Abstract
Aim
It is unknown whether the halotolerant bacterium Halomonas sp. produces a range of secondary metabolites with antimicrobial qualities. In the past few years, there has been a growing interest in the biotechnological capability of halophilic bacteria for the production of antimicrobial compounds.
Materials and methods
The current review intended to assess the antibacterial and antifungal properties of microbial metabolites, explicitly those produced as secondary metabolites by a putative halophilic bacterium. First, phenotypic and genotypic identification were used to identify and confirm the obtained potent halophilic bacterium as Halomonas sp., and its antioxidant properties and biological compatibility were studied.
Results
The extracellular metabolites that were obtained exhibit a moderation zone of inhibition against 11 mm of Staphylococcus aureus, 12 mm of Pseudomonas aeruginosa, and 11 mm of Candida albicans. The optimal inhibitory concentration for S. aureus and P. aeruginosa is 256 µg/mL, while the minimum inhibitory concentration (MIC) for C. albicans is 128 µg/mL. The antioxidant property of crude metabolites indicates that 100% scavenging at 512 µg/mL, and the blow at 256 µg/mL, are not reasonable levels of antioxidant activity.
Conclusion
Secondary metabolites appear to be highly biologically compatible, as there is no hemolytic activity at any of the tested concentrations. According to the study, Halomonas sp.'s secondary metabolites could be a source for the synthesis of novel antimicrobial compounds.
Keywords: antimicrobial activity, anti-oxidant potential, gc-ms analysis, halomonas sp, secondary metabolites
Introduction
Bacteria classified as halophilic grow best in environments with high concentrations of salt. Extreme environments, such as saltwater ponds, saline lakes, hypersaline soils, arid soils, and preserved food, can all support the survival and growth of these bacteria [1]. The development and metabolic activity of halophilic bacteria depend heavily on their capacity to endure a range of environments [2]. The rise in antibiotic resistance has made it harder to find antibiotics from different sources in recent years. Therefore, according to Deepalaxmi and Gayathri (2018) [3], halophilic bacteria may offer an alternative source of antimicrobial compounds. According to Gasperotti et al. (2018) [4], halophilic bacteria such as Halomonas sp. are known to flourish in exceptionally salinized conditions and have the capacity to produce stable secondary metabolites with antimicrobial qualities like 1-acetyl-β-carboline, diketopiperazines (DKPs) cyclo(2-OHPro-Phe), macrolactin and succinoyl macrolactones. These secondary metabolites have demonstrated a significant role in the fields of biotechnology and pharmaceuticals. They include compatible solutes, enzymes, carotenoid pigments, and biopolymers [5,6]. There hasn't been much research done on the antimicrobial qualities of the extracellular and intracellular metabolites that Halomonas denitrificans produces. On the other hand, recent research has demonstrated that antimicrobial metabolites are produced by halotolerant Bacillus licheniformis strains that were isolated from saline and hypersaline environments [7]. Both halophilic and non-halophilic bacteria are members of the Gammaproteobacteria class, which includes the Halomonadaceae. This includes halophilic and halotolerant species like Modicisalibacter, Halomonas, Cobetia, Chromohalobacter, and Kushneria. Conversely, non-halophilic bacteria are present in closely related halophilic members of the family, such as Zymobacter, Carnimonas, and Halotalea [8].
The fields of microbial biotechnology and related research have opened up new avenues for the industrial use of ambient microbes that generate bioactive compounds and secondary metabolites. Microbes are widely distributed and beneficial to sustainable development in many fields, such as pharmaceuticals, industry, agriculture, and other related areas where pathogen control is needed. The rod-shaped, gram-negative, non-spore-forming Halomonas organisms are mostly dependent on respiratory metabolism. They take electrons from either oxygen or nitrate; some species are denitrifiers, and some have fermentative metabolism. While some species of Halomonas may be considered halotolerant, most species show a preference for mild salinities. Microbial secondary metabolites are substances that microorganisms produce. Rather than being necessary for their basic metabolic processes, such as growth and reproduction, these metabolites are vital to their survival and ability to adapt to various environmental conditions [9]. The microbial cells of Halomonas sp. produce organic compounds known as intracellular metabolites, which are indicative of biological activity in the organism. These metabolites can be produced by a variety of metabolic pathways and have roles in the cell [10]. They are crucial for a number of cellular functions, including the synthesis of macromolecules, the production of energy, and the preservation of cellular homeostasis [11]. Additionally, the intracellular metabolites from halophilic bacteria also exhibit antimicrobial properties [12]. Bacillus halophilus sp. BS3 and Kocuria marina BS-15 produce polymeric biosurfactants with anticancer properties, while other halophilic bacteria produce lipopeptide-like biosurfactants [13,14]. Therefore, the promising field for drug discovery and development may be the antimicrobial qualities of the extracellular, intracellular, and lipopeptides of halophilic bacteria produced by H. denitrificans. Additional investigation into these characteristics may result in the creation of novel antibiotics and all-natural preservatives, in addition to other biotechnological uses. In general, microbial metabolites are significant for clinical and therapeutic uses. Therefore, investigating these microbial metabolites from environmental microorganisms, such as halophilic bacteria, may have a wide range of applications.
