Abstract
Background
Triple-negative breast cancer is a breast cancer subtype characterized by its challenging prognosis, and establishing prognostic models aids its clinical treatment. PANoptosis, a recently identified type of programmed cell death, influences tumor growth and patient outcomes. Nonetheless, the precise impact of PANoptosis-related genes on the prognosis of triple-negative breast cancer has yet to be determined.
Methods
Clinical information for the triple-negative breast cancer samples was collected from the Gene Expression Omnibus and The Cancer Genome Atlas databases, while 19 PANoptosis-related genes were sourced from previous studies. We first categorized PANoptosis-related subtypes and determined the differentially expressed genes between them. Subsequently, we developed and validated a PANoptosis-associated predictive model using LASSO and Cox multivariate regression analyses. Statistical evaluations were conducted using R software, and the mRNA expression levels of the genes were quantified using real-time PCR.
Results
Using consensus clustering analysis, we divided triple-negative breast cancer patients into two clusters based on PANoptosis-related genes and identified 1054 differentially expressed genes between these clusters. Prognostic-related genes were subsequently selected to re-cluster patients, validating their predictive ability. A prognostic model was then constructed based on four genes: BTN2A2, CACNA1H, PIGR, and S100B. The expression and enriched cell types of these genes were examined and the expression levels were validated in vitro. Furthermore, the model was validated, and a nomogram was created to enhance personalized risk assessment. The risk score, proven to be an independent prognostic indicator for triple-negative breast cancer, showed a positive correlation with both age and disease stage. Immune infiltration and drug sensitivity analyses suggested appropriate therapies for different risk groups. Mutation profiles and pathway enrichment were analyzed, providing insights into potential therapeutic targets.
Conclusion
A PANoptosis-related prognostic model was successfully developed for triple-negative breast cancer, offering a novel approach for predicting patient prognosis and guiding treatment strategies.
Keywords: PANoptosis, Tumor microenvironment, Triple-negtive breast cancer, Prognosis, Immunotherapy
1. Introduction
Globally, breast cancer has the highest incidence rate and ranks as the fifth leading cause of cancer mortality [1]. Triple-negative breast cancer (TNBC) comprises about 15–20 % of all breast cancer cases and poses a significant clinical challenge because of its higher recurrence risks and lower prognostic outcomes [2]. Because these tumors lack key receptors such as estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER-2), TNBC is not amenable to endocrine therapy or HER-2 targeted treatments, making it a particularly challenging form of breast cancer to treat [3]. The dearth of effective therapies underscores the increasing importance of identifying novel biomarkers for TNBC, which are crucial for enabling personalized prognostic assessments and developing therapeutic strategies.
Although cell death is an essential mechanism vital for human well-being that oversees cell proliferation, organ formation, and tissue homeostasis, its disruption or circumvention can promote the transformation of cells into malignant states and propel the onset of tumorigenesis [4]. Pyroptosis, necroptosis, and apoptosis are among the most thoroughly characterized forms of programmed cell death identified to date, each governed by intricate molecular machinery crucial for initiating, transmitting, and executing cell death [5]. However, recent studies have revealed that these seemingly independent pathways exhibit substantial cross-talk, demonstrating a complex interplay among their molecular components. Consequently, PANoptosis was newly introduced by Malireddi et al. in 2019. Although it integrates the key features of pyroptosis, apoptosis, and necroptosis, it eludes conventional categorization into any single one of these pathways [6].
Similar to the pivotal roles of the apoptosome, inflammasome, and necrosome in regulating apoptosis, pyroptosis, and necroptosis, the PANoptosome is essential in orchestrating PANoptosis [7]. Crucial upstream molecules of the PANoptosome, responsible for initiating its assembly and inducing PANoptosis, include ZBP1, AIM2, and RIPK1 [8]. Additionally, various other targets related to apoptosis (such as Caspase-6, Caspase-7, and PARP), necroptosis (such as MLKL, RIPK1, and RIPK3), and pyroptosis (such as Caspase-1, Caspase-3, and GSDMD) were also involved in this novel form of cell death [[9], [10], [11], [12]]. PANoptosis is essential in various diseases, such as infections, inflammatory conditions, and cancer. In cancer, PANoptosis is associated with the growth and proliferation of cancer cells. The induction of PANoptosis triggers strong inflammatory responses, thereby enhancing tumor immunotherapy [13]. Additionally, it presents a promising strategy for overcoming drug resistance typically associated with conventional cell death pathways [14]. The potential of PANoptosis in the prognostic processes of cancer merits further exploration.
Recently, multiple studies have reported that PANoptosis is involved in tumorigenesis and prognosis among numerous cancer types, including pancreatic cancer, hepatocellular carcinoma, and colon cancer [[15], [16], [17]]. Prognostic models based on PANoptosis-related targets have been successfully established for these cancers. These models contribute to enhanced accuracy in predicting outcomes, enabling clinicians to provide more personalized treatment guidance. In TNBC, some prognostic models related to necroptosis, cuproptosis, and autophagy have already been established [[18], [19], [20]]. However, prognostic models specifically related to PANoptosis have not yet been developed, indicating a potential area for future research in TNBC prognosis. As a result, in this investigation, we discovered PANoptosis-related genes (PRGs) and effectively built a predictive model for TNBC patients using these genes. Furthermore, we conducted extensive bioinformatics studies and experimental validations to support our model, which provides novel insights into prognostic prognosis and precise treatment options. Ultimately, our approach aims to enhance the prognosis for patients with TNBC.
2. Materials and methods
2.1. Data collection from GEO and TCGA databases
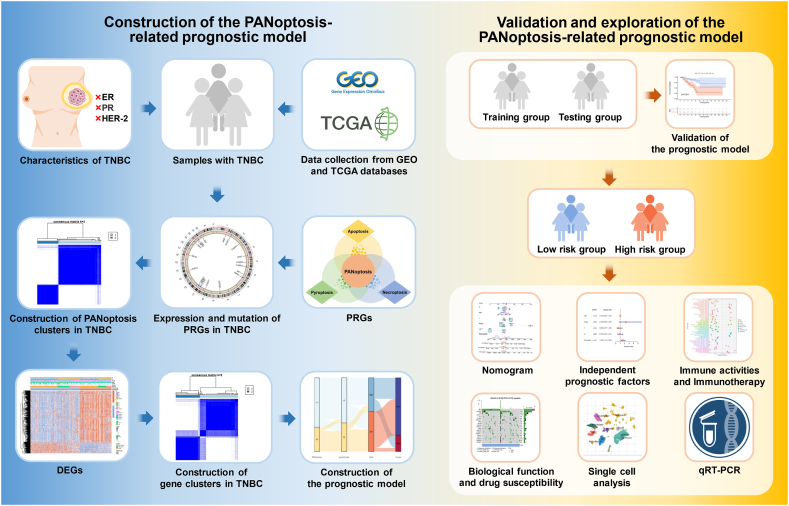
The study's procedure is shown in Fig. 1. The samples of patients were collected from the Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/) and The Cancer Genome Atlas (TCGA) (https://portal.gdc.cancer.gov/) databases. We acquired gene expressions, mutations, copy number variations (CNVs), and clinical information for breast cancer from the TCGA database. The TNBC samples with negative expression of ER, PR, and HER-2 were specifically chosen. The R software (version 4.3.1) was subsequently employed to convert fragments per kilobase million (FPKM) into transcripts per million (TPM) for subsequent analysis. Having excluded records with incomplete data, a total of 240 samples, including 123 TNBC samples with 123 tumor tissues and 22 normal tissues, and 117 breast cancer samples with normal tissues, were obtained for further study from the TCGA database (Table S1). Additionally, datasets GSE58812 (comprising 107 samples) and GSE135565 (comprising 83 samples) were retrieved from the GEO database. Afterward, we utilized the "limma” package in R language to iteratively filter intersecting genes obtained from both the GEO and TCGA databases. Subsequently, we removed normal samples from the TCGA data and checked whether the data had been log2-transformed. If not, logarithmic transformation was applied. Following the removal of batch effects, the two datasets were merged, resulting in a total of 430 samples selected for constructing the PANoptosis-related model. PRGs were identified based on previous research, and detailed information is provided in Table S2 [[21], [22], [23], [24]].
Fig. 1.
The workflow of PANoptosis-related model construction in TNBC.
2.2. Differential expressions, mutations, and CNVs of PRGs
To explore the clinical features of PRGs in TNBC, we examined the expression differences in normal and tumor samples obtained from the TCGA database. Additionally, we utilized the “maftools” and “RCircos” packages to identify and visualize genetic mutations and chromosome locations of the CNVs in PRGs on 23 chromosomes. Subsequently, we assessed the survival status with diverse PRG expression levels via the “survival” and “survminer” packages. A prognostic network analysis was also conducted among the 19 PRGs.
2.3. Consensus clustering analysis
Based on the levels of PRG expression, we conducted cluster analysis using the “ConsensusClusterPlus” package, using the k-means algorithm and determined the optimal number of clusters within the range of k = 2–9 through 1000 tests. The differential expression of PRGs between these clusters was visualized using the “pheatmap” package, and TNBC samples were categorized by principal component analysis (PCA) according to PANoptosis clusters. Furthermore, Kaplan-Meier (KM) curves were generated to assess differences in overall survival (OS) among distinct clusters. To assess the biological functions of these clusters, gene set variation analysis (GSVA) was conducted using “c2.cp.kegg_legacy.v2023.2.Hs.symbols.gmt” and “c5.go.v2023.2.Hs.symbols.gmt” datasets sourced from the MSigDB database (https://www.gsea-msigdb.org/gsea/msigdb). Additionally, Single Sample Gene Set Enrichment Analysis (ssGSEA) was used to evaluate the immune infiltration.
