Abstract
Background and Aims
Primary sclerosing cholangitis (PSC) is a chronic inflammatory liver disorder without effective medical treatment which is characterized by inflammation and fibrotic structures around the bile ducts. Biliary epithelial cells (cholangiocytes) are the target and potential disease drivers in PSC, yet little is known if cholangiocytes from PSC patients differ from non-PSC controls. To characterize cholangiocytes at early rather than end-stage disease, cholangiocyte organoids (COs) were derived from diseased bile ducts of PSC patients and compared to organoids generated from disease controls.
Methods
Cholangiocytes were obtained during endoscopic retrograde cholangiopancreatography (ERCP) brushing of diseased bile duct areas and expanded as organoids using previously established culture methods. Stable CO lines were analyzed for cell type identity, basic cholangiocyte function, and transcriptomic signature.
Results
We demonstrate that cholangiocytes, derived from the damaged area within the bile ducts of PSC patients, can be expanded in culture without displaying functional or genetic disease-related features. We further show that COs from patients who later were diagnosed with dysplasia exhibit higher expression of the cancer-associated genes PGC, FXYD2, MIR4435-2HG, and HES1.
Conclusions
Our results demonstrate that PSC organoids are largely similar to control organoids after culture and highlight the significance of COs as a tool for regenerative medicine approaches as well as their potential for discovering new potential biomarkers for diagnosing cholangiocarcinoma.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10620-024-08570-y.
Keywords: Cholangiopathies, In vitro modeling, Single-cell sequencing, Cholangiocarcinoma, Autoimmune liver disorders, Biliary epithelial cells
Introduction
Primary sclerosing cholangitis (PSC) is a progressive hepatobiliary disorder characterized by chronic biliary inflammation, cholestasis, and periductal fibrosis, which often progresses to end-stage liver disease with cirrhosis [1]. There are no available medical treatments altering PSC prognosis, and at present, liver transplantation is the only available treatment option [2, 3]. In addition to the progressive nature of the disease leading to liver failure, the ongoing chronic inflammation within the bile ducts puts PSC patients at a high risk of developing cholangiocarcinoma (CCA) that represents a major contributor to PSC-related mortality [4]. There is a clear contribution of genetic factors predisposing to the disease and genetic findings from PSC patients indicate altered immune cell pathways, trafficking and immune tolerance, as well as distinct T-cell phenotypes [5, 6] and antibody production [7] in the liver of PSC patients. These findings clearly position PSC as an immune-mediated disease, but there is still insufficient knowledge on other predisposing risk factors and the exact cellular events and interactions driving disease progression.
Biliary epithelial cells (cholangiocytes) are a heterogeneous group of biliary cells lining the bile ducts which modify the content of bile as it is transported along the biliary tract [8]. In PSC, impaired cholangiocyte function, resulting from a combination of biliary insults, leads to destruction of the biliary epithelial cell barrier and, eventually, to leakage of toxic bile into hepatic tissue, resulting in sequential biliary inflammation and development of liver fibrosis. It is still unclear whether specific disease-related cholangiocyte features are intrinsic or occur due to changes within the biliary environment upon disease onset [9]. Specifically impaired functions of biliary epithelial cells, such as cellular senescence or increased proliferation and ongoing secretion of proinflammatory modulators by biliary epithelial cells, have been suggested to promote disease development [10–12]. Clarification of cholangiocyte biology and the pathophysiology in PSC has been hampered by inadequate disease models and challenges in culturing and expanding cholangiocytes isolated from bile ducts of PSC patients [13, 14]. To that end, methods based on organoid technology for culturing patient-derived cholangiocytes have been developed, greatly simplifying the in vitro expansion and study of cholangiocytes derived from patients compared to previous methods [15–17]. These organoids resemble primary adult cholangiocytes and importantly retain key functions and characteristics of their tissue of origin, thereby offering a unique potential for mechanistic studies but also open up for use for biliary tissue repair in regenerative medicine approaches [16]. It is still unclear whether these in vitro cultured cells retain features of disease imprint after extended culture, and whether this is reflected in their transcriptomic profile and can be related to the disease phenotype observed in the patients. These issues are crucial when using cholangiocyte organoids (COs) as a tool for analyzing cellular disease mechanisms ongoing during disease progression and have ramifications for the use of such cells in regenerative medicine.
Only few studies have described the isolation and culture of cholangiocytes from PSC patients largely due to difficulties in collecting sufficient patient material for research [18–20]. Previous studies have utilized cholangiocytes either from bile or explanted end-stage liver tissue which may not capture the relevant cellular types at the site of injury at earlier disease stages. Herein, we directly isolated cholangiocytes from diseased bile ducts of PSC patients and non-PSC controls undergoing endoscopic retrograde cholangiopancreatography (ERCP). Collected cells were expanded into organoid cell lines followed by single-cell RNA sequencing (scRNAseq) analysis.
Methods
Patient Samples
Cholangiocytes were obtained from the bile ducts of patients undergoing ERCP. PSC and control patients were recruited from the Section of Gastroenterology at the Department of Transplantation Medicine, Oslo University Hospital Rikshospitalet. Written informed consent was obtained prior to ERCP and ethics approval for the use of cells from human livers and bile ducts was approved by the Regional Ethics Committee (2012-286/2016-1540). Patient inclusion in the study was done independent of gender.
Isolation of Cholangiocytes from Patient Bile Ducts
ERCP brush samples were placed into William’s E + (WE +) medium (Gibco Inc., Waltham, Massachusetts, USA) with 50 ng/ml of epidermal growth factor (EGF) (R&D Systems Inc., Minneapolis, Minnesota, USA) and 10 μM Rho-associated kinase inhibitor Y-27632 (Selleck Chemicals LLC., Houston, Texas, USA) on ice and carefully washed to collect patient material. Isolated ERCP material was centrifuged at 400 g for 4 min, supernatant was discarded, and the cells were seeded onto 24-well cell culture plates in 50 μl droplets of supplemented WE + culture medium [21] and Matrigel (Corning Inc., New York, New York, USA) mixed at a 1:3 ratio for in vitro 3D expansion [17].
Expansion and Maintenance of Cholangiocyte Organoid Lines
Cell culture of COs was performed as previously described [17, 21]. Briefly, cells were cultured with a solubilized basement membrane matrix (Matrigel) and WE + medium, supplemented with a combination of EGF (Bio-Techne Co., Minneapolis, Minnesota, USA), R-spondin 1 (Bio-Techne Co.), and Dickkopf-related protein 1 (DKK-1) (Bio-Techne Co.) to promote the expansion of cholangiocytes in the form of organoids [21]. Organoid media was exchanged every 48 h and the organoid lines were split every 5–7 days depending on organoid quality and quantity. After split 4, all organoid lines were transferred to liquid nitrogen and stored for scRNAseq and further analysis. For scRNAseq, all organoid lines were simultaneously revived from liquid nitrogen storage and synchronized in culture. All experiments were performed using passage 6 organoids unless otherwise stated.
Immunofluorescence Staining of Cholangiocyte Organoids
For staining, organoids were fixed with 4% paraformaldehyde (PFA) in PBS for 20 min. Fixation solution was removed and the organoids were washed twice in PBS for 10 min each. Permeabilization and blocking were performed by adding 0.1% Triton X-100 and 5% donkey serum in PBS for 1 h. Organoids were stained with primary antibodies (Supplementary Table 1) overnight at 4 °C. Samples were washed 3 times in PBS for 45 min each and secondary antibody staining (Supplementary Table 1) was performed for 1 h at room temperature, followed by incubation with Hoechst 33258 in PBS for 20 min. Organoids were washed 3 times in PBS for 45 min each and then imaged using fluorescence microscopy. All immunofluorescence images were acquired using a Zeiss Axiovert 200 M inverted microscope (Zeiss Group, Oberkochen, Germany) or a Zeiss LSM 700 (LSM 710) confocal microscope. The ImageJ 1.48 k software [22] was used for image processing. Immunofluorescence images are representative of at least 3 different CO lines.
Flow Cytometry Analysis
COs were dissociated from Matrigel using Cell Recovery Solution (Corning Inc.) for 30 min at 4 °C and were then centrifuged at 444 g for 4 min and dissociated to single cells using TrypLE Express (Gibco Inc.). Cells were stained for 30 min on ice, washed twice in flow buffer (PBS containing 5% fetal bovine serum and 0.09% sodium azide), and measured and analyzed using a BD FACSVerse Flow Cytometer and the BD FACSuite software (BD Life Sciences comp., Franklin Lakes, New Jersey, USA). A complete list of the antibodies and dilutions used is provided in Supplementary Table 2.
