Abstract
The Nef protein of the simian and human immunodeficiency viruses is known to directly bind and downregulate the CD4 receptor. Although the molecular mechanism is well understood, direct binding of Nef and CD4 is difficult to demonstrate and is believed to be of low affinity. Applying nuclear magnetic resonance and fluorescence spectroscopy, we biophysically reevaluated the CD4-Nef complex and found the dissociation constant to be in the submicromolar range. We conclude that additional, so far disregarded residues in the N terminus of Nef are important for interaction with CD4.
Human immunodeficiency virus type 1 (HIV-1) Nef is a protein containing roughly 200 amino acid residues. It is a membrane-associated protein that is produced at the earliest stage of viral gene expression (13) and is a component of viral particles (39). Nef has been reported to have diverse effects on cellular signal transduction pathways. It interacts with various cellular protein kinases and acts both as a kinase substrate and as a modulator of kinase activity (7, 19, 22). In addition, Nef has been demonstrated to downregulate cell surface receptors CD4 and major histocompatibility complex class I (3, 9, 15, 23, 30, 31, 35). Nef-mediated downmodulation of CD4 is well understood now and appears to involve a whole set of factors. At least two distinct motifs in a long loop region of the protein were found to bind adaptins (AP 1, 2, and 3) (10–12, 18, 28). One of these motifs was additionally reported to interact with the regulatory unit of a vacuolar proton pump also involved in CD4 downregulation (29). The β-subunit of COPI coatomers was shown to bind Nef subsequently to adaptins and seems to direct CD4 to a degradation pathway (8, 34). From mutational analysis it is known that residues 407 to 418 in the cytoplasmic tail of CD4 are necessary and sufficient for downregulation of CD4 by Nef (1, 4, 14, 37). Three-dimensional structures are known from both the CD4 cytoplasmic domain (40, 41) and Nef proteins with N-terminal and, in some cases, additional deletions (5, 20, 26). Nuclear magnetic resonance (NMR) investigations into the interaction between CD4 and Nef using a 13-residue peptide of CD4 (residues 407 to 419) and several Nef mutants (NefΔ2–39 and NefΔ2–39, Δ159–173) elucidated residues in these Nef deletion mutants that were affected by binding of CD4(407–419) (21). The dissociation constant (KD) of this complex, however, was found to be only in the range of 1 mM. Although N-terminal amino acid sequences among Nef proteins are not conserved, some residues therein are known to be essential for downregulation of CD4 expression (2, 24, 25). Moreover, a study employing the yeast two-hybrid system suggests that residues important for CD4 binding are scattered all over the Nef sequence (36).
We focused our investigations on direct in vitro binding between essentially complete binding partners. In particular, we used a chemically synthesized peptide comprising all 31 C-terminal residues (403 to 433) of human CD4 with a fluorescein label at its N terminus and a recombinantly expressed full-length Nef protein from HIV-1 strain SF2. The CD4 peptide (403 to 433) and other peptides for controls (Fig. 1A) were purchased as reversed-phase high-performance liquid chromatography-purified products (Interactiva Biotechnologie, Ulm, Germany, and Jerini Biotools, Berlin, Germany). Identity was confirmed by matrix assisted laser desorption ionization time of flight mass spectrometry. Nef was overproduced as a polyhistidine-tagged fusion protein in Escherichia coli BL21(DE3) harboring the Nef gene from the HIV-1 isolate SF2 in a pET15b vector (Novagen) and the plasmid pUBS520 encoding the tRNA argU (27). Bacteria were grown aerobically at 37°C in Luria broth or M9 medium (38) containing ampicillin (200 μg/ml) and kanamycin (100 μg/ml). Cells were induced at an optical density at 600 nm of about 0.9 with 1 mM isopropyl-β-d-1-thiogalactopyranoside