2.4. Construction of the PANoptosis-based prognostic model
Differentially expressed genes (DEGs) were identified in specific clusters using the “limma” package, with the criteria of false discovery rate (FDR) < 0.05 and |log2FC| < 0.585. The Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) analyses were utilized to examine the biological signatures associated with these DEGs (P < 0.05). Using univariate Cox regression, prognostic-related genes were further retrieved from the DEGs (P < 0.05). Following that, samples were classified into distinct gene subtypes in accordance with the expression of these prognostic-related DEGs through cluster analysis. Additionally, the clinical characteristics of the different gene clusters were also assessed.
Based on prognostic-related DEGs, A risk model was developed using R packages such as “caret”, “glmnet”, “survival”, “survminer”, and “timeROC”. Initially, the samples were randomly divided into two equal sets: one served as the training group for establishing the PANoptosis-related risk model, and the other as the testing group for validating the constructed risk model. To reduce overfitting in the genetic data, least absolute shrinkage and selection operator (LASSO) regression analysis was utilized, followed by cross-validation. Ultimately, we conducted the multivariate Cox regression analysis to pinpoint candidate genes for the development for constructing the PANoptosis based model. To compute individual risk scores, a formula was defined as follows: risk score = ∑(Expi ∗ coefi). In this equation, coefi represents the regression coefficient, and Expi represents the gene expression value for each gene, respectively.
To delve deeper into the clinical features of risk groups, risk scores were depicted for different PANoptosis clusters and gene clusters. Subsequently, using the midpoint value from the training group, all samples were classified into higher and lower risk score groups, and different clinical characteristics of these risk groups were compared. A sankey diagram was created by the “dplyr” package to visualize the distribution of PANoptosis-related clusters and gene clusters among risk groups and survival status.
To further validate the constructed risk model, we developed a nomogram incorporating four clinicopathological characteristics: age, stage, stage T, and stage N. Receiver operating characteristic (ROC) curves for these unique attributes were created to assess the effectiveness of the nomogram alongside all clinical characteristics. Finally, independent prognostic analyses were conducted on the risk model, utilizing both univariate and multivariate Cox analyses.
2.5. Assessment of immune correlation with risk levels
The “infiltration_estimation_for_tcga.csv” dataset was obtained from the TIMER2.0 database (http://timer.cistrome.org/) to assess the infiltration of immune cells in two risk groups. Using the CIBERSORT method, we investigated correlations among immune cells, immune functions, and risk scores within these groups. Furthermore, differential analyses of the tumor microenvironment (TME) scores, including immune, stromal, and ESTIMATE scores, were conducted across different risk levels through the ESTIMATE algorithm. Moreover, we examined the association between stemness scores and risk scores. We further analyzed the immune checkpoint gene profiles across higher and lower risk score groups. Finally, we explored the response to immune checkpoint inhibitor treatment in two risk groups within TNBC, utilizing the Tumor Immune Dysfunction and Exclusion (TIDE) database (http://tide.dfci.harvard.edu/).
2.6. Assessment of mutations, biological functions, and chemotherapeutics between risk groups
Aiming to examine gene mutations in patients with TNBC within two distinct risk groups, we used the “maftools” package. The tumor mutation burden (TMB) scores were calculated by Spearman's method. After that, we conducted gene set enrichment analysis (GSEA) to identify distinct physiological roles in different risk levels. Lastly, we employed the “pRophetic” tool to assess the effectiveness of frequently used chemotherapeutic agents by calculating the half maximal inhibitory concentration (IC50) values.
2.7. Expression and distribution of genes in the model
We used the Human Protein Atlas (HPA) platform (https://www.proteinatlas.org) to examine the differential expression of the genes utilized in model construction at the protein level, comparing tumor and normal tissues. The Tumor Immune Single-cell Hub (TISCH) database (http://tisch.compgenomics.org) is used to evaluate the expression patterns of genes related to risk model construction in various immune cell subpopulations of breast cancer at the single-cell level. The differential expression levels of these genes in patients with TNBC were also assessed between normal and tumor tissues.
2.8. Cell lines and quantitative real-time polymerase chain reaction PCR (qRT-PCR)
The TNBC cell lines MDA-MB-231 and MDA-MB-468 and the normal breast cell line MCF-10A were acquired from the American Type Culture Collection (ATCC, Manassas, VA, USA). These cell lines were maintained at 37 °C in Dulbecco's Modified Eagle Medium (DMEM), enriched with 10 % heat-inactivated fetal bovine serum (FBS) and 1 % penicillin/streptomycin mixture, in a moisture-controlled incubator with 5 % CO2. Total RNA from both cell lines was isolated using TRIzol reagent (Ambion), and cDNA was synthesized using the RevertAid First Strand cDNA Synthesis kit (Thermo). The relative mRNA expression was determined via qRT-PCR using the Tip Green qPCR SuperMix kit (TransStart), with GAPDH serving as the normalization standard. Primer sequences were provided in Table S3.
2.9. Statistical analysis
All the evaluations were executed using R tools (version 4.3.1) along with its auxiliary packages, setting a significance level at P < 0.05.
3. Results
3.1. The expression and mutation characteristics of PRGs in TNBC
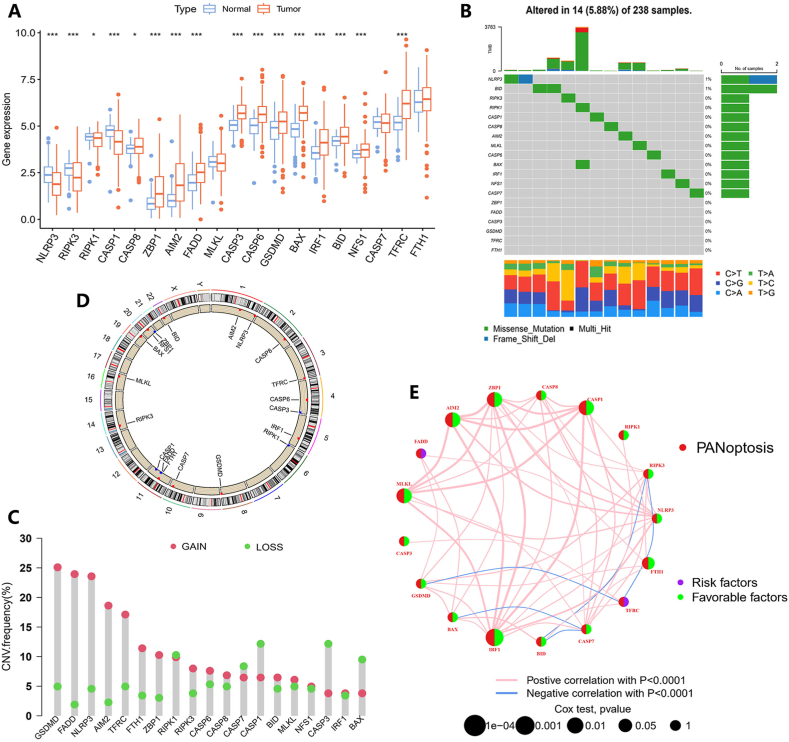
Starting with 19 PRGs identified in previous research, we initially performed analyses of gene expression differences between normal and TNBC samples within the TCGA dataset. Thirteen PRGs, including RIPK1, CASP8, ZBP1, AIM2, FADD, CASP3, CASP6, GSDMD, BAX, IRF1, BID, NFS1, and TFRC, were significantly up-regulated, while three PRGs were significantly down-regulated in TNBC patients (Fig. 2A). Then, we analyzed somatic mutations in 19 PRGs, revealing that there were 14 (5.88 %) mutations in PRGs among 238 TNBC samples. NLRP3 and BID exhibited mutation frequencies of 1 % each, while six PRGs (ZBP1, FADD, CASP3, GSDMD, TFRC, and FTH1) showed no mutations (Fig. 2B). Next, we analyzed the CNVs in 19 PRGs. Most CNVs in these genes showed an increase, such as GSDMD, FADD, NLRP3, and AIM2, while CNVs in BAX, CASP1, CASP3, and CASP7 showed an apparent decrease (Fig. 2C). We also visualized the locations of 19 CNVs on 23 human chromosomes (Fig. 2D and Table S4). To investigate the correlation among these 19 PRGs in TNBC, a network was conducted based on their interactions and prognostic effects (Fig. 2E). Cox univariate regression analysis was employed to determine whether these PRGs impact OS in patients with TNBC. Results with a significance level of P < 0.05 were shown using KM curves (Fig. S1 and Table S5).
Fig. 2.
Gene expression and mutation characteristics of PRGs in TNBC. (A) Expression levels of PRGs in TNBC compared to normal samples. (∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.) (B) Mutation frequency of 19 PRGs in TNBC patients. (C) The distribution of CNV changes in 19 PRGs across 23 chromosomes. (D) Incidence of CNV amplification and deletion in PRGs. (E) Development of a predictive network using PRGs.