γ-Glutamyl Transferase Activity
γ-glutamyl transferase (GGT) activity was measured in triplicate using the MaxDiscovery GGT Enzymatic Assay Kit (BioScientific, Avondale, Arizona, USA) based on the manufacturer's instructions. Absorbance units refer to light absorbance at a wavelength of 405 nm.
Alkaline Phosphatase Staining
Alkaline phosphatase (ALP) staining was performed using the BCIP/NBT Color Development Substrate (5-bromo-4-chloro-3-indolyl-phosphate/nitro blue tetrazolium) (Promega Co., Madison, Wisconsin, USA) according to the manufacturer’s instructions.
REAL-TIME Quantitative Polymerase Chain Reaction
RNA extraction and quantitative polymerase chain reaction (qPCR) were performed as previously described [23]. A complete list of the primers used is provided in Supplementary Table 3. All qPCR data are presented as the median, interquartile range (IQR), and range (minimum to maximum) of four independent CO lines unless otherwise stated. Values were normalized to the housekeeping gene hydroxymethylbilane synthase (HMBS).
Rhodamine 123 Assay
Organoids were initially incubated for 30 min in the presence or absence of 50 µM verapamil (Merck and Co Inc., New York, New York, USA). Organoids were then incubated with 100 µM rhodamine 123 (Merck and Co Inc.) and 1 µg/ml Hoechst 33342 in supplemented WE + culture medium for 5 min. Organoids were washed 3 times in WE + medium and then incubated for 40 min in fresh supplemented WE + culture medium. Organoids were imaged using a Zeiss LSM 710 confocal microscope. Images were analyzed using ImageJ [22] with a mean intensity measurement taken for the interior of each organoid. The experiment was run in triplicate with 40 organoids quantified in total per condition.
Cytokine-Stimulation Experiments
COs were stimulated with 50 ng/ml tumor necrosis factor α (TNF-α) (Bio-Techne Co.), transforming growth factor β (TGF-β) (Bio-Techne Co.) or interleukin 17 (IL-17) (Bio-Techne Co.) in supplemented WE + culture medium for 5 days. Medium and stimulation cytokines were exchanged every 48 h. Senescence-associated-β-galactosidase (SA-β-Gal) stain assay (Abcam Limited, Cambridge, UK) of unstimulated and stimulated organoids was performed following the manufacturer’s instructions. Cell culture supernatant was analyzed by Luminex Discovery Assay (R&D Systems Inc.), following the manufacturer’s instructions. The experiment was repeated in n > 3 independent stimulation experiments with individual patients.
Single-Cell RNA Sequencing and Data Analysis
COs were harvested after the sixth passage and processed for scRNAseq using the 10X Genomics scRNAseq platform and Chromium Next GEM Single Cell 3’ Gel Bead Kit v3.1 (10X Genomics Inc., Pleasanton, California, USA). Raw sequencing reads were delivered in FASTQ format.
Initial quality checks, alignment, and transcript quantification were performed using Cell Ranger version 3.1.0. The alignment was performed against the GRCh38-3.0.0 H. sapiens reference genome; a total 41,954 cells were initially called on default acceptance thresholds.
Further quality control checks and filtering were performed, including cells with fewer than 2500 features (genes) were discarded; the threshold was determined based on the nfeature distribution (Supplementary Figs. 1, 2, 3). In addition, cells with > 15% reads mapping to mitochondrial genes, > 40% reads mapping to ribosomal genes, or > 100,000 unique molecular identifiers (UMIs) were removed; the latter corresponded to doublets (Supplementary Figs. 2, 3). Once the cells were filtered, mitochondrial, ribosomal, and Y chromosome genes were excluded from the expression matrix, prior to normalization. Cell-cycle was assessed using standard sets of cell-cycle genes, using dedicated Seurat functions; no significant batch effect was noted; the cell-cycle genes were subsequently excluded from expression matrix. 15,564 cells passed all the quality control filters, across samples; post-filtering, the expression levels were normalized using SCTransform [24].
To evaluate the clustering stability and optimize hyper-parameters, we employed ClustAssess [25]; parameters such as the feature set (most abundant or highly variable genes, and the number of genes not affected by random noise variation), the resolution, and number of clusters were all assessed. The tested number of genes were 500, 1000, 1500, and 2000, in line with the nfeature distributions across samples. The stability is evaluated using Element-Centric similarity; the consistency of outputs on random iterations was summarized using Element-Centric Consistency (ECC) [26], on 100 iterations on distinct random seeds. We identified the transition point from signal to noise at the 1500 most variable features, which we subsequently used for downstream clustering (Supplementary Figs. 4, 5).
Clustering was performed with Seurat v4.3.0. Initially, a Principal Component Analysis (PCA) was performed on the 1500 most variable features, retaining the first 30 principal components. The Louvain clustering algorithm was applied on the PCs, with a resolution of 0.8, revealing 13 distinct clusters. Differential Expression (DE) analysis was performed using the ROC test, considering genes expressed in > 25% of the cells, and a log2FC > 0.25.
scRNA data were visualized using the ShinyCell package from R. Heatmap generation for group comparisons was performed using the webtool ClustVis [27].
Spatial Transcriptomics and Sequencing
Spatial transcriptomics and sequencing have previously been described for the dataset used [28]. Biliary regions were defined by expression of the cholangiocyte markers KRT7 or EPCAM (KRT7 > 2 counts per spot, EPCAM > 1 counts per spot).
Gene expression levels were visualized within pre-defined biliary regions in Loupe Browser software (10X Genomics Inc.).
Statistical Analysis
All statistical analysis for non-scRNAseq data was performed using GraphPad Prism 9 (GraphPad Software, La Jolla, California, USA). Double sided Student’s t test was used to evaluate statistical significance. P values below 0.05 were considered significant. The number of replicates refers to organoid lines unless otherwise stated.
Results
Generation of PSC Cholangiocyte Organoids from ERCP Brushes
To determine if CO cultures could be established directly from the bile ducts of PSC patients, we collected brush samples from 13 patients undergoing ERCP with an age ranging from 29 to 74 years; of these, 10 patients had a diagnose of PSC, while 3 were non-PSC patients who were included as controls (full clinical characteristics in Table 1). Six of the PSC patients had a diagnosis of inflammatory bowel disease (IBD) (four patients with ulcerative colitis (UC) and two patients with Morbus Crohn). The most common indication for ERCP was bile duct obstruction (Table 2) and the parallel brush samples taken for clinical diagnostics showed normal cytological findings in approximately 50% of the cases (Table 2).
Table 1.
Clinical characteristics at the time of ERCP procedure
| Patient | Diagnosis | Gender | Age (Years) | IBD | Biliary malignancy | ALP (U/L) | ALT (U/L) | AST (U/L) | G-GT (U/L) | Bilirubin (U/L) |
|---|---|---|---|---|---|---|---|---|---|---|
| PSC 1 | PSC | M | 30 | UC | None | 133 | 66 | 47 | 162 | 7 |
| PSC 2 | PSC | M | 52 | None | None | 642 | 116 | 136 | 1070 | 25 |
| PSC 3 | PSC | M | 51 | None | None | 45 | 28 | 31 | 42 | 5 |
| PSC 4 | PSC* | M | 64 | UC | None | 106 | 17 | 19 | 219 | 9 |
| PSC 5 | PSC | F | 45 | Mb Crohn | CCA | 593 | 67 | 89 | 194 | 91 |
| PSC 6 | PSC | F | 20 | Mb Crohn | None | 99 | 23 | 30 | 39 | 7 |
| PSC 7 | PSC | M | 44 | UC | None | 405 | 194 | 114 | 687 | 22 |
| PSC 8 | PSC | M | 42 | None | BIL-IN II** | 141 | 101 | 67 | 192 | 22 |
| BIL-IN I*** | ||||||||||
| PSC 9 | PSC/AIH | M | 45 | UC | BIL-IN I**** | 556 | 349 | 286 | 556 | 24 |
| PSC 10 | PSC | M | 29 | None | None | 224 | 46 | 41 | 281 | 27 |
| Control 1 | Chronic pancreatitis | F | 65 | None | None | 64 | 15 | 20 | 27 | 3 |
| Control 2 | Pancreatic cancer | F | 69 | None | None | 709 | 365 | 254 | 794 | 10 |
| Control 3 | Cholecystitis | F | 74 | None | None | 191 | 112 | 62 | 328 | 11 |
Control, non-PSC patient
AIH autoimmune hepatitis, IBD Inflammatory bowel disease, Mb Crohn Morbus Crohn, UC ulcerative colitis, CCA cholangiocarcinoma, BIL-IN biliary intraepithelial neoplasia, ERCP endoscopic retrograde cholangiopancreatography, ALP alkaline phosphatase, ALT alanine transaminase, AST aspartate transaminase, G-GT gamma-glutamyl transferase
*Small duct PSC
**Vesica biliaris
***Ductus choledochus
****Ductus hepaticus communis
Table 2.
