Abstract
We have previously shown for the paramyxovirus simian virus 5 (SV5) that a functional promoter for RNA replication requires proper spacing between two discontinuous elements: a 19-base segment at the 3′ terminus (conserved region I [CRI]) and an 18-base internal region (CRII) that is contained within the coding region of the L protein gene. In the work described here, we have used a reverse-genetics system to determine if the 53-base segment between CRI and CRII contains additional sequence-specific signals required for optimal replication or if this segment functions solely as a sequence-independent spacer region. A series of copyback defective interfering minigenome analogs were constructed to contain substitutions of nonviral sequences in place of bases 21 to 72 of the antigenomic promoter, and the relative level of RNA replication was measured by Northern blot analysis. The results from our mutational analysis indicate that in addition to CRI and CRII, optimal replication from the SV5 antigenomic promoter requires a third sequence-dependent element located 51 to 66 bases from the 3′ end of the RNA. Minigenome RNA replication was not affected by changes in the either the position of this element in relation to CRI and CRII or the predicted hexamer phase of NP encapsidation. Thus, optimal RNA replication from the SV5 antigenomic promoter requires three sequence-dependent elements, CRI, CRII and bases 51 to 66.
The paramyxoviruses are a diverse family of nonsegmented negative-sense RNA viruses. Sendai virus (SeV), measles virus, and respiratory syncytial virus are prototypes of the respiro-, morbilli-, and pneumoviruses, respectively. Simian virus 5 (SV5) is the prototype of the Rubulavirus genus, that also includes mumps virus, SV41, and human parainfluenza virus type 2 (HPIV2). While many general aspects of viral RNA replication are shared by these virus groups, significant differences in their requirements for cis-acting sequences have become evident through sequence analysis and the use of reverse genetics (reviewed in references 3 and 4). For the nonsegmented negative-strand RNA viruses, the 3′ termini of the genomic and antigenomic RNAs contain signals directing viral RNA replication (1, 3, 9).
Three factors have been previously identified that can influence the level of RNA synthesized from the paramyxovirus promoters: the number of nucleotides in the viral RNA template, the primary nucleotide sequences near the termini of the viral RNA, and proper spacing between discontinuous segments of the viral promoter (11). The length of a paramyxovirus RNA template can be a major factor determining the level of RNA replication, with genome replication being most efficient when the total number of nucleotides is an even multiple of six (2). Based on the finding that the SeV NP binds six nucleotides (6), this “rule of six” could reflect a requirement that the 3′-end nucleotides of the template may need to be precisely encapsidated by NP. Alternatively, optimum replication may require that critical cis-acting sequences must be recognized in the correct positions with their encapsidating NP monomers (i.e., phasing; 8, 18). The degree to which replication of a particular paramyxovirus genome adheres to the rule-of-six requirement differs among viruses. For SeV, the rule of six is an apparent strict requirement for RNA replication (2), although expression of the SeV C protein can influence the stringency (24). By contrast, there is no replicative advantage to respiratory syncytial virus genome analogs having any of the integer genome lengths tested (20). SV5 and HPIV3 are intermediate in this regard, since RNA replication is optimal for a genome whose length is a multiple of six, but this is not an absolute requirement for RNA replication (5, 10).
A second major factor directing the efficiency of paramyxovirus RNA replication is the primary sequence near the termini of the viral RNA. For SV5, a previous analysis has shown that the antigenomic promoter contains two essential sequence-specific elements, conserved region I (CRI) and CRII. CRI and CRII were identified by a sequence alignment of rubulavirus genomes (12). CRI is composed of the 3′-terminal 19 nucleotides. CRII was identified by mutational analysis as an internal promoter element within the coding region of the L protein gene located between 72 and 90 bases from the 3′ end of the genome (12). The sequences within CRII that are important for optimal SV5 RNA replication have been mapped by saturation mutagenesis (11), and they consist of a CG motif located in the first two positions of three sequential hexamers of nucleotides (5′-CGGGAUCGAUGGCGAGAA-3′, template sense; Fig. 1A).