3.2. Development and feature analysis of PANoptosis clusters in TNBC
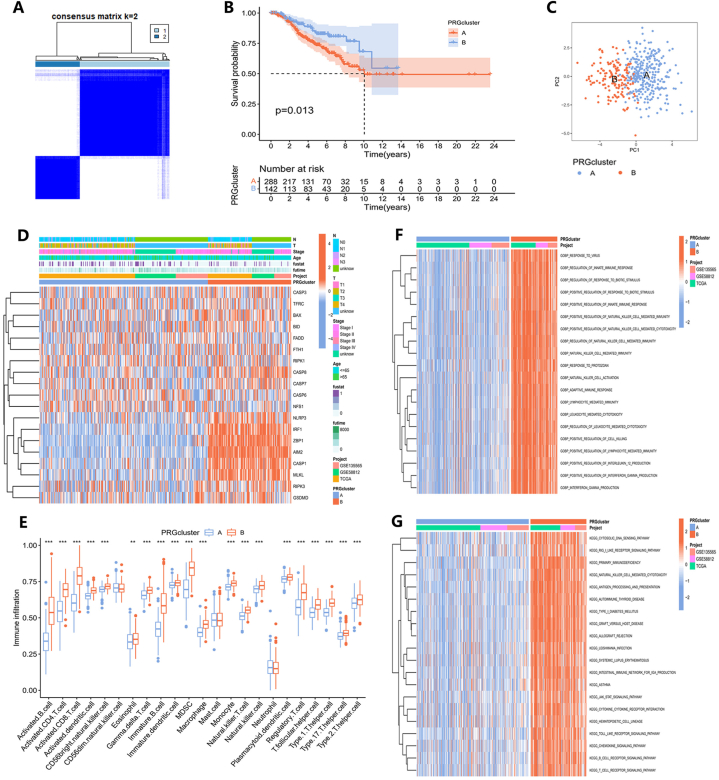
We conducted a consensus clustering analysis to identify different clusters of TNBC based on the differential expression of 19 PRGs. The optimal threshold for segmenting the entire sample into different clusters was determined to be k = 2 (Fig. 3A). The KM curve in Fig. 3B showed that cluster A exhibited superior OS compared to cluster B (P = 0.013). Furthermore, the PCA result suggests that, based on the expression of PRGs, a significant distinction is observed between the two clusters (Fig. 3C). Furthermore, we carried out a comparative study to analyze the expression patterns of these 19 PRGs in the two identified clusters. A heatmap has been generated to elucidate the specific details of the relationships between gene expression, clinical characteristics, and different clusters (Fig. 3D).
Fig. 3.
Construction and feature analysis of PANoptosis clusters. (A) Based on consensus clustering analysis, two PANoptosis clusters were identified. (B) The KM survival curves of PANoptosis clusters. (C) the PCA result of PANoptosis clusters. (D) Variations in the expression of PRGs between PANoptosis clusters. (E) Immune cell infiltrate differences between PANoptosis clusters. (∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.) (F) The GO and (G) KEGG analysis of PANoptosis clusters.
Based on the ssGSEA algorithm, we also evaluated the correlation between the two clusters and 23 immune cell subpopulations separately (Fig. 3E). Most of these immune cell subpopulations exhibited higher infiltration in cluster B compared to cluster A. Only CD56dim natural killer cells, mast cells, and neutrophils demonstrated no significant differences between cluster A and cluster B. We used GSVA enrichment analysis to examine the differences in physiological traits between the two groups and to determine the relevant pathways and biological functions. The GO and KEGG enrichment results for each cluster are depicted in distinct heatmaps (Fig. 3F and G; Tables S6 and S7).
3.3. Construction of gene clusters in TNBC
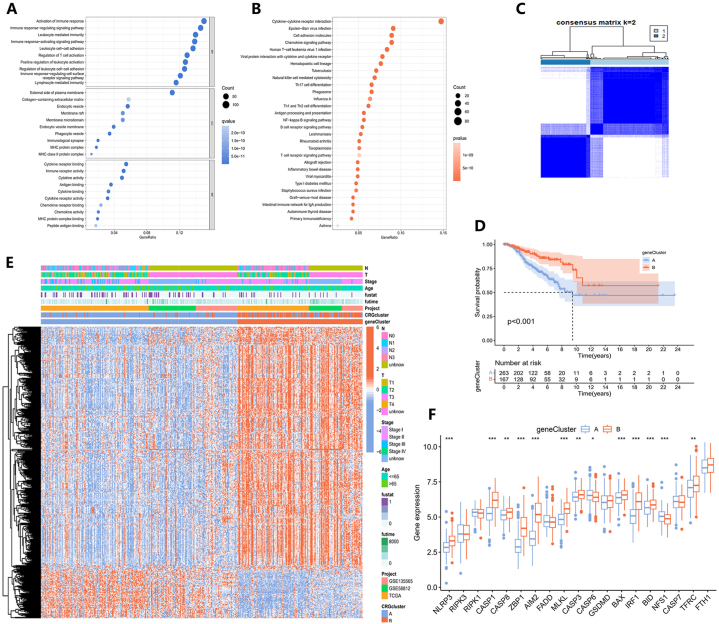
Based on the two PANoptosis clusters we constructed, we identified 1054 DEGs for subsequent analysis (Table S8). To explore the enrichment of biological functions and pathways in these DEGs, we performed both GO and KEGG analyses (Tables S9 and S10). We presented the top 30 enriched pathways in KEGG analysis and the top 10 functions in each of the BP, CC, and MF terms in GO analysis, ranked by P-values (Fig. 4A and B). To assess the prognostic significance of these DEGs, we conducted a univariate Cox analysis, identifying 473 prognostic DEGs with P < 0.05 for further analysis (Table S11). Using the consensus clustering algorithm, all TNBC samples were then classified into two gene clusters based on the expression levels of DEGs (Fig. 4C). The KM survival plot depicted that the OS between the two gene clusters was significantly different (P < 0.001), and patients in gene cluster A were more prone to a worse prognosis (Fig. 4D). The differential expression and clinicopathological characteristics of prognostic DEGs were revealed in a heatmap (Fig. 4E). Additionally, the expression levels of PRGs were also analyzed across these two gene clusters, unveiling significant differences (Fig. 4F).
Fig. 4.
Construction of gene clusters utilizing DEGs between PANoptosis clusters. (A) The GO and (B) KEGG analyses of DEGs between PANoptosis clusters. (C) Identification of two gene clusters based on DEGs. (D) The KM survival curves of the two clusters. (E) Differential expression of DEGs between two gene clusters. (F) PRG expression levels in gene clusters. (∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.)
3.4. Development of the PANpotosis-related model
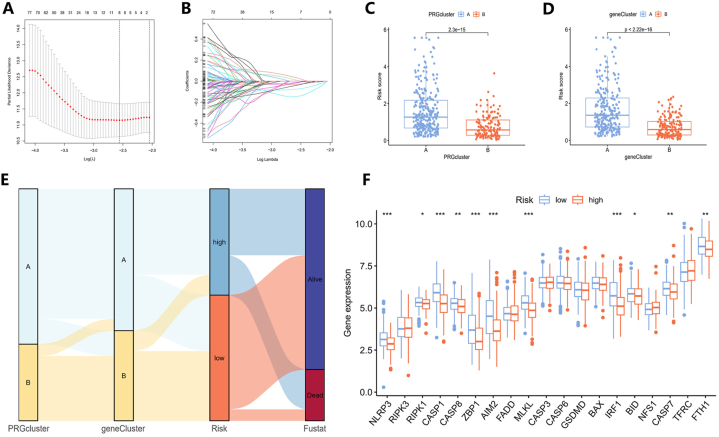
After determining the prognosis-related DEGs, we created a risk model related to PANoptosis using these genes and randomly assigned all samples to training and testing groups. Optimal prognostic factors were determined through the use of LASSO regression and Cox multivariate regression analysis (Fig. 5A and B). Eight prognosis-related genes were singled out via minimum partial likelihood deviance using Lasso regression and used for subsequent Cox multivariate regression analysis. The output of multivariate Cox regression analysis only includes four genes (BTN2A2, CACNA1H, PIGR and S100B) ultimately selected for constructing the prognostic model and their corresponding coef values. According to this model, a patient's risk score is calculated by summing the products of each gene's expression and its corresponding coefficient (Table 1).
Fig. 5.
Construction of the PANoptosis-related prognostic model. (A) LASSO regression analysis and (B) cross-validation for identifying key prognostic genes and assessing partial likelihood deviance. (C) The risk scores between PANoptosis clusters and (D) gene clusters. (E) The connections among PANoptosis clusters, gene clusters, risk score groups, and survival statuses were depicted in a Sankey diagram. (F) PRG expression levels in different risk groups. (∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.)
Table 1.
The result of Cox multivariate regression analysis based on eight prognosis-related genes.
| id | coef |
|---|---|
| BTN2A2 | −0.505696739094738 |
| CACNA1H | 0.200767206653681 |
| PIGR | −0.223659461925913 |
| S100B | −0.178945869507183 |
Upon determining individual risk scores, notable differences were found between different gene clusters and PANoptosis clusters (Fig. 5C and D). By utilizing the median risk assessment value from the training cohort as a cutoff, the patients were separated into two distinct groups: a low-risk group with 233 samples and a high-risk group containing 197 samples. To depict the connections among PANoptosis clusters, gene clusters, risk score groups, and survival status, a Sankey diagram was generated (Fig. 5E). Furthermore, the differential expressions of PANoptosis-related genes (PRGs) between the two risk categories were also highlighted. Analysis indicated that eleven PRGs had significant expression differences, with all exhibiting elevated expression in patients with lower risk scores (Fig. 5F).
