Abstract
Bluetongue virus (BTV) is an arthropod-borne virus transmitted by Culicoides species to vertebrate hosts. The double-capsid virion is infectious for Culicoides vector and mammalian cells, while the inner core is infectious for only Culicoides-derived cells. The recently determined crystal structure of the BTV core has revealed an accessible RGD motif between amino acids 168 to 170 of the outer core protein VP7, whose structure and position would be consistent with a role in cell entry. To delineate the biological role of the RGD sequence within VP7, we have introduced point mutations in the RGD tripeptide and generated three recombinant baculoviruses, each expressing a mutant derivative of VP7 (VP7-AGD, VP7-ADL, and VP7-AGQ). Each expressed mutant protein was purified, and the oligomeric nature and secondary structure of each was compared with those of the wild-type (wt) VP7 molecule. Each mutant VP7 protein was used to generate empty core-like particles (CLPs) and were shown to be biochemically and morphologically identical to those of wt CLPs. However, when mutant CLPs were used in an in vitro cell binding assay, each showed reduced binding to Culicoides cells compared to wt CLPs. Twelve monoclonal antibodies (MAbs) was generated using purified VP7 or CLPs as a source of antigen and were utilized for epitope mapping with available chimeric VP7 molecules and the RGD mutants. Several MAbs bound to the RGD motif on the core, as shown by immunogold labeling and cryoelectron microscopy. RGD-specific MAb H1.5, but not those directed to other regions of the core, inhibited the binding activity of CLPs to the Culicoides cell surface. Together, these data indicate that the RGD motif present on BTV VP7 is responsible for Culicoides cell binding activity.
Orbiviruses (within the family Reoviridae), are vectored to vertebrate species by arthropods (e.g., gnats, mosquitoes and ticks) and are able to replicate in both hosts. Bluetongue virus (BTV) is the prototype virus of the genus and is transmitted by gnats (Culicoides species), causing diseases of economic importance in ruminants in many parts of the world. Vector-virus interactions play a crucial role in vector-borne disease epidemiology. The spread of Culicoides species from BTV-endemic to non-BTV (or related African horsesickness virus, AHSV, and epizootic hemorrhagic disease virus EHDV, of deer) regions of the world in the past highlights the concern that these viruses are a threat to regions of the world that are presently free from them.
The initiation of a virus infection involves virus binding to ligands on the cell surface prior to cell entry by a number of mechanisms (depending on the virus). Like many other viruses, BTV appears to utilize a protein molecule(s) of mammalian cells as a receptor (20); however, it is also possible that alternative receptors may be utilized in different tissues and in different species and as accessory molecules.
BTV has a genome composed of 10 segments of double-stranded RNA packaged within a double icosahedral capsid. The outer capsid layer, which is lost at an early stage of the infection process, is composed of two major structural proteins (VP2 and VP5). These proteins are involved in host cell attachment and penetration during the initial stages of infection (22). After entry into the cells, the virus is uncoated (by removal of VP2 and VP5) to yield a transcriptionally active core particle which is composed of two major proteins (VP7 and VP3) and three minor proteins (VP1, VP2, and VP3) in addition to the double-stranded RNA genome (28, 55, 56). Since BTV and other orbiviruses are transmitted between their mammalian hosts by the bite of insect vectors, the viruses must remain infectious in the insect gut, an environment which can remove the BTV outer layers. This implies that BTV particles, either lacking the complete outer capsid proteins or with modified outer capsid proteins, are infectious for the insect vector. Indeed, Mertens and coworkers have demonstrated that BTV cores are highly infectious for the Culicoides vector and Culicoides-derived cell culture (KC cells) (42–44) and that the specific infectivity of in vitro-generated cores for the KC cells was directly comparable to that of the intact virus particles. Cores were similarly infectious when oral infectivity studies were carried out using Culicoides species. The high level of core-associated infectivity for KC cells suggests that the initial stages of core-cell interaction and entry use an alternate entry mechanism to that used by complete particles.
The outermost BTV core protein, VP7, is the most accessible protein of the BTV core and suggests that it may participate in vector cell entry (67). VP7 has an arginine-glycine-aspartate (RGD) tripeptide motif present at amino acid residues 168 to 170, one of the ligand sites recognized by host proteins that belong to the integrin family, such as fibronectin, vitronectin, and fibrinogen (29, 57, 58). From X-ray crystallographic structures, the RGD motif in BTV VP7 is located on the upper domain of the two domain molecule (1, 19) and appears to be accessible on the surface. The RGD motif has a conformation similar to that seen in the RGD motif of the VP1 protein of foot-and-mouth disease virus (FMDV) and γ-crystallin, which attaches to αV β3 integrin (6, 19, 20, 32, 38, 50, 51, 66). It is plausible, therefore, that RGD-integrin binding is an initial step of BTV core attachment to insect cells.
In this study we have evaluated the role of the VP7 RGD sequence in cell attachment activity by taking advantage of an established biological assay system which allows synthesis and purification of high-yield recombinant core-like particles (CLPs) from Sf9 insect cells (17). Not only are the CLPs composed of VP7 and VP3 antigenically equivalent to the BTV core but also both have similar three-dimensional (3D) structures when analyzed by cryoelectron microscopy (cryo-EM) (23–25). Thus, CLPs mimic the authentic core and could be used as a surrogate of authentic cores in cell binding studies. We have introduced point mutations within the RGD tripeptide of the VP7 protein to generate VP7 with an altered RGD motif. Three such RGD-targeted VP7 mutants were expressed using the baculovirus expression system, and their antigenic and biochemical properties were characterized. Each mutant derivative also assembled into CLPs in the presence of the VP3 protein, implying that the RGD site is accessible and is not involved in protein-protein interactions during core assembly. In addition, a panel of 12 monoclonal antibodies (MAbs) was raised against the VP7 protein and characterized. An antigenic site corresponding to the RGD sequence of VP7 was subsequently mapped by reacting the antibodies with the VP7 mutants in enzyme-linked immunosorbent assay (ELISA) based tests and by using immunogold labeling of CLPs. The general location of antibody binding was confirmed by cryo-EM studies. When these mutant CLPs were assessed for their binding activities in Culicoides cells, each preparation showed a decreased level of binding in comparison to the wild-type (wt) CLPs. Together, the data presented here demonstrate that the VP7 RGD motif is involved in the binding of the BTV core into Culicoides cells.
MATERIALS AND METHODS
Viruses and cells.
Spodoptera frugiperda (Sf9) cells were grown in suspension or monolayer cultures at 28°C in TC100 medium supplemented with 10% fetal calf serum. Derivatives of Autographa californica nuclear polyhedrosis virus containing the wt BTV-10 VP7 or BTV-17 VP3 gene (Ac10BTV7 and Ac17BTV3) and the BTV-10 VP7 mutants were plaque purified and propagated as described previously (17). The KC cell line, derived from the embryos of AK colony insects (63), was kindly provided by Sally Wechsler, US Department of Agriculture Center, Laramie, Wyo., and were grown at 28°C in Schneider's medium (Sigma) supplemented with 10% FCS.
Construction of recombinant transfer vectors and isolation of recombinant baculoviruses expressing mutant VP7 proteins.
Mutations in VP7 were made in the baculovirus transfer vector pAcCL29 (37), using synthetic oligonucleotides and the method described by Kunkel et al. (35). The wild-type BTV-10 VP7 was derived from the transfer vector pAcYM1.10BTV7 (48). The oligonucleotides used for mutagenesis and the resulting amino acid changes are shown in Table 1. All the oligonucleotides represent the coding-strand complement, with mutated anticodons underlined. The arginine residue (Arg-168) in the BTV-10 VP7 gene was mutated to alanine to create pAcCL29BTV10.7R168A. A second mutation was also introduced to convert the aspartic residue (Asp-170) to glutamine or leucine in pAcCL29BTV10.7RD170AQ and pAcCL29BTV10.7RD170AL, respectively. The alterations in the recombinant plasmids were verified by sequence analyses (59). The Lipofection technique was used to cotransfect monolayers of Sf9 cells with recombinant transfer vectors and Bsu36I triple-cut A. californica nuclear polyhedrosis virus DNA (34). Recombinant baculoviruses were selected on the basis of their LacZ-negative phenotypes, plaque purified, and propagated as described elsewhere (34). Three recombinant viruses, AGD, AGQ and AGL, were selected for further investigation.
