Abstract
Enzymatic reaction-mediated microbial transformation has emerged as a promising technology with significant potential in various industries. These technologies offer the ability to produce enzymes on a large scale, optimize their functionality, and enable sustainable production processes. By utilizing microbial hosts and manipulating their genetic makeup, enzymes can be synthesized efficiently and tailored to meet specific industrial requirements. This leads to enhanced enzyme performance and selectivity, facilitating the development of novel processes and the production of valuable compounds. Moreover, microbial transformation and biosynthesis offer sustainable alternatives to traditional chemical methods, reducing environmental impact and promoting greener production practices. Microbial transformations enrich drug candidate diversity and enhance active ingredient potency, benefiting the pharmaceutical industry. Continued advancements in genetic engineering and bioprocess optimization drive further innovation and application development in Enzymatic reaction-mediated microbial transformation. The integration of AI for predicting enzymatic reactions and optimizing pathways marks a promising direction for future research. In summary, these technologies have the potential to revolutionize several industries by providing cost-effective, sustainable solutions.
Keywords: Use of microorganisms, Five chemical reactions, Drug development
1. Introduction
Enzymatic reaction-mediated microbial transformation are two important processes used in the field of biotechnology.
Microbial transformation, involves the use of microorganisms to catalyze chemical reactions for the production of useful compounds. This technology utilizes bacteria, fungi, and yeasts to convert compounds or substances. There are numerous types and a wide range of microorganisms, each with strong substrate specificity. Microbial transformation offers mild reaction conditions, simple operation, high purity products, and minimal environmental pollution [1]. The earliest microbial transformations were conducted using wild microorganisms found in fermented wine [2]. This technique has found applications in various fields such as pharmaceuticals, agriculture, and bioremediation [3]. Microbial transformation technology originated from the observation of fermentation processes in the 19th century, initially laid the foundation for Louis Pasteur's research on yeast fermentation. Over time, this field has evolved into a highly specialized technology involving the intersection of microbiology, biochemistry, and genetic engineering. Microbial transformation can be utilized to modify existing drugs, enhancing their solubility, stability, and bioavailability. In the field of agriculture, microbial transformation is employed to produce natural products such as insecticides, herbicides, and fungicides, which are often environmentally friendly and have fewer side effects compared to synthetic pesticides. Moreover, microbial transformation is crucial in bioremediation to eliminate or alter pollutants in the environment. Microorganisms have the ability to break down complex organic compounds into simpler, less harmful substances, which can be easily eliminated or metabolized by other organisms [4].
Enzyme biosynthesis is the natural process through which living cells produce enzymes. Biosynthesis, which is related to microbial transformation, can be altered using genetic engineering techniques to create enzymes with specific properties or functions [5]. As a result, various new enzymes with desired properties have been developed.
Microorganisms such as fungi, yeast, and bacteria have the ability to convert simple starting materials into structurally diverse molecules with desired properties, often achieving high yields [6]. This makes them appealing as industrial hosts for large-scale production [7].
In the context of enzymes, microbial transformation is the process of using microorganisms to create or modify enzymes for various applications. It is an important tool for producing enzymes like amylase, cellulase, protease, lipase, etc. [8].Scientists create these wide varieties of enzymes with different properties and functions, which can be used in various industries including food, pharmaceuticals, agriculture and energy. For example, enzymes produced by these processes are used in the production of detergents, textiles, and paper and pulp industries [9].
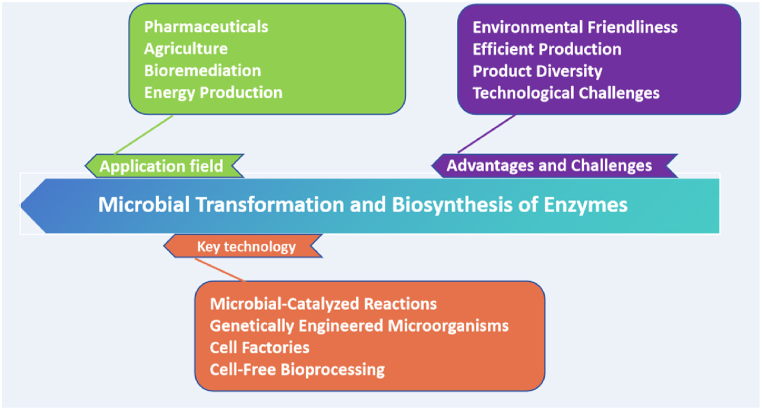
In this article, we will explore enzyme-catalyzed microbial transformation technologies, which represent a significant process with extensive applications in the field of biotechnology. This review aims to comprehensively analyze the latest advancements in enzyme-catalyzed microbial transformation technologies within the field of biotechnology and to explore their potential applications in the pharmaceutical industry. We focus on the role of microbial transformation in enhancing drug diversity, increasing the efficacy of active ingredients, and reducing drug toxicity (Fig. 1). By evaluating the developmental trends and challenges in this field, this review will provide a clear roadmap for future research directions.
Fig. 1.
The graphical abstract of this article.
2. The main reaction types of microbial transformation and enzymatic reactions
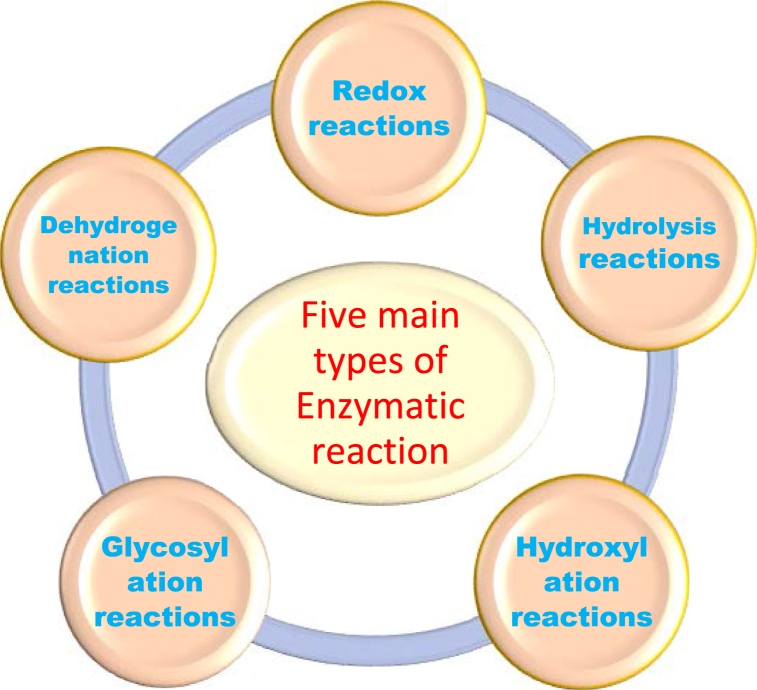
There are five common chemical reactions that depend on microorganisms (Fig. 2). The following will be described one by one.
Fig. 2.
The five main reaction types of enzymatic reaction.
2.1. Redox reactions
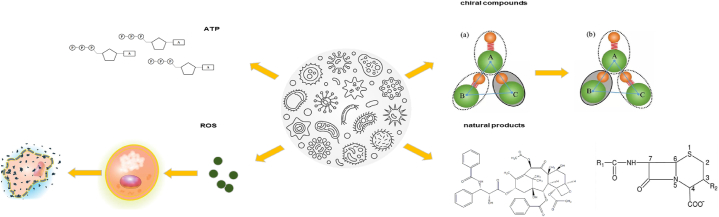
Redox reactions play a critical role in microbial transformation processes. These reactions involve the transfer of electrons from one molecule to another [10,11], and they are essential for the metabolism of microorganisms. During microbial transformation, microorganisms can either oxidize or reduce organic compounds to produce energy or to break down complex molecules. It shows the process of generating ATP and microbial fermentation to produce organic compounds (Fig. 3).
Fig. 3.
The oxidation reaction involved in microorganisms in the conversion of microorganisms has several important significances (1) Production of ATP; (2) Causes the formation of active oxygen, which causes oxidation stress and cell damage; (3) The oxidation reaction can also be used to produce valuable compounds through microbial fermentation or biological conversion process.