FIG. 1.
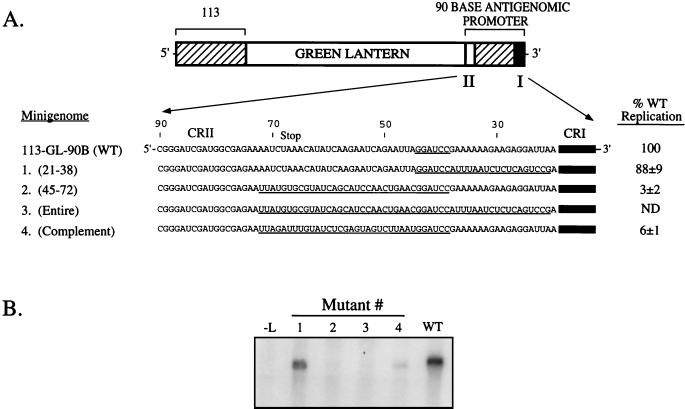
The antigenomic promoter contains a sequence-dependent element in the intervening region between CRI and CRII. (A) Structures of the copyback 113-GL-90B minigenome and substitution mutants. The GL sequence is flanked on the 5′ end by 113 bases of trailer-L gene sequence and on the 3′ end by the 90-base antigenomic promoter. CRI, CRII, and the intervening region are represented by filled, stippled, and cross-hatched boxes, respectively. The 90-base antigenomic promoter is expanded to show the sequence of CRII and the intervening region. The L protein stop codon and the distance from the 3′ end are indicated. For minigenome mutants 1 to 4, the locations of foreign sequence substitutions are underlined. Relative replication for each mutant is expressed as a percentage of the level determined for the WT, which was analyzed in parallel (average ± standard deviation). A representative Northern blot of replication for mutants 1 to 4 is shown in panel B. Lane −L, control from transfected cells in which the L plasmid was omitted. ND, none detected.
For SV5, we have shown that proper spacing between CRI and CRII is a third factor that can influence the level of RNA replication from the antigenomic promoter (12). Six-base insertions or deletions in the RNA segment separating the 3′-terminal CRI element from the internal CRII element can lead to dramatic reductions in RNA replication. This finding suggests that SV5 RNA replication has a requirement for spacing between CRI and CRII which is independent of the rule-of-six requirement. We have proposed that the sensitivity of SV5 RNA replication to changes in the length of the region between CRI and CRII reflects a requirement for these two RNA segments to align to the same face of the helical nucleocapsid template (12). The purpose of the study reported here was to determine if the 53-base intervening region between CRI and CRII contains additional cis-acting signals required for optimal replication or if it serves solely as a sequence-independent spacer between CRI and CRII.
We have previously developed an in vivo assay (10) with which to measure RNA replication from a copyback defective interfering minigenome analog. The 113-GL-90 minigenome (12) encodes a 966-base copyback defective interfering analog with 113 bases from the 5′ terminus of the SV5 genome and the 3′ 90-base antigenomic promoter (Fig. 1A). These viral sequences flank the green lantern (GL) open reading frame, which serves as a nonviral reporter sequence for Northern blotting. cDNA encoding the minigenome analogs were flanked on the 5′ and 3′ ends by the T7 polymerase promoter and DNA encoding the hepatitis delta virus ribozyme, respectively (10, 15, 16). The parental 113-GL-90B plasmid contains a BamHI site engineered into SV5 bases 39 to 44 to facilitate cloning of mutant genome sequences. We have previously shown that this alteration does not affect minigenome replication (12). RNA replication of all mutants was expressed as a percentage of the level determined for 113-GL-90B. Using 113-GL-90B as a template, PCR was used to introduce mutations into the intervening region of the 3′ antigenomic promoter between CRI and CRII as described previously (14). The resulting products were digested with StuI and SphI and used to replace the corresponding DNA fragment in the wild-type (WT) p113-GL-90B plasmid. All mutations were verified by DNA sequencing.