TABLE 1.
Oligonucleotide primers used in site-directed mutagenesis
| Mutant name | Nucleotide sequencea |
|---|---|
| BTV-10.7 | 5′-TACATCTCCTCTGCGAGG-3′ |
| R168A | 5′-TACATCTCCTGCCGCTCCAGC-3′ |
| RD170AQ | 5′-CTGTTGTACTTCTCCTGCCGCTCCAGC-3′ |
| RD170AL | 5′-CTGTTGTACAAGTCCTGCCGCTCCAGC-3′ |
The oligonucleotides used for site-directed mutagenesis represent the complement of the coding strand of VP7 gene. The underlined triplets represent the altered sequences.
SDS-PAGE and Western blot analyses.
The protein samples were boiled in protein dissociation buffer (10 mM Tris-HCl [pH 6.8], 10% [vol/vol] β-mercaptoethanol, 10% [wt/vol] sodium dodecyl sulfate [SDS], 25% [vol/vol] glycerol, 0.02% [wt/vol] bromophenol blue) at 100°C for 10 min and resolved by SDS-polyacrylamide gel electrophoresis (PAGE) (10% polyacrylamide) followed by staining with Coomassie Brilliant blue. For Western blot analyses proteins were transferred from gels onto a polyvinylidene difluoride membrane by standard blotting procedures. The membranes were incubated with an anti-BTV-10 antiserum diluted 1:1,000 in blocking buffer containing 5% (wt/vol) skimmed milk and 0.05% (vol/vol) Tween 20 in phosphate-buffered saline (PBS) and then with a secondary antibody conjugated with alkaline phosphatase and developed with the alkaline phosphatase substrate (NBT-BCIP [GIBCO-BRL] in 0.1 M Tris-HCl [pH 8.5]–0.1 M NaCl–0.05M MgCl2).
Purification of recombinant VP7 protein and BTV CLPs.
The wt and mutant BTV-10 VP7 proteins were purified as described previously (1). The infected Sf9 cells were infected with either AcBTV10.7 or one of the three recombinant viruses (AGD, AGQ, and AGL) at a multiplicity of infection of 5 PFU per cell, harvested at 48 h postinfection, washed in PBS, resuspended in cold TNN (100 mM NaCl, 50 mM Tris-HCl [pH 8.0], 0.5% Nonidet P-40), and homogenized at 4°C as described above. Cell debris and nuclei were precipitated by centrifugation (10 min at 16,000 × g). Ice-cold saturated ammonium sulfate in 100 mM Tris-HCl (pH 7.5) was added to the cytoplasmic cell extracts to a final concentration of 20%. The protein was precipitated by centrifugation, resuspended in 10 mM Tris HCl (pH 8.5), isolated from the insoluble material following removal by pelleting at 10,000 × g for 10 min, and then dialyzed against the same buffer at 4°C overnight. The extract was stored at −20°C prior to analysis.
Baculovirus-expressed CLPs were purified as described previously (17). In brief, Sf9 cells were coinfected with AcBTV17.3 virus and the mutant VP7 baculoviruses at a multiplicity of infection of 5 PFU per cell. After incubation at 28°C for 48 h, the cells were harvested and the baculovirus-expressed CLPs were purified by a sucrose gradient (66%, [wt/vol] and 30% [wt/vol]) centrifugation. The recovered CLPs were analyzed for the presence of VP3 and VP7 proteins by SDS-PAGE (10% polyacrylamide) and Western blot analysis using anti-BTV-10 polyclonal antibodies, and the morphology was verified by EM.
Production of MAbs against BTV VP7.
Female BALB/c mice were inoculated intraperitoneally three times, at 2-week intervals, with 100 μl of either sucrose gradient-purified baculovirus-expressed recombinant wt CLP or partially purified baculovirus-expressed recombinant VP7 at a concentration of ∼500 μg/ml. The animals were rested for 3 weeks before intravenous inoculation with a further 100 μl of the same protein. Their spleens were removed 3 days later. Fusions were carried out by using polyethylene glycol 1500 (BCL) and the myeloma cell line SP2/o (16). VP7-specific antibody production was detected and verified by indirect ELISA, indirect immunofluorescence, and Western blotting using recombinant VP7 antigen and other baculovirus recombinant proteins as a control (18). Antibody-positive clones were isolated and maintained in RPMI 1640 (GIBCO) with 15% Myoclone Super Plus fetal calf serum (GIBCO). Peritoneal macrophages were added to these cultures to assist growth of the clones. Mouse ascites were induced by intraperitoneal inoculation of 106 to 107 hybridoma cells in BALB/c mice pretreated with 0.5 ml of pristane (2,6,10,14-tetramethylpentadecane) 3 to 6 days prior to the inoculation.
Antibody purification.
Anti-BTV-10 VP7 antibodies in ascitic fluid (AF) were purified using an affinity chromatography kit (Pharmacia). AF was diluted 1:1 with binding buffer and centrifuged at 1,000 × g for 5 min to remove large protein aggregates. After sterilization using a 0.45-μm-pore-size Millipore filter, the AF was purified by affinity chromatography using a Pharmacia Biotech Mab Trap G II kit. The column was equilibrated with 3 ml of binding buffer (0.2 M sodium phosphate [pH 7.0]). The sample was loaded to bind the immunoglobulin G (IgG) to the protein G in the column via the Fc region. Contaminants were removed from the column by washing with 5 ml of binding buffer. The bound IgG was eluted with 3 ml of elution buffer (1.0 M glycine-HCl [pH 2.7]) and immediately neutralized with 75 μl of 1.0 M Tris-HCl (pH 9.0) per ml of purified IgG.
Production of Fab fragments.
A 100-μl volume of each purified antibody was incubated at 37°C for 2 h with 100 μl of 0.02-mg/ml papain attached to beaded agarose (Sigma). The reaction was stopped by addition of 20 μl of 300 mM iodoacetamide. Complete fractionation was monitored by SDS-PAGE (10% polyacrylamide) examination of the product.
ELISA.
Conventional ELISA procedures were performed as described previously (9). In brief, Micro ELISA plates were coated overnight with an optimal concentration of antigen (10 μg/well) in carbonate coating buffer (15 mM Na2CO3, 36 mM NaHCO3 [pH 9.6]) and subsequently blocked with PBS supplemented with 2% (wt/vol) skim milk. Incubation of primary antibody in serial two or fivefold dilutions was followed by incubation with alkaline phosphatase-conjugated secondary antibody (anti-mouse IgG [Sigma] diluted 1:500 in PBS). The reaction was developed by adding the substrate (1 mg/ml of disodium-ρ-nitrophenyl phosphate [pNPP; Sigma] in 100 mM glycine–1 mM MgCl2–1 mM ZnCl2 [pH 10.4]), and stopped with 3 N NaOH; the optical density was read at 405 nm.
EM and immunogold labeling.
Purified wt and mutant CLPs were resuspended in water, and 10-μl drops of CLPs were absorbed onto carbon-coated copper 400 mesh EM grids for 15 min, washed with water, and negatively stained with 2% (wt/vol) uranyl acetate. The grids were examined in a JEOL 100CX electron microscope at 100 kV.
For immunogold labeling, grids with absorbed CLPs were floated first on a blocking solution of 0.1% (wt/vol) bovine serum albumin BSA in PBS for 15 min and then processed as follows: (i) 15 min on a droplet of the appropriate VP7 monoclonal antibody (AF diluted in 1:500 PBS containing 0.1% BSA), followed by three quick washes in PBS; (2) 15 min on a droplet of gold particles (diameter, 5 nm) conjugated to goat anti-mouse IgG and IgM antibody (British Biocell International) diluted 1:20 in PBS containing 0.1% BSA, followed by three quick washes in PBS; and (iii) 40 s in 2% uranyl acetate. The grid was then blotted and air dried. In between each stage, the grids were carefully blotted onto filter paper by holding the grid perpendicular to the paper. In each experiment, negative control grids with no VP7 antibody and a nonspecific antibody were also prepared. For a positive control, mouse anti-CLP polyclonal antibody was used. The grids were examined as above, and images were obtained on Agfa Scientia Electron Microscope film and developed in Kodak D19 for 3.5 min.
Cryo-EM.
The Fab-CLP complexes were made by incubating the CLPs with Fab for 1 h at room temperature with regular vortexing. The complexes were then incubated at 4°C overnight. For complete decoration of all VP7 epitopes, a twofold excess of Fab was added for each VP7 molecule. Specimens were prepared for cryo-EM using standard procedures (13). A 4-μl aliquot of the specimen was applied to one side of a holey carbon grid. This grid was then blotted and plunged into a bath of liquid ethane (−180°C). The frozen-hydrated sample was transferred to a precooled low-temperature cryoholder and observed in either a JEOL 1200 or Philips CM120 transmission electron microscope. Regions of interest were imaged at ×30,000 and ×28,000 magnification, respectively with an electron dose of 5 electrons/Å2. From each region, a focal pair was recorded with intended defocus values of 1.2 and 2.4 μm. The electron images were recorded with a 1-s exposure on Kodak SO-163 films. The films were developed in Kodak D-19 developer for 12 min at 21°C and fixed in Kodak fixer for 10 min.
3D structural analysis.
Micrographs were selected based on particle concentration, quality of ice, and appropriate defocus. The images were digitized on a Zeiss SCAI microdensitometer (Carl Zeiss, Inc., Englewood, Colo.) using a 7-μm step size. The pixels were then averaged to give a 14-μm step size, which corresponded to 4.67 Å/pixel in the object for the JEOL microscope or 5.0 Å/pixel in the object for the Philips microscope. CLP-antibody complexes were extracted with a suitable box size. The determination of the orientational parameters, their refinement, and the 3D reconstructions were produced by ICOS Toolkit software suite using standard procedures (11). Three-dimensional reconstruction, from a set of particles, which adequately represented the icosahedral asymmetric unit, was computed using cylindrical expansion methods. The further-from-focus micrograph in each focal pair was processed first to obtain a low-resolution reconstruction, and this was then used to assist in finding correct orientations for the particles imaged in the corresponding closer-to-focus micrograph.
The reconstructions of the various particles were computed to a resolution within the first zero of the contrast transfer function of the corresponding micrograph. The reconstructions were viewed on a Silicon Graphics Workstation using IRIS Explorer v3.5 (Numerical Algorithms Group, Inc.).
Direct binding of CLPs onto KC cells.
Confluent monolayers of KC cells (63) were prepared in 96-well culture plates. KC cells were seeded at 106 cells per well and grown overnight at 28°C in Schneider medium supplemented with 5% FCS. Before the cells were used for binding to CLPs, the cell medium was removed from each well. The KC cells were washed with prechilled PBS–0.1% sodium azide to a final concentration of 10 μg/ml. To each well, 50 μl of the diluted CLPs were added and the absorption was allowed to continue for 30 min at room temperature. The cells were washed three times with prechilled PBS before being fixed with prechilled 90% ethanol–10% PBS for 10 min and subsequently washed three times with prechilled PBS. To each well of fixed cells, 100 μl of blocking buffer (1% skim milk in PBS) was added, and the cells were incubated for 1 h at 37°C and washed three times with PBS before detection of CLP binding. CLPs bound to the cells were detected by two types of antibodies, the polyclonal BTV-10 antibody and MAb H1.4 diluted with PBS containing 0.5% skim milk and 0.05% Tween 20 to final dilutions of 1:1,000 and 1:5,000, respectively. CLP-adsorbed cells in each well were incubated with 50 μl of the respective antibody for 1 h at 37°C and washed three times with PBS prior to the addition 50 μl of anti-mouse antibody–alkaline phosphatase conjugate. Antibody binding on adsorbed CLPs was detected similarly to the detection used in the normal ELISA procedure described above. Each experiment was repeated at least three times.
RESULTS
Mutagenesis of the unique RGD motif of VP7 does not alter the overall antigenic and biochemical characteristics of VP7.
RGD domains are involved in the binding of viruses to the cell membrane. The X-ray structural analysis of VP7 and BTV cores has revealed that an RGD motif is accessible on the surface of the 38-kDa VP7 molecule (Fig. 1) and likely to be antigenic to determine the antigenic nature and to assess the role of the RGD motif in cell binding, mutations targeting this tripeptide were designed that did not perturb either the VP7 monomer-monomer or the trimer-trimer interactions in the core assembly. In one, the RGD site was changed to AGD by changing arginine at 168 position to alanine. In one of the two remaining double mutants, arginine 168 is replaced by alanine and aspartate 170 is replaced by glutamine, converting the RGD site to AGQ, and in the other, arginine 168 is replaced by alanine and aspartate is replaced by leucine generating the sequence AGL. The rationale for creating the mutant motifs is that the AGQ motif is found at the equivalent position in the closely related AHSV-4 VP7 (2), suggesting that it should support cell entry, whilse the AGL mutant has precedent. Mutant VP7 DNAs were used to generate three recombinant baculoviruses, VP7-AGD, VP7-ADQ, and VP7-AGL, each with an altered VP7 molecule. To demonstrate that the recombinant RGD-mutated VP7 proteins were expressed in insect cells, extracts of Sf9 cells infected with the different baculoviruses were analyzed by SDS-PAGE. The expression of mutant VP7 molecules was examined by SDS-PAGE and compared with the baculovirus-expressed wt VP7 (48). As shown in Fig. 2A, a protein band of 38 kDa was identified for each of the RGD mutants (lanes 2 to 4). Both the levels of expression and sizes of the mutant proteins were similar to those of VP7 (lane 1). The authenticity of each mutant VP7 was further confirmed by Western blot analyses using an available polyclonal antiserum raised to BTV10-VP7. As shown in Fig. 2B, all three mutant proteins (lanes 2 to 4) reacted with the polyclonal antibody as well as with wt-VP7 (lane 1), indicating that these mutants retained the overall molecular and antigenic properties of the VP7 protein.
FIG. 1.
Alpha carbon trace representation of BTV-10 VP7 with RGD site indicated. The VP7 trimer is made up of three monomers (purple, grey, and yellow), and the position of the RGD triplet within a short beta strand (cyan) is shown on the grey monomer. Detail of RGD triplet and the surrounding sequence are shown in exploded view as a ball-and-stick model.
FIG. 2.
Expression of wt and mutated VP7 proteins. Sf9 cells were infected with Ac10BTV.7, expressing the wt (lane 1) or mutated (VP7-AGD [lane 2], VP7-AGQ [lane 3], and VP7-AGL [lane 4]) VP7 proteins. (A) The infected-cell lysates were analyzed by SDS-PAGE (10% polyacrylamide) and stained with Coomassie brilliant blue. (B) The proteins were electroblotted onto a membrane and reacted with BTV-10 polyclonal antisera. The arrow indicates the size of wt and mutated VP7 proteins. The protein mass markers are indicated on the left.
One of the striking features of BTV VP7 is that it oligomerizes readily into a trimeric form both in solution and in crystals (1), which are the building blocks for core assembly (36). Since the RGD residues are located in a loop on the VP7 upper domain, not at the interface between VP7 monomers, the mutated VP7 proteins should not perturb the monomer-monomer interactions. To confirm that the VP7 mutants still retained the ability to form trimers, we purified each expressed VP7 mutant and investigated their oligomeric nature using an assay system that we developed previously (36). All three mutants formed trimers which could be detected by SDS-PAGE (data not shown), confirming that substitution within the RGD motif did not alter the interface regions of the VP7 mutants. Similarly, circular dichroism analysis of the purified recombinant proteins showed no discernible change in the spectra of the RGD site-mutagenized VP7 proteins compared to spectra of unmodified VP7, indicating that the RGD mutations had no overall effect on the secondary elements (αhelix or βstrand) of VP7 (data not shown).
