogenicity of the mutant, a nonmutagenized control plasmid was generated in a parallel reaction. The nef+ mutant pBS-3′LWn+ and the nef− control pBS-3′LWn− plasmids were both digested with SalI and XbaI and gel purified. HXB/LW proviral plasmid DNA was digested with SalI and XhoI and gel purified. The digested proviral plasmid DNA was partially ligated at the SalI site with either the 3′-LWn+ or the 3′-LWn− insert. A293T cells were transfected with the partially ligated DNA using Effectine reagent (Qiagen, Inc.), and the viral supernatant was collected after 48 h and amplified in phytohemagglutinin (PHA)-activated PBMCs. The HXB/LWn+ proviral DNA was used directly as a template for sequencing with primers from either side of the insert based on sequences from HXB2. Sequences were confirmed from both strands.
HIV-1 replication in PBMCs and viral supernatant production.
Proviral DNA (0.8 μg) was transfected into A293T cells, and supernatants were used to infect PHA-activated PBMCs as described previously (23, 39). Supernatants were collected and analyzed by a multinuclear activation of a galactosidase indicator (MAGI) assay performed as previously described using U373-MAGI-CXCR4CEM glioblastoma cells (41). Supernatants with titers of greater than 5 × 104 infectious units (IU)/ml were stored as viral stocks for infection.
Western blot analysis of nef expression.
Western blot analysis was performed with total cell extracts from transfected A293T cells or infected H9 T cells as described previously (23). The anti-nef polyclonal antibody (kindly provided by R. Swanstrom, University of North Carolina, Chapel Hill) was used to detect the nef protein. NL4–3 virus-infected H9 cells were used as positive controls. An antitubulin monoclonal antibody (MAb) was used to monitor protein levels.
Infection of SCID-hu Thy/Liv mice or HF-TOC.
Animal transplantation procedures for SCID-hu Thy/Liv construction have been described elsewhere (32). Infection of SCID-Thy/Liv mice was performed as previously described (39). Briefly, SCID-hu Thy/Liv mice were infected with supernatants collected from PHA-activated PBMCs containing no HIV-1 (mock) or 4 × 104 IU of HIV-1/ml. Fifty microliters (∼2,000 IU) was injected into each thymus graft. Biopsy specimens were obtained from Thy/Liv organs at various times, and thymocytes were analyzed for p24 and proviral DNA. Thymocyte subsets were analyzed with a fluorescence-activated cell sorter (FACS) as described below.
The HF-TOC procedures were modified slightly from those described previously (4). Briefly, human fetal thymi (19 to 24 gestational weeks) were dissected into ∼2-mm3 fragments containing at least three to five intact thymic lobules under a dissecting microscope. These fragments were transferred onto sterile organ culture membranes (Millipore) floating on medium (RPMI medium, 10% fetal calf serum, 50 μg of streptomycin/ml, 50 U of penicillin G/ml, minimal essential medium vitamin solution [Gibco-BRL], insulin-transferrin-sodium selenite medium supplement [Sigma]) in six-well tissue culture plates. Equal amounts of virus (∼800 IU) in 20 μl of supernatant from infected PHA-activated PBMCs or 20 μl of control supernatant from mock-infected PHA-activated PBMCs was added to each fragment. The fragments were then cultured at 37°C in 5% CO2 for 7 to 12 days with daily changes of culture medium. Thymocytes were teased out of the fragments and analyzed as described above.
Viral replication assays.
p24 production (picograms/106 thymocytes) was measured using a Vironostica p24 enzyme-linked immunosorbent assay (ELISA) kit (Organon Teknika Corp., Durham, N.C.) and cell lysate in phosphate-buffered saline (PBS)–1% Triton X-100. Semiquantitative DNA PCR analysis was performed as described previously (23, 39). Briefly, human thymocytes from HIV-1- or mock-infected Thy/Liv grafts were assayed by 10-fold dilution of infected cells into uninfected human cells. Genomic DNA was prepared from the mixed cells. ACH2 cells (one HIV-1 genome per cell) were used as standard controls.
Immunohistochemistry.
At 6 weeks postinfection, SCID-hu Thy/Liv organs were fixed in PBS–4% paraformaldehyde, frozen, and sectioned. They were then stained with human anti-HIV-1 serum as described previously (3).
Flow cytometric analyses.
Thymocytes isolated from SCID-hu Thy/Liv organs or HF-TOC fragments were stained with phycoerythrin-CD4 and tricolor-CD8 (Caltag) in PBS–2% fetal bovine serum, washed, and resuspended in PBS–1% formaldehyde as previously described (23, 39).
RESULTS
HXB/LW and NL4–3 infect the human thymus with similar replication kinetics.
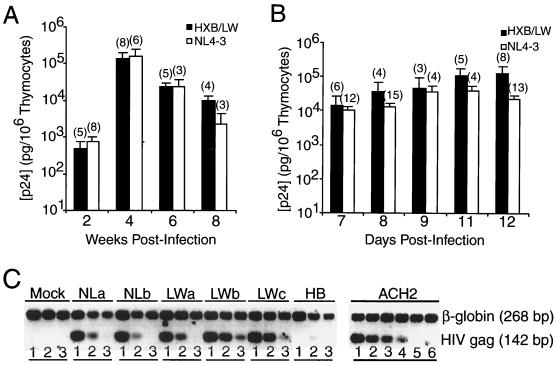
NL4–3 and HXB/LW, as well as HXB2, are derived from the Lai/IIIB isolate (Table 1). They all replicate efficiently in PHA-activated PBMCs (40). HIV-1 replication in the SCID-hu Thy/Liv model was assessed by a p24 antigen ELISA and by DNA PCR analysis. As previously reported (40), no significant HXB2 replication was detected up to 6 weeks postinoculation. In contrast, challenge with the recombinant HXB/LW virus was associated at 3 to 6 weeks postinfection with high levels of viral replication, comparable to that seen in NL4–3-infected Thy/Liv organs (Fig. 1A). Similar results were obtained after infection with the same HIV-1 clones in the HF-TOC model (4, 40). Analysis of data from 18 independent experiments demonstrated that NL4–3 and HXB/LW replicated to similar levels, with similar kinetics, in the HF-TOC model (Fig. 1B). Quantitation of proviral DNA confirmed that about 10% of human thymocytes were infected with HXB/LW, comparable to the infection levels seen with NL4–3 (Fig. 1C) or primary isolates (39, 40).