Materials and methods
Halophilic isolate
The Department of Microbiology, Saveetha Medical College and Hospital, Chennai, India, provided the strong halophilic bacterial isolate, which was obtained from the Bio-control and Microbial Product Lab. It was kept in a nutrient agar slant medium with 8% NaCl and maintained at 7°C for additional analysis.
Clinical pathogens
The antimicrobial activity assay involved the use of five distinct pathogenic bacteria: Candida albicans, Pseudomonas aeruginosa, methicillin-resistant Staphylococcus aureus (MRSA), Escherichia coli, and Enterococcus faecalis. For 30 days, the pathogenic organisms were regularly cultivated and sub-cultured in Mueller-Hinton Agar (MHA) medium (HIMEDIA M173-500G; HiMedia, Thane, Maharashtra, India) to maintain cell viability, while C. albicans was kept in Sabourad Dextrose Agar (SDA) medium (HiMedia).
Bacterial DNA isolation and amplification of 16s rRNA
The DNA isolation and 16S rRNA sequencing of potent halophilic bacteria was performed at Eurofins Genomics (Bangalore, India). The halophilic bacteria were cultured in Nutrient Broth (NB) medium. Following a 24-hour growth period, the bacterial cells were cleaned with phosphate-buffered saline (PBS). Lysis buffer was then added to the bacterial cells to facilitate the chemical lysis process that released the DNA. After adding the binding buffer to the lysate to denature the proteins and safeguard the DNA, the DNA was bound to silica. It was added to a spin column with a silica membrane; in the presence of a high concentration of salt, DNA will bind to the membrane. To eliminate impurities, the silica membrane was cleaned using several washing buffers. Water was utilized as the elution buffer to remove the DNA from the silica membrane, and agarose gel electrophoresis was used to examine the resultant DNA sample. Until its next use, the extracted DNA was stored at -20°C or -80°C.
Amplification of DNA
Using universal primers, the extracted DNA from the bacterial sample was amplified by polymerase chain reaction (PCR) to produce a copy of the 16S rRNA gene region. The conserved region was intensified using primers 27F (5'-AGAGTTTGATCMTGGCTCAG-3') and 1492R (5'-GGTTACCTTGTTACGACTT-3'). The PCR mixture is first denatured at 95°C for five minutes. After that, there are 35 cycles of denaturation (at 95°C for 30 seconds), annealing (at 55-60°C for 30 seconds), and extension (at 72°C for 60 seconds), culminating in a final extension of 10 minutes at 72°C.
Amplification and genomic analysis of 16S rRNA
Sequencing
The refined PCR result was sent to the European Sequencing Center in Bangalore, a sequencing service provider, for Sanger sequencing. The File Alignment Sequence Transfer (FAST) format sequencing data was obtained through the use of the European Molecular Biology Open Software Suite (EMBOSS) Merger, which merged the forward and reverse primers (bioinformatics.nl). The obtained nucleotides were run through the National Center for Biotechnology Information's (NCBI's) Basic Local Alignment Search Tool (BLAST) analysis. Nucleotide BLAST analysis was used for data analysis, and the data was downloaded as a Fast-All (FASTA) file. Neighbor-Joining analysis of the evolutionary history was performed [15]. The evolutionary history of the taxa was examined using the bootstrap consensus (1000 replicates) method, as described by Felsenstein (1985) [16]. Using the Likelihood method, as outlined by Tamura et al. (2004) [17], branches corresponding to evolutionary distances were calculated, and evolutionary analyses were carried out in Molecular Evolutionary Genetics Analysis 11 (MEGA11) [18].
Synthesis and recovery of secondary metabolites
The production of microbial metabolites was carried out in an NB medium with 8% NaCl. The obtained bacterial culture was cultivated for nine days in 250 milliliters of NB medium, or until a change in the growth medium's color was noticed. Centrifugation was used to gather the cell-free supernatant after growth and filtration followed. After adding 1/2 of the ethyl acetate to the obtained culture filtrate and thoroughly mixing it in a separating funnel, the ethyl acetate was separated, collected, and used to conduct the crude metabolites before being stored for additional research.
Antimicrobial test: well diffusion technique
The secondary metabolites of Halomonas sp. were assessed using the well diffusion assay against the following five pathogens: C. albicans, P. aeruginosa, E. coli, MRSA, and E. faecalis. The pathogens were grown as a mother inoculum using sterile Mueller-Hinton Broth (MHB), and their optical density (OD) was adjusted to 0.4 at 600 nm. On sterile MHA medium plates, the pathogens were subsequently dispersed using lawn culture techniques. After making wells with a cork borer, 100 µL of crude metabolites were added to the MHA plates. Using a zone scale, the zones of inhibition were identified after a 16-hour incubation period at 37°C. The experiments were conducted in three sets.