VP7 molecules with mutations at the RGD site interact with VP3 and assemble into CLPs that are morphologically indistinguishable from normal CLPs.
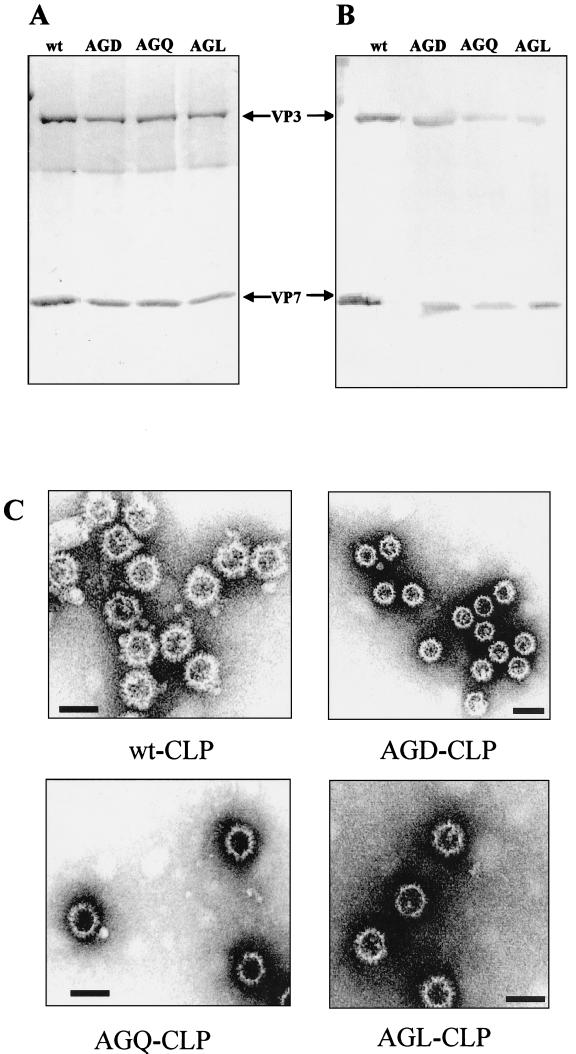
Since the RGD site is located on the upper domain of the protein and is not in contact with the underlying scaffolding VP3 layer, the mutated VP7 molecules should still assemble into CLPs in the presence of the VP3 molecule. Therefore, to determine this, we used a coexpression system in which assembly of CLPs was assessed by coinfection of insect cells with the recombinant virus expressing either wt VP7 or one of the three VP7 mutants and a recombinant virus expressing authentic VP3 molecules. The assembled CLPs were purified through a sucrose gradient and analyzed by SDS-PAGE. As shown in Fig. 3A, two protein bands at 120 and 38 kDa, representing VP3 and wt (lane1) or RGD-mutated (lanes 2 to 4) VP7s, respectively, were observed for each CLP preparation. Both protein bands reacted specifically in the immunoblot with polyclonal antiserum raised to BTV-10 virus (Fig. 3B). This indicates that the mutated VP7 proteins were still able to assemble into CLPs when coexpressed with VP3. The morphology of the CLP prepared by each mutant VP7 was further analyzed by EM, which revealed that the RGD-mutated CLPs formed from VP7-AGD, VP7-AGQ, and VP7-AGL exhibited the same size and morphological features as did the wt CLPs formed with unmodified VP7 (Fig. 3C). The RGD-mutated CLPs appeared highly stable upon storage at 4°C in 0.2 M Tris (pH 8.0) for prolonged periods (more than 2 months).
FIG. 3.
CLP formation by VP3 and wt or mutated VP7 proteins. Sf9 cells were coinfected with baculoviruses expressing VP3 with either the wt VP7 or various VP7 mutants. The CLPs were purified as described in Materials and Methods. (A and B) The CLPs were analyzed by SDS-PAGE (10% polyacrylaride) and stained with Coomassie brilliant blue (A) or immunoblotted with polyclonal Ab to BTV-10 (B). The arrows indicate the sizes of VP3 and mutated VP7 proteins. (C) CLP formation was confirmed by electron microscopy. Bars, 100 nm.
These data indicate that the mutations at Arg-168 and Asp-170 do not interfere with the interaction with VP3 and could form stable CLPs that are structurally similar to the native CLPs.
Identification of MAbs targeted to the RGD site of VP7 molecule.
To determine further the accessibility of the RGD motif in the BTV core and to map the antigenic region of VP7, a panel of 12 VP7 MAbs (MAbs H1.2 to H1.11 and CLPIII) that reacted specifically with native VP7 antigen as well as with CLPs were generated as described in Materials and Methods. Further, to determine whether these MAbs are targeted to the upper domain of VP7, their reactivities with two previously generated domain-switched VP7 chimeric constructs (45) were assessed by ELISA and Western blot analysis. One of these consists of only the upper domain of BTV VP7, while the lower domain is derived from the AHSV (ABA) VP7 molecule, and the second is a reverse construct (BAB) in which the upper domain is replaced by the fragment of AHSV VP7 but the lower is BTV VP7. The antibodies showed no cross-reactivity with either the purified AHSV VP7 protein or the purified BAB VP7 but reacted with ABA VP7 protein as strongly as with the wt BTV VP7 (Fig. 4A), demonstrating that all MAbs are targeted to the upper domain of VP7 molecule.
FIG. 4.
ELISA of the wt, chimeric, and mutant VP7 proteins. Micro-ELISA plates were coated with the purified recombinant proteins. A panel of six VP7 MAbs (H1.2 to H1.6 AND CLPIII) was tested with all the antigens, using 1:1,000 to 1:24,000 dilutions of sera. The optical density (OD) reading at 405 nm of a 1:10,000 dilution of each MAb is shown. (A) The micro-ELISA plates were coated with either the wt (■) or one of the two domain-switched chimeric, ABA ( ), BAB (▨) VP7. (B) Micro-ELISA plates were coated with either the wt (■) or the RGD mutants, AGD (
), BAB (▨) VP7. (B) Micro-ELISA plates were coated with either the wt (■) or the RGD mutants, AGD ( ), AGL (□), AGQ (▨).
), AGL (□), AGQ (▨).
To identify whether any of these MAbs specifically mapped to the RGD site of the VP7 molecule, each MAb was subsequently assessed for its binding ability to each of the purified RGD substitution mutants is an ELISA. As shown in Fig. 4B, 3 of the 12 MAbs (H1.5, H1.6, and CLPIII) did not bind to the RGD mutated VP7 proteins, although all the other MAbs bound to some degree; this suggests that at least three MAbs are specifically targeted to the region of the RGD site. This is probably because this site is particularly immunogenic.
To evaluate the accessibility of the RGD motif in the BTV core and to confirm further that H1.5 and CLPIII indeed recognize the RGD site of the molecule, CLPs generated with each of the RGD mutants as well as with native VP7 molecule were examined for their binding ability to these MAbs by immuno-EM. As shown in Fig. 5, immunogold staining of the mutants with these two MAbs produced similar results. There was no binding of antibodies of H1.5 or CLPIII to the CLPs generated with the VP7-AGQ mutant (Fig. 5A). However, immunogold-labeled MAb H1.4 bound successfully to ADQ-CLP (Fig. 5B). This indicates that MAbs H1.5 and CLPIII, but not H1.4, are targeted to the RGD site in VP7. Similar results were obtained with the other two VP7-AGD and VP7-ADL mutants (data not shown). Further, ELISA using CLPs assembled with each VP7 mutant molecule confirmed that H1.5 and CLPIII do not bind to the CLPs that were generated with the three mutants but bound to the wt CLPs strongly (data not shown).