TABLE 1.
Summary of HIV-1 isolates and clones
| HIV-1 | PBMCa | Thy-Repb | Presence (+) or absence (−) of the following HIV-1 gene:
|
||
|---|---|---|---|---|---|
| nef | vpr | vpu | |||
| Lai/IIIB | + | − | − | − | − |
| HXB2 | + | − | − | − | − |
| NL4-3 | + | + | + | + | + |
| HXB/LW | + | + | − | − | + |
Ability to replicate in activated human PBMCs.
Ability to replicate in human thymus.
FIG. 1.
Similar replication kinetics of HXB/LW and NL4–3 in SCID-hu Thy/Liv mouse and HF-TOC model systems. (A) Replication of HXB/LW and NL4–3 in SCID-hu Thy/Liv mice. Levels of p24 capsid protein associated with 106 cells are shown on the y axis. Each column represents the average values for each virus (numbers in parentheses are numbers of animals), along with standard error bars. (B) Replication of HXB/LW and NL4–3 in HF-TOC. Thymus fragments were infected with equivalent infectious units, and thymocytes were harvested at various times postinfection. Levels of p24 capsid protein associated with 106 cells are shown on the y axis. Data represent the average values at the indicated times (numbers in parentheses are numbers of donors), along with standard error bars. (C) Quantitation of proviral DNA. Human thymocytes from SCID-hu Thy/Liv mice infected with NL4–3 (two SCID-hu Thy/Liv mice; panels NLa and NLb), HXB/LW (three SCID-hu Thy/Liv mice; panels LWa to LWc), or HXB2 (panel HB) were assayed by 10-fold dilution of infected cells into uninfected human cells. Lanes 1, 10,000 sample cells; lanes 2, 1,000 sample cells plus 9,000 normal human cells; lanes 3, 100 sample cells plus 9,900 normal human cells. ACH2 cells (one HIV-1 genome per cell) were used as standard controls (lane 4, 10 ACH2 cells plus 9,990 normal human cells; lane 5, 1 ACH2 cell plus 9,999 normal human cells; lane 6, 10,000 normal human cells). β-Globin primers were used as internal controls.
HXB/LW replication was uncoupled from pathogenicity in the human thymus.
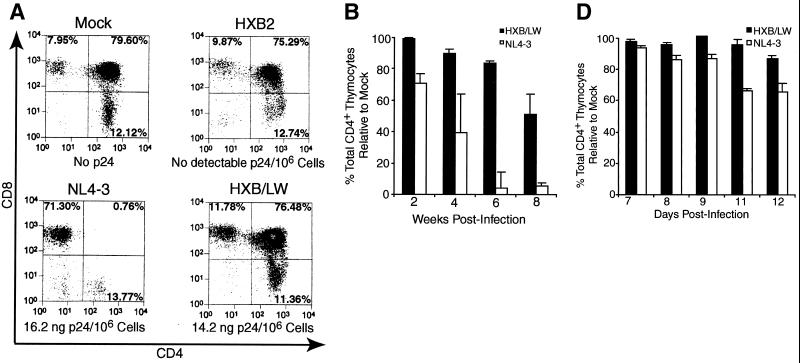
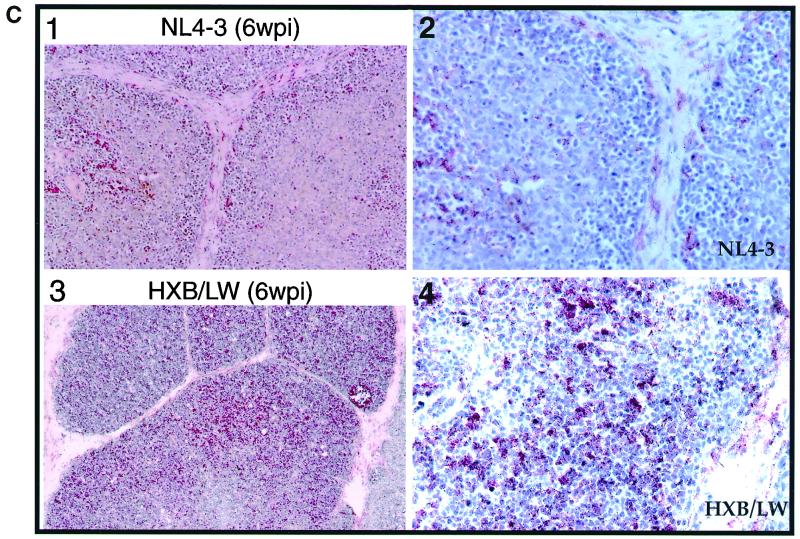
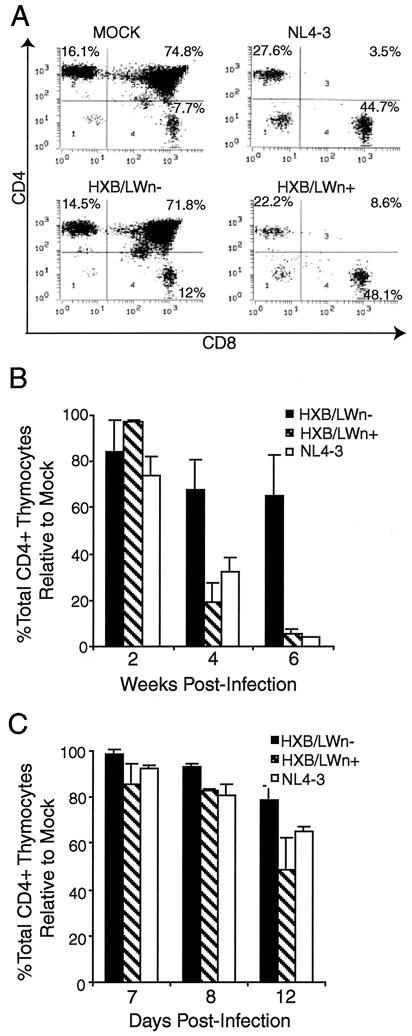
The HXB2 clone failed to replicate and thus showed no detectable p24 capsid protein or thymocyte depletion (Fig. 2A). NL4–3 infection of SCID-hu Thy/Liv organs led to significant depletion (up to 90% of total CD4+ thymocytes relative to the results obtained for mock-infected Thy/Liv organs) of CD4+ thymocytes at 3 to 8 weeks postinfection (Fig. 2A and B). In contrast, HXB/LW, with a level of replication similar to that of NL4–3, showed no significant depletion of thymocytes at up to 6 weeks postinfection (Fig. 2A and B). Immunohistochemical detection of infected cells in SCID-hu Thy/Liv organs infected with HXB/LW revealed large numbers of thymocytes expressing HIV antigens within well-defined cortex and medulla of intact thymic lobules (Fig. 2C, lower panels). As previously reported (3), NL4–3 infected Thy/Liv organs were dramatically disrupted, and thymocytes were depleted (Fig. 2C, upper panels).