for 2 to 3 h, harvested by centrifugation (3,000 × g; 4°C) and sonicated in phosphate-buffered saline (PBS) buffer (8.8 mM Na2HPO4, 0.94 mM KH2PO4[pH 7.4], 150 mM NaCl). The extract was clarified by centrifugation for 30 min at 20,000 × g at 4°C and applied to a TALON metal affinity column (Clontech). After being washed with 50 mM potassium phosphate buffer (pH 7.5) containing 300 mM NaCl, the protein was eluted with 150 mM imidazol in the same buffer. Fractions containing the fusion protein, as determined by denaturing sodium dodecyl sulfate-polyacrylamide gel electrophoresis, were pooled, desalted, and applied to a butyl Sepharose column (Pharmacia, Uppsala, Sweden) in PBS containing 8.4% (by weight) ammonium sulfate. The bound protein was eluted with a gradient from 8.4% to 0% (by weight) ammonium sulfate in PBS buffer. Fractions containing the fusion protein were pooled, desalted, and concentrated (Macrosep; molecular weight cutoff, 10 kDa; Pall Filtron). The purified fusion protein was cleaved for 3 to 4 h at room temperature with 0.3 U of biotinylated thrombin (Pharmacia) per mg of protein, yielding the full-length Nef protein from HIV-1 strain SF2 (MGGKWSKRSMGGWSAIRERMRRAEPRAEPAADGVGAVSRDLEKHGAITSSNTAATNADCAWLEAQEEEEVGFPVRPQVPLRPMTYKAALDISHFLKEKGGLEGLIWSQRRQEILDLWIYHTQGYFPDWQNYTPGPGIRYPLTFGWCFKLVPVEPEKVEEANEGENNSLLHPMSLHGMEDAEKEVLVWRFDSKLAFHHMARELHPEYYKDC) with an additional eight amino acids at its N terminus (GSHMLEDP). Thrombin was separated from Nef by treatment with 15 μl of streptavidin agarose (Novagen) per unit. After centrifugation, the supernatant was applied to a HiLoad Superdex 75 prep grade column (Pharmacia) and eluted with PBS. All buffers used in this study were degassed and supplemented with 14 mM 2-mercaptoethanol. Fractions containing Nef protein were pooled and concentrated. The identity of the protein was confirmed by matrix assisted laser desorption ionization time of flight mass spectrometry and N-terminal amino acid sequence analysis.
FIG. 1.
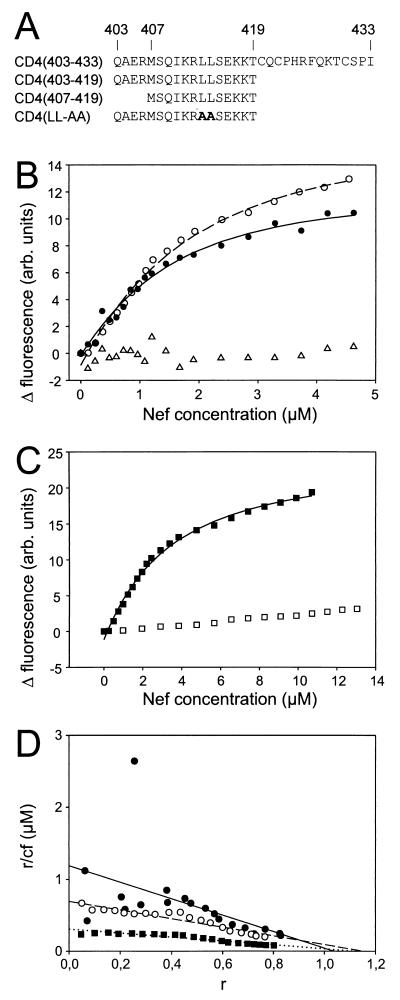
(A) Overview of N-terminal fluoresceinylated CD4 peptides used for Nef binding studies. Amino acid sequences are given using the one-letter-code for the CD4 peptides named on the left. In addition, the residue numbers corresponding to the respective sequence positions in CD4 are shown in the top line. (B) Fluorescence titrations of 1.1 μM fluoresceinylated CD4(403–433) (●), 1.0 μM fluoresceinylated CD4(403–419) (○), and as a control, 1.1 μM FITC isomer I (▵) as a function of Nef concentration. Values result from differences in the fluorescences of CD4 peptides or FITC with PBS buffer-solved Nef protein and PBS buffer without Nef. Assuming a simple bimolecular reaction between Nef and the CD4 peptide, analysis by nonlinear curve fitting yielded KD values of 0.87 and 1.4 μM for CD4(403–433) (solid line) and CD4(403–419) (dashed line), respectively. (C) Fluorescence titrations of 1.0 μM fluoresceinylated CD4(407–419) (■) and 1.0 μM fluoresceinylated CD4(LL-AA) (□) as a function of the Nef concentration. Nonlinear curve fitting yielded a KD value of 3.3 μM for CD4(407–419) (solid line). (D) Scatchard plot of the data points shown in panels B and C for binding of HIV-1 Nef to CD4(403–433) (●), CD4(403–419) (○), and CD4(407–419) (■) peptides. The x axis (r) corresponds to the ratio of bound Nef to the total CD4 peptide concentration, and r/cf (y axis) is the ratio of r and unbound Nef. Linear regression analysis (solid, dashed, and dotted lines) yielded dissociation constants (−1/KD, slope) and numbers of Nef binding sites per CD4 molecule (x axis intercept), the latter being close to 1 in all cases. It can easily be seen that most data points are very close to the respective fit curves. Some values for CD4(403–433) are more scattered than data points derived from other peptides. These experimental difficulties may be due to the presence of three cysteine residues in CD4(403–433).
