Highlights
-
•
Developed a ConvNeXtv2 Fusion based Mask R-CNN model for automatic segmentation of bone tumors from CT scans.
-
•
Utilized data from two hospitals to ensure robustness and generalizability of the model.
-
•
Implemented advanced AI techniques to enhance diagnostic accuracy in bone metastases.
-
•
Facilitated personalized treatment strategies by accurately identifying lung cancer metastasis.
-
•
Demonstrated potential for reducing the need for extensive and costly diagnostic procedures.
Abstract
Lung cancer, which is a leading cause of cancer-related deaths worldwide, frequently metastasizes to the bones, significantly diminishing patients’ quality of life and complicating treatment strategies. This study aims to develop an advanced 3D Mask R-CNN model, enhanced with the ConvNeXt-V2 backbone, for the automatic segmentation of bone tumors and identification of lung cancer metastasis to support personalized treatment planning. Data were collected from two hospitals: Center A (106 patients) and Center B (265 patients). The data from Center B were used for training, while Center A’s dataset served as an independent external validation set. High-resolution CT scans with 1 mm slice thickness and no inter-slice gaps were utilized, and the regions of interest (ROIs) were manually segmented and validated by two experienced radiologists. The 3D Mask R-CNN model achieved a Dice Similarity Coefficient (DSC) of 0.856, a sensitivity of 0.921, and a specificity of 0.961 on the training set. On the test set, it achieved a DSC of 0.849, a sensitivity of 0.911, and a specificity of 0.931. For the classification task, the model attained an AUC of 0.865, an accuracy of 0.866, a sensitivity of 0.875, and a specificity of 0.835 on the training set, while achieving an AUC of 0.842, an accuracy of 0.836, a sensitivity of 0.847, and a specificity of 0.819 on the test set. These results highlight the model’s potential in improving the accuracy of bone tumor segmentation and lung cancer metastasis detection, paving the way for enhanced diagnostic workflows and personalized treatment strategies in clinical oncology.
1. Introduction
Lung cancer remains a leading cause of cancer mortality globally, with an estimated 1.8 million deaths annually, accounting for nearly one-fifth of all cancer-related deaths [1], [2], [3]. Its aggressive nature and propensity for early metastasis to distant organs pose significant challenges in clinical management. One of the most devastating complications of advanced lung cancer is skeletal metastasis, which occurs in approximately 30–40 % of patients [4]. The presence of bone metastases is often indicative of a poor prognosis, with a median survival time of less than one year following diagnosis [5]. These metastases can lead to severe pain, pathological fractures, spinal cord compression, and hypercalcemia, all of which substantially reduce a patient’s quality of life and increase the complexity of treatment [6], [7]. Furthermore, skeletal-related events (SREs) associated with bone metastases necessitate intensive medical intervention, including radiotherapy, surgery, and the use of bone-modifying agents, thereby imposing a significant economic burden on healthcare systems [8], [9]. The early and accurate detection of bone metastases is crucial for timely intervention and improved clinical outcomes in lung cancer patients [10], [11].
Patients with distal bone metastases often undergo extensive and specialized diagnostic procedures to determine the primary source of the metastatic tumors, especially in cases where lung cancer is suspected [2]. These diagnostic protocols typically involve a combination of imaging studies, such as X-rays, CT scans, MRI, and PET scans, as well as bone biopsies and various laboratory tests [12]. While these procedures are essential for accurate diagnosis, they can be time-consuming, invasive, and costly, placing a significant burden on both patients and healthcare systems [7], [13], [14]. Consequently, there is a critical need for more efficient and cost-effective methods to swiftly and accurately identify the primary origin of bone metastases, particularly for lung cancer [15], [16]. Rapid identification of the metastatic source is pivotal for implementing precise and timely treatment strategies, which can significantly improve patient outcomes and quality of life [17], [18], [19].
Recent advancements in artificial intelligence (AI) and machine learning are revolutionizing medical diagnostics, particularly in oncology [20]. Automated segmentation and analysis of bone tumors using advanced models like 3D Mask R-CNN have shown a remarkable ability to enhance diagnostic precision. In recent studies, the integration of the ConvNeXt-V2 backbone has significantly improved feature extraction and segmentation performance, especially in complex medical imaging tasks [21], [22], [23]. ConvNeXt-V2, as demonstrated in recent research, enables the extraction of more enriched and semantically significant feature maps, improving the identification of metastatic regions within 3D CT scans. These advances reduce the need for multiple, time-consuming diagnostic tests, offering a faster and more accurate means of assessing bone metastases [24]. The integration of AI in diagnostic workflows not only optimizes resource utilization but also accelerates decision-making processes, ultimately contributing to better clinical management of patients with bone metastases.
The accurate prediction of whether bone metastases originate from lung cancer is crucial for creating personalized treatment plans. With AI-driven models, such predictions can be achieved more efficiently, reducing unnecessary procedures and enabling clinicians to focus on tailored therapeutic strategies. The implementation of AI into diagnostic workflows not only improves resource utilization but also significantly accelerates decision-making processes. This ultimately leads to better clinical outcomes and more effective management of bone metastases, a critical issue for lung cancer patients, as bone metastases occur in approximately 30–40 % of cases This study aims to further develop a deep learning model based on the 3D Mask R-CNN framework, incorporating the ConvNeXt-V2 backbone for enhanced performance. The model will focus on the automatic segmentation of bone tumors and the determination of whether metastasis originates from lung cancer. By integrating this advanced technology, we seek to improve the treatment and management of bone metastases, ensuring timely and precise interventions that ultimately enhance patient outcomes.
2. Methodology
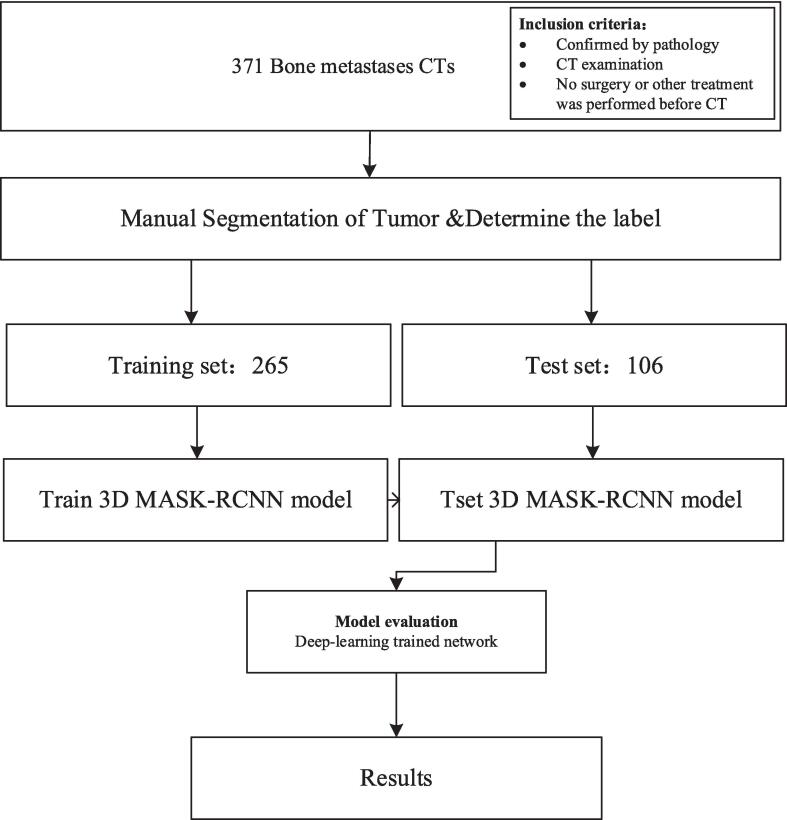
The flow chart of the study design is displayed in Fig. 1.
Fig. 1.
The flow chart of our study design.
2.1. Data collection
This study is designed as a dual-center experiment, involving patients with bone metastases from two hospitals, referred to as Center A and Center B. The data collection process was meticulously planned to ensure robust and scientifically sound results.
Patient Cohort:
-
•
Center A: A total of 106 patients with bone metastases were collected. Among these, 56 patients had metastases originating from lung cancer, while the remaining 50 patients had bone metastases from other primary cancers.
-
•
Center B: A larger cohort of 265 patients with bone metastases was included. Out of these, 89 patients had lung cancer as the primary source of their metastases, and the remaining 176 patients had bone metastases from other types of cancers.
Data Utilization: To ensure the scientific rigor and validity of our experimental results, we adopted a strategic approach in data utilization:
-
•Training Set: Data from Center B was used as the training set. This set included 265 patients, allowing the model to learn from a diverse and extensive dataset. The breakdown of this dataset is as follows:
-
•Lung cancer metastases: 89 patients
-
•Other cancer metastases: 176 patients
-
•
-
•External Validation Set: Data from Center A was utilized as the external validation set. This cohort, comprising 106 patients, was entirely independent of the training data to ensure unbiased evaluation of the model's performance. The breakdown is:
-
•Lung cancer metastases: 56 patients
-
•Other cancer metastases: 50 patients
-
•
Rationale: Using data from Center B for training and data from Center A for external validation enhances the robustness of the model by:
-
•
Preventing Overfitting: Ensuring the model is trained on one dataset and validated on a separate dataset reduces the risk of overfitting and enhances the generalizability of the model.
-
•
Ensuring Scientific Validity: The external validation using data from a different center provides a more rigorous test of the model's performance in real-world settings, reflecting its potential clinical applicability.
Data Types and Preprocessing: The collected data consists of CT scan images with a slice thickness of 1 mm and no inter-slice gap. Preprocessing steps involved:
-
•
Normalizing the CT imaging data to ensure consistency across different scans.
-
•
Augmenting the dataset with techniques such as rotation, scaling, and flipping to increase the diversity of training samples.
-
•
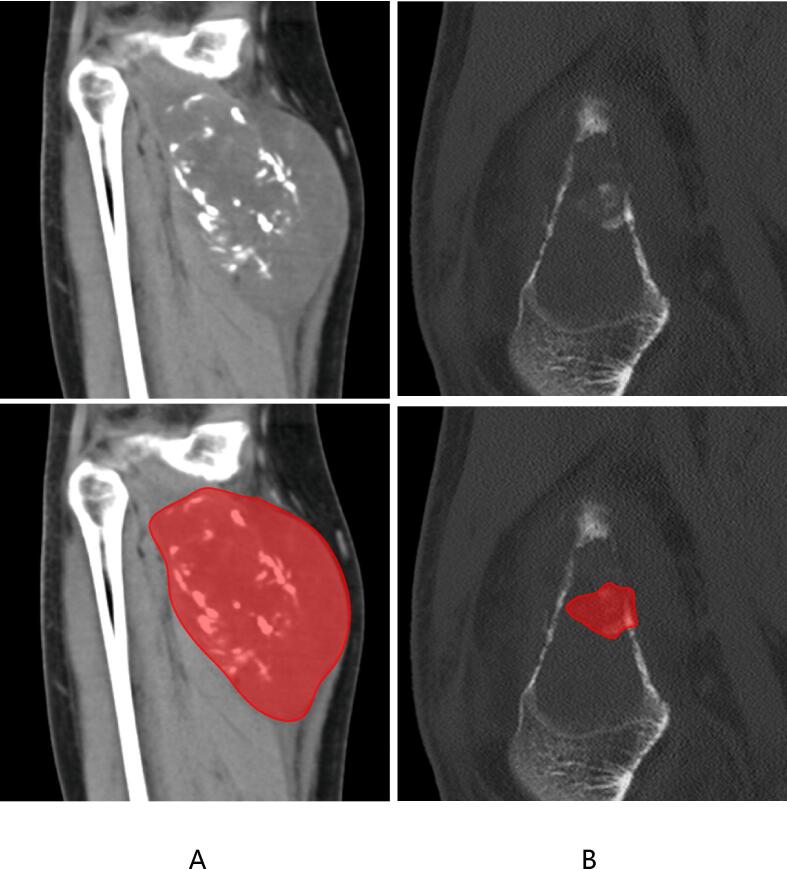
Segmenting the images to isolate the regions of interest (ROIs) for bone metastases. The segmentation was meticulously performed using specialized software by two orthopedic radiologists, each with over 15 years of experience in bone imaging diagnosis. To ensure accuracy, the segmentation masks were mutually validated by these experts, ensuring high-quality and precise annotations. Shown in Fig. 2.
Fig. 2.
ROI segmentation of CT. The diagram in A shows the segmentation of lung cancer metastasis, while B shows the segmentation of other cancer metastasis.
By adopting this dual-center approach and leveraging advanced preprocessing techniques, we aim to develop a robust deep learning model capable of accurately segmenting bone tumors and predicting whether the metastasis originates from lung cancer. This methodology ensures that the model is both scientifically valid and clinically applicable.
2.2. Model development
In this study, we utilized a 3D Mask R-CNN model to achieve automatic segmentation of bone tumors and predict whether the metastasis originates from lung cancer. The 3D Mask R-CNN extends the original Mask R-CNN to handle volumetric data, making it ideal for processing the spatial and contextual information inherent in CT scan volumes. This model architecture effectively manages the complexities of 3D medical imaging. The 3D Mask R-CNN model incorporates a 3D convolutional neural network within its Region Proposal Network (RPN) component to extract features from the CT scans, which is critical for generating region proposals. These features are then used to create anchor boxes with varying aspect ratios and scales, enhancing the accuracy of region proposals. The RPN assigns two values to each anchor box: the probability that the box contains an object, and the offset values required to adjust the box to accurately fit the object. To eliminate redundant region proposals, the RPN applies a non-maximum suppression algorithm, ensuring the selection of the most relevant proposals based on the extracted features and predefined anchor boxes. This approach allows the model to effectively identify and segment regions of interest in the 3D medical images, facilitating accurate diagnosis and treatment planning for patients with bone metastases. [25], [26].
After generating region proposals, the Mask R-CNN framework is responsible for classifying and segmenting the proposed regions. The RoI Align layer plays a crucial role by aligning the region proposals with the feature maps, ensuring spatial accuracy and reducing misalignment issues that can arise during the pooling process. This step significantly enhances the precision of the model, particularly when handling the fine details of 3D medical images.
Once aligned, the feature maps are processed by two parallel networks: a classifier and a segmentation network. The classifier is tasked with predicting the probability that a given region contains the target object (such as a tumor), allowing the model to filter out irrelevant areas. Simultaneously, the segmentation network generates a binary mask for each region, identifying the precise location of the object within the CT scan volume.
This dual-stage process—classification and segmentation—ensures both the accurate identification of tumors and their detailed segmentation. By combining these two tasks, the 3D Mask R-CNN model achieves robust and highly accurate detection and segmentation of tumors in CT scans. This integrated approach allows for a more reliable and comprehensive analysis, which is critical in clinical applications for diagnosing and treating bone metastases [26], [27].
2.3. Model architecture
Backbone Network (ConvNeXt-V2): The backbone of our improved 3D Mask R-CNN model is a ConvNeXt-V2, replacing the traditional ResNet-50. ConvNeXt-V2 is designed to enhance feature extraction by leveraging deeper convolutional layers and the Global Response Normalization (GRN) layer, which reduces redundant activations and increases feature diversity during training. Pre-trained on large-scale medical imaging datasets, this backbone captures spatial and contextual details in CT scans more effectively than ResNet-50, thus improving segmentation accuracy and model generalization.
Region Proposal Network (RPN): The RPN identifies regions of interest (ROIs) by sliding a small network over the feature map output generated by ConvNeXt-V2. It proposes candidate bounding boxes, which are refined in subsequent stages. Key RPN parameters include:
-
•
Anchor Scales: [4, 8, 16, 32, 64] for improved detection of objects at varying sizes.
-
•
Anchor Ratios: [0.5, 1, 2], allowing the model to account for different object shapes.
-
•
IoU Threshold: 0.95, optimized for balancing precision and recall during positive anchor classification.
RoI Align Layer: The RoI Align layer ensures precise spatial alignment of features by extracting fixed-size feature maps from the region proposals. This minimizes any potential misalignment and is crucial for maintaining high segmentation accuracy.
Fully Connected Layers: Two fully connected layers process the fixed-size feature maps to predict class scores and bounding box adjustments for each region proposal. These layers use ReLU activation functions to maintain computational efficiency while improving accuracy.
Mask Head: The mask branch is responsible for segmentation, consisting of several convolutional layers that generate binary masks for each region of interest. These masks pinpoint the precise location of tumors within the bounding boxes, ensuring detailed and accurate segmentation.
Training Parameters:
-
•
Optimizer: AdamW optimizer, chosen for its superior performance in handling large datasets and reducing overfitting.
-
•
Learning Rate: 1e-4 with a decay rate of 0.05 per epoch to gradually reduce the learning rate as training progresses.
-
•
Batch Size: 8, to ensure efficient utilization of GPU memory without compromising on performance.
-
•
Loss Functions: A combination of classification loss (cross-entropy), bounding box regression loss (smooth L1), and mask prediction loss (binary cross-entropy) ensures the model accurately predicts the class, bounding box, and segmentation mask for each ROI.
Data Augmentation: To improve the generalization of the model, a range of data augmentation techniques are applied:
-
•
Random Rotation: Up to 15 degrees to simulate variations in tumor orientation.
-
•
Random Scaling: Between 0.8 and 1.2 times the original size, ensuring robustness across different image sizes.
-
•
Random Flipping: Applied along the sagittal, coronal, and axial planes, reflecting the 3D nature of the medical images.
Implementation Details: The model was developed using the PyTorch deep learning framework, leveraging high-performance NVIDIA GPUs for training. By adopting the 3D Mask R-CNN architecture with the ConvNeXt-V2 backbone, our model aims to provide precise and robust segmentation of bone tumors. It can also predict whether the metastasis originated from lung cancer, thereby facilitating more accurate diagnoses and aiding in the development of personalized treatment plans (Fig. 3).
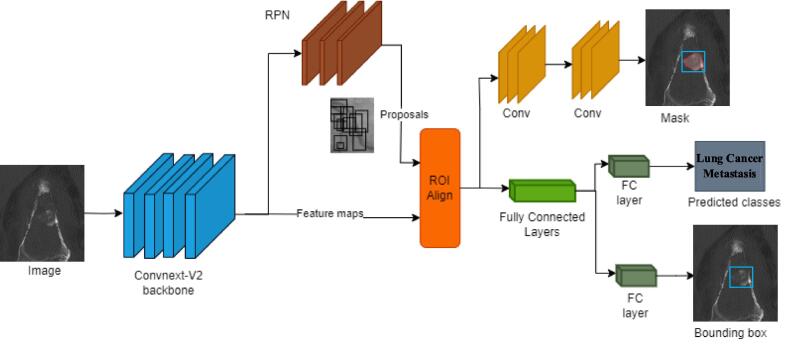
Fig. 3.
Architecture of 3D Mask RCNN. The architecture illustrates the key components of the 3D Mask R-CNN model with the ConvNeXt-V2 backbone. Input CT images are processed through the ConvNeXt-V2 backbone, extracting detailed feature maps. These maps are passed through the Region Proposal Network (RPN) to generate object proposals. The RoI Align layer ensures accurate spatial alignment of features, which are then fed into two branches: the segmentation branch, which generates binary masks to localize tumor regions, and the classification branch, which predicts the class (e.g., lung cancer metastasis) and refines the bounding box for precise localization.
2.4. Model training
To train our enhanced 3D Mask R-CNN model for bone tumor detection, we utilized a dataset comprising 265 CT scans, collected from patients with confirmed bone metastases. Prior to training, we standardized the resolution of all scans to a uniform voxel size of 1 mm × 1 mm × 1 mm, ensuring consistency across the dataset. Intensity normalization was applied to bring all pixel values to a zero mean and unit variance, which is crucial for minimizing variations in image quality that could affect model performance.
The dataset was randomly divided into training, validation, and testing sets in a 70:15:15 ratio. This allocation ensures sufficient data for model training while reserving a validation set for tuning hyperparameters and a testing set for evaluating model generalization. To further enhance the diversity of the training data and prevent overfitting, we implemented a series of 3D augmentation techniques, including random rotations (up to 15 degrees), translations, and scaling (ranging from 0.8 to 1.2 times the original size). Additionally, random flipping along the sagittal, coronal, and axial planes was employed to simulate variations in tumor positioning.
We employed the AdamW optimizer due to its improved performance in managing large datasets and reducing overfitting. The initial learning rate was set to 1e-4, and a decay schedule was applied, reducing the learning rate by a factor of 0.1 after 40 epochs to fine-tune the model as it approached convergence. The model was trained for 50 epochs, with early stopping applied based on validation loss to prevent overfitting and ensure optimal performance.
Model optimization was achieved using a composite loss function, combining classification loss and segmentation loss. The classification loss, computed using cross-entropy, measures the difference between the predicted and true class probabilities. The segmentation loss, based on binary cross-entropy, evaluates the discrepancy between the predicted binary masks and the ground truth masks, ensuring precise tumor segmentation. This multi-component loss function enabled the model to accurately predict both the tumor’s class (lung cancer metastasis or not) and its exact location within the CT scan.
By leveraging these techniques, our 3D Mask R-CNN model demonstrates robust performance in both detecting and segmenting bone tumors from CT scans, providing a valuable tool for clinical applications [28], [29].
2.5. Model evaluation
To assess the performance of our 3D Mask R-CNN model on the test dataset, we employed a comprehensive set of evaluation metrics [33], including the Dice Similarity Coefficient (DSC), sensitivity, specificity, and accuracy, which together provide a thorough understanding of the model's effectiveness in segmenting bone tumors and predicting their source.
Dice Similarity Coefficient (DSC): The DSC is a crucial metric for evaluating the overlap between the predicted segmentation masks and the ground truth. It measures spatial accuracy, with values ranging from 0 (no overlap) to 1 (perfect overlap). The DSC is particularly useful in medical imaging, as it quantifies the similarity between the predicted and actual tumor regions, directly reflecting the segmentation quality.
Sensitivity (True Positive Rate): Sensitivity measures the proportion of actual positive cases (e.g., tumors) that were correctly identified by the model. A high sensitivity indicates that the model is adept at detecting tumors, ensuring that few metastatic regions are missed during diagnosis.
Specificity (True Negative Rate): Specificity reflects the proportion of true negative cases (e.g., non-tumor regions) that the model correctly identified. High specificity ensures that healthy tissue is not mistakenly labeled as tumor, thus minimizing false positives and reducing unnecessary interventions.
Accuracy: Accuracy provides an overall assessment of the model’s performance by measuring the proportion of correct predictions (both true positives and true negatives) relative to all predictions made. It balances the detection of tumors with the correct identification of non-tumor regions, making it a comprehensive indicator of model reliability.
In addition to these core metrics, we used the Receiver Operating Characteristic (ROC) curve and Area Under the Curve (AUC) to evaluate the model’s classification performance. The ROC curve visualizes the trade-off between sensitivity and specificity across various decision thresholds, offering insight into the model’s ability to distinguish between different classes (tumor vs. non-tumor). The AUC quantifies the overall performance of the model, with values closer to 1 indicating near-perfect classification performance. A higher AUC signifies that the model is proficient at distinguishing between metastatic bone tumors and other regions, even under varying thresholds.
By utilizing these metrics collectively, we can assess not only the model’s segmentation precision but also its predictive capability in determining the origin of bone metastases. This multi-dimensional evaluation ensures a robust and reliable analysis of the model's performance in both segmentation and classification tasks, making it a valuable tool for clinical applications in detecting and managing bone tumors.
3. Results
Segmentation Performance
Our 3D Mask R-CNN model achieved high accuracy in segmenting bone tumors from CT scans, with strong performance observed across both the training and testing sets. Specifically, the Dice Similarity Coefficient (DSC), a metric commonly used to evaluate segmentation overlap between the predicted masks and ground truth, was 0.956 on the training set and 0.929 on the testing set (see Table 1). These DSC values indicate a high degree of precision in the model’s ability to identify and segment tumor regions from the CT scans, showing the model’s capacity to generalize well from training data to unseen test data.
Table 1.
Details of model segmentation performance evaluation parameters.
| Process | DSC |
|---|---|
| Training | 0.956 |
| Testing | 0.929 |
Classification Performance
In addition to segmentation, the model's performance in distinguishing between bone metastases originating from lung cancer and other sources was evaluated using several metrics, including Area Under the Curve (AUC), accuracy, sensitivity, and specificity. The model demonstrated solid classification results, particularly when utilizing the ConvNeXt-V2 backbone. On the training set, the ConvNeXt-V2-based model achieved an AUC of 0.991, an accuracy of 0.951, a sensitivity of 0.925, and a specificity of 0.980. On the test set, the model showed robust performance, achieving an AUC of 0.955, an accuracy of 0.886, a sensitivity of 0.808, and a specificity of 1.000 (see Table 2). These results underscore the effectiveness of the model in distinguishing between lung cancer metastases and other cancer types.
Table 2.
Details of model classification performance evaluation parameters.
| Architecture | Accuracy | AUC | 95 % CI | Sensitivity | Specificity | F1 | Task |
|---|---|---|---|---|---|---|---|
| Resnet-50 | 0.853 | 0.931 | 0.885–0.977 | 0.830 | 0.878 | 0.854 | label-train |