Minimum inhibitory concentration (MIC)
Regulations from the European Committee on Antimicrobial Susceptibility Testing (EUCAST) and the Clinical and Laboratory Standards Institute (CLSI) were followed in determining the MIC. The first 10 wells of microtiter test plates were filled with crude metabolites, Ci, at concentrations of Ci = (512, 256, 128, 64, 32, 16, 8, 4, 2, 1) mg/L. Furthermore, Pi - which stands for 5 μL of human pathogens - was added to every well, with the exception of the negative control. The plates were incubated at 37°C for 16 hours. Following incubation, the volume of newly made MTT (VMTT) was added, with VMTT equal to 10 μL and the concentration of MTT (CMTT) equal to 5 mg/mL. After covering the wells with aluminum foil, they were left alone for the time for MTT incubation (tMTT) = 1 hour. After that, a volume of dimethyl sulfoxide (VDMSO) (solubilization solution) was added; VDMSO = 100 μL; the mixture was then left for 15 to 30 minutes to fully solubilize. In an enzyme-linked immunosorbent assay (ELISA) reader, the OD at a wavelength of λ = 595 nm was used to calculate the percentage of cell death (PD) using the following formula.
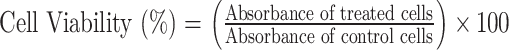
The results observed were recorded and contrasted with the effectiveness of commercially available antibiotics, as described by Rajagopal et al. (2023) [19].
Assay for antioxidant activity using 2,2-diphenyl-1-picrylhydrazyl (DPPH)
The DPPH assay method, described by Malterud (1993) [20], was used to measure the DPPH scavenging activity (SA) of crude metabolites from Halomonas sp. In the beginning, the concentration range of the crude metabolites, Ci, was diluted to include Ci = (512, 256, 128, 64, 32, 16, 8, 4, 2, 1) mg/L. After adding 100 μL of DPPH, the mixture was left to incubate for 20 minutes at room temperature in the dark. Next, a negative control was added, with Vmethanol = 100 μL, containing 99% methyl alcohol. Serving as the positive control was ascorbic acid. At λ = 517 nm, the absorbance (A) was measured. The percentage (%) of DPPH radical SA was determined using the formula:
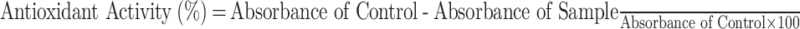
The results were evaluated and contrasted using the methodology of Ralte et al. (2022) [21].
Hemolytic activity
Human blood cells were used to test the biological compatibility of secondary metabolites from Halomonas sp., specifically the recently extracted human blood cells from the participants, followed by three PBS washes. PBS was used as the negative control, and 1% Triton X-100 was used as the positive control. The various concentrations of secondary metabolites (512, 256, 128, 64, 32, 16, 8, 4, 2, and 1 μg/mL) were diluted in 800 µL of PBS solution, and 200 μL of blood sample was added to each microcentrifuge tube separately. The tubes were centrifuged at 5000 rpm for seven minutes and allowed to incubate for an hour at 37°C. The amount of hemoglobin released was calculated using absorbance measurements made at 570 nm [22].
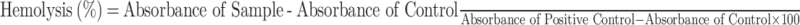
These investigations were done twice and results are expressed by mean ± standard deviation (±SD).
Gas chromatography-mass spectrometry (GC-MS) examination of halophilic bacteria's secondary metabolites
A study on the molecules found in Halomonas sp. secondary metabolites was carried out using the GC Ultra and DSQ II model mass spectrometers. The engine vacuum strain (P), connection point temperature (T_connection), source temperature (T_source), and injector port temperature (T_injector) were all set on the instrument to 250°C, 200°C, and 250 psi, respectively. The oven temperature (T_oven) was programmable, with changes specified as follows. The DB-35 MS non-polar section measured 0.25 μm in inner diameter and 0.25 mm in outer diameter. Helium was used as the carrier gas at a rate of 1 mL/min. The mass spectrometer was set up to find fragments between 50 Da and 650 Da in mass. The ionization energy (IE) of the MS was set at -70 eV, and it included a pre-filter to remove neutral particles. The results were contrasted with the library's reference data.
Statistical analysis
Every experiment was conducted thrice, and the mean ± SDs were used to express the results using Prism software (GraphPad Software Inc., San Diego, CA, USA).
Results
Morphological and phenotypic characterization of halophilic bacteria
In nutrient agar medium with 8% NaCl, the halophilic bacteria's morphology revealed pale yellow, round-shaped, flat, and translucent colonies. A gram-negative rod arrangement was observed under light microscopy following gram staining (Figure 1).