In the process of microbial metabolism, organic compounds serve as electron donors [12], while electron acceptors can be inorganic compounds such as oxygen, nitrate, sulfate, and carbon dioxide [13]. These electron acceptors can also be organic compounds, such as aromatic compounds, which serve as terminal electron acceptors in anaerobic respiration [14].
The redox reactions involved in microbial transformation have several important implications. Firstly, they are responsible for the production of ATP, which is the primary energy currency in living organisms. Secondly, they can lead to the formation of reactive oxygen species, which can cause oxidative stress and damage to cells. Finally, redox reactions can also be used to produce valuable compounds [15], such as pharmaceuticals or biofuels [16], through microbial fermentation or Microbial transformation processes.
Microbial redox reactions are used to synthesize chiral compounds by transforming a precursor compound to a chiral molecule through a series of oxidation and reduction reactions. For example, in the synthesis of the anti-inflammatory drug ibuprofen, a microbial reduction reaction is used to convert the precursor compound α-methylstyrene to (S)-ibuprofen [17], the active form of the drug that has higher potency. The reduction reaction is carried out using enzymes produced by microbes belonging to the genus Candida.
In another case, microorganisms have been exploited to catalyze specific redox reactions that lead to the production of various natural products, such as the synthesis of the powerful anticancer drug paclitaxel and the antibiotic cephalosporins. Erythromycin is primarily derived from the bacterium Saccharopolyspora erythraea, which undergoes fermentation to synthesize the antibiotic [18]. The fermentation process involves a series of redox reactions, where the bacteria utilize carbon sources (such as sugars) to generate energy and reduce various organic molecules. These redox reactions result in the production of the antibiotic erythromycin, which can then be isolated and used in pharmaceutical applications. The microbial fermentation process enables the efficient and scalable production of antibiotics [19] and other pharmaceutical compounds.
2.2. Hydrolysis reactions
Hydrolysis reactions are used to break down complex carbohydrates, such as cellulose and hemicellulose, from plant biomass into simple sugars [20], which can then be fermented by microbes to produce biofuels such as ethanol or butanol [21,22]. One example is the use of fungi, specifically Aspergillus niger, to produce xylanase enzymes that degrade the plant polymer xylan to release hemicellulosic sugars for industrial purposes [23]. The use of xylanase reduces energy consumption and enhances chemical recovery due to increased dissolution rate of pretreatment residues.
Beta-lactamase enzymes, produced by certain bacteria like Escherichia coli and Staphylococcus aureus [24], are responsible for the hydrolysis of beta-lactam antibiotics, such as penicillins and cephalosporins. These enzymes cleave the beta-lactam ring in the antibiotic molecule through a hydrolysis reaction, rendering the antibiotic ineffective [25]. This microbial hydrolysis of antibiotics represents a mechanism of antibiotic resistance in bacteria, as the hydrolysis by beta-lactamase enzymes protects the bacteria from the antimicrobial activity of these drugs. Understanding and studying these hydrolysis reactions can aid in the development of strategies to combat antibiotic resistance and improve the effectiveness of antibiotic therapies.
2.3. Hydroxylation reactions
The hydroxylation reaction is influenced by a variety of factors, including the structure of the substrate, the type of hydroxylase enzyme, and the environmental conditions. Microbial hydroxylation has become an important tool in biotechnology for the production of pharmaceuticals and other valuable compounds.
Hydroxylases can be used to add hydroxyl groups to steroid molecules to produce drugs such as cortisone and prednisone [26], which are used to treat inflammation and autoimmune disorders.
Hydroxylation can also be used in bioremediation to break down toxic organic compounds. Microorganisms can use hydroxylation to add a hydroxyl group to pollutants such as polychlorinated biphenyls (PCBs) and polycyclic aromatic hydrocarbons (PAHs), making them more soluble [27] and easier to degrade. This process can help to reduce the environmental impact of these pollutants and promote the remediation of contaminated sites.
2.4. Glycosylation reactions
In microbial transformation, glycosylation can be used to produce glycoproteins and other glycoconjugates with specific glycan structures and functions [28]. For example, microorganisms can be engineered to produce recombinant proteins with human-like glycosylation patterns, which can improve the therapeutic efficacy and reduce the immunogenicity of these proteins.
Glycosylation reactions can also be used in bio-catalysis to synthesize complex glycan structures with high efficiency and specificity. Microorganisms can be engineered to express glycosyltransferases and other enzymes involved in glycan biosynthesis, allowing for the production of custom glycans for various industrial and biomedical applications.
An example of a microbial transformation involving a glycosylation reaction is the production of teichoic acid in Bacillus subtilis [29]. Teichoic acid is a glycopeptide polymer composed of glycerol phosphate chains attached to serine residues in the peptidoglycan layer of the cell wall of gram-positive bacteria including Bacillus subtilis [30]. During its production, an alpha-(1,3)-glucan chain is transferred to a lipid carrier molecule, undecaprenol, through the action of enzymes called glycosyl transferases (GTs). Subsequently, the GTs extend the glucose polymer by adding additional glucopyranose moieties through condensation reactions, ultimately leading to the formation of teichoic acid polysaccharides.
2.5. Dehydrogenation reactions
Microorganisms, including bacteria, fungi, and yeast, have been found to possess enzymes capable of catalyzing dehydrogenation reactions [31]. These enzymes, known as dehydrogenases, are involved in diverse metabolic pathways, including energy production, biosynthesis, and detoxification processes [32]. They exhibit high specificity towards particular substrates, enabling the selective dehydrogenation of specific molecules.
In the field of microbial biotechnology, dehydrogenation reactions have found applications in the production of various industrially valuable compounds [33]. For example, the microbial synthesis of fine chemicals and pharmaceutical intermediates often involves dehydrogenation steps to introduce specific functional groups or modify chemical structures.
One area where dehydrogenation reactions in microbial transformations have shown promise is in the production of biofuels. Microorganisms have been engineered to carry out dehydrogenation reactions on lignocellulosic biomass, converting sugars and other organic compounds into biofuels like ethanol and butanol [34]. This process involves the dehydrogenation of sugars to produce aldehydes, which are subsequently reduced to alcohols [35]. By carefully selecting and engineering microbial strains, researchers aim to improve the efficiency and yield of these dehydrogenation reactions, thereby advancing the viability of biofuel production.
3. Application of the technology in Medicine
Given the central role of microbial transformation technologies in biotechnology, we now turn to discuss their application in the pharmaceutical sector for enhancing drug diversity.
3.1. Increase drug diversity
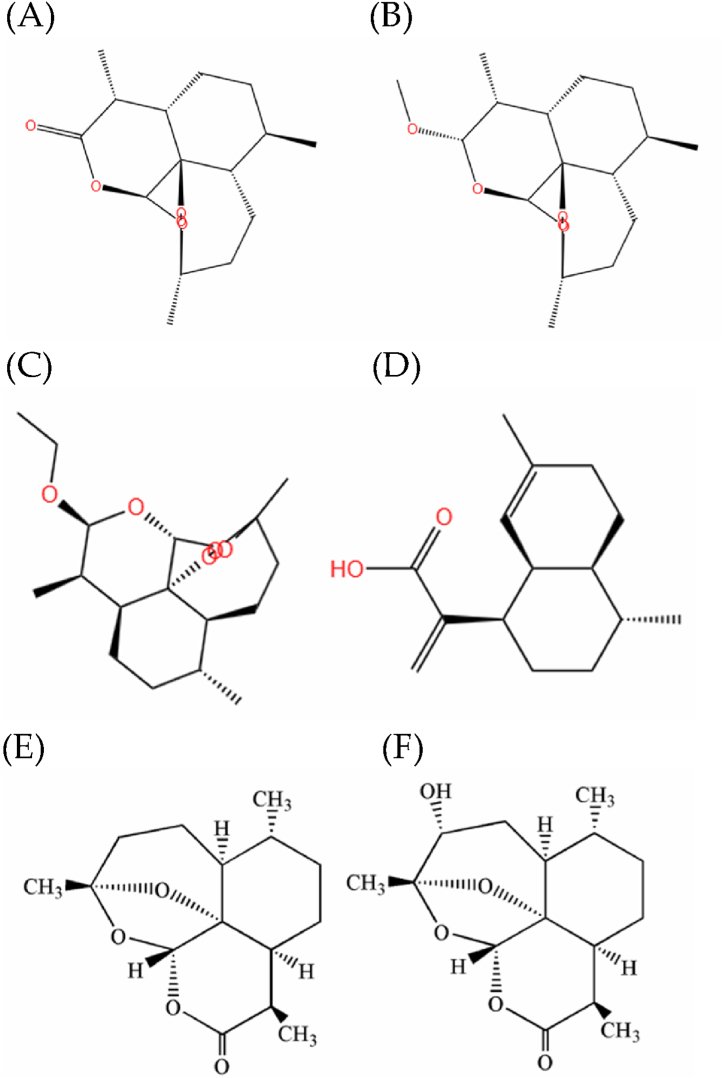
Microbial transformation has been recognized as a valuable technique, which increases the diversity of drug candidates [36]. By utilizing the enzymatic activities of microorganisms, researchers can introduce structural modifications to existing drug molecules, leading to the synthesis of new derivatives [37]. For example, microbial transformation of different artemisinin compounds leads to different products (Table 1). This approach allows for the exploration of chemical variations that may enhance the pharmacological properties, such as potency, selectivity, and solubility, of the original compound. Fig. 4 shows the structural format of several common penicillin derivatives.
Table 1.
Some examples of Microbial transformation of artemisinin derivatives.
| substrate | microbes involved in the reaction | site of chemical reaction | main product |
|---|---|---|---|
| Artemisinin | Penicillium simplissimum | C-3 | 3β-acetoxy artemisinin [38] |
| Rhizopus stolonifera | C-10 | 10β-hydroxy artemisinin [39] | |
| Artemether | Streptomyces lavendulae | C-14 | 14-hydroxy artemether |
| Cunninghamella elegans | C-9 | 9β-hydroxy artemther | |
| Streptomyces lavendulae | C-9 | 9α-hydroxy artemether | |
| Arteether | Beauveria sulfurescens | C-7 | 7β-hydroxy-β-arteether |
| Cunninghamella elegans | C-7 | 7β-hydroxy-β-arteether | |
| Streptomyces lavendulae | C-14 | 14-hydroxy-β-arteether | |
| artemisinic acid | Mucor mucedo | C-3 | 3β-hydroxyartemisinic acids |
| Trichotheciumroseum | C-3 | 3β-hydroxyartemisinic acids |
Fig. 4.
Chemical structural formulas of several artemisinin derivatives (A) Artemisinin; (B) Artemether; (C) Arteether; (D) Artemisinic acid; (E) 1-deoxyartemisinin; (F) 4α-hydroxy-1-deoxyartemisinin [40].
One of the main advantages of microbial transformation is its ability to introduce regioselective and stereoselective modifications to drug molecules [41]. Microorganisms possess a diverse range of enzymes, which can selectively modify specific functional groups or chiral centers in a molecule. This enzymatic selectivity enables the generation of derivatives with specific structural features that can be important for optimizing drug potency, reducing side effects, or improving metabolic stability [42].
Microbial transformation also offers a practical and cost-effective approach for generating drug derivatives compared to traditional chemical synthesis. The use of microorganisms as biocatalysts eliminates the need for complex and expensive synthetic methodologies, as the enzymatic reactions occur under mild conditions and in aqueous environments. This reduces the requirement for hazardous reagents and simplifies the purification process, making microbial transformation a more sustainable and environmentally friendly approach [43].
Recent studies have demonstrated the effectiveness of microbial transformation in generating new drug derivatives. For example, microbial transformation has been employed to modify natural products, such as alkaloids, terpenoids, and polyketides [44], resulting in the synthesis of structurally diverse derivatives with improved pharmacological profiles. Additionally, microbial transformation has been used to functionalize small molecules, such as aromatic compounds and heterocycles, leading to the generation of drug-like molecules with enhanced drug-likeness and improved physicochemical properties.
The microbial transformation represents a powerful approach to increase the types of derivatives available for drug research. This technique allows for the introduction of regioselective and stereoselective modifications, provides a cost-effective alternative to traditional chemical synthesis [45,46], and enables the synthesis of compounds that are difficult to access by other means.
3.2. Improve the active ingredient of the drug
Microbial transformation technology has emerged as a powerful tool in drug research for increasing the content of active ingredients or enhancing their availability [47]. This approach utilizes microorganisms to modify drug molecules, leading to improved potency, bioavailability, and therapeutic efficacy [48].
Microbial transformation technology offers several advantages in increasing the content of active ingredients in drugs [49]. Microorganisms possess a wide array of enzymatic activities that can modify drug molecule. These enzymatic transformations can enhance the structural complexity and diversity of drug compounds, leading to increased content of active ingredients [50].
One way the technology can enhance the content of active ingredients is by increasing the yield of specific compounds during the production process [51]. Microorganisms can be genetically engineered or selected for their ability to efficiently produce target metabolites [52]. By optimizing the enzymatic pathways or enhancing the expression of specific enzymes, researchers can improve the biosynthetic capacity of microorganisms, resulting in higher yields of desired active ingredients. This can be particularly advantageous for natural product-based drugs where the active ingredient is derived from microbial fermentation or microbial transformation processes [53].
In addition to increasing the content of active ingredients, microbial transformation technology can also enhance the availability of drug molecules in the body. One challenge in drug development is the low bioavailability of certain compounds, which limits their therapeutic effectiveness [54]. Microorganisms can be used to modify drug molecules and improve their pharmacokinetic properties, such as solubility, stability, and absorption [40].
Microbial transformation technology can be employed to introduce specific functional groups or modifications that enhance the water solubility of drugs. This can improve their dissolution rate, bioavailability, and systemic exposure, thereby increasing their therapeutic efficacy. Microorganisms can also be utilized to metabolically convert prodrugs into their active form, facilitating drug delivery and enhancing drug availability [55]. For example, the antibiotic drug sulfasalazine is commonly used in the treatment of inflammatory bowel disease. However, sulfasalazine has limited oral bioavailability due to poor absorption in the gastrointestinal tract. The bacterium Escherichia coli possesses an enzyme called azoreductase, which can efficiently reduce the azo bond present in sulfasalazine, resulting in the formation of 5-ASA. By utilizing microbial transformation technology, the prodrug sulfasalazine can be bioactivated into its active form inside the body, leading to improved therapeutic effectiveness.
Recent advancements in microbial transformation technology have demonstrated its effectiveness in increasing the content and availability of active ingredients in drug research. Studies have shown the successful application of microbial transformation in enhancing the yield of specific metabolites and improving the pharmacokinetic properties of drug molecules. These advancements highlight the potential of microbial transformation technology as a valuable tool in drug development, allowing for the optimization of active ingredient content and improved therapeutic outcomes [56].
3.3. Reduce drug toxicity
Microbial transformation technology has emerged as a promising approach in drug research for reducing drug toxicity. This innovative technique utilizes microorganisms to modify drug molecules [57], leading to structural alterations that can mitigate adverse effects and enhance drug safety or act as a detoxifier [58].
Microbial transformation technology offers several strategies to address drug toxicity concerns. One approach involves the enzymatic modification of drug molecules to enhance their metabolic stability [59,60]. By introducing specific structural modifications through microbial transformation, researchers can create derivatives that are less susceptible to metabolic breakdown by host enzymes, reducing the formation of toxic metabolites and enhancing drug safety.
Scientists often employ the technology to modify the chemical structure of drugs, aiming to reduce their inherent toxicity [61,62]. Microorganisms can enzymatically modify functional groups or introduce specific substitutions, leading to structural alterations that can alter drug-target interactions or metabolic pathways. These modifications can enhance the selectivity of drugs towards their intended targets, minimizing off-target effects and reducing the potential for toxicity.
In addition, microbial transformation can be utilized to convert prodrugs into their active forms, thereby improving the therapeutic index and reducing toxicity. Prodrugs are inactive or less active drug forms that are metabolically converted into the active drug within the body. Microorganisms possess enzymes capable of catalyzing these conversions, enabling the targeted release of active drugs while minimizing systemic toxicity [63].
Moreover, we use the technology to optimize the pharmacokinetic properties of drugs, contributing to reduced toxicity. Through enhancing drug distribution, absorption, and elimination [64], scientists achieve better control of drug exposure and minimizing toxic effects associated with high systemic concentrations.
Recent studies have demonstrated the efficacy of microbial transformation technology in reducing drug toxicity. For example, Microbial transformation has been used to modify chemotherapeutic drugs or their products in combination with chemotherapeutic drugs, enhancing their selectivity towards cancer cells while reducing their toxicity towards healthy tissues [65]. In another example, microbial transformation has been utilized to modify drug candidates with hepatotoxic potential, resulting in derivatives with improved safety profiles. Furthermore, microbial transformation and biosynthesis technology have been instrumental in the production of antibody-drug conjugates (ADCs). ADCs are a class of targeted therapies that combine the specificity of monoclonal antibodies with the cytotoxic effects of chemotherapeutic drugs. Through microbial biosynthesis, researchers can precisely attach the cytotoxic drug to the antibody, ensuring targeted delivery to cancer cells while minimizing systemic toxicity. This approach has shown promising results in reducing the side effects associated with traditional chemotherapy.
The study by Koppel et al. highlights the contribution of the human gut microbiota in the metabolism of xenobiotics, including pharmaceutical preparations. The gut microbiota has the ability to transform ingested compounds into metabolites with altered activities, toxicities, and lifetimes within the body. This microbial transformation can lead to the inactivation or detoxification of drugs, thereby reducing their toxicity [66]. One specific example is that the researchers investigated the ability of the human gut bacterium Eggerthella lenta to inactivate the cardiac drug digoxin and they found that this bacterium possesses a specific cytochrome enzyme that can metabolize digoxin, leading to its inactivation. This microbial-mediated inactivation of digoxin has implications for drug efficacy and toxicity. Understanding the role of gut bacteria in drug metabolism can help identify potential interactions and optimize drug dosing to reduce toxicity.
Beyond its applications in the pharmaceutical field, microbial transformation technology also demonstrates significant potential in the chemical, agricultural, and energy sectors. In the chemical industry, bio-polymers and bio-plastics synthesized through microbial transformation are gradually replacing traditional petroleum-based plastics, providing new pathways for achieving a circular economy and sustainable development. In agriculture, microbial transformation technology is used to produce bio-pesticides and bio-fertilizers, which have a smaller environmental impact and help improve crop yield and quality.
4. Genetic engineering and microbial transformation
Recognizing the potential of microbial transformation in drug development, we further investigate how genetic engineering can be integrated with microbial transformation to promote the sustainable production of chemicals and fuels. Researchers have focused on harnessing the power of microbial autotrophy, developing sustainable alternatives for chemical and fuel production, and enhancing the production of valuable molecules through metabolic engineering.
One area of research that has gained significant attention is the engineering of autotrophic microorganisms. Autotrophic microorganisms have the ability to convert carbon dioxide into biomass by deriving energy from light or inorganic electron donors. This metabolic capability has attracted interest in the development of sustainable bioprocesses for the production of fuels, chemicals, and other valuable compounds. Genetic and metabolic engineering approaches have been employed to optimize the performance of autotrophic microorganisms and enhance their productivity [67].
In the field of green chemistry, researchers have focused on developing C1-gas fermenting acetogenic chassis organisms. These organisms have the ability to convert carbon dioxide, carbon monoxide, and acetates into valuable chemicals and fuels. Genetic and metabolic engineering efforts have been directed towards improving the efficiency and yield of these processes, with the aim of developing sustainable alternatives for chemical production [68].
Metabolic engineering has also played a crucial role in the production and derivatization of valuable molecules. Ursolic acid (UA), a promising molecule with various biological activities, has been the subject of intense research. Advances in metabolic engineering have enabled the enhancement of UA production through the manipulation of biosynthetic pathways and optimization of cellular metabolism. These developments have the potential to contribute to the development of new drugs and therapies.
Microbial interactions with plants have also been a focus of research in the past three years. Fungal interactions with plants, both beneficial and detrimental, have been studied to gain insights into plant diseases and develop strategies for crop propickion [69]. Innovative methods and new insights into plant-fungal interactions have been explored, providing a foundation for the development of sustainable agricultural practice.
In the field of agriculture, engineered microbes have shown great potential for enhancing agricultural production and addressing food security challenges. Biotechnological developments in microbial engineering have enabled the manipulation of microbial traits to improve crop yield, nutrient uptake, and disease resistance. These advancements have the potential to contribute to sustainable agriculture and ensure food security for the growing world population.
5. Microbial transformation and cell engineering
As advancements in genetic engineering continue, research in cell engineering has also made significant progress, particularly in constructing microbial cell factories for the production of various chemicals and materials. The field of metabolic engineering has seen significant advancements, with the development of microbial cell factories for the production of various chemicals and materials. Metabolic engineering involves the modification of cellular metabolism to enhance the production of desired compounds [70]. This approach has been applied to a wide range of microorganisms, including bacteria, yeast, and filamentous fungi.
One area of research in cell engineering is the use of electroactive microbial biofilms. Cyclic voltammetry, an electrochemical technique, has been employed to investigate the behavior of these biofilms. This technique allows for the measurement of the electrochemical activity of the biofilm, providing insights into its metabolic processes. The study of electroactive microbial biofilms has applications in areas such as wastewater treatment and bio-electrochemical systems.
Another area of research is the engineering of microbial cells for the production of specific compounds. For example, microbial cell factories have been developed for the production of L-valine, an amino acid with various industrial applications [71]. Metabolic engineering strategies have been employed to optimize the production of L-valine in microorganisms such as Corynebacterium glutamicum, Escherichia coli, Bacillus subtilis, and yeast strains. These efforts involve the manipulation of metabolic pathways and the introduction of genetic modifications to enhance the production of the target compound.
In addition to metabolic engineering, synthetic biology has also played a significant role in cell engineering and microbial transformation. Synthetic biology involves the design and construction of biological systems with novel functions [72]. This field has been applied to the development of microbial consortia, which are communities of different microorganisms that work together to perform specific tasks. Synthetic microbial consortia have been designed for various applications, including the production of biofuels and the degradation of environmental pollutants [73].
The field of cell engineering and microbial transformation has seen rapid progress in recent years, with advancements in metabolic engineering, synthetic biology, and electrochemical techniques. These developments have enabled the design and construction of microbial cell factories for the production of a wide range of chemicals and materials. Future research in this field is likely to focus on further optimizing metabolic pathways, improving the efficiency of microbial cell factories, and exploring new applications for cell engineering and microbial transformation.
6. Microbial transformation, biosynthesis of enzymes and AI
The application of artificial intelligence (AI) in microbial transformation has emerged as a pivotal area of research, offering innovative solutions and insights into the integration of AI in microbiology and biotechnology. In the realm of microbiology, a paradigm shift towards next-generation microbiology has been observed, emphasizing the evolving landscape of metagenomics and metaproteomics in microbial research [74]. Furthermore, the role of AI-2, a well-studied autoinducer, in inhibiting Candida albicans biofilm formation has been explored, shedding light on the molecular mechanisms underlying the interaction between bacteria and fungi [75].
The utilization of machine learning in predicting extensive enzymatic reactions has been a focus of recent research, demonstrating the potential of comprehensive machine learning models in predicting enzymatic reactions [76]. Additionally, global analysis of adenylate-forming enzymes has revealed the β-lactone biosynthesis pathway in pathogenic Nocardia, showcasing the diverse applications of AI in understanding biosynthesis pathways in bacteria [77].
The engineering of the substrate specificity of toluene-degrading enzyme XylM using biosensor XylS and machine learning has expanded the versatility of machine learning in enzyme engineering, showcasing its potential in biocatalysis and metabolic engineering [78]. Furthermore, advances in AI-based microbiome for postmortem interval estimation have been explored, emphasizing the potential of AI in estimating postmortem intervals based on microbiome data [79].
In the context of microbial transformation, the integration of AI has the potential to revolutionize the understanding and manipulation of microbial ecosystems. The utilization of AI in predicting microbial interactions and ecological networks has the potential to provide valuable insights into the complex relationships within microbial communities. Moreover, AI-based approaches can enhance the understanding of microbial transformation processes, enabling the identification of optimal conditions for the production of specific compounds through microbial synthesis. The development of AI-powered tools for the analysis of microbial genomic and metagenomic data has the potential to significantly advance the understanding of microbial transformation processes and the discovery of novel biocatalysts.
Furthermore, the integration of AI in microbial transformation has the potential to streamline the identification and characterization of microbial enzymes involved in transformation processes. AI-based approaches can facilitate the prediction of enzyme-substrate interactions and the design of novel enzymes with enhanced catalytic properties, thereby accelerating the development of biocatalysts for microbial transformation. Additionally, AI-powered platforms for high-throughput screening of microbial strains and enzymatic activities can significantly expedite the discovery and optimization of microbial transformation processes.
The integration of AI in microbial transformation holds great promise for advancing the understanding and application of microbial transformation processes. The utilization of machine learning and AI-based approaches has the potential to revolutionize the discovery, optimization, and application of microbial biocatalysts, thereby contributing to the development of sustainable and efficient biotechnological processes.
The integration of diverse disciplines is pivotal for the advancement of microbial transformation technologies. Collaborations between AI specialists and microbiologists, for instance, can lead to the development of intelligent systems capable of predicting enzyme activities and optimizing metabolic pathways. Synthetic biology, with its focus on designing and engineering new biological functions, can contribute by creating tailored microbial strains for specific industrial applications. Moreover, the intersection of material science and microbial transformation may pave the way for the production of novel biomaterials with unique properties. Cross-disciplinary initiatives not only foster innovation but also ensure a comprehensive approach to problem-solving in the field.
7. Cell-free bioprocessing and cascade enzymatic biocatalysis
Following the successful application of AI technologies in microbial transformation, cell-free bioprocessing technologies have also demonstrated their potential in the production of bioactive compounds. Cell-free bioprocessing has evolved through the integration of synthetic biology principles, enhancing the efficiency of bio-machinery interfacing with synthetic environments. This integration has enabled the production of bioactive compounds with improved properties.
Innovative transformation strategies and optimization techniques have been a focal point of recent research to further enhance the efficiency and selectivity of bioprocessing methods. Studies have explored advanced tools such as vibrational spectroscopy for rapid screening of microbial cells and the development of new on-line parameters derived from automated flow cytometry for process monitoring and control. Additionally, investigations into trace organic contaminant transfer and transformation in bioretention cells have provided insights into the fate of contaminants during runoff events, emphasizing the significance of environmental bioprocessing applications [80,81].
Some researchers have highlighted the intricate relationship between the two. Studies have shown that advancements in microbial fuel cells (MFCs) have enabled micropower generation in small-scale applications, although concerns about their practical large-scale viability have been raise. Additionally, research on sediment microbial fuel cells has illustrated the effectiveness of data-driven modeling in predicting system performance. These findings emphasize the importance of comprehending the dynamics of microbial processes in bioprocessing applications.
Recent medical research has highlighted the intricate relationship between cascade enzymatic biocatalysis and microbial transformation. Studies have explored strategies to optimize in vitro multi-enzymatic reactions, emphasizing that each additional reaction step expands the accessible product range while increasing overall complexity. Furthermore, research has focused on the development of plasmid designs for tunable two-enzyme co-expression to promote whole-cell production of cellobiose, underlining the importance of precise balancing between enzyme activities for efficient flux [82].
Investigations into photo-biocatalytic cascades have shown the potential of combining chemical and enzymatic transformations fueled by light, with challenges related to enzyme-photocatalyst compatibility noted [83]. Advancements in flow biocatalysis have been highlighted as a challenging alternative for synthesizing active pharmaceutical ingredients and natural compounds, leveraging recent biotechnological and protein engineering advances to significantly expand the synthetic enzymatic toolbox [84]. The integration of heterogeneous catalysis and biocatalysis in tandem reactions has been proposed as a promising approach to control product selectivity, showing the potential to merge enzymatic transformations into existing chemical process.
The combination of cell-free bioprocessing technology with microbial transformation offers a new, efficient pathway for industrial production. For example, in the production of high-value pharmaceutical intermediates and active ingredients, cell-free bioprocessing technology enables a faster and more flexible production process, while reducing raw material requirements and production costs. Furthermore, tandem enzyme catalysis technology, by precisely regulating enzyme activity and reaction conditions, can enhance the yield and purity of specific compounds, which is particularly important for the fine chemical and pharmaceutical industries.
8. Advantages and disadvantages of enzymatic reaction-mediated microbial transformation
Despite the numerous innovations and applications brought about by microbial transformation technologies, it is crucial to consider their advantages and disadvantages when assessing their suitability for specific applications.
8.1. Advantages
Advancements in microbial transformation technology have also been instrumental in reducing the toxicity of natural product-based drugs. This approach enables the production of safer derivatives derived from natural sources, offering potential benefits in drug discovery and development.
-
1)
Versatility: Microbial transformation can enable genetic engineering techniques that involve adding, removing, altering or replacing specific traits, allowing versatile modifications of living organisms [85].
One example is gene cloning in yeasts Saccharomyces cerevisiae [86], which can be employed for protein overproduction, by exploiting their amenability to homologous recombination [87], leading to stable cell lines ideal for large scale protein production.
-
2)
Efficiency: Microbial transformation methods allow high efficiency transferring plasmids containing desired genes or gene combinations into target hosts, as observed during the introduction of chloramphenicol resistance markers, lacZ reporter gene constructs, fluorescent proteins, etc., enabling facile selection and tracking of transformed colonies. Research reports that during the treatment of organic waste metal compounds, Shewanella oneidensis MR-1 combined with activated carbon (AC) enhanced the microbial reduction of hydrated iron, and the maximum reduction rate of hydrated iron increased by 1.7–8.2 times.
-
3)
Accessibility: Compared to other species, eukaryotic microbes offer higher ease-of-use concerning competence control for natural transformations at room temperature under non-permissive conditions [88]. In particular, S. cerevisiae displays exceptional adaptability towards plasmid uptake through various mechanisms.
-
4)
Environmentally Beneficial Applications: By introducing carbon fixing genes into cyanobacteria, we potentially generate self-renewing biofuels or fertilizers, producing organic compounds requiring no direct sunlight usage while simultaneously removing CO2 from the atmosphere [39].Other examples include using algae for photobioreactors that photosynthesize and remove excess greenhouse gases from industrial emissions.
8.2. Disadvantages
Microbial transformation, while having its advantages, also comes with certain disadvantages that need to be considered and to be avoid in practice. Here, we will explore these drawbacks.
-
1)
Limited substrate specificity: Microbes often have a narrow range of substrates they can transform. This limitation restricts the applicability of microbial transformation to specific compounds with compatible chemical structures. As a result, not all drugs or chemical compounds can be effectively modified through microbial transformation.
-
2)
Contamination risks: Microbial transformation processes are susceptible to contamination from unwanted microorganisms. Contaminating microorganisms can compete with the desired strains, negatively impacting the transformation efficiency and product quality. Strict quality control measures and sterile conditions are necessary to prevent contamination during fermentation and transformation steps.
-
3)
Biohazards: One of the main concerns is the transmission of pathogens through microbial transformations. Limited data exist on the specific routes of transmission from microbial sources [89]. This lack of information makes it challenging to fully assess and mitigate the risks associated with biohazards. Furthermore, individuals with compromised immune systems may be more susceptible to infections caused by the introduction of live microorganisms.
-
4)
Bacteriophage attacks: Bacteriophages can specifically target and infect bacteria that are involved in microbial transformation processes, leading to the disruption or inhibition of these processes [90]. This can result in a decrease in microbial activity and diversity, as well as a change in the composition of microbial communities.
In addition, certain other limitations in practical applications warrant attention. For instance, the substrate specificity of microbial transformation is limited, restricting its applicability to compounds with incompatible chemical structures. To overcome this limitation, researchers can explore genetic engineering to modify microorganisms, thereby broadening their substrate range, or develop new microbial strains specifically targeting certain types of compounds.
Furthermore, microbial transformation processes are susceptible to contamination by competing microorganisms, which can affect transformation efficiency and product quality. To mitigate this risk, stricter quality control measures can be implemented, including the use of sterile techniques during fermentation and transformation steps, as well as regular monitoring and control of the microbial composition in the fermentation environment.
Biosafety issues are also a critical aspect of microbial transformation. The use of live microorganisms may pose the risk of pathogen transmission. To alleviate this risk, thorough biosafety assessments of the microorganisms used should be conducted, and all operations should be performed within biosafety cabinets to prevent the spread of pathogens.
9. Prospects for the field
Although microbial transformation technology exhibits tremendous potential, several challenges need to be overcome before achieving widespread application. For instance, the substrate specificity limitations in the microbial transformation process restrict its applicability to a wide range of compounds. To address this challenge, future research could focus on the genetic engineering of microorganisms to broaden their substrate range or the development of new microbial strains specifically targeting certain types of compounds. Furthermore, the efficiency and product yield of the microbial transformation process still require further improvement to meet the demands of industrial-scale production. This may involve a deeper understanding and optimization of microbial metabolic pathways, as well as the development of more efficient bioreactors and fermentation conditions. Microbial transformation technology may also face regulatory and ethical issues in practical applications, particularly when involving the release of genetically modified microorganisms into the environment. Therefore, the establishment of a robust safety assessment and regulatory framework will be crucial to ensure the safe application of these technologies. Finally, to achieve the sustainable development of microbial transformation technology, further research and development are needed to explore environmentally friendly production methods, reduce the dependence on natural resources, and minimize waste and pollutant emissions during the production process.
As we reflect on the advancements in enzymatic reaction-mediated microbial transformation, it is imperative to gaze into the horizon and outline the trajectory of future research. A pivotal direction for future research lies in the engineering of microbial strains with broadened substrate specificity and enhanced catalytic efficiency. Synthetic biology offers a powerful platform for the design of novel enzymes and pathways, enabling the harnessing of previously intractable biochemical reactions. Cross-disciplinary collaborations are encouraged to unlock the full potential of microbial transformation. Synergies with material science, nanotechnology, and computational biology could lead to the development of innovative biocatalysts and biomaterials, as well as novel solutions for environmental remediation.
10. Conclusions
In conclusion, the advancements in enzymatic reaction-mediated microbial transformation have significantly reshaped the landscape of industrial biotechnology and drug development. The ability to tailor enzymes for specific functions and to produce novel compounds through microbial biosynthesis has opened up new horizons for creating more effective and sustainable industrial processes. Notably, the integration of AI in predicting and optimizing these Microbial transformation pathways has been a game-changer, offering unprecedented precision and efficiency in the design of microbial cell factories.
However, alongside these strides, challenges such as substrate specificity, contamination risks, and biohazard concerns demand attention. Future research should focus on developing robust protocols and advanced genetic engineering techniques to mitigate these issues, ensuring the safe and efficient application of microbial transformation technology.
Looking ahead, the convergence of microbial transformation with cutting-edge fields like synthetic biology and AI holds immense promise. It is expected to further amplify the potential of this technology, leading to groundbreaking innovations in the production of biofuels, pharmaceuticals, and other valuable chemicals. With continued advancements, enzymatic reaction-mediated microbial transformation is poised to make a substantial impact on the road to sustainable and green biotechnology.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Availability of data and materials
This is a review, all data and materials are references.
Disclaimer/Publisher's note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of the editors. Editors disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
Funding
This research was supported by Project supported by Beijing Ji-shui-tan Research Funding (code: QN-202302).
CRediT authorship contribution statement
Cheng-chao Zheng: Writing – review & editing, Methodology, Funding acquisition. Liang Gao: Methodology, Investigation, Formal analysis, Data curation. Hao Sun: Resources, Project administration, Methodology, Investigation. Xin-Yu Zhao: Supervision, Project administration. Zhu-qing Gao: Validation, Supervision, Funding acquisition. Jie Liu: Visualization, Validation, Supervision. Wei Guo: Writing – review & editing, Writing – original draft, Data curation, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Huo C., Han F., Xiao Y., Kim H.J., Lee I.S. Microbial transformation of yakuchinone A and cytotoxicity evaluation of its metabolites. Int. J. Mol. Sci. 2022;23(7) doi: 10.3390/ijms23073992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balland P.A., et al. The new paradigm of economic complexity. Res Policy. 2022;51(3) doi: 10.1016/j.respol.2021.104450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma S., et al. Inhibitory effect of fermented flammulina velutipes polysaccharides on mice intestinal inflammation. Front. Nutr. 2022;9 doi: 10.3389/fnut.2022.934073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou L., et al. Amiloride ameliorates muscle wasting in cancer cachexia through inhibiting tumor-derived exosome release. Skelet Muscle. 2021;11(1):17. doi: 10.1186/s13395-021-00274-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu J., et al. Conformationally engineering flexible peptides on silver nanoparticles. iScience. 2022;25(6) doi: 10.1016/j.isci.2022.104324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lim H.G., et al. Vibrio sp. dhg as a platform for the biorefinery of brown macroalgae. Nat. Commun. 2019;10(1):2486. doi: 10.1038/s41467-019-10371-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Linde D., et al. Structural characterization of two short unspecific peroxygenases: two different dimeric arrangements. Antioxidants. 2022;11(5) doi: 10.3390/antiox11050891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scott I.M., Zhu H., Schieck K., Follick A., Reynolds L.B., Menassa R. Non-target effects of hyperthermostable α-amylase transgenic nicotiana tabacum in the laboratory and the field. Front. Plant Sci. 2019;10:878. doi: 10.3389/fpls.2019.00878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Schaick G., et al. Native liquid chromatography and mass spectrometry to structurally and functionally characterize endo-xylanase proteoforms. Int. J. Mol. Sci. 2022;23(3) doi: 10.3390/ijms23031307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang Y., et al. Trisulfide bond-mediated doxorubicin dimeric prodrug nanoassemblies with high drug loading, high self-assembly stability, and high tumor selectivity. Sci. Adv. 2020;6(45) doi: 10.1126/sciadv.abc1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Brien E., Silva R.M., Barton J.K. Redox signaling through DNA. Isr. J. Chem. 2016;56(9–10):705–723. doi: 10.1002/ijch.201600022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kushkevych I., Kotrsová V., Dordević D., Buňková L., Vítězová M., Amedei A. Hydrogen sulfide effects on the survival of lactobacilli with emphasis on the development of inflammatory bowel diseases. Biomolecules. 2019;9(12) doi: 10.3390/biom9120752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhuang X., Wang S., Wu S. Electron transfer in the biogeochemical sulfur cycle. Life. 2024;14(5):591. doi: 10.3390/life14050591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Narsaria A.K., et al. Distortion-controlled redshift of organic dye molecules. Chemistry (Weinheim an der Bergstrasse, Germany) 2020;26(9):2080–2093. doi: 10.1002/chem.201905355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernandes A.S. Redox-active molecules as therapeutic agents. Antioxidants. 2022;11(5):1004. doi: 10.3390/antiox11051004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ansari I., et al. Wild halophytic phragmites karka biomass saccharification by bacterial enzyme cocktail. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.714940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heine T., et al. On the enigma of glutathione-dependent styrene degradation in gordonia rubripertincta CWB2. Appl. Environ. Microbiol. 2018;84(9) doi: 10.1128/AEM.00154-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yun K., et al. Droplet-microfluidic-based promoter engineering and expression fine-tuning for improved erythromycin production in Saccharopolyspora erythraea NRRL 23338. Front. Bioeng. Biotechnol. 2022;10 doi: 10.3389/fbioe.2022.864977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song M.W., Park J.Y., Lee H.S., Kim K.T., Paik H.D. Co-fermentation by lactobacillus brevis B7 improves the antioxidant and immunomodulatory activities of hydroponic ginseng-fortified yogurt. Antioxidants. 2021;10(9) doi: 10.3390/antiox10091447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Y., et al. Integrated microbiome and metabolome analysis reveals a novel interplay between commensal bacteria and metabolites in colorectal cancer. Theranostics. 2019;9(14):4101–4114. doi: 10.7150/thno.35186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang Y., et al. Quantitative proteomic analysis to reveal expression differences for butanol production from glycerol and glucose by Clostridium sp. strain CT7. Microb. Cell Fact. 2021;20(1):12. doi: 10.1186/s12934-021-01508-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang B., Nie X., Xiao Y., Gu Y., Jiang W., Yang C. Ferrous-iron-activated transcriptional factor AdhR regulates redox homeostasis in Clostridium beijerinckii. Appl. Environ. Microbiol. 2020;86(7) doi: 10.1128/AEM.02782-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miao T., et al. Improved production of xylanase in Pichia pastoris and its application in xylose production from xylan. Front. Bioeng. Biotechnol. 2021;9 doi: 10.3389/fbioe.2021.690702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miltgen G., et al. One Health compartmental analysis of ESBL-producing Escherichia coli on Reunion Island reveals partitioning between humans and livestock. J. Antimicrob. Chemother. 2022;77(5):1254–1262. doi: 10.1093/jac/dkac054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amanatidou E., Matthews A.C., Kuhlicke U., Neu T.R., McEvoy J.P., Raymond B. Biofilms facilitate cheating and social exploitation of β-lactam resistance in Escherichia coli. NPJ Biofilms Microbiomes. 2019;5(1):36. doi: 10.1038/s41522-019-0109-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Donova M. Microbial steroid production technologies: current trends and prospects. Microorganisms. 2021;10(1):53. doi: 10.3390/microorganisms10010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saktrakulkla P., Li X., Martinez A., Lehmler H.J., Hornbuckle K.C. Hydroxylated polychlorinated biphenyls are emerging legacy pollutants in contaminated sediments. Environ. Sci. Technol. 2022;56(4):2269–2278. doi: 10.1021/acs.est.1c04780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harten I.A., et al. The synthesis and secretion of versican isoform V3 by mammalian cells: a role for N-linked glycosylation. Matrix Biol. 2020;89:27–42. doi: 10.1016/j.matbio.2020.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang X., et al. Transcriptional regulator AcrR increases ethanol tolerance through regulation of fatty acid synthesis in lactobacillus plantarum. Appl. Environ. Microbiol. 2019;85(22) doi: 10.1128/AEM.01690-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guyet A., Alofi A., Daniel R.A. Insights into the roles of lipoteichoic acids and MprF in Bacillus subtilis. mBio. 2023;14(1) doi: 10.1128/mbio.02667-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu Y., et al. iTRAQ-based quantitative proteomic analysis of arthrobacter simplex in response to cortisone acetate and its mutants with improved Δ(1)-dehydrogenation efficiency. J. Agric. Food Chem. 2023;71(16):6376–6388. doi: 10.1021/acs.jafc.3c00417. [DOI] [PubMed] [Google Scholar]
- 32.Sackett J.D., Kamble N., Leach E., Schuelke T., Wilbanks E., Rowe A.R. Genome-scale mutational analysis of cathode-oxidizing thioclava electrotropha ElOx9(T) Front. Microbiol. 2022;13 doi: 10.3389/fmicb.2022.909824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiao Y., Han F., Lee I.S. Biotransformation of the phenolic constituents from licorice and cytotoxicity evaluation of their metabolites. Int. J. Mol. Sci. 2021;22(18) doi: 10.3390/ijms221810109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pang J., Liu Z., Zhang Q., Lu X., Qi Q. Systematic analysis of Escherichia coli isolates from sheep and cattle suggests adaption to the rumen niche. Appl. Environ. Microbiol. 2020;86(20) doi: 10.1128/AEM.01417-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang W., Tian X., Jiao H., Jackstell R., Beller M. Iridium-catalyzed domino hydroformylation/Hydrogenation of Olefins to alcohols: synergy of two ligands. Chemistry (Weinheim an der Bergstrasse, Germany) 2022;28(9) doi: 10.1002/chem.202104012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiao Y., Lee I.S. Effects of microbial transformation on the biological activities of prenylated chalcones from angelica keiskei. Foods. 2022;11(4) doi: 10.3390/foods11040543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cao L., Shcherbin E., Mohimani H. A metabolome- and metagenome-wide association network reveals microbial natural products and microbial biotransformation products from the human microbiota. mSystems. 2019;4(4) doi: 10.1128/mSystems.00387-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goswami A., et al. Bio-transformation of artemisinin using soil microbe: direct C-acetoxylation of artemisinin at C-9 by Penicillium simplissimum. Bioorg. Med. Chem. Lett. 2010;20(1):359–361. doi: 10.1016/j.bmcl.2009.10.097. [DOI] [PubMed] [Google Scholar]
- 39.Bai W., Ranaivoarisoa T.O., Singh R., Rengasamy K., Bose A. n-Butanol production by Rhodopseudomonas palustris TIE-1. Commun. Biol. 2021;4(1):1257. doi: 10.1038/s42003-021-02781-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo J., et al. Biotransformation of artemisinin to a novel derivative via ring rearrangement by Aspergillus Niger. Appl. Microbiol. Biotechnol. 2022;106(7):2433–2444. doi: 10.1007/s00253-022-11888-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zu H., Gu J., Zhang H., Fan A., Nie Y., Xu Y. Highly enantioselective synthesis of (R)-1,3-butanediol via deracemization of the corresponding racemate by a whole-cell stereoinverting cascade system. Microb. Cell Fact. 2020;19(1):125. doi: 10.1186/s12934-020-01384-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tromp A.T., et al. Host-receptor post-translational modifications refine staphylococcal leukocidin cytotoxicity. Toxins. 2020;12(2) doi: 10.3390/toxins12020106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou L., et al. Evaluation of metabolic engineering strategies on 2-ketoisovalerate production by Escherichia coli. Appl. Environ. Microbiol. 2022;88(17) doi: 10.1128/aem.00976-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dong L., et al. Anti-inflammatory effects of metabolites from antarctic fungal strain pleosporales sp. SF-7343 in HaCaT human keratinocytes. Int. J. Mol. Sci. 2021;22(18) doi: 10.3390/ijms22189674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lv Y., et al. Highly efficient preparation of cyclic dinucleotides via engineering of dinucleotide cyclases in Escherichia coli. Front. Microbiol. 2019;10:2111. doi: 10.3389/fmicb.2019.02111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiao S., et al. Comprehensive metabolic profiling of euphorbiasteroid in rats by integrating UPLC-Q/TOF-MS and NMR as well as microbial biotransformation. Metabolites. 2022;12(9) doi: 10.3390/metabo12090830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang R., et al. Efficient one-step biocatalytic multienzyme cascade strategy for direct conversion of phytosterol to C-17-Hydroxylated steroids. Appl. Environ. Microbiol. 2021;87(24) doi: 10.1128/AEM.00321-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Łyczko P., Panek A., Ceremuga I., Świzdor A. The catalytic activity of mycelial fungi towards 7-oxo-DHEA - an endogenous derivative of steroidal hormone dehydroepiandrosterone. Microb. Biotechnol. 2021;14(5):2187–2198. doi: 10.1111/1751-7915.13903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tao A., Feng X., Sheng Y., Song Z. Optimization of the artemisia polysaccharide fermentation process by Aspergillus Niger. Front. Nutr. 2022;9 doi: 10.3389/fnut.2022.842766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao Y.Q., et al. One-pot biosynthesis of 7β-hydroxyandrost-4-ene-3,17-dione from phytosterols by cofactor regeneration system in engineered mycolicibacterium neoaurum. Microb. Cell Fact. 2022;21(1):59. doi: 10.1186/s12934-022-01786-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu A., et al. Use of non-Saccharomyces yeast Co-fermentation with Saccharomyces cerevisiae to improve the polyphenol and volatile aroma compound contents in nanfeng tangerine wines. Journal of fungi (Basel, Switzerland) 2022;8(2) doi: 10.3390/jof8020128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moon J.H., Lee K., Lee J.H., Lee P.C. Redesign and reconstruction of a steviol-biosynthetic pathway for enhanced production of steviol in Escherichia coli. Microb. Cell Fact. 2020;19(1):20. doi: 10.1186/s12934-020-1291-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Choi Y., Bose S., Shin N.R., Song E.J., Nam Y.D., Kim H. Lactate-fortified puerariae radix fermented by bifidobacterium breve improved diet-induced metabolic dysregulation via alteration of gut microbial communities. Nutrients. 2020;12(2) doi: 10.3390/nu12020276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu W.Y., et al. Doxorubicin cardiomyopathy is ameliorated by acacetin via Sirt1-mediated activation of AMPK/Nrf2 signal molecules. J. Cell Mol. Med. 2020;24(20):12141–12153. doi: 10.1111/jcmm.15859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang J., et al. 4-Hydroxyisoleucine alleviates macrophage-related chronic inflammation and metabolic syndrome in mice fed a high-fat diet. Front. Pharmacol. 2020;11 doi: 10.3389/fphar.2020.606514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yan X.T., et al. In vitro anti-obesity effect of shenheling extract (SHLE) fermented with lactobacillus fermentum grx08. Foods. 2022;11(9) doi: 10.3390/foods11091221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ma P.Y., et al. Native endophytes of tripterygium wilfordii-mediated biotransformation reduces toxicity of celastrol. Front. Microbiol. 2022;13 doi: 10.3389/fmicb.2022.810565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kolawole O., et al. Comparative in vitro assessment of a range of commercial feed additives with multiple mycotoxin binding claims. Toxins. 2019;11(11) doi: 10.3390/toxins11110659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Luo Y., Zhou T. Connecting the dots: targeting the microbiome in drug toxicity. Med. Res. Rev. 2022;42(1):83–111. doi: 10.1002/med.21805. [DOI] [PubMed] [Google Scholar]
- 60.Peng Y., et al. Microbial biotransformation to obtain stilbene methylglucoside with GPR119 agonistic activity. Front. Microbiol. 2023;14 doi: 10.3389/fmicb.2023.1148513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Clarke G., Sandhu K.V., Griffin B.T., Dinan T.G., Cryan J.F., Hyland N.P. Gut reactions: breaking down xenobiotic-microbiome interactions. Pharmacol. Rev. 2019;71(2):198–224. doi: 10.1124/pr.118.015768. [DOI] [PubMed] [Google Scholar]
- 62.Farouk F., Shamma R. Chemical structure modifications and nano-technology applications for improving ADME-Tox properties, a review. Arch. Pharm. (Weinheim) 2019;352(2) doi: 10.1002/ardp.201800213. [DOI] [PubMed] [Google Scholar]
- 63.Cai C., et al. Deoxynivalenol degradation by various microbial communities and its impacts on different bacterial flora. Toxins. 2022;14(8) doi: 10.3390/toxins14080537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zimmermann-Kogadeeva M., Zimmermann M., Goodman A.L. Insights from pharmacokinetic models of host-microbiome drug metabolism. Gut Microb. 2020;11(3):587–596. doi: 10.1080/19490976.2019.1667724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Binder P., et al. Pak2 regulation of Nrf2 serves as a novel signaling nexus linking ER stress response and oxidative stress in the heart. Front Cardiovasc Med. 2022;9 doi: 10.3389/fcvm.2022.851419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Linke J.A., Rayat A., Ward J.M. Production of indigo by recombinant bacteria. Bioresour Bioprocess. 2023;10(1):20. doi: 10.1186/s40643-023-00626-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang S., Zhao F., Yang M., Lin Y., Han S. Metabolic engineering of Saccharomyces cerevisiae for the synthesis of valuable chemicals. Crit. Rev. Biotechnol. 2024;44(2):163–190. doi: 10.1080/07388551.2022.2153008. [DOI] [PubMed] [Google Scholar]
- 68.Ibitoye S.E., Jen T.C., Mahamood R.M., Akinlabi E.T. Densification of agro-residues for sustainable energy generation: an overview. Bioresour Bioprocess. 2021;8(1):75. doi: 10.1186/s40643-021-00427-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim S.H., Vujanovic V. Early transcriptomic response of the mycoparasite Sphaerodes mycoparasitica to the mycotoxigenic Fusarium graminearum 3-ADON, the cause of Fusarium head blight. Bioresour Bioprocess. 2021;8(1):127. doi: 10.1186/s40643-021-00479-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ko Y.S., et al. Tools and strategies of systems metabolic engineering for the development of microbial cell factories for chemical production. Chem. Soc. Rev. 2020;49(14):4615–4636. doi: 10.1039/d0cs00155d. [DOI] [PubMed] [Google Scholar]
- 71.Gao H., Tuyishime P., Zhang X., Yang T., Xu M., Rao Z. Engineering of microbial cells for L-valine production: challenges and opportunities. Microb. Cell Fact. 2021;20(1):172. doi: 10.1186/s12934-021-01665-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Goold H.D., Wright P., Hailstones D. Emerging opportunities for synthetic biology in agriculture. Genes. 2018;9(7):341. doi: 10.3390/genes9070341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang B., Wang Y., Shi S., Li F., Song H. [Design and applications of synthetic electroactive microbial consortia. Sheng wu gong cheng xue bao = Chinese journal of biotechnology. 2023;39(3):858–880. doi: 10.13345/j.cjb.220773. [DOI] [PubMed] [Google Scholar]
- 74.Nam N.N., Do H., Loan Trinh K.T., Lee N.Y. Metagenomics: an effective approach for exploring microbial diversity and functions. Foods. 2023;12(11):2140. doi: 10.3390/foods12112140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Angehrn Z., et al. Artificial intelligence and machine learning applied at the point of care. Front. Pharmacol. 2020;11:759. doi: 10.3389/fphar.2020.00759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Scheetz J., et al. A survey of clinicians on the use of artificial intelligence in ophthalmology, dermatology, radiology and radiation oncology. Sci. Rep. 2021;11(1):5193. doi: 10.1038/s41598-021-84698-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Boillat T., Nawaz F.A., Rivas H. Readiness to embrace artificial intelligence among medical doctors and students: questionnaire-based study. JMIR Medical Education. 2022;8(2) doi: 10.2196/34973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ye J., et al. Formation mechanism of microbial diversity in artificial intelligence devices due to intermediate disturbance by low-dose UV radiation for complementary medicine. Evid Based Complement Alternat Med. 2022;2022 doi: 10.1155/2022/2874835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu R., Wang Y., Zou J. Research on the transformation from financial accounting to management accounting based on drools rule engine. Comput. Intell. Neurosci. 2022;2022 doi: 10.1155/2022/9445776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jasinska A.J., Apetrei C., Pandrea I. Walk on the wild side: SIV infection in African non-human primate hosts-from the field to the laboratory. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.1060985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sheehan S.A., Retzbach E.P., Shen Y., Krishnan H., Goldberg G.S. Heterocellular N-cadherin junctions enable nontransformed cells to inhibit the growth of adjacent transformed cells. Cell Commun. Signal. 2022;20(1):19. doi: 10.1186/s12964-021-00817-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schwaiger K.N., Voit A., Dobiašová H., Luley C., Wiltschi B., Nidetzky B. Plasmid design for tunable two-enzyme Co-expression promotes whole-cell production of cellobiose. Biotechnol. J. 2020;15(11) doi: 10.1002/biot.202000063. [DOI] [PubMed] [Google Scholar]
- 83.Özgen F.F., Runda M.E., Schmidt S. Photo-biocatalytic cascades: combining chemical and enzymatic transformations fueled by light. Chembiochem. 2021;22(5):790–806. doi: 10.1002/cbic.202000587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Santi M., et al. Flow biocatalysis: a challenging alternative for the synthesis of APIs and natural compounds. Int. J. Mol. Sci. 2021;22(3):990. doi: 10.3390/ijms22030990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nethery M.A., Hidalgo-Cantabrana C., Roberts A., Barrangou R. CRISPR-based engineering of phages for in situ bacterial base editing. Proc. Natl. Acad. Sci. U.S.A. 2022;119(46) doi: 10.1073/pnas.2206744119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Strawn G., et al. Genome-wide screen identifies new set of genes for improved heterologous laccase expression in Saccharomyces cerevisiae. Microb. Cell Fact. 2024;23(1):36. doi: 10.1186/s12934-024-02298-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhou J., Li S.M. Conversion of viridicatic acid to crustosic acid by cytochrome P450 enzyme-catalysed hydroxylation and spontaneous cyclisation. Appl. Microbiol. Biotechnol. 2021;105(24):9181–9189. doi: 10.1007/s00253-021-11674-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dai P., Lv Y., Gao Y., Gong X., Zhang Y., Zhang X. ZafA gene is important for trichophyton mentagrophytes growth and pathogenicity. Int. J. Mol. Sci. 2019;20(4) doi: 10.3390/ijms20040848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kurushima J. [Mechanism of high-frequent horizontal gene transfer in Gram positive bacterial pathogens], Nihon saikingaku zasshi. Jpn. J. Bacteriol. 2023;78(4):179–187. doi: 10.3412/jsb.78.179. [DOI] [PubMed] [Google Scholar]
- 90.Cycoń M., Mrozik A., Piotrowska-Seget Z. Antibiotics in the soil environment-degradation and their impact on microbial activity and diversity. Front. Microbiol. 2019;10:338. doi: 10.3389/fmicb.2019.00338. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This is a review, all data and materials are references.