Abstract
Background
Tuberculous meningitis (TBM) is a severe central nervous system (CNS) infection with a challenging diagnosis due to inadequate detection methods. This study evaluated current clinical detection methods and their applicability.
Methods
A cohort of 514 CNS infection patients from 2018 to 2020 was studied. Data on general demographics, Cerebrospinal Fluid (CSF) analysis, epidemiology, and clinical outcomes were collected. TBM patients were identified, and the sensitivities of mmetagenomic next-generation sequencing (NGS), GeneXpert, and microbial culture were compared. Kappa statistic assessed the consistency between methods.
Results
Among the patients involved, TBM (29%) and neurosyphilis (25%) were the two most prevalent CNS infections. CSF analysis indicated that 76% of patients had leukocytosis, suggesting a potential CNS inflammation. In TBM cases, 92.5% had elevated CSF protein and leukocyte counts. Moreover, the percentage of positive mNGS results was 55.6%. GeneXpert and MTB cultures alone had lower sensitivity, but combined use resulted in a 53.4% positive rate.
Conclusions
This study highlights the high sensitivity of mNGS, comparable to GeneXpert and MTB culture. The combined methods are cost-effective and straightforward, and can partially substitute for mNGS, offering valuable alternatives for TBM diagnosis and providing insights into multiple diagnostic strategies in clinical practice.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12879-024-10037-4.
Keywords: TBM, CNS infection, mNGS, GeneXpert, MTB culture
Introduction
Tuberculous meningitis (TBM) is an important CNS infectious disease caused by Mycobacterium tuberculosis (MTB). It occurs when MTB spreads through the bloodstream, crosses the blood-brain barrier, and invades the meninges, submembrane, or ependyma surface, leading to meningitis [1]. The diagnosis of TBM relies on the medical history of the patients, clinical manifestations, imaging findings, and subsequent laboratory examinations. However, the conventional diagnostic methods for TBM are complicated and lack specificity. Accordingly, rapid and accurate diagnostic tools for MTB are insufficient. In addition, approximately 50% of patients with CNS infections cannot be identified for the cause of meningitis due to the limited availability of CNS samples [2–4]. Delayed diagnosis of TBM might cause poor clinical outcomes, increase the stress of patients and their families, and markedly increase healthcare costs [5]. Therefore, more precise diagnostic tools are urgently needed.
Conventional approaches for detecting CNS infections in clinical practice include polymerase chain reaction (PCR)-based examinations of cerebrospinal fluid (CSF) specimens [6–9]. The GeneXpert system is a tool for the rapid detection of tuberculosis infections recommended by the World Health Organization (WHO) [10]. GeneXpert can be used to explore the expression of genes of known pathogens and may serve as the gold standard for clinical diagnosis. The mainstream approaches for the clinical detection of TBM include GeneXpert and MTB culture [11]. However, these methods can identify only known pathogens; thus, rapidly identifying unknown or rare pathogenic microorganisms is challenging. Thus, there is an urgent need for rapid and high-throughput detection tools. mNGS, also known as high-throughput sequencing, allows for prompt sequencing of large numbers of DNA molecules [12–14]. mNGS has gradually been used in the clinical setting, particularly in the diagnosis of knotty infectious diseases [12, 15–17]. The advantages of NGS include the ability to analyze a range of potential microorganisms in a single test [18, 19]. However, studies comparing the efficiency of mNGS with that of other diagnostic tools are insufficient.
This study aimed to compare the clinical performance and effectiveness of mNGS with those of other conventional microbiological tests. We attempted to identify an optimal diagnostic strategy for TBM in the clinical setting.
Methods
Patients
This study included 933 case reports from January 1, 2018, to December 31, 2020, in our hospital. After excluding duplicate and incomplete records, 827 cases remained. Following imaging, clinical signs, and laboratory tests, 514 patients were diagnosed by clinicians as having characteristics of CNS infection and were included in the subsequent analysis of this study. This study strictly adhered to the Helsinki Declaration (Brazil, October 2013) [20]. The study’s protocols and ethics were approved by the Scientific Research Ethics Committee of Shenzhen Third People’s Hospital (approval number: 2020002). All patients enrolled in this study provided signed informed consent.