3.5. Evaluation of the PANoptosis-based prognostic model
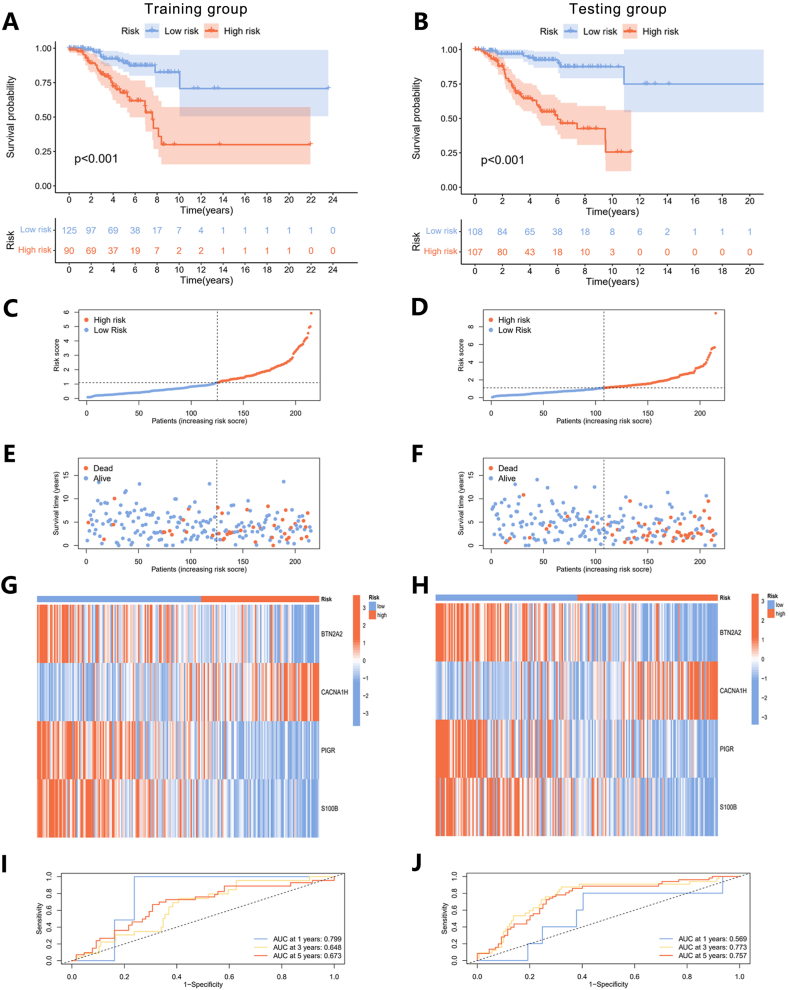
To showcase the clinical applicability of the risk model, we plotted concordance index curves for all samples (Fig. S2), the training group, and the testing group, respectively. The Concordance index of risk scores surpassed age, stage, stage T, and stage N in both groups, confirming the suitability of risk scores for evaluating the model's construction (Fig. S3). Next, we evaluated the variations in risk scores, survival conditions, and the expression profiles of genes incorporated into the model among distinct samples. In both the training and testing groups, patients with higher risk scores showed a stronger association with poorer prognoses (Fig. 6A and B). Additionally, as the disease progressed and the risk scores increased, they exhibited increased susceptibility to mortality (Fig. 6C, D, 6E, and 6F). The expression levels of BTN2A2, PIGR, and S100B were found to decrease in association with higher risk scores, while the expression of CACNA1H showed an increase across both sets (Fig. 6G and H). We generated ROC curves while calculating the area under the ROC curve (AUC) for different years to assess the performance of the risk model (Fig. 6I and J). At the first year, the AUC values were 0.569 for the training group and 0.799 for the testing group. At 3 years, the values were 0.773 and 0.648, and at 5 years, they were 0.757 and 0.673. These results indicate that the risk model demonstrated good prognostic accuracy and was well-constructed for further studies.
Fig. 6.
Validation of the prognostic model. (A) The KM survival curves of different risk groups in training and (B) testing groups. (C) The ROC curves for the training and (D) testing groups. (E–J) Ranked dot plots (E and F), scatter plots (G and H), and heatmaps (I and J) showed the distribution of risk score, survival status, and gene expression in training and testing groups.
3.6. Establishment and validation of the nomogram
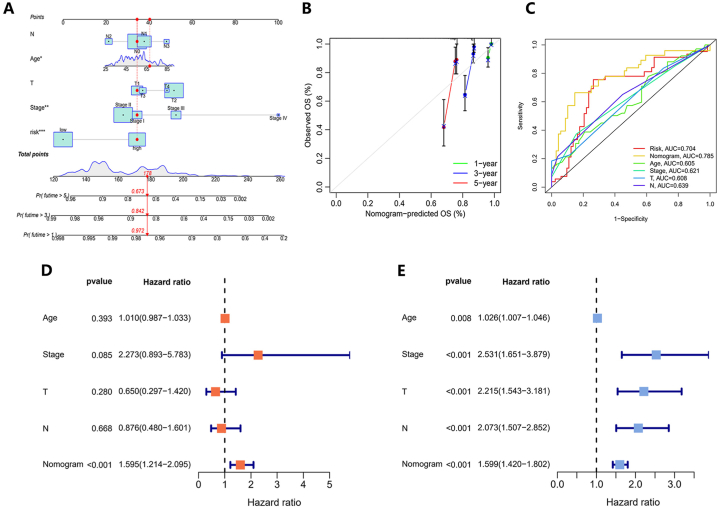
Utilizing five clinical features including age, stage, stage T, and stage N, we developed a personalized score nomogram to perform comprehensive risk assessments for patients based on the constructed model. The clinical prognosis worsened with an elevation in the cumulative score for each prognostic index (Fig. 7A). Furthermore, the calibration plot suggested that the nomogram had the capability to make precise predictions (Fig. 7B). Based on the nomogram, ROC curves of each parameter were established and the AUC values were calculated (Fig. 7C). The AUC scores of risk score, nomogram, age, stage, stage T, and stage N were 0.704, 0.785, 0.605, 0.621, 0.608, and 0.639 respectively. The AUC score of the nomogram was the highest, implying that based on this prognostic model, using this nomogram to predict OS is better than any other factor. Additionally, the result revealed that the nomogram serves as a more effective independent prognostic marker for patients compared to factors such as age, overall stage, T stage, and N stage, as determined by both univariate and multivariate Cox analyses (Fig. 7D and E).
Fig. 7.
Development and confirmation of the nomogram. (A) The nomogram was designed to estimate the survival outcomes of TNBC patients utilizing risk score and clinical factors. (B) The calibration plot showed the capability of the nomogram to make precise predictions. (C) ROC curve analysis for the nomogram and additional clinical characteristics. (D) The multivariate and (E) univariateCox analyses indicated that the nomogram could be independently utilized for predicting patient outcomes.
3.7. Independent prognostic factors of the risk model
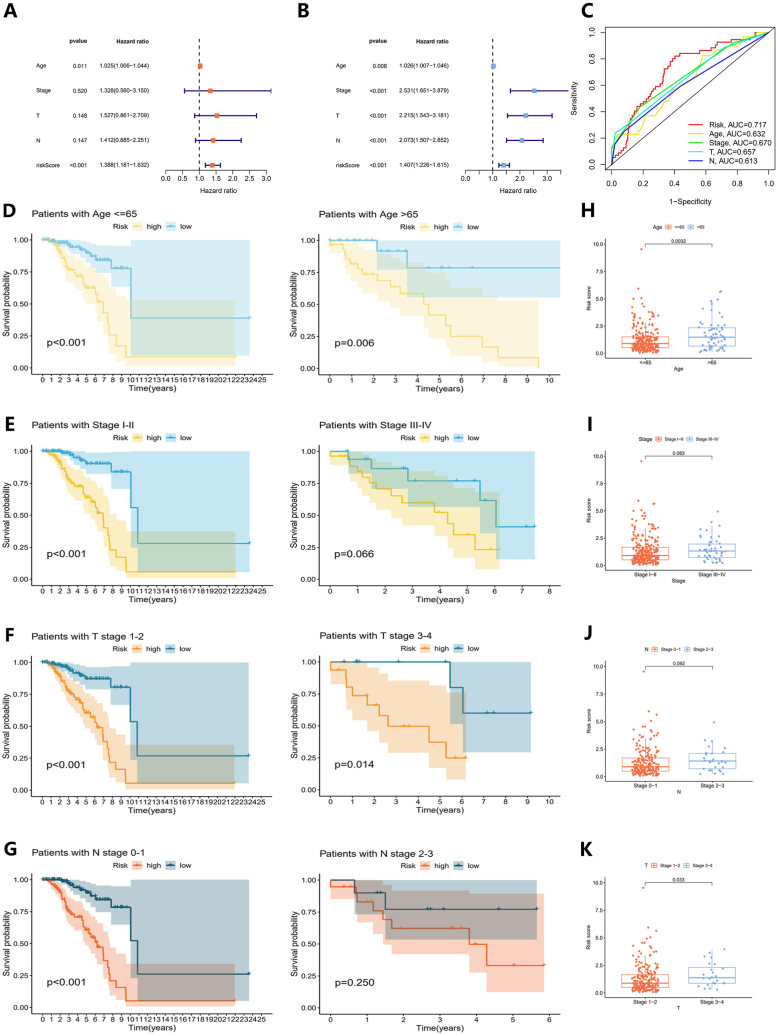
We further identified clinical features that could be considered as standalone predictors of prognosis. Based on the results from both univariate and multivariate analyses, age and the risk score were identified as significant independent prognostic factors (P < 0.05) (Fig. 8A and B). The AUC scores of risk score, age, stage, stage T, and stage N were 0.717, 0.632, 0.670, 0.657, and 0.613 respectively (Fig. 8C). To assess the model's general capability, stratification analyses were also performed. These showed that patients with lower risk scores had notably longer OS times compared to those with higher risk scores, across various age groups and stages of TNBC (Fig. 8D, E, 8F, and 8G). Additionally, a series of boxplots revealed positive correlations between tumor progression, patient age, and increasing risk scores (Fig. 8H, I, 8J, and 8K).
Fig. 8.
Independent prognostic analysis of the prognostic model. (A) The multivariate and (B) univariate Cox analyses of clinical features such as the risk score, age, stage, stage T, and stage N. (C) The ROC curves for these clinical features. (D-G) Differences in OS between high and low-risk groups in (D) age groups, (E) stage groups, (F) T stage groups, and (G) N stage groups. (H-K) The distribution of patient risk scores in (H) age groups, (I) stage groups, (J) T stage groups, and (K) N stage groups.