We used fluorescence titrations to determine the dissociation constant (KD) values of full-length Nef and several CD4 peptides (Fig. 1A). Fluorescence measurements were carried out at 298K on an SLM Aminco Fourier Transform Spectrofluorometer 48000 MHF (SLM Instruments Inc.) and an LS50B (Perkin-Elmer) in standard fluorescence cells of SUPRASIL quartz glass (10 by 10 mm; Hellma, Müllheim, Germany). Fluorescence was measured with permanent stirring in PBS buffer using excitation and emission wavelengths of 495 and 520 nm, respectively. Appropriate amounts of Nef were added to the fluorescent component solved in PBS buffer. As a control the same titrations were performed with buffer devoid of Nef. Figure 1B shows the change of fluoresceinyl-CD4(403–433) relative fluorescence as a function of the Nef concentration. Assuming a simple bimolecular reaction between Nef and CD4, analysis by nonlinear curve fitting (32) yielded a KD value of 0.87 ± 0.19 μM. As additional parameters, minimum (−0.13 ± 0.26) and maximum (12.6 ± 0.74) fluorescences were fitted from the experimental data. Both values are in accordance with the experimental data and indicate the fit to be correct. Fluorescein isothiocyanate (FITC-I) (Sigma) as a control did not bind to Nef (Fig. 1B). An independent evaluation employing a Scatchard plot analysis with a linear regression analysis (Fig. 1D) confirmed the KD value to be 0.84 μM.
The observed KD value for binding of full-length Nef to CD4(403–433) is about 1,000-fold lower than that observed for Nef mutants NefΔ2–39 and NefΔ2–39,Δ159–173 and CD4(407–419) (21). Differences in both studies are in the length of the CD4 peptide and the completeness of the Nef protein. The possibility that the C-terminal tail of the CD4 cytoplasmic domain is involved in Nef binding can be excluded based on all mutation experiments (1, 4, 14, 36, 37). To be sure that our in vitro assay was in accordance with published data derived from assays in cell cultures, we determined the dissociation constant of a CD4 peptide C-terminally truncated at exactly the same position as the peptide used by Grzesiek and colleagues (21) in their study. The dissociation constant of 1.4 μM (Fig. 1B and D) obtained for Nef and CD4(403–419) suggests only a minor role of residues 420 to 433 in CD4 for Nef binding.
In the present study, a CD4 peptide was used that starts at Gln-403, which builds the N-terminal cap of an α-helix (40). This helix cap could not form in the CD4 peptide (residues 407 to 419) used by Grzesiek and colleagues. Gratton and colleagues (17) concluded based on their mutational studies that a correlation exists between the presence of this α-helix in CD4 and susceptibility to downregulation by Nef. All these data suggest that the existence of a preformed α-helix in CD4 supports binding to Nef. To measure the contribution of the four residues forming the helix cap, we determined the dissociation constant of CD4 peptide (residues 407 to 419) and full-length Nef to be 3.3 μM (Fig. 1C and D). Thus, the presence of residues 403 to 406 forming the helix cap increases the affinity of CD4 to Nef by a factor of roughly 2. The CD4 peptide (407 to 419) yielding a KD of 3.3 μM for Nef binding has exactly the same sequence as that used in earlier studies reporting a KD of 1 mM (21). However, the amino acid sequence of the Nef protein used in the present study was complete, in contrast to that used in earlier studies, which lacked 38 N-terminal residues (21). This strongly suggests that an intact N-terminal region of Nef is important for high-affinity binding to CD4.