Figure 1. Colony morphology of halophilic bacteria.
A) Colony morphology of potent halophilic bacterium; B) Gram's staning of halophilic bacterium
The biochemical tests show that urease is positive, hydrogen sulfide production is negative, oxidase and catalase are positive, and indole is negative.
Molecular identification of bacterial isolates
The GenBank NCBI database assigned the DNA sequences of the halophilic bacterial isolates through the use of the BLAST 2.0 program for identification. The halophilic organism most closely resembled H. denitrificans, according to the results of the BLAST run. It was followed by Halomonas shengliensis, Halomonas huangheensis, Halomonas binhaiensis, Halomonas cupid, Halomonas stenophila, and Halomonas pacifica. It was confirmed that the potent bacterium belonged to the Halomonas sp. (Figure 2).
Figure 2. Phylogenetic tree construction of halophilic bacterium.
Antimicrobial activity of microbial metabolites
A 14 mm zone of inhibition was observed by the extracellular metabolites of Halomonas sp. against both S. aureus and C. albicans. However, it was unable to demonstrate any antimicrobial activity against E. coli and E. faecalis. In contrast, S. aureus, P. aeruginosa, and E. coli were all effectively inhibited by the positive control, which consisted of commercially available antibiotics (Figure 3; Table 1).
Table 1. Antimicrobial activity of extracellular metabolites produced by Halomonas sp.
ECM: Extracellular metabolites; PC: Positive control
| Zone of inhibition (mg/mL) | |||||||||
| Gram-positive bacterium | Gram-negative bacterium | Fungal pathogen | |||||||
| Staphylococcus aureus | Enterococcus faecalis | Escherichia coli | Pseudomonas aeruginosa | Candida albicans | |||||
| ECM | PC | ECM | PC | ECM | PC | ECM | PC | ECM | PC |
| 14 | 25 | - | - | - | 27 | 12 | 27 | 13 | - |
Figure 3. Antimicrobial property of secondary metabolites from Halomonas sp.
A) Staphylococcus aureus; B) Enterococcus faecalis; C) Escherichia coli; D) Pseudomonas aeruginosa; E) Candida albicans
MIC
It was established what Halomonas sp.'s MIC was for each of the five clinical pathogens. It was found that the extracellular metabolites from Halomonas sp. had a maximum concentration of growth inhibition of 128 mg/L and 256 mg/L, respectively, against S. aureus, C. albicans, and P. aeruginosa. Table 2 shows that no growth inhibition was observed against E. coli or E. faecalis.
Table 2. Minimum inhibitory concentration of extracellular metabolites produced by Halomonas sp.
ECM: Extracellular metabolites; PC: Positive control; ND: Not determined
| Minimum inhibitory concentration (MIC) (mg/L) | |||||||||
| Gram-positive bacterium | Gram-negative bacterium | Fungal pathogen | |||||||
| Staphylococcus aureus | Enterococcus faecalis | Escherichia coli | Pseudomonas aeruginosa | Candida albicans | |||||
| ECM | PC | ECM | PC | ECM | PC | ECM | PC | ECM | PC |
| 128 | 2 | ND | ND | ND | 1 | 256 | 2 | 128 | ND |
Antioxidant activity of microbial metabolites
The extracellular metabolites of Halomonas sp. were found to have antioxidant activity that was able to reduce free radicals at a concentration of 512 mg/mL but did not reduce DPPH SA at a concentration of 256 mg/mL (Figure 4).
Figure 4. Anti-oxidant activity of secondary metabolites form Halomonas sp.
PC: Positive control; NC: Negative control
Hemolytic activity of the microbial metabolites
The effects on red blood cells of varying concentrations of secondary metabolites were evaluated. This information provides a quantitative assessment of hemolytic activity and includes measurements of hemoglobin release or alterations in cell integrity. According to a comparison of the results, there is no hemolytic activity at any of the tested concentrations of the secondary metabolites (Figure 5).
Figure 5. Hemolytic activity of secondary metabolites from Halomonas denitrificans.
PC: Positive control; NC: Negative control
GC-MS analysis
A complex mixture of compounds was discovered by GC-MS analysis of crude metabolites from Halomonas sp. (Figure 6).
Figure 6. Chromatogram of GC-MS analysis of secondary metabolites.