FIG. 2.
Reduced pathogenicity of HXB/LW in human thymus. (A) Replication and pathogenic activities of NL4–3, HXB2, and HXB2/LW in SCID-hu Thy/Liv mice. SCID-hu Thy/Liv mice were injected with equivalent infectious units of each virus, and thymocytes were analyzed at 4 weeks postinfection. Levels of cell-associated p24 antigen in 106 thymocytes were measured. (B) Comparison of CD4+ thymocyte depletion in SCID-hu Thy/Liv mice infected with HXB/LW or NL4–3. Thymocytes from the infected SCID-hu Thy/Liv organs shown in Fig. 1A were analyzed by FACS for CD4 and CD8. The total percentages of CD4+ thymocytes (CD4+ CD8− and CD4+ CD8+) are shown on the y axis. The percentage of total CD4+ cells in mock-infected animals at each time point in each experiment was set to 100%. The results shown are the average values, along with standard error bars. (C) Immunohistochemical staining of NL4–3- and HXB/LW-infected SCID-hu Thy/Liv organs. Frozen sections of NL4–3-infected (upper panels) and HXB/LW-infected (lower panels) SCID-hu Thy/Liv organs (6 weeks postinfection [wpi]) were stained with human anti-HIV serum and hematoxylin-eosin to detect HIV antigens and thymocytes, respectively. Magnifications: ×10 for left panels and ×60 for right panels. (D) Comparison of CD4+ thymocyte depletion in HF-TOC infected with HXB/LW or NL4–3. Thymocytes isolated from the infected HF-TOC fragments shown in Fig. 1B were analyzed by FACS for CD4 and CD8. The total percentages of CD4+ thymocytes (CD4+ CD8− and CD4+ CD8+) are shown on the y axis. The percentage of total CD4+ cells in mock-infected tissues at each time point in each experiment was set to 100%. The results shown are the average values, along with standard error bars.
Similar pathogenesis was observed with the HF-TOC system (4). Only NL4–3-infected HF-TOC fragments showed significant thymocyte depletion relative to that in mock-infected fragments by 12 days postinfection (Fig. 2D).
The results of the replication and pathogenesis studies clearly established a difference in the pathogenic activities of HXB/LW and NL4–3. The replication activity of HXB/LW could be separated from its pathogenic activity in both models of thymic HIV-1 infection. This uncoupling of replication from pathogenicity enabled us to use the HXB/LW virus to map replication-independent genomic determinants of pathogenicity in vivo.
Restoration of the nef ORF in HXB/LW restored thymocyte depletion in the human thymus.
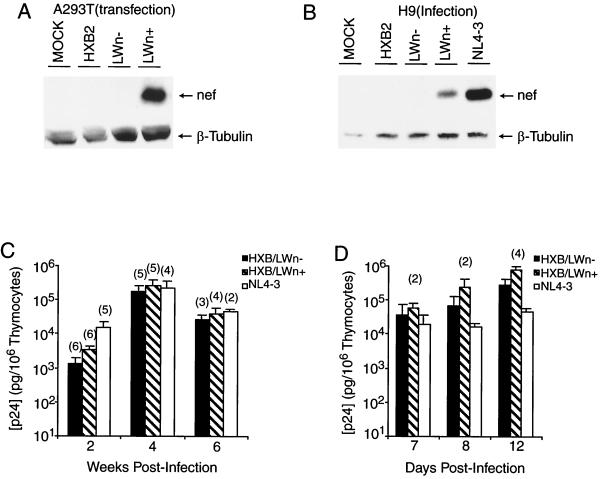
Like the LW12.3 clone isolated from the infected laboratory worker, HXB/LW encoded defective nef and vpr genes (Table 1) (40). Since it has been reported that vpr is not required for the replication and pathogenicity of HIV-1 in the SCID-hu Thy/Liv model (2, 18), we tested whether nef could enhance the pathogenicity of HXB/LW in the in vivo model. HXB/LW with a restored nef ORF (HXB/LWn+) was generated by repairing the premature stop codon in the HXB/LW nef gene (see Materials and Methods). An unmutagenized control virus (HXB/LWn−) was simultaneously generated with similar procedures. The HXB/LWn− control virus was phenotypically indistinguishable from HXB/LW in T-cell lines, in the HF-TOC system, and in SCID-hu Thy/Liv mice (data not shown). As expected, HXB/LWn+, but not HXB2 or HXB/LWn− (or HXB/LW), expressed a full-length nef protein in both transfected A293T cells and infected human T cells (Fig. 3A and B).
FIG. 3.
Repaired nef gene did not alter HXB/LW replication kinetics. (A) The expression of nef in A293T cells transfected with proviruses encoding HXB2, HXB/LWn−, and HXB/LWn+ was detected by Western blot analysis. The same blot was probed with an antitubulin MAb to monitor total protein levels. (B) The expression of nef in H9 T cells infected with HXB2, HXB/LWn−, and HXB/LWn+ was detected by Western blot analysis. NL4–3-infected H9 cells were included as positive controls. The same blot was probed with an antitubulin MAb to monitor total protein levels. (C) Similar levels of HXB/LWn+, HXB/LWn−, and NL4–3 replication in SCID-hu Thy/Liv mice. SCID-hu Thy/Liv organs were infected with equivalent infectious units, and thymocytes were harvested at various times. Levels of p24 capsid protein associated with 106 cells are shown on the y axis. Each column represents the average of three independent experiments (numbers in parentheses are numbers of animals), along with calculated standard error bars. (D) Similar levels of HXB/LWn+, HXB/LWn−, and NL4–3 replication in HF-TOC. Fetal thymus fragments were infected with equivalent infectious units, and thymocytes were harvested at various times. Levels of p24 capsid protein associated with 106 cells are shown on the y axis. Values at each time point represent averages from four independent experiments (numbers in parentheses are numbers of donors), along with calculated standard error bars.
HXB/LWn+ and HXB/LWn− showed similar replication activities in SCID-hu Thy/Liv mice (Fig. 3C) and HF-TOC (Fig. 3D). However, when pathogenic activity was analyzed in these experiments, infection by HXB/LWn+ led to thymocyte depletion at levels similar to those observed in NL4–3 infections (Fig. 4A), whereas HXB/LWn−-infected tissues showed no significant thymocyte depletion. Kinetic analysis of thymocyte depletion showed that HXB/LWn+ was similar to NL4–3 in pathogenicity, while HXB/LWn− showed no significant pathogenic activity until late times postinfection (Table 2 and Fig. 4B and C). These results clearly demonstrated that repairing the premature stop codon of the HXB/LWn− nef gene was sufficient to restore pathogenicity to this virus, without significantly altering its replication kinetics.
FIG. 4.
Repaired nef gene enhanced pathogenesis in HXB/LW-infected thymus. (A) Similar levels of thymocyte depletion in HXB/LWn+- and NL4–3-infected SCID-hu Thy/Liv mice. SCID-hu Thy/Liv mice were injected with equivalent infectious units of virus, and thymocytes were harvested at 4 weeks postinfection and analyzed by FACS. The thymocyte subpopulations were expressed as a percentage of the events in the live gate. The levels of p24 capsid protein associated with 106 thymocytes in each SCID-hu Thy/Liv mouse were as follows: LWn+, 490 ng; LWn− (HXB/LW), 670 ng; and NL4–3, 67 ng. (B) Pathogenesis kinetics in SCID-hu Thy/Liv mice infected with HXB/LWn+, HXB/LWn−, and NL4–3. Thymocytes from the infected SCID-hu Thy/Liv organs shown in Fig. 3C were analyzed by FACS at the times shown on the x axis. The total percentage of CD4+ thymocytes relative to that in mock infections is shown on the y axis. The percentage of total CD4+ cells in mock-infected animals at each time in each experiment was set to 100%. The results shown are the average values, along with standard error bars. (C) Pathogenesis kinetics in HF-TOC infected with HXB/LWn+, HXB/LWn−, and NL4–3. Thymocytes from the infected HF-TOC fragments shown in Fig. 3D were analyzed by FACS at the times shown on the x axis. The total percentage of CD4+ thymocytes relative to that in mock infections is shown on the y axis. The percentage of total CD4+ cells in mock-infected animals at each time in each experiment was set to 100%. The results shown are the average values, along with standard error bars.
TABLE 2.
Summary of SCID-hu Thy/Liv mouse model viral replication and pathogenesis
| Wk postinfection | Virus | No. of animals | Mean % of cells (SD) that were
|
Mean ng of p24 (SD)/106 cells | |||
|---|---|---|---|---|---|---|---|
| Livea | CD4+ | CD4+ CD8+ | CD8+ | ||||
| 2 | Mock | 5 | 72.9 (10.8) | 13.3 (3.7) | 77.1 (5.7) | 5.6 (1.0) | 0 |
| HXB/LWn− | 6 | 76.2 (10.4) | 12.2 (3.7) | 67.0 (29.9) | 6.6 (0.6) | 1.3 (1.4) | |
| HXB/LWn+ | 6 | 82.6 (4.6) | 10.9 (1.5) | 80.2 (2.3) | 7.6 (1.5) | 3.3 (2.7) | |
| NL4-3 | 5 | 61.8 (11.3) | 19.8 (11.0) | 49.5 (11.8) | 16.8 (5.7) | 15.6 (14.1) | |
| 4 | Mock | 4 | 50.9 (14.5) | 16.7 (7.5) | 72.4 (8.0) | 9.1 (1.2) | 0 |
| NXB/LWn− | 5 | 45.0 (29.7) | 15.6 (3.1) | 55 (24.0) | 22.1 (14.1) | 210.9 (191.2) | |
| HXB/LWn+ | 5 | 6.7 (5.0) | 12 (10.8) | 5.4 (5.7) | 49.9 (17.7) | 261.2 (313.2) | |
| NL4-3 | 4 | 12.1 (4.2) | 16.0 (8.6) | 3.4 (1.8) | 49 (15.1) | 229.7 (240.5) | |
| 6 | Mock | 2 | 75.1 (3.9) | 9.6 (1.8) | 86.7 (2.1) | 3.2 (0.4) | 0 |
| HXB/LWn− | 2 | 19.2 (15.4) | 22 (8.0) | 57.3 (18.5) | 14.3 (4.5) | 37.1 (4.6) | |
| HXB/LWn+ | 2 | 7.1 (0.4) | 3.9 (4.1) | 4.4 (1.2) | 78.4 (2.0) | 72.3 (23.2) | |
| NL4-3 | 2 | 10.1 (3.6) | 1.9 (1.0) | 1.8 (0.5) | 82.7 (6.9) | 45.5 (12.3) | |
Percentage of cells in live gate.
The replication of HXB/LW was genetically separable from thymocyte depletion in SCID-hu Thy/Liv organs.
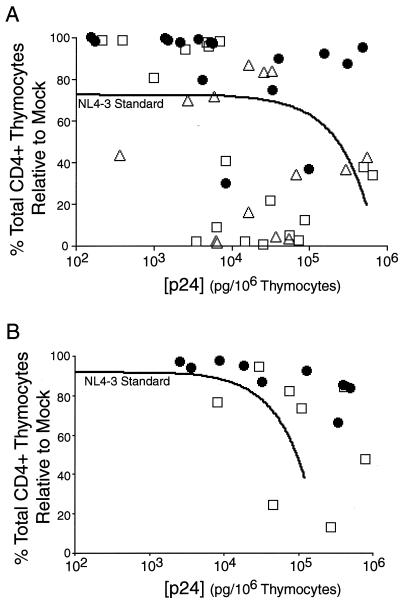
To analyze the relative replication and pathogenic activities of HXB/LW (with or without a functional nef gene) and NL4–3, we plotted the pathogenicity of each virus against its replication as described previously (5). In comparison to NL4–3 standard curves generated from 10 independent SCID-hu Thy/Liv experiments (32 SCID-hu Thy/Liv mice infected with NL4–3) (Fig. 5A) or 26 independent HF-TOC experiments (100 HF-TOC fragments infected with NL4–3) (Fig. 5B), HXB/LWn− showed significantly reduced pathogenic activity at similar viral loads. In both model systems, all but two of the data points for HXB/LWn− remained above the NL4–3 standard curves (Fig. 5), demonstrating the reduced pathogenicity of this virus. In contrast, HXB/LWn+ and NL4–3 from the same experiments showed similar pathogenic activities in both the SCID-hu Thy/Liv and the HF-TOC models (Fig. 5). The high R2 value (0.76) for the HF-TOC NL4–3 standard indicates that the data points fit with the nonlinear regression model (5). However, the low value (R2 = 0.17) for the SCID-hu Thy/Liv NL4–3 standard suggests that some data points did not fit the regression line as well. This lack of fit was probably due to greater thymocyte depletion at late times after infection with NL4–3 in Thy/Liv organs (Fig. 4B) that led to reduced cell-associated p24 levels (3, 39). The HF-TOC cultures did not reach the same level of depletion (Fig. 4C).
FIG. 5.
Relative pathogenicity of HXB/LWn+, HXB/LWn−, and NL4–3 correlated to the presence of an intact nef ORF. (A) Pathogenic plot for SCID-hu Thy/Liv mice. p24 capsid protein levels (x axis) were plotted against the total percentage of CD4+ thymocytes relative to that in mock infections (y axis) for the time points from 2 to 6 weeks postinfection from the experiments shown in Fig. 3C. The NL4–3 standard regression curve was generated from 32 SCID-hu Thy/Liv mice infected with the NL4–3 virus from 10 different experiments (R2 = 0.17). Numbers of mice were as follows: HXB/LWn−, n = 16 (●); HXB/LWn+, n = 18 (□); NL4–3, n = 14 (▵); and NL4–3 standard, n = 32. (B) Pathogenicity plot for HF-TOC. p24 capsid protein levels (x axis) were plotted against the total percentage of CD4+ thymocytes (y axis) for all time points from the HF-TOC experiments shown in Fig. 3D. The NL4–3 standard regression curve was generated from 100 HF-TOC assays of 26 different donor thymus tissues infected with the NL4–3 virus (R2 = 0.79). Numbers of tissues were as follows: HXB/LWn−, n = 9 (●); HXB/LWn+, n = 8 (□); and NL4–3 standard, n = 100.
Collectively, our results demonstrate that HXB/LWn− has the ability to replicate in vivo, but with significantly reduced pathogenic activity. Furthermore, an intact nef ORF can restore its pathogenic activity, with no significant effect on its replication in the thymus.
DISCUSSION
From a laboratory worker accidentally exposed to an attenuated HIV-1 isolate, viruses have been recovered that have reverted to replicate in vivo (40, 43). Using SCID-hu Thy/Liv mice as an in vivo model for HIV-1 replication and pathogenesis, we showed that the replication activity of a recombinant HIV-1 clone derived from the laboratory worker isolate was separable from its pathogenicity in vivo.
For other lentiviruses, single point mutations have been shown to convert an attenuated virus to a pathogenic one in vivo. However, pathogenic activity always has been correlated with enhanced replication activity. For example, lower viral loads were associated with reduced pathogenicity of SIV with a nef deletion in monkeys (20) or of HIV-1 in SCID-hu Thy/Liv mice (17). In a recent study of several CCR5-dependent HIV-1 isolates, Scoggins et al. also correlated replication efficiency in SCID-hu Thy/Liv mice with pathogenicity (37). The present study documented the first example of unique structural determinants in HIV-1 that appear to enhance infectivity but not pathogenicity in vivo.
Interestingly, the SalI-BamHI fragment of HXB2 that was replaced by that of LW12.3 in HXB/LW also showed in vivo defects in replication and pathogenicity in the SHIV genome (26, 35). In one report, SHIV-HXB2c demonstrated greatly reduced replication in infected monkeys compared with SHIV-89.6 (a primary HIV-1 isolate), although both chimeric viruses replicated efficiently in simian peripheral blood lymphocytes in vitro (35). Both reports indicated that laboratory-adapted HIV-1 isolates may have accumulated mutations in important genes for in vivo replication and transmission. Even SHIV-89.6 showed reduced replication and pathogenicity in monkeys. After several passages in vivo, SHIV variants with enhanced replication and pathogenicity accumulated (19). Thus, multiple determinants appear to be involved in both replication and pathogenesis or uniquely in pathogenicity independent of replication.
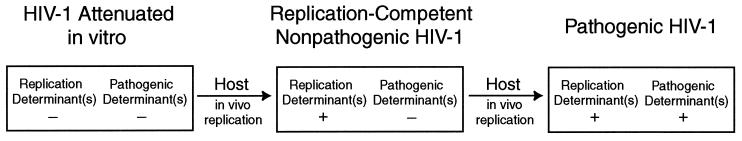
Our results suggest that while the laboratory-attenuated Lai/IIIB virus had recovered the ability to replicate in vivo through passage in the infected laboratory worker, it had not regained corresponding pathogenic activity. In the recombinant HXB/LW, the nef gene can function as a pathogenic factor independent of replication activity. We have shown that reversion of the replication determinants in LW12.3 env was not sufficient for pathogenicity in the human thymus. Restoration of the nef gene was required for HIV-1 pathogenic activity in vivo. As both env and nef are believed to be involved in replication as well as pathogenicity, we propose that Lai/IIIB has been attenuated by mutations in multiple genetic determinants required for replication and pathogenesis in vivo (Fig. 6). As the virus was passaged in vivo, we propose that replication determinants in env reverted to allow it to replicate more efficiently in the infected laboratory worker. However, the attenuated pathogenic determinants had not yet reverted in the LW12.3 isolate from which the recombinant HXB/LW was derived. Additional pathogenic determinants are also implicated in the HXB/LW virus because, at late times postinfection, HXB/LW-infected Thy/Liv organs also showed low, but significant, levels of thymocyte depletion (Fig. 2 and 4 and Table 2). The delayed or attenuated pathogenic activity may be due to the attenuated pathogenic LW12.3 env gene or to other HIV-1 pathogenic factors.
FIG. 6.
Model of in vivo reversion of HIV-1 attenuated in vitro. Tissue culture-attenuated viruses carry mutations in multiple genetic determinants of replication and pathogenicity, rendering them unable to replicate efficiently in vivo. After a short passage in vivo, reversions of a replication determinant(s) generate replication-competent isolates that remain attenuated for a pathogenic determinant(s). After further passage in vivo, reversion of a pathogenic determinant(s) results in a pathogenic virus.
Etemad-Moghadam et al. recently reported that env mutations in in vivo-passaged SHIV were associated with increased resistance to neutralizing antibody and that these mutations exerted a negative effect on the pathogenic potential of the virus in some cell lines (12). Consistent with this report, one of the sequence changes in the env V3 loop common to HXB/LW (40) and other isolates from the infected laboratory worker (25, 43) is associated with escape from antibody neutralization (10). Furthermore, this mutation interfered with the in vitro pathogenicity of the virus (10). The CD4+ T-cell count of the infected laboratory worker has decreased in recent years, suggesting the emergence of pathogenic revertants. Unfortunately, no recent HIV-1 isolates from this patient are available for analysis. However, based on our model (Fig. 6), we predict that additional changes in genes such as env or nef resulting in enhanced pathogenicity will be observed in later-stage laboratory worker isolates.
Many studies have demonstrated the importance of the nef gene in the replication and pathogenicity of HIV-1. Alterations and deletions in the nef gene have been associated with long-term survivors (9, 21, 28, 36). nef has multiple effects on HIV-1-infected cells: it down-regulates the CD4 receptor and major histocompatibility complex class I molecules from the infected cell surface and alters multiple T-cell signaling pathways (reviewed in references 8 and 33). These activities of nef have been mapped to distinct functional domains of the protein (7, 16, 27). The HXB/LW virus provides a valuable system for determining which functional domain of the nef protein is required specifically for in vivo HIV-1 pathogenicity in the human thymus. In addition, since the mechanism by which nef exerts its pathogenic activity in vivo is largely unknown, the HXB/LW virus may also be used to further examine mechanisms of nef-mediated pathogenesis in vivo.
ACKNOWLEDGMENTS
We are grateful to J. Harton for critical reading of the manuscript and to R. Swanstrom, M. Bonyhadi, J. M. McCune, and H. Kaneshima for helpful discussions. We thank the members of the S. Fiscus laboratory for providing PHA-activated PBMCs and for assistance with the p24 ELISA. The following reagent was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, National Institutes of Health (NIH): U373-MAGI-CXCR4CEM cells (Michael Emerman).
This work was supported by NIH grant AI41356 (to L.S.). K.M.D. is supported by a fellowship from the Irvington Institute of Immunological Research and Toys-R-Us, Inc. E.D.M. was funded in part by a Lineberger Comprehensive Cancer Center postdoctoral training grant (CA09156) and by the American Foundation for AIDS Research (amfAR 70520–28-RFI).
Footnotes
This paper is dedicated to the memory of Eric D. Miller.
REFERENCES
- 1.Aldrovandi G M, Gao L, Bristol G, Zack J A. Regions of human immunodeficiency virus type 1 nef required for function in vivo. J Virol. 1998;72:7032–7039. doi: 10.1128/jvi.72.9.7032-7039.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aldrovandi G M, Zack J A. Replication and pathogenicity of human immunodeficiency virus type 1 accessory gene mutants in SCID-hu mice. J Virol. 1996;70:1505–1511. doi: 10.1128/jvi.70.3.1505-1511.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonyhadi L M, Rabin L, Salimi S, Brown D A, Kosek J, McCune J M, Kaneshima H. HIV induces thymus depletion in vivo. Nature. 1993;363:728–732. doi: 10.1038/363728a0. [DOI] [PubMed] [Google Scholar]
- 4.Bonyhadi L M, Su L, Auten J, McCune J M, Kaneshima H. Development of a human thymic organ culture model for the study of HIV pathogenesis. AIDS Res Hum Retrovir. 1995;11:1073–1080. doi: 10.1089/aid.1995.11.1073. [DOI] [PubMed] [Google Scholar]
- 5.Camerini D, Su H P, Gamez-Torre G, Johnson M L, Zack J A, Chen I S. Human immunodeficiency virus type 1 pathogenesis in SCID-hu mice correlates with syncytium-inducing phenotype and viral replication. J Virol. 2000;74:3196–3204. doi: 10.1128/jvi.74.7.3196-3204.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang S P, Bowman B H, Weiss J B, Garcia R E, White T J. The origin of HIV-1 isolate HTLV-IIIB. Nature. 1993;363:466–469. doi: 10.1038/363466a0. [DOI] [PubMed] [Google Scholar]
- 7.Cohen G B, Rangan V S, Chen B K, Smith S, Baltimore D. The human thioesterase II protein binds to a site on HIV-1 nef critical for CD4 down-regulation. J Biol Chem. 2000;275:23097–23105. doi: 10.1074/jbc.M000536200. [DOI] [PubMed] [Google Scholar]
- 8.Cullen B R. HIV-1 auxiliary proteins: making connections in a dying cell. Cell. 1998;93:685–692. doi: 10.1016/s0092-8674(00)81431-2. [DOI] [PubMed] [Google Scholar]
- 9.Deacon N J, Tsykin A, Solomon A, Smith K, Ludford-Menting M, Hooker D J, McPhee D A, Greenway A L, Ellett A, Chatfield C, et al. Genomic structure of an attenuated quasi species of HIV-1 from a blood transfusion donor and recipients. Science. 1995;270:988–991. doi: 10.1126/science.270.5238.988. [DOI] [PubMed] [Google Scholar]
- 10.Di Marzo Veronese F, Reitz M S, Gupta G, Robert-Guroff M, Boyer-Thompson C, Louie A, Gallo R C, Lusso P. Loss of a neutralizing epitope by a spontaneous point mutation in the V3 loop of HIV-1 isolated from an infected laboratory worker. J Biol Chem. 1993;268:25894–25901. [PubMed] [Google Scholar]
- 11.Duus K M, Hatfield C, Grose C. Cell surface expression and fusion by the varicella-zoster virus gH:gL glycoprotein complex: analysis by laser scanning confocal microscopy. Virology. 1995;210:429–440. doi: 10.1006/viro.1995.1359. [DOI] [PubMed] [Google Scholar]
- 12.Etemad-Moghadam B, Sun Y, Nicholson E K, Fernandes M, Liou K, Gomila R, Lee J, Sodroski J. Envelope glycoprotein determinants of increased fusogenicity in a pathogenic simian-human immunodeficiency virus (SHIV-KB9) passaged in vivo. J Virol. 2000;74:4433–4440. doi: 10.1128/jvi.74.9.4433-4440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fauci A S. Multifactorial nature of human immunodeficiency virus disease: implications for therapy. Science. 1993;262:1011–1018. doi: 10.1126/science.8235617. [DOI] [PubMed] [Google Scholar]
- 14.Hahn B H, Shaw G M, Arya S K, Popovic M, Gallo R C, Wong S F. Molecular cloning and characterization of the HTLV-III virus associated with AIDS. Nature. 1984;312:166–169. doi: 10.1038/312166a0. [DOI] [PubMed] [Google Scholar]
- 15.Hanna Z, Kay D G, Rebai N, Guimond A, Jothy S, Jolicoeur P. Nef harbors a major determinant of pathogenicity for an AIDS-like disease induced by HIV-1 in transgenic mice. Cell. 1998;95:163–175. doi: 10.1016/s0092-8674(00)81748-1. [DOI] [PubMed] [Google Scholar]
- 16.Iafrate A J, Bronson S, Skowronski J. Separable functions of Nef disrupt two aspects of T cell receptor machinery: CD4 expression and CD3 signaling. EMBO J. 1997;16:673–684. doi: 10.1093/emboj/16.4.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jamieson B D, Aldrovandi G M, Planelles V, Jowett J B M, Gao L, Bloch L M, Chen I S Y, Zack J A. Requirement of human immunodeficiency virus type 1 nef for in vivo replication and pathogenicity. J Virol. 1994;68:3478–3485. doi: 10.1128/jvi.68.6.3478-3485.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jamieson B D, Zack J A. In vivo pathogenesis of a human immunodeficiency virus type 1 reporter virus. J Virol. 1998;72:6520–6526. doi: 10.1128/jvi.72.8.6520-6526.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karlsson G B, Halloran M, Li J, Park I W, Gomila R, Reimann K A, Axthelm M K, Iliff S A, Letvin L, Sodroski J. Characterization of molecularly cloned simian-human immunodeficiency viruses causing rapid CD4+ lymphocyte depletion in rhesus monkeys. J Virol. 1997;71:4218–4225. doi: 10.1128/jvi.71.6.4218-4225.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kestler H W D, Ringler D J, Mori K, Panicali D L, Sehgal P K, Daniel M D, Desrosiers R C. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell. 1991;65:651–662. doi: 10.1016/0092-8674(91)90097-i. [DOI] [PubMed] [Google Scholar]
- 21.Kirchhoff F, Easterbrook P J, Douglas N, Troop M, Greenough T C, Weber J, Carl S, Sullivan J L, Daniels R S. Sequence variations in human immunodeficiency virus type 1 Nef are associated with different stages of disease. J Virol. 1999;73:5497–5508. doi: 10.1128/jvi.73.7.5497-5508.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kong L I, Taylor M E, Waters D, Blattner W A, Hahn B H, Shaw G M. Genetic analysis of sequential HIV-1 isolates from an infected lab worker. Int Conf AIDS. 1989;5:518. [Google Scholar]
- 23.Kovalev G, Duus K, Wang L, Lee R, Bonyhadi M, Ho D, McCune J M, Kaneshima H, Su L. Induction of MHC class I expression on immature thymocytes in HIV-1-infected SCID-hu Thy/Liv mice: evidence of indirect mechanisms. J Immunol. 1999;162:7555–7562. [PMC free article] [PubMed] [Google Scholar]
- 24.Levy J A. Pathogenesis of human immunodeficiency virus infection. Microbiol Rev. 1993;57:183–289. doi: 10.1128/mr.57.1.183-289.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lori F, Hall L, Lusso P, Popovic M, Markham P, Franchini G, Reitz M J. Effect of reciprocal complementation of two defective human immunodeficiency virus type 1 (HIV-1) molecular clones on HIV-1 cell tropism and virulence. J Virol. 1992;66:5553–5560. doi: 10.1128/jvi.66.9.5553-5560.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu Y, Brosio P, Lafaile M, Li J, Collman R G, Sodroski J, Miller C J. Vaginal transmission of chimeric simian-human immunodeficiency viruses in rhesus macaques. J Virol. 1996;70:3045–3050. doi: 10.1128/jvi.70.5.3045-3050.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mangasarian A, Piguet V, Wang J K, Chen Y L, Trono D. Nef-induced CD4 and major histocompatibility complex class I (MHC-I) down-regulation are governed by distinct determinants: N-terminal alpha helix and proline repeat of Nef selectively regulate MHC-I trafficking. J Virol. 1999;73:1964–1973. doi: 10.1128/jvi.73.3.1964-1973.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mariani R, Kirchhoff F, Greenough T C, Sullivan J L, Desrosiers R C, Skowronski J. High frequency of defective nef alleles in a long-term survivor with nonprogressive human immunodeficiency virus type 1 infection. J Virol. 1996;70:7752–7764. doi: 10.1128/jvi.70.11.7752-7764.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCune J, Kaneshima H, Krowka J, Namikawa R, Outzen H, Peault B, Rabin L, Shih C C, Yee E, Lieberman M, et al. The SCID-hu mouse: a small animal model for HIV infection and pathogenesis. Annu Rev Immunol. 1991;9:399–429. doi: 10.1146/annurev.iy.09.040191.002151. [DOI] [PubMed] [Google Scholar]
- 30.McCune J M. HIV-1: the infective process in vivo. Cell. 1991;64:351–363. doi: 10.1016/0092-8674(91)90644-e. [DOI] [PubMed] [Google Scholar]
- 31.McCune J M, Namikawa R, Kaneshima H, Shultz L D, Lieberman M, Weissman I L. The SCID-hu mouse: a model for the analysis of human hematolymphoid differentiation and function. Science. 1988;241:1632–1639. doi: 10.1126/science.241.4873.1632. [DOI] [PubMed] [Google Scholar]
- 32.Namikawa R, Weilbaecher K N, Kaneshima H, Yee E J, McCune J M. Long-term human hematopoiesis in the SCID-hu mouse. J Exp Med. 1990;172:1055–1063. doi: 10.1084/jem.172.4.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peter F. HIV Nef: the mother of all evil? Immunity. 1998;9:433–437. doi: 10.1016/s1074-7613(00)80626-3. [DOI] [PubMed] [Google Scholar]
- 34.Popovic M, Sarngadharan M G, Read E, Gallo R C. Detection, isolation, and continuous production of cytopathic retroviruses (HTLV-III) from patients with AIDS and pre-AIDS. Science. 1984;224:497–500. doi: 10.1126/science.6200935. [DOI] [PubMed] [Google Scholar]
- 35.Reimann K, Li J, Voss G, Lekutis C, Tenner-Racz K, Racz P, Lin W, Montefiori D, Lee-Parritz D, Lu Y, Collman R, Sodroski J, Letvin N L. An env gene derived from a primary human immunodeficiency virus type 1 isolate confers high in vivo replicative capacity to a chimeric simian-human immunodeficiency virus in rhesus monkeys. J Virol. 1996;70:3198–3206. doi: 10.1128/jvi.70.5.3198-3206.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salvi R, Garbuglia A R, Di Caro A, Pulciani S, Montella F, Benedetto A. Grossly defective nef gene sequences in a human immunodeficiency virus type 1-seropositive long-term nonprogressor. J Virol. 1998;72:3646–3657. doi: 10.1128/jvi.72.5.3646-3657.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scoggins R M, Taylor J R, Jr, Patrie J, van't Wout A B, Schuitemaker H, Camerini D. Pathogenesis of primary R5 human immunodeficiency virus type 1 clones in SCID-hu mice. J Virol. 2000;74:3205–3216. doi: 10.1128/jvi.74.7.3205-3216.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shaw G M, Hahn B H, Arya S K, Groopman J E, Gallo R C, Wong-Staal F. Molecular characterization of human T-cell leukemia (lymphotropic) virus type III in the acquired immune deficiency syndrome. Science. 1984;226:1165–1171. doi: 10.1126/science.6095449. [DOI] [PubMed] [Google Scholar]
- 39.Su L, Kaneshima H, Bonyhadi M, Salimi S, Kraft D, Rabin L, McCune J M. HIV-1 induced thymocyte depletion is associated with indirect cytopathicity and infection of progenitor cells in vivo. Immunity. 1995;2:25–36. doi: 10.1016/1074-7613(95)90076-4. [DOI] [PubMed] [Google Scholar]
- 40.Su L, Kaneshima H, Bonyhadi M L, Lee R, Auten J, Wolf A, Du B, Rabin L, Hahn B H, Terwilliger E, McCune J M. Identification of HIV-1 determinants for replication in vivo. Virology. 1997;227:46–52. doi: 10.1006/viro.1996.8338. [DOI] [PubMed] [Google Scholar]
- 41.Vodicka A M, Goh W C, Wu L I, Rogel M E, Bartz S R, Schweickart V L, Raport C J, Emerman M. Indicator cell lines for detection of primary strains of human and simian immunodeficiency viruses. Virology. 1997;233:193–198. doi: 10.1006/viro.1997.8606. [DOI] [PubMed] [Google Scholar]
- 42.Weiss R A. How does HIV cause AIDS? Science. 1993;260:1273–1279. doi: 10.1126/science.8493571. [DOI] [PubMed] [Google Scholar]
- 43.Weiss S H, Goedert J J, Gartner S, Popovic M, Waters D, Markham P, di Marzo Veronese F, Gail M H, Barkley W E, Gibbons J, Gill F, Leuther M, Shaw G M, Gallo R C, Blattner W A. Risk of human immunodeficiency virus (HIV-1) infection among laboratory workers. Science. 1988;239:68–71. doi: 10.1126/science.3336776. [DOI] [PubMed] [Google Scholar]
- 44.Yao Z, Jones D H, Grose C. Site-directed mutagenesis of herpesvirus glycoprotein phosphorylation sites by recombination polymerase chain reaction. PCR Methods Appl. 1992;1:205–207. doi: 10.1101/gr.1.3.205. [DOI] [PubMed] [Google Scholar]