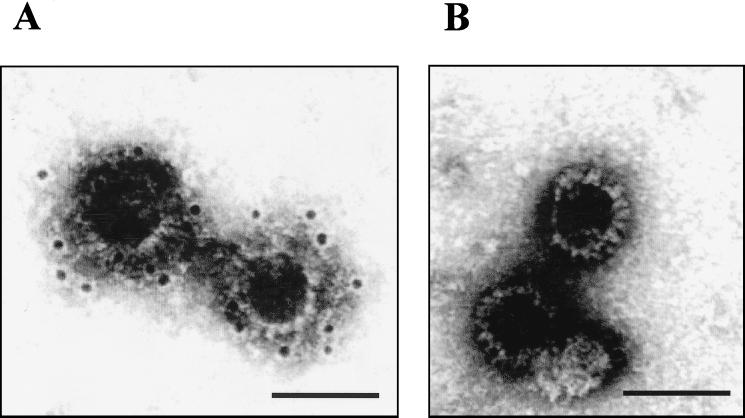
FIG. 5.
Immunogold labeling of CLPs with VP7 MAb H1.5. CLPs formed with VP3 and wt VP7 or VP7-AGQ were immunogold labeled with MAb, followed by the secondary antibody (goat anti-mouse) coupled with 10-nm-diameter gold particles. The complexes were visualized by EM. Bars, 100 nm. (A) wt CLPs showing the gold particles bound to the surface of CLPs; (B) CLPs formed by the VP7-AGQ, with no gold particles bound.
Visualization of CLP-antibody complexes by high-resolution cryo-EM.
The immunogold labeling studies showed that CLPIII and H1.5 had reduced binding to the CLPs containing VP7 with mutated RGD sequence compared with binding to the wt CLPs. This suggested, therefore, that these antibodies bound to a site which included the RGD epitope of VP7. Several methods can provide information concerning the region of antibody binding to the particles, including cryo-EM and 3D image reconstruction, which allow visualization of the antigen-antibody complexes at relatively high resolution (26). Therefore, to gain some insight into the antibody binding location on CLPs, we have generated CLP-H1.5/CLPIII Fab complexes as described in Materials and Methods and have examined these by cryo-EM and 3D image reconstruction. Sufficient Fabs were added to the CLPs that optimal antibody decoration of the VP7 molecules could be achieved without a large excess of Fab, which could interfere with the reconstruction. The decorated particles were well separated in ice and were homogeneous in appearance, as shown in Fig. 6A. As expected, the decorated particles had a different appearance from undecorated CLP. A halo of even thickness surrounding each particle (Fig. 6A) was observed that completely obscured the VP7 trimers. The VP3 subcore layer could be seen as a dark ring in the particle images. The decorated particles were ∼140 Å larger in diameter than the undecorated particles, resulting in a total diameter of ∼840 Å. These particles were used for 3D image reconstruction. The radial-density plot (data not shown) was calculated from the 3D reconstruction to represent the protein components at various radii within the 3D structure. By comparing this radial-density plot with that of the WT CLP reconstruction, it was evident that attached to the VP7 layer of the CLP was a layer of extra density which extended to a radius of ∼420 Å. This density has been attributed to the bound antibody molecules. It is clear in Fig. 6B that the extra density, attributed to Fab (yellow), sits on the surface of the CLP. An equatorial slice of the reconstruction reveals that the Fab is attached to the upper domain of the VP7 (blue) (Fig. 6C). Density due to the Fab was seen extending from all trimers in the particle. The removal of radial sections corresponding to Fab density revealed the VP7 directly beneath the antibody layer. This layer of protein was largely unchanged in conformation or position (Fig. 6B).
FIG. 6.
Cryon-EM of the CLP-H1.5 complex. (A) Micrograph of the CLP-H1.5 complex. An arrow indicates the diffuse electron density surrounding the particle. The arrowhead indicates a ring due to the smooth inner shell formed by the VP3 layer. The inset shows the CLPs without the MAb. (B) Surface representation of CLP-H1.5 complex viewed down the threefold axis. The decorated particle has a radius of ∼420 Å. The antibody fragments are shown in yellow. The VP7 trimers to which the antibodies attach are colored blue. The VP3 layer is exposed to the surface of the particle at the fivefold axes and is colored green. (C) Equatorial slice of the reconstruction (∼47 Å thick). Note that the antibody fragments (yellow) are bound to the upper domain of the VP7 (blue).
RGD mutations inhibit CLP binding to KC cells, and MAbs targeted to RGD compete with CLP binding.
To assess whether the RGD site plays a role in core entry, we have developed an assay system to investigate the cell binding ability of CLPs as representatives of BTV cores. In this assay, Culicoides-derived KC cells, which are permissive for BTV core infection, are used as an immobilized layer and the binding of both wt and mutant CLPs is assessed following incubation by an ELISA using either polyclonal BTV antibody (Fig. 7A) or MAb H1.4, a MAb (Fig. 7B) whose binding is not affected by the RGD mutations. Using this assay format, wt CLPs were found to bind to KC cells in a dose-dependent manner (Fig. 7) irrespective of the serum used for detection. In contrast to wt CLPs however, CLPs generated with the VP7-RGD mutants showed only a low-level binding to the cell surface and failed to show a significant dose-response curve. These data are consistent with a role for the RGD sequence of VP7 in cell binding.
FIG. 7.
Binding of wt and RGD-mutated CLPs to KC cells in binding ELISA. KC cells were grown in 96-well plates, and wt and RGD-mutated CLPs were allowed to bind. The CLP attachment was detected as described in Materials and Methods. The amount of CLP bound is expressed as the optical density (OD) at 405 nm and is plotted against increasing CLP concentrations (0 to 250 μg), and the bound CLP was detected by polyclonal BTV-10 antibody (A) or MAb H1.4 (B). Note the high level of binding for the wt CLP (⧫) with the increasing concentration of CLPs but no increase of binding with the same concentrations of CLPs for VP7-AGD (▴), VP7-AGQ (□), and VP7-AGL (○). The data plotted represent the average of three experiments performed in parallel. The standard deviation at a given point was no more than 7% of the value shown.
To confirm a role for the VP7 RGD motif, a second series of cell binding assays were carried out. In these, the binding activity of CLPs to KC cells was determined in the presence of MAb H1.5, a MAb whose epitope includes the RGD site of VP7. Following incubation of a fixed concentration of CLPs with different amounts of H1.5, residual CLP binding to the cell surface was detected by ELISA using a BTV polyclonal antibody. The addition of MAb H1.5 competed with the cell binding ability of CLPs, which increased significantly as the concentration of MAb decreased (Fig. 8). The addition of MAb to RGD mutant CLPs had no effect on the low level of cell binding observed. The data obtained with MAb H1.5 competition are consistent with a role for the RGD motif of VP7 in CLP binding to the KC cell surface and, by implication, with the entry of BTV cores into Culicoides cells.
FIG. 8.
Binding of wt and RGD-mutated CLPs to KC cells in the presence of MAb H1.5. KC cells were grown in 96-well plates, 100 μg of either wt or RGD-mutated CLPs was bound in the presence of increasing dilutions of MAb H1.5 (1:500 to 1:10,000), and the bound CLPs were detected using polyclonal BTV-10 antibody as described in Materials and Methods. The experiment were carried out three times for each preparation. The average amount of CLP bound in three experiments is expressed as the optical density (OD) at 405 nm and is plotted against decreasing MAb dilutions. The bound CLPs are indicated as follows: ⧫, wt; ●, KC cells; ▴, VP7-AGD; □, VP7-AGQ; ○, VP7-AGL.
DISCUSSION
BTV-VP3 and VP7 proteins self-assemble into empty CLP when coexpressed in insect cells by using recombinant baculoviruses (17). Assembled CLPs mimic the authentic virus cores and provide an in vitro model for direct structure-functional studies. In this study we examined the effect of point mutations introduced into VP7 on virus-host protein interactions. The expression of recombinant capsid proteins and their assembly into virus-like particles have been similarly used to study virus-host interaction in binding and internalization of bovine papillomavirus (46), Norwalk virus (64), and rotaviruses (15).
Comparative sequence data and structural studies have implicated the RGD tripeptide sequence in BTV-VP7 (amino acid residues 168 to 170) in virus attachment to Culicoides host cells. To date, however, little direct evidence has been available to test this hypothesis for BTV even though RGD is critical for other virus-receptor interactions. To provide direct evidence for VP7 RGD tripeptide involvement in virus attachment, we introduced point mutations at amino acid residues 168 and 170, converting RGD to either AGD, AGQ, or AGL. The coexpression of RGD-mutated proteins with BTV-VP3 in the baculovirus system resulted in the assembly of CLPs and showed no gross change in the overall conformation by the introduction of mutations in this region. The assembled CLPs formed by these RGD-mutated VP7s appeared equivalent to authentic BTV cores in morphological, biochemical, and overall immunological characteristics. This view is supported by three sets of experimental data: (i) RGD-mutated VP7 proteins reacted with anti-BTV serum in immunoblot assays as did the wt; (2) RGD-mutated VP7 proteins were structurally similar to wt VP7, as indicated by the similarity in secondary structures from CD analysis and their ability to oligomerization into trimers, a feature of the VP7 molecule; and (iii) the assembled CLPs were structurally and functionally equivalent to the authentic CLPs. Since no gross change of structure occurred by mutation of RGD, the role of the sequence in host cell binding was assessed. A direct approach, in which CLPs were assayed by indirect ELISA for binding to the surface of Culicoides (KC) cells, indicated that alteration of the RGD sequence led to a reduced level of CLP binding. An indirect approach, in which MAbs raised against VP7 were used as competitor for CLP binding, showed that only MAbs that mapped directly to the RGD motif gave effective competition. MAbs that bound the upper domain of VP7 but were not affected by mutations at RGD did not compete for KC cell surface binding. Together, these data indicate that the RGD motif of BTV VP7 is accessible in the context of the CLP (MAb generation and cryo-EM decoration) and is responsible for cell surface binding. The RGD tripeptide is conserved for all BTV serotypes of VP7s as well as for EHDV, despite the location of the motif in an exposed position, where antibody-selected variation might be expected to occur (31, 54, 68). The RGD motif of AHSV is located in a slightly different position from that of BTV but also on a highly flexible and exposed loop (2). Moreover, the structure of the integrin insertion region, domain I, can be easily docked onto the RGD motif of both BTV and AHSV VP7 proteins (3).
There is accumulating evidence for the involvement of an RGD or an RGD-like tripeptide amino acid sequence in virus binding to cellular integrins during infection by a number of viruses. The role played by these tripeptide-containing ligands could be indirect, as is the case for adenovirus (4, 12, 47, 65), or direct, as for rotaviruses (10, 21, 27), papillomoviruses (14), and members of the Picornaviridae family, including echoviruses 1, 8, and 9 (5, 30, 49, 60, 70); coxsackievirus A9 (7, 8, 53, 61); FMDV (6, 32, 50, 51); and human parechovirus (62). It has been shown conclusively that the RGD motif in FMDV is essential for virus entry, since mutations in this sequence, including the conservative change to RGE, resulted in noninfectious virus particles that failed to enter cells (39–41). Recent extensive analysis of the mechanism of rotavirus cell entry has shown that such tripeptides do indeed act as ligands for integrins and that peptides containing these sequences and MAbs against integrins block the entry and infectivity of rotavirus (21, 27, 69). Using CLPs as a model system for cell binding, VP7 mutants were assessed for their cell attachment capability and the data obtained demonstrated clearly that mutated CLPs either did not bind to Culicoides-derived cells at all or bound at much reduced level compared to the wt CLPs, suggesting a role for RGD in core uptake by the insect cells.
MAbs specific for integrins that recognize the RGD sequences of viral proteins block virus attachment and infectivity of other viruses (33, 52, 61, 62). In our study we have used the VP7 MAbs that are RGD site specific (e.g., H1.5 and CLPIII) to block the CLP attachment and have demonstrated that such MAbs interfere with CLP binding to KC cells. Cryo-EM analysis has shown that Fab fragments of these MAbs decorate the surface of the CLPs effectively and that FAbs bind to a region which includes the RGD site. Together, the data support the hypothesis that the VP7 RGD motif, as presented in associated with VP3, is at least part of the receptor binding sequence of BTV cores.
ACKNOWLEDGMENTS
We thank T. Booth, IVEM, Oxford, United Kingdom, and D. Stuart, Oxford University, Oxford, United Kingdom, for their support and advice on structural work, and we are grateful to B. V. V. Prasad, Baylor College, Houston, Tex., for providing the cryo-EM facility. We also thank M. Gross, New Chemistry Laboratory, University of Oxford, for assistance in circular dichroism analysis.
This work was partly funded by grants from the BBSRC (United Kingdom) and NIH (United States).
REFERENCES
- 1.Basak A K, Stuart D I, Roy P. Preliminary crystallographic study of bluetongue virus capsid protein, VP7. J Mol Biol. 1992;228:687–689. doi: 10.1016/0022-2836(92)90850-j. [DOI] [PubMed] [Google Scholar]
- 2.Basak A K, Gouet P, Grimes J, Roy P, Stuart D. Crystal structure of the top domain of African horsesickness virus VP7: comparisons with bluetongue virus VP7. J Virol. 1996;70:3797–3806. doi: 10.1128/jvi.70.6.3797-3806.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basak A K, Grimes J, Gouet P, Roy P, Stuart D I. Structures of orbivirus VP7: implications for the role of this protein in the viral life cycle. Structure. 1997;5:871–883. doi: 10.1016/s0969-2126(97)00242-6. [DOI] [PubMed] [Google Scholar]
- 4.Belin M T, Boulanger P. Involvement of cellular adhesion sequences in the attachment of adenovirus to the HeLa cell surface. J Gen Virol. 1993;74:1485–1497. doi: 10.1099/0022-1317-74-8-1485. [DOI] [PubMed] [Google Scholar]
- 5.Bergelson J M, Shepley B M C, Chan C, Hemler M E, Finberg R W. Identification of integrin VLA-2 as a receptor for echovirus 1. Science. 1992;255:1718–1720. doi: 10.1126/science.1553561. [DOI] [PubMed] [Google Scholar]
- 6.Berinstein A, Roivainen M, Hovi T, Mason P W, Baxt B. Antibodies to the vitronectin receptor (integrin αvβ3 inhibit binding and infection of foot and mouth disease virus to cultured cells. J Virol. 1995;69:2664–2666. doi: 10.1128/jvi.69.4.2664-2666.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang K H P, Auvinen P, Hyypiä T, Stanway G. The nucleotide sequence of coxsackievirus A9: implications for receptor binding and entero-virus classification. J Gen Virol. 1989;70:3269–3280. doi: 10.1099/0022-1317-70-12-3269. [DOI] [PubMed] [Google Scholar]
- 8.Chang K H, Day C, Walker J, Stanway G. The nucleotide sequences of wild type coxsackievirus A9 strains imply that the RGD motif in VP1 is functionally significant. J Gen Virol. 1992;73:621–626. doi: 10.1099/0022-1317-73-3-621. [DOI] [PubMed] [Google Scholar]
- 9.Chuma T, Le Blois H, Sanchez-Vizcaino J M, Diaz-Laviada M, Roy P. Expression of the major core antigen VP7 of African horsesickness virus by a recombinant baculovirus and its use as a group-specific diagnostic reagent. J Gen Virol. 1992;73:925–931. doi: 10.1099/0022-1317-73-4-925. [DOI] [PubMed] [Google Scholar]
