Abstract
Stem cell therapies for Parkinson’s disease are at an exciting time of development, and several clinical trials have recently been initiated. Human pluripotent stem cells are differentiated into transplantable dopamine (DA) progenitors which are proliferative at the time of grafting and undergo terminal differentiation and maturation in vivo. While the progenitors are homogeneous at the time of transplantation, they give rise to heterogeneous grafts composed not only of therapeutic DA neurons but also of other mature cell types. The mechanisms for graft diversification are unclear. We used single-nucleus RNA-seq and ATAC-seq to profile DA progenitors before transplantation combined with molecular barcode-based tracing to determine origin and shared lineages of the mature cell types in the grafts. Our data demonstrate that astrocytes, vascular leptomeningeal cells, and DA neurons are the main component of the DAergic grafts, originating from a common progenitor that is tripotent at the time of transplantation.
Transplanted dopamine progenitors give rise to multiple cell types in the mature graft.
INTRODUCTION
Parkinson’s disease (PD) is the second most common neurodegenerative disease and an ideal candidate for stem cell therapy as it is primarily one neuronal cell type, the dopamine (DA) neurons of the substantia nigra pars compacta, that is affected. Replacing this missing cellular component with new DA neurons is expected to restore DA neurotransmission in the DA-depleted parts of the striatum. Clinical proof-of-concept studies in patients with PD have involved intrastriatal transplantation of subdissected tissue from the human fetal ventral mesencephalon, which is the area where DA neurons are born during development. These studies have shown that the transplanted cells can provide long-term motor improvements in a subset of patients but that the use of fetal tissue is both ethically and logistically challenging (1). For better consistency, quality control, and cell availability, manufactured cell products based on pluripotent stem cells have been developed by several teams, including our own, and are now being investigated in clinical trials (2–4). In these efforts, DA progenitors of high purity are generated by patterning human embryonic stem cells (hESCs) or human-induced pluripotent stem cells into a ventral midbrain (VM) fate and subsequently grafting these to the brain, where the cells will undergo their last cell divisions, differentiation, and functional maturation.
The preclinical studies leading up to these trials have excluded the presence of remaining pluripotent cells, as well as efficiently minimized the presence of insufficiently or incorrectly specified off-target cells (2–4). Thus, DA progenitors of high purity are transplanted, yet the cells give rise to heterogeneous grafts composed not only of DA neurons but also of different mature cell types such as astrocytes (3, 5), oligodendrocytes (5), and vascular leptomeningeal cells (VLMCs) (5, 6). In some cases, grafts also gave rise to choroid plexus cells, but these were later determined to arise from a contaminating progenitor cell population caused by insufficient bone morphogenetic protein inhibition present at the time of grafting, which could be eliminated by culture modification (4). However, the presence of the other non-DA cell types does not appear to be associated with insufficient patterning or presence of contaminating off-target progenitor populations at the time of grafting. This is further supported by a recent experimental study performed by Xu et al. (6) showing that while cell sorting for a highly specific double-positive EN1+/LMX1A+ DA progenitor can remove off-target contaminants, these purified DA progenitors still give rise to mature grafts containing astrocytes, neurons, and VLMCs in equal proportion as the unsorted grafts (6). These findings raise intriguing questions on how posttransplantation diversification occurs, and if control of the diversification process can be harnessed to produce grafts with predefined, optimal cellular compositions.
In this study, we used a single-nucleus assay for sequencing transposase-accessible chromatin (snATAC-seq) and mRNA expression in paired maps to assess the homogeneity of progenitors at the time of transplantation. This was followed by combined single-cell transcriptomics and barcoding lineage tracing of the stem cell–derived DA progenitors to experimentally address graft diversification and computationally reconstruct the cellular origin of the different cell types. The transcriptional analysis revealed three major cell types present in the grafts: DA neurons, astrocytes, and VLMCs. Barcode retrieval and mapping to individual cells identified shared barcodes among all the different cell types within the grafts, indicating a shared cellular origin. To pinpoint the time point when lineage diversification occurs, we also barcode-tagged the transplanted cells in vivo 1 month after grafting and found that the majority of the cells that remained proliferative at this time point also maintained their multipotent differentiation potential. Together, single-cell multiomics and barcode-based lineage tracing revealed that the different subtypes of graft-derived cell types originate from a common cellular progenitor. We also identified a subset of genes whose combined expression was associated with a bias of the progenitors toward a DAergic fate.
RESULTS
Multiomic characterization reveals homogeneity of hESC-derived DA progenitors
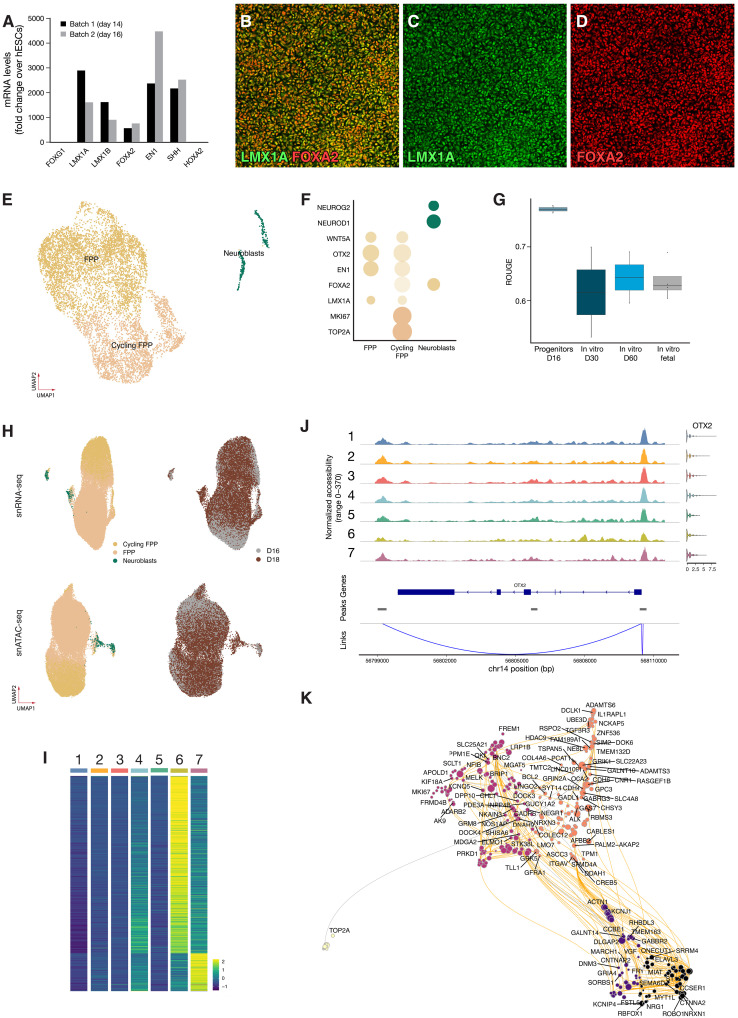
Extrinsic patterning signals were used to guide stem cell differentiation into a caudal VM fate to obtain highly pure populations of DA progenitors according to the same protocol as was used to produce guanosine monophosphate–grade cells in our clinical trial STEM-PD (NCT05635409) (7). The differentiations resulted in a homogeneous population of progenitors with high expression of LMX1A, EN1, FOXA2, and OTX2 at days 14 and 16 [quantitative reverse transcription polymerase chain reaction (PCR); Fig. 1A] and confirmed by confocal microscopy (Fig. 1, B to D, and fig. S1, A and B). The progenitor cells were analyzed at day 16 of differentiation using single-cell RNA sequencing (scRNA-seq), and after quality control and filtering (see Materials and Methods), we obtained transcriptomes from a total of 8166 cells with a median of 30,566 unique molecular identifiers (UMIs) and 5354 detected genes per cell. The progenitors were clustered and annotated on the basis of literature-curated marker genes (Fig. 1, E and F). This revealed one major cluster of midbrain floor plate progenitors (FPPs; 96% of the cells, expressing LMX1A, EN1, WNT5A, OTX2, and FOXA2). The FPP cluster could be further divided into a proliferating and a nonproliferating subcluster, where the main difference was expression of genes associated with cell division (i.e., MKI67 and TOP2A). In total, 58% of the cells in the progenitor population was predicted to be actively cycling. In addition to FPPs, we also identified a small postmitotic, neuroblast population (4% of cells) expressing proneuronal genes (NEUROD1 and NEUROG2). To quantify the homogeneity of the progenitors, we applied Ratio of Global Unshifted Entropy (ROGUE) (8) statistics to determine the purity of each single-cell sample while accounting for sample to sample differences and compared it to samples differentiated in vitro for up to 60 days (9). As expected, hESC-derived progenitors showed a very high ROGUE score of ~0.8 (Fig. 1G) with a decreasing value as the samples were differentiated in vitro (declining to 0.64 at 60 days). We also compared hESC-derived progenitors to fetal VM samples that are known to contain a higher number of different cell types (10), and we also confirmed using the ROGUE analysis that hESC-derived progenitors were more homogeneous than dissected fetal midbrain (post-conception weeks 8 to 11, 0.62; Fig. 1G). In conclusion, scRNA-seq confirmed high transcriptional homogeneity of hESC-derived VM progenitors at day 16, which is a standard time point for transplantation in preclinical as well as in clinical studies (2, 3, 11).
Fig. 1. Multiomic characterization of VM progenitors.
(A) Quantitative PCR quantification of midbrain markers in progenitors at days 14 and 16 (D16). (B to D) Representative confocal images of DA progenitors of high homogeneity coexpressing FOXA2 and LMX1A. (E) UMAP visualization of scRNA-seq data from progenitors before grafting (n = 8166) colored by cell type. (F) Dot plot of key progenitor markers expression per cell type. (G) Boxplot showing cell purity for each dataset as assessed by ROGUE (n = 2 to 4). (H) UMAP visualization of integrated snATAC- and snRNA-seq of progenitors from days 16 and 18 (n = 2 replicates per time point) colored by predicted cell type and day. (I) Differentially accessible peaks (false discovery rate adjusted P < 0.05) for any subcluster; see also fig. S1 [(C) and (D)]. (J) Linked peaks with transcription of OTX2 across clusters show no differential accessibility across clusters. (K) UMAP embedding of the inferred Pando-based TF network in DA progenitors.