TBM diagnostic criteria
Diagnostic guidelines specified at an international seminar on sharing TBM diagnosis and treatment practices held in Cape Town, South Africa, in 2009 were used as a reference in our definition of TBM diagnostic criteria [18, 19, 21]. Patients were classified into three categories based on the scoring system (Appendix Table 1): those with a score ≥ 12 or ≥ 10 (when imaging examination was not possible) were considered highly indicative of TBM, those with a score of 6–11 or 6–9 (when imaging examination was not possible) were considered possibly diagnostic for TBM, and those with a score of less than 6 were considered indicative of a non-TBM-infected patient [18, 19, 21]. In the final diagnosis process, patients with a high frequency of TBM infection and those who tested positive for MTB were defined as confirmed TBM patients.
Etiologic tests
In this study, all tests were conducted on established detection platforms. Appendix Table 2 provides an overview of the conventional methods used for the clinical detection of pathogens in the brain parenchyma or CSF. These methods include microbial culture, antigen tests, and serological tests.
MTB culture and GeneXpert
Colombian blood agar and Haemophilus chocolate agar plates, as well as Sabouraud agar plates, were inoculated with CSF. Bacterial growth in blood cultures and CSF was identified using standard microbiological methods, including colony morphology, Gram staining, biochemical analysis, and the IVD MALDI Biotyper System, an automated rapid mass spectrometry detection system (Bruker Daltonik GmbH, Germany) for colony identification. Gram staining and India ink staining microscopy were used to examine cerebrospinal fluid smears, and acid-fast bacilli staining was used to evaluate cerebrospinal fluid smears.
CSF specimens were inoculated into BBL MGIT tubes (7 ml, BD, America), and a fully automated BACTEC MGIT 960 System mycobacterial detection system (BD, America) was used to culture MTB for six weeks. The GeneXpert Dx System (Cepheid, America) was used to identify MTB and detect rifampin and isoniazid resistance.
mNGS
Sample processing and RNA extraction
One.5 − 3 mL of fresh CSF was collected from patients according to the standard sample collection procedure. A total of 450 µL of CSF was taken to extract RNA following the manufacturer’s operational manual using a TIANMicrobe magnetic bead method and a pathogenic microorganism DNA/RNA extraction kit (NG550-01).
RNA enrichment
After mixing 33 µl of the extracted nucleic acid sample with 7 µl of the enrichment reaction mixture, the mixture was incubated on a PCR machine at 37 °C for 10 min, after which magnetic bead purification was performed to remove DNA from the nucleic acids, thereby increasing the concentration of RNA.
Reverse transcription and two-strand synthesis
The unenriched nucleic acid or the enriched nucleic acid is subjected to fragmentation, one-strand synthesis and two-strand synthesis to form double-stranded DNA nucleic acid; then, the DNA is purified by magnetic beads, and the purified DNA is used for DNA library construction.
Construction of DNA libraries and sequencing
Then, DNA libraries were constructed through DNA fragmentation, end repair, adapter ligation and PCR amplification. An Agilent 2100 was used for quality control of the DNA libraries. Quality-qualified libraries were pooled, and DNA Nanoball (DNB) was generated and sequenced on the BGISEQ-50/MGISEQ-2000 platform.
Bioinformatic analysis
High-quality sequencing data were generated by removing low-quality reads, followed by computational subtraction of human host sequences mapped to the human reference genome (hg19) using Burrows–Wheeler Alignment [22]. The remaining data obtained by removing low-complexity reads were classified by simultaneously aligning them to the Pathogens Metagenomic Database (PMDB), which consists of bacteria, fungi, viruses and parasites. The classification reference databases were downloaded from NCBI (ftp://ftp.ncbi.nlm.nih.gov/genomes/).