- 10.Coulson B, Lodrigan S L, Lee D J. Rotavirus contains integrin ligand sequences and a disintegrin-like domain that are implicated in virus cell entry into cells. Proc Natl Acad Sci USA. 1997;94:5389–5394. doi: 10.1073/pnas.94.10.5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crowther R A. Procedures for three-dimensional reconstruction of spherical viruses by Fourier synthesis from electron micrographs. Philos Trans R Soc London Ser B. 1971;261:221–230. doi: 10.1098/rstb.1971.0054. [DOI] [PubMed] [Google Scholar]
- 12.Defer C, Belin M T, Caillet Boudin M L, Boulanger P. Human adenovirus-host cell interactions: comparative study with members of subgroups B and C. J Virol. 1990;64:3661–3673. doi: 10.1128/jvi.64.8.3661-3673.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dubochet J, Adrian M, Chang J J, Homo J C, Lepault J, McDowell A W, Schultz P. Cryo-electron microscopy of vitrified specimens. Q Rev Biophys. 1988;21:129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- 14.Evander M, Frazer I, Payne E, Qi Y, Hengst K, McMillan N. Identification of the α6 integrin as a candidate receptor for papillomaviruses. J Virol. 1997;69:2449–2456. doi: 10.1128/jvi.71.3.2449-2456.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Falconer M M, Gilbert J M, Roper A M, Greenberg H B, Gavora J S. Rotavirus-induced fusion from without in tissue culture cells. J Virol. 1995;69:5582–5591. doi: 10.1128/jvi.69.9.5582-5591.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fazekas de St. Groth S, Scheidegger D. Production of monoclonal antibodies. Strategy and tactics. J Immunol Methods. 1980;35:1–21. doi: 10.1016/0022-1759(80)90146-5. [DOI] [PubMed] [Google Scholar]
- 17.French T J, Roy P. Synthesis of bluetongue virus (BTV) core-like particles by a recombinant baculovirus expressing the two major structural core proteins of BTV. J Virol. 1990;64:1530–1536. doi: 10.1128/jvi.64.4.1530-1536.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gould E A, Buckley A, Cammack N. Use of the biotin-streptavidin interaction to improve flavivirus detection by immunofluorescence and ELISA tests. J Virol Methods. 1985;11:41–48. doi: 10.1016/0166-0934(85)90123-5. [DOI] [PubMed] [Google Scholar]
- 19.Grimes J, Basak A K, Roy P, Stuart D. The crystal structure of bluetongue virus VP7. Nature. 1995;373:167–170. doi: 10.1038/373167a0. [DOI] [PubMed] [Google Scholar]
- 20.Grimes J M, Burroughs J N, Gouet P, Diprose J M, Malby R, Zientara S, Mertens P P C, Stuart D I. The atomic structure of the bluetongue virus core. Nature. 1998;395:470–478. doi: 10.1038/26694. [DOI] [PubMed] [Google Scholar]
- 21.Guerrero C A, Mendez E, Zarate S, Isa P, Lopez S, Arias C F. Integrin alpha v beta 3 mediates rotavirus cell entry. Proc Natl Acad Sci USA. 2000;97:14644–14649. doi: 10.1073/pnas.250299897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hassan S, Roy P. Expression and functional characterization of bluetongue virus VP2 protein. Role in cell entry. J Virol. 2000;73:9832–9842. doi: 10.1128/jvi.73.12.9832-9842.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hewat E A, Booth T F, Roy P. Structure of bluetongue virus particles by cryo-electron microscopy. J Struct Biol. 1992;109:61–69. doi: 10.1016/1047-8477(92)90068-l. [DOI] [PubMed] [Google Scholar]
- 24.Hewat E A, Booth T F, Loudon P T, Roy P. 3D reconstruction of bluetongue virus core-like particles by cryo-electron microscopy. Virology. 1992;189:10–20. doi: 10.1016/0042-6822(92)90676-g. [DOI] [PubMed] [Google Scholar]
- 25.Hewat E A, Booth T F, Roy P. Structure of correctly self-assembled bluetongue virus-like particles. J Struct Biol. 1994;112:183–191. doi: 10.1006/jsbi.1994.1019. [DOI] [PubMed] [Google Scholar]
- 26.Hewat E A, Verdaguer N, Fita I, Blakemore W, Brookes S, King A, Newman J, Domingo E, Mateu M G, Stuart D I. Structure of the complex of an Fab fragment of a neutralizing antibody with foot-and-mouth disease virus: positioning of a highly mobile antigenic loop. EMBO J. 1997;16:1492–1500. doi: 10.1093/emboj/16.7.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hewish M J, Takada Y, Coulson B S. Integrins α2β1 and α4β1 can mediate SA11 rotavirus attachment and entry into cells. J Virol. 2000;71:6749–6756. doi: 10.1128/jvi.74.1.228-236.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huismans H, Van Dijk A A, Els H J. Uncoating of parental bluetongue virus to core and subcore particles in infected L cells. Virology. 1987;157:180–188. doi: 10.1016/0042-6822(87)90327-8. [DOI] [PubMed] [Google Scholar]
- 29.Hynes R O. Integrins: versatility, modulation, and signalling in cell adhesion. Cell. 1992;69:11–25. doi: 10.1016/0092-8674(92)90115-s. [DOI] [PubMed] [Google Scholar]
- 30.Hyypiä T, Horsnell C, Maaronen M, Khan M, Kalkkinen N, Auvinen P, Kinnunen L, Stanway G. A distinct picornavirus group identified by sequence analysis. Proc Natl Acad Sci USA. 1992;89:8847–8851. doi: 10.1073/pnas.89.18.8847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iwata H, Yamagawa M, Roy P. Evolutionary relationships among the gnat-transmitted orbiviruses that cause African horsesickness, bluetongue and epizootic haemorrhagic disease as evidenced by their capsid protein sequences. Virology. 1992;191:251–261. doi: 10.1016/0042-6822(92)90187-t. [DOI] [PubMed] [Google Scholar]
- 32.Jackson T, Sharma A, Ghazaleh R A, Blakemore W E, Ellard F M, Simmons D L, Stuart D I, Newman J W, King A M. Arginine-glycine-aspartic acid specific binding by foot-and-mouth disease viruses to the purified integrin receptor, alpha(v)beta3, in vitro. J Virol. 1997;71:8357–8361. doi: 10.1128/jvi.71.11.8357-8361.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kalter D C, Gendelman H E, Meltzer M S. Inhibition of human-immunodeficiency-virus infection in monocytes by monoclonal-antibodies against leukocyte adhesion molecules. Immunol Lett. 1991;30:219–228. doi: 10.1016/0165-2478(91)90029-a. [DOI] [PubMed] [Google Scholar]
- 34.Kitts P A, Possee R D. A method for producing recombinant baculovirus expression vectors at high efficiency. BioTechniques. 1993;14:810–817. [PubMed] [Google Scholar]
- 35.Kunkel T A, Roberts J D, Zakuor R A. Rapid and efficient method of site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 36.Limn C-H, Staeuber N, Monastyrskaya K, Gouet P, Roy P. Functional dissection of the major structural protein of bluetongue virus: identification of key residues within VP7 essential for capsid assembly. J Virol. 2000;74:8658–8669. doi: 10.1128/jvi.74.18.8658-8669.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Livingstone C, Jones I M. Baculovirus expression vector with single-strand capability. Nucleic Acids Res. 1989;17:2366. doi: 10.1093/nar/17.6.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Logan D, Abu-Ghazaleh R, Blakemore W, Curry S, Jackson T, King A, Lea S, Lewis R, Newman J, Parry N, Rowlands D, Stuart D, Fry E. Structure of a major immunogenic site on foot-and-mouth disease virus. Nature. 1993;362:566–568. doi: 10.1038/362566a0. [DOI] [PubMed] [Google Scholar]