In a standard replication assay, dishes of A549 human lung cells were infected with recombinant vaccinia virus expressing T7 RNA polymerase (vTF7.3; 7) for 1 h at a multiplicity of infection of ∼5 and then cotransfected with the SV5 minigenome analog (1 μg), pGEM3-L (2 μg), pGEM2-P (0.2 μg), and pUC19-NP (3 μg) as described previously (10). After 36 to 48 h, total intracellular RNA was harvested and analyzed by Northern blotting using a 32P-labeled minus-sense GL-specific riboprobe (12). This riboprobe is the same sense as the GL sequence found in the primary T7-derived transcript in order to monitor conversion of the SV5 GL RNA to the complement strand by the SV5 P, L, and NP proteins. RNA levels were quantitated using Phosphorlmager analysis and are expressed as percentages of the 113-GL-90B (WT) level, which was analyzed in parallel. In our assays, we have considered a threefold (33% of the WT level) or greater alteration in RNA replication a significant change. To determine if the RNA replication products are resistant to nuclease digestion, extracts from transfected cells were prepared by resuspension in lysis buffer (10 mM Tris [pH 7.5], 10 mM NaCl, 1 mM CaCl2, 1% Triton X-100, 0.5% sodium deoxycholate; 17) and gently mixed. After addition of micrococcal nuclease (Boehringer Mannheim; 1 μg/ml), samples were incubated at 30°C for 30 min and treated with Trizol and the resulting RNA was analyzed by Northern blotting.
To determine if the region between CRI and CRII contains important cis-acting sequences, four SV5 GL minigenomes were constructed such that portions of the intervening region were replaced with nonviral sequences. Substitution of nonviral sequences for bases 21 to 38 from the 3′ end of the SV5 RNA created a minigenome template that replicated to WT levels (mutant 1, Fig. 1A and B), as shown previously (12). By contrast, substitution of foreign sequence for bases 45 to 72 created a minigenome which replicated to ∼3% of WT levels (mutant 2). This result indicated that bases 45 to 72 of the intervening region contain a sequence necessary for optimal RNA replication. This result was confirmed in the case of mutant 3 (Fig. 1A and B), in which no replication products were detected when the entire intervening region between CRI and CRII (bases 21 to 72) was replaced with a combination of the nonviral sequences in mutants 1 and 2.
The foreign sequences chosen to substitute for the intervening region were random and were not biased in their nucleotide composition. However, our previous results with a transcribing minigenome have shown that substitutions with certain foreign sequences can inhibit polymerase function (19). To test the possibility that an inhibitory sequence had been inadvertently added to create replication-defective mutant 2, a fourth mutant was constructed to contain a different nonviral sequence (mutant 4, Fig. 1A). Mutant 4 was engineered such that the intervening region between bases 45 and 72 was replaced with the complement of the WT sequence. Mutant 4 was similar to mutants 2 and 3 in replicating to levels that were ∼6% of the WT level (Fig. 1A and B). These results indicate that the decreased replication for mutants 2, 3, and 4 was due not to the addition of an inhibitory sequence but rather to the removal of a positive-acting sequence required for optimal RNA replication. Taken together, these results suggest that, in addition to serving as a spacer to align CRI and CRII, the intervening region contains a sequence-dependent element required for optimal RNA replication.