As a final control, we carried out a binding study with Nef and a CD4 peptide (403 to 419) in which leucines 413 and 414 were replaced with alanines (Fig. 1C). This mutation is reported to render CD4 refractory to Nef-induced downregulation (1). No dissociation constant could be determined from the data points measured within the Nef concentration range between 0 and more than 13 μM, suggesting that mutation of leucines 413 and 414 to alanines drastically reduces the affinity of CD4 to Nef. This observation is in perfect accordance with published data (1, 21). It is known from mutational analysis that residues 407 to 418 of CD4 are necessary and sufficient for downregulation of CD4 by Nef (1, 4, 14, 37). Our in vitro studies basically confirm these observations. Residues N- and C-terminal to this part of CD4, however, seem to have a minor but significant contribution to Nef binding.
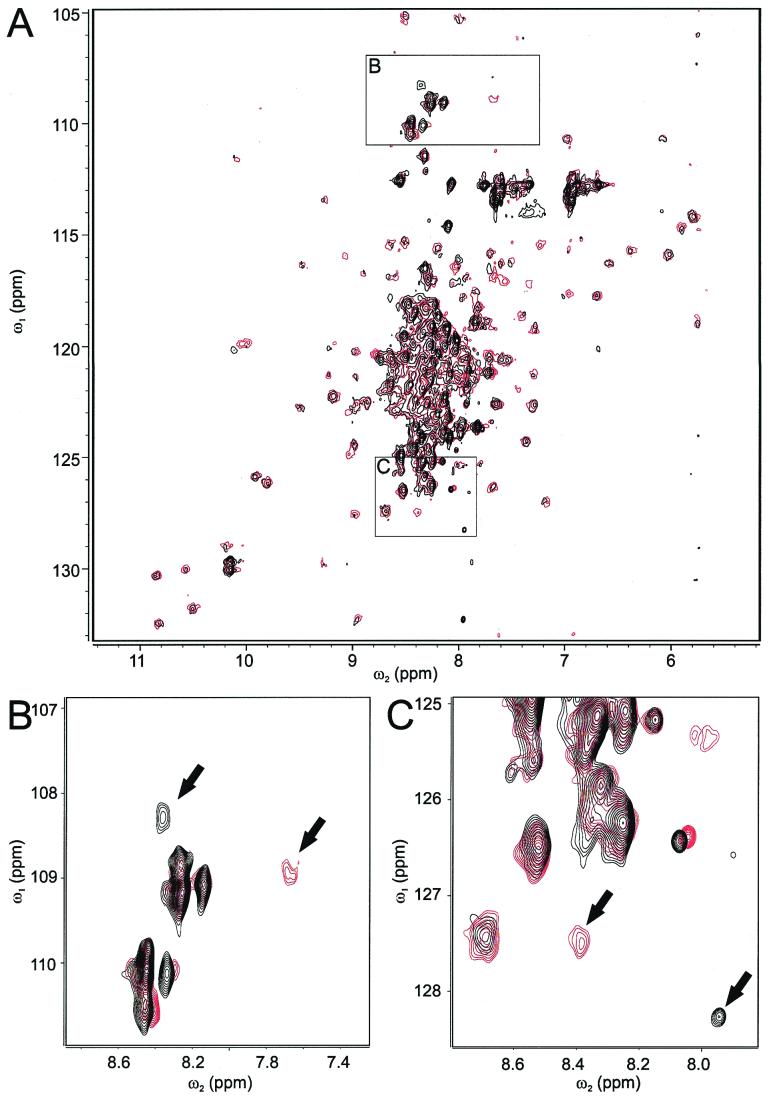
To ultimately elucidate details of binding of Nef to CD4, knowledge of the three-dimensional complex structure is desired. To pursue this long-term goal and to confirm high-affinity binding of Nef and CD4 by an additional method, we employed NMR spectroscopy. Observation of chemical shift changes in a protein upon ligand binding is a sensitive method for measuring the strength of an interaction and for defining the protein's interaction surface (16, 33). Especially useful and sensitive are, for example, heteronuclear single quantum coherence (HSQC) spectra. Thus, 1H, 15N HSQC spectra of 15N-labeled full-length Nef protein with increasing amounts of CD4(403–433) peptide were recorded. Titrations were performed in quarter steps in respect to the molar ratio of the Nef protein and the CD4 peptide. From a KD of about 1 μM or even below, dissociation rates of less than 100 Hz can be expected even in the case of diffusion-controlled association. Thus, exchange between free and CD4-bound Nef should be slow on the NMR chemical shift time scale at least for some of the 1H, 15N amide cross resonances. Indeed, intensities of some of the amide resonances in the 1H, 15N HSQC spectra decreased without shifting while new resonances appeared with increasing intensities during ongoing titration with CD4 peptide (Fig. 2). Assuming that the resonance pairs shown in Fig. 2B and C belong to the same amide cross-resonances of Nef in the presence and absence of the CD4 peptide, their proton chemical shift distances of 510 and 330 Hz, respectively, indicate that exchange between bound and unbound Nef is significantly slower than 300 Hz. A number of other resonances (one can be seen in Fig. 2C) shifted during titration with the CD4 peptide up to 30 Hz, suggesting the dissociation of the complex to be fast compared to this time scale. Both observations confirm that the dissociation rate of the complex is about 100 Hz and, given that the association