Statistical analysis
R software V.3.5.1 was used for statistical analysis, and a two-tailed p value < 0.05 was used to indicate statistical significance. Categorical variables are expressed as frequencies (N) and percentages (%), while continuous variables are expressed as medians and interquartile ranges (IQRs). The Kruskal–Wallis sum test, Pearson chi-square test, and Fisher’s exact test were used to compare categorical variables. Additionally, SPSS v.19.0 was used to calculate the kappa coefficient to evaluate the consistency of the different detection methods, with the interference of chance factors eliminated. A kappa ≥ 0.75 suggested substantial agreement; 0.4 < kappa < 0.75 indicated moderate agreement; and a kappa ≤ 0.4 indicated poor agreement. Moreover, statistical significance was indicated by a p value < 0.05. All the statistical data were processed using GraphPad Prism v.8.0.1, and some tables were generated using Microsoft Office Excel 2020.
Results
Patient characteristics
Among all patients included in the study, 514 patients were diagnosed with CNS infection. Analysis of the final discharge diagnoses revealed that out of these 514 patients, 146 (29%) were diagnosed with TBM, 128 (25%) with neurosyphilis, 65 (13%) with viral meningitis, 27 (5%) with bacterial meningitis, 38 (7%) with fungal meningitis, 22 (4%) with HIV-related encephalopathy, 16 (3%) with parasitic meningitis, and 72 (14%) with unconfirmed CNS infections.
The median age of the patients was 37 years, and there was a significantly greater incidence of CNS infections in males than in females. Patients diagnosed with neurosyphilis were predominantly in the age range of 51 years or older, while other types of CNS infections predominantly occurred among individuals aged between 21 and 35 years.
Headache (52.3%, 269/514) and fever (51.4%, 264/514) were identified as the most prevalent signs and symptoms. More than half of the enrolled patients (57.4%, 295/514) had received prior antibiotic treatment before undergoing lumbar puncture, and 264 individuals (51.4%, 264/514) presented with either a fever or documented history of fever upon admission.
The demographic and clinical characteristics of the 514 CNS infection patients are listed in Table 1. Routine detection of cerebrospinal fluid (CSF) biochemical indices was performed for 499 patients (499/514), revealing a significant increase in the CSF leukocyte count in 76% of the patients (379/499). CSF protein concentrations increased substantially (> 0.45 g/L) in 481 patients (73.2% of individuals; 352/481). These two phenomena were most prevalent in patients with TBM (92.5% and 92.5%, respectively). In addition, there was a significant decrease in CSF glucose levels in 51.4% (75/146) of patients with TBM and an increase in peripheral blood C-reactive protein levels in 62% of patients (91/146).
Table 1.
Demographic and clinical characteristics of 514 CNS infection patients at Shenzhen Third People’s Hospital, China
| Total | Bacterial | Viral | MTB | Fungal | Parasite | Neurosyphilis | Others | P Value | |
|---|---|---|---|---|---|---|---|---|---|
| Characteristics | 514 | 27 | 65 | 146 | 38 | 16 | 128 | 94 | |
| Age, n (%) | |||||||||
| Mean ± SD | 39.76 ± 15.97 | 36.00 ± 17.58 | 33.95 ± 14.50 | 36.71 ± 17.22 | 39.92 ± 11.61 | 33.56 ± 9.08 | 47.78 ± 13.71 | 39.53 ± 16.09 | < 0.001 |
| 1–20 | 40(7.8) | 4(14.8) | 12(18.5) | 14(9.6) | 1(2.6) | 0(0.0) | 0(0.0) | 8(8.5) | |
| 21–35 | 199(38.7) | 11(40.7) | 26(40.0) | 70(47.9) | 17(44.7) | 12(75.0) | 30(23.4) | 33(35.1) | |
| 36–50 | 140(27.2) | 8(29.6) | 18(27.7) | 25(17.1) | 12(31.6) | 3(18.8) | 40(31.3) | 35(37.2) | |
| ≥51 | 135(26.3) | 4(14.8) | 9(13.8) | 37(25.3) | 8(21.1) | 1(6.3) | 58(45.3) | 18(19.1) | |
| Gender, n (%) | 0.249 | ||||||||
