Abstract
The potential roles of an amino acid deletion at codon 67 (Δ67) and a Thr-to-Gly change at codon 69 (T69G) in the reverse transcriptase of human immunodeficiency virus (HIV) type 1 in drug sensitivity and relative replication fitness were studied. Our results suggest that the Δ67 and T69G changes can be categorized as mutations associated with multidrug resistance. The combination of both mutations with an L74I change (Δ67+T69G/L74I) leads to a novel 3′-azido-3′-deoxythymidine resistance motif and compensates for impaired HIV replication.
Current analyses of genotypic drug resistance patterns have revealed the presence of stable rearrangements, including amino acid insertions and deletions in the β3-β4 hairpin loop of the finger domain of the reverse transcriptase (RT) of human immunodeficiency virus type 1 (HIV-1) (3, 18a, 19). The emergence of these stable rearrangements is associated with failures of combination multiple antiretroviral therapy (3, 7, 11, 13, 16, 18, 18a, 19). A two-amino-acid insertion between codons 69 and 70 in the RT has been shown to confer resistance to multiple nucleoside RT inhibitors (NRTIs) (11, 13, 18). We have recently identified an amino acid deletion at codon 67 (Δ67) and a Thr-to-Gly change at codon 69 (T69G) in the RT of HIV-1 from a patient who had failed combination therapy with 3′-azido-3′-deoxythymidine (AZT) and 2′,3′-dideoxyinosine (ddI) (7). The emergence of this same combination of mutations in the RT has also recently been reported under the selection pressure of 2′,3′-didehydro-3′-deoxythymidine (D4T)–(−)β-l-2′,3′-dideoxy-3′-thiacytidine (3TC)–indinavir (14). Therefore, it appears that the emergence of the Δ67 deletion and the T69G change is not the result of any single treatment regimen but rather may be due to an unusual set of host and/or viral characteristics.
We have demonstrated that high-level resistance to AZT (up to 1,810-fold) was seen in the setting of the Δ67 and T69G changes in association with the AZT resistance mutations K70R, T215F, and K219Q and the non-NRTI (NNRTI) resistance mutations L74I and K103N (7). Further study has revealed that the emergence of the T69G mutation confers drug resistance at the expense of fitness (8). Subsequently, the development of the Δ67 deletion led to a virus with improved replication and high-level AZT resistance (8). The purpose of the present study was to determine the overall profiles of resistance to licensed NRTIs and NNRTIs in viruses containing the Δ67 deletion and the T69G mutation.
A series of HIV-1 variants were created in a cloned proviral DNA pNL4.3 (1) backbone using a Quickchange site mutagenesis kit (Stratagene, La Jolla, Calif.) (8). Drug resistance assays were carried out using MT-2 cells (4, 5) following previously described methods (8). Sensitivities to each drug were reported as the concentrations of the drugs that inhibited p24 production by 50% (IC50s) in tissue culture systems (8, 20). Significant differences between HIV-1 variants in drug sensitivity were calculated by using the unpaired t test of the StatView program (Abacus Concepts, Berkeley, Calif.).
Constructs containing either the Δ67 or the T69G change demonstrated increases in the IC50s of 3′-dideoxycytidine (ddCi Sigma, St. Louis, Mo.) (P < 0.01), ddI (Sigma) (P < 0.01), D4T (P < 0.05 for Δ67, P < 0.01 for T69G) (Sigma), 3TC (P < 0.01), and abacavir (P < 0.01) (Table 1). They were sensitive to AZT (Sigma), nevirapine, and delavirdine. Interestingly, the Δ67 or T69G change increased sensitivity to efavirenz (P < 0.01). A mutant containing both the Δ67 and T69G changes (Δ67+T69G) was sensitive to AZT, ddI, nevirapine, delavirdine, and efavirenz. The Δ67 and T69G changes increased the ICs50 of ddC (P < 0.05), 3TC (P < 0.01), D4T (P < 0.05), and abacavir (P < 0.05) (Table 1). Ross et al. recently reported that the Δ67 mutation in the HXB2 backbone did not lead to high-level resistance to NRTIs (14). Differences in assay methodology or subtle difference in the HIV genotypic backbone may explain this difference between our results and theirs.