Epigenetic changes are known to foreshadow changes in gene expression (12), suggesting that the earliest events for cell fate choice are unlikely to be captured using gene expression alone. Thus, to get an ever deeper analysis of potential progenitor heterogeneity, we also performed multiome single-nucleus RNA and ATAC sequencing using the Chromium 10X platform comprehensively characterizing the full cellular transcriptome and chromatin accessibility. After quality control and filtering, we retained 15,605 paired profiles for day 16 and 27,014 for day 18 progenitors. Consistent with the scRNA-seq analysis, the integrated data partitioned into one major cluster of FPPs and one small cluster of postmitotic neurons (Fig. 1H). A consensus set of 329,315 accessible peaks representing potential cis-regulatory elements were identified which showed a similar pattern when visualized on Uniform Manifold Approximation and Projection (UMAP) embeddings separately (Fig. 1H). Differential abundance analysis between the different clusters using a higher resolution factor identified seven subclusters (fig. S1, C and D) and 621 differentially abundant peaks; a great majority of which were more abundant in the postmitotic clusters and with very few differentially abundant peaks in the progenitor subclusters (Fig. 1I). We used the joint profiles obtained from VM progenitors to detect cis-regulatory interactions and identified Domains Of Regulatory Chromatin (referred to as DORCs; fig. S1E) (13), which exhibit substantial overlap with super-enhancers. This did not differ between the clusters but could be further investigated for potential role in DA differentiation. Next, we mapped the linked expression of the VM markers LMX1A, FOXA2, EN1, and OTX2 (Fig. 1J) with putative cis-regulatory elements across the different clusters. None of these showed any evidence of differential expression nor differential promotor usage, confirming the finding that the progenitors were transcriptionally and epigenetically homogeneous at this stage in terms of both chromatin accessibility and gene activity of DA-related transcription factors (TFs).
We next used the Pando algorithm (14) to infer the gene regulatory networks (GRNs) underlying progenitor development and potential. This algorithm uses multimodal single-cell genomic measurements to model gene expression through TF-peak interactions. We used a general linear model to explore the relationships between each gene, its target TF, and the binding site accessibility, resulting in a list of regulated genes (gene modules) and regulatory motifs for each TF. In total, 86 TF modules were identified, comprising 375 unique TFs and 595 target genes (table S1). To visualize the GRNs, we used UMAP embedding and performed k-means clustering on the resulting graph, leading to the identification of five distinct clusters related to specific aspects of DA progenitor biology (Fig. 1K), including one cluster of TFs driving a neurogenesis program and another driving cell cycle. Gene Ontology enrichment analysis of gene modules provided further insights into the biological processes and functions associated with each cluster, with detailed data included in table S2. Scoring the gene modules across different cell clusters enabled us to map these modules to specific cell types, enhancing our understanding of the distinct biological roles within the DA progenitor population. For example, gene module 1 includes genes such as EBF3, MYT1L, ONECUT2, ROBO1, and SEMA6D, which are pivotal for midbrain progenitor biology and neural connectivity (fig. S1F). In addition, gene module 2, relevant across all subclusters, includes CREB5, GFRA1, and ITGAV, highlighting their roles in neural differentiation, survival, and midbrain structure organization (fig. S1G). These findings confirmed the preclinical data and snRNA-seq data presented here that the DA progenitors were highly homogeneous at the time point of transplantation, both at the level of the transcriptome as well as chromatin accessibility and gene activity of DA-related TFs. We therefore concluded that graft diversification is likely to take place after delivery to the brain and suggest a yet undefined multilineage differentiation potential of the DA progenitors.
Lineage tracing of DA progenitors through lentiviral barcoding and snRNA-seq
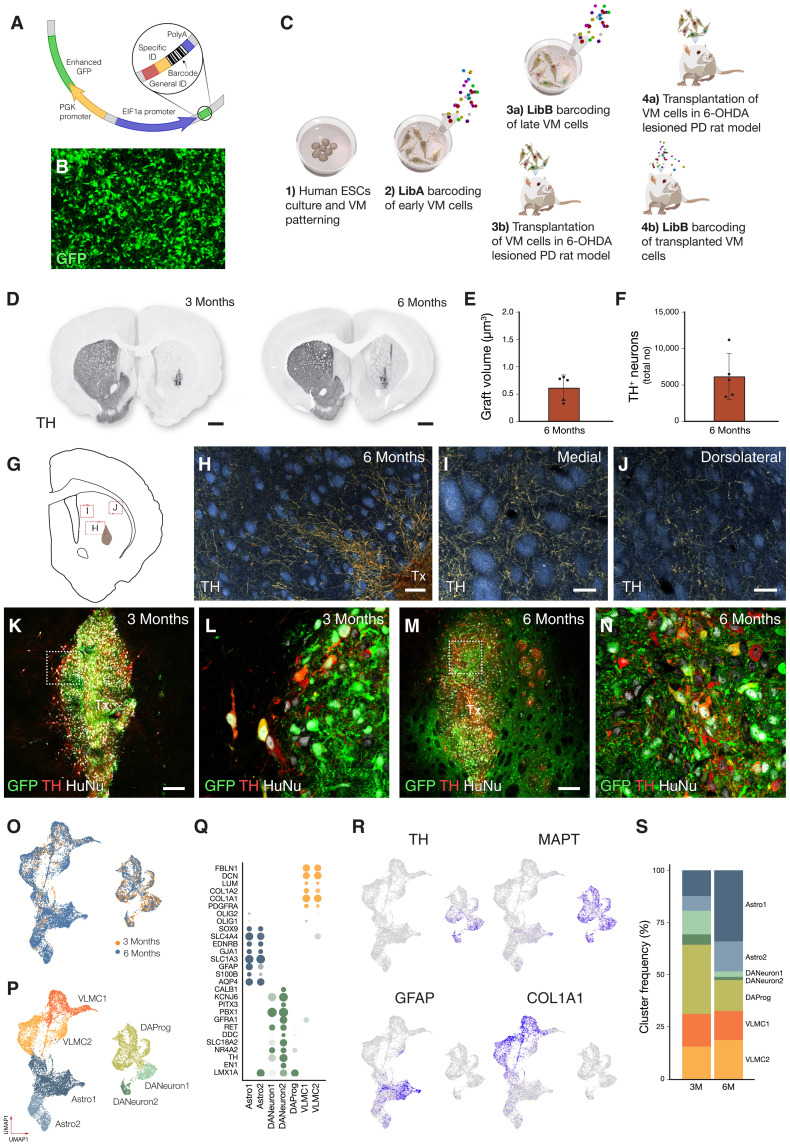
To experimentally address whether lineage diversification occurs after transplantation, we used pooled lentiviral libraries to tag the DA progenitors at different time points of differentiation with unique and heritable genetic barcodes. We made use of two libraries (LibA and LibB) that each contained 20-bp long, random nucleotide sequences (molecular barcodes) followed by a short library identification sequence and a polyadenylation signal, all driven by the eIF1α promoter (Fig. 2A). In addition, the vector also contained green fluorescent protein (GFP) for detection of transduction (Fig. 2B and fig. S1H). The libraries had a bottlenecked diversity of around 1 M sequencing-validated molecular barcodes each. In the first study, the two different pooled lentiviral barcode libraries were used to tag DA progenitors before transplantation at days 11 and 16 in vitro, followed by transplantation into a preclinical xenograft rat model of PD at day 18 (n = 16 rats; Fig. 2C). In the second experiment, LibA was delivered at day 11 in vitro and transplanted at day 18 (n = 16 rats). These animals received intracerebral injections of LibB in vivo 1 month after grafting (n = 16 rat; Fig. 2C).
Fig. 2. Succesful grafting and snRNA-seq characterization of midbrain grafts.
(A) Schematic of lentiviral vector used for lineage tracing. (B) Representative confocal image of progenitors expressing GFP, indicative of successful transduction. (C) Experimental design for combined clonal tracing and transcriptomics in neural grafts. (D) Neuron and TH-rich grafts were analyzed 3 and 6 months after transplant (representative images from n = 8 for each time point). (E and F) Quantification of graft volume and TH+ neurons. (G) Schematic of the striatal region from where panels (H) to (J) were captured. (H to J) Dark field microscopy of TH staining confirming extensive DA innervation. Tx, transplant. (K to N) In grafts from cells that received LibA and LibB in vitro, there is a complete colocalization of GFP and human grafted cells. Inset shows GFP+ TH+ cells. (L) In grafts where LibB was delivered to the grafted cell 1 month after transplant, GFP can be detected both in graft and host parenchyma. (O and P) UMAP visualization of stem cell–derived cells (n = 14,414) from 12 rats colored by graft age (O) and cell type (P). (Q) Dot plot of key marker genes showing % of cells expressing and expression level of indicated genes. (R) Featureplots of key genes to distinguish DA neurons [TH and microtubule associated protein tau (MAPT)], astrocytes (GFAP), and VLMC (COL1A1). (S) Bar plot of % of identified cell types in short-term (3 months, n = 6) and long-term (6 months, n = 6) grafts.