| Male | 369(71.8) | 17(63.0) | 45(69.2) | 97(66.4) | 33(86.8) | 12(75.0) | 96(75.0) | 69(73.4) | |
| Female | 145(28.2) | 10(37.0) | 20(30.8) | 49(33.6) | 5(13.2) | 4(25.0) | 32(25.0) | 25(26.6) | |
| History, n (%) | |||||||||
| HIV positive | 129(25.1) | 7(25.9) | 15(23.1) | 13(8.9) | 25(65.8) | 9(56.3) | 23(18.0) | 37(39.4) | < 0.001 |
| Antibiotics prior to admission | 295(57.4) | 16(59.3) | 26(40.0) | 95(65.1) | 22(57.9) | 6(37.5) | 77(60.2) | 53(56.4) | 0.030 |
| Clinical signs and symptoms | |||||||||
| Fever(T > 38℃) | 264(51.4) | 22(81.5) | 39(60.0) | 117(80.1) | 23(60.5) | 7(43.8) | 10(7.8) | 46(48.9) | < 0.001 |
| Headache | 269(52.3) | 21(77.8) | 47(72.3) | 104(71.2) | 25(65.8) | 8(50.0) | 15(11.7) | 49(52.1) | < 0.001 |
| Altered mental status | 248(48.2) | 13(48.1) | 29(44.6) | 75(51.4) | 13(34.2) | 7(43.8) | 71(55.5) | 40(42.6) | 0.2 |
| Focal neurologic signs | 150(29.2) | 10(37.0) | 17(26.2) | 44(30.1) | 8(21.1) | 6(37.5) | 42(32.8) | 23(24.5) | NA |
| CSF examination | |||||||||
| Elevated white cell count, n = 499 | 379(76.0) | 22(81.5) | 51(78.5) | 135(92.5) | 35(92.1) | 10(62.5) | 76(59.4) | 50(53.2) | < 0.001 |
| Elevated neutrophil count, n = 499 | 209(41.9) | 19(70.4) | 25(38.5) | 97(66.4) | 23(60.5) | 3(18.8) | 18(14.1) | 24(25.5) | < 0.001 |
| Elevated protein, n = 481 | 352(73.2) | 20(74.1) | 38(58.5) | 135(92.5) | 26(68.4) | 11(68.8) | 73(57.0) | 49(52.1) | < 0.001 |
| Decreased glucose, n = 491 | 115(23.4) | 7(25.9) | 4(6.2) | 75(51.4) | 14(36.8) | 3(18.8) | 2(1.6) | 10(10.6) | < 0.001 |
| Peripheral blood examination | |||||||||
| Elevated white cell count, n = 511 | 127(24.9) | 15(55.6) | 13(20.0) | 49(33.6) | 6(16.2) | 2(12.5) | 18(14.1) | 24(25.5) | < 0.001 |
| Low white cell count, n = 511 | 34(6.7) | 1(3.7) | 4(6.2) | 5(3.4) | 7(18.9) | 1(6.3) | 3(2.3) | 13(13.8) | < 0.001 |
| Elevated lymphocyte count, n = 511 | 28(5.5) | 2(7.4) | 5(7.7) | 7(4.8) | 0(0.0) | 0(0.0) | 5(3.9) | 9(9.6) | 0.3 |
| Thrombocytopenia, n = 511 | 19(3.7) | 2(7.4) | 2(3.1) | 4(2.7) | 3(7.9) | 1(6.3) | 0(0.0) | 7(7.4) | 0.007 |
| Elevated C-reactive protein, n = 486 | 245(50.4) | 19(70.4) | 26(40.0) | 91(62.3) | 27(71.1) | 12(75.0) | 25(19.5) | 45(47.9) | < 0.001 |
Note Categorical variables are represented as the frequency (N) and percentage (%); chi-square tests or Fisher’s exact tests were used to compare categorical variables; IQR: interquartile range; NA: not available. Elevated or decreased in white blood cell count refers to white blood cell count > 9.5 × 109/L or < 3.5 × 109/L; Elevated neutrophil count refers to the number of neutrophils > 6.8 × 109/L; Elevated protein refers to protein concentration > 0.45 g/L; Decreased glucose refers to sugar < 2.2 mmol/L or less than 50% of blood sugar; Elevated C-reactive protein refers to the concentration of C-reactive protein > 6.3 mg/L
Laboratory Test results for TBM patients based on CSF samples
Among the 146 patients diagnosed with TBM, 58 (39.7%, 58/146) were unable to yield positive results for MTB infection through laboratory culture, mNGS, or GeneXpert. However, based on their clinical symptoms and other diagnostic criteria, they were still diagnosed with TBM. Of the 70 patients (47.9%, 70/146) diagnosed with TBM by GeneXpert and microbial culture, 37 patients tested positive by both MTB culture and GeneXpert, 18 patients tested positive only by GeneXpert, and 15 patients tested positive solely through MTB culture. Additionally, this study included the mNGS test results of 18 TBM patients (Appendix Table 3); among them, ten were positive for MTB using mNGS alone, five were positive only by mNGS but negative by conventional approaches, three were positive by both mNGS and GeneXpert, one was identified as positive via mNGS and MTB culture simultaneously, and one was identified as positive across all detection methods.