ERCP indication, region, and result including clinical course and follow-up time
| Patient | Indication ERCP | Region of bile ducts | Result brush cytology | Clinical course | Follow-up time |
|---|---|---|---|---|---|
| PSC 1 | Dominant stricture | Dominant stricture ductus choledochus | Regular cylindrical epithelium | Recurrent cholangitis | 28 months |
| PSC 2 | Anastomotic stricture | HJRY anastomosis | Acellular material | LTX eleven years before ERCP/recurrent PSC/retransplant two years after ERCP | 23 months |
| PSC 3 | Control inconclusive brush | Ductus choledochus | Regular cylindrical epithelium | No relevant clinical events | 60 months |
| PSC 4 | Strictures and Ca19-9 > 2500 | Dominant stricture left ductus choledochus | Acellular material | No relevant clinical events | 60 months |
| PSC 5 | Pruritus | Left and right ductus choledochus | Regular cylindrical epithelium | Died, CCA diagnosis 5 months after ERCP | 16 months |
| PSC 6 | Strictures/pain | Dominant stricture ductus choledochus | Regular cylindrical epithelium | No relevant clinical events | 59 months |
| PSC 7 | Unsuccessful MRCP | Dominant stricture ductus hepaticus | Regular cylindrical epithelium | No relevant clinical events | 58 months |
| PSC 8 | Polysomy FISH | Extrahepatic bile duct | Inconclusive regarding dysplasia | LTX five months after ERCP | 5 months |
| PSC 9 | Pruritus, jaundice | Ductus hepaticus communis | Low grade dysplasia | LTX two months after ERCP | 2 months |
| PSC 10 | Pruritus, pain | Ductus hepaticus communis and left ductus choledochus | Inflammation | Progressive disease, multiple endoscopic procedures | 52 months |
| Control 1 | Pancreatic stones | Unknown | No information | Died | 26 months |
| Control 2 | Dilated bile ducts | Ampulla Vateri | Regular cylindrical epithelium | Died | 27 months |
| Control 3 | Investigate papillary pathology | Left ductus choledochus | No information | No relevant clinical events | 52 months |
LTX Liver transplantation, HJRY hepaticojejunostomy, MRCP magnetic resonance cholangiopancreatography, FISH fluorescence in situ hybridization, ERCP endoscopic retrograde cholangiopancreatography, CA19-9 cancer antigen 19–9
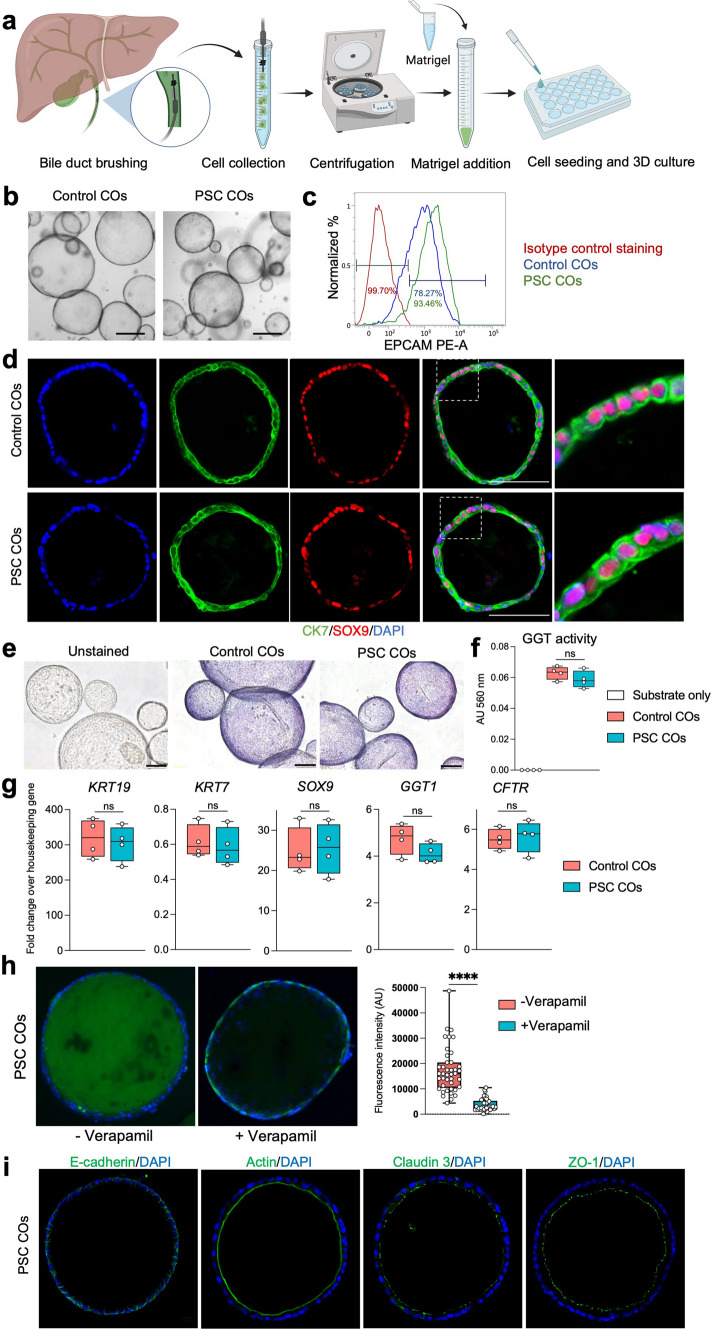
To generate COs from ERCP brushings, cells from the collected brushes were mixed with Matrigel and supplemented WE + medium and seeded as droplets into cell culture dishes (Fig. 1A). First organoids were observed as early as 3–4 days in 3D Matrigel culture (Supplementary Fig. 6). The organoid cultures underwent splitting after 6–8 days to remove dead cells and debris and pure organoid lines were obtained after approximately two splits and 14 days in culture (Fig. 1B). The COs exhibited stable expression of the key biliary markers epithelial cell adhesion molecule (EPCAM) by flow cytometry and cytokeratin 7 (CK7) as well as SRY-related high mobility group box transcription factor 9 (SOX9) by immunofluorescence staining (Fig. 1C, D), confirming the cholangiocyte identity of the expanded cells. The organoid lines derived from PSC patients and non-PSC controls showed similar morphology, size, and expression of biliary markers (Fig. 1B–D). To determine if organoids retained biliary-associated markers and function, ALP and GGT activity were measured. COs from PSC and control patients demonstrated comparable levels of ALP and GGT activity (Fig. 1E, F) and comparable levels of the biliary markers keratin 19 (KRT19), KRT7, SOX9, GGT, and cystic fibrosis transmembrane conductance regulator (CFTR) on a transcriptional level as assessed by qPCR (Fig. 1G).
Fig. 1.
Characterization of cholangiocyte organoids generated from ERCP brushings. a Overview of cholangiocyte organoid (CO) derivation from ERCP brushing. b Representative brightfield images showing COs from a control patient and a patient affected by PSC. Scale bars 100 μm. c Representative flow cytometry histograms demonstrating expression of the key biliary marker EPCAM in control and PSC COs. d Expression of biliary markers in control and PSC COs by immunofluorescence. Scale bars 100 μm. e Representative images of COs from control and PSC patients demonstrating alkaline phosphatase (ALP) activity. Scale bars 100 μm. f COs from control and PSC patients demonstrate gamma-glutamyl transferase (GGT) activity at comparable levels. N = 4; ns, non-significant. g Expression of the biliary markers KRT19, KRT7, SOX9, GGT1, and CFTR by control and PSC COs, detected by quantitative-PCR. Housekeeping gene, HMBS; n = 4; ns, non-significant. h Rhodamine 123 assay demonstrating intact MDR1 transporter function in PSC COs. n = 40 organoids. Error bars represent s.d. i Correct localization of main cytoskeleton and tight junction markers in PSC COs, demonstrated by immunofluorescence. Scale bar 50 μm. CK7 cytokeratin 7, KRT19 keratin 19, SOX9 SRY-related high mobility group box transcription factor 9, GGT gamma-glutamyltransferase 1, CFTR cystic fibrosis transmembrane conductance regulator, ZO-1 zonula occludens-1 protein, MDR1 multidrug resistance protein 1, **** P < 0.0001
To assess functionality of the PSC-derived organoids, we additionally assessed secretory capacity by a Rhodamine 123 assay and evaluated the expression and localization of the key tight junction and cytoskeleton proteins E-cadherin, actin, claudin 3, and zonula occludens-1 (ZO-1) that are important for establishing intact barrier function. PSC COs demonstrated active secretion through the multidrug-resistance-receptor 1 (MDR1) and correct expression and localization of key tight junction and cytoskeleton proteins (Fig. 1H, I). As a part of the functional assessment, we further addressed the reactivity of PSC COs to disease-relevant stimuli (TNF-α, TGF-β and IL-17) by immunohistochemistry, immunofluorescence, and multiplex enzyme-linked immunosorbent assay (ELISA). PSC-derived organoids reacted to external stimulation by an upregulation of key markers for cellular senescence (SA-β-Gal and protein 21 (p21)) and by increased secretion of mediators that are relevant for immune-modulation and tissue remodeling (Supplementary Fig. 7).
