Abstract
As do human herpesvirus 6 variants A and B (HHV-6A and -6B), HHV-7 encodes a homolog of the alphaherpesvirus origin binding protein (OBP), which binds at sites in the origin of lytic replication (oriLyt) to initiate DNA replication. In this study, we sought to characterize the interaction of the HHV-7 OBP (OBPH7) with its cognate sites in the 600-bp HHV-7 oriLyt. We expressed the carboxyl-terminal domain of OBPH7 and found that amino acids 484 to 787 of OBPH7 were sufficient for DNA binding activity by electrophoretic mobility shift analysis. OBPH7 has one high-affinity binding site (OBP-2) located on one flank of an AT-rich spacer element and a low-affinity site (OBP-1) on the other. This is in contrast to the HHV-6B OBP (OBPH6B), which binds with similar affinity to its two cognate OBP sites in the HHV-6B oriLyt. The minimal recognition element of the OBP-2 site was mapped to a 14-bp sequence. The OBPH7 consensus recognition sequence of the 9-bp core, BRTYCWCCT (where B is a T, G, or C; R is a G or A; Y is a T or C; and W is a T or A), overlaps with the OBPH6B consensus YGWYCWCCY and establishes YCWCC as the roseolovirus OBP core recognition sequence. Heteroduplex analysis suggests that OBPH7 interacts along one face of the DNA helix, with the major groove, as do OBPH6B and herpes simplex virus type 1 OBP. Together, these results illustrate both conserved and divergent DNA binding properties between OBPH7 and OBPH6B.
Human herpesvirus 7 (HHV-7) is a widely prevalent betaherpesvirus with an in vitro tropism for CD4+ T lymphocytes (reviewed in reference 5). HHV-7 is usually acquired during early childhood after HHV-6 infection and is likely transmitted via saliva (29). HHV-7 has been associated with febrile illnesses in children, neurological manifestations during primary infection, and clinical complications in organ transplant patients.
The betaherpesvirus subfamily consists of the cytomegaloviruses and the roseoloviruses (HHV-7 and HHV-6 variants A and B [HHV-6A and HHV-6B]). Their genomes are genetically colinear, with origins of lytic replication (oriLyts) in analogous positions upstream from U41, the gene encoding the major DNA binding protein (1, 7, 11, 23, 27). However, sequence features of their oriLyt regions indicate substantial differences between cytomegaloviruses and roseoloviruses in their mechanisms for initiating viral replication. The minimal oriLyt regions of the cytomegaloviruses are long (>1.3-kb) complex structures consisting of multiple inverted and direct repeats, in addition to numerous transcription factor recognition sites that are believed to mediate activation of the replication origin (2, 20, 21, 28). Unlike the cytomegaloviruses, oriLyts of HHV-6A and HHV-6B have features in common with the replication origins of alphaherpesviruses; these origins are less complex and are centered around binding sites for a virus-encoded replication initiator protein, the OBP.
Roseoloviruses each encode a homolog of the alphaherpesvirus OBP (14, 16, 23), which has no homolog in the cytomegaloviruses. The most extensively characterized alphaherpesvirus OBP, herpes simplex virus type 1 (HSV-1) OBP (OBPH1), binds as a dimer at each of two sites in its origins of replication (Box sites I and II in each oriS and two Box I sites in oriL) (17). Homodimer interactions at each box site are believed to lead to local DNA bending of the intervening AT-rich element, unwinding of the region in association with the single-stranded DNA binding protein, and recruitment of the viral replication machinery (6).
Like OBPH1, the OBP of HHV-6B (OBPH6B) binds two sites (OBP-1 and OBP-2) that flank an AT-rich spacer element in the minimal core of the oriLyt region (14). The consensus recognition sequence differs between OBPH6B and OBPH1, and they are unable to bind each other's OBP site (15). OBPH7 and OBPH6B, encoded by the U73 gene in each virus, share 58% amino acid identity, in contrast to their respective 32 and 31% identities with the more distantly related OBPH1 (23). Although the HHV-7 genome is generally well conserved with respect to HHV-6A and HHV-6B, there is little sequence similarity in the origin regions (reference 27 and data not shown). Nonetheless, van Loon et al. demonstrated that an HHV-6 oriLyt-containing plasmid could replicate in HHV-7-infected cells (27). In addition, they found that mutation of the only site in the HHV-7 oriLyt region that matches the OBPH6B consensus recognition sequence resulted in the loss of transient plasmid replication in HHV-7-infected cells. Although these results indicate that OBPH7 may recognize sequences similar to those in the HHV-6 oriLyt, they suggest that there are OBP sites in the HHV-7 oriLyt that do not conform to the OBPH6B consensus. Recognizing the limits of relying on the consensus sequence of a distinct, albeit closely related, herpesvirus to predict OBPH7-binding sites, we have used a direct biochemical approach for their identification.
In this study, we characterized the interaction of OBPH7 with its cognate oriLyt to better understand the initiation mechanism of HHV-7 DNA replication. We found that in the HHV-7 oriLyt, OBPH7 binds to two sites that flank an AT-rich spacer, and we studied its sequence and spatial binding requirements.
MATERIALS AND METHODS
RNA preparation and RT-PCR analysis.
For mRNA studies, HHV-7(SB) was propagated in SupT1 cells as previously described (4). Total RNA was prepared from 106 SupT1 cells 7 days postinfection with HHV-7(SB) with a High Pure RNA isolation kit (Roche, Indianapolis, Ind.) according to the manufacturer's protocol. One microgram of RNA (extracted from approximately 1.5 × 105 cells) was treated with DNase (Roche) at 37°C for 30 min according to the manufacturer's recommendations. Twenty nanograms of DNase-treated RNA was reverse transcribed from oligo(dT) primers in a 40-μl reaction mixture by using a GeneAmp RNA PCR kit (PE Biosystems, Foster City, Calif.) according to the manufacturer's protocol. Ten microliters of the reverse transcriptase (RT) reaction mixture was then used for PCR analysis using primers specific for HHV-7 U73 (CCGAATCAAAACATTTACTCTA and AATCCGCTCTAATAGATTCTGCTA) and for the cellular gene glyceraldehyde-3′-phosphate dehydrogenase (GAPDH) (Stratagene, La Jolla, Calif.). Thermal cycling conditions were as follows: 35 cycles of 96°C for 15 s, 60°C for 30 s, and 68°C for 30 s with an increased extension of 5 s per cycle.
Plasmid construction.
U73 fragments encoding the carboxyl-terminal portions of OBPH7 were amplified from HHV-7(SB) nucleocapsid DNA using a proofreading DNA polymerase (Pfx; GIBCO BRL, Rockville, Md.). Primers used were as follows: for the construct without a Kozak sequence, CGCGGATCCATGAACGGAGAATTCTA and CCGGATATCCGTTATGAACGCAATA; and for constructs containing a Kozak consensus sequence surrounding the ATG, CGCATCACGGGATCCGCCACCATGGACGGAGAATTCTA and CCGGATATCCGTTATGAACGCAATA (restriction endonuclease sites used for cloning are italicized, residues in boldface indicate U73-derived nucleotides, and underlined ATG sequences were inserted for translation initiation). Using standard cloning techniques, these amplimers were digested, purified, and then ligated into the EcoRV site of pcDNA3 (Invitrogen, Carlsbad, Calif.) for the non-Kozak primer set or the BamHI and EcoRV sites of pcDNA3 and pCMV-Tag 2B (Stratagene) for the Kozak-containing primer set. pCMV-Tag 2B provides a FLAG epitope tag at the amino terminus of the protein. A negative control pcDNA3-derived construct was also generated that contained U73 in the reverse orientation.
OBPH7 in vitro expression.
OBP was expressed in coupled in vitro transcription-translation (IVTT) reactions (Promega, Madison, Wis.) as previously described with 2 μg of input DNA and 55% reticulocyte lysate (14). Negative control lysate was generated by programming the reactions with a plasmid containing U73 in the reverse orientation. 35S-labeled in vitro-translated products were mixed with loading buffer and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis as previously described (14).