Comparisons of different detection methods for TBM-positivity
The individual test results presented in Table 2 demonstrated that metagenomic next-generation sequencing yielded the highest positivity rate (55.6%), followed by GeneXpert (43.7%) and microbial culture (38.6%). When considering combined test results, the highest positivity rate was observed with GeneXpert + microbial cultures, at a rate of 53.4%, followed closely by combinations involving either GeneXpert + mNGS or mNGS + microbial cultures, which both had a positivity rate of 52.9%. No significant difference was observed among these three methods.
Table 2.
Comparison of positive rates of clinical testing for tuberculous meningitis
| Total number of people(n) | Positive(n) | positive rate (%) | |
|---|---|---|---|
| Total | 146 | 80 | 54.8 |
| mNGS | 18 | 10 | 55.6 |
| GeneXpert | 135 | 59 | 43.7 |
| Culture | 140 | 54 | 38.6 |
| GeneXpert + Culture | 133 | 71 | 53.4 |
| GeneXpert + mNGS | 17 | 9 | 52.9 |
| Culture + mNGS | 17 | 9 | 52.9 |
Consistency of mNGS, microbial cultures, and GeneXpert in TBM diagnosis
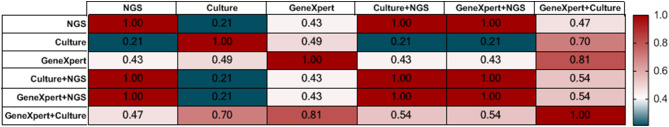
To evaluate the performance of mNGS for diagnosing TBM, we examined the consistency of the mNGS results and other conventional approaches. Kappa statistics (Fig. 1) indicated perfect agreement among mNGS alone or mNGS plus microbial culture or GeneXpert (kappa = 1, P < 0.0001). Furthermore, there was perfect agreement between mNGS combined with GeneXpert and mNGS combined with microbial culture (kappa = 1, P < 0.0001). Additionally, there was substantial agreement between the GeneXpert and GeneXpert + microbial culture results (kappa = 0.81, p < 0.0001). Moderate agreement was detected between GeneXpert and microbial cultures (kappa = 0.49, P < 0.0001), GeneXpert and mNGS (kappa = 0.43, P = 0.031), and mNGS and GeneXpert + microbial cultures (kappa = 0.47, p = 0.019). However, poor agreement was noted between mNGS and microbial culture results (kappa = 0.21, p = 0.156).
Fig. 1.
Comparison of results consistency of TBM detection methods. Kappa = 1 indicates perfect agreement, kappa ≥ 0.75 indicates substantial agreement, 0.4 < kappa < 0.75 indicates moderate agreement, and kappa ≤ 0.4 indicates poor agreement
Discussion
Early diagnosis of TBM can facilitate effective treatment. This study explored the characteristics of patients with TBM and patients whose CNS was infected with other etiologic agents. The study clearly demonstrated that while high leukocyte counts and protein concentrations in CSF are often observed in TBM-infected patients, they can also be observed in other central nervous system infections. Therefore, it is important to consider other factors, such as clinical symptoms and imaging results, when diagnosing TBM, as routine testing alone is not sufficient for diagnosis.