GC-MS: Gas chromatography-mass spectrometry
Propanoic acid, 3-chloro-, 4-formylphenyl ester, with a peak area of 1.31% and a lower probability of 38.75%, is among the substances that have been identified. On the other hand, 2-piperidinone has been detected with a relative peak area of 1.42% and a high identification probability of 84.95%. Another component detected had a probability of 35.27% and a peak area of 0.36%. 2,4-Di-tert-butylphenol was also present, with a probability of 41.1% and a peak area of 0.54%. Many piperazine derivatives, including 3-methyl-6-(1-methylethyl)- and 2,5-piperazinedione, as well as their structural analogs, showed peak areas of less than 1% and probabilities exceeding 70%. Pyrrolo[1,2-a]pyrazine-1,4-dione, hexahydro-, which represented the highest peak area at 20.06% with a 94.37% probability, was one of the noteworthy findings. Other noteworthy compounds included pyrimidine-2(1H)-thione, 4,4,6-trimethyl-1-(1-phenylethyl)-, with a peak area of 2.76% and a 20.61% identification probability, and cyclo, with a peak area of 9.31% and a high probability of 94.78%. Each metabolite found in the Japan Pharmacopeia-Excipients for Microorganisms (JP-ECMs) is thoroughly profiled by the analysis, and Table 3 shows the molecular name, molecular formula, and molecular weight of each metabolite.
Table 3. GC-MS analysis of secondary metabolites from Halomonas sp.
GC-MS: Gas chromatography-mass spectrometry
| Peak | Reaction time | Area % | Name | Probability % | Formula | Mol. wt |
| 1 | 7.525 | 1.42 | 2-piperidinone | 84.95 | C5H9NO | 99.13 |
| 2 | 12.052 | 1.31 | Propanoic acid, 3-chloro-, 4-formylphenyl ester | 38.75 | C10H9ClO3 | 212.02 |
| 3 | 12.795 | 0.36 | Benzeneacetamide | 35.27 | C8HNO | 135.07 |
| 4 | 15.667 | 0.54 | 2,4-Di-tert-butylphenol | 41.1 | C14H22O | 206.17 |
| 5 | 18.340 | 0.42 | 2,5-piperazinedione, 3-methyl-6-(1-methylethyl)- | 87.16 | C8H14N2O2 | 170.11 |
| 6 | 19.965 | 3.17 | 1,4-diazabicyclo[4.3.0]nonan-2,5-dione, 3-methyl | 90.99 | C8H12N2O2 | 168.09 |
| 7 | 20.364 | 1.07 | dl-Alanyl-dl-leucine | 80.77 | C9H18N2O3 | 202.13 |
| 8 | 20.490 | 1.69 | (3S,6S)-3-butyl-6-methylpiperazine-2,5-dione | 46.12 | C9H16N2O2 | 184.12 |
| 9 | 20.861 | 5.32 | Pyrrolo[1,2-a]pyrazine-1,4-dione, hexahydro- | 81.2 | C9H10N2O2 | 154.07 |
| 10 | 21.054 | 0.50 | 3-Isobutyl-2,5-piperazinedione | 73.31 | C8H14N2O2 | 170.11 |
| 11 | 22.257 | 9.31 | Cyclo(L-prolyl-L-valine) | 94.78 | C10H16N2O2 | 196.12 |
| 12 | 25.309 | 20.06 | Pyrrolo[1,2-a]pyrazine-1,4-dione, hexahydro-3-(2-methylpropyl)- | 94.37 | C11H18N2O2 | 210.04 |
| 13 | 30.420 | 0.95 | 2,5-piperazinedione, 3-methyl-6-(phenylmethyl)- | 72.01 | C12H14N2O2 | 218.11 |
| 14 | 30.976 | 28.53 | 2,5-piperazinedione, 3,6-bis(2-methylpropyl) | 71.98 | C12H22N2O2 | 226.17 |
| 15 | 32.788 | 0.67 | 2,5-piperazinedione, 3-benzyl-6-isopropyl- | 83.72 | C14H18N2O2 | 246.14 |
| 16 | 34.955 | 13.18 | Pyrrolo[1,2-a]pyrazine-1,4-dione, hexahydro-3-(phenylmethyl)- | 86.66 | C14H16N2O2 | 244.12 |
| 17 | 38.174 | 2.76 | Pyrimidine-2(1H)-thione, 4,4,6-trimethyl-1-(1-phenylethyl)- | 20.61 | C15H19N2S | 259.13 |
| 18 | 38.995 | 6.75 | Cyclo-(l-leucyl-l-phenylalanyl) | 11.98 | C15H20N2O2 | 260.15 |
| 19 | 39.917 | 1.97 | p-(methylthio)benzyl alcohol | 19.23 | C8H10SO | 154.05 |
Discussion
The genus Halomonas was first described by Vreeland et al. in 1980 [23]. It belongs to the phylum Proteobacteria, class Gammaproteobacteria, order Oceanospirillales, and family Halomonadaceae. Among the species represented in the genus Halomonas are H. denitrificans, Halomonas salaria sp. nov, Halomonas gomseomensis sp. nov, and Halomonas janggokensis sp. nov [24]. Halomonas, also known as H. huangheensis [25], Halomonas nitroreducens, according to González-Domenech et al. (2008) [26]; Halomonas litopenaei was identified by Xue et al. (2018) [27], while Halomonas urmiana was identified by Khan et al. (2020) [28]. Most of them are halophilic and halotolerant bacteria that can survive at concentrations of 7-12% NaCl. Halomonas sp. are aerobic, gram-negative, non-spore-forming rods that measure 0.6-0.8 × 1.2-1.6 μm. Their colonies are smooth, translucent, round with complete edges, and brownish-yellow in color. These cells test positive for the enzymes catalase and oxidase, and they are motile because they have peritrichous flagella. Their growth is noted at pH levels between 7 and 10, with pH 8 to 9 being the ideal range, and temperatures between 5 and 50°C (with an optimum at 25 to 35°C). Furthermore, salinities between 2% and 20% NaCl are favorable for Halomonas sp. growth (the ideal range is between 8% and 10% NaCl). The Voges-Proskauer test produces negative results, and they neither produce indole nor hydrogen sulfide (H2S). Additionally, they can lower nitrite and nitrate [24].