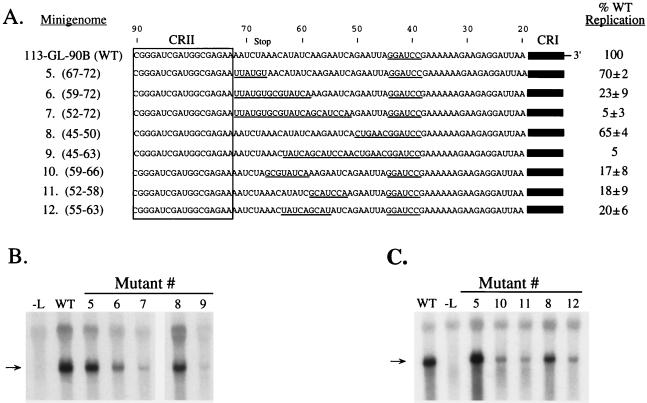
A series of mutants were constructed to map the locations of the cis-acting sequences between bases 45 and 72 (Fig. 2). Mutants 5 to 9 were constructed to contain progressive 5′-to-3′ (mutants 5 to 7) or 3′-to-5′ (mutants 8 and 9) substitutions with the same nonviral sequences used in the initial mutagenesis shown in Fig. 1. Mutant 5, which contains a substitution of bases 67 to 72, replicated to levels slightly lower than the WT minigenome. However, a significant decrease in RNA replication was seen for mutant 6, in which the progressive 5′-to-3′ substitutions extended to base 59. RNA replication was reduced even further in the case of mutant 7, in which the 5′-to-3′ substitutions extended to base 52. Likewise, in the case of the 3′-to-5′ substitutions, replication was slightly reduced for mutant 8, which contains substitutions in bases 45 to 50, but there was a significant reduction in RNA synthesis with mutant 9, in which the mutations extended to base 63. Thus, mutants 5 and 8 define the 5′ and 3′ borders, respectively, of a signal for optimal SV5 RNA replication that is located between bases 51 and 66 from the 3′ of the antigenome.
FIG. 2.
SV5 replication requires a sequence-dependent element located between bases 51 and 66 of the antigenomic promoter. (A) Structures of mutant minigenomes. Underlined sequences indicate progressive 5′-to-3′ (mutants 5 to 7) or 3′-to-5′ (mutants 8 and 9) substitutions of nonviral sequences introduced into the 113-GL-90B template. Numbers in parentheses indicate bases replaced with nonviral sequences. Mutants 10 to 12 contain six to nine base substitutions between bases 52 and 66. The replication level of each mutant is expressed as a percentage of the level determined for the WT template, which was analyzed in parallel (average ± standard deviation). Representative Northern blots of replication products from mutants 5 to 9 and mutants 5, 8, and 10 to 12 are shown in panels B and C, respectively. Lane −L, transfected cells in which the L plasmid was omitted. The arrows indicate replication products.
As an alternative to the progressive substitutions in mutants 5 to 9, a set of mutants were constructed such that six to nine nonviral-sequence base substitutions were engineered into the minigenome template in place of bases 52 to 66. For each of these mutants, the six to nine base substitutions (mutants 10 to 12) reduced replication to ∼20% of the WT level (Fig. 2A and C), indicating that important sequences are distributed throughout this region. Taken together, these data indicate that, in addition to CRI and CRII, the SV5 antigenomic promoter contains a third sequence-dependent element located between bases 51 and 66 that is required for optimal RNA replication. Like CRII (12), this intervening region element is also an internal segment found in the L gene, with bases 51 to 66 located within sequences corresponding to the 3′ noncoding region of the L gene.
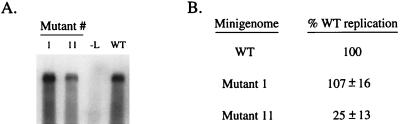
The viral RNA contained within a paramyxovirus nucleocapsid is resistant to nuclease degradation, due to encapsidation by NP. To determine if the RNA synthesized from the mutant templates was encapsidated, replication products were treated with micrococcal nuclease. Cells infected with vTF7.3 were transfected with plasmids encoding the NP, P, and L proteins, along with plasmids encoding mutant minigenome 1 (bases 21 to 38 replaced), which synthesizes WT levels of replication products, or mutant minigenome 11 (bases 52 to 58 replaced), which produces about fivefold less RNA. Lysates were prepared and then treated with micrococcal nuclease as described above. RNA protected from digestion was then analyzed by Northern blotting. As shown in Fig. 3, the levels of nuclease-resistant RNA synthesized by mutants 1 and 11 were 107 ± 16 and 25 ± 13% of that seen for the WT template, respectively. These values are very similar to that seen when total RNA was analyzed as shown in Fig. 1 and 2. Thus, these data indicate that substitutions in the antigenomic promoter result in mutant templates that synthesize reduced levels of RNA, but the viral RNA products are still encapsidated into nucleocapsid-like structures.