Electrophoretic mobility shift analysis (EMSA).
Oligonucleotides were annealed, labeled, and purified as described previously (14). DNA was incubated at room temperature for 20 min in 10 μl of either reaction buffer A or B with 2 μl of programmed IVTT lysate. Reaction buffer A consisted of 12 mM HEPES-NaOH (pH 7.6), 4 mM Tris-HCl (pH 7.6), 125 mM NaCl, 1 mM EDTA, 5 mM MgCl2, 1 mM dithiothreitol, 120 μg of bovine serum albumin per ml, 12% glycerol, 5 μg of salmon testes DNA per ml, and a cocktail of protease inhibitors (Complete Mini EDTA-free; Roche). Buffer B (14) was similar to buffer A except that it contained 50 mM NaCl and no MgCl2. DNA-protein complexes were separated in 5% polyacrylamide gels (60:1 acrylamide:bis-acrylamide) by using a low-ionic-strength electrophoresis buffer (14) at 4°C.
For competitive EMSA, reactions were set up as described above except that unlabeled competitor DNA was incubated with the lysate for 10 min at room temperature before the addition of the labeled target DNA. The percentage of binding inhibition was calculated by dividing the difference between the amount of signal in the absence and presence of competitor oligonucleotides by the amount of signal without competitor and then multiplying by 100. The percentage of wild-type inhibition was calculated by dividing the percentage of binding inhibition of the mutated competitor oligonucleotides by the percentage of binding inhibition of the wild-type oligonucleotide and then multiplying by 100. The percentage of oligonucleotide 7–2, L6R7, or 7–2B inhibition was calculated similarly. For supershift experiments, monoclonal antibodies (MAbs) against the FLAG (Stratagene) or the Xpress (Invitrogen) epitopes were incubated with the protein-DNA complexes for an additional 10 min at room temperature.
RESULTS
The HHV-7 oriLyt region.
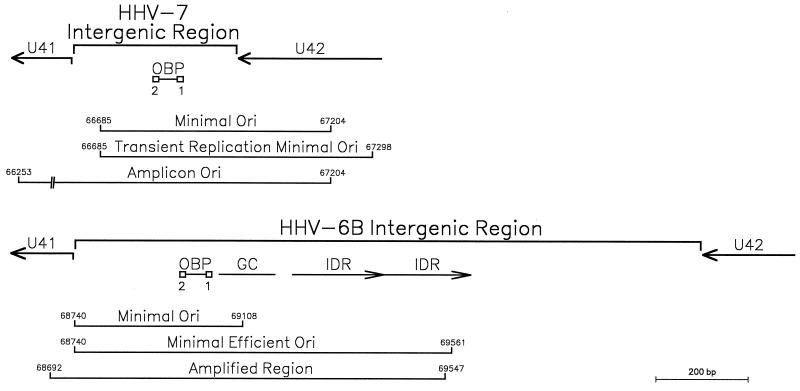
As for the whole genome, the HHV-7 U41-U42 intergenic region is more compact than those of HHV-6A and HHV-6B (Fig. 1). The approximately 600-bp HHV-7 oriLyt region mapped in transient replication assays is composed of nearly all of the U41-U42 intergenic region and extends 300 bases into U42 (27). HHV-7 amplicons constructed by Romi et al. (24) utilized an approximately 1-kb oriLyt region that extended only 200 bases into U42; this suggests that the minimal oriLyt may be as short as 500 bp.
FIG. 1.
The oriLyt regions of HHV-7 and HHV-6B. The HHV-7 minimal origin as defined from plasmid (27) and amplicon (24) constructs includes two OBP sites flanking an AT-rich spacer element (described in this report). The HHV-6B minimal efficient origin determined by transient replication analysis (7) and a region found amplified in some cell culture passages of HHV-6B(Z29) (26) contains a G+C-rich region and IDRs to the right of the OBP sites. Coordinates are derived from HHV-7(RK) and HHV-6B(Z29).
The 500-bp minimal HHV-7 oriLyt shares little sequence similarity with the HHV-6 oriLyts; it is one of the most divergent regions between the genomes of these viruses (data not shown). As illustrated in Fig. 1, within the 817-bp HHV-6B oriLyt region is a 400-bp minimal essential domain that contains two OBP-binding sites flanking an AT-rich spacer element. Replication efficiency is enhanced by adjacent sequences that include a GC-rich segment and a putative DNA unwinding element within the AT-rich imperfect direct repeats (IDRs) of 187 and 192 bp (7, 18, 26). The HHV-6A oriLyt region has a similar structure with an approximate 91% sequence identity to the homologous HHV-6B region, except there are three copies of an IDR that is related to, but shorter than, that in HHV-6B (8, 11). The minimal HHV-7 oriLyt is not marked by the striking GC- and AT-rich regions of the HHV-6 oriLyts and has no structures that correspond to the long IDRs or putative DNA unwinding elements found in the HHV-6A and HHV-6B oriLyt regions (8, 27). Within the otherwise divergent HHV-7 oriLyt, one site (OBP-2) is identical to the HHV-6 OBP-2 site and a second site (OBP-1) contains seven of nine matches to the OBPH6 consensus recognition sequence (15); these sites flank a 50-bp AT-rich element in the HHV-7 oriLyt (Fig. 1) and have an overall structure that is similar to the lytic origin regions of HHV-6A, HHV-6B, and most alphaherpesviruses. These are the OBPH7-binding sites identified biochemically in this work.
U73 expression in HHV-7(SB)-infected cells.
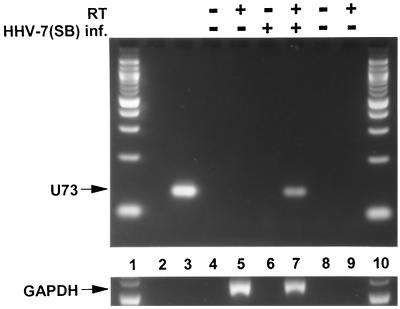
RT-PCR was done to confirm that the gene encoding OBPH7 (U73) is expressed during lytic replication. A 133-bp amplimer was generated from RNA extracted from HHV-7(SB)-infected SupT1 cells (Fig. 2, lane 7) but not uninfected cells (lane 5). A 550-bp GAPDH product was amplified in reactions containing both uninfected and infected SupT1 RNA only upon the addition of RT (lanes 5 and 7), demonstrating the absence of DNA contamination and the presence of amplifiable RNA. The U73 primers were specific for HHV-7, since they did not produce a product with HHV-6B(Z29) DNA (lane 2).
FIG. 2.
RT-PCR detection of HHV-7(SB) U73 (OBPH7) transcripts. Lanes 1 and 10 contain a 100-bp DNA ladder (New England Biolabs, Inc., Beverly, Mass.). Lanes 2 and 3 contain HHV-6B(Z29) and HHV-7(SB) viral nucleocapsid DNA, respectively. Lanes 4 and 5 and lanes 6 and 7 contain the PCR products of reactions with uninfected SupT1 RNA and HHV-7(SB)-infected SupT1 RNA, respectively. Lanes 8 and 9 are no-template negative controls. RT was added to lanes 5, 7, and 9. The upper and lower panels contain PCR products obtained using U73 and GAPDH primers, respectively.
In vitro expression of truncated OBPH7.