| 0.864 | 0.859 | 0.713–1.000 | 0.846 | 0.889 | 0.880 | label-test | |
| Convnext-Base | 0.892 | 0.939 | 0.891–0.986 | 0.906 | 0.878 | 0.897 | label-train |
| 0.864 | 0.925 | 0.844–1.000 | 0.846 | 0.889 | 0.880 | label-test | |
| Resnet-101 | 0.912 | 0.976 | 0.952–0.998 | 0.868 | 0.959 | 0.911 | label-train |
| 0.864 | 0.940 | 0.876–1.000 | 0.769 | 1.000 | 0.870 | label-test | |
| Convnext-V2-Base | 0.951 | 0.991 | 0.979–1.000 | 0.925 | 0.980 | 0.951 | label-train |
| 0.886 | 0.955 | 0.901–1.000 | 0.808 | 1.000 | 0.894 | label-test | |
| YOLO-V8 | 0.853 | 0.929 | 0.881–0.976 | 0.736 | 0.980 | 0.839 | label-train |
| 0.841 | 0.850 | 0.706–0.994 | 0.885 | 0.778 | 0.868 | label-test | |
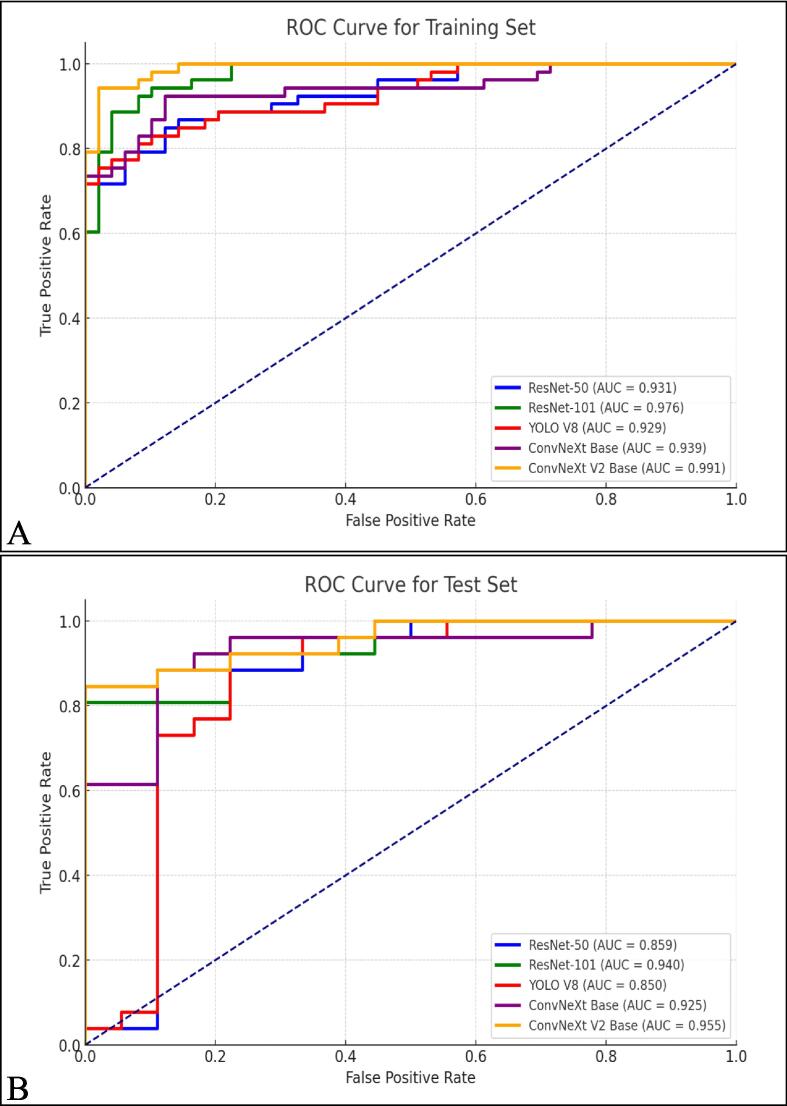
The ROC curves for the training and testing sets, shown in Fig. 4, further illustrate the model's classification capabilities. For the training set (Fig. 4A), the ConvNeXt-V2 backbone outperformed other architectures with an AUC of 0.991, closely followed by ResNet-101 with an AUC of 0.976. On the testing set (Fig. 4B), the ConvNeXt-V2 backbone continued to show strong performance, achieving an AUC of 0.955, indicating its superior ability to distinguish between lung cancer metastasis and other metastases, even in unseen data. These results confirm that the 3D Mask R-CNN model, particularly when using the ConvNeXt-V2 backbone, is highly effective in both segmentation and classification tasks. This performance is critical for real-world clinical applications, where precise tumor identification and accurate classification are essential for effective treatment planning.
Fig. 4.
ROC of automatic discrimination of bone metastases from lung cancer (A) and other cancers (B).
4. Discussion
The findings of this study underscore the significant clinical potential of our 3D Mask R-CNN model, especially when enhanced with the ConvNeXt-V2 backbone, in the segmentation and classification of bone metastases originating from lung cancer. Bone metastases, particularly from lung cancer, are associated with severe complications such as pathological fractures, spinal cord compression, and hypercalcemia, all of which severely impact a patient’s quality of life. Early and precise detection of metastatic sites is crucial for effective treatment planning, enabling the timely implementation of radiation therapy, surgery, or systemic treatments [30].
Our model’s ability to accurately identify metastatic regions through automated segmentation is a major step toward improving clinical outcomes. Manual segmentation of tumors is labor-intensive, prone to inter-observer variability, and time-consuming. By automating this process with a robust AI-driven model, clinicians can focus more on treatment planning and patient care. Additionally, automated segmentation allows for the consistent monitoring of tumor progression or regression during treatment, providing a reliable method for evaluating the efficacy of therapeutic interventions [10].
Furthermore, the model’s classification capabilities in distinguishing lung cancer metastases from other types of cancer metastases are clinically valuable. Accurate identification of the primary source of metastasis is critical for guiding personalized treatment strategies, such as targeted therapies or immunotherapies specific to the cancer type. By integrating this model into clinical workflows, unnecessary biopsies or additional imaging tests could be reduced, streamlining the diagnostic process and potentially lowering healthcare costs.
The integration of the ConvNeXt-V2 backbone into the 3D Mask R-CNN architecture is a significant technological innovation in the realm of medical imaging. ConvNeXt-V2, with its deeper convolutional layers and Global Response Normalization (GRN), allows the model to extract more semantically rich and nuanced features from the CT scan images. This is particularly important for the detection of small or complex metastatic lesions that may be missed by more traditional models, improving the overall sensitivity and accuracy of the system [19].
Moreover, the transition from 2D to 3D imaging in the Mask R-CNN model marks a substantial leap forward in medical imaging technology. CT scans produce volumetric data, and the ability of the 3D Mask R-CNN model to process this data while maintaining spatial coherence significantly enhances the precision of segmentation. This dual-stage approach—combining region proposal generation with segmentation and classification—enables the model to not only identify tumor regions but also differentiate them based on their primary cancer source, thus offering more holistic insights into patient diagnosis and treatment [26].