Five rats per time point and condition were perfused and processed for histology at 3- and 6-month posttransplantation, confirming formation of DA-rich grafts (Fig. 2, D to N). The graft volume was relatively small (Fig. 2E). The total number of tyrosine hydroxylase (TH) cells per graft was 6384 ± 1366 (Fig. 2F) with a yield of 2.1% which is in range of similar studies from our group (2). Dark field microscopy of the TH staining confirmed extensive DAergic innervation with TH+ fibers extending from the graft core (Fig. 2H) into the medial (Fig. 2I) and dorsolateral striatum (Fig. 2J). In animals from group 1, where both barcode libraries were delivered in vitro before grafting, GFP, HuNu, and TH-expressing cells were all contained within the graft (Fig. 2, K and L). In contrast, in group 2 where LibA was delivered in vitro and LibB was delivered in vivo, transduction with GFP was detected in both the graft and the surrounding host tissue, as was expected (Fig. 2, M and N). The remaining grafts were analyzed using snRNA-seq 3 and 6 months after grafting, enabling the identification of cell identity based on their transcriptional profile (n = 3 per condition, total of 12 animals). From these animals, a total of 14,414 human nuclei were retained after quality filtering (details in Materials and Methods and fig. S2A). These were integrated using Seurat canonical correlation analysis (15), revealing a similar cell composition at 3 and 6 months after grafting (Fig. 2O). On the basis of marker gene analysis and comparisons with primary reference atlases (see Materials and Methods), three major cell types, DA neurons, astrocytes, and VLMCs, were detected in the grafts in roughly equal proportions (Fig. 2, P to S). On the basis of silhouette analysis, DA neurons could be further subdivided into a cluster of postmitotic DA precursors (SOX6+/LMX1A+) and two more mature DA neurons cluster with high expression of TH and KCNJ6 (GIRK2). Similarly, two astrocytic clusters were identified: Astro-2 showed expression of GINS3, indicating a growth and reparative phenotype (16), whereas Astro-1 was more immature (VIM+/NES+). While the number of captured DA neurons and VLMCs was similar at 3 and 6 months, the number of astrocytes increased over time from 19 to 38% (Fig. 2S). This is in line with our previous observations on graft composition using scRNA-seq (5). However, in contrast to our previous study based on scRNA-seq, snRNA-seq as used in this current study captured 10-fold more neurons. Analysis of each graft separately was also performed and showed similar graft outcomes in all animals analyzed (fig. S2B) validating the experimental setup and confirming the robustness of the approach. In the subsequent analysis, 3- and 6-month grafts were merged and analyzed together.
After profiling by snRNA-seq, one or more LibA barcodes could be robustly detected in 70% of the mature cells in the graft (76% of the neurons, 67% of astrocytes, and 69% of VLMCs contained barcodes; Fig. 3A) and in 53% of cells for LibB (63, 46, and 53% for neurons, astrocytes, and VLMCs, respectively) with comparable sensitivity across animals (fig. S3A). This sensitivity is in line with other similar lineage tracing reports (17, 18) with transgene promotor silencing and inefficient capture on bead oligo-dT reverse transcription primers as likely contributors to loss of barcode detection of LibA, aggravated by interference of viral superinfection for LibB (19). Using the observed barcode diversity (fig. S3B) and frequency from each specific library, we modeled the rate of barcode “collision”—the event in which two different cells are independently labeled with the same barcode at the time of transduction—and found that our library setup could be used to label more than 100,000 cells grafted nuclei before reaching an estimated barcode collision rate of 1%. In addition, when analyzing the presence of specific barcodes in the animals where the barcode (LibB) was delivered in vivo, we found that individual barcode sequences were virtually never detected in more than one animal (see fig. S3C), confirming high barcode calling specificity. Thus, a shared barcode between cells can with high certainty be interpreted as a shared cellular origin.
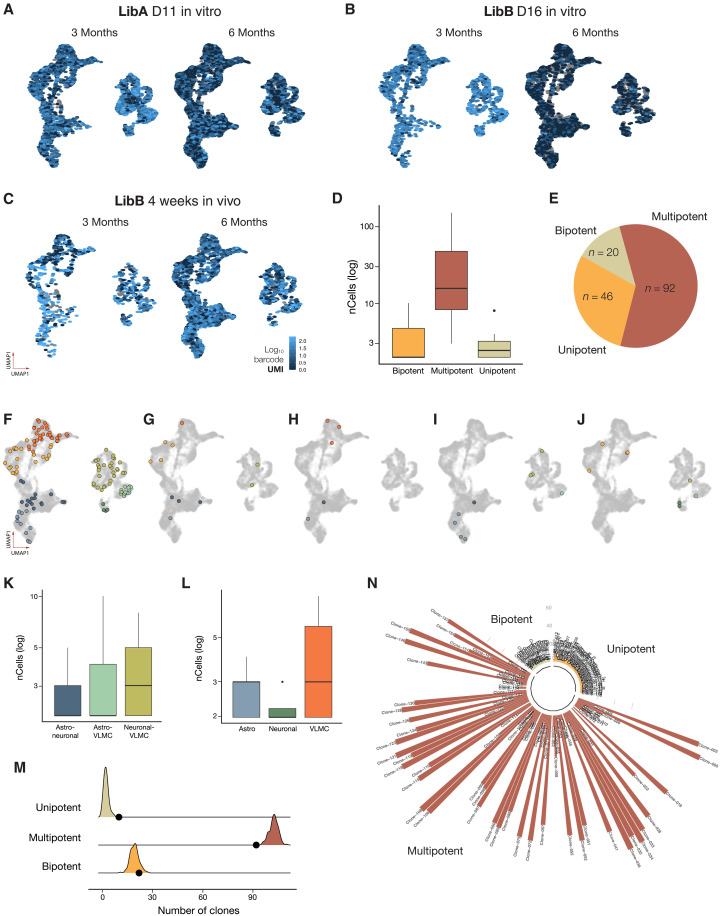
Fig. 3. Clonal dynamics derived from progenitors barcoded at day 11.
(A to C) Expression level of dominant viral barcode derived from LibA and LibB delivered at day 11, day 16, and week 4, respectively (n = 6 rats for LibA and n = 3 for each of LibB day 16 and LibB week 4). (D) Clone size per potency after barcode delivery at day 11. Lower and upper hinges correspond to the first and third quartiles, and the whisker extends from the hinge to the largest value no further than |1.5 * IQR| from the hinge (where IQR is the interquartile range or the distance between the first and third quartiles). (E) Proportion of clone types identified. (F and G) Representative multipotent clones containing all three major cell types. (H to J) Representative clone plots of bipotent clones. (K) Clone sizes of bipotent clone pairs detected after delivery of barcode library at day 11. (L) Clone sizes of unipotent clones detected after delivery of barcode library at day 11. (M) Simulation results when generating 10,000 random clones without any fate bias. Shaded areas indicate result of simulation, and black dots indicate results from the actual data. (N) Manhattan plot of clone sizes for day 11 clones colored by potency.
DA progenitors are multipotent at day 11
To determine the lineage relationship of the mature cells present in the graft, we first projected all cells/clones with unique barcodes from LibA (delivered at day 11) onto a UMAP manifold of the transcriptome (Fig. 3, A to C). This mapping was based on barcode transcripts captured by snRNA-seq and then specifically amplified from the cDNA pool for retrieval of cell barcode–viral barcode pairs (supported by at least two UMIs per cell; see details in Materials and Methods). In this mapping, we found 158 clones of different sizes ranging from 2 to 150 cells with a median clone size of 9.5 (Fig. 3D and fig. S3D). Further analysis showed that most clones (68%) contained all three cell types (neurons, astrocytes, and VLMCs), but we also detected clones containing only two cell types (bipotent) and few clones with only one cell type (unipotent; Fig. 3E). The multipotent clones were different in size ranging from large clones (see example in Fig. 3F) to clones with very few cells of each type (see example in Fig. 3G). The bipotent clones contained neurons/astrocytes (Fig. 3H), neurons/VLMCs (Fig. 3I), and VLMCs/astrocytes (Fig. 3J), with no significant difference in the contribution to each combination (Fig. 3K; P = 0.34). The few unipotent clones detected were typically smaller in size and without bias for specific cell types (Fig. 3L). To determine whether the detected bi- and unipotent clones could be explained by sampling bias, we performed a simulation assay generating 10,000 sets of 158 random clones with size and cell type proportion matching those of the real data but with no fate bias. This analysis indicated that detected bipotent clones were at the proportion expected if there was no fate bias (n = 19 versus 20 ± 3; table S3), whereas multipotent clones were slightly underrepresented (n = 92 versus 103 ± 20) and unipotent were slightly overrepresented (n = 10 versus 2 ± 1). Thus, the majority of the clones, including those identified as uni-/bipotent, are likely to be multipotent (Fig. 3, M and N) since their appearance is within one SD of random sampling. Together, this dataset indicates a shared origin of virtually all cell types in the graft, from which can be inferred that DA progenitors at day 11 have the potential to give rise to all three mature cell types in the graft.
Progressive fate restriction occurs primarily after transplantation
We next sought to investigate whether and when lineage diversification occurred during DA neuron differentiation by analyzing the barcodes from LibB that were delivered at later stages of differentiation [day 16 in vitro (Fig. 4A) or 1 month after grafting in vivo (Fig. 4B)].