TABLE 1.
Susceptibilities of recombinant viruses to RT inhibitors
| Virusb | Mean IC50 ± SE (nM)a
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| AZT | ddC | ddI | 3TC | D4T | Abacavir | Nevirapine | Delavirdine | Efavirenz | |
| WT (NL4.3) | 15 ± 6.5 (1.0) | 38 ± 7.1 (1.0) | 297 ± 54 (1.0) | 103 ± 23 (1.0) | 53 ± 3.5 (1.0) | 18 ± 5.2 (1.0) | 17 ± 1.7 (1.0) | 6.2 ± 1.7 (1.0) | 1.9 ± 0.3 (1.0) |
| Δ67 | 20 ± 7.0 (1.3) | 494 ± 102 (13)c | 1087 ± 119 (3.7)c | 1379 ± 257 (13)c | 312 ± 105 (5.9)d | 322 ± 56 (18)c | 25 ± 10 (1.5) | 6.7 ± 1.2 (1.1) | 0.4 ± 0.2 (0.2)c |
| T69G | 23 ± 5.5 (1.5) | 282 ± 60 (7.4)c | 2564 ± 821 (8.6)c | 477 ± 123 (4.6)c | 266 ± 45 (6.3)c | 130 ± 24 (7.2)c | 14 ± 3.8 (0.8) | 3.8 ± 1.1 (0.6) | 0.5 ± 0.1 (0.3)c |
| Δ67+T69G | 26 ± 10 (1.7) | 83 ± 19 (2.2)d | 256 ± 55 (0.9) | 394 ± 49 (3.8)c | 500 ± 153 (9.4)d | 214 ± 67 (12)d | 16 ± 3.5 (0.9) | 16 ± 6.1 (2.6) | 1.8 ± 0.3 (0.9) |
| K103N | 24 ± 7.8 (1.6) | 49 ± 6.1 (1.3) | 400 ± 58 (1.3) | 178 ± 36 (1.7) | 190 ± 70 (3.6) | 32 ± 6.0 (1.8) | 2,436 ± 379 (143)c | 1,263 ± 187 (204)c | 34 ± 2.4 (18)c |
| Δ67+K103N | 17 ± 6.5 (1.1) | 199 ± 70 (5.2)d | 1,040 ± 181 (3.5)c | 287 ± 72 (2.8)d | 378 ± 112 (7.1)∗d | 268 ± 67 (15)d | 2,082 ± 362 (122)c | 633 ± 186 (102)d | 7.3 ± 2.8 (3.8)c |
| T69G+K103N | 21 ± 6.5 (1.4) | 163 ± 53 (4.3)d | 633 ± 233 (2.1) | 381 ± 147 (3.7) | 543 ± 178 (10)d | 149 ± 26 (8.3)c | 577 ± 91 (34)c | 452 ± 61 (73)c | 8.0 ± 1.2 (4.2)c |
| Δ67+T69G+K103N | 22 ± 6.6 (1.5) | 97 ± 28 (2.6)d | 467 ± 88 (1.6) | 121 ± 15 (1.2) | 219 ± 57 (4.1)d | 178 ± 22 (9.9)c | 1,811 ± 651 (106)c | 224 ± 42 (36)c | 11 ± 3.0 (7.3)c |
The IC50 of each drug was calculated. All assays were independently performed at least three times. The values in parentheses show fold differences from the WT.
Resistant mutations. Δ67, deletion of the amino acid at RT codon 67.
P < 0.01.
P < 0.05.
Since the Δ67 or T69G change increased sensitivity to efavirenz, further phenotypic studies were performed to assess the impact of these mutations on NNRTI-resistant variants of HIV-1. As described by other groups, the variant containing the K103N change showed resistance to nevirapine, delavirdine, and efavirenz (15). Addition of either the Δ67 or T69G mutation to the K103N mutant diminished resistance to efavirenz. In contrast, the presence of both the Δ67 and T69G changes in the K103N mutant partially decreased resistance to all NNRTIs (Table 1).
We have previously reported that the combination of an L74I change, an NNRTI resistance mutation with the Δ67 deletion, and a T69G change (Δ67+T69G/L74I) in the RT resulted in AZT resistance. The IC50 of AZT for the Δ67+T69G/L74I mutant was fivefold higher than that for the wild type (7). The further addition of a K103N change to the Δ67+T69G/L74I motif led to a further increase (16-fold) in the IC50 of AZT (7).
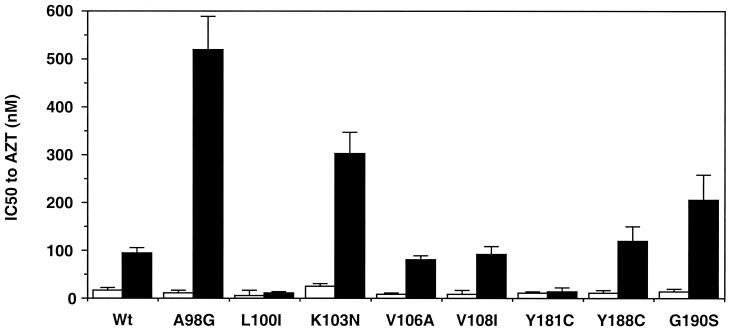
To define the combined effects of the Δ67+T69G/L74I motif and other NNRTI resistance mutations on AZT resistance, a series of constructs containing the well-described NNRTI resistance mutations were created by site-directed mutagenesis (8). Addition of an A98G change to the Δ67+T69G/L74I motif also led to an increase in the IC50 (520 ± 70 nM; P < 0.01) of AZT. No significant changes in AZT resistance were seen upon addition of V106A (92 ± 20 nM), Y108A (100 ± 20 nM), Y188C (164 ± 48 nM), or G190S (205 ± 53 nM; P < 0.1). The Y181C and L100I changes can lead to suppression of AZT resistance (2, 10). Addition of the Y181C or L100I change diminished the IC50 of AZT for the Δ67+T69G/L74I motif-containing mutant to 10 ± 1.7 or 12 ± 8 nM, respectively (Fig. 1). An M184V change in the RT confers resistance to 3TC and suppresses AZT resistance (15). Addition of an M184V mutation also downregulated AZT resistance in the Δ67+T69G/L74I construct (24 ± 4.5 nM).
FIG. 1.
Impact of the Δ67+T69G/L74I motif in combination with known NNRTI resistance mutations on AZT sensitivity. Recombinant mutants were constructed by site-directed mutagenesis. The Δ67+T69G/L74I motif in the RT gene of NL4.3 was induced in each variant containing a single NNRTI resistance mutation. Recombinant viruses were assessed for AZT susceptibility with the drug resistance assay (7, 20). Open bars show the nanomolar IC50s of AZT for mutants without the Δ67+T69G/L74I mutations, and closed bars show the IC50s of AZT for mutants with the Δ67+T69G/L74I mutations. The data shown are means ± standard errors. Each construct was independently tested at least three times.
Mutations M41L, D67N, K70R, T215F/Y, and K219Q/E are associated with well-described AZT resistance (15). Codons 41, 67, and 70 are present in the β3-β4 loop region of the finger domain of the RT and separated by approximately 25 Å from codons 215 and 219 in the β11a-β11b loop. Meanwhile, codons 98 and 103 are present in the β5-β6 loop of the NNRTI binding site and separated by approximately 40 Å from the finger domain (17). Despite this distance, the combination of the Δ67 deletion and the T69G and L74I changes with A98G or K103N led to AZT resistance (Fig. 1). The crystal structure of a covalently trapped catalytic complex of HIV-1 RT has been reported (6). Binding of the template-primer and a deoxynucleoside triphosphate to the DNA-binding cleft between the finger and palm domains of the RT has been shown to induce the outer part of the finger domain, the fingertip, to bend inward toward the palm domain (6). Recently, Winters et al. have reported that the Δ67 deletion in the β3-β4 loop and the T69G mutation in HIV-1 are associated with nucleoside analogs and have proposed a model to explain their findings (18a). In this model, these mutations lead to changes in the three-dimensional structure of the β3-β4 loop and alpha helices C and E in both RT-DNA open (17) and RT-DNA-dTTP closed structures (6). These changes lead to the loss of hydrogen bonds between the RT and dTTP (18a). Therefore, the changes in these regions may lead to an inefficient RT and then allow a novel interaction between the RT and the template-primer-deoxynucleoside triphosphate, subsequently leading to a novel drug resistance.
In the absence of L74I, AZT resistance was not seen in the Δ67+T69G/K103N construct (data not shown). Therefore, the L74I mutation or the combination of Δ67+T69G/L74I changes in the finger domain might facilitate long-distance “communication” between the finger and palm domains through the template-primer, as first reported by Kleim et al. (9).
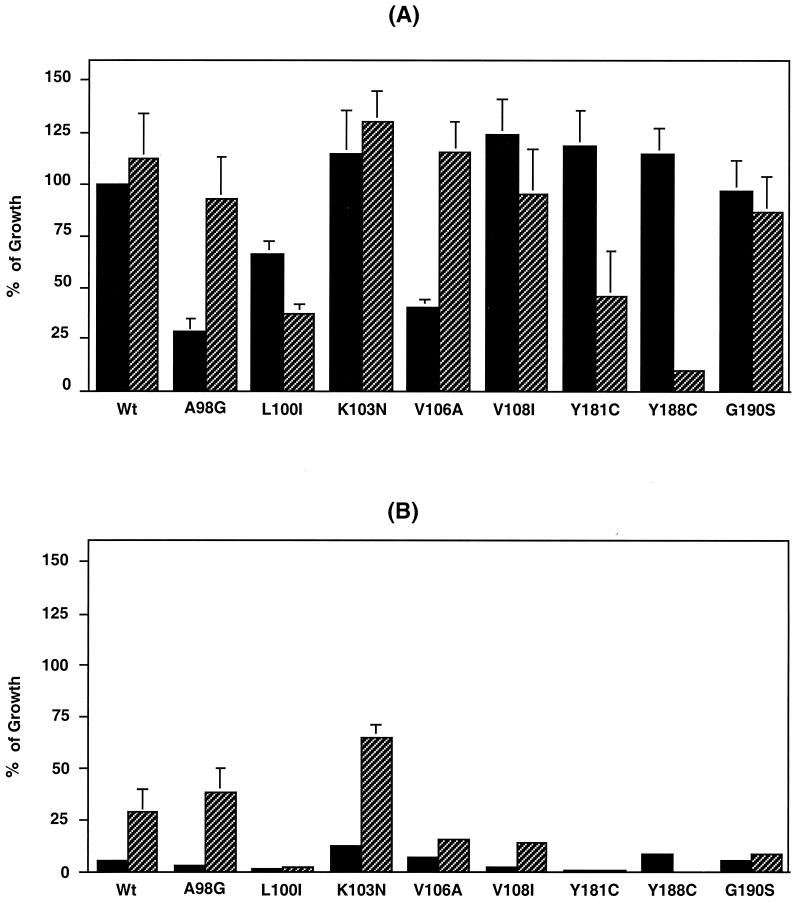
We have previously reported that the T69G mutation led to ddI resistance in an AZT-resistant backbone and was associated with impaired HIV replication (8). Emergence of the Δ67 deletion and the L74I change in the RT compensated for this decrease in HIV replication, even though the Δ67 deletion alone led to impaired HIV replication (8). To characterize the effect of the Δ67+T69G/L74I motif on the replication capability of HIV containing NNRTI resistance mutations, relative replication fitness assays were performed in the presence or absence of 1 μM AZT. Infected MT-2 cells were cultured at 0.1 × 106/ml for 7 days, and p24 levels in day 7 supernatants were measured by a p24 antigen capture kit (8) (Fig. 2). In the absence of AZT (Fig. 2A), a K103N, V108I, Y181C, Y188C, or G190S change in the RT did not lead to a significant difference in growth compared with the wild-type (WT) virus (P > 0.05). In contrast, a A98G, L100I, or V106A change, decreased virus replication to 28 ± 7.0, 66 ± 6.5, or 40 ± 4.8% of that of the WT, respectively. The impaired HIV replication induced by the A98G or V106A change was compensated for by the addition of the Δ67+T69G/L74I motif to the mutant. Meanwhile, addition of the Δ67+T69G/L74I motif to the L100I, Y181C, or Y188C change significantly decreased HIV replication to 37 ± 4.4, 46 ± 21, or 10 ± 2.0% of that of the WT, respectively (P < 0.01). In the presence of 1 μM AZT (Fig. 2B), all variants without the Δ67+T69G/L74I motif were sensitive to AZT. Addition of the motif to the WT or A98G or K103N mutant resulted in partial restoration of replication to 28 ± 11, 38 ± 11, and 65 ± 6.3%, respectively. Archer et al. demonstrated that NNRTI resistance mutations could alter the rate of RNase H cleavage, which is correlated with HIV replication (1a). Since addition of the Δ67+T69G/L74I motif to variants containing mutations associated with resistance to NNRTI affected their replication capability in this study (Fig. 2A), the motif might also influence RNase H cleavage.
FIG. 2.
Replication properties of recombinant HIV-1 variants. Recombinant HIV-1 mutants were constructed by site-directed mutagenesis. MT-2 cells were infected with 1,250 50% tissue culture-infective loses/3 × 106 cells and cultured for 7 days. To determine virus growth properties, levels of p24 antigen in day 7 culture supernatant were measured with a p24 antigen capture kit (8). The Δ67+T69G/L74I motif in the RT gene of NL4.3 was induced in each variant containing a single NNRTI resistance mutation. Viruses were cultured in the absence (A) or presence (B) of 1 μM AZT. Closed bars show results for variants without the Δ67+T69G/L74I mutations, and hatched bars show results for variants with the Δ67+T69G/L74I mutations. Results are expressed as the mean percentage of growth ± the standard error of the mean compared to WT growth in the absence of AZT from three independent experiments. In this experiment, the WT p24 concentration was 713 ± 53 ng/ml.
To define the novel AZT resistance mechanism, a virion-associated RT inhibition assay was also attempted using serial twofold dilutions of AZT-TTP (DuPont NEN, Boston, Mass.) and a commercially available RT assay kit (Roche Molecular Biochemical, Indianapolis, Ind.). The virion-associated RT activity of the Δ67+T69G/L74I motif did not show resistance to AZT-TTP (data not shown). Further biochemical study of this Δ67+T69G/L74I motif is necessary to understand the precise mechanism(s) of the novel AZT resistance motif, as well as the compensated replication fitness. Recently, Meyer et al. demonstrated a novel mechanism of AZT resistance due to an increase in primer-unblocking activity (12). It is hoped that such knowledge will lead to the development of better therapies and salvage therapies.
Acknowledgments
We thank Hiroaki Mitsuya at the NCI for kindly providing 3TC and abacavir. Nevirapine, delavirdine, and efavirenz were provided by Boehringer Ingelheim (Ridgefield, Conn.), Pharmacia & Upjohn (Kalamazoo, Mich.), and DuPont Pharmaceutical Company (Wilmington, Del.), respectively. MT-2 cells and pNL4.3 were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH.
This study was supported by the National Institute of Allergy and Infectious Diseases under contract N01-CO-56000 with SAIC—Frederick.