3.8. Evaluation of immune activities and immunotherapy between risk groups
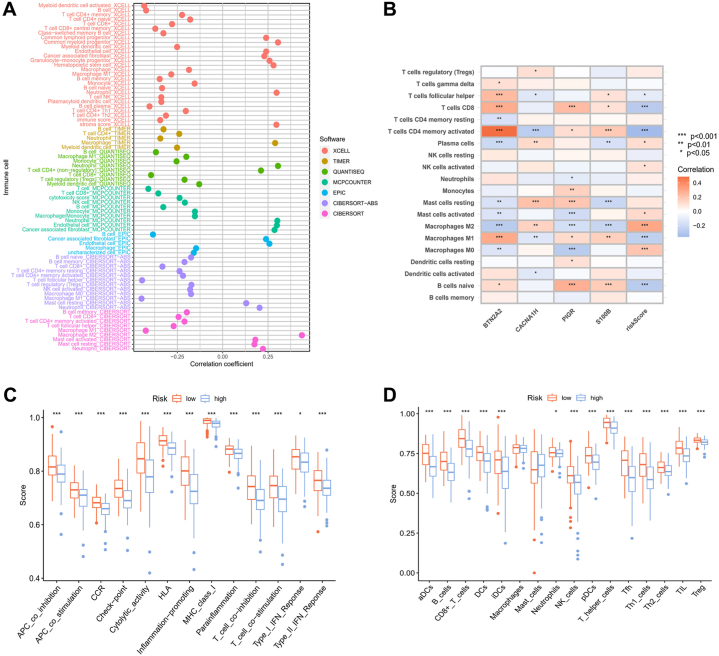
Using several computational techniques, we studied the impact of risk levels on tumor-infiltrating immune cells (Figs. 9A and S4). The high-risk group displayed a more significant association with endothelial cells, mast cells, neutrophils, and cancer-associated fibroblasts. Furthermore, through correlation analyses of the expression levels of BTN2A2, PIGR, CACNA1H, and S100B with immune cells, we revealed significant associations between particular genes and specific immune cell types (Fig. 9B). To illustrate the relationship between risk groups and immune cells along with their functions, the ssGSEA approach was used to calculate enrichment scores (Fig. 9C and D). The results indicated that the lower risk group exhibited higher scores in immune-associated activities and greater abundance of immune cells.
Fig. 9.
Immune activities between two risk groups. (A) The relationship between risk scores and populations of immune cells was assessed through multiple algorithms. (B) The connection between immune cell quantities and the expression patterns of four genes. (C) Variations in immune functions and (D) the infiltration of immune cells in different risk groups. (∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.)
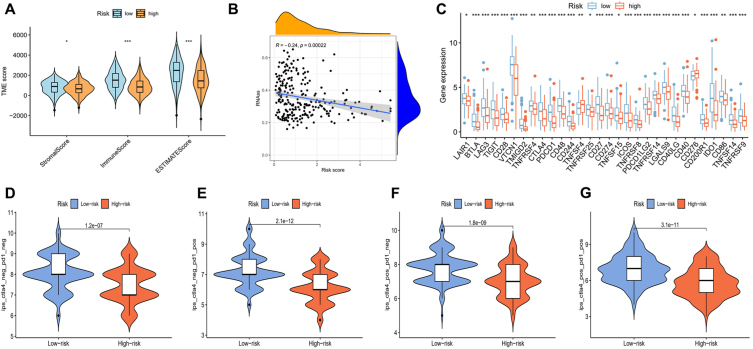
TME is a multifaceted and dynamic system where immune cells, inflammatory cells, and immune checkpoints engage with one another, influencing tumor growth, metastasis, and treatment response [25]. TME scores were calculated for the two risk groups, unveiling markedly elevated stromal, immune, and ESTIMATE scores associated with lower risk scores (Fig. 10A). The stemness features of cancer are influenced by the tumor immune microenvironment, contributing to diverse bio-functions in tumor cells [26]. In TNBC, the correlation between RNA stemness scores (RNAss) and risk scores showed a significant negative association, suggesting that patients with lower risk scores exhibit more significant stem cell characteristics (Fig. 10B). Additionally, we performed an analysis of the expression levels of checkpoint-related genes within the two risk categories, noting an increased expression for most of these genes in the low-risk group (Fig. 10C). Through immune checkpoint pathways, cancer cells can masquerade as normal cells within the human body [27]. Therefore, blocking these pathways is a highly promising approach for achieving anti-cancer immunity. We explored the immune checkpoint inhibitors treatment response represented by cytotoxic T-lymphocyte associated protein 4 (CTLA-4)/programmed cell death protein 1 (PD-1) inhibitors. The results indicated significantly better immunotherapeutic effects in patients with low-risk scores (Fig. 10D, E, 10F, and 10G).
Fig. 10.
Cancer immunotherapy efficacy between two risk groups. (A) TME scores between different risk groups. (∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.) (B) Stemness scores between different risk groups. (C) Differential expression of checkpoint-related genes between different risk groups. (∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.) (D, E, F, and G) The correlation between treatment using immune checkpoint inhibitors and distinct risk groups.
3.9. The mutation, biological function, and drug susceptibility analysis between risk groups
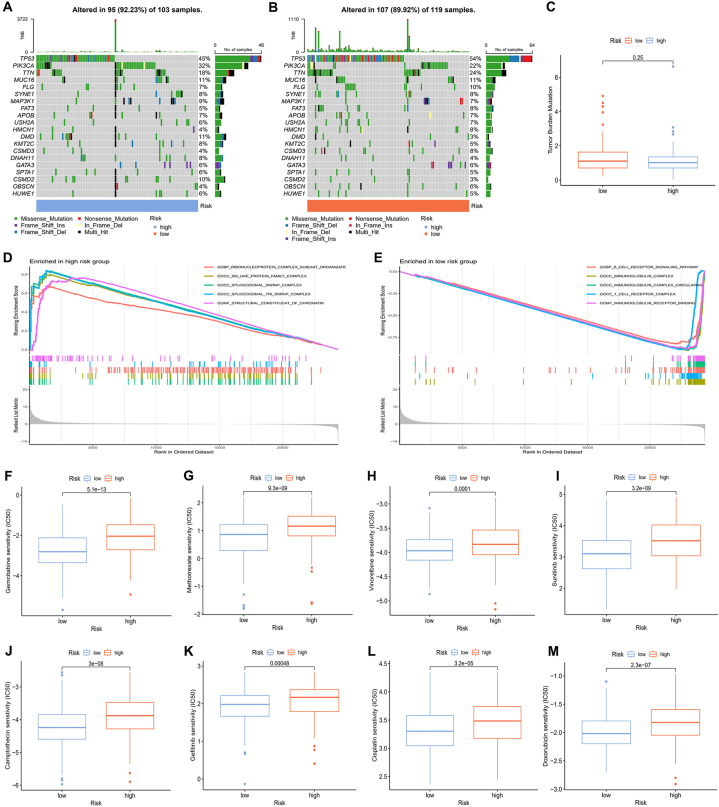
The analysis of somatic mutations was conducted for the two different risk score groups (Fig. 11A and B). The mutation frequency of TP53 was notably the highest, especially in patients with lower risk levels. Furthermore, genes such as TTN and FLG displayed increased mutation rates in patients with lower risk scores, whereas PIK3CA, MAP3K1, and DMD were more frequently mutated in patients with higher risk scores. TMB was also calculated in these two groups, showing no significant associations with risk scores (Fig. 11C). To further explore the enrichment of the biological functions in different risk groups, the GSEA analysis has been performed (Fig. 11D and E; Tables S12 and S13). Based on normalized enrichment scores and P-values, we selected the top five most enriched biological functions in each group respectively. The majority of functions enriched in the lower risk cohort were associated with immune activities, whereas those in the higher risk cohort were primarily linked to chromatin structures. We also investigated the relationship between drug sensitivity and different risk levels. The results of comparing IC50 values between the two risk groups suggested that chemotherapy drugs, including camptothecin, cisplatin, doxorubicin, gefitinib, gemcitabine, methotrexate, sunitinib, and vinorelbine, were all more effective for patients with lower risk scores (Fig. 11F, G, 11H, 11I, 11J, 11K, 11L, and 11M).
Fig. 11.
Differences in mutations, biological functions, and response to medication between risk groups. (A) Frequency of mutations in DEGs within patients with high-risk scores and (B) low-risk scores. (C) The correlation between TMB scores and risk groups. (D) The GO enrichment analyses in high- and (B) low-risk samples. (F-M) Sensitivity to (F) gemcitabine, (G) methotrexate, (H) vinorelbine, (I) sunitinib, (J) camptothecin, (K) gefitinib, (L) cisplatin, and (M) doxorubicin in the different risk groups.