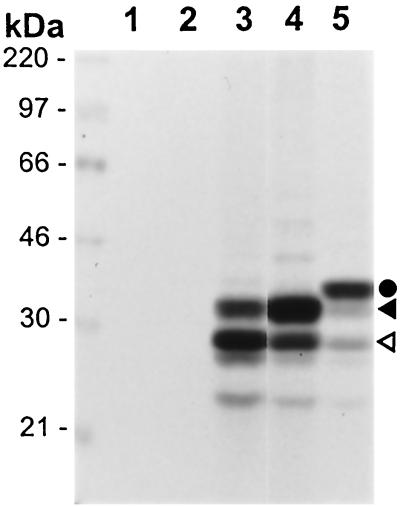
Unlike OBPH1, baculovirus and bacterial expression of full-length OBPH6B has yielded insoluble protein (15). As for OBPH6B (14), full-length OBPH7 protein expressed in IVTT formed high-molecular-weight aggregates that had little binding activity by EMSA (data not shown). The carboxyl-terminal portion of OBPH7 (amino acids [aa] 484 to 787) closely corresponds to the smallest region of OBPH6B (aa 482 to 770) required for DNA binding activity as measured by EMSA (15). The carboxyl-terminal portion of OBPH7 was expressed by IVTT from three plasmid constructs. From an OBP expression construct lacking a Kozak translation initiation sequence surrounding the initiating methionine, 33- and 29-kDa products were detected (Fig. 3, lane 3). The presence of a Kozak sequence resulted in higher levels of the 33-kDa protein that closely approximated the expected molecular mass of 35.4 kDa (Fig. 3, lane 4). This suggests that the smaller products initiated from internal AUG sequences. An amino-terminal FLAG-tagged OBPH7 fusion protein of 37 kDa was also generated that was consistent with the predicted size of 36.9 kDa (Fig. 4, lane 5). Truncated OBPH7 and FLAG-tagged OBPH7 generated from constructs containing a Kozak sequence were used in the remainder of the experiments described in this report.
FIG. 3.
In vitro expression of truncated OBPH7. IVTT mixtures programmed with pCMV-Tag 2B vector DNA (lane 1), pcDNA3 with truncated U73 in the reverse orientation (lane 2), or plasmids containing the carboxyl-terminal region (aa 484 to 787) of HHV-7 U73 (lanes 3 to 5) were incubated in the presence of [35S]methionine and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis as described previously (14). OBPH7 was expressed from pcDNA3 constructs either lacking (lane 3) or containing (lane 4) a Kozak translation initiation sequence. Lane 5 contains N-terminally FLAG-tagged OBPH7 expressed from pCMV-Tag 2B. The circle indicates the 37-kDa FLAG-tagged OBPH7 fusion protein. The solid and open triangles indicate the 33- and 29-kDA products generated from constructs containing the carboxyl-terminal portion of HHV-7 U73.
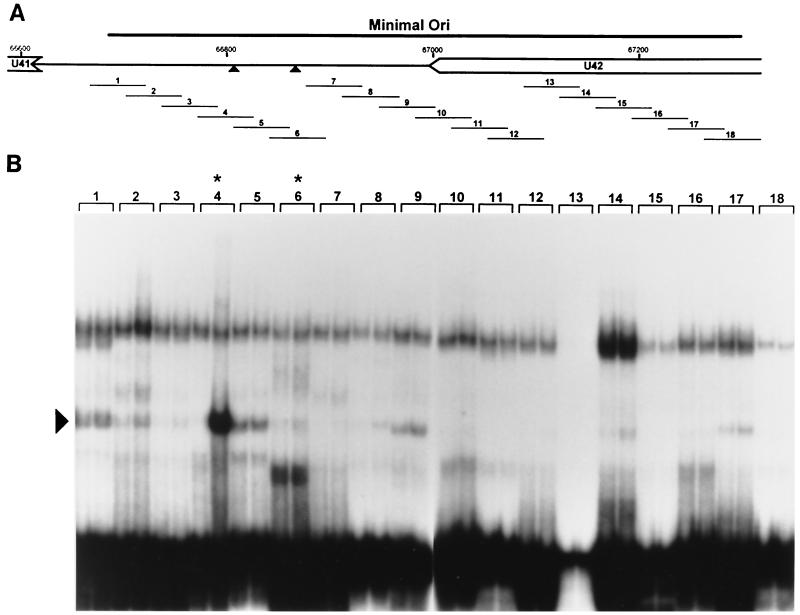
FIG. 4.
Identification of an OBPH7-binding site in the HHV-7 oriLyt. (A) Schematic diagram of the genomic location of the 600-bp HHV-7 minimal oriLyt region (27). The triangles indicate the putative HHV-7 OBP-2 and OBP-1 sites. (B) The 18 double-stranded 55-bp oligonucleotides (1 to 18) shown in panel A were used for EMSA. 32P-labeled oligonucleotides were reacted with IVTT lysate programmed with truncated U73 in the reverse orientation (negative control, left lane for each oligonucleotide) or with truncated U73 in the correct forward orientation (right lane for each oligonucleotide). Asterisks indicate oligonucleotides with the putative OBP-2 and OBP-1 sites. Oligonucleotide 13 labeled inefficiently to lower specific activity; no specific binding was detected upon long exposure or by PhosphorImager analysis (data not shown). The arrowhead indicates the complex generated in the presence of oligonucleotide 4 and OBPH7-containing IVTT.
The high-affinity OBPH7 site in the HHV-7 oriLyt is identical to the HHV-6 OBP-2 site.
In preliminary EMSA experiments, we found that OBPH7 recognized an oligonucleotide containing the putative HHV-7 OBP-2 site (data not shown). The reaction buffer used in the initial experiments (buffer B) was previously identified as optimal for OBPH6B (14). Subsequent titrations indicated that higher NaCl and MgCl2 concentrations (125 mM NaCl and 5 mM MgCl2) were required for optimal OBPH7 binding (buffer A); these binding conditions were used in the following series of experiments.
Twenty-one sites in the minimal HHV-7 oriLyt have at least seven of nine matches to the OPBH6B consensus sequence, YGWYCWCCY. To perform an unbiased biochemical search for all OBP sites, 18 overlapping 55-bp DNA duplexes that span the 600-bp oriLyt region were examined for OBPH7 binding (Fig. 4A). Since the IVTT expression system contains a multitude of reticulocyte proteins that may interact with the DNA targets, we included a negative control lysate in parallel with the OBPH7-containing lysate for each oligonucleotide examined. The experiment was performed in duplicate, and shifts were quantitated by PhosphorImager analysis. As shown in Fig. 4B, oligonucleotide 4 was the only target that reproducibly generated a >2-fold difference in any shift pattern between the OBPH7-containing lane and the negative control lane. Oligonucleotide 4 contains a 9-bp sequence identical to the HHV-6 OBP-2 site. Maintenance of this sequence is required for HHV-7 oriLyt-mediated plasmid replication (27). No OBPH7-specific binding was detected to oligonucleotide 6, which contains a sequence with seven of nine matches to the OBPH6B consensus recognition sequence and is located on the flank of an AT-rich element opposite from the OBP-2 site. Because of the correspondence of sequence and context between this site and the HHV-6 OBP-1 site, we had expected recognition of oligonucleotide 6 by OBPH7 in the direct binding analysis. We sought to confirm the results from oligonucleotides 4 and 6 in a competition analysis.
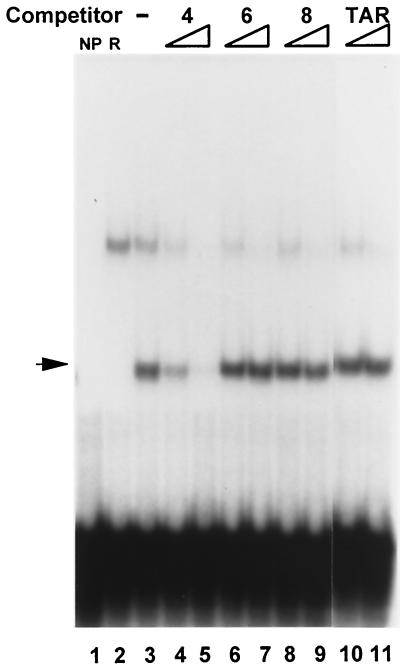
As shown in Fig. 5, only oligonucleotide 4, which contains the putative OBP-2 site, competed for binding to OBPH7 with labeled oligonucleotide 4. There was no evidence for recognition of oligonucleotide 6 (which contains the putative OBP-1 site) or oligonucleotide 8 and TAR (no OBP site similarity). These results confirm the specificity of the OBP-2 site interaction and demonstrate that there is a single high-affinity site for OBPH7 (designated OBP-2) in the HHV-7 oriLyt.