rate is diffusion controlled (<108 Hz M−1), the resulting dissociation constant is 1 μM or less, which is in perfect agreement with the results from the fluorescence titration. Most of the amide resonances in the HSQC spectra did not show significant changes (Fig. 2A), indicating that the overall three-dimensional structure of Nef does not change dramatically upon binding of CD4. Because assignment of resonances of the Nef variant (SF2) used in this study was not sequence specific, it is not possible at the present stage to directly identify Nef residues involved in CD4 binding.
FIG. 2.
Overview (A) and two different zooms (B and C) of superimposed 1H, 15N HSQC spectra of HIV-1 Nef in the absence (red contour lines) and presence (black contour lines) of an equimolar concentration of the CD4(403–433) peptide. Note that for reasons of clarity, contour levels are not identical in panels A, B, and C. During titration the red-colored peaks, indicated by arrows, did not shift, but their intensities decreased with ongoing titration (data not shown), and new peaks (black-colored, indicated by arrows) appeared. NMR samples contained 180 μM uniformly 15N-labeled full-length Nef protein in PBS buffer with 10% D2O. All NMR spectra were recorded at 298K on a Varian Unity INOVA spectrometer working at a proton frequency of 750 MHz and equipped with a Varian XYZ-PFG-1H{13C, 15N} probe. Spectra were processed using the Varian VNMR software and analyzed using XEASY (6).
It is worth mentioning that our study was carried out with the CD4 peptide and Nef protein not anchored on the same side of a membrane as would be the case in vivo. Thus, in a living cell, binding affinity between Nef and CD4 can be expected to be even greater than observed in our in vitro system due to a much more favorable entropic term of the binding energy.
Employing a synthetic CD4(403–433) peptide and the recombinant full-length Nef protein purified by a novel highly native procedure under reducing and oxygen-excluding conditions, we were able to show high-affinity binding between HIV Nef and CD4. The resulting complex may be suitable for high-resolution structure determination by NMR spectroscopy or X-ray crystallography. Although this will take significant additional efforts, it will yield better insight into interference with host signal transduction proteins by viral proteins. Exploring new target proteins in HIV infection, aside from reverse transcriptase and protease, is becoming increasingly important. The fluorescence assay for measurement of CD4-Nef binding described in our study is possibly suitable to be employed directly or in a modified form for high-throughput screening assays to find substances that interfere with CD4-Nef interaction.
Acknowledgments
This work was supported by grants from the Deutsche Forschungsgemeinschaft (DFG) to D.W. and A.S.B. (SFB 466, A4, and B1).
We are grateful to R. Mattes (Institut für Industrielle Genetik, Universität Stuttgart) for providing the plasmid pUBS520. We thank K.-H. Gührs and B. Schlott (Institut für Molekulare Biotechnologie, Jena, Germany) for carrying out mass spectroscopy and N-terminal amino acid sequencing.
REFERENCES
- 1.Aiken C, Konner J, Landau N R, Lenburg M E, Trono D. Nef induces CD4 endocytosis: requirement for a critical dileucine motif in the membrane-proximal CD4 cytoplasmic domain. Cell. 1994;76:853–864. doi: 10.1016/0092-8674(94)90360-3. [DOI] [PubMed] [Google Scholar]
- 2.Aiken C, Krause L, Chen Y L, Trono D. Mutational analysis of HIV-1 Nef: identification of two mutants that are temperature-sensitive for CD4 downregulation. Virology. 1996;217:293–300. doi: 10.1006/viro.1996.0116. [DOI] [PubMed] [Google Scholar]
- 3.Anderson S, Shugars D C, Swanstrom R, Garcia J V. Nef from primary isolates of human immunodeficiency virus type 1 suppresses surface CD4 expression in human and mouse T cells. J Virol. 1993;67:4923–4931. doi: 10.1128/jvi.67.8.4923-4931.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson S J, Lenburg M, Landau N R, Garcia J V. The cytoplasmic domain of CD4 is sufficient for its down-regulation from the cell surface by human immunodeficiency virus type 1 Nef. J Virol. 1994;68:3092–3101. doi: 10.1128/jvi.68.5.3092-3101.1994. . (Erratum, 68:4705.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arold S, Franken P, Strub M P, Hoh F, Benichou S, Benarous R, Dumas C. The crystal structure of HIV-1 Nef protein bound to the Fyn kinase SH3 domain suggests a role for this complex in altered T cell receptor signaling. Structure. 1997;5:1361–1372. doi: 10.1016/s0969-2126(97)00286-4. [DOI] [PubMed] [Google Scholar]
- 6.Bartels C, Xia T, Billeter M, Güntert P, Wüthrich K. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J Biomol NMR. 1995;6:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- 7.Baur A S, Sass G, Laffert B, Willbold D, Cheng Mayer C, Peterlin B M. The N-terminus of Nef from HIV-1/SIV associates with a protein complex containing Lck and a serine kinase. Immunity. 1997;6:283–291. doi: 10.1016/s1074-7613(00)80331-3. [DOI] [PubMed] [Google Scholar]
- 8.Benichou S, Bomsel M, Bodeus M, Durand H, Doute M, Letourneur F, Camonis J, Benarous R. Physical interaction of the HIV-1 Nef protein with beta-COP, a component of non-clathrin-coated vesicles essential for membrane traffic. J Biol Chem. 1994;269:30073–30076. [PubMed] [Google Scholar]
- 9.Benson R E, Sanfridson A, Ottinger J S, Doyle C, Cullen B R. Downregulation of cell-surface CD4 expression by simian immunodeficiency virus Nef prevents viral super infection. J Exp Med. 1993;177:1561–1566. doi: 10.1084/jem.177.6.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bresnahan P A, Yonemoto W, Ferrell S, Williams-Herman D, Geleziunas R, Greene W C. A dileucine motif in HIV-1 Nef acts as an internalization signal for CD4 downregulation and binds the AP-1 clathrin adaptor. Curr Biol. 1998;8:1235–1238. doi: 10.1016/s0960-9822(07)00517-9. [DOI] [PubMed] [Google Scholar]
- 11.Craig H M, Pandori M W, Guatelli J C. Interaction of HIV-1 Nef with the cellular dileucine-based sorting pathway is required for CD4 down-regulation and optimal viral infectivity. Proc Natl Acad Sci USA. 1998;95:11229–11234. doi: 10.1073/pnas.95.19.11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Craig H M, Reddy T R, Riggs N L, Dao P P, Guatelli J C. Interactions of HIV-1 nef with the mu subunits of adaptor protein complexes 1, 2, and 3: role of the dileucine-based sorting motif. Virology. 2000;271:9–17. doi: 10.1006/viro.2000.0277. [DOI] [PubMed] [Google Scholar]
- 13.Cullen B R. The role of Nef in the replication cycle of the human and simian immunodeficiency viruses. Virology. 1994;205:1–6. doi: 10.1006/viro.1994.1613. [DOI] [PubMed] [Google Scholar]
- 14.Garcia J V, Alfano J, Miller A D. The negative effect of human immunodeficiency virus type 1 Nef on cell surface CD4 expression is not species specific and requires the cytoplasmic domain of CD4. J Virol. 1993;67:1511–1516. doi: 10.1128/jvi.67.3.1511-1516.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia J V, Miller A D. Serine phosphorylation-independent downregulation of cell-surface CD4 by nef. Nature. 1991;350:508–511. doi: 10.1038/350508a0. [DOI] [PubMed] [Google Scholar]
- 16.Görlach M, Wittekind M, Beckman R A, Mueller L, Dreyfuss G. Interaction of the RNA-binding domain of the hnRNP C proteins with RNA. EMBO J. 1992;11:3289–3295. doi: 10.1002/j.1460-2075.1992.tb05407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gratton S, Yao X J, Venkatesan S, Cohen E A, Sekaly R P. Molecular analysis of the cytoplasmic domain of CD4: overlapping but noncompetitive requirement for lck association and down-regulation by Nef. J Immunol. 1996;157:3305–3311. [PubMed] [Google Scholar]
- 18.Greenberg M E, Bronson S, Lock M, Neumann M, Pavlakis G N, Skowronski J. Co-localization of HIV-1 Nef with the AP-2 adaptor protein complex correlates with Nef-induced CD4 down-regulation. EMBO J. 1997;16:6964–6976. doi: 10.1093/emboj/16.23.6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greenway A, Azad A, Mills J, McPhee D. Human immunodeficiency virus type 1 Nef binds directly to Lck and mitogen-activated protein kinase, inhibiting kinase activity. J Virol. 1996;70:6701–6708. doi: 10.1128/jvi.70.10.6701-6708.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grzesiek S, Bax A, Clore G M, Gronenborn A M, Hu J S, Kaufman J, Palmer I, Stahl S J, Wingfield P T. The solution structure of HIV-1 Nef reveals an unexpected fold and permits delineation of the binding surface for the SH3 domain of Hck tyrosine protein kinase. Nat Struct Biol. 1996;3:340–345. doi: 10.1038/nsb0496-340. [DOI] [PubMed] [Google Scholar]
- 21.Grzesiek S, Stahl S J, Wingfield P T, Bax A. The CD4 determinant for downregulation by HIV-1 Nef directly binds to Nef. Mapping of the Nef binding surface by NMR. Biochemistry. 1996;35:10256–10261. doi: 10.1021/bi9611164. [DOI] [PubMed] [Google Scholar]
- 22.Harris M. HIV: a new role for Nef in the spread of HIV. Curr Biol. 1999;9:R459–R461. doi: 10.1016/s0960-9822(99)80282-6. [DOI] [PubMed] [Google Scholar]
- 23.Harris M, Coates K. Identification of cellular proteins that bind to the human immunodeficiency virus type 1 nef gene product in vitro: a role for myristylation. J Gen Virol. 1993;74:1581–1589. doi: 10.1099/0022-1317-74-8-1581. [DOI] [PubMed] [Google Scholar]
- 24.Hua J, Blair W, Truant R, Cullen B R. Identification of regions in HIV-1 Nef required for efficient downregulation of cell surface CD4. Virology. 1997;231:231–238. doi: 10.1006/viro.1997.8517. [DOI] [PubMed] [Google Scholar]
- 25.Iafrate A J, Bronson S, Skowronski J. Separable functions of Nef disrupt two aspects of T cell receptor machinery: CD4 expression and CD3 signaling. EMBO J. 1997;16:673–684. doi: 10.1093/emboj/16.4.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee C H, Saksela K, Mirza U A, Chait B T, Kuriyan J. Crystal structure of the conserved core of HIV-1 Nef complexed with a Src family SH3 domain. Cell. 1996;85:931–942. doi: 10.1016/s0092-8674(00)81276-3. [DOI] [PubMed] [Google Scholar]
- 27.Lindsey D F, Mullin D A, Walker J R. Characterization of the cryptic lambdoid prophage DLP12 of Escherichia coli and overlap of the DLP12 integrase gene with the tRNA gene argU. J Bacteriol. 1989;171:6197–6205. doi: 10.1128/jb.171.11.6197-6205.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lock M, Greenberg M E, Iafrate A J, Swigut T, Muench J, Kirchhoff F, Shohdy N, Skowronski J. Two elements target SIV Nef to the AP-2 clathrin adaptor complex, but only one is required for the induction of CD4 endocytosis. EMBO J. 1999;18:2722–2733. doi: 10.1093/emboj/18.10.2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu X, Yu H, Liu S H, Brodsky F M, Peterlin B M. Interactions between HIV1 Nef and vacuolar ATPase facilitate the internalization of CD4. Immunity. 1998;8:647–656. doi: 10.1016/s1074-7613(00)80569-5. [DOI] [PubMed] [Google Scholar]
- 30.Mariani R, Skowronski J. CD4 down-regulation by nef alleles isolated from human immunodeficiency virus type 1-infected individuals. Proc Natl Acad Sci USA. 1993;90:5549–5553. doi: 10.1073/pnas.90.12.5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marsh J W. The numerous effector functions of Nef. Arch Biochem Biophys. 1999;365:192–198. doi: 10.1006/abbi.1999.1208. [DOI] [PubMed] [Google Scholar]
- 32.Müller B, Restle T, Reinstein J, Goody R S. Interaction of fluorescently labeled dideoxynucleotides with HIV-1 reverse transcriptase. Biochemistry. 1991;30:3709–3715. doi: 10.1021/bi00229a017. [DOI] [PubMed] [Google Scholar]
- 33.Otting G, Qian Y Q, Billeter M, Müller M, Affolter M, Gehring W J, Wüthrich K. Protein-DNA contacts in the structure of a homeodomain-DNA complex determined by nuclear magnetic resonance spectroscopy in solution. EMBO J. 1990;9:3085–3092. doi: 10.1002/j.1460-2075.1990.tb07505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piguet V, Gu F, Foti M, Demaurex N, Gruenberg J, Carpentier J L, Trono D. Nef-induced CD4 degradation: a diacidic-based motif in Nef functions as a lysosomal targeting signal through the binding of beta-COP in endosomes. Cell. 1999;97:63–73. doi: 10.1016/s0092-8674(00)80715-1. [DOI] [PubMed] [Google Scholar]
- 35.Renkema H G, Saksela K. Interactions of HIV-1 NEF with cellular signal transducing proteins. Front Biosci. 2000;5:D268–D283. doi: 10.2741/renkema. [DOI] [PubMed] [Google Scholar]
- 36.Rossi F, Gallina A, Milanesi G. Nef-CD4 physical interaction sensed with the yeast two-hybrid system. Virology. 1996;217:397–403. doi: 10.1006/viro.1996.0130. [DOI] [PubMed] [Google Scholar]
- 37.Salghetti S, Mariani R, Skowronski J. Human immunodeficiency virus type 1 Nef and p56lck protein-tyrosine kinase interact with a common element in CD4 cytoplasmic tail. Proc Natl Acad Sci USA. 1995;92:349–353. doi: 10.1073/pnas.92.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 39.Welker R, Kottler H, Kalbitzer H R, Kräusslich H G. Human immunodeficiency virus type 1 Nef protein is incorporated into virus particles and specifically cleaved by the viral proteinase. Virology. 1996;219:228–236. doi: 10.1006/viro.1996.0240. [DOI] [PubMed] [Google Scholar]
- 40.Willbold D, Rösch P. Solution structure of the human CD4 (403–419) receptor peptide. J Biomed Sci. 1996;3:435–441. doi: 10.1007/BF02258047. [DOI] [PubMed] [Google Scholar]
- 41.Wray V, Mertins D, Kiess M, Henklein P, Trowitzsch-Kienast W, Schubert U. Solution structure of the cytoplasmic domain of the human CD4 glycoprotein by CD and 1H NMR spectroscopy: implications for biological functions. Biochemistry. 1998;37:8527–8538. doi: 10.1021/bi9723111. [DOI] [PubMed] [Google Scholar]