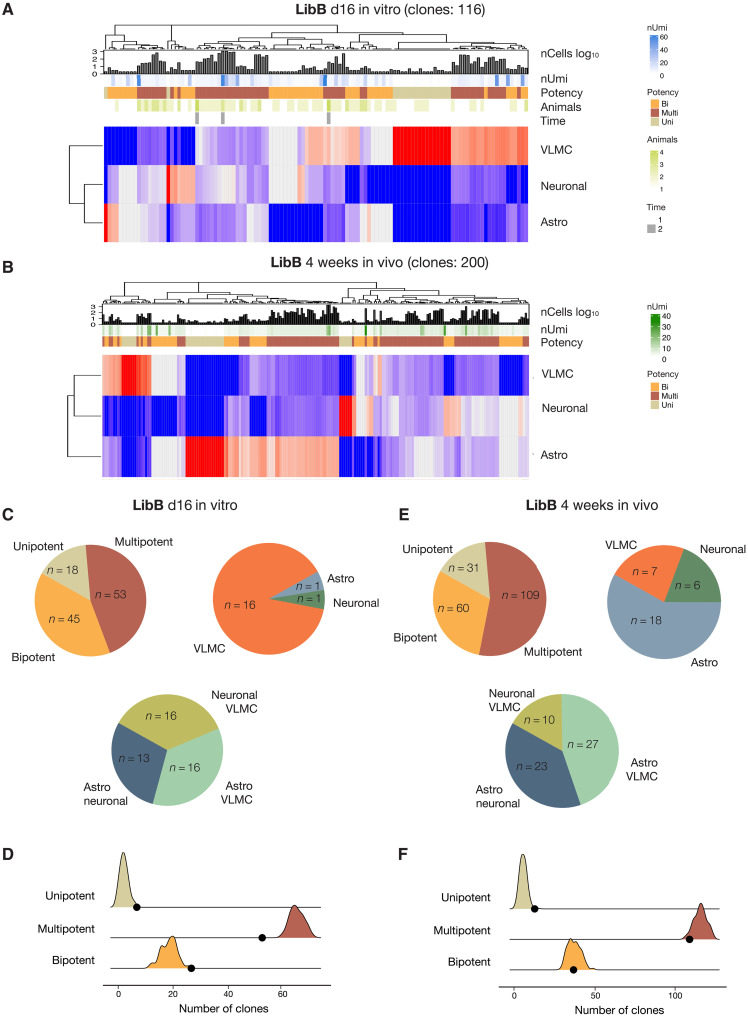
Fig. 4. Progressive fate restrictions of day 16 and week 4 in vivo barcoded progenitors.
(A and B) Heatmap of cell distribution in clones present in the mature graft following barcode delivery at day 16 (A) and week 4 in vivo (B) where each row corresponds to one clone. Red indicates high number of cells, and blue indicates low number of cells. Row annotation gives nUmi = average expression level of viral transcript; nCellslog10 = clone size; potency = uni/bi/multipotency; animals = number of animals where the clone was detected; and time whether the clone was detected in both short- and long-term experiment. (C and E) Proportion of multipotent, bipotent pairs, and unipotent clones detected after barcode delivery at day 16. (D and F) Simulation results of unbiased clones compared to real data. See also Fig. 3D.
In clones arising from cells labeled at day 16 in vitro, we identified 116 multicellular clones (Fig. 4A). These clones were typically smaller (median size, 6) than the clones arising from day 11 progenitors (fig. S3D) with a slightly higher proportion of bi- (39%) and unipotent (15%) clones but again with no significant difference in bipotent pair frequency (Fig. 4C). Simulation-based testing performed on cells labeled on day 16 indicated that unipotent clone was within range expected by chance, while bipotent clones were statistically overrepresented in the true data whereas multipotent were underrepresented (Fig. 4D and table S3), suggesting that a small proportion of progenitors with a more restricted potency may have appeared by day 16.
Next, looking at clones arising from cells labeled in vivo by intracerebral delivery of LibB 1 month after grafting, i.e., at a time point when a population of dividing cells are still present in the graft (3, 20), we identified 200 multicellular clones with a median size of seven cells per clone (Fig. 4B). The majority of them were not only multipotent (n = 109) but also bipotent and unipotent clones were detected (Fig. 4E). Simulation for this dataset indicated that multi- and bipotent clones were within range expected, whereas unipotent clones were slightly overrepresented (n = 13 versus 6 ± 2; Fig. 4F and table S3). Thus, even 6 weeks after initiation of differentiation where the maturation for the last 4 weeks has taken place in vivo within the brain parenchyma, many progenitors retain their multipotency.
Relationship between clones of cells in the mature graft and the transcriptional profile of the progenitor cell of origin
To trace back individual clones to their progenitor cell of origin, we analyzed the progenitors that were targeted with barcodes at days 11 and day 16 in vitro (Fig. 1, A to C) at the time of transplantation (day 18). We detected barcodes in 4056 progenitors (Fig. 5A), and for 506 of these, we could detect the same unique combination of barcodes as where present in the clones at 3 and/or 6 months after grafting, thereby inferring a direct lineage relationship between a progenitor and its differentiated progeny. First, we wanted to investigate whether the progenitors that gave rise to uni-, bi-, or multipotent clones formed defined clusters already at the progenitor stage. To determine this, we mapped the barcodes from each specific clone detected in the mature graft back to the progenitor population and found no enrichment of any clone type (multi-, bi-, or unipotent) in any of the defined progenitor clusters at day 18 (Figs. 1E and 5B; P > 0.5). Nor did any of the clone types map to any specific region of the UMAP, indicating that there was no bias in the progenitors in the contribution to multi-, bi-, or unipotent clones, respectively. This was also the case when we not only mapped cells based on potency but also took into account the cell type composition in the uni- and bipotent clones (Fig. 5C). This supports the conclusion that most, if not all, mature clones in the graft share a common cellular origin.
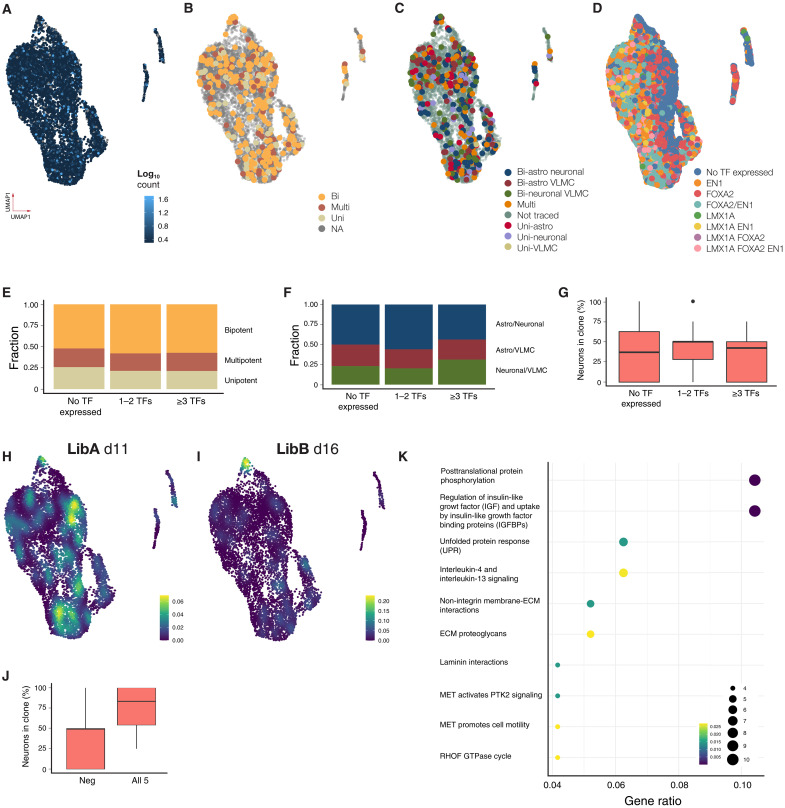
Fig. 5. Clonal capacity and transcriptional profile of day 18 progenitors pretransplantation.
(A to D) UMAP visualization of scRNA-seq from barcoded progenitors (n = 8166, two replicates) colored by barcode transcription level (A), their major clonal (LibA) (B), subclonal capacity (LibA) (C), and VM TF expression (D). NA, not available. (E to G) Fate of progenitors expressing no, one, two, or at least three VM TFs in terms of major clonal fraction (E), bipotent fraction (F), and fraction neurons (G). (H and I) Density plot of progenitors giving rise to neuronal biased clones barcoded at day 11 (H) or day 16 (I). (J) Fraction of neurons in clones expressing top five genes associated with neuronal fate compared to the rest. (K) Pathway analysis of genes associated with neuron biased progenitors at day 16. ECM, extracellular matrix; MET, metanephrine; GTPase, guanosine triphosphatase; PTK2, Protein Tyrosine Kinase 2; RHOF, Ras Homolog Family Member F.
To investigate this further, we next took a progenitor centered approach, mapping the progenitor’s expression of the most commonly used TFs for the developing VM and/or DA progenitors: FOXA2, LMX1A, OTX2, and EN1 forward onto their in vivo clones to determine whether expression of an individual TF/combination of TFs could be linked to a specific cell fate in clones appearing after transplantation. OTX2 and FOXA2 showed homogeneous expression in the progenitor cluster, and LMX1A and EN1 showed signs of coexpression with highest expression in the more differentiated cell clusters (Fig. 5D). We then grouped progenitors expressing either 1 or 2 midbrain TFs at high levels (n = 102), ≥3 (n = 28) compared to rest (n = 50) and their ability to generate neuron rich clones. We could not detect any potency bias linked to a specific TF expression either in terms of potency (Fig. 5E, P > 0.1), type of bipotency (Fig. 5F, P > 0.1), or fraction of neurons generated (Fig. 5G, P > 0.1). In addition, we examined cell cycle phases in progenitors using the phase predicted from the scRNA-seq data to see whether it influenced cell fate acquisition. Most progenitors were actively cycling (58%) with no correlation between cell cycle phase and differentiation outcomes, indicating that the majority of progenitors are cycling at transplantation. This suggests that while forced cell cycle exit could affect postgrafting cell fate, intrinsic cycling does not predict differentiation potential.
We also investigated expression of key gliogeneic TFs (21), including nuclear factor 1 A-type (NFIA) and activating transcription factor 3, as well as other gliogenic candidates, and found no significant differences in accessibility or expression at the progenitor stage (fig. S4A). Nor did they correlate with progenitor potency indicating that canonical gliogeneic TFs do not dictate fate at this stage. This might be explained with the timing of late gliogenesis, where such markers are typically not yet regulated. To find genes expressed in early VLMCs, we reanalyzed data from a previous study (22) and identified 27 TFs associated with early VLMC genesis. None of the identified VLMC genes showed differential expression or accessibility across any clusters of progenitors at the time of transplantation, as shown in the enclosed featureplot of MEOX2 and PTTG1 (fig. S4B).