In a similar vein, the powerful bacteria form round, translucent, flat colonies that are pale yellow in color, and gram-negative rods are arranged in light microscopy following gram staining. The biochemical tests show that urease is positive, hydrogen sulfide production is negative, oxidase and catalase are positive, and indole is negative. The powerful halophilic bacterium was identified as Halomonas sp. After molecular identification, the bacterium was found to have high similarities with H. denitrificans, H. shengliensis, H. huangheensis, H. binhaiensis, H. cupid, H. stenophila, and H. pacifica. Poly(3-hydroxybutyrate) (PHB), a metabolite linked to PHB accumulation during various log phases of bacterial growth, is produced by the Halomonas sp. These results imply that the utilization of Halomonas sp. KM-1 in the production of PHB exhibits multicomponent and phase-specific mechanisms [29]. El-Garawani et al. (2020) [6] reported that a recently isolated strain of Halomonas sp. (HA1) exhibits anticancer potential by inducing apoptosis and G2/M arrest in hepatocellular carcinoma (HepG2) cell line. According to a study by Youssif et al. (2020) [30], extracellular polymeric substances (EPS) were produced by the marine microorganism namely Halomonas sp. non-alcoholic steatohepatitis (NASH) in a medium containing 4M NaCl, pH 9, and a 7% initial inoculum. Additionally, the EPS demonstrated antimicrobial activity against a range of pathogens, with the greatest antibacterial activity demonstrated against P. aeruginosa and S. aureus, as well as Bacillus subtilis, E. coli, P. aeruginosa, and C. albicans. According to Youssif et al. (2020) [30], the EPS demonstrated antioxidant and anti-inflammatory activity, indicating the significance of using it for the treatment of chronic diseases where oxidative stress and inflammation play important roles. El-Garawani et al.'s recent study from 2020 [6] demonstrates that the marine isolate of Halomonas sp. exhibited anticancer activity against HepG2 liver cancer cells. Halomonas sp. produces a variety of metabolites, such as di-peptides and biosurfactants, which have anticancer properties. In particular, El-Garawani et al. (2020) [6] identified two biosurfactants as the most potent compounds: Surfactin C14 and Surfactin C15. The secondary metabolites produced by S. aureus, P. aeruginosa, and C. albicans exhibit moderate inhibition in the current study. These metabolites also exhibit antioxidant properties at higher concentrations and have high biological compatibility with human blood samples. However, more research is needed to fully understand the pharmacological activity and to optimize the culture media for the production of secondary metabolites, as well as for their purification and characterization.
Limitations of the study
While this study shows promising results, it's important to acknowledge some limitations. The observed antimicrobial activity of the secondary metabolites was moderate, and further research is needed to enhance their potency. Additionally, the study focused on a limited number of clinical pathogens, so evaluating their efficacy against a wider range of microorganisms is crucial. Analyzing the crude extract limits the understanding of individual compound activities, making it essential to isolate and characterize these compounds. Furthermore, the study's in vitro nature necessitates in vivo studies to assess the metabolites' true efficacy and safety. Finally, the potential long-term effects, such as resistance development or toxicity, remain unexplored and require further investigation. Addressing these limitations will provide a more comprehensive understanding of the therapeutic potential of these secondary metabolites.
Conclusions
Halomonas sp. was found to be the potent bacterium isolated from Saltpan, Tamil Nadu, India. It exhibits moderate inhibition against S. aureus and E. coli for both the well diffusion and MIC assays, as well as moderate antioxidant properties for both intracellular and extracellular metabolites. Blood samples and the metabolites generated by Halomonas sp. are highly compatible. It yields 23 distinct compounds, of which seven are major; however, based on 96% similarity, one molecule may be identical to its name. Therefore, additional mass production is needed to purify and analyze the molecule responsible for this microbial and antioxidant activity and to validate its efficacy against various pathogens and its mode of action, based on the microbial activity of secondary metabolites and compounds produced by this bacterium.
Disclosures
Human subjects: All authors have confirmed that this study did not involve human participants or tissue.
Animal subjects: All authors have confirmed that this study did not involve animal subjects or tissue.
Conflicts of interest: In compliance with the ICMJE uniform disclosure form, all authors declare the following:
Payment/services info: All authors have declared that no financial support was received from any organization for the submitted work.
Financial relationships: All authors have declared that they have no financial relationships at present or within the previous three years with any organizations that might have an interest in the submitted work.
Other relationships: All authors have declared that there are no other relationships or activities that could appear to have influenced the submitted work.
Author Contributions
Concept and design: Manivannan Nandhagopal, Keerthana Perumal, Jayaprakash Seenuvasan
Acquisition, analysis, or interpretation of data: Manivannan Nandhagopal, Keerthana Perumal, Jayaprakash Seenuvasan
Drafting of the manuscript: Manivannan Nandhagopal, Keerthana Perumal, Jayaprakash Seenuvasan
Critical review of the manuscript for important intellectual content: Manivannan Nandhagopal, Keerthana Perumal, Jayaprakash Seenuvasan
Supervision: Manivannan Nandhagopal
References
- 1.Isolation, identification and enzymatic activity of halotolerant and halophilic fungi from the Great Sebkha of Oran in northwestern of Algeria. Chamekh R, Deniel F, Donot C, Jany JL, Nodet P, Belabid L. Mycobiology. 2019;47:230–241. doi: 10.1080/12298093.2019.1623979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diversity of prokaryotic microorganisms in alkaline saline soil of the Qarhan Salt Lake area in the Qinghai-Tibet Plateau. Wang Y, Bao G. Res Sq. 2021;12:3365. doi: 10.1038/s41598-022-07311-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Screening of bioactive compound, antimicrobial activity producing halophilic isolates from the saltpans of Thoothukudi district. Deepalaxmi RK, Gayathri C. Afr J Microbiol Res. 2018;12:338–344. [Google Scholar]
- 4.Identification of two different chemosensory pathways in representatives of the genus Halomonas. Gasperotti AF, Revuelta MV, Studdert CA, Herrera Seitz MK. BMC Genomics. 2018;19:266. doi: 10.1186/s12864-018-4655-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Microbial diversity of saline environments: searching for cytotoxic activities. Díaz-Cárdenas C, Cantillo A, Rojas LY, et al. AMB Express. 2017;7:223. doi: 10.1186/s13568-017-0527-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.A newly isolated strain of Halomonas sp. (HA1) exerts anticancer potential via induction of apoptosis and G(2)/M arrest in hepatocellular carcinoma (HepG2) cell line. El-Garawani IM, El-Sabbagh SM, Abbas NH, et al. Sci Rep. 2020;10:14076. doi: 10.1038/s41598-020-70945-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.A comparative GC-MS analysis of bioactive secondary metabolites produced by halotolerant Bacillus spp. isolated from the Great Sebkha of Oran. Nas F, Aissaoui N, Mahjoubi M, et al. Int Microbiol. 2021;24:455–470. doi: 10.1007/s10123-021-00185-x. [DOI] [PubMed] [Google Scholar]
- 8.Denitrification as an important taxonomic marker within the genus Halomonas. González-Domenech CM, Martínez-Checa F, Béjar V, Quesada E. Syst Appl Microbiol. 2010;33:85–93. doi: 10.1016/j.syapm.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 9.Antimicrobial and antibiofilm activities of bacterial strain P-6B from Segara Anakan against MRSA 2983. Asnani A, Dinda AP, Nursyamsi AR, Anjarwati DU. MOLEKUL. 2022;17:116–124. [Google Scholar]
- 10.Impact of the cold shock phenomenon on quantification of intracellular metabolites in bacteria. Wittmann C, Krömer JO, Kiefer P, Binz T, Heinzle E. Anal Biochem. 2004;327:135–139. doi: 10.1016/j.ab.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 11.Determination of microbial maintenance in acetogenesis and methanogenesis by experimental and modeling techniques. Bonk F, Popp D, Weinrich S, et al. Front Microbiol. 2019;10:166. doi: 10.3389/fmicb.2019.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Intracellular Staphylococcus aureus elicits the production of host very long-chain saturated fatty acids with antimicrobial activity. Bravo-Santano N, Ellis JK, Calle Y, Keun HC, Behrends V, Letek M. Metabolites. 2019;9:148. doi: 10.3390/metabo9070148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Isolation and characterization of halophilic Bacillus sp. BS3 able to produce pharmacologically important biosurfactants. Donio MBS, Ronica SFA, Viji VT, Velmurugan S, Jenifer JA, Michaelbabu M, Citarasu T. Asian Pac J Trop Med. 2013;6:876–883. doi: 10.1016/S1995-7645(13)60156-X. [DOI] [PubMed] [Google Scholar]