FIG. 3.
The replication products made in the in vivo replication assay are resistant to nuclease degradation. A replication assay was carried out using WT 113-GL-90B, mutant minigenome 1 (bases 21 to 38 replaced), and mutant minigenome 11 (bases 52 to 58 replaced) templates. Lysates were prepared and treated with micrococcal nuclease, and samples were analyzed by Northern blotting as described in the text. The replication level of each mutant is expressed as a percentage of WT (average ± standard deviation). Lane −L, transfected cells in which the L plasmid was omitted.
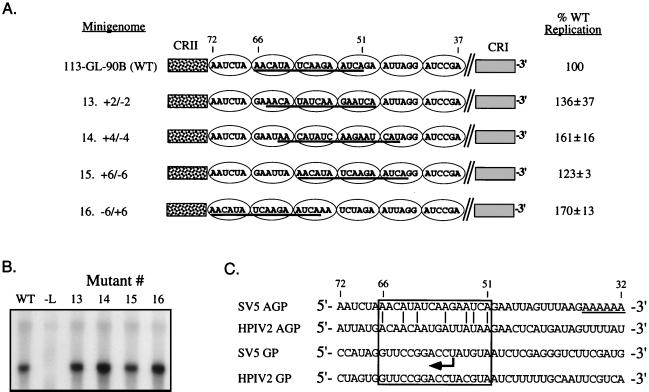
SV5 RNA replication is sensitive to changes in both the spacing between CRI and CRII and the overall length of the RNA template (rule of six; 10, 12). A set of mutant minigenomes was constructed to determine if the position relative to CRI and CRII or the phase of encapsidation of bases 51 to 66 within an NP monomer is an important factor for optimal RNA replication. As shown in Fig. 4A, mutant minigenomes were constructed such that two, four, or six bases were individually removed from the 3′ side of the intervening region segment and inserted on the 5′ side. For example, in mutant 13, the GA dinucleotide flanking the 3′ side of the intervening region element was removed and inserted into the 5′-flanking side. For mutant 16, this resulted in the intervening region element being moved six nucleotides closer to CRII (Fig. 4A). When analyzed in our replication assay, each of the position mutants synthesized RNA to levels slightly higher than the WT level (Fig. 4A and B). From these data, we concluded that changes in the position and the hexamer phase of bases 51 to 66 do not decrease RNA replication from the antigenomic promoter.
FIG. 4.
Alterations in the distance from the 3′ end of the genome and the hexamer phase of bases 51 to 66 do not affect RNA replication or encapsidation. (A) Structures of mutant templates with phase changes in bases 51 to 66. The sequence of bases 37 to 72 is grouped by ovals which represent NP monomers binding six nucleotides. In minigenomes 13 to 15, bases 51 to 66 (underlined) were shifted 3′ toward CRI by 2, 4, or 6 bases, respectively. The total number of bases remained unchanged by engineering an insertion and a deletion flanking bases 51 to 66. In mutant 16, bases 51 to 66 were moved 5′ by six bases toward CRII. Extracts from transfected cells were prepared by lysis in a resuspension buffer and treated with 10 μg of micrococcal nuclease per ml. The replication level of each mutant is expressed as a percentage (average ± standard deviation) of WT replication which was analyzed in parallel. A representative Northern blot of mutants 13 to 16 is shown in panel B. Lane −L, transfected cells in which the L plasmid was omitted. (C) Sequence alignment of bases 32 to 72 from the SV5 and HPIV-2 genomic (GP) and antigenomic (AGP) promoters. The site for polyadenylation of the SV5 L protein gene is underlined. The arrow indicates the transcription start site of the SV5 NP gene.
In summary, we have used mutational analysis to show that the intervening region between CRI and CRII contains an additional sequence-specific element required for optimal replication. Thus, our data demonstrate that the SV5 antigenomic promoter consists of three noncontinuous sequence-dependent RNA segments: CRI, CRII, and a third element located 51 to 66 bases from the 3′ end. These cis-acting signals are separated by sequence-independent spacer regions.
The three sequence-dependent elements we have identified in the SV5 antigenomic promoter vary significantly in their conservation between members of the Rubulavirus genus. The 19-base CRI element located at the 3′ end of the genomic and antigenomic RNAs is the most highly conserved region among the Rubulavirus genomes, and substitutions in this element can lead to a dramatic loss of replication (12). The three copies of the 5′-CGNNNN-3′ motif within SV5 internal element CRII are highly conserved among the rubulaviruses and are found 72 to 90 bases from the 3′ end of the genomic and the antigenomic RNAs (11, 12). By contrast, the third promoter element identified here shows no apparent sequence identity among Rubulavirus antigenomic promoters (12). In addition, our data indicate that replication is dramatically affected only when the majority of the cis-acting sequence is replaced, while smaller substitutions have lesser effects. Thus, the third promoter element may be a functionally redundant signal that is distributed across bases 51 to 66. We have previously shown that a minigenome containing the 3′-terminal 90 bases of the HPIV-2 antigenomic promoter can be replicated by the SV5 polymerase to levels matching that of the WT SV5 minigenome (12). However, HPIV2 and SV5 antigenomic promoters show no extensive sequence identity between bases 51 and 66, other than a series of conserved A residues that are spaced in every third position (Fig. 4C). Further mutational analyses is required to define in more detail the features of bases 51 to 66 that are important for optimal RNA replication.
For the rhabdovirus vesicular stomatitis virus, recent data indicate that the level of viral RNA replication can be influenced by the extent of terminal complementarity in the promoter region (26). By contrast, it has been reported that terminal complementarity is not a factor dictating SeV minigenome RNA replication (23). Several lines of evidence are inconsistent with SV5 RNA replication being influenced by terminal complementarity beyond that found in the 3′-terminal CRI element. First, we have previously shown that a chimeric minigenome containing 113 SV5-specific 5′-terminal bases and the 98-base HPIV-2 antigenomic promoter at the 3′ end is able to direct RNA replication by the SV5 polymerase to levels matching that of the WT minigenome (12). Second, mutant 1 minigenome that has bases 21 to 38 replaced with nonviral sequences (Fig. 1) can direct very efficient RNA replication. In both of these minigenome templates, the extent of terminal complementarity has been disrupted by HPIV-2-specific sequences or nonviral substitutions with very little reduction in RNA replication. Taken together, we conclude that disruption of terminal complementarity by the intervening-region substitutions described here cannot account for the decreases in RNA replication.
The cis-acting promoter element between bases 51 and 66 could function as a polymerase binding site on the viral RNA. For a number of RNA viruses, such as Qβ phage (21), turnip crinkle virus satellite C RNA (22), and influenza virus (25), RNA synthesis requires a discontinuous segment of the genome in addition to a cis-acting 3′-end sequence. These 3′-distal discontinuous promoter elements may act as a scaffold for directing the viral polymerase to initiate at the correct site on the viral RNA. We have previously proposed that the internal CRII element serves as part of a binding site for the SV5 polymerase to position the enzyme near the 3′ end of the viral RNA (11). This is based on our model which predicts that CRI and CRII are positioned on the same face of the helical nucleocapsid template (12). In our model for the antigenomic promoter, bases 51 to 66 would be located on the side of the nucleocapsid template opposite that of the CRI-CRII face. This could reflect an additional binding site for host or viral proteins involved in RNA replication. It has been postulated that the paramyxovirus nucleocapsid exists in distinct forms (6), and it is possible that these structures reposition bases 51 to 66 in relation to CRI and CRII to differentially modulate RNA replication.