The high performance of the model, demonstrated by the high Dice Similarity Coefficient (DSC) values (0.956 for training and 0.929 for testing) and strong AUC values (0.991 for training and 0.955 for testing), places this model among the best in the current landscape of medical imaging technologies for cancer diagnosis. The use of the ConvNeXt-V2 backbone addresses some of the challenges faced by traditional convolutional neural networks (CNNs), such as feature loss in deeper layers, allowing for better generalization in both segmentation and classification tasks.
This study contributes to the field in several important ways. First, by successfully integrating ConvNeXt-V2 into the 3D Mask R-CNN framework, we have demonstrated an improvement in both segmentation and classification tasks for bone metastases detection. This novel combination enhances the ability to handle complex 3D data [31], [32] and offers a scalable solution for integrating AI into clinical workflows.
Secondly, the dual-center study design adds robustness to the model’s performance. By training and testing the model on independent datasets from two different hospitals, we ensured that the model was not overfitted to a specific dataset, enhancing its generalizability across different clinical settings. This aspect is crucial for developing AI models that are capable of being deployed in diverse healthcare environments.
Lastly, this study provides a pathway for future research in personalized cancer treatment. The combination of accurate segmentation with classification of the metastatic source enables a more tailored therapeutic approach, which can lead to improved patient outcomes and quality of life. This model also opens the door for further integration of AI in cancer diagnostics, not only for bone metastases but potentially for other cancer types and metastatic sites as well.
5. Conclusion
In summary, this study demonstrates the feasibility and effectiveness of using a 3D Mask R-CNN model, enhanced by the ConvNeXt-V2 backbone, for the automatic segmentation and classification of bone metastases from lung cancer. The high accuracy, sensitivity, and specificity achieved in both segmentation and classification tasks highlight the potential of AI-driven models to significantly improve diagnostic workflows in oncology. Moving forward, expanding the dataset and integrating additional clinical data, such as genetic or laboratory information, will further enhance the performance and clinical applicability of the model. This study lays the groundwork for broader adoption of AI in cancer diagnostics, with the ultimate goal of improving patient care and outcomes.
Funding support
Supported by Medicine Plus Program of Shenzhen University, No.2024YG011; Shenzhen Health Elite Talents, No.2021XKQ193; Science and Technology Program of Nanshan District, Shenzhen, No.NS2023128; Education Reform Project of Guangdong Province, No.2021JD082.
CRediT authorship contribution statement
Ketong Zhao: Writing – original draft, Data curation, Writing – review & editing. Ping Dai: Data curation. Ping Xiao: Data curation. Yuhang Pan: Methodology. Litao Liao: Resources. Junru Liu: Software. Xuemei Yang: Supervision. Zhenxing Li: Formal analysis. Yanjun Ma: Formal analysis. Jianxi Liu: Software. Zhengbo Zhang: Data curation. Shupeng Li: Methodology. Hailong Zhang: Methodology. Sheng Chen: Investigation. Feiyue Cai: Writing – review & editing. Zhen Tan: Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Contributor Information
Feiyue Cai, Email: caifeiyue@ocibe.com.
Zhen Tan, Email: tanzhen@szu.edu.cn.
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2020. CA Cancer J. Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Coleman R.E. Clinical features of metastatic bone disease and risk of skeletal morbidity. Clin. Cancer Res. 2006;12:6243s–6249s. doi: 10.1158/1078-0432.Ccr-06-0931. [DOI] [PubMed] [Google Scholar]
- 3.Coletti C., Acosta G.F., Keslacy S., Coletti D. Exercise-mediated reinnervation of skeletal muscle in elderly people: an update. Eur. J. Transl. Myol. 2022;32 doi: 10.4081/ejtm.2022.10416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsuya A., Kurata T., Tamura K., Fukuoka M. Skeletal metastases in non-small cell lung cancer: a retrospective study. Lung Cancer. 2007;57:229–232. doi: 10.1016/j.lungcan.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 5.Medeiros B., Allan A.L. Molecular mechanisms of breast cancer metastasis to the lung: clinical and experimental perspectives. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20092272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riihimäki M., et al. Metastatic sites and survival in lung cancer. Lung Cancer. 2014;86:78–84. doi: 10.1016/j.lungcan.2014.07.020. [DOI] [PubMed] [Google Scholar]
- 7.Body J.J., et al. Bone health in the elderly cancer patient: a SIOG position paper. Cancer Treat. Rev. 2016;51:46–53. doi: 10.1016/j.ctrv.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 8.Salamanna F., Borsari V., Contartese D., Nicoli Aldini N., Fini M. Link between estrogen deficiency osteoporosis and susceptibility to bone metastases: a way towards precision medicine in cancer patients. Breast. 2018;41:42–50. doi: 10.1016/j.breast.2018.06.013. [DOI] [PubMed] [Google Scholar]