3.10. Evaluation of the four model genes
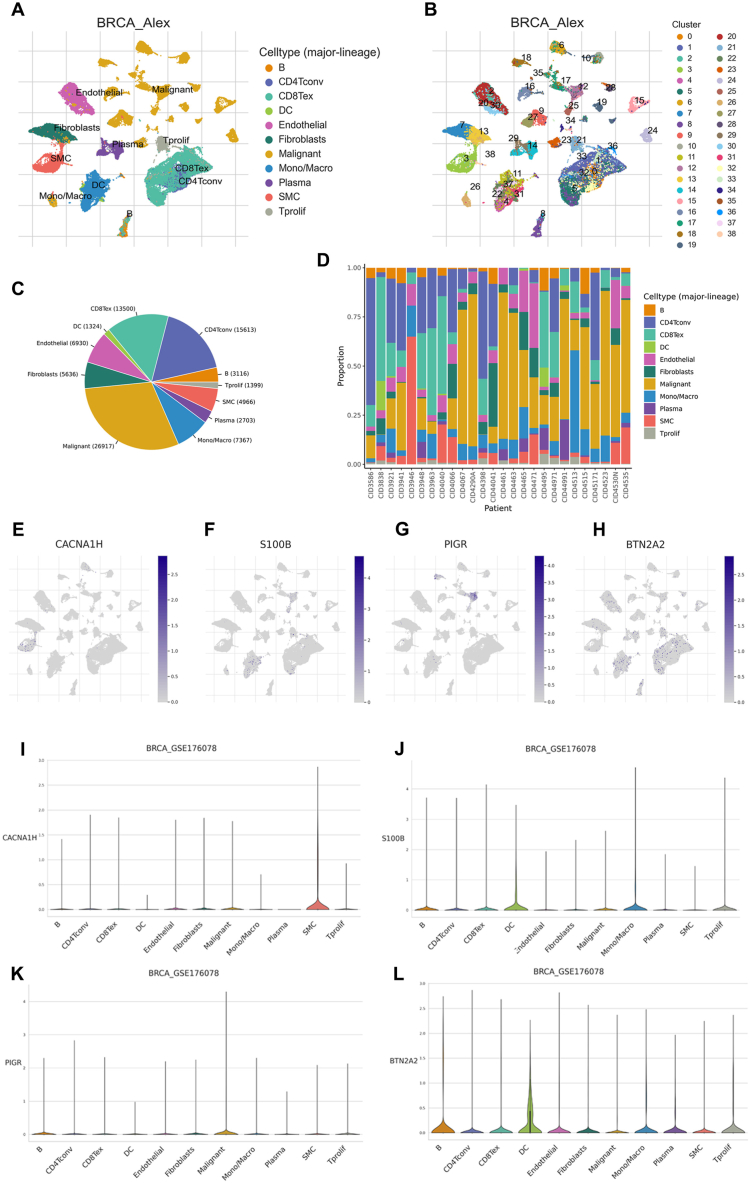
We employed the GSE176078 dataset from the TISCH platform to investigate the expression characteristics of the four genes included in the prognostic model across different cell clusters in breast cancer. Within the GSE176078 dataset, 11 principal cell types and 39 major cell populations have been identified (Fig. 12A and B). The ratios of various cell types in both aggregate samples and individual patients were also depicted (Fig. 12C and D). Results showed that S100B and BTN2A2 were predominantly expressed in dendritic cells (DC), PIGR exhibited main expression in malignant cells, and CACNA1H was primarily expressed in smooth muscle cells (SMC) (Fig. 12E, F, 12G, 12H, 12I, 12J, 12K, and 12L).
Fig. 12.
Analysis at the single-cell level of genes used for model development. (A) Cell types and (B) cell populations in the GSE176078 dataset. (C) The ratios of various cell types in both aggregate samples and (D) individual patients. (E-L) The distribution of CACNA1H (E and I), S100B (F and J), PIGR (G and K), and BTN2A2(H and L) in different cell types.
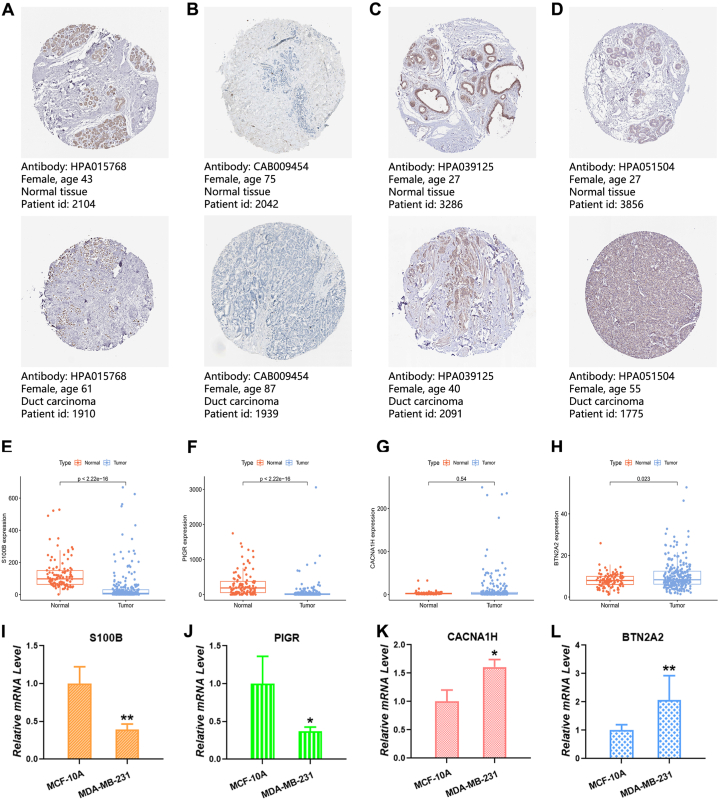
Moreover, we delved into the immunohistochemical data for these four genes in breast cancer using the HPA database. S100B (Fig. 13A) and PIGR (Fig. 13B) exhibited heightened expression in normal tissues, while BTN2A2 (Fig. 13C) and CACNA1H (Fig. 13D) exhibited heightened expression in tumor tissues. Examining these genes in TNBC patients from TCGA and GEO databases revealed increased levels of S100B (Fig. 13E) and PIGR (Fig. 13F) in normal samples, with BTN2A2 (Fig. 13G) displaying increased levels in tumor samples. Although CACNA1H (Fig. 13H) also exhibited increased expression in tumor samples, it did not reach statistical significance. Lastly, we assessed the relative mRNA levels of these four genes using qRT-PCR, indicating higher mRNA levels of S100B and PIGR in MCF-10A cells and higher mRNA levels of BTN2A2 and CACNA1H in MDA-MB-231 and MDA-MB-468 cells (Fig. 13I, J, 13K, and 13L).
Fig. 13.
Evaluation of genes in normal and tumor samples. (A-D) Immunohistochemical analysis of (A) S100B, (B) PIGR, (C) CACNA1H, and (D) BTN2A2 in normal and tumor tissues. (E-H) Variations in gene expression between normal and TNBC tissues. (I-L) Analysis of the relative mRNA levels of the four genes through qRT-PCR. (∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.)
4. Discussion
TNBC represents a breast cancer subtype renowned for its unfavorable prognostic outcome, characterized by low survival rates, high recurrence rates, significant metastatic potential, and limited therapeutic options [28]. Consequently, numerous research endeavors have been devoted to developing prognostic models, aiming to provide solutions for disease management and risk assessment in TNBC. However, as our comprehension of the pathogenesis and progression of this disease deepens, there arises a need for further investigation into the role and predictive potential of emerging mechanisms in TNBC prognosis. PANoptosis, an innovative form of cell death recently identified, incorporates key elements of apoptosis, pyroptosis, and necroptosis. Recent reports have highlighted the importance of this cellular death mechanism due to its pivotal involvement in both cancer progression and the effectiveness of anti-tumor therapies. Notably, several targets within PANoptosis exhibit close ties to breast cancer, suggesting its potential as a mechanistic pathway for both therapeutic interventions and prognostic assessments in this specific cancer subtype [29]. However, the specific role of PANoptosis in TNBC prognosis necessitates further investigation.
In this study, we established a PANoptosis-related prognostic model in TNBC, utilizing 19 PRGs sourced from recent studies. Our initial step aimed to comprehend the variations of these PRGs in TNBC, involving the evaluation of their differential expression and genetic mutation profiles. By classifying all samples into two separate PANoptosis clusters according to the expression levels of these 19 PRGs, we further investigated disparities in immune infiltration and biological functions between these clusters. The findings revealed predominant upregulation of immune infiltration and biological functions in cluster B, with a significantly superior OS compared to cluster A. Additionally, we identified 1054 DEGs between these clusters and conducted KEGG and GO analyses on these genes. To comprehend the relationship between PANoptosis and these DEGs, we categorized the patients into two gene clusters according to these genes. The statistically significant OS difference between the two gene clusters revealed a strong relationship between PANoptosis and gene clusters. As a result, we developed a predictive model using these DEGs to perform risk stratification and predict the OS in TNBC patients. The final selection for model construction included four genes: BTN2A2, CACNA1H, PIGR, and S100B.
Utilizing the HPA database, we identified that S100B and PIGR displayed lower expression levels in breast cancer tumor tissues, while BTN2A2 and CACNA1H exhibited higher expression. These findings align coherently with the results of our further analyses conducted using the TCGA and GEO databases, and they are supported by our PCR experiment validations of the expression of these four genes in TNBC. Furthermore, we identified the distribution of the four genes across various cell types, including dendritic cells, malignant cells, and smooth muscle cells, which are particularly associated with immune responses and tumor metastasis [30,31]. Recent studies have highlighted the significance of both PIGR and BTN2A2 as integral components of the immune system. PIGR, part of the immunoglobulin superfamily, is vital for mucosal defense by mediating transcytosis. Recognized as a prognostic marker, PIGR contributes to an improved breast cancer prognosis by enhancing tumor immunity and exerting tumor-suppressive effects [32]. BTN2A2 is thought to act as a co-inhibitory molecule, regulating T cell-mediated immunity by dampening T cell activation [33,34]. Its expression level is strongly associated with the prognosis of breast cancer, with its functions potentially related to tumor immune infiltration, as well as the migratory and proliferative abilities of cancer cells [35,36]. The presence of metastasis typically signifies an advanced cancer stage and is often linked with a worse prognosis. S100 proteins vary in expression across different cancers and are strongly linked to tumor progression. Among the S100 family, S100B is notably associated with cancer metastasis and serves as a predictive marker for metastatic development in malignant melanoma, breast cancer, and lung cancer [[37], [38], [39]]. CACNA1H encodes the T-type calcium channel, which is involved in various cellular processes, such as cell proliferation and apoptotic cell death [40]. In breast cancer, the expression of the CACNA1H gene is strongly correlated with genes involved in epithelial-mesenchymal transition, indicating its potential role in tumor metastasis [41]. In summary, the four genes chosen for model construction are vital in breast cancer progression and prognosis, particularly concerning tumor immunity and metastasis.