FIG. 5.
Specificity of the OBPH7 protein–OBP-2 DNA interaction by competitive EMSA. OBPH7 binding to oligonucleotides 4, 6, and 8 (Fig. 4) was analyzed in competition analysis. Sixteen- and 80-fold molar excesses of the oligonucleotides were added as competitors for binding to labeled oligonucleotide 4. Oligonucleotide 8 and TAR are double-stranded oligonucleotides with no sequence similarity to an OBP site. NP, no protein added to the binding reaction; R, binding reaction contained protein from an IVTT reaction mixture programmed with a plasmid containing HHV-7 U73 in the reverse orientation; −, no competitor present; arrow, the specific shift with OBPH7.
OBPH7 is present in the specific shifted complex.
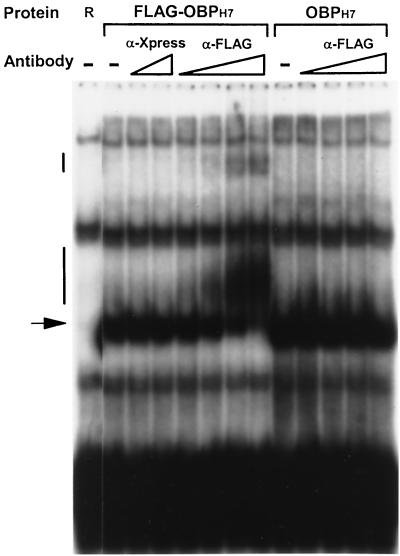
To verify that the specific shifted complex contained OBPH7, we did an antibody supershift experiment. A truncated OBPH7 protein with an amino-terminal FLAG epitope was reacted with oligonucleotide 7–2 (see Fig. 7C) in the presence of specific and nonspecific antibodies. The 50-bp 7–2 is similar to the 55-bp oligonucleotide 4 except the OBP-2 site is in a slightly different location in relation to the flanking sequence. As shown in Fig. 6, in the absence of added antibodies, both FLAG-OBPH7 and OBPH7 generated protein-DNA complexes with similar mobilities. DNA-protein complexes containing FLAG-OBPH7 were recognized by the antibody against the FLAG epitope to generate tertiary supershifted complexes. This antibody did not affect the mobility of DNA complexes containing OBPH7 protein that lacked the FLAG epitope. The specificity of this interaction was further demonstrated by the absence of a higher-mobility product upon addition of antibody of the same immunoglobulin G (IgG) subclass (IgG1) against an epitope (Xpress) not present in the OBP proteins. This experiment also demonstrates that the other shifts seen are due to interactions with other proteins of the reticulocyte lysate and are not the specific shifts of interest involving OBPH7.
FIG. 7.
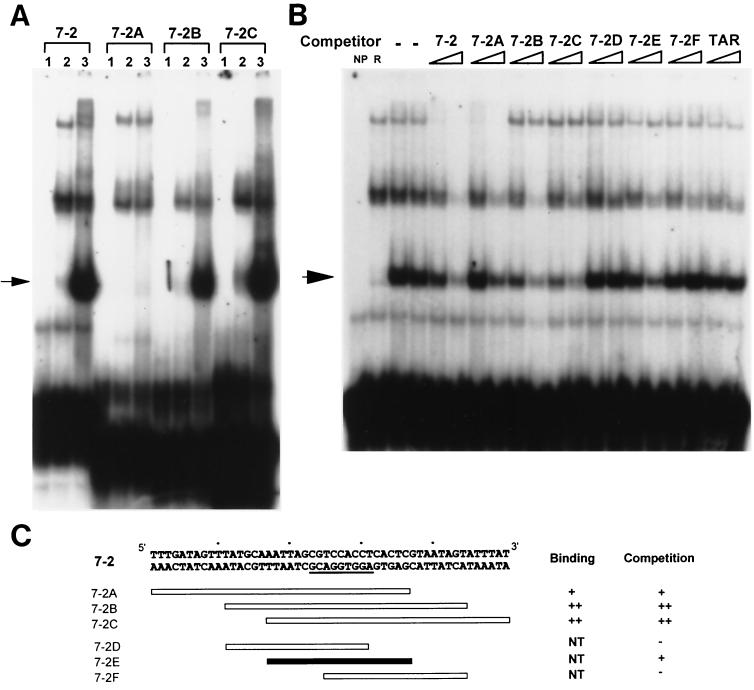
Mapping the OBP-2-binding site. (A) Oligonucleotides that span the 50-bp 7–2 oligonucleotide that contains the OBP-2 site were tested for their ability to be directly bound by OBPH7. For each oligonucleotide target, lane 1 contains a binding reaction mixture without any reticulocyte lysate, lane 2 contains reverse negative control lysate, and lane 3 contains lysate with truncated OBPH7 protein. (B) Competition EMSA with unlabeled oligonucleotides at 16- and 80-fold molar excess. −, no competitor was added. NP and R are negative controls as described in the Fig. 5 legend. In panels A and B, arrows point to the specific shifts. (C) Schematic diagram of oligonucleotides used and summary of binding and competition assay results shown in panels A and B. NT, not tested. ++ indicates a stronger degree of interaction of an oligonucleotide with OBPH7 than +; −, no competition observed.
FIG. 6.
Specificity of the OBPH7 protein–OBP-2 DNA interaction by supershift. OBPH7 was incubated with increasing amounts of MAbs against the Xpress (Invitrogen) or FLAG (Stratagene) epitope before the addition of the labeled 7–2 oligonucleotide (see Fig. 7C) containing the HHV-7 OBP-2 site. The triangles below α-Xpress indicate 97.6 pg and 6.25 ng of input IgG MAb. The triangles below α-FLAG indicate the addition of 97.6 pg, 0.391 ng, 1.58 ng, and 6.25 ng of IgG MAb. −, no antibody added; R, negative control lysate; arrow, the specific shift with OBPH7 protein; vertical bars, the supershifted complexes.
The OBPH7 minimal recognition element.
The following series of experiments was intended to define the boundaries of the minimal OBPH7 recognition element. Three sets of shorter oligonucleotides derived from the 50-bp 7–2 oligonucleotide were generated and examined for their ability to bind OBPH7. The first set of overlapping 34-bp oligonucleotides (7–2A, -B, and -C) (Fig. 7C) collectively spans the 50-bp region. In both direct binding and competition experiments, 7–2B and 7–2C were strongly bound by OBPH7 while 7–2A reacted only weakly (Fig. 7A and B). These results indicate that the 8-bp sequence to the right of the 7–2A boundary is not essential for recognition but does influence binding efficiency. A second set of overlapping 20-bp DNA duplexes that span 7–2B was then tested for competition against labeled 7–2 (Fig. 7B); as summarized in Fig. 7C, only 7–2E competed.
Finally, a third panel of oligonucleotides was generated that contained variable amounts of native sequence flanking either the left or right side of the 9-bp HHV-7 OBP-2 sequence that is identical to the HHV-6B OBP-2 site. These oligonucleotides were tested for their ability to compete with labeled 7–2B for binding to OBPH7. In these experiments, the amount of radioactivity in each shifted band was quantitated and is presented as the percentage of binding inhibition relative to that of a full-length control, oligonucleotide L6R7, as described in Materials and Methods. The oligonucleotides and competition results are summarized in Table 1. The shortest oligonucleotide that competed for binding was the 14-bp L0R5 oligonucleotide. This defines the minimal recognition sequence as a 14-bp element consisting of the 9-bp OBP-2 sequence plus five bases flanking to its right.
TABLE 1.