REFERENCES
- 1.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Archer R H, Dykes C, Gerondelis P, Lloyd A, Fay P, Reichman R C, Bambara R A, Demeter L M. Mutants of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase resistant to nonnucleoside reverse transcriptase inhibitors demonstrate altered rates of RNase H cleavage that correlate with HIV-1 replication fitness in cell culture. J Virol. 2000;74:8390–8401. doi: 10.1128/jvi.74.18.8390-8401.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Byrnes V W, Emini E A, Schleif W A, Condra J H, Schneider C L, Long W J, Wolfgang J A, Graham D J, Gotlib L, Schlabach A J, Wolanski B S, Blahy O M, Quintero J C, Rhodes A, Roth E, Titus D L, Sardana V V. Susceptibilities of human immunodeficiency virus type 1 enzyme and viral variants expressing multiple resistance-engendering amino acid substitutions to reverse transcriptase inhibitors. Antimicrob Agents Chemother. 1994;38:1404–1407. doi: 10.1128/aac.38.6.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Jong J J, Goudsmit J, Lukashov V V, Hillebrand M E, Baan E, Huismans R, Danner S A, ten Veen J H, de Wolf F, Jurriaans S. Insertion of two amino acids combined with changes in reverse transcriptase containing tyrosine-215 of HIV-1 resistant to multiple nucleoside analogs. AIDS. 1999;13:75–80. doi: 10.1097/00002030-199901140-00010. [DOI] [PubMed] [Google Scholar]
- 4.Haertle T, Carrera C J, Wasson D B, Sowers L C, Richman D D, Carson D A. Metabolism and anti-human immunodeficiency virus-1 activity of 2-halo-29,39-dideoxyadenosine derivatives. J Biol Chem. 1988;263:5870–5875. [PubMed] [Google Scholar]
- 5.Harada S, Koyanagi Y, Yamamoto N. Infection of HTLV-III/LAV in HTLV-carrying cells MT-2 and MT-4 and application in a plaque assay. Science. 1985;229:563–566. doi: 10.1126/science.2992081. [DOI] [PubMed] [Google Scholar]
- 6.Huang H, Chopra R, Verdine G L, Harrison S C. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implication for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 7.Imamichi T, Sinha T, Imamichi H, Zhang Y M, Metcalf J A, Falloon J, Lane H C. High-level resistance to 3′-azido-3′-deoxythimidine due to a deletion in the reverse transcriptase gene of human immunodeficiency virus type 1. J Virol. 2000;74:1023–1028. doi: 10.1128/jvi.74.2.1023-1028.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Imamichi T, Berg S C, Imamichi H, Lopez J C, Metcalf J A, Falloon J, Lane H C. Relative replication fitness of a high-level 3′-azido-3′-deoxythimidine-resistant variant of human Immunodeficiency virus type 1 possessing an amino acid deletion at codon 67 and a novel substitution (Thr→Gly) at codon 69. J Virol. 2000;74:10958–10964. doi: 10.1128/jvi.74.23.10958-10964.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kleim J P, Rosner M, Winkler I, Paessens A, Kirsch R, Hsiou V, Arnold E, Riess G. Selective pressure of a quinoxaline non-nucleoside inhibitor of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) on HIV-1 replication results in the emergence of nucleoside RT-inhibitor-specific (RT Leu-743Val or Ile and Val-753Leu or Ile) HIV-1 mutants. Proc Natl Acad Sci USA. 1996;93:34–38. doi: 10.1073/pnas.93.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larder B A. 3′-Azido-3′-deoxythymidine resistance suppressed by a mutation conferring human immunodeficiency virus type 1 resistance to nonnucleoside reverse transcriptase inhibitors. Antimicrob Agents Chemother. 