For the nonsegmented negative-strand viruses, RNA replication is tightly coupled to encapsidation by the viral nucleocapsid protein NP (1). Thus, bases 51 to 66 could alternatively function as a signal for efficient encapsidation by the NP protein. A current model for the control of replication versus transcription is based on antitermination of RNA synthesis at or near the junction between the leader and the NP start site (reviewed in reference 9). It is noteworthy that the cis-acting sequences described here are located ∼55 bases from the 3′ end of the viral antigenomic RNA, a position analogous to the leader-NP junction in the genomic promoter (Fig. 4C). Thus, bases 51 to 66 could form a signal for encapsidation of nascent RNA as it emerges from the viral polymerase. It might be predicted that this strong encapsidation signal would not be present in the genomic promoter and thus make it a weaker promoter for replication. We are currently testing the role of the third promoter element in differential RNA replication from the genomic and antigenomic promoters.
Acknowledgments
We thank Liz Wansley and Doug Lyles for helpful comments on the manuscript.
This work was supported by NIH grant AI42023.
REFERENCES
- 1.Banerjee A K, Barik S. Gene expression of vesicular stomatitis virus genome RNA. Virology. 1992;188:417–428. doi: 10.1016/0042-6822(92)90495-b. [DOI] [PubMed] [Google Scholar]
- 2.Calain P, Roux L. The rule of six, a basic feature for efficient replication of Sendai virus defective interfering RNA. J Virol. 1993;67:4822–4830. doi: 10.1128/jvi.67.8.4822-4830.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collins P, Chanock R M, McIntosh K. Parainfluenza viruses. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. New York, N.Y: Lippincott-Raven Publishers; 1996. pp. 1205–1241. [Google Scholar]
- 4.Conzelmann K-K. Genetic manipulation of non-segmented negative-strand RNA viruses. J Gen Virol. 1996;77:381–389. doi: 10.1099/0022-1317-77-3-381. [DOI] [PubMed] [Google Scholar]
- 5.Durbin A P, Siew J W, Murphy B R, Collins P L. Minimum protein requirements for transcription and RNA replication of a minigenome of human parainfluenza virus type 3 and evaluation of the rule of six. Virology. 1997;234:74–83. doi: 10.1006/viro.1997.8633. [DOI] [PubMed] [Google Scholar]
- 6.Egelman E, Wu S, Amrein M, Portner A, Murti G. The Sendai virus nucleocapsid exists in at least four different helical states. J Virol. 1989;63:2233–2243. doi: 10.1128/jvi.63.5.2233-2243.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;85:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kolakofsky D, Pelet T, Garcin D, Hausmann S, Curran J, Roux L. Paramyxovirus RNA synthesis and the requirement for hexamer genome length: the rule of six revisited. J Virol. 1998;72:891–899. doi: 10.1128/jvi.72.2.891-899.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lamb R A, Kolakofsky D. Paramyxoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. New York, N.Y: Lippincott-Raven Publishers; 1996. pp. 1177–1204. [Google Scholar]
- 10.Murphy S K, Parks G D. Genome nucleotide lengths that are divisible by six are not essential but enhance replication of defective interfering RNAs of the paramyxovirus simian virus 5. Virology. 1997;232:145–157. doi: 10.1006/viro.1997.8530. [DOI] [PubMed] [Google Scholar]
- 11.Murphy S K, Parks G D. RNA replication for the paramyxovirus simian virus 5 requires an internal repeated (CGNNNN) sequence motif. J Virol. 1999;73:805–809. doi: 10.1128/jvi.73.1.805-809.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murphy S K, Ito Y, Parks G D. A functional antigenomic promoter for the paramyxovirus simian virus 5 requires proper spacing between an essential internal segment and the 3′ terminus. J Virol. 1998;72:10–19. doi: 10.1128/jvi.72.1.10-19.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parks G D, Ward C D, Lamb R A. Molecular cloning of the NP and L genes of simian virus 5: identification of highly conserved domains in paramyxovirus NP and L proteins. Virus Res. 1992;22:259–279. doi: 10.1016/0168-1702(92)90057-g. [DOI] [PubMed] [Google Scholar]