Oligonucleotides used for EMSA
| Competitor | Sequence | Length (bp) | Binding inhibition (%)a |
|---|---|---|---|
| AATTAGCGTCCACCTCACTCGT | |||
| L6R7 | ********************** | 22 | 100 |
| L5R6 | ******************** | 20 | 100 |
| L4R5 | ****************** | 18 | 89 |
| L3R4 | **************** | 16 | 0 |
| L2R3 | ************** | 14 | 0 |
| L1R2 | ************ | 12 | 0 |
| L0R1 | ********** | 10 | 0 |
| L3R6 | ****************** | 18 | 100 |
| L3R5 | ***************** | 17 | 100 |
| L2R5 | **************** | 16 | 100 |
| L1R5 | *************** | 15 | 100 |
| L0R5b | ************** | 14 | 76 |
| L5R4 | ****************** | 18 | 47 |
| L4R4 | ***************** | 17 | 28 |
| L4R3 | **************** | 16 | 0 |
| L4R2 | *************** | 15 | 0 |
| L4R1 | ************** | 14 | 0 |
The percentage inhibition of radiolabeled oligonucleotide 7–2B binding to truncated OBPH7, relative to the inhibition from oligonucleotide L6R7.
L0R5 is the smallest oligonucleotide that competes for OBPH7 recognition.
The OBPH7 and the OBPH6B consensus recognition sequences are closely related.
The consensus recognition sequence for a DNA binding protein is a powerful tool for understanding the basis of sequence specificity for a protein-DNA interaction. In addition, comparison of the consensus sequences among OBPs enables an evaluation of their evolutionary divergence. To determine the OBPH7 consensus recognition sequence, we used the saturation mutagenesis technique described previously for OBPH6B (15) and OBPH1 (13). We designed a panel of double-stranded oligonucleotides that contained all possible single base pair substitutions at each position of the 14-bp minimal recognition element; 60-fold molar excesses of these oligonucleotides were then tested for their ability to compete with 7–2B for binding to OBPH7. The effect of each substitution was determined by quantifying the radioactivity of the shifted band and expressing this value as the percentage of binding inhibition relative to that of the wild-type 7–2E or 7–2Eext oligonucleotide.
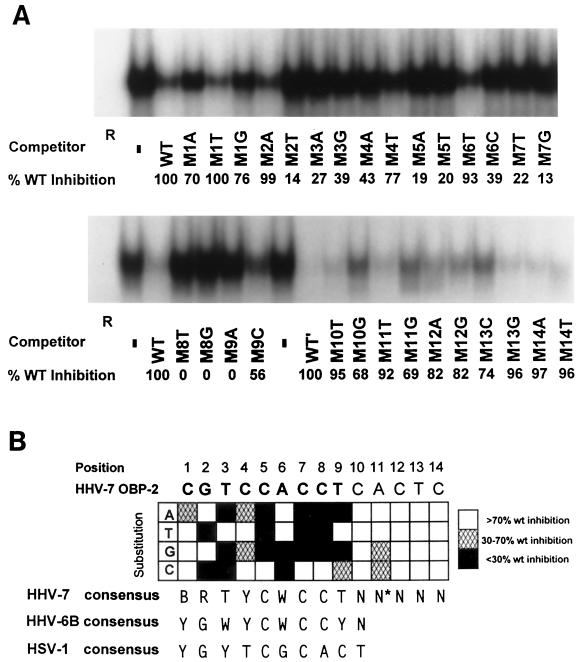
A representative set of the competition experiments is presented in Fig. 8A. Mutated oligonucleotides such as M2T (substitution with a thymidine at position 2), which contained a substitution that resulted in <70% inhibition of binding compared to the wild-type oligonucleotide, were considered to have a nonpermissive substitution. Mutated oligonucleotides such as M2A (Fig. 8A), which retained its ability to compete for binding (>70%) as compared to that of the wild-type oligonucleotide, contained a permissive base pair. Designation as a permissive or nonpermissive change was based on quantitative trends observed in at least three independent experiments. The results of these experiments are summarized in Fig. 8B and allow deduction of the OBPH7 consensus sequence, BRTYCWCCT.
FIG. 8.
Effect of substitutions in the 14-bp core OBP-2 sequence and the resulting OBPH7 consensus recognition sequence. (A) Saturation mutagenesis of the minimal OBPH7 recognition sequence (Table 1, oligonucleotide L0R5). At each position of the 14-bp minimal recognition sequence, oligonucleotides were generated that contained changes to the other three possible base pairs. The label beneath each lane of the gel indicates the position and change made in the competitor used. Sixty-fold molar excesses of unlabeled mutated oligonucleotides were used in competitive EMSA against 32P-labeled 7–2B. The amount of residual shifted radioactivity after competition was quantified by PhosphorImager analysis (Molecular Dynamics). The inhibition of binding of 32P-labeled 7–2B DNA to truncated OBPH7 by each competitor DNA duplex is shown beneath the gel as the percent inhibition relative to the wild-type DNA duplex (WT). The other competitor DNA duplexes shown in panel B were analyzed similarly (data not shown). Positions 10 to 14 were analyzed in the context of the sequence of the longer 7–2Eext oligonucleotide, AATTAGCGTCCACCTCACTCGTAATAGT (WT′), to avoid effects that might be more dependent on proximity to the end of the oligonucleotide than sequence alone. −, no competitor was added. (B) Summary of saturation mutagenesis and the resulting consensus recognition sequence. Open blocks indicate that the given alteration at that position did not result in loss of recognition (competition was at least 70% of WT) and is therefore a permissive change. Hatched and solid blocks indicate that the alteration reduced the binding ability partially or severely, respectively. These designations were based on quantitative trends observed in at least three independent experiments. The asterisk on the N at position 11 indicates that although two of the changes were slightly below the cutoff for recognition, the slight gradient in recognition of all alterations at this position did not enable identification of a clearly preferred sequence. In the consensus sequence, B is a T, G, or C; R is a G or A; Y is a T or C; W is a T or A; and N is any nucleotide.
OBPH7 tolerates less sequence variation from positions 2 through 9 (Fig. 8B). While there was some degree of sequence preference at positions 10 through 14, the gradient was so slight that a consensus sequence could not be determined. In comparison with the OBPH6B consensus at positions 1 and 2, the OBPH7 consensus recognition sequence is more permissive. At positions 3 and 9 there was overlap with the OBPH6B consensus, but OBPH7 is less permissive than OBPH6B. From the consensus sequences for OBPH6B and OBPH7, YCWCC was identified as the central core sequence for roseolovirus OBP recognition, in comparison to the C—C core shared with OBPH1 and the other alphaherpesviruses. The OBP-2 site and the putative OBP-1 site are the only sites in the HHV-7 oriLyt that exactly match the experimentally derived OBPH7 consensus sequence.
Identification of OBP-1 as a low-affinity binding site.
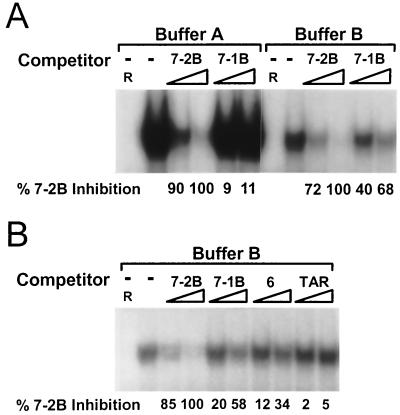
As described above, the only sites in the HHV-7 oriLyt that exactly match the OBPH7 consensus sequence are the OBP-2 site we identified in the previous mapping experiments and the putative OBP-1 site, which is in a position and context that suggest its possible function as an OBP-binding site. Because oligonucleotides containing the putative OBP-1 site were not recognized in any of the preceding experiments, we examined the effect of different buffer conditions on DNA binding activity. As measured by signal intensity in the absence of competitor, the ability of OBPH7 to bind an oligonucleotide that contains the OBP-2 site, 7–2B, was reduced 10-fold in buffer B (the buffer optimized for OBPH6B) (14) compared to buffer A (the buffer optimized for OBPH7), but the protein-DNA interactions were concomitantly altered to allow binding to the 34-bp 7–1B oligonucleotide that contained the OBP-1 sequence (Fig. 9). As shown in Fig. 9B, a second oligonucleotide that contained the OBP-1 sequence, the 55-bp oligonucleotide 6 (Fig. 4A), was also bound by OBPH7 in buffer B. This reactivity was not due to a loss of specificity in buffer B conditions, since the nonspecific TAR oligonucleotide does not compete for binding to OBPH7 (Fig. 9B) and the oligonucleotide M3A, which contains a single nonpermissive mutation, was not recognized (data not shown). OBPH7 had a higher affinity for oligonucleotides containing the OBP-2 site than for those containing the OBP-1 site in both binding conditions.