1992;36:2664–2669. doi: 10.1128/aac.36.12.2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Larder B A, Bloor S, Kemp S D, Hertogs K, Desmet R L, Miller V, Sturmer M, Staszewski S, Ren J, Stammers D K, Stuart D I, Pauwels R. A family of insertion mutations between codons 67 and 70 of human immunodeficiency virus type 1 reverse transcriptase confer multinucleoside analog resistance. Antimicrob Agents Chemother. 1999;43:1961–1967. doi: 10.1128/aac.43.8.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer P R, Matsuura S E, Mian A M, So A G, Scott W A. A mechanism of AZT resistance: an increase in nucleotide-dependent primer unblocking by mutant HIV-1 reverse transcriptase. Mol Cell. 1999;4:35–43. doi: 10.1016/s1097-2765(00)80185-9. [DOI] [PubMed] [Google Scholar]
- 13.Ross L, Johnson M, Graham N, Shaefer M, St. Clair M. The reverse transcriptase codon 69 insertion is observed in nucleoside reverse transcriptase inhibitor-experienced HIV-1-infected individuals, including those without prior or concurrent zidovudine therapy. J Hum Virol. 1999;2:290–295. [PubMed] [Google Scholar]
- 14.Ross L, Johnson M, Ferris R G, Short S A, Boone L R, Melby T E, Lanier R, Shaefer M, St. Clair M. Deletions in the beta3-beta4 hairpin loop of HIV-1 reverse transcriptase are observed in HIV-1 isolated from subjects during long-term antiretroviral therapy. J Hum Virol. 2000;3:144–149. [PubMed] [Google Scholar]
- 15.Schinazi R F, Larder B A, Mellors J W. Mutations in retroviral genes associated with drug resistance: 1999–2000 update. Int Antiviral News. 2000;8:65–91. [Google Scholar]
- 16.Tamalet C, Izopet J, Koch N, Fantini J, Yahi N. Stable rearrangements of the β3-β4 hairpin loop of HIV-1 reverse transcriptase in plasma viruses from patients receiving combination therapy. AIDS. 1998;12:F161–F166. doi: 10.1097/00002030-199814000-00003. [DOI] [PubMed] [Google Scholar]
- 17.Tantillo C, Ding J, Jacobo-Molina A, Nanni R G, Boyer P L, Hughes S H, Pauwels R, Andries K, Janssen P A J, Arnold E. Locations of anti-AIDS drug binding sites and resistance mutations in the three dimensional structure of HIV-1 RT. J Mol Biol. 1994;243:369–387. doi: 10.1006/jmbi.1994.1665. [DOI] [PubMed] [Google Scholar]
- 18.Winters M A, Coolley K L, Girard Y A, Levee D J, Hamdan H, Shafer R W, Katzenstein D A, Merigan T C. A 6-basepair insert in the reverse transcriptase gene of human immunodeficiency virus type 1 confers resistance to multiple nucleoside inhibitors. J Clin Investig. 1998;102:1769–1775. doi: 10.1172/JCI4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18a.Winters M A, Coolley K L, Cheng P, Girard Y A, Hamdan H, Kovari L C, Merigan T C. Genotypic, phenotypic, and modeling studies of a deletion in the beta3-beta4 region of the human immunodeficiency virus type 1 reverse transcriptase gene that is associated with resistance to nucleoside reverse transcriptase inhibitors. J Virol. 2000;74:10707–10711. doi: 10.1128/jvi.74.22.10707-10713.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yahi N, Tamalet C, Tourres C, Tivoli N, Ariasi F, Volt F, Gastaut J A, Gallais H, Moreau H J, Fantini J. Mutation pattern of the reverse transcriptase and protease genes in human immunodeficiency virus type 1-infected patients undergoing combination therapy: survey of 787 sequences. J Clin Microbiol. 1999;37:4099–4106. doi: 10.1128/jcm.37.12.4099-4106.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y-M, Imamichi H, Imamichi T, Lane H C, Falloon J, Vasudevachari M B, Salzman N P. Drug resistance during indinavir therapy is caused by mutations in the protease gene and in its Gag substrate cleavage sites. J Virol. 1997;71:6662–6670. doi: 10.1128/jvi.71.9.6662-6670.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]