- 39.Mason P W, Reider E, Baxt B. RGD sequence of foot and mouth disease virus is essential for infecting cells via the natural receptor but can be bypassed by an antibody-dependent enhancement pathway. Proc Natl Acad Sci USA. 1994;91:1932–1936. doi: 10.1073/pnas.91.5.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mateu M G, Valero M L, Andreu D, Domingo E. Systematic replacement of amino acid residues within the Arg-Gly-Asp, containing loop of foot and mouth disease virus and effect on cell recognition. J Biol Chem. 1996;271:12814–12819. doi: 10.1074/jbc.271.22.12814. [DOI] [PubMed] [Google Scholar]
- 41.McKenna T S C, Lubroth J, Rieder E, Baxt B, Mason P W. Receptor binding site-deleted foot and mouth (FMD) virus protects cattle from FMD. J Virol. 1995;69:5787–5790. doi: 10.1128/jvi.69.9.5787-5790.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mellor P S. The replication of bluetongue virus in Culicoides vectors. Curr Top Microbiol Immunol. 1990;162:143–161. doi: 10.1007/978-3-642-75247-6_6. [DOI] [PubMed] [Google Scholar]
- 43.Mertens P P C, Burroughs J N, Anderson J. Purification and properties of virus particles, infectious subviral particles, and cores of bluetongue virus serotypes 1 and 4. Virology. 1987;157:375–86. doi: 10.1016/0042-6822(87)90280-7. [DOI] [PubMed] [Google Scholar]
- 44.Mertens P P C, Burroughs J N, Fu H, Wellby M P, Jennings D M, O'Hara R S, Walton A, Mellor P S. Enhanced infectivity of modified bluetongue virus particles for Culicoides vectors and to insect cell lines. Virology. 1996;217:582–893. doi: 10.1006/viro.1996.0153. [DOI] [PubMed] [Google Scholar]
- 45.Monastyrskaya K, Staeuber N, Roy P. Effects of domain-switching and site-directed mutagenesis on the properties and functions of the VP7 proteins of two orbiviruses. Virology. 1997;237:217–227. doi: 10.1006/viro.1997.8776. [DOI] [PubMed] [Google Scholar]
- 46.Muller M, Gissman L, Cristiano R J, Sun X-Y, Frazer I H, Jenson A B, Alonso A, Zentgraf H, Zhou J. Papillomavirus capsid binding and uptake by cells from different tissues and species. J Virol. 1995;69:948–954. doi: 10.1128/jvi.69.2.948-954.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nemerow G R, Cheresh D A, Wickham T J. Adenovirus entry into host cells: A role for alpha-v integrins. Trends Cell Biol. 1994;4:52–55. doi: 10.1016/0962-8924(94)90010-8. [DOI] [PubMed] [Google Scholar]
- 48.Oldfield S, Adachi A, Urakawa T, Hirasawa T, Roy P. Purification and characterization of the major group-specific core antigen VP7 of bluetongue virus synthesized by a recombinant baculovirus. J Gen Virol. 1990;71:2649–2656. doi: 10.1099/0022-1317-71-11-2649. [DOI] [PubMed] [Google Scholar]
- 49.Pulli T, Koivunen E, Hyypiä T. Cell-surface Interactions of echovirus 22. J Biol Chem. 1997;272:21126–21180. doi: 10.1074/jbc.272.34.21176. [DOI] [PubMed] [Google Scholar]
- 50.Rieder E, Baxt B, Lubroth J, Mason P W. Vaccines prepared from chimeras of foot-and-mouth disease virus (FMDV) induce neutralizing antibodies and protective immunity to multiple serotypes of FMDV. J Virol. 1994;68:7092–7098. doi: 10.1128/jvi.68.11.7092-7098.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rieder E, Baxt B, Lubroth J, Mason P W. Animal-derived antigenic variants of foot-and-mouth disease virus (FMDV) type A-12 have low affinity for cells in culture. J Virol. 1994;68:5296–5299. doi: 10.1128/jvi.68.8.5296-5299.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rieder E, Berinstein A, Baxt B, Kang A, Mason P W. Propagation of an attenuated virus by design: engineering a novel receptor for a noninfectious foot-and-mouth disease virus. Proc Natl Acad Sci USA. 1996;93:10428–10433. doi: 10.1073/pnas.93.19.10428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roivainen M, Piirainen L, Hovi T, Virtanen I, Rikonen T, Heino J, Hyypiä T. Entry of coxsackievirus A9 into host cells: specific interactions with αvβ3 integrin, the vitronectin receptor. Virology. 1994;203:357–365. doi: 10.1006/viro.1994.1494. [DOI] [PubMed] [Google Scholar]
- 54.Roy P, Hirasawa T, Fernandez M, Blinov V M, Sanchez-Vizcaino Rodrique J M. The complete sequence of the group-specific antigen, VP7, of African horsesickness virus serotype 4 reveals a close relationship to bluetongue virus. J Gen Virol. 1991;72:1237–1241. doi: 10.1099/0022-1317-72-6-1237. [DOI] [PubMed] [Google Scholar]
- 55.Roy P. Bluetongue virus proteins. J Gen Virol. 1992;73:3051–3064. doi: 10.1099/0022-1317-73-12-3051. [DOI] [PubMed] [Google Scholar]
- 56.Roy P. Orbivirus structure and assembly. Virology. 1996;216:1–11. doi: 10.1006/viro.1996.0028. [DOI] [PubMed] [Google Scholar]
- 57.Ruoslahti E, Pierschbacher M D. New perspectives in cell adhesion: RGD and integrins. Science. 1987;238:491–496. doi: 10.1126/science.2821619. [DOI] [PubMed] [Google Scholar]
- 58.Ruoslahti E. Integrins. J Clin Investig. 1991;87:1–7. doi: 10.1172/JCI114957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stanway G, Kalkkinen N, Roivainen M, Ghazi F, Khan M, Smyth M, Meurman O, Hyypia T. Molecular and biological characteristics of echovirus 22, a representative of a new picornavirus group. J Virol. 1994;68:8232–8238. doi: 10.1128/jvi.68.12.8232-8238.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Triantafilou M, Triantafilou K, Wilson K M, Takada Y, Fernandez N, Stanway G. Involvement of β2-microglobulin and integrin αvβ3 molecules in the coxsackievirus A9 virus infectious cycle. J Gen Virol. 2000;80:2591–2600. doi: 10.1099/0022-1317-80-10-2591. [DOI] [PubMed] [Google Scholar]
- 62.Triantafilou M, Triantafilou K, Takada Y, Fernandez N. Human parechovirus 1 utilizes integrins αvβ3 and αvβ1 as receptors. J Virol. 2000;74:5856–5862. doi: 10.1128/jvi.74.13.5856-5862.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wechsler S J, McHolland L E, Wilson W C. A RNA virus in cells from Culicoides variipennis. J Invertebs Pathol. 1991;57:200–205. doi: 10.1016/0022-2011(91)90117-9. [DOI] [PubMed] [Google Scholar]
- 64.White L J, Ball J M, Hardy M E, Tanaka T N, Kitamoto N, Estes M. Attachment and entry of recombinant Norwalk virus capsids to cultured human and animal cell lines. J Virol. 1996;70:6589–6597. doi: 10.1128/jvi.70.10.6589-6597.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wickham T J, Mathias P, Cheresh D A, Nemerow G R. Integrins alpha v beta 3 and alpha v beta 5 promote adenovirus internalization but not virus attachment. Cell. 1993;73:309–319. doi: 10.1016/0092-8674(93)90231-e. [DOI] [PubMed] [Google Scholar]
- 66.Wistow G, Turnell B, Summers L, Slingsby C, Moss D, Miller L, Lindley P, Blundell T. X-ray analysis of the eye lens protein gamma-II crystallin at 1.9 A resolution. J Mol Biol. 1983;170:175–202. doi: 10.1016/s0022-2836(83)80232-0. [DOI] [PubMed] [Google Scholar]
- 67.Xu G, Wilson W, Mecham J, Murphy K, Zhou E-M, Tabachnick W. VP7: an attachment protein of bluetongue virus for cellular receptors in Culicoides variipennis. J Gen Virol. 1997;78:1617–1623. doi: 10.1099/0022-1317-78-7-1617. [DOI] [PubMed] [Google Scholar]
- 68.Yu Y, Fukusho A, Ritter D G, Roy P. Complete nucleotide sequence of the group-reactive antigen VP7 gene of bluetongue virus. Nucleic Acids Res. 1988;16:1620. doi: 10.1093/nar/16.4.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zárate S, Espinosa R, Romero P, Guerrero C A, Arias F, López S. Integrin α2β1 mediates the cell attachment of the rotavirus neuraminidase-resistant variant nar3. Virology. 2000;287:50–54. doi: 10.1006/viro.2000.0660. [DOI] [PubMed] [Google Scholar]
- 70.Zimmermann H, Eggers H J, Nelsen-Salz B. Cell attachment of mouse virulence of echovirus 9 correlate with an RGD motif in the capsid protein VP1. Virology. 1997;233:149–156. doi: 10.1006/viro.1997.8601. [DOI] [PubMed] [Google Scholar]