In addition, we evaluated the clinical performance of mNGS in overcoming the challenges in diagnosing TBM in some patients with TBM. A comparison of the results showed that mNGS yielded higher positivity rates than did conventional GeneXpert and microbial culture methods. The highest positivity rates were achieved by combining mNGS with routine tests, including serological tests and tests of specimens other than CSF. Furthermore, mNGS analysis identified more potential etiologic agents than did conventional direct testing of CSF specimens. Eleven infected patients were diagnosed only by mNGS. These findings demonstrated false-negative results for some patients with TBM according to routine tests, indicating the potential use of mNGS in the clinical diagnosis of TBM.
The diagnosis and treatment of TBM are challenging due to the similarity of symptoms to those of other CNS infections and the possibility of multiple etiologic agents [10, 20, 21]. GeneXpert and microbial culture methods have limitations in detecting unknown or variant etiologic agents [6]. However, mNGS has the potential to overcome these limitations by simultaneously detecting known and unknown genes of all pathogenic microorganisms in the database [22]. As databases improve and machine learning approaches are adopted, mNGS will become more widely used in the future.
A single diagnostic method often fails to provide accurate results. Combining multiple diagnostic approaches for TBM patients tends to yield more precise outcomes. Our results showed that mNGS yielded higher positivity rates than did GeneXpert and microbial culture methods when only one test was performed. However, there was no significant difference in the percentage of positive cells when the two test methods were combined. Interestingly, the percentage of positive mNGS + GeneXpert and mNGS + microbial culture results was notably greater than that of GeneXpert or microbial culture alone. Relevant studies have also shown significantly greater positivity rates with mNGS than with other conventional methods for detecting CNS infections [12, 23–25]. According to the results of CSF samples from patients with CNS infections on the same sequencing platform during the same period, the detection specificity of mNGS reached 94.2% [26], indicating that mNGS is a highly sensitive and specific diagnostic method suitable for clinical diagnosis. In conclusion, we recommend adopting mNGS as a preferred clinical diagnostic approach for patients with suspected TBM or combining mNGS with other methods for TBM diagnosis.
Although the application prospects of mNGS are very promising, there are still certain limitations in its current diagnostic processes. In the TBM patient data we collected, the sensitivity of mNGS was less than 60%. Factors that may affect MTB detection include contamination of CSF samples during collection, low MTB concentration, and insufficient sequencing depth [27]. Therefore, for the clinical diagnosis of TBM and other CNS diseases, it is necessary not only to improve bioinformatic alignment but also to optimize sampling procedures and technical aspects.
Considering the higher cost of mNGS, patients with lower incomes can substitute mNGS with GeneXpert + MTB culture. However, longer growth cycles of MTB (2–3 weeks on average) are unconducive to early diagnosis and effective treatment by clinicians. This approach allows for accurate and timely diagnosis in critically ill patients, which can lead to better outcomes and potentially save lives. In patients with mild illness, cost-effective GeneXpert + microbial culture can still provide reliable results while minimizing unnecessary expenses. Overall, a tailored approach to diagnostic testing based on the severity of illness can optimize patient care and resource utilization.
This study has several limitations. Although we confirmed the advantages of mNGS in diagnosing TBM, its disadvantages might still hinder its wider clinical application. First, this study did not determine the titer of etiologic agents during mNGS, which may lead to false-negative, false-positive, or low-abundance results for high-priority etiologic agents obscured by other high-abundance agents. Second, to follow the wishes of patients, we cannot ask all patients to undergo other tests, including mNGS. This led to the limited number of patients who underwent mNGS in this study, which may have affected the evaluation of the efficacy of mNGS. Third, due to the limitations of available diagnostic methods and the restricted accuracy of traditional diagnostic approaches, we cannot rule out the possibility that a small number of patients with milder symptoms may have been misdiagnosed with TBM in our study. Finally, although we have made every effort to collect as many CNS patient samples as possible over a three-year period and conducted thorough testing, the case information and test results we obtained are still limited due to the single-center nature of our study. In future research, we will collaborate with multiple centers to conduct larger-scale testing in order to optimize the diagnosis of CNS infection patients.