Upon comparing the survival rates and risk scores among the testing group, training group, and the entire patient cohort, it became evident that patients categorized as higher risk consistently exhibited lower survival rates. Furthermore, the risk associated with the disease was observed to escalate with its progression. These findings provide robust support for the efficacy of our constructed model. With the model, we constructed a nomogram to provide a more straightforward approach for assessing patient risk. Additionally, we identified independent prognostic factors significantly impacting patients with TNBC. The results demonstrate that the nomogram we developed proves to be a more effective prognostic tool compared to other individual factors for forecasting outcomes. Meanwhile, age and risk score were considered as potential key prognostic factors in TNBC, offering guidance for clinical decisions.
Immunotherapy aims to overcome tumor-induced immunosuppression, enabling the immune system to identify and destroy cancer cells more effectively [42]. Increasing research indicates that the TME is pivotal in modulating immunotherapy outcomes by suppressing or enhancing immune responses [43]. Our findings indicated a significantly elevated TME score in patients with lower risk scores, implying that they may have a more robust immune response and be better suited for immunotherapy. The immune cells within the TME are crucial in tumorigenesis, capable of either hindering or facilitating tumor progression. Our data indicate a negative association among the majority of analyzed immune cells and risk scores in TNBC, implying that an increase and activation of immune cells may be associated with a decreased risk. The risk score showed an inverse relationship with T cells, naive B cells, and M1 macrophages (M1-TAMs), and a positive correlation with plasma cells, mast cells, NK cells, and M2 macrophages (M2-TAMs). T cells are essential for targeting and eliminating tumor cells, their recruitment and activation in tumors are critical for potent antitumor responses [44]. Although naive B cells do not directly participate in immune responses, they can be activated and differentiated under appropriate conditions to contribute to tumor suppression [45]. Mast cells contribute to tumor development and metastasis by the secretion of growth factors and pro-inflammatory cytokines, thereby promoting tumor growth, angiogenesis, and tissue remodeling [46]. In most cases, NK cells exert antitumor effects. However, the function of NK cells may be suppressed in the TME, leading to a reduction in their cell toxicity and consequently resulting in adverse prognoses [47]. Plasma cells might additionally foster tumor growth through the release of immunosuppressive cytokines that inhibit or misdirect the response of effector T cells. Furthermore, different phenotypes of TAMs have unique functions in modulating tumour growth [48]. Recent research in TNBC revealed a favorable prognosis associated with M1-TAMs, while an unfavorable prognosis is correlated with M2-TAMs [49]. These findings align with our conclusions, providing additional evidence for the robustness and precision of our constructed model. Immune checkpoints are crucial in immunotherapy, with checkpoint blockade emerging as an important anti-cancer strategy. We examined the relationship between risk levels and immune checkpoint gene expression, finding that patients with lower risk exhibited significantly higher expression compared to those at higher risk. This suggests that individuals with lower risk scores possess increased sensitivity to cancer immunotherapy targeting immune checkpoints. Among immune checkpoints, CTLA-4 and PD-1 have been extensively explored and are known to negatively regulate T-cell immune function at various stages of T-cell activation [50]. Our study findings reveal an inverse correlation between prognostic risk and CTLA-4 and PD-1 levels, indicating the potential effectiveness of CTLA-4 and PD-1 blockade as strategies for treating TNBC.
Cancer cell stemness plays a critical role in driving tumor development and often serves as the foundation for its survival [51]. Typically, cancer stemness correlates positively with poor survival, metastasis, and drug resistance [[52], [53], [54]]. However, our research suggests a negative correlation between RNAss and risk scores in TNBC patients. Several factors could account for this finding. Firstly, treatment received by our selected samples might play a role. Patients with higher risk scores may undergo more aggressive treatments, impacting the population and activity of stem cells. Conversely, those with lower risk scores often undergo lighter treatment regimens, potentially enhancing the survival of stem cells. This disparity may also be ascribed to the restricted sample size in our research.
In the context of cancer, TMB represents the mutation frequency within tumor cells. A greater number of mutations increases the probability of the immune system detecting and eliminating the tumor [55]. Our analysis suggested that the TMB in low-risk patients was slightly elevated, yet this difference did not demonstrate statistical significance. This may be ascribed to the restricted sample size in our research. TP53 stands out as the most frequently mutated gene in TNBC. Its mutation potentially enhanced immunogenic activity, leading to adverse prognostic outcomes [56]. Our research revealed that TP53 mutations were the most common in both the two risk score levels, with the mutation rate significantly elevated in patients with higher risk scores, consistent with findings reported in earlier studies. Furthermore, we conducted the GSEA analysis to clarify the biological roles related to the risk evaluation. The analysis revealed that lower risk scores correlated with various immune signaling pathways, underscoring the potential importance of immune-based therapies for TNBC patients with lower risk. Ultimately, an investigation into the therapeutic efficacy of different chemotherapy agents across distinct risk groups was conducted, with the objective of offering more tailored treatment options for TNBC patients.
Our findings revealed a noteworthy correlation between the PANoptosis prognostic model, immune function, and responses to immunotherapy. Despite the successful development of the PANoptosis-based prognostic model, our study has areas for improvement. Firstly, the limited data source with a small sample size may have overlooked some crucial prognostic factors. Secondly, our current research heavily relies on the exploration of existing data from public databases, necessitating additional experiments to further validate our research findings.
5. Conclusion
Our study analyzed the expression and mutation characteristics of PNGs, employing these genes to initially formulate a model specific to PANoptosis in TNBC. Subsequently, we affirmed the reliability of our constructed prognostic model and developed a nomogram to enhance risk assessment. Following the successful development and verification of the PANoptosis-based predictive model, we investigated the relationships between risk stratification and immunotherapy, gene mutations, biological functions, and responsiveness to chemotherapy drugs. These discoveries may offer more precise guidance for tailoring personalized treatment strategies in patients with TNBC.
Funding
This research was funded by Innovation and Technology Fund-Mainland-Hong Kong Joint Funding Scheme (MHP/010/20), Shenzhen Science and Technology Innovation Commission (JCYJ20220531090802006), Research Centre for Chinese Medicine Innovation of The Hong Kong Polytechnic University (E-ABCT-BBBB-1 and E-ABCT-BBBB-3), The Hong Kong Polytechnic University Shenzhen Research Institute Life Science Research Start-up Fund (I2023A008), Traditional Chinese Medicine Bureau of Guangdong Province (202404101916049020).
Availability of data and materials
Clinical data for this research were obtained from the GEO (https://www.ncbi.nlm.nih.gov/geo/) and TCGA (https://portal.gdc.cancer.gov/) databases and can be downloaded from the provided websites. Datasets and custom scripts are available upon request.
CRediT authorship contribution statement
Jia-Wen Chen: Writing – original draft, Visualization, Validation, Software, Formal analysis, Data curation, Conceptualization. Rui-Hong Gong: Writing – original draft, Validation, Investigation, Data curation. Chi Teng: Investigation, Data curation, Conceptualization. Yu-Shan Lin: Validation, Investigation, Data curation. Li-Sha Shen: Investigation. Zesi Lin: Investigation. Sibao Chen: Writing – review & editing, Validation, Supervision, Investigation, Funding acquisition. Guo-Qing Chen: Writing – original draft, Visualization, Validation, Investigation, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e38732.
Contributor Information
Sibao Chen, Email: sibao.chen@polyu.edu.hk.
Guo-Qing Chen, Email: guoqing.chen@polyu.edu.hk.