Single-Cell RNA Sequencing Demonstrates Transcriptomic Overlap Between Organoid Lines Derived from Individual Patients
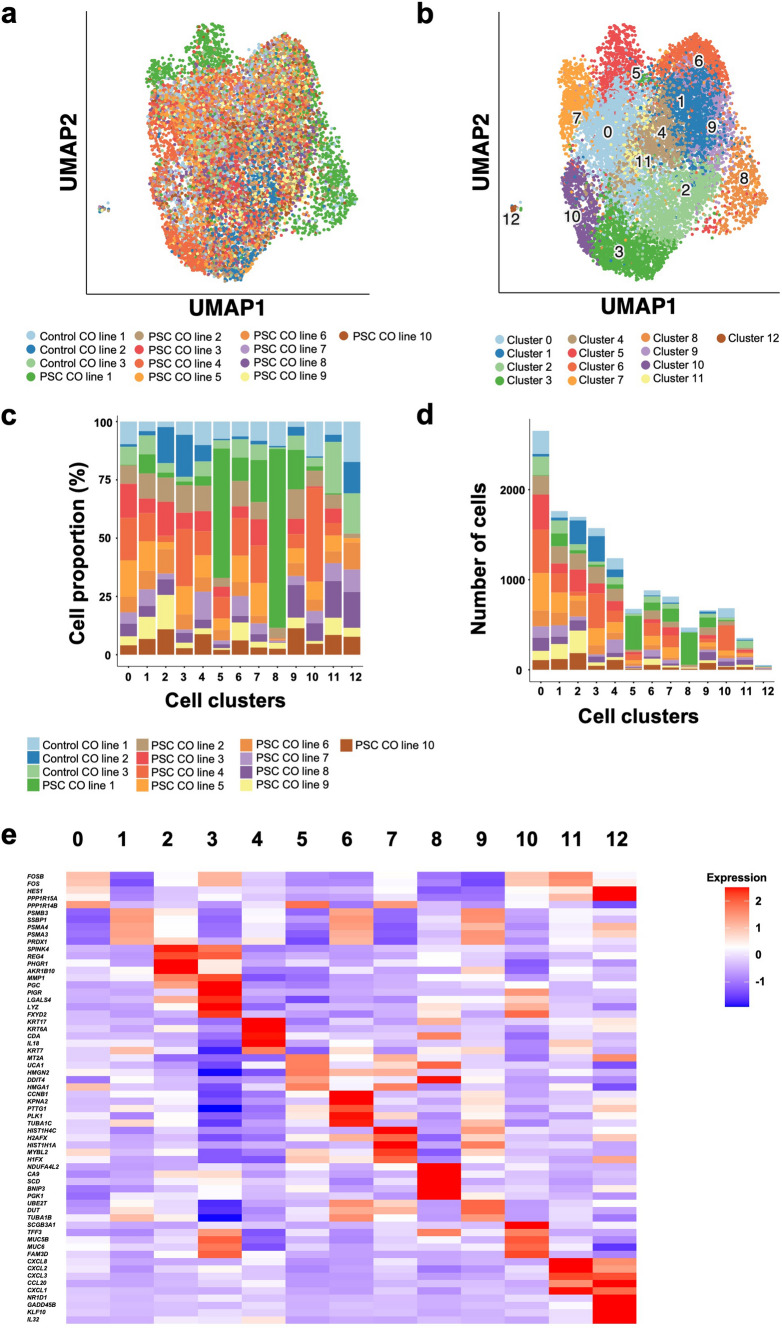
To assess potential patient-specific transcriptomic signature differences between the individual CO lines, we expanded the organoid lines until passage 6, homogenized the COs to single-cell suspensions, and then performed scRNAseq to compare the transcriptional profile of all samples (n = 13). Cluster analysis of scRNAseq transcriptomes revealed close similarity between all cultured CO lines (Fig. 2A). Seurat [29] clustering, with hyper-parameters optimized on data characteristics using ClustAssess [25], revealed 13 clusters when all CO lines were evaluated together (Fig. 2B). No patient-specific clusters were identified, although the cell proportions within each cluster varied between the patients (Fig. 2C). Clusters 0–11 grouped closely to each other. Cluster 12, which consisted of a small subpopulation of cells, clustered furthest away from the other clusters (Fig. 2B, D). Cluster-specific differentially expressed genes (DEG) were identified; focusing on positive markers, we identified genes that are associated with cellular stress responses such as apoptosis (KLF10, GADD45B) and inflammatory processes (NR1D1, CXCL1, IL32) to be strongly upregulated in cluster 12 (Fig. 2E), indicating that cluster 12 consists of a small subpopulation of dying and inflammatory active cells. Cluster 0, the largest subpopulation of cells (Fig. 2D), differentiated from the remaining clusters by an upregulation of genes associated to the regulation of cell proliferation and differentiation (FOS, FOSB, PPP1R15A) (Fig. 2E). DEG analysis revealed that clusters 0–11 were defined by subtle changes in functional gene groups, indicating functional subgroups of cholangiocytes within the CO lines that differ from each other in their cell-cycle state (cluster 1, 6, and 7), metabolic state (cluster 8—upregulated glycolysis/gluconeogenesis), secretion level (Cluster 3 and 10—upregulated secretion/mucus layer), inflammatory response (Cluster 11—upregulated chemotaxis/chemokine activity), DNA repair and chromatin remodeling (Cluster 5), and intermediate filament assembly (Cluster 4) (Fig. 2E, Supplementary Data 1).
Fig. 2.