- 14.Kocuria marina BS-15 a biosurfactant producing halophilic bacteria isolated from solar salt works in India. Sarafin Y, Donio MB, Velmurugan S, Michaelbabu M, Citarasu T. Saudi J Biol Sci. 2014;21:511–519. doi: 10.1016/j.sjbs.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.The neighbor-joining method: a new method for reconstructing phylogenetic trees. Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 16.Confidence limits on phylogenies: an approach using the bootstrap. Felsenstein J. Evolution. 1985;39:783. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 17.Prospects for inferring very large phylogenies by using the neighbor-joining method. Tamura K, Nei M, Kumar S. Proc Natl Acad Sci U S A. 2004;101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Molecular evolutionary genetics analysis version 11. Tamura K, Stecher G, Kumar S. Mol Biol Evol. 2021;38:3022–3027. doi: 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Study on new series of bis-benzimidazole derivatives synthesis, characterization, single crystal XRD, biological activity and molecular docking. Rajagopal K, Dhandayutham S, Nandhagopal M, Narayanasamy M. J Mol Struct. 2023;1283:135253. [Google Scholar]
- 20.Shared understanding of the qualitative research process. Guidelines for the medical researcher. Malterud K. Fam Pract. 1993;10:201–206. doi: 10.1093/fampra/10.2.201. [DOI] [PubMed] [Google Scholar]
- 21.GC-MS and molecular docking analyses of phytochemicals from the underutilized plant, Parkia timoriana revealed candidate anti-cancerous and anti-inflammatory agents. Ralte L, Khiangte L, Thangjam NM, Kumar A, Singh YT. Sci Rep. 2022;12:3395. doi: 10.1038/s41598-022-07320-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Preparation and characterization of cross-linked chitosan/palladium nanocomposites for catalytic and antibacterial activity. Dhanavel S, Manivannan N, Mathivanan N, Gupta VK, Narayanan V, Stephen A. J Mol Liq. 2018;257:32–41. [Google Scholar]
- 23.Halomonas elongata, a new genus and species of extremely salt-tolerant bacteria. Vreeland RH, Litchfield CD, Martin EL, Elliot E. Int J Syst Bacteriol. 1980;30:485–495. [Google Scholar]
- 24.Halomonas gomseomensis sp. nov., Halomonas janggokensis sp. nov., Halomonas salaria sp. nov. and Halomonas denitrificans sp. nov., moderately halophilic bacteria isolated from saline water. Kim KK, Jin L, Yang HC, Lee ST. Int J Syst Evol Microbiol. 2007;57:675–681. doi: 10.1099/ijs.0.64767-0. [DOI] [PubMed] [Google Scholar]
- 25.Halomonas huangheensis sp. nov., a moderately halophilic bacterium isolated from a saline-alkali soil. Miao C, Jia F, Wan Y, Zhang W, Lin M, Jin W. Int J Syst Evol Microbiol. 2014;64:915–920. doi: 10.1099/ijs.0.056556-0. [DOI] [PubMed] [Google Scholar]
- 26.Halomonas nitroreducens sp. nov., a novel nitrate- and nitrite-reducing species. González-Domenech CM, Béjar V, Martínez-Checa F, Quesada E. Int J Syst Evol Microbiol. 2008;58:872–876. doi: 10.1099/ijs.0.65415-0. [DOI] [PubMed] [Google Scholar]
- 27.Halomonas litopenaei sp. nov., a moderately halophilic, exopolysaccharide-producing bacterium isolated from a shrimp hatchery. Xue M, Wen CQ, Liu L, et al. Int J Syst Evol Microbiol. 2018;68:3914–3921. doi: 10.1099/ijsem.0.003090. [DOI] [PubMed] [Google Scholar]
- 28.Halomonas urmiana sp. nov., a moderately halophilic bacterium isolated from Urmia Lake in Iran. Khan SA, Zununi VS, Forouhandeh H, et al. Int J Syst Evol Microbiol. 2020;70:2254–2260. doi: 10.1099/ijsem.0.004005. [DOI] [PubMed] [Google Scholar]
- 29.Metabolomics-based component profiling of Halomonas sp. KM-1 during different growth phases in poly(3-hydroxybutyrate) production. Jin YX, Shi LH, Kawata Y. Bioresour Technol. 2013;140:73–79. doi: 10.1016/j.biortech.2013.04.059. [DOI] [PubMed] [Google Scholar]
- 30.Production and characterization of extracellular polymeric substances by marine Halomonas sp. NASH isolated from Wadi El-Natroun. Youssif AM, Hamed MM, Abdrabo MAA. J Pure Appl Microbiol. 2020;14:2745–2756. [Google Scholar]