We next took a less stringent approach and classified neuron-biased clones, defined as either unipotent clones with only neurons or bipotent/multipotent with >50% of the cells in the clones being neurons. When these were mapped back to day 11 barcoded progenitors, we did not identify any statistically significant enrichment of neuron-biased progenitors within the UMAP (Fig. 5H), nor did we detect any genes significantly enriched among those progenitors. However, when mapping back to day 16 barcoded progenitors, we found a strong enrichment of neuron-biased progenitors located within the apical part of the progenitor cluster in the UMAP (P < 0.01; Fig. 5I, yellow). By performing differential gene expression analysis among progenitors giving rise to different clones regardless of cluster identity, we identified 411 genes with an adjusted P value < 0.01 for any of the possible potencies (fig. S4C) and revealed 76 genes enriched in neuronal biased progenitors (day 16; Fig. 5I). Using the top five genes enriched in neuron biased progenitors, we could identify a signature that generated clones with 73.6 ± 31% neurons compared to 39 ± 29% in progenitors not expressing these genes (Fig. 5J, P = 0.0039). Pathway analysis of genes enriched in progenitors identified a significant neuron-biased activation of the IGF pathway (Fig. 5K), together with the RNA binding protein TOB2, TFs BCLAF1 and SPEN, as well as the glutamine transporter SLC38A2 as significantly enriched genes.
DISCUSSION
Single-cell technologies are rapidly emerging as new tools in regenerative medicine and have been used to profile the developmental trajectory from pluripotency to different types of therapeutic cells such as pancreatic islets, retinal cells, and neurons in vitro (9, 22–26) and to provide a better understanding of normal development of the specific cell types (27–32). More recently, single-cell transcriptional profiling has been used to determine the cell composition and maturation in vivo after delivery in disease-relevant animal models for PD and diabetes (33, 34). To date, only very few studies of this kind are published, but they have had a large impact on the field since they provide a comprehensive and unbiased profiling of cells formed after transplantation. They have also revealed that transcriptionally homogeneous cell preparations give rise to heterogeneous outcome after terminal differentiation in vivo in both PD models (5) and in type 1 diabetes models (34).
Our study focuses on stem cell therapy in PD, where scRNA-seq previously has been used to map DA neuron development (32, 33) and to profile stem cell–derived progenitor and neurons in vitro (9, 22) as well as in vivo (5, 6, 35). These, as well as additional studies based on histology and/or bulk RNA-seq, have revealed that despite the ability to generate transcriptionally homogeneous DA progenitor cells for grafting, only a fraction of the resulting transplant will be composed of mature DA neurons. Other types of neurons, astrocytes, oligodendrocytes, VLMCs, and choroid plexus have been reported to also be present in the mature stem cell–derived grafts (6, 7, 36–41). This raises a fundamental question in stem cell biology and the use of stem cells for regenerative medicine: How does a heterogeneous outcome from a homogeneous progenitor preparation arise? Likely explanations for this can be either that a small proportion of non-DA progenitors are present in the cultures at the time of transplantation, and these subsequently expand in vivo or that the progenitors used for transplantation are multipotent and generate all the different cell types in the mature graft. Some mature cells reported to be present in DA grafts, like off-target contaminants or choroid plexus cells, have been determined to arise from contaminating progenitor populations present at time of grafting. These progenitors can be removed via modifications to culture conditions or fluorescence-activated cell sorting (FACS) before grafting (4, 6). However, the origin of DA neurons, astrocytes, and VLMCs in the mature grafts remains to be determined which was the focus of this study.
We report a multiome analysis of DA progenitors where both the transcriptome and chromatin accessibility are measured on the same cell. These datasets provide a valuable resource to further investigate the gene regulatory programs driving human DA progenitors and their GRNs. Moreover, this analysis confirms previous transcriptional studies (6, 33, 42) and extends them by showing that the DA progenitors are also highly homogeneous in terms of chromatin profile and gene accessibility. This in turn supports the hypothesis that even progenitors that are transcriptionally and epigenetically homogeneous at the time of transplantation eventually give rise to different mature cell types in the graft. To experimentally prove this, we developed a barcode-based molecular tracing system where pooled libraries of heritable barcodes were delivered via lentiviral vectors early in the trajectory from stem cell to DA neuron differentiation, coupled to snRNA-seq of mature grafts 3 and 6 months after transplantation to detect mature cell types and their origin. This type of orthogonal lentiviral barcoding libraries provides a much higher efficacy, fidelity, and diversity than, for example, the iTracer system, recently reported in brain organoids (18). The iTracer system exploits CRISPR-Cas9–induced scar formation to provide the second unique temporal identity, which is necessary to map lineage relation of cells that undergo protracted differentiation over extended time periods, where it is not possible to create a continuous “state map” in transcriptomic space which is a commonly used approach to map lineage in cells that are developmentally close in time (43).
The transcriptional analysis was performed on nuclei isolated from the mature grafts and revealed the presence of DA neurons, astrocytes, and VLMCs similar to what have been reported by us and others previously using scRNA-seq of grafted tissue (5, 6). The main advantage of switching from scRNA-seq to snRNA-seq as we did here is that we capture many more neurons. The large number of grafted neurons captured allowed to determine neuronal subtype specificity and showed that in principle the only mature neurons present in the graft are the DA neurons. By mapping unique barcodes on different cell types, we found that most clones contained cells of all mature identities, indicating a shared cellular origin of DA neurons, astrocytes, and VLMCs from a common progenitor. This observation supports previous findings that FACS-purified DA progenitors from broader VM differentiations not only enrich for DA neurons but also give rise to VLMCs (6). An interesting and unexpected observation in this study was that the proliferative cells that were targeted with barcodes 1 month after grafting also remained multipotent with a capacity to generate clones with shared barcodes in all mature cell types in the graft.
As graft heterogeneity can arise from a mixed progenitor composition, we have taken significant steps to confirm that this is not the case for astrocytes, VLMCs, or DA neurons. This confidence is based on two key observations: first, the profiling of both the transcriptome and chromatin accessibility at the single-cell level, which includes a quantification of homogeneity compared to other datasets (Fig. 1G). Second, the use of molecular barcodes shows that almost all progenitors have the ability to generate all cell types in the mature graft (Fig. 3). These findings together support our conclusion that the progenitors are homogeneous, both at the molecular level and in their differentiation potential, and that graft diversification does not arise from different progenitors being present at time of grafting but rather that the graft heterogeneity arise from transplantation of homogeneous progenitors which are multipotent and have the potential to give rise to all mature cell types in the graft. Together, these observations show a common cellular origin of graft-derived cells and that final fate specification occurs during the last cell divisions.
Subsequent sequencing of the pretransplantation progenitors and their barcodes allowed us to link a certain progenitor profile with its differentiation potential. While we were not able to detect any potency bias linked to a specific progenitor population or gene expression status, we could identify neuron-biased progenitors appearing at day 16 and identified a set of genes (TOB2, BCLAF1, SPEN, and SCL38A2) whose expression conferred a neuronal bias. Our data show that cell cycle phase does not predict cell fate acquisition. Most progenitor cells are actively cycling at transplantation, with no correlation between cell cycle phase and differentiation outcomes in terms of cell types or clone size. However, forced cell cycle exit could still affect cell fate postgrafting (44). A better understanding of how and when lineage diversification occur and how it can be affected could be further exploited to guide further refinement of current therapeutic cell preparations toward a more effective composition.
Our study experimentally address the mechanism for neural graft diversification at the molecular level, and this has high translational relevance. The data reveal that the midbrain-patterned DA progenitors have the potential to give rise to different mature cell types after grafting. This in turn implies that while cell sorting can successfully be used to remove contaminating and incorrectly specified off-target cells in the progenitor population before transplantation (4, 6), a heterogeneous cell composition in the resulting grafts cannot be avoided by cell sorting at the progenitor stage. To obtain pure grafts to conduct experiments to determine each cell type contribution to the therapeutic effect, cells would need to be cultured for longer, sorted, and transplanted at the postmitotic stage when their final identity is set. This has been tested (45), but both the sorting procedure and the grafting of mature cells compromise graft survival and graft outgrowth, leading to suboptimal grafts. With newer and more gentle methods for cell sorting as well as developing bioengineering solutions for better graft survival of mature cells on the horizon, these types of experiments will likely soon be possible to perform (46). Alternatively, cells can be genetically modified to avoid or promote a particular developmental path or to induce cell death in specific cell populations once formed. These types of studies will serve as an important basis for experimentally addressing the role of different cell types in the grafts. The data from such studies would in turn guide the translational path forward in terms of determining optimal graft composition and ultimately controlling the diversification of grafted cells with a composition designed for an optimal therapeutic effect, and methods to obtain this using clinically compatible strategies.
MATERIALS AND METHODS
Lentiviral barcode library generation
A third-generation lentiviral backbone pCCLsin.cPPT.hPGK.eGFP.WPRE was digested upstream of the phosphoglycerate kinase 1 (PGK) promoter using the Xho I restriction site. A second expression cassette EF1a promoter and synthetic polyA sequence were inserted in trans with the two promoters separated by a Conserved Translational Element insulator sequence. Barcodes were ordered as 20 nucleotides long, High Purity Salt Free purified oligos (Eurofins Genomics), and defined as ambiguity nucleotides by using the sequence V-H-D-B (IUPAC ambiguity code) repeated five times. Barcodes were flanked by a static 12-bp sequence containing a library ID that allows for identification of the origin. A 40-μl Gibson assembly reaction (New England Biolabs) was performed to insert barcoded oligos into 50 ng of the vector, using a molar ratio of 1:2. The reaction was incubated for 1 hour at 50°C, purified using DNA Clean and Concentrator-5 (Zymo Research), and eluted in 5 μl. One microliter of purified Gibson assembly product was transformed into 20 μl of MegaX DH10B T1R Electrocomp (Thermo Fisher Scientific) cells according to the manufacturer’s protocol. Five individual transformations were performed and pooled into one tube. A small fraction of the transformed bacteria was plated on agar plates to validate the transformation efficacy. The none plated transformed bacteria were grown overnight as a maxi prep and purified using the ZymoPURE Plasmid Maxiprep Kit (Zymo Research). Two independent libraries (with separate library IDs) were generated, libraries A and B. To determine the library diversities, the barcodes were amplified using a Nextera indexing PCR and sequenced together on an Illumina MiSeq. Using a barcode analysis pipeline previously described (47), a barcode whitelist was determined for each library, and the diversity was determined to be 938,837 and 1,071,126 unique barcodes for libraries A and B, respectively [determined by non-unique counts of valid barcodes from plasmid sequencing after Starcode all-pairs clustering to remove sequencing errors (48)].