Cholangiocyte organoids from individual patients overlap in their transcriptomic profile. a UMAP plots illustrate a general transcriptomic overlap between individual COs from PSC (n = 10) and non-PSC patients (n = 3) after stable culture. b Identification of 12 separate clusters across all CO lines. c Proportion of cells from each patient contributing to each cholangiocyte cluster. d Number of cells per patient in each cell cluster. e Top 5 upregulated genes per cluster. UMAP Uniform Manifold Approximation and Projection, CO cholangiocyte organoid
Cholangiocyte Organoids Derived from PSC Patients Do Not Demonstrate Specific Disease-Relevant Features in their Transcriptional Profile Compared to Controls
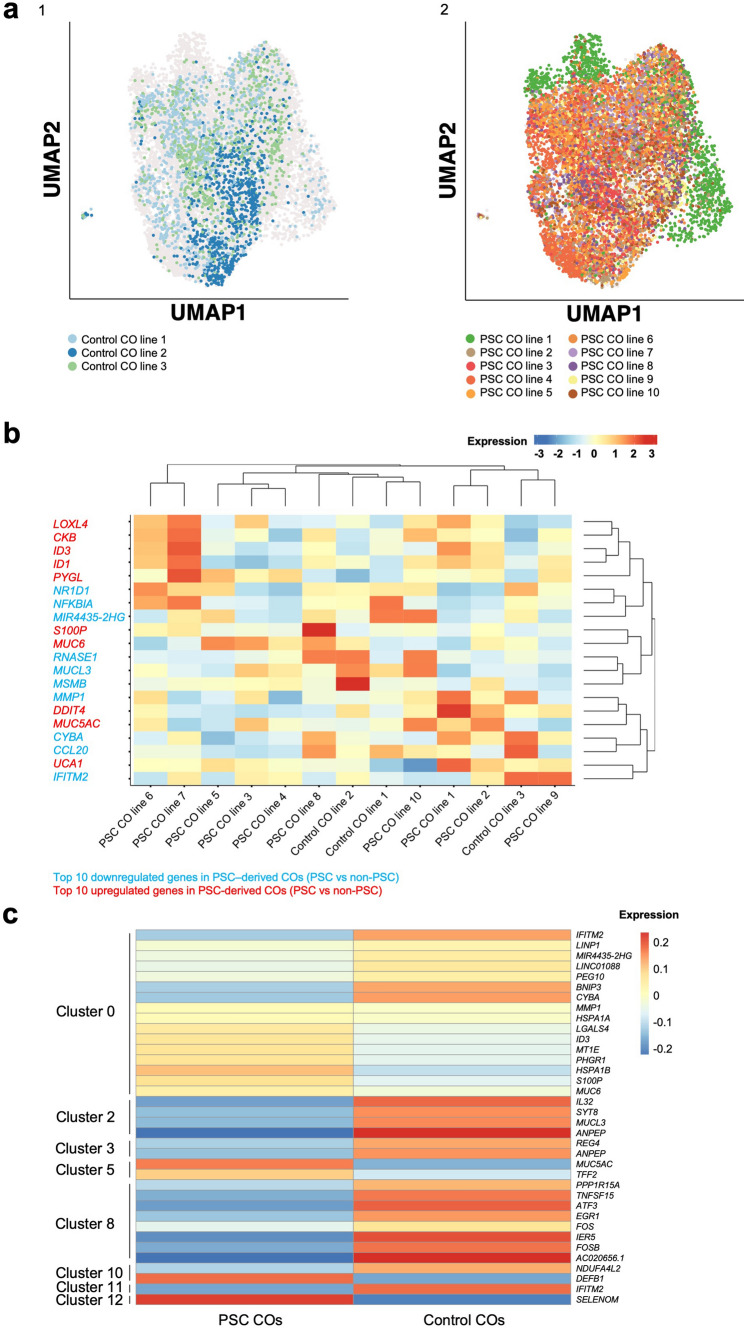
Organoid lines from different patients shared a core transcriptional profile; cluster analysis revealed differences in the cell proportions of each individual patient within the different functional subgroups (clusters). We therefore hypothesized that cholangiocytes from individual patients may retain distinct transcriptome signatures reflecting disease etiology. scRNAseq revealed close similarity between lines derived from PSC patients and lines derived from non-PSC control patients (Fig. 3A). Global analysis identified 132 significantly DEG when comparing PSC and non-PSC CO lines (Supplementary Data 2). The most differentially expressed genes were REG4 (average log2 fold change (avg-log2FC) of -1.02), MUCL3 (avg-log2FC of -0.78) and IL32 (avg-log2FC of -0.73), all of which were downregulated in PSC-derived CO lines (Supplementary Data 2). PSC-derived organoids further showed significant downregulation of MSMB (avg-log2FC of -0.67), NFKBIA (avg-log2FC of -0.58), and MMP1 (avg-log2FC of -0.56) and an increased expression of MUC6 (avg-log2FC of 0.68), ID1 (avg-log2FC of 0.68), and ID3 (avg-log2FC of 0.67) (Supplementary Data 2, Fig. 3B). Significant DEGs were patient-specific rather than global changes relating to disease etiology (Fig. 3B).
Fig. 3.
Transcriptional profiling reveals a strong overlap between cholangiocyte organoids from PSC patients and organoids from non-PSC patients. a UMAP plots show a general transcriptomic overlap between individual samples from (1) non-PSC (n = 3) and (2) PSC patients (n = 10) after organoid culture. b Heatmap demonstrating the top 10 up- and downregulated genes in PSC compared to the non-PSC group. c Most differential expressed genes between the PSC and non-PSC patient groups per cluster (log2FC > 0.5). UMAP Uniform Manifold Approximation and Projection, CO cholangiocyte organoid
Since the cluster analysis revealed patient/line-specific proportions of different functional subgroups within the individual patients (Fig. 2C and 3B), we next analyzed scRNAseq for changes between PSC and non-PSC CO lines within each functional cluster. Subcluster DEG analysis revealed minor changes in gene expression that could not specifically be related to disease etiology (avg-log2FC values < 0.94, Supplementary Data 3). The most upregulated genes in PSC-derived CO lines were MUC5AC and TFF2 (Cluster 5), DEFB1 (Cluster 10), and SELENOM (Cluster 12). Downregulated genes in the PSC group were among others ANPEP and IL32 (Cluster 2), TNFSF15, ATF3, IER5, FOSB, and AC020656.1 (Cluster 8) and IFITM2.1 (Cluster 11) (Fig. 3C).
Additionally, we performed in situ spatial transcriptomic analysis of liver tissue samples from PSC and non-PSC control samples and confirmed the expression of SELENOM, MUC6, ID1, ID3, and DEFB1 in relevant biliary regions in PSC tissue (n = 4) and non-PSC tissue (n = 2) (Supplementary Figs. 8, 9).
Transcriptional Profiling Reveals an Upregulation of Cancer-Related Genes in Cholangiocyte Organoids Derived from a Patient Who Went on to Develop CCA
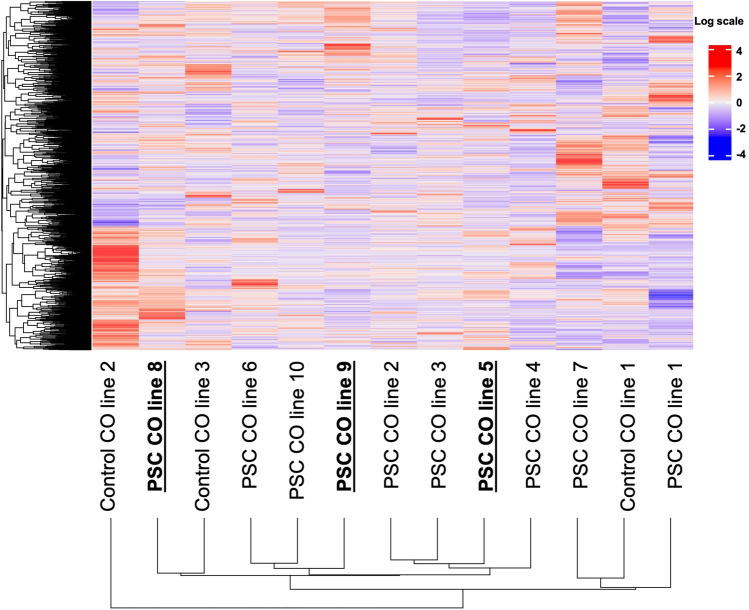
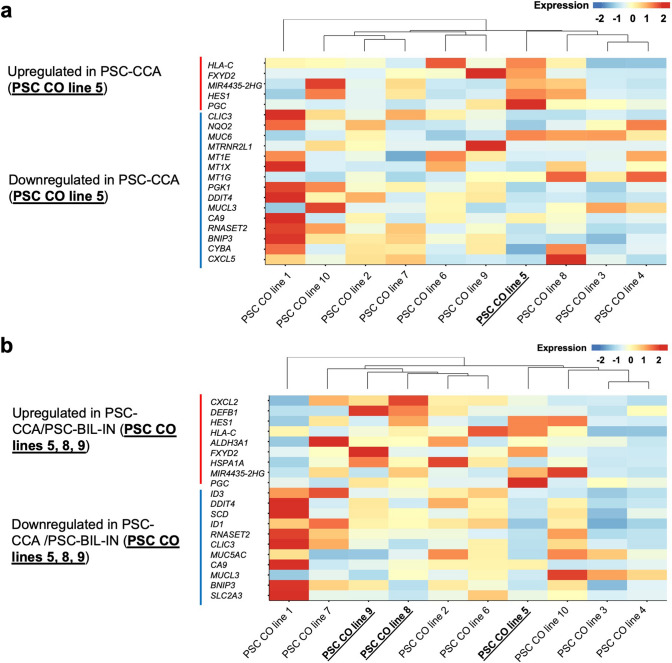
During follow-up after the ERCP procedure (2–60 months; mean 36 months), two of the included PSC patients were diagnosed with biliary intraepithelial neoplasia stage I/II (BIL-IN I/II) (after 5 and 2 months) and one PSC patient was diagnosed with CCA (after 5 months). To investigate whether a possible cancer-related disease imprint could be seen in the organoid culture based on cells collected before the diagnosis, we next compared the complete transcriptomes of all CO lines to each other (Fig. 4). DEG and clustering of the individual CO lines did not show a significant overlap between CO lines that were generated from patients with a putative malignant background on transcriptome level (Fig. 4). We next compared DEG between the CO line derived from a patient with later CCA diagnosis and the remaining PSC-derived samples (Fig. 5A) and between the groups PSC (no dysplasia) and CCA + BIL-IN I/II (diagnosis after ERCP procedure) (Fig. 5B). DEG revealed an upregulation of previously described cancer-related genes (PGC, FXYD2, MIR4435-2HG, and HES1) in CCA compared to non-cancer control samples (Fig. 5A) and an upregulation of the potential tumor suppressor DEFB1 in samples associated with the development of CCA or BIL-IN (Fig. 5B), highlighting a potential of COs as a possible prognostic tool for diagnosing CCA in the future.
Fig. 4.
Transcriptomic similarities and clustering between the individual CO lines. Heatmap shows the complete transcriptomes of all CO samples. Differentially expressed genes are color coded according to the log scale. CO lines from patients who were later diagnosed with biliary malignancy are marked in bold. CO cholangiocyte organoid
Fig. 5.
Transcriptional profiling reveals higher expression of the potential pan-cancer regulator PGC in cholangiocyte organoids derived from a CCA patient. a Top 20 differentiated genes in a CO sample generated from a patient who later was diagnosed with CCA (PSC CO line 5). b Top 20 differentiated genes in a PSC patient group diagnosed with CCA or biliary intraepithelial neoplasia stage I/II (BIL-IN) (n = 3) (diagnosis after cell isolation) compared to PSC patients without biliary malignancy (n = 7). CO cholangiocyte organoid
Discussion
Understanding the pathogenesis of PSC would require the detailed analysis of affected tissue and diseased target cells, which together constitute the disease-specific microenvironment. Most studies evaluating cholangiocytes in PSC have used cells collected from bile or explanted end-stage diseased liver tissue [18, 19], both sources which may not completely represent the cholangiocyte subtypes at the site of chronic inflammation. To assess the phenotype of cholangiocytes originating from the diseased area of the bile ducts, we isolated primary cholangiocytes from ERCP brushes, expanded the collected cells into organoids, and performed high-resolution scRNAseq to establish a comprehensive overview of CO lines derived from PSC and non-PSC patients.
We show that isolation of cholangiocytes directly from the disease-affected common and hepatic bile ducts of patients undergoing ERCP offers a robust and reproducible method for obtaining primary patient cells for organoid expansion. CO lines were successfully established from 13 brush samples and the resulting organoids were easily expanded and expressed key lineage markers characteristic of mature primary cholangiocytes. Importantly, we did not observe morphologic or functional differences between CO lines derived from PSC patients and those derived from non-PSC control patients, suggesting our method may be a valuable tool for future regenerative medicine approaches aiming to treat PSC patients using autologous organoids [16, 30].
A previous study comparing scRNAseq data from fresh human cholangiocytes to cholangiocyte organoids demonstrated a loss of transcriptional diversity after prolonged cell culture [16]. In line with those published data on cholangiocytes from non-PSC patients, we observed that primary cholangiocytes from PSC patients expanded in organoid culture media expressed a common transcriptome profile irrespective of disease etiology. Moderate changes in gene expression seemed to be related to cell line-specific gene signatures (cell-cycle status, culture-dependent stress features) rather than to disease etiology. Contrary to previous studies suggesting that chronic cholangiocyte activation in PSC can initiate non-reversible cellular senescence and induce the transition of cholangiocytes into a strongly proinflammatory senescence-associated secretory (SASP) phenotype [10, 12, 31], we observed that genes associated with an inflammatory response (CCL20, MMP1, IL32, CXCL2, and CXCL8) were slightly downregulated in PSC organoids cultured under non-inflammatory conditions. Although we did not observe any differences in proliferation relative to disease etiology, we saw an upregulation of genes that are thought to promote cellular senescence [32, 33] (ID1 and ID3) in PSC CO lines. Regarding the small avg-log2FC values for those genes, the biological significance of those results remains to be determined. Staining for the key senescence markers SA-β-Gal and p21 further indicated low levels of senescence in PSC COs. External stimulation of PSC-derived organoids with disease-relevant cytokines demonstrated upregulation of both SA-β-Gal and p21, accompanied by an increased secretion of immune-modulating and tissue remodeling mediators that might be of relevance for PSC pathogenesis.
Taken together, these results clearly suggest that the senescent-like phenotype and SASP of PSC cholangiocytes could be specific to the proinflammatory biliary environment during ongoing disease rather than intrinsic and can be reversed when cholangiocytes are expanded in culture.
Of note, future studies should also address potential functional differences between intra- and extrahepatic cholangiocytes in PSC given the known differences between cholangiocytes in these two compartments [34], as our current study comprises the study of extrahepatic cholangiocyte organoids solely.
New diagnostic tools delivering confident results for CCA diagnosis remain the largest unmet need in PSC patients [35]. To evaluate the potential of biliary-derived organoids in this regard, we performed a targeted transcriptome assessment of COs expanded from 3 patients who were later diagnosed with CCA or BIL-IN I/II. In CCA and BIL-IN I/II organoids, we saw a significant upregulation of pepsinogen C (PGC) which has been described as a pan-cancer regulator and has previously been shown to be upregulated in CCA [36]. PGC is thought to regulate the K-RAS signaling pathway and other cancer-related pathways and is a potential prognostic cancer biomarker, but its definitive role in tumor microenvironments is still unclear [36]. We further observed a cancer-related upregulation of FXYD2, which has been shown to be upregulated in CCA tissues [37], as well as an upregulation of the potential pan-cancer biomarker MIR4435-2HG and the potential CCA-driver HES1 [38, 39]. Upregulation of these markers in CCA/BIL-IN I/II-derived organoids after prolonged culture suggests that COs could be used as a predictive tool to triage patients at higher risk of developing CCA. This approach holds potential for establishing a new diagnostic test for CCA based on organoid expansion and sequencing. As the results described herein are limited to 3 CCA/BIL-IN I/II samples, a larger patient group is needed for replication and extension of these results.
In summary, our study demonstrates that primary cholangiocytes can be isolated and expanded into functional COs from ERCP brushes taken from the site of injury of PSC and non-PSC patients. scRNAseq profiling shows PSC and non-PSC COs are largely similar; however, detailed assessment of organoids generated from PSC patients who later developed dysplasia indicates that COs may retain certain disease-relevant features despite their adaption toward the in vitro environment and suggest a future potential application of COs as a diagnostic tool for CCA. Future studies investigating the phenotype of those PSC-derived COs under inflammatory culture conditions may further unlock the full potential of those cells for future regenerative medical approaches aiming to treat PSC patients and/or future mechanistic advances unraveling disease mechanisms and developing new medicinal approaches for the treatment of PSC.
Supplementary Information
Below is the link to the electronic supplementary material.Below is the link to the electronic supplementary material.
Supplementary Figure 1: Number of features per sample. (a) Proportion plot showing the number of features (nFeature) per sample. (b) Saturation plot (nFeature vs nCount scatter plot), demonstrating the logarithmic relationship between the number of counts (nCount, sequencing depth per cell) and number of features (quantified genes) per cell. PSC.Org1 and PSC.ORG1.1 correspond to the same biological sample (batch effect assessment). ORG1-3, non-PSC samples; PSC.ORG1-10, PSC samples. (JPG 201 KB)
Supplementary Figure 2: Violin plots summarising the number of features, number of counts, ribosomal protein percentage and percentage of mitochondrial content across cells, per sample. (a) Violin plots displaying the distribution of the number of features detected in each cell, aggregated by sample. (b) Violin plots showing the distribution of the number of counts. (c) Violin plots displaying the distribution of the ribosomal protein (rp) percentage. (d) Violin plots showing the distribution of percentage of mitochondrial content (mt) detected in each cell, aggregated by sample. We note the consistent bi-modal distribution, which prompted a filtering of cells with nfeature values less than 2500. PSC.Org1 and PSC.ORG1.1 correspond to the same biological sample (batch effect assessment). ORG1-3, non-PSC samples; PSC.ORG1-10, PSC samples. (JPG 729 KB)
Supplementary Figure 3: UMAPs showing the number of features, counts, RP percentage and percentage of mitochondrial content across cells. (a) UMAP displaying the number of features. (b) UMAP displaying the number of counts. (c) UMAP displaying the ribosomal protein (rp) percentage detected in each cell. (d) UMAP displaying the percentage of mitochondrial content (mt). UMAP, Uniform Manifold Approximation and Projection. (JPG 628 KB)
Supplementary Figure 4: Evaluation of cluster stability and output consistency. Boxplots showing the Element-Centric Consistency (ECC) and Element Centric Similarity (ECS) for each feature set. UMAPs showing the stable clusters per gene set and seurat clusters vs stability of chosen gene set. These UMAPs illustrate the correspondence between the clusters obtained by seurat for the analysis, against the stability of the selected configuration obtained by ClustAssess. UMAP, Uniform Manifold Approximation and Projection. (JPG 806 KB)
Supplementary Figure 5: Stability analysis linking the number of clusters (k, y-axis) and resolution (x-axis). This plot assesses the effect/impact of different resolution values on the number of clusters and subsequently the stability of each partition. Different clustering methods are explored; the stable configurations are those with a high ECC (yellow shade) across iterations; the size of each data point reflects the frequency of the most prevalent partition when resolution and the number of clusters are held constant. Element-Centric Consistency, ECC. (JPG 36 KB)
Supplementary file 6: Expansion of ERCP-derived cholangiocyte organoids in Matrigel. First organoid structures are typically visible 3-4 days after the start of culture. Depending on size and morphology of the organoids, cells are split every 6-8 days. Magnification: 4x. (JPG 151 KB)
Supplementary Figure 7: PSC cholangiocyte organoids demonstrate disease relevant responses to external stimuli by upregulation of senescence-associated markers and by secreting immune-modulating and tissue-remodulating mediators. (a) Representative images and senescence staining of stimulated COs derived from PSC patients. 1) Phase contrast images of unstimulated and stimulated organoids. 2) SA-β-Gal stain of unstimulated and stimulated organoids. Scale bars: 500 μm. 3) Representative confocal images of p21 stained organoids with and without stimulation. Scale bars: 50 μm. Treatment dose: 50 ng/mL, 5 days. (b) Secretion of inflammatory and fibrogenic mediators into the cell culture supernatant after stimulation. Data points from one representative stimulation experiment are shown (2 independent cell culture wells). n>3 independent stimulation experiments with individual patients. Treatment dose: 50 ng/mL, 5 days. CO, cholangiocyte organoid; SA-β-Gal, senescence-associated- β-galactosidase; p21, protein 21. (JPG 789 KB)
Supplementary Figure 8: Definition and characterization of biliary regions in human tissue samples using spatial transcriptomics. (a) Biliary regions defined by expression of the cholangiocyte markers KRT7 or EPCAM (KRT7 >2 counts per spot, EPCAM >1 counts per spot). (b) Log-normalized expression levels of the key cholangiocyte markers KRT19, SOX9, CFTR, SCTR within the defined biliary regions. (JPG 624 KB)
Supplementary Figure 9: Spatial transcriptomics demonstrates the presence of specific genes detected by scRNAseq in PSC COs also in relevant biliary regions within PSC tissue samples. Spatial UMAPs indicate the presence of (a) SELENOM (b) MUC6 (c) ID1, (d) ID3 and (e) DEFB1 in relevant pre-defined biliary regions in PSC samples (n=4) and non-PSC disease controls (n=2). Violin plots show log-normalized expression levels. (JPG 560 KB)
Author’s contribution
A.K.F., B.K.C., and E.M. wrote the main manuscript text. A.K.F., T.B, H.W.H., and F. S. performed the experimental work. A.K.F., I.M., and M.L.L.N. performed the primary data analysis. J.Ø., B.K.C., L.K.E., J.H., I.M., and M.L.L.N. contributed to data analysis and interpretation. L.K.E. collected patient-information. E.M., F.S., T.H.K., and L.V. coordinated study design, analysis, and final presentation of the data and gave financial support to this study. All authors reviewed the manuscript.
Funding
Open access funding provided by University of Oslo (incl Oslo University Hospital). This work was supported by the Norwegian PSC Research Center and the Scientia Fellowship European Union’s Horizon 2020 research and innovation program under the Marie Sklodowska-Curie grant agreement No. 801133 and the Norwegian Research council Grant Nos. 315399 and 325435. The work completed in Cambridge was supported by the European Research Council Grant New-Chol.
Data availability
Data transparency Raw data/processed data described in this study are available upon written request to the last author.
Declarations
Conflict of interest
The authors declare no competing interests.
Ethical approval
This study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by the Regional Ethics Committee (2012-286/2016-1540).
Consent to participate
Written informed consent was obtained from all individual participants in this study.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Karlsen TH, Folseraas T, Thorburn D, Vesterhus M. Primary sclerosing cholangitis—a comprehensive review. J. Hepatol. 2017;67:1298–1323. [DOI] [PubMed] [Google Scholar]
- 2.European Association for the Study of the Liver. EASL Clinical Practice Guidelines: management of cholestatic liver diseases. J. Hepatol. 2009;51:237–267. [DOI] [PubMed] [Google Scholar]
- 3.Chapman R, Fevery J, Kalloo A et al. Diagnosis and management of primary sclerosing cholangitis. Hepatology 2010;51:660–678. [DOI] [PubMed] [Google Scholar]
- 4.Boonstra K, Weersma RK, van Erpecum KJ et al. Population-based epidemiology, malignancy risk, and outcome of primary sclerosing cholangitis. Hepatology 2013;58:2045–2055. [DOI] [PubMed] [Google Scholar]
- 5.Poch T, Krause J, Casar C et al. Single-cell atlas of hepatic T cells reveals expansion of liver-resident naive-like CD4+ T cells in primary sclerosing cholangitis. J. Hepatol. 2021;75:414–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liaskou E, Klemsdal Henriksen EK, Holm K et al. High-throughput T-cell receptor sequencing across chronic liver diseases reveals distinct disease-associated repertoires. Hepatology 2016;63:1608–1619. [DOI] [PubMed] [Google Scholar]
- 7.Chung BK, Guevel BT, Reynolds GM et al. Phenotyping and auto-antibody production by liver-infiltrating B cells in primary sclerosing cholangitis and primary biliary cholangitis. J. Autoimmun. 2017;77:45–54. [DOI] [PubMed] [Google Scholar]
- 8.Tabibian JH, Masyuk AI, Masyuk TV, O’Hara SP, LaRusso NF. Physiology of cholangiocytes. Compr. Physiol. 2013;3:541–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jalan-Sakrikar N, De Assuncao TM, Navarro-Corcuera A et al. Induced pluripotent stem cells from subjects with primary sclerosing cholangitis develop a senescence phenotype following biliary differentiation. Hepatol. Commun. 2022;6:345–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tabibian JH, O’Hara SP, Splinter PL, Trussoni CE, LaRusso NF. Cholangiocyte senescence by way of N-ras activation is a characteristic of primary sclerosing cholangitis. Hepatology 2014;59:2263–2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meng L, Quezada M, Levine P et al. Functional role of cellular senescence in biliary injury. Am. J. Pathol. 2015;185:602–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guicciardi ME, Trussoni CE, LaRusso NF, Gores GJ. The spectrum of reactive cholangiocytes in primary sclerosing cholangitis. Hepatology 2020;71:741–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sato K, Glaser S, Kennedy L et al. Preclinical insights into cholangiopathies: disease modeling and emerging therapeutic targets. Expert Opin. Ther. Targets 2019;23:461–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pollheimer MJ, Trauner M, Fickert P. Will we ever model PSC?—“it’s hard to be a PSC model!” Clin. Res. Hepatol. Gastroenterol. 2011;35:792–804. [DOI] [PubMed] [Google Scholar]
- 15.Sato K, Zhang W, Safarikia S et al. Organoids and spheroids as models for studying cholestatic liver injury and cholangiocarcinoma. Hepatology 2021;74:491–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sampaziotis F, Muraro D, Tysoe OC et al. Cholangiocyte organoids can repair bile ducts after transplantation in the human liver. Science 2021;371:839–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tysoe OC, Justin AW, Brevini T et al. Isolation and propagation of primary human cholangiocyte organoids for the generation of bioengineered biliary tissue. Nat. Protoc. 2019;14:1884–1925. [DOI] [PubMed] [Google Scholar]
- 18.Soroka CJ, Assis DN, Alrabadi LS et al. Bile-derived organoids from patients with primary sclerosing cholangitis recapitulate their inflammatory immune profile. Hepatology. 2019;70:871–882. [DOI] [PubMed] [Google Scholar]
- 19.Tabibian JH, Trussoni CE, O’Hara SP, Splinter PL, Heimbach JK, LaRusso NF. Characterization of cultured cholangiocytes isolated from livers of patients with primary sclerosing cholangitis. Lab. Invest. 2014;94:1126–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loarca L, De Assuncao TM, Jalan-Sakrikar N et al. Development and characterization of cholangioids from normal and diseased human cholangiocytes as an in vitro model to study primary sclerosing cholangitis. Lab. Invest. 2017;97:1385–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sampaziotis F, de Brito MC, Geti I, Bertero A, Hannan NR, Vallier L. Directed differentiation of human induced pluripotent stem cells into functional cholangiocyte-like cells. Nat. Protoc. 2017;12:814–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schneider CA, Rasband WS, Eliceiri KW. NIH image to ImageJ: 25 years of image analysis. Nat. Methods 2012;9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dianat N, Dubois-Pot-Schneider H, Steichen C et al. Generation of functional cholangiocyte-like cells from human pluripotent stem cells and HepaRG cells. Hepatology 2014;60:700–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hafemeister C, Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019;20:296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arash S, Munteanu A, Mohorianu I. ClustAssess: tools for assessing the robustness of single-cell clustering. bioRxiv. 2022; 2022–01.