- 9.Wallington-Beddoe C.T., Mynott R.L. Prognostic and predictive biomarker developments in multiple myeloma. J. Hematol. Oncol. 2021;14:151. doi: 10.1186/s13045-021-01162-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lipton A., Costa L., Ali S., Demers L. Use of markers of bone turnover for monitoring bone metastases and the response to therapy. Semin. Oncol. 2001;28:54–59. doi: 10.1016/s0093-7754(01)90233-7. [DOI] [PubMed] [Google Scholar]
- 11.Nielsen O.S., Munro A.J., Tannock I.F. Bone metastases: pathophysiology and management policy. J. Clin. Oncol. 1991;9:509–524. doi: 10.1200/jco.1991.9.3.509. [DOI] [PubMed] [Google Scholar]
- 12.Vanel D., Bittoun J., Tardivon A. MRI of bone metastases. Eur. Radiol. 1998;8:1345–1351. doi: 10.1007/s003300050549. [DOI] [PubMed] [Google Scholar]
- 13.Deberne M., et al. Inaugural bone metastases in non-small cell lung cancer: a specific prognostic entity? BMC Cancer. 2014;14:416. doi: 10.1186/1471-2407-14-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y., et al. ESA-UNet for assisted diagnosis of cardiac magnetic resonance image based on the semantic segmentation of the heart. Front. Cardiovasc. Med. 2022;9:1012450. doi: 10.3389/fcvm.2022.1012450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niu Y., et al. Risk factors for bone metastasis in patients with primary lung cancer: a systematic review. Medicine (Baltimore) 2019;98:e14084. doi: 10.1097/MD.0000000000014084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Z.Q., et al. Bone age recognition based on mask R-CNN using xception regression model. Front. Physiol. 2023;14:1062034. doi: 10.3389/fphys.2023.1062034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saiki M., et al. Characterization of computed tomography imaging of rearranged during transfection-rearranged lung cancer. Clin. Lung Cancer. 2018;19:435–440.e431. doi: 10.1016/j.cllc.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 18.Gentile M., et al. Application of “omics” sciences to the prediction of bone metastases from breast cancer: state of the art. J. Bone Oncol. 2021;26 doi: 10.1016/j.jbo.2020.100337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y.Z., et al. Automated meniscus segmentation and tear detection of knee MRI with a 3D mask-RCNN. Eur. J. Med. Res. 2022;27:247. doi: 10.1186/s40001-022-00883-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tomasian A., Jennings J.W. Bone metastases: state of the art in minimally invasive interventional oncology. Radiographics. 2021;41:1475–1492. doi: 10.1148/rg.2021210007. [DOI] [PubMed] [Google Scholar]
- 21.Spadea M.F., et al. Deep convolution neural network (DCNN) multiplane approach to synthetic CT generation from MR images-application in brain proton. Therapy. Int. J. Radiat. Oncol. Biol. Phys. 2019;105:495–503. doi: 10.1016/j.ijrobp.2019.06.2535. [DOI] [PubMed] [Google Scholar]
- 22.Chen S., et al. MR-based synthetic CT image for intensity-modulated proton treatment planning of nasopharyngeal carcinoma patients. Acta Oncol. 2022;61:1417–1424. doi: 10.1080/0284186x.2022.2140017. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y., Liu C., Zhang X., Deng W. Synthetic CT generation based on T2 weighted MRI of nasopharyngeal carcinoma (NPC) using a deep convolutional neural network (DCNN) Front. Oncol. 2019;9:1333. doi: 10.3389/fonc.2019.01333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang H., et al. Application of deep convolutional neural networks for discriminating benign, borderline, and malignant serous ovarian tumors from ultrasound images. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.770683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen Y., Ji R., Wang C., Li X., Li X. Weakly supervised object detection via object-specific pixel gradient. IEEE Trans. Neural Netw. Learn. Syst. 2018;29:5960–5970. doi: 10.1109/tnnls.2018.2816021. [DOI] [PubMed] [Google Scholar]
- 26.Li X., et al. Generalized focal loss: towards efficient representation learning for dense object detection. IEEE Trans. Pattern Anal. Mach. Intell. 2023;45:3139–3153. doi: 10.1109/tpami.2022.3180392. [DOI] [PubMed] [Google Scholar]
- 27.Lin T.Y., Goyal P., Girshick R., He K., Dollar P. Focal loss for dense object detection. IEEE Trans. Pattern Anal. Mach. Intell. 2020;42:318–327. doi: 10.1109/tpami.2018.2858826. [DOI] [PubMed] [Google Scholar]
- 28.Szentimrey Z., de Ribaupierre S., Fenster A., Ukwatta E. Automated 3D U-net based segmentation of neonatal cerebral ventricles from 3D ultrasound images. Med. Phys. 2022;49:1034–1046. doi: 10.1002/mp.15432. [DOI] [PubMed] [Google Scholar]
- 29.Nodirov J., Abdusalomov A.B., Whangbo T.K. Attention 3D U-net with multiple skip connections for segmentation of brain tumor images. Sensors (basel) 2022;22 doi: 10.3390/s22176501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsuya A., Kurata T., Tamura K., Fukuoka M. Skeletal metastases in non-small cell lung cancer: a retrospective study. Lung Cancer. 2007;57 doi: 10.1016/j.lungcan.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 31.Shao Y., Yin Y., Shichang Du., Xia T., Xi L. Leakage monitoring in static sealing interface based on three dimensional surface topography indicator. ASME Trans. Manufact. Sci. Eng. 2018;140(10) [Google Scholar]
- 32.Zhao C., Lv J., Shichang Du. Geometrical deviation modeling and monitoring of 3D surface based on multi-output Gaussian process. Measurement. 2022;199 [Google Scholar]
- 33.K.K.L. Wong, Cybernetical Intelligence: Engineering Cybernetics with Machine Intelligence, John Wiley & Sons, Inc., Hoboken, New Jersey, ISBN: 9781394217489, 2023.