Neural cell differentiation and lentiviral barcode library delivery
RC17 hESCs (Roslin Cells) were cultured at 37°C and 5% CO2 and used for all transplantation experiments. Regional patterning of hESCs toward a VM fate was performed via dual SMAD inhibition to induce neuralization, GSK3 inhibitor–mediated caudalization, and SHH-mediated ventralization, as described in detail in Nolbrant et al. (7). VM cells were genetically tagged with lentiviral barcode library, which carries GFP reporter, on day 11 for library A and subsequently tagged with library B on day 16 during cell differentiation culture or 4 weeks later postcell transplantation. Each lentiviral transduction was performed with a multiplicity of infection of 1. Transduction successful rate was evaluated by flow cytometry analysis on the GFP expression.
Cell transplantation
Lentiviral barcoded VM cells were transplanted into striatal brain region of Parkinsonism rat, including Sprague-Dawley rats (n = 6) as well as athymic nude rats (n = 6). Lesion of the nigrostriatal pathway in rats was induced by unilateral injection of 6-hydroxydopamine into the right medial forebrain bundle, with a volume of 3 μl at a freebase concentration of 3.5 μg/μl to the following coordinates relative to bregma: Anterior/Posterior (A/P), −4.4; Medial/Lateral (M/L), −1.1; Dorsal/Ventral (DV) (from dura), −7.8; adjusted to flat head. Lesion severity was measured 4 weeks later by amphetamine-induced rotations [intraperitoneal injection of amphetamine (2.5 mg/kg); Apoteksbolaget, Sweden] and recorded over 90 min using an automated system (Omnitech Electronics). After this, animals were grafted to the striatum with VM-patterned progenitor cells. Rats received 300,000 cells into two deposits in striatum, at a total volume of 4 μl, at a concentration 75,000 cells/μl, at a rate of 1 μl/min and diffusion time of 2 min to the following coordinates relative to bregma: A/P, +1.0; M/L, −2.6; D/V (from dura), −4.5/−3.5; adjusted to flat head. Sprague-Dawley rats were immunosuppressed with daily injections of cyclosporine (10 mg/kg per day, intraperitoneally; Apoteksbolaget, Sweden), starting 2 days before transplantation, until the end of the experiment, to prevent graft rejection. Animals were perfused 3 or 6 months after cell transplantation. All experimental procedures followed the guidelines and recommendations of Swedish animal protection legislation and were approved by Lund Animal Ethics board under the approval number 5.8.18-08579/2017.
Histology
Before perfusion, rats were given terminal anesthesia with a lethal dose of sodium pentobarbitone (60 mg/kg) injected intraperitoneally (Apoteksbolaget, Sweden). The animals were transcardially perfused with physiological saline solution followed by ice-cold 4% paraformaldehyde. Brains were postfixed overnight in 4% paraformaldehyde in the fridge, transferred to 25% sucrose for 24 hours, and sectioned at a thickness of 35 μm (1:8 series) using a freezing microtome. For DAB-developed immunohistochemistry, free floating sections were incubated with tris-EDTA (pH 9.0) for 15 min at 80°C for antigen retrieval. Immunohistochemistry was performed on free floating sections that were incubated with primary antibodies overnight in 0.1 M KPBS solution containing 0.25% Triton X-100 and 5% serum for the species specific to the secondary antibody. Sections were then incubated with fluorophore-conjugated (fluorescent detection) or biotin-coupled (DAB detection) secondary antibodies for 1 hour in the same solution. All stained sections were mounted on gelatin-coated microscope slides. Fluorescent sections were coverslipped using polyvinyl alcohol mounting medium with DABCO (Sigma-Aldrich). DAB-developed sections were dehydrated in an ascending series of alcohols, cleared with xylene, coverslipped using DPX mountant, and left to dry overnight at room temperature. The primary antibodies used were the following: mouse anti-HuNu (1:200; Merck Millipore, catalog no. MAB1281), mouse anti-hNCAM (human neural cell adhesion molecule 1; 1:1000; Santa Cruz Biotechnology, catalog no. sc-106), rabbit anti-TH (1:1000; Merck Millipore, catalog no. AB152), sheep anti-hCOL1A1 (1:200; R&D, catalog no. AF6220), rabbit anti-hGFAP (1:200; R&D, catalog no. MAB25941), and chicken anti-GFP (1:1000; Abcam, catalog no. ab13970).
Nuclei isolation, snRNA-seq, and snATAC-seq
Grafted VM-patterned hESCs were dissected from rat striatum (n = 6 for VM intrastriatal grafts at 3-month posttransplantation and n = 6 for hESC intrastriatal grafts at 6-month posttransplantation) and transferred to a glass douncer containing 1 ml of nuclei lysis buffer [consisted of sucrose, CaCl2, MgAc, EDTA, tril-HCl, dithiothreitol, Triton X-100, ribonuclease (RNase) inhibitor, and RNase-free water] on ice. Tissue was completely minced and homogenized to nuclei suspension by sample grinding with Dounce homogenizers with 10 strokes with loose pestle and then 10 strokes with tight pestle. The nuclei suspension was filtered by loading through a 35-μm-diameter filter and followed by centrifuging for 10 min at 900g and 4°C. The nuclei pellet was collected and washed with cold wash buffer, which consisted of the following reagents: 1× phosphate-buffered saline, 0.5% bovine serum albumin, and RNase inhibitor (0.4 U/μl) for three times. After removing the supernatant from the last wash, nuclei were resuspended in 200 μl of DRAQ5 containing resuspension buffer. Nuclei were sorted on the basis of size to remove cell debris and fractured nuclei using the BD FACSAria III Cell Sorter. For RNA-seq, 7000 to 10,000 nuclei per sample with a maximum of 45 μl of total solution volume were collected for snRNA-seq library generation. Single nuclei were captured using Chromium (10X Genomics, PN-120233), a droplet-based scRNA-seq device according to the manufacturer’s recommendations. Briefly, after single-nucleus encapsulation with barcoded beads, cell lysis and reverse transcription were performed in a droplet reaction with polyT primers containing cell-specific barcodes, UMI, and sequencing adaptor sequences. After pooling and a first round of amplification, the library was split in half. The first half was fragmented and processed into a conventional snRNA-seq library using the manufacturer’s protocols. We used the second, unfragmented half to amplify barcodes reads by two rounds of PCR with the first introducing the TruSeq Read 2 sequence 3′ of the barcode (primer: 5′-GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTCCATTTCAGGTGTCGTGA-3′ matched with a P5 primer: 5′-CTACACGACGCTCTTCCGATCT-3′). In the second PCR, an i7 index and the P7 sequence were added to the TruSeq Read 2 overhang.
Successful library preparation was confirmed by a bioanalyzer (DNA HS kit, Agilent) and sequenced on Illumina NovaSeq 5000 2 × 100 bp (transcriptome libraries) and 2 × 50 bp (barcodes). For ATAC-seq, VM-patterned hESCs were sorted on the basis of GFP reporter, and then the nuclei preparation was performed according to the Chromium Next GEM Single-Cell Multiome ATAC + Gene Expression (GEX) protocol (CG000338).
Alignment, data processing, and analysis of transcriptomic data from grafts and progenitors
Cell Ranger (version 3.0, 10X Genomics) was used to demultiplex base call files to FastQ files and align reads. Default alignment parameters were used together with a combined human/rat reference (both version 93 from Ensembl). Seurat (version 4) was applied to snRNA-seq data for preprocessing and downstream analysis. Ribosomal protein genes and pseudogenes were excluded from analysis. Nuclei with more than 9000 or less than 200 detected genes as well as those with a mitochondrial transcript proportion greater than 10% were excluded. After normalization and integration using the “SCTransform” approach, based on regularized negative binomial regression, 5000 highly variable genes were identified and integrated using the corresponding functions in the Seurat package. Data were dimensionally reduced by principal components analysis (n = 30) and used for graph-based clustering (shared nearest-neighbor graph calculation and clustering using Louvain). Cell cycle scores were calculated for each cell but were of limited importance for overall data structure and hence not regressed out. After determining differentially expressed genes, we manually assigned major cell classes to each cluster (neurons, astrocytes, and VLMCs) using a combination of top differentially expressed genes and canonical markers. In addition, SingleR (49) with the LaManno brain data reference was used to confirm cell types. Pathway analysis was performed using clusterProfiler (50) with the Biocarta pathway database and P value adjustment using Benjami-Hochberg method. Multiome snATAC/snRNA-seq data were aligned and counted using Cell Ranger ARC (version 2.0) and further processed using the Signac workflow (51). Briefly, cells with fewer than 1000 or more than 100,000 ATAC fragments or <1000/>50,000 counts were filtered, as were cells with a nucleosomal enrichment of >2 or a transcriptional start site enrichment of <1. Peaks within each dataset were called using MACS2 (52). Dimensional reduction was performed with latent semantic indexing (LSI) followed by identification of top variable features and singular value decomposition. UMAP projection was performed using LSI components 2 to 50. Differentially accessible peaks were identified using a logistic regression model with “nFeature_peaks” as a latent variable. Peaks with an adjusted P value < 0.05 were considered significant.