FIG. 9.
Identification of a second, lower-affinity OBP site (OBP-1) by competitive EMSA. (A) Competition with 16- and 80-fold molar excesses of unlabeled DNA duplexes containing HHV-7 OBP-2 (7–2B) and OBP-1 (7–1B, GGAGGGTTCATTGATCCTCCTTGCCTGCAATTCT) with 32P-labeled 7–2 for binding to OBPH7 in buffers A and B (see Materials and Methods). The amount of residual shifted target was quantitated by PhosphorImager analysis. The percentage of inhibition of binding to 32P-labeled 7–2 DNA relative to 7–2B at 80-fold molar excesses by each competitor DNA duplex is shown beneath the gel. −, no competitor present; R, negative control lysate. (B) Conditions are the same as described for panel A, except that binding buffer B was used. Oligonucleotide 6 contains the OBP-1 site and is described in the legend to Fig. 4. TAR is described in the legend for Fig. 5.
Heteroduplex analysis suggests that OBPH7 interacts with one face of the DNA helix.
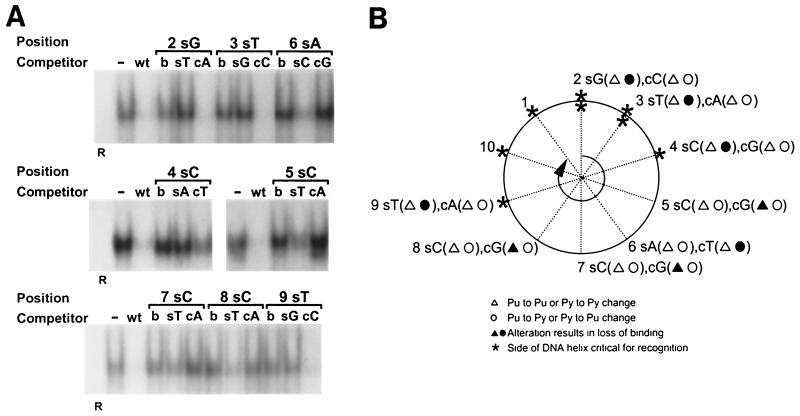
To identify the strand-specific base interactions required for OBPH7 recognition of the high-affinity OBP-2 site, we generated heteroduplex oligonucleotides derived from the 7–2E sequence (Fig. 7C) that contain single mismatched base pairs at positions 2 through 9. The heteroduplex oligonucleotides and corresponding oligonucleotides with complementary substitutions on both strands (nonheteroduplex) were compared for their ability to compete for binding against the wild-type sequence in oligonucleotide 7–2E. Sense strand alterations at positions 2, 3, 4, and 9 as well as complementary strand alterations at positions 5 through 8 resulted in loss of recognition by OBPH7 at levels similar to or greater than alterations on both strands at these positions (Fig. 10A). Changes on the opposite strand at these positions resulted in at least a threefold-greater inhibition of binding than the alteration on both strands, indicating that the principal interaction is not on this strand at this position. For example, when both strands (Fig. 10A, lane b) at position 2 were changed from a GC to a TA base pair, the resulting oligonucleotide did not compete for binding. A sense strand changed to a T at position 2 (lane sT) was also unable to compete; this is in contrast to the effect of a complementary strand change to an A (lane cA), which competes as well as the wild-type sequence. This indicates that the base on the sense strand of the oligonucleotide at position 2 is a possible contact point for OBPH7.
FIG. 10.
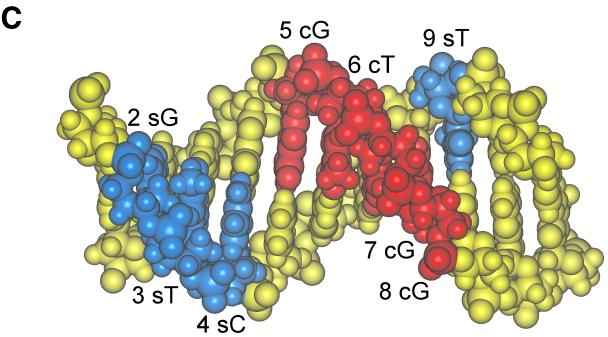
EMSA to measure recognition of heteroduplexes. (A) Heteroduplex oligonucleotides containing single mismatched base pairs at positions 2 through 9 were created by annealing a wild-type 7–2Eext oligonucleotide to complementary oligonucleotides containing single nucleotide substitutions. For positions 2 through 9, 60-fold molar excesses of heteroduplex oligonucleotides with a mutation on the 5′-to-3′ sense strand (s) or a mutation of the 3′-to-5′ complementary strand (c) were compared for their ability to compete for binding with truncated OBPH7 against oligonucleotides containing the mutation on both strands (b). −, no competitor; WT, wild-type 7–2E is the competitor; R, negative control protein lysate. (B) Helical wheel representation of heteroduplex analysis. The 9-bp OBP-2 core sequence is arranged on a helical wheel to approximate the 10.4 base residues per turn of a B-form DNA helix. For each position, two heteroduplex oligonucleotides were synthesized with changes on the sense strand (first set of parentheses) and two were made with changes on the complementary strand (second set of parentheses). Purine-to-purine and pyrimidine-to-pyrimidine changes are indicated by triangles. Purine-to-pyrimidine and pyrimidine-to-purine changes are indicated as circles. Solid symbols indicate that strand-specific alteration is responsible for loss of recognition at that position in the core sequence. Asterisks indicate the side of the DNA helix that is critical for recognition; thus, a change on the complementary strand that affects recognition is indicated by an asterisk on the opposite side of the helix (e.g., the T at position 6). (C) A B-form DNA model of the OBP-2 site, CGTCCACCTCA, was produced using Insight II, Release 2000 (Molecular Simulations, Incorporated, San Diego, Calif.). Labeled positions indicate strand mismatches that resulted in the loss of recognition by OBPH7.
The strand-specific alterations that resulted in loss of recognition and thus are considered possible points of contact with OBPH7 are summarized in Fig. 10B and C. In the helical wheel representation of the OBP-2 sequence, the side of the DNA helix that is critical for recognition is found along one face (Fig. 10B). In a simulation of the OBP-2 site as B-form DNA, 7 of the 8 potential points of contact in the core recognition element examined in Fig. 10A are present in the major groove (Fig. 10C). These results indicate that OBPH7 may interact with the core of its OBP site in the major groove along one face of the DNA helix. Position 9 lies outside the major groove, but along the same face, and may reflect possible phosphate interactions that are disrupted by a strand mismatch.
DISCUSSION
We have characterized the interaction of OBPH7 with the HHV-7 oriLyt. As with alphaherpesviruses, this is likely the first step in a cascade of events required for initiation of viral replication. Specifically, we determined that (i) the carboxyl-terminal 304 aa are sufficient for DNA binding activity, (ii) there are two binding sites with markedly different affinities for OBPH7 in the HHV-7 oriLyt, (iii) BRTYCWCCT is the OBPH7 consensus recognition sequence, and (iv) strand-specific interactions in the 9-bp OBP core occur along one face of the DNA helix.
OBPH7 sequence recognition.