- 26.Gates AJ, Wood IB, Hetrick WP, Ahn YY. Element-centric clustering comparison unifies overlaps and hierarchy. Sci. Rep. 2019;9:8574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Metsalu T, Vilo J. ClustVis: a web tool for visualizing clustering of multivariate data using principal component analysis and heatmap. Nucleic Acids Res. 2015;43:W566–W570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chung BK, Øgaard J, Reims HM, Karlsen TH, Melum E. Spatial transcriptomics identifies enriched gene expression and cell types in human liver fibrosis. Hepatol. Commun. 2022;6:2538–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sampaziotis F, Justin AW, Tysoe OC et al. Reconstruction of the mouse extrahepatic biliary tree using primary human extrahepatic cholangiocyte organoids. Nat. Med. 2017;23:954–963. [DOI] [PubMed] [Google Scholar]
- 31.Carpino G, Cardinale V, Renzi A et al. Activation of biliary tree stem cells within peribiliary glands in primary sclerosing cholangitis. J. Hepatol. 2015;63:1220–1228. [DOI] [PubMed] [Google Scholar]
- 32.Alani RM, Young AZ, Shifflett CB. Id1 regulation of cellular senescence through transcriptional repression of p16/Ink4a. Proc. Natl. Acad. Sci. USA 2001;98:7812–7816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zebedee Z, Hara E. Id proteins in cell cycle control and cellular senescence. Oncogene 2001;20:8317–8325. [DOI] [PubMed] [Google Scholar]
- 34.Rimland CA, Tilson SG, Morell CM et al. Regional differences in human biliary tissues and corresponding in vitro-derived organoids. Hepatology 2021;73:247–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lang SA, Bednarsch J, Joechle K et al. Prognostic biomarkers for cholangiocarcinoma (CCA): state of the art. Expert Rev. Gastroenterol. Hepatol. 2021;15:497–510. [DOI] [PubMed] [Google Scholar]
- 36.Shen S, Li H, Liu J, Sun L, Yuan Y. The panoramic picture of pepsinogen gene family with pan-cancer. Cancer Med. 2020;9:9064–9080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Z, Tang Y, Li L et al. Downregulation of FXYD2 Is associated with poor prognosis and increased regulatory T cell infiltration in clear cell renal cell carcinoma. J. Immunol. Res. 2022;2022:4946197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhong C, Xie Z, Zeng LH, Yuan C, Duan S. MIR4435-2HG is a potential pan-cancer biomarker for diagnosis and prognosis. Front. Immunol. 2022;13:855078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsumori T, Kodama Y, Takai A et al. Hes1 Is essential in proliferating ductal cell-mediated development of intrahepatic cholangiocarcinoma. Cancer Res. 2020;80:5305–5316. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Number of features per sample. (a) Proportion plot showing the number of features (nFeature) per sample. (b) Saturation plot (nFeature vs nCount scatter plot), demonstrating the logarithmic relationship between the number of counts (nCount, sequencing depth per cell) and number of features (quantified genes) per cell. PSC.Org1 and PSC.ORG1.1 correspond to the same biological sample (batch effect assessment). ORG1-3, non-PSC samples; PSC.ORG1-10, PSC samples. (JPG 201 KB)
Supplementary Figure 2: Violin plots summarising the number of features, number of counts, ribosomal protein percentage and percentage of mitochondrial content across cells, per sample. (a) Violin plots displaying the distribution of the number of features detected in each cell, aggregated by sample. (b) Violin plots showing the distribution of the number of counts. (c) Violin plots displaying the distribution of the ribosomal protein (rp) percentage. (d) Violin plots showing the distribution of percentage of mitochondrial content (mt) detected in each cell, aggregated by sample. We note the consistent bi-modal distribution, which prompted a filtering of cells with nfeature values less than 2500. PSC.Org1 and PSC.ORG1.1 correspond to the same biological sample (batch effect assessment). ORG1-3, non-PSC samples; PSC.ORG1-10, PSC samples. (JPG 729 KB)
Supplementary Figure 3: UMAPs showing the number of features, counts, RP percentage and percentage of mitochondrial content across cells. (a) UMAP displaying the number of features. (b) UMAP displaying the number of counts. (c) UMAP displaying the ribosomal protein (rp) percentage detected in each cell. (d) UMAP displaying the percentage of mitochondrial content (mt). UMAP, Uniform Manifold Approximation and Projection. (JPG 628 KB)
Supplementary Figure 4: Evaluation of cluster stability and output consistency. Boxplots showing the Element-Centric Consistency (ECC) and Element Centric Similarity (ECS) for each feature set. UMAPs showing the stable clusters per gene set and seurat clusters vs stability of chosen gene set. These UMAPs illustrate the correspondence between the clusters obtained by seurat for the analysis, against the stability of the selected configuration obtained by ClustAssess. UMAP, Uniform Manifold Approximation and Projection. (JPG 806 KB)
Supplementary Figure 5: Stability analysis linking the number of clusters (k, y-axis) and resolution (x-axis). This plot assesses the effect/impact of different resolution values on the number of clusters and subsequently the stability of each partition. Different clustering methods are explored; the stable configurations are those with a high ECC (yellow shade) across iterations; the size of each data point reflects the frequency of the most prevalent partition when resolution and the number of clusters are held constant. Element-Centric Consistency, ECC. (JPG 36 KB)
Supplementary file 6: Expansion of ERCP-derived cholangiocyte organoids in Matrigel. First organoid structures are typically visible 3-4 days after the start of culture. Depending on size and morphology of the organoids, cells are split every 6-8 days. Magnification: 4x. (JPG 151 KB)
Supplementary Figure 7: PSC cholangiocyte organoids demonstrate disease relevant responses to external stimuli by upregulation of senescence-associated markers and by secreting immune-modulating and tissue-remodulating mediators. (a) Representative images and senescence staining of stimulated COs derived from PSC patients. 1) Phase contrast images of unstimulated and stimulated organoids. 2) SA-β-Gal stain of unstimulated and stimulated organoids. Scale bars: 500 μm. 3) Representative confocal images of p21 stained organoids with and without stimulation. Scale bars: 50 μm. Treatment dose: 50 ng/mL, 5 days. (b) Secretion of inflammatory and fibrogenic mediators into the cell culture supernatant after stimulation. Data points from one representative stimulation experiment are shown (2 independent cell culture wells). n>3 independent stimulation experiments with individual patients. Treatment dose: 50 ng/mL, 5 days. CO, cholangiocyte organoid; SA-β-Gal, senescence-associated- β-galactosidase; p21, protein 21. (JPG 789 KB)
Supplementary Figure 8: Definition and characterization of biliary regions in human tissue samples using spatial transcriptomics. (a) Biliary regions defined by expression of the cholangiocyte markers KRT7 or EPCAM (KRT7 >2 counts per spot, EPCAM >1 counts per spot). (b) Log-normalized expression levels of the key cholangiocyte markers KRT19, SOX9, CFTR, SCTR within the defined biliary regions. (JPG 624 KB)
Supplementary Figure 9: Spatial transcriptomics demonstrates the presence of specific genes detected by scRNAseq in PSC COs also in relevant biliary regions within PSC tissue samples. Spatial UMAPs indicate the presence of (a) SELENOM (b) MUC6 (c) ID1, (d) ID3 and (e) DEFB1 in relevant pre-defined biliary regions in PSC samples (n=4) and non-PSC disease controls (n=2). Violin plots show log-normalized expression levels. (JPG 560 KB)
Data Availability Statement
Data transparency Raw data/processed data described in this study are available upon written request to the last author.