Barcode demultiplexing and clonal analysis
Reads containing the barcode sequence were extracted from the FastQ files after demultiplexing with bcl2fastq (v 2.19). Reads that contained the library motif were identified using a custom perl script with the main function of extracting reads matching GTCGTGA[ACTG]{20}CTCGAC for LibA and GTCGTGA[ACTG]{20}CTCGAC for LibB outputting the read ID, 10× cell barcode, UMI, library barcode sequence, and library ID. Parsed output was then used to construct a cell barcode × viral lib matrix. An error correction step was then performed to adjust sequencing errors using Starcode where viral barcodes one edit distance apart were collapsed on a cell-by-cell basis. The resulting matrix was then filtered to remove any cell barcodes not found in the filtered Cell Ranger output or not present in the pool from a prior produced whitelist of barcodes.
Statistics
All statistical analyses were performed using the R software package (version 4.2.0). Sample sizes were not predetermined but were instead similar to previous publications (33). Hypothesis testing was done using Wilcoxon rank sum test unless otherwise specified. To determine whether clone bias were due to differences in potency or merely an effect of incomplete sampling, we performed a simulation assay. Ten thousand sets of random clones with size and cell type proportion matching those of the real but without any fate bias were generated (null hypothesis) and compared to those observed in the data. To assess barcode collisions, we used a simulation approach in R. We leveraged the measured relative barcode abundances and the estimated total barcode library size. Using base R functions, we simulated labeling a starting cell population with sizes ranging from 1 to 100 M cells. Each simulation was repeated 10,000 times. We then compared the expected number of unique barcodes (based on library size) to the average number of unique barcodes assigned in each simulated population. This discrepancy between expected and observed unique barcodes represents the estimated number of barcode collisions at a given cell population size.
Acknowledgments
We thank U. Jarl, B. Mattson, J. G. Johansson, M. Sparrenius, E. Sozzi, Y. Sharma, and A. Björklund for excellent technical assistance and scientific input. We would also like to acknowledge Clinical Genomics Lund, SciLifeLab, and Center for Translational Genomics (CTG), Lund University for providing sequencing service.
Funding: This work was supported by funding to M.P. from the New York Stem Cell Foundation, European Research Council (ERC) under ERC Grant Agreement 771427, European Union–funded project NSC-Reconstruct (European Union, H2020, GA no 874758, 2020-23), Swedish Research Council (2021-00661), Swedish Parkinson Foundation (Parkinsonfonden), Swedish Brain Foundation, Strategic Research Area at Lund University Multipark, the Novo Nordisk Foundation (NNF21CC0073729), and Knut and Alice Wallenberg Stiftelse (KAW 2018-0040). This work was also supported by funding to P.S. from the Segerfalk foundation.
Author contributions: P.S.: Writing—original draft, conceptualization, writing—review and editing, methodology, data curation, validation, formal analysis, software, and visualization. Y.Z.: Conceptualization, data curation, writing—review and editing, and methodology. F.N.: Investigation, writing—review and editing, and methodology. A.F.: Investigation, writing—review and editing, and methodology. N.K., M.Å., M.D., and J.Y.: Methodology and resources. A.K. and T.B.: Writing—original draft, conceptualization, and supervision. MP: Writing—original draft, conceptualization, supervision, and funding acquisition.
Competing interests: M.P. is the owner of Parmar Cells, and A.K. is the owner of Kirkeby Cell Therapy APS. M.P. and A.K. are coinventors on patents related to this work filed by Biolamina AB and Lund University (WO2016162747A2/A3, filed 08 April 2016, published 13 October 2016 and 05 January 2017). Both performed paid consultancy to Novo Nordisk. M.P. is member of SAB for Arbor Biotechnologies. T.B. and M.D. are inventors of multiple patents related to gene therapy, and they are also founders and directors of Brave Bioscience AB. T.B. is a cofounder and SAB member of Dyno Therapeutics. The other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. All unique reagents generated in this study are available from the lead contact with a completed material transfer agreement. Sequencing data are available at GEO as noted below. The barcoding vectors can be provided by M.P. pending scientific review and a completed material transfer agreement. Requests for the material should be submitted to M.P. (malin.parmar@med.lu.se). The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (53) and are accessible through GEO series accession number GSE200610 (https://ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE200610).
Supplementary Materials
The PDF file includes:
Figs. S1 to S4
Legends for tables S1 to S3
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S3
REFERENCES AND NOTES
- 1.Barker R. A., Drouin-Ouellet J., Parmar M., Cell-based therapies for Parkinson disease—Past insights and future potential. Nat. Rev. Neurol. 11, 492–503 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Kirkeby A., Nelander J., Hoban D. B., Rogelius N., Bjartmarz H., Storm P., Fiorenzano A., Adler A. F., Vale S., Mudannayake J., Zhang Y., Cardoso T., Mattsson B., Landau A. M., Glud A. N., Sørensen J. C., Lillethorup T. P., Lowdell M., Carvalho C., Bain O., van Vliet T., Lindvall O., Björklund A., Harry B., Cutting E., Widner H., Paul G., Barker R. A., Parmar M., Preclinical quality, safety, and efficacy of a human embryonic stem cell-derived product for the treatment of Parkinson’s disease, STEM-PD. Cell Stem Cell 30, 1299–1314 (2023). [DOI] [PubMed] [Google Scholar]
- 3.Piao J., Zabierowski S., Dubose B. N., Hill E. J., Navare M., Claros N., Rosen S., Ramnarine K., Horn C., Fredrickson C., Wong K., Safford B., Kriks S., El Maarouf A., Rutishauser U., Henchcliffe C., Wang Y., Riviere I., Mann S., Bermudez V., Irion S., Studer L., Tomishima M., Tabar V., Preclinical efficacy and safety of a human embryonic stem cell-derived midbrain dopamine progenitor product, MSK-DA01. Cell Stem Cell 28, 217–229.e7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doi D., Magotani H., Kikuchi T., Ikeda M., Hiramatsu S., Yoshida K., Amano N., Nomura M., Umekage M., Morizane A., Takahashi J., Pre-clinical study of induced pluripotent stem cell-derived dopaminergic progenitor cells for Parkinson’s disease. Nat. Commun. 11, 3369 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tiklová K., Nolbrant S., Fiorenzano A., Björklund Å. K., Sharma Y., Heuer A., Gillberg L., Hoban D. B., Cardoso T., Adler A. F., Birtele M., Lundén-Miguel H., Volakakis N., Kirkeby A., Perlmann T., Parmar M., Single cell transcriptomics identifies stem cell-derived graft composition in a model of Parkinson’s disease. Nat. Commun. 11, 2434 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu P., He H., Gao Q., Zhou Y., Wu Z., Zhang X., Sun L., Hu G., Guan Q., You Z., Zhang X., Zheng W., Xiong M., Chen Y., Human midbrain dopaminergic neuronal differentiation markers predict cell therapy outcomes in a Parkinson’s disease model. J. Clin. Invest. 132, e156768 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nolbrant S., Heuer A., Parmar M., Kirkeby A., Generation of high-purity human ventral midbrain dopaminergic progenitors for in vitro maturation and intracerebral transplantation. Nat. Protoc. 12, 1962–1979 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Liu B., Li C., Li Z., Wang D., Ren X., Zhang Z., An entropy-based metric for assessing the purity of single cell populations. Nat. Commun. 11, 3155 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nilsson F., Storm P., Sozzi E., Hidalgo Gil D., Birtele M., Sharma Y., Parmar M., Fiorenzano A., Single-cell profiling of coding and noncoding genes in human dopamine neuron differentiation. Cells 10, 137 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Birtele M., Storm P., Sharma Y., Kajtez J., Wahlestedt J. N., Sozzi E., Nilsson F., Stott S., He X. L., Mattsson B., Single-cell transcriptional and functional analysis of dopaminergic neurons in organoid-like cultures derived from human fetal midbrain. Development 149, dev200504 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hiller B. M., Marmion D. J., Thompson C. A., Elliott N. A., Federoff H., Brundin P., Mattis V. B., McMahon C. W., Kordower J. H., Optimizing maturity and dose of iPSC-derived dopamine progenitor cell therapy for Parkinson’s disease. NPJ Regen. Med. 7, 24 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernstein B. E., Mikkelsen T. S., Xie X., Kamal M., Huebert D. J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Jaenisch R., Wagschal A., Feil R., Schreiber S. L., Lander E. S., A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Ma S., Zhang B., LaFave L. M., Earl A. S., Chiang Z., Hu Y., Ding J., Brack A., Kartha V. K., Tay T., Law T., Lareau C., Hsu Y.-C., Regev A., Buenrostro J. D., Chromatin potential identified by shared single-cell profiling of RNA and chromatin. Cell 183, 1103–1116.e20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fleck J. S., Jansen S. M. J., Wollny D., Zenk F., Seimiya M., Jain A., Okamoto R., Santel M., He Z., Camp J. G., Treutlein B., Inferring and perturbing cell fate regulomes in human brain organoids. Nature 621, 365–372 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W. M., Hao Y., Stoeckius M., Smibert P., Satija R., Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Agarwal D., Sandor C., Volpato V., Caffrey T. M., Monzón-Sandoval J., Bowden R., Alegre-Abarrategui J., Wade-Martins R., Webber C., A single-cell atlas of the human substantia nigra reveals cell-specific pathways associated with neurological disorders. Nat. Commun. 11, 4183 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weinreb C., Rodriguez-Fraticelli A., Camargo F. D., Klein A. M., Lineage tracing on transcriptional landscapes links state to fate during differentiation. Science 367, eaaw3381 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He Z., Maynard A., Jain A., Gerber T., Petri R., Lin H.-C., Santel M., Ly K., Dupré J.