The minimal OBPH7 recognition element is 14 bp in length, similar to the 13-bp minimal element of OBPH6B (15) and the 15-bp element of OBPH1 (13). For both OBPH6B and OBPH7, additional sequence was required on the right flank of the 9-bp core recognition sequence, suggesting that the flanking sequence is required to stabilize the protein-DNA interaction. The roseolovirus OBPs have identical sequence specificity in the central 5-bp region of the 9-bp recognition core (YCWCC); outside the 5-bp region, their specificity diverges. This difference in specificity does not appear to have been substantially influenced by differences in the buffers used during determination of the consensus sequences (data not shown). For both viruses, the consensus sequences are consistent with the authentic OBP-1 sites. Ultimate proof would require evaluation of these sequences under physiologic conditions such as transient replication assays.
Using the OBPH7 consensus recognition sequence to search the genomes of HHV-7 strains JI (23) and RK (22) for exact matches, we identified 11 potential OBP sites in addition to the 2 identified in the HHV-7 oriLyt in both genomes. Three are present in each copy of the genome-bounding direct repeat elements (a total of six copies per genome), with the remaining sites scattered in the unique segment. None of the sequences is linked to another in a manner suggestive of an authentic origin region.
Like OBPH6B (15) and OBPH1 (13, 25), OBPH7 required strand-specific base interactions within the OBP recognition core that align along one face of the DNA helix in the major groove. These results are consistent with the higher-resolution model for OBPH1, in which an alpha helix in its carboxyl-terminal DNA binding domain fits into the major groove to mediate specific base contacts at an OBP site (9, 13, 25).
Unlike OBPH6B, which recognizes both OBP sites in the HHV-6 oriLyt with comparable affinities, OBPH7 has a higher affinity for OBP-2 than OBP-1. The OBP-1 site fits the OBPH7 consensus sequence, yet there are three differences in the 9-bp core between OBP-1 (GATCCTCCT) and OBP-2 (CGTCCACCT). In competition experiments utilizing a buffer (buffer A) in which the OBP-2 site is bound strongly while OBP-1 is not detectably bound, there was a progressive decrease in binding to oligonucleotides that contained various combinations of the OBP-1-derived sequence (data not shown). This indicates that the differences in the 9-bp recognition core compound to create an overall low-affinity OBP-1 site.
Mutated OBPH1 binding sites that are not individually bound in vitro have less of an impact on binding by full-length OBPH1 in the context of the entire oriS (9). This cooperativity is ablated upon disruption of the amino-terminal multimerization domain of OBPH1 (12). Truncated OBPH7 recognized OBP-1 and OBP-2 with markedly different affinities, suggesting that cooperative interactions through multimerization of full-length OBPH7 may be required at the OBP sites in the HHV-7 oriLyt. Protein-protein interactions with OBPH7 bound to the high-affinity OBP-2 site may increase the stability of an OBP complex at the low-affinity OBP-1 site. Unlike OBPH7, OBPH6B binds its two sites with comparable affinities. This indicates that OBP protein-DNA interactions differ between OBPHB6 and OBPH7.
Comparison of the OBP-binding regions of roseoloviruses and alphaherpesviruses.
The HHV-7 oriLyt has features conserved in the lytic origin regions of alphaherpesviruses and of HHV-6A and HHV-6B (Fig. 11). Like HHV-6A and HHV-6B, the HHV-7 AT-rich spacer region is longer than in alphaherpesvirus ori regions and has imperfect dyad symmetry. Unlike the other origin regions shown here, the AT-rich spacer element in HHV-7 contains few AT repeats.
FIG. 11.
Comparison of the OBP sites in the ori regions of alphaherpesviruses and roseoloviruses. OBP-binding regions of the oriLyts of alphaherpesviruses and roseoloviruses are shown. HSV1 L, HSV-1 oriL; HSV1 S, HSV-1 oriS; HSV2 L, HSV-2 oriL; HSV2 S, HSV-2 oriS; EHV1 S, equine herpesvirus 1 oriS; PRV, pseudorabies virus; VZV, varicella-zoster virus; MDV2, Marek's disease virus 2; I, II, and III, Box I, II, and III of HSV-1 oriS. Residues that match the HHV-7 consensus are shaded. Horizontal arrows indicate a structure with dyad symmetry in the HHV-7 sequence. The HHV-7 Box III-like sequence is underlined.
Interestingly, the HHV-7 oriLyt contains a sequence that has a 9-of-12 match with the HSV Box III element and a 6-of-9 match with the OBPH7 consensus recognition sequence; a similar sequence is not present in the HHV-6A and HHV-6B oriLyts. This element is found in a position in HHV-7 that corresponds to its location in HSV, that is, immediately to the left of the Box I site. Mutations in Box III decrease the replication of an oriS-containing plasmid in transient replication assays (19). However, the HSV Box III site is detectably bound only in the context of cooperative binding of full-length OBPH1 to an oriS segment containing the Box I and II sites (9, 10). Recent data suggest that Box I and III may interact to form a secondary structure through a 6-bp region of dyad symmetry in oriS that is required for stable interaction with OBPH1 (3). There is also an element of dyad symmetry (5 bp) between HHV-7 OBP-1 and its Box III-like site.
The OPB-1 site-containing oligonucleotides 6 and 7–1B both had comparable affinities for OBPH7, but only oligonucleotide 6 contains the Box III-like site. Further analysis of the interaction of full-length OBPH7 with oligonucleotides containing mutations in the Box III site is required to determine what impact it may have on recognition of the flanking OBP-1 site. HHV-7-infected cells replicated an HHV-6 oriLyt-containing plasmid (27), and the HHV-6 oriLyt region lacks the 12-bp HSV Box III-like site found in the HHV-7 oriLyt. This suggests that the Box III-like element is probably not essential for possible cooperative interactions of OBPH7 at the origin of replication by other HHV-7 replication factors.
Differences in the interaction of roseolovirus OBPs with their oriLyts.
OBPH7 and OBPH6B have similarities in their DNA binding properties, namely, that their consensus recognition sequences overlap and they recognize an identical 9-bp element in their oriLyt regions. However, in transient replication-infection assays, the HHV-7 replication machinery replicated an HHV-6 oriLyt-containing plasmid while an HHV-7 oriLyt-containing plasmid did not replicate in HHV-6B-infected cells (27). The basis for the lack of reciprocity may simply lie in differences in sequence recognition between OBPH6B and OBPH7: the 9-bp core of the HHV-6B OBP-1 site differs by a single base from the OBPH7 consensus recognition sequence while the HHV-7 OBP-1 site and OBPH6B consensus sequence differ at two positions. This interaction could be further affected by differences in proximal flanking sequences. In addition, as described above, the HHV-7 oriLyt lacks the long IDRs and putative DNA unwinding element of the HHV-6 oriLyts but contains a putative Box III-like element. The sum of these differences within and around the OBP sites is likely to affect OBPH6B and OBPH7 activities in the oriLyts.
Much can be learned about the mechanism of DNA replication initiation of roseoloviruses from a comparative functional analysis of their closely related, yet distinct, OBP proteins. We are presently studying reciprocity at the level of DNA binding between the OBPs and OBP sites of HHV-6 and HHV-7.
ACKNOWLEDGMENTS
L.T.K. was supported in part through an appointment to the Research Participation Program at the Centers for Disease Control and Prevention administered by the Oak Ridge Institute for Science and Education.
We thank Elizabeth Neuhaus for constructing the DNA model, Felicia Stamey for sequencing the OBP constructs, the CDC Biotechnology Core Facility for oligonucleotide synthesis, and James Gathany for photography.