- 14.Parks G D. Mapping of a region of the paramyxovirus L protein required for the formation of a stable complex with the viral phosphoprotein P. J Virol. 1994;68:4862–4872. doi: 10.1128/jvi.68.8.4862-4872.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pattnaik A K, Wertz G W. Replication and amplification of defective interfering particle RNAs of vesicular stomatitis virus in cells expressing viral proteins from vectors containing cloned cDNAs. J Virol. 1990;64:2948–2957. doi: 10.1128/jvi.64.6.2948-2957.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pattnaik A, Ball A, Legrone A, Wertz G. Infectious defective interfering particles of VSV from transcripts of a cDNA clone. Cell. 1992;69:1011–1020. doi: 10.1016/0092-8674(92)90619-n. [DOI] [PubMed] [Google Scholar]
- 17.Peeples M E, Collins P L. Mutations in the 5′ trailer region of a respiratory syncytial virus minigenome which limit RNA replication to one step. J Virol. 2000;74:146–155. doi: 10.1128/jvi.74.1.146-155.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pelet T, Delenda C, Gubbay O, Garcin D, Kolakofsky D. Partial characterization of a Sendai virus replication promoter and the rule of six. Virology. 1996;224:405–414. doi: 10.1006/viro.1996.0547. [DOI] [PubMed] [Google Scholar]
- 19.Rassa J C, Parks G D. Highly diverse intergenic regions of the paramyxovirus simian virus 5 cooperate with the gene end U tract in viral transcription termination and can influence reinitiation at a downstream gene. J Virol. 1999;73:3904–3912. doi: 10.1128/jvi.73.5.3904-3912.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Samal S, Collins P. RNA replication by a respiratory syncytial virus analog does not obey the rule of six and retains a nonviral trinucleotide extension at the leader end. J Virol. 1996;70:5075–5082. doi: 10.1128/jvi.70.8.5075-5082.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schuppli D, Miranda G, Qiu S, Weber H. A branched stem-loop structure in the M-site of bacteriophage qbeta RNA is important for template recognition by qbeta replicase holoenzyme. J Mol Biol. 1998;283:585–593. doi: 10.1006/jmbi.1998.2123. [DOI] [PubMed] [Google Scholar]
- 22.Song C, Simon A. Requirement of a 3′-terminal stem-loop in in vitro transcription by an RNA dependent RNA polymerase. J Mol Biol. 1995;254:6–14. doi: 10.1006/jmbi.1995.0594. [DOI] [PubMed] [Google Scholar]
- 23.Tapparel C, Roux L. The efficiency of Sendai virus genome replication: the importance of the RNA primary sequence independent of terminal complementarity. Virology. 1996;225:163–167. doi: 10.1006/viro.1996.0584. [DOI] [PubMed] [Google Scholar]
- 24.Tapparel C, Hausmann S, Pelet T, Curran J, Kolakofsky D, Roux L. Inhibition of Sendai virus genome replication due to promoter-increased selectivity: a possible role for the accessory C proteins. J Virol. 1997;71:9588–9599. doi: 10.1128/jvi.71.12.9588-9599.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tiley L S, Hagen M, Matthews J T, Krystal M. Sequence-specific binding of the influenza virus RNA polymerase to sequences located at the 5′ ends of the viral RNAs. J Virol. 1994;68:5108–5116. doi: 10.1128/jvi.68.8.5108-5116.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wertz G W, Whelan S, LeGrone A, Ball L A. Extent of terminal complementarity modulates the balance between transcription and replication of vesicular stomatitis virus RNA. Proc Natl Acad Sci USA. 1994;91:8587–8591. doi: 10.1073/pnas.91.18.8587. [DOI] [PMC free article] [PubMed] [Google Scholar]