-S., Sidow L., Sanchis Calleja F., Jansen S. M. J., Riesenberg S., Camp J. G., Treutlein B., Lineage recording in human cerebral organoids. Nat. Methods 19, 90–99 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Volsky D. J., Simm M., Shahabuddin M., Li G., Chao W., Potash M. J., Interference to human immunodeficiency virus type 1 infection in the absence of downmodulation of the principal virus receptor, CD4. J. Virol. 70, 3823–3833 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirkeby A., Grealish S., Wolf D. A., Nelander J., Wood J., Lundblad M., Lindvall O., Parmar M., Generation of regionally specified neural progenitors and functional neurons from human embryonic stem cells under defined conditions. Cell Rep. 1, 703–714 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Tiwari N., Pataskar A., Péron S., Thakurela S., Sahu S. K., Figueres-Oñate M., Marichal N., López-Mascaraque L., Tiwari V. K., Berninger B., Stage-specific transcription factors drive astrogliogenesis by remodeling gene regulatory landscapes. Cell Stem Cell 23, 557–571.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fiorenzano A., Sozzi E., Birtele M., Kajtez J., Giacomoni J., Nilsson F., Bruzelius A., Sharma Y., Zhang Y., Mattsson B., Emnéus J., Ottosson D. R., Storm P., Parmar M., Single-cell transcriptomics captures features of human midbrain development and dopamine neuron diversity in brain organoids. Nat. Commun. 12, 7302 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Veres A., Faust A. L., Bushnell H. L., Engquist E. N., Kenty J. H.-R., Harb G., Poh Y.-C., Sintov E., Gürtler M., Pagliuca F. W., Peterson Q. P., Melton D. A., Charting cellular identity during human in vitro β-cell differentiation. Nature 569, 368–373 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharon N., Chawla R., Mueller J., Vanderhooft J., Whitehorn L. J., Rosenthal B., Gürtler M., Estanboulieh R. R., Shvartsman D., Gifford D. K., Trapnell C., Melton D., A peninsular structure coordinates asynchronous differentiation with morphogenesis to generate pancreatic islets. Cell 176, 790–804.e13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collin J., Queen R., Zerti D., Dorgau B., Hussain R., Coxhead J., Cockell S., Lako M., Deconstructing retinal organoids: Single cell RNA-seq reveals the cellular components of human pluripotent stem cell-derived retina. Stem Cells 37, 593–598 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fiorenzano A., Sozzi E., Parmar M., Storm P., Dopamine neuron diversity: Recent advances and current challenges in human stem cell models and single cell sequencing. Cells 10, 1366 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu Y., Wang X., Hu B., Mao Y., Chen Y., Yan L., Yong J., Dong J., Wei Y., Wang W., Wen L., Qiao J., Tang F., Dissecting the transcriptome landscape of the human fetal neural retina and retinal pigment epithelium by single-cell RNA-seq analysis. PLOS Biol. 17, e3000365 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu Z., Liao X., Li N., Zhou H., Li H., Zhang Q., Hu K., Yang P., Hou S., A single-cell transcriptome atlas of the human retinal pigment epithelium. Front. Cell Dev. Biol. 9, 802457 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hendley A. M., Rao A. A., Leonhardt L., Ashe S., Smith J. A., Giacometti S., Peng X. L., Jiang H., Berrios D. I., Pawlak M., Li L. Y., Lee J., Collisson E. A., Anderson M. S., Fragiadakis G. K., Yeh J. J., Ye C. J., Kim G. E., Weaver V. M., Hebrok M., Single-cell transcriptome analysis defines heterogeneity of the murine pancreatic ductal tree. eLife 10, e67776 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu X.-X., Qiu W.-L., Yang L., Zhang Y., He M.-Y., Li L.-C., Xu C.-R., Defining multistep cell fate decision pathways during pancreatic development at single-cell resolution. EMBO J. 38, e100164 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kee N., Volakakis N., Kirkeby A., Dahl L., Storvall H., Nolbrant S., Lahti L., Björklund Å. K., Gillberg L., Joodmardi E., Sandberg R., Parmar M., Perlmann T., Single-cell analysis reveals a close relationship between differentiating dopamine and subthalamic nucleus neuronal lineages. Cell Stem Cell 20, 29–40 (2017). [DOI] [PubMed] [Google Scholar]
- 32.La Manno G., Gyllborg D., Codeluppi S., Nishimura K., Salto C., Zeisel A., Borm L. E., Stott S. R. W., Toledo E. M., Villaescusa J. C., Lönnerberg P., Ryge J., Barker R. A., Arenas E., Linnarsson S., Molecular diversity of midbrain development in mouse, human, and stem cells. Cell 167, 566–580.e19 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tiklová K., Björklund Å. K., Lahti L., Fiorenzano A., Nolbrant S., Gillberg L., Volakakis N., Yokota C., Hilscher M. M., Hauling T., Holmström F., Joodmardi E., Nilsson M., Parmar M., Perlmann T., Single-cell RNA sequencing reveals midbrain dopamine neuron diversity emerging during mouse brain development. Nat. Commun. 10, 581 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Augsornworawat P., Maxwell K. G., Velazco-Cruz L., Millman J. R., Single-cell transcriptome profiling reveals β cell maturation in stem cell-derived islets after transplantation. Cell Rep. 32, 108067 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rájová J., Davidsson M., Avallone M., Hartnor M., Aldrin-Kirk P., Cardoso T., Nolbrant S., Mollbrink A., Storm P., Heuer A., Parmar M., Björklund T., Deconvolution of spatial sequencing provides accurate characterization of hESC-derived DA transplants in vivo. Mol. Ther. Methods Clin. Dev. 29, 381–394 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kirkeby A., Parmar M., Building authentic midbrain dopaminergic neurons from stem cells - lessons from development. Transl. Neurosci. 3, 314–319 (2012). [Google Scholar]
- 37.Kriks S., Shim J.-W., Piao J., Ganat Y. M., Wakeman D. R., Xie Z., Carrillo-Reid L., Auyeung G., Antonacci C., Buch A., Yang L., Beal M. F., Surmeier D. J., Kordower J. H., Tabar V., Studer L., Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson’s disease. Nature 480, 547–551 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim T. W., Piao J., Koo S. Y., Kriks S., Chung S. Y., Betel D., Socci N. D., Choi S. J., Zabierowski S., Dubose B. N., Hill E. J., Mosharov E. V., Irion S., Tomishima M. J., Tabar V., Studer L., Biphasic activation of WNT signaling facilitates the derivation of midbrain dopamine neurons from hESCs for translational use. Cell Stem Cell 28, 343–355.e5 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bye C. R., Penna V., de Luzy I. R., Gantner C. W., Hunt C. P. J., Thompson L. H., Parish C. L., Transcriptional profiling of xenogeneic transplants: Examining human pluripotent stem cell-derived grafts in the rodent brain. Stem Cell Rep. 13, 877–890 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gantner C. W., de Luzy I. R., Kauhausen J. A., Moriarty N., Niclis J. C., Bye C. R., Penna V., Hunt C. P. J., Ermine C. M., Pouton C. W., Kirik D., Thompson L. H., Parish C. L., Viral delivery of GDNF promotes functional integration of human stem cell grafts in parkinson’s disease. Cell Stem Cell 26, 511–526.e5 (2020). [DOI] [PubMed] [Google Scholar]
- 41.Niclis J. C., Gantner C. W., Hunt C. P. J., Kauhausen J. A., Durnall J. C., Haynes J. M., Pouton C. W., Parish C. L., Thompson L. H., A PITX3-EGFP reporter line reveals connectivity of dopamine and non-dopamine neuronal subtypes in grafts generated from human embryonic stem cells. Stem Cell Rep. 9, 868–882 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kikuchi T., Morizane A., Doi D., Magotani H., Onoe H., Hayashi T., Mizuma H., Takara S., Takahashi R., Inoue H., Morita S., Yamamoto M., Okita K., Nakagawa M., Parmar M., Takahashi J., Human iPS cell-derived dopaminergic neurons function in a primate Parkinson’s disease model. Nature 548, 592–596 (2017). [DOI] [PubMed] [Google Scholar]
- 43.VanHorn S., Morris S. A., Next-generation lineage tracing and fate mapping to interrogate development. Dev. Cell 56, 7–21 (2021). [DOI] [PubMed] [Google Scholar]
- 44.Hiller B. M., Marmion D. J., Gross R. M., Thompson C. A., Chavez C. A., Brundin P., Wakeman D. R., McMahon C. W., Kordower J. H., Mitomycin-C treatment during differentiation of induced pluripotent stem cell-derived dopamine neurons reduces proliferation without compromising survival or function in vivo. Stem Cells Transl. Med. 10, 278–290 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Luzy I. R., Niclis J. C., Gantner C. W., Kauhausen J. A., Hunt C. P. J., Ermine C., Pouton C. W., Thompson L. H., Parish C. L., Isolation of LMX1a ventral midbrain progenitors improves the safety and predictability of human pluripotent stem cell-derived neural transplants in parkinsonian disease. J. Neurosci. Off. J. Soc. Neurosci. 39, 9521–9531 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moriarty N., Pandit A., Dowd E., Encapsulation of primary dopaminergic neurons in a GDNF-loaded collagen hydrogel increases their survival, re-innervation and function after intra-striatal transplantation. Sci. Rep. 7, 1–14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davidsson M., Diaz-Fernandez P., Schwich O. D., Torroba M., Wang G., Björklund T., A novel process of viral vector barcoding and library preparation enables high-diversity library generation and recombination-free paired-end sequencing. Sci. Rep. 6, 37563 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zorita E., Cuscó P., Filion G. J., Starcode: Sequence clustering based on all-pairs search. Bioinforma. Oxf. Engl. 31, 1913–1919 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aran D., Looney A. P., Liu L., Wu E., Fong V., Hsu A., Chak S., Naikawadi R. P., Wolters P. J., Abate A. R., Butte A. J., Bhattacharya M., Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 20, 163–172 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu T., Hu E., Xu S., Chen M., Guo P., Dai Z., Feng T., Zhou L., Tang W., Zhan L., Fu X., Liu S., Bo X., Yu G., clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. The Innovation 2, 100141 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.T. Stuart, A. Srivastava, C. Lareau, R. Satija, Multimodal single-cell chromatin analysis with Signac. bioRxiv [Preprint] (2020); https://doi.org/10.1101/2020.11.09.373613.
- 52.Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nusbaum C., Myers R. M., Brown M., Li W., Liu X. S., Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Edgar R., Domrachev M., Lash A. E., Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S4
Legends for tables S1 to S3
Tables S1 to S3