REFERENCES
- 1.Anders D G, Kacica M A, Pari G, Punturieri S M. Boundaries and structure of human cytomegalovirus oriLyt, a complex origin for lytic-phase DNA replication. J Virol. 1992;66:3373–3384. doi: 10.1128/jvi.66.6.3373-3384.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anders D G, Punturieri S M. Multicomponent origin of cytomegalovirus lytic-phase DNA replication. J Virol. 1991;65:931–937. doi: 10.1128/jvi.65.2.931-937.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aslani A, Simonsson S, Elias P. A novel conformation of the herpes simplex virus origin of DNA replication recognized by the origin binding protein. J Biol Chem. 2000;275:5880–5887. doi: 10.1074/jbc.275.8.5880. [DOI] [PubMed] [Google Scholar]
- 4.Black J B, Burns D A, Goldsmith C S, Feorino P M, Kite-Powell K, Schinazi R F, Krug P W, Pellett P E. Biologic properties of human herpesvirus 7 strain SB. Virus Res. 1997;52:25–41. doi: 10.1016/s0168-1702(97)00102-0. [DOI] [PubMed] [Google Scholar]
- 5.Black J B, Pellett P E. Human herpesvirus 7. Rev Med Virol. 1999;9:245–262. doi: 10.1002/(sici)1099-1654(199910/12)9:4<245::aid-rmv253>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 6.Boehmer P E, Lehman I R. Herpes simplex virus DNA replication. Annu Rev Biochem. 1997;66:347–384. doi: 10.1146/annurev.biochem.66.1.347. [DOI] [PubMed] [Google Scholar]
- 7.Dewhurst S, Dollard S C, Pellett P E, Dambaugh T R. Identification of a lytic-phase origin of DNA replication in human herpesvirus 6B strain Z29. J Virol. 1993;67:7680–7683. doi: 10.1128/jvi.67.12.7680-7683.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dewhurst S, Krenitsky D M, Dykes C. Human herpesvirus 6B origin: sequence diversity, requirement for two binding sites for origin-binding protein, and enhanced replication from origin multimers. J Virol. 1994;68:6799–6803. doi: 10.1128/jvi.68.10.6799-6803.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elias P, Gustafsson C M, Hammarsten O. The origin binding protein of herpes simplex virus 1 binds cooperatively to the viral origin of replication oris. J Biol Chem. 1990;265:17167–17173. [PubMed] [Google Scholar]
- 10.Elias P, Gustafsson C M, Hammarsten O, Stow N D. Structural elements required for the cooperative binding of the herpes simplex virus origin binding protein to oriS reside in the N-terminal part of the protein. J Biol Chem. 1992;267:17424–17429. [PubMed] [Google Scholar]
- 11.Gompels U A, Nicholas J, Lawrence G, Jones M, Thomson B J, Martin M E, Efstathiou S, Craxton M, Macaulay H A. The DNA sequence of human herpesvirus-6: structure, coding content, and genome evolution. Virology. 1995;209:29–51. doi: 10.1006/viro.1995.1228. [DOI] [PubMed] [Google Scholar]
- 12.Hazuda D J, Perry H C, McClements W L. Cooperative interactions between replication origin-bound molecules of herpes simplex virus origin-binding protein are mediated via the amino terminus of the protein. J Biol Chem. 1992;267:14309–14315. [PubMed] [Google Scholar]
- 13.Hazuda D J, Perry H C, Naylor A M, McClements W L. Characterization of the herpes simplex virus origin binding protein interaction with oris. J Biol Chem. 1991;266:24621–24626. [PubMed] [Google Scholar]
- 14.Inoue N, Dambaugh T R, Rapp J C, Pellett P E. Alphaherpesvirus origin-binding protein homolog encoded by human herpesvirus 6B, a betaherpesvirus, binds to nucleotide sequences that are similar to ori regions of alphaherpesviruses. J Virol. 1994;68:4126–4136. doi: 10.1128/jvi.68.7.4126-4136.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inoue N, Pellett P E. Human herpesvirus 6B origin-binding protein: DNA-binding domain and consensus binding sequence. J Virol. 1995;69:4619–4627. doi: 10.1128/jvi.69.8.4619-4627.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lawrence G L, Nicholas J, Barrell B G. Human herpesvirus 6 (strain U1102) encodes homologues of the conserved herpesvirus glycoprotein gM and the alphaherpesvirus origin-binding protein. J Gen Virol. 1995;76:147–152. doi: 10.1099/0022-1317-76-1-147. [DOI] [PubMed] [Google Scholar]
- 17.Lee S S, Lehman I R. The interaction of herpes simplex type 1 virus origin-binding protein (UL9 protein) with Box I, the high affinity element of the viral origin of DNA replication. J Biol Chem. 1999;274:18613–18617. doi: 10.1074/jbc.274.26.18613. [DOI] [PubMed] [Google Scholar]
- 18.Lindquester G J, O'Brian J J, Anton E D, Greenamoyer C A, Pellett P E, Dambaugh T R. Genetic content of a 20.9 kb segment of human herpesvirus 6B strain Z29 spanning the homologs of human herpesvirus 6A genes U40–57 and containing the origin of DNA replication. Arch Virol. 1997;142:103–123. doi: 10.1007/s007050050062. [DOI] [PubMed] [Google Scholar]
- 19.Martin D W, Deb S P, Klauer J S, Deb S. Analysis of the herpes simplex virus type 1 OriS sequence: mapping of functional domains. J Virol. 1991;65:4359–4369. doi: 10.1128/jvi.65.8.4359-4369.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masse M J, Karlin S, Schachtel G A, Mocarski E S. Human cytomegalovirus origin of DNA replication (oriLyt) resides within a highly complex repetitive region. Proc Natl Acad Sci USA. 1992;89:5246–5250. doi: 10.1073/pnas.89.12.5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masse M J O, Messerle M, Mocarski E. The location and sequence composition of the murine cytomegalovirus replicator (oriLyt) Virology. 1997;230:350–360. doi: 10.1006/viro.1997.8473. [DOI] [PubMed] [Google Scholar]
- 22.Megaw A G, Rapaport D, Avidor B, Frenkel N, Davison A J. The DNA sequence of the RK strain of human herpesvirus 7. Virology. 1998;244:119–132. doi: 10.1006/viro.1998.9105. [DOI] [PubMed] [Google Scholar]
- 23.Nicholas J. Determination and analysis of the complete nucleotide sequence of human herpesvirus 7. J Virol. 1996;70:5975–5989. doi: 10.1128/jvi.70.9.5975-5989.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Romi H, Singer O, Rapaport D, Frenkel N. Tamplicon-7, a novel T-lymphotropic vector derived from human herpesvirus 7. J Virol. 1999;73:7001–7007. doi: 10.1128/jvi.73.8.7001-7007.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simonsson S, Samuelsson T, Elias P. The herpes simplex virus type 1 origin binding protein. Specific recognition of phosphates and methyl groups defines the interacting surface for a monomeric DNA binding domain in the major groove of DNA. J Biol Chem. 1998;273:24633–24639. doi: 10.1074/jbc.273.38.24633. [DOI] [PubMed] [Google Scholar]
- 26.Stamey F R, Dominguez G, Black J B, Dambaugh T R, Pellett P E. Intragenomic linear amplification of human herpesvirus 6B oriLyt suggests acquisition of oriLyt by transposition. J Virol. 1995;69:589–596. doi: 10.1128/jvi.69.1.589-596.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Loon N, Dykes C, Deng H, Dominguez G, Nicholas J, Dewhurst S. Identification and analysis of a lytic-phase origin of DNA replication in human herpesvirus 7. J Virol. 1997;71:3279–3284. doi: 10.1128/jvi.71.4.3279-3284.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vink C, Beuken E, Bruggeman C A. Cloning and functional characterization of the origin of lytic-phase DNA replication of rat cytomegalovirus. J Gen Virol. 1997;78:2963–2973. doi: 10.1099/0022-1317-78-11-2963. [DOI] [PubMed] [Google Scholar]
- 29.Wyatt L S, Frenkel N. Human herpesvirus 7 is a constitutive inhabitant of adult human saliva. J Virol. 1992;66:3206–3209. doi: 10.1128/jvi.66.5.3206-3209.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]