Conclusions
This study compared the sensitivity and consistency of th + ree different detection methods used individually and in combination for diagnosing TBM. Among the three methods, mNGS has the highest sensitivity. GeneXpert combined with MTB culture offers similar sensitivity to mNGS. Considering the short detection time and high sensitivity, for critical patients, we recommend mNGS for TBM diagnosis. In less developed regions, a combination of GeneXpert and MTB culture is a viable alternative.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We would like to thank the patients who participated in this study. We acknowledge the contributions of all the healthcare providers from Shenzhen Third People’s Hospital.
Abbreviations
- TBM
Tuberculous meningitis
- CNS
Central nervous system
- mNGS
Metagenomic next-generation sequencing
- CSF
cerebrospinal fluid
- MTB
Mycobacterium tuberculosis
- AFB
Acid-fast bacillus
- PCR
Polymerase chain reaction
- WHO
World Health Organization
- CMV
Cytomegalovirus
- EBV
Epstein‒Barr virus
- HSV-1/2
Herpes simplex virus-1/2
Author contributions
MC, LW, and GL conceived the study; SZ, YT, and WX collected clinical information; XZ, DL, FT, DW, YW, and FW analyzed the data; and ZL, LW, MC, GL, and XZ were major contributors to the writing of the manuscript. All the authors read and agreed with the content.
Funding
This study was funded by grants from Shenzhen High-level Hospital Construction Fund (No. G22244G2001), Guangdong Provincial Department of Science and Technology(2023A1515220104), Shenzhen University High Level Clinical Special Project - Shenzhen Neurosurgery Specialist Training Pilot Base Promotion Project (LCXKB202415), and Shenzhen Clinical Research Center for Tuberculosis (20210617141509001). The funders played no role in the study design, data collection, data analysis, data interpretation, or writing of the report.
Data availability
All the data involved in this study were included in this published article. The original data can be reasonably obtained after consulting the corresponding author by email.
Declarations
Ethics approval and consent to participate
All the study protocols and ethics protocols used were approved by the Scientific Research Ethics Committee of Shenzhen Third People’s Hospital (approval number/ID: 2020002), and all the patients enrolled in this study provided written informed consent. The final clinical diagnosis of patients completing the study was reviewed by certified medical practitioners and researchers (Shengkun Zhang). All the results and charts were reviewed and approved by Xujie Zhu.
Consent for publication
Not Applicable.
Competing interests
The authors declare no competing interests.
Footnotes
The original online version of this article was revised: Following publication of the original article, we were notified that the author affiliations were incorrectly reported.
Zengchen Liu and Xujie Zhu are co-first authors.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
11/8/2024
A Correction to this paper has been published: 10.1186/s12879-024-10172-y
Contributor Information
Guobao Li, Email: l3gb06@aliyun.com.
Lanlan Wei, Email: weilanlan@mail.sustech.edu.cn.
Ming Chu, Email: chuming@mail.sustech.edu.cn.
References
- 1.Manyelo CM et al. Tuberculous meningitis: Pathogenesis, Immune responses, Diagnostic challenges, and the potential of biomarker-based approaches. J Clin Microbiol, 2021. 59(3). [DOI] [PMC free article] [PubMed]
- 2.Glaser CA, et al. In search of encephalitis etiologies: diagnostic challenges in the California Encephalitis Project, 1998–2000. Clin Infect Dis. 2003;36(6):731–42. [DOI] [PubMed] [Google Scholar]
- 3.Glaser CA, et al. Beyond viruses: clinical profiles and etiologies associated with encephalitis. Clin Infect Dis. 2006;43(12):1565–77. [DOI] [PubMed] [Google Scholar]
- 4.Granerod J, et al. Causes of encephalitis and differences in their clinical presentations in England: a multicentre, population-based prospective study. Lancet Infect Dis. 2010;10(12):835–44. [DOI] [PubMed] [Google Scholar]
- 5.Khetsuriani N, Holman RC, Anderson LJ. Burden of encephalitis-associated hospitalizations in the United States, 1988–1997. Clin Infect Dis. 2002;35(2):175–82. [DOI] [PubMed] [Google Scholar]
- 6.Mechal Y, et al. Evaluation of GeneXpert MTB/RIF system performances in the diagnosis of extrapulmonary tuberculosis. BMC Infect Dis. 2019;19(1):1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zubair AS, et al. A critically ill patient with Central Nervous System Tuberculosis and negative initial workup. Front Neurol. 2020;11:430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singhi P, Saini AG. Fungal and parasitic CNS infections. Indian J Pediatr. 2019;86(1):83–90. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y, et al. Clinical application and evaluation of metagenomic next-generation sequencing in suspected adult central nervous system infection. J Transl Med. 2020;18(1):199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Organization WH. Global Tuberculosis Report 2023.. 2023; https://www.who.int/publications/i/item/9789240083851
- 11.Méchaï F, Bouchaud O. Tuberculous meningitis: challenges in diagnosis and management. Rev Neurol (Paris). 2019;175(7–8):451–7. [DOI] [PubMed] [Google Scholar]
- 12.Wilson MR, et al. Clinical metagenomic sequencing for diagnosis of Meningitis and Encephalitis. N Engl J Med. 2019;380(24):2327–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aly SM, Sabri DM. Next generation sequencing (NGS): a golden tool in forensic toolkit. Arch Med Sadowej Kryminol. 2015;65(4):260–71. [DOI] [PubMed] [Google Scholar]
- 14.Yoshinaga Y, et al. Genome Sequencing Methods Mol Biol. 2018;1775:37–52. [DOI] [PubMed] [Google Scholar]
- 15.Deurenberg RH, et al. Application of next generation sequencing in clinical microbiology and infection prevention. J Biotechnol. 2017;243:16–24. [DOI] [PubMed] [Google Scholar]
- 16.Behjati S, Tarpey PS. What is next generation sequencing? Arch Dis Child Educ Pract Ed. 2013;98(6):236–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xuan J, et al. Next-generation sequencing in the clinic: promises and challenges. Cancer Lett. 2013;340(2):284–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forbes JD, et al. Metagenomics: the Next Culture-Independent game changer. Front Microbiol. 2017;8:1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldberg B, et al. Making the Leap from Research Laboratory to Clinic: challenges and opportunities for Next-Generation sequencing in Infectious Disease Diagnostics. mBio. 2015;6(6):e01888–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen X, et al. Immunological network analysis in HPV associated head and neck squamous cancer and implications for disease prognosis. Mol Immunol. 2018;96:28–36. [DOI] [PubMed] [Google Scholar]
- 21.Marais S, et al. Tuberculous meningitis: a uniform case definition for use in clinical research. Lancet Infect Dis. 2010;10(11):803–12. [DOI] [PubMed] [Google Scholar]
- 22.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Zeggeren IE, et al. Diagnostic accuracy of VIDISCA-NGS in patients with suspected central nervous system infections. Clin Microbiol Infect. 2021;27(4):631. .e7-631.e12. [DOI] [PubMed] [Google Scholar]
- 24.Hasan MR, et al. A metagenomics-based diagnostic approach for central nervous system infections in hospital acute care setting. Sci Rep. 2020;10(1):11194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xing XW, et al. Evaluation of Next-Generation sequencing for the diagnosis of infections of the Central Nervous System caused by the neurotropic herpes viruses: a pilot study. Eur Neurol. 2018;80(5–6):283–8. [DOI] [PubMed] [Google Scholar]
- 26.Fan S et al. Metagenomic Next-generation Sequencing of Cerebrospinal Fluid for the Diagnosis of Central Nervous System Infections: A Multicentre Prospective Study. bioRxiv, 2019: p. 658047.
- 27.Chiu CY, Miller SA. Clinical metagenomics. Nat Rev Genet. 2019;20(6):341–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data involved in this study were included in this published article. The original data can be reasonably obtained after consulting the corresponding author by email.