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
References
- 1.Chhikara B.S., Parang K. Global Cancer Statistics 2022: the trends projection analysis. Chemical Biology Letters. 2023;10 451-451. [Google Scholar]
- 2.Bou Zerdan M., Ghorayeb T., Saliba F., Allam S., Bou Zerdan M., Yaghi M., Bilani N., Jaafar R., Nahleh Z. Triple negative breast cancer: updates on classification and treatment in 2021. Cancers. 2022;14:1253. doi: 10.3390/cancers14051253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tong L., Yu X., Wang S., Chen L., Wu Y. Research progress on molecular subtyping and modern treatment of triple-negative breast cancer. Breast Cancer. 2023:647–658. doi: 10.2147/BCTT.S426121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen J.-W., Chen S., Chen G.-Q. 2023. Recent Advances in Natural Compounds Inducing Non-apoptotic Cell Death for Anticancer Drug Resistance. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galluzzi L., Vitale I., Aaronson S.A., Abrams J.M., Adam D., Agostinis P., Alnemri E.S., Altucci L., Amelio I., Andrews D.W. Molecular mechanisms of cell death: recommendations of the nomenclature committee on cell death 2018. Cell Death Differ. 2018;25:486–541. doi: 10.1038/s41418-017-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malireddi R.S., Kesavardhana S., Kanneganti T.-D. ZBP1 and TAK1: master regulators of NLRP3 inflammasome/pyroptosis, apoptosis, and necroptosis (PAN-optosis) Front. Cell. Infect. Microbiol. 2019;9:406. doi: 10.3389/fcimb.2019.00406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun X., Yang Y., Meng X., Li J., Liu X., Liu H. PANoptosis: mechanisms, biology, and role in disease. Immunol. Rev. 2024;321:246–262. doi: 10.1111/imr.13279. [DOI] [PubMed] [Google Scholar]
- 8.Cai H., Lv M., Wang T. PANoptosis in cancer, the triangle of cell death. Cancer Med. 2023 doi: 10.1002/cam4.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inoue S., Browne G., Melino G., Cohen G. Ordering of caspases in cells undergoing apoptosis by the intrinsic pathway. Cell Death Differ. 2009;16:1053–1061. doi: 10.1038/cdd.2009.29. [DOI] [PubMed] [Google Scholar]
- 10.Karlowitz R., van Wijk S.J. Surviving death: emerging concepts of RIPK3 and MLKL ubiquitination in the regulation of necroptosis. FEBS J. 2023;290:37–54. doi: 10.1111/febs.16255. [DOI] [PubMed] [Google Scholar]
- 11.Zhang R., Gou W., Yi P., Qin Z., Zhu D., Jia J., Liu L., Jiang X., Feng J. Tetracaine hydrochloride induces macrophage pyroptosis through caspase-1/11-GSDMD signaling pathways. Exp. Ther. Med. 2023;26:1–13. doi: 10.3892/etm.2023.12127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cai H., Lv M., Wang T. PANoptosis in cancer, the triangle of cell death. Cancer Med. 2023;12:22206–22223. doi: 10.1002/cam4.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao J., Xiong A., Liu J., Li X., Wang J., Zhang L., Liu Y., Xiong Y., Li G., He X. PANoptosis: bridging apoptosis, pyroptosis, and necroptosis in cancer progression and treatment. Cancer Gene Ther. 2024:1–14. doi: 10.1038/s41417-024-00765-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiong Y. The emerging role of PANoptosis in cancer treatment. Biomed. Pharmacother. 2023;168 doi: 10.1016/j.biopha.2023.115696. [DOI] [PubMed] [Google Scholar]
- 15.Wang X., Sun R., Chan S., Meng L., Xu Y., Zuo X., Wang Z., Hu X., Han Q., Dai L., Bai T., Yu Z., Wang M., Yang W., Zhang H., Chen W. PANoptosis-based molecular clustering and prognostic signature predicts patient survival and immune landscape in colon cancer. Front. Genet. 2022;13 doi: 10.3389/fgene.2022.955355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang B., Huang B., Zhang X., Li S., Zhu J., Chen X., Song H., Shang D. PANoptosis-related molecular subtype and prognostic model associated with the immune microenvironment and individualized therapy in pancreatic cancer. Front. Oncol. 2023;13 doi: 10.3389/fonc.2023.1217654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Z., Zhang F., Pang P., Li Y., Chen X., Sun S., Bian Y. Identification of PANoptosis-relevant subgroups to evaluate the prognosis and immune landscape of patients with liver hepatocellular carcinoma. Front. Cell Dev. Biol. 2023;11 doi: 10.3389/fcell.2023.1210456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pu S., Zhou Y., Xie P., Gao X., Liu Y., Ren Y., He J., Hao N. Identification of necroptosis-related subtypes and prognosis model in triple negative breast cancer. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.964118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sha S., Si L., Wu X., Chen Y., Xiong H., Xu Y., Liu W., Mei H., Wang T., Li M. Prognostic analysis of cuproptosis-related gene in triple-negative breast cancer. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.922780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan C., Liu Q., Jia R. Construction and validation of a prognostic risk model for triple-negative breast cancer based on autophagy-related genes. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.829045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu P., Ke Z.R., Chen J.X., Li S.J., Ma T.L., Fan X.L. Advances in mechanism and regulation of PANoptosis: prospects in disease treatment. Front. Immunol. 2023;14 doi: 10.3389/fimmu.2023.1120034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin J.F., Hu P.S., Wang Y.Y., Tan Y.T., Yu K., Liao K., Wu Q.N., Li T., Meng Q., Lin J.Z., Liu Z.X., Pu H.Y., Ju H.Q., Xu R.H., Qiu M.Z. Phosphorylated NFS1 weakens oxaliplatin-based chemosensitivity of colorectal cancer by preventing PANoptosis. Signal Transduct. Targeted Ther. 2022;7:54. doi: 10.1038/s41392-022-00889-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pandian N., Kanneganti T.D. PANoptosis: a unique innate immune inflammatory cell death modality. J. Immunol. 2022;209:1625–1633. doi: 10.4049/jimmunol.2200508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Samir P., Malireddi R.K.S., Kanneganti T.D. The PANoptosome: a deadly protein complex driving pyroptosis, apoptosis, and necroptosis (PANoptosis) Front. Cell. Infect. Microbiol. 2020;10:238. doi: 10.3389/fcimb.2020.00238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bilotta M.T., Antignani A., Fitzgerald D.J. Managing the TME to improve the efficacy of cancer therapy. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.954992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aponte P.M., Caicedo A. Stemness in cancer: stem cells, cancer stem cells, and their microenvironment. Stem Cell. Int. 2017;2017 doi: 10.1155/2017/5619472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li B., Chan H.L., Chen P. Immune checkpoint inhibitors: basics and challenges. Curr. Med. Chem. 2019;26:3009–3025. doi: 10.2174/0929867324666170804143706. [DOI] [PubMed] [Google Scholar]
- 28.de Ruijter T.C., Veeck J., de Hoon J.P., van Engeland M., Tjan-Heijnen V.C. Characteristics of triple-negative breast cancer. J. Cancer Res. Clin. Oncol. 2011;137:183–192. doi: 10.1007/s00432-010-0957-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu X., Miao M., Sun J., Wu J., Qin X. PANoptosis: a potential new target for programmed cell death in breast cancer treatment and prognosis. Apoptosis. 2023 doi: 10.1007/s10495-023-01904-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pittet M.J., Di Pilato M., Garris C., Mempel T.R. Dendritic cells as shepherds of T cell immunity in cancer. Immunity. 2023 doi: 10.1016/j.immuni.2023.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeClerck Y.A., Yean T.-D., Chan D., Shimada H., Langley K.E. Inhibition of tumor invasion of smooth muscle cell layers by recombinant human metalloproteinase inhibitor. Cancer Res. 1991;51:2151–2157. [PubMed] [Google Scholar]
- 32.Sun Z., Tan B., Dong K., Pu Q., Gao H., Li P., Zhao G. 2023. PIGR Predicts Good Clinical Outcomes and Plays a Tumor Suppressor Role in the Development of Breast Cancer via Enhancing Tumor Immunity. [Google Scholar]
- 33.Smith I.A., Knezevic B.R., Ammann J.U., Rhodes D.A., Aw D., Palmer D.B., Mather I.H., Trowsdale J. BTN1A1, the mammary gland butyrophilin, and BTN2A2 are both inhibitors of T cell activation. J. Immunol. 2010;184:3514–3525. doi: 10.4049/jimmunol.0900416. [DOI] [PubMed] [Google Scholar]
- 34.Ren H., Li S., Liu X., Li W., Hao J., Zhao N. Multi-omics analysis of the expression and prognostic value of the butyrophilins in breast cancer. J. Leukoc. Biol. 2021;110:1181–1195. doi: 10.1002/JLB.5MA0321-158RR. [DOI] [PubMed] [Google Scholar]
- 35.Wang H., Pu S., Xu H., Yang L., Shao L., Chen X., Huang X., Pu J. BTN2A2, a new biomarker and therapeutic target for glioma. Aging. 2023;15:10996–11011. doi: 10.18632/aging.205039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y., Zhao N., Zhang X., Li Z., Liang Z., Yang J., Liu X., Wu Y., Chen K., Gao Y. Bibliometrics analysis of butyrophilins as immune regulators [1992–2019] and implications for cancer prognosis. Front. Immunol. 2020;11:1187. doi: 10.3389/fimmu.2020.01187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yen M.-C., Huang Y.-C., Kan J.-Y., Kuo P.-L., Hou M.-F., Hsu Y.-L. S100B expression in breast cancer as a predictive marker for cancer metastasis. Int. J. Oncol. 2018;52:433–440. doi: 10.3892/ijo.2017.4226. [DOI] [PubMed] [Google Scholar]
- 38.Harpio R., Einarsson R. S100 proteins as cancer biomarkers with focus on S100B in malignant melanoma. Clin. Biochem. 2004;37:512–518. doi: 10.1016/j.clinbiochem.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 39.Chen L., Hu X., Wu H., Jia Y., Liu J., Mu X., Wu H., Zhao Y. Over-expression of S100B protein as a serum marker of brain metastasis in non-small cell lung cancer and its prognostic value. Pathol. Res. Pract. 2019;215:427–432. doi: 10.1016/j.prp.2018.11.011. [DOI] [PubMed] [Google Scholar]
- 40.Sharma S., Wu S.-Y., Jimenez H., Xing F., Zhu D., Liu Y., Wu K., Tyagi A., Zhao D., Lo H.-W. Ca2+ and CACNA1H mediate targeted suppression of breast cancer brain metastasis by AM RF EMF. EBioMedicine. 2019;44:194–208. doi: 10.1016/j.ebiom.2019.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ibrahim F.A.R., Hussein Z.U.N., Yousef A.I., Abd El Moneim N.A., Hussein A.M., Ahmed A.F.M., Ragab N.M., Al-Masry O. Insights on possible interplay between epithelial-mesenchymal transition and T-type voltage gated calcium channels genes in metastatic breast carcinoma. Heliyon. 2022;8 doi: 10.1016/j.heliyon.2022.e10160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ye F., Dewanjee S., Li Y., Jha N.K., Chen Z.-S., Kumar A., Vishakha, Behl T., Jha S.K., Tang H. Advancements in clinical aspects of targeted therapy and immunotherapy in breast cancer. Mol. Cancer. 2023;22:105. doi: 10.1186/s12943-023-01805-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang H., Qiao J., Fu Y.-X. Immunotherapy and tumor microenvironment. Cancer Lett. 2016;370:85–90. doi: 10.1016/j.canlet.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Molon B., Calì B., Viola A. T cells and cancer: how metabolism shapes immunity. Front. Immunol. 2016;7:20. doi: 10.3389/fimmu.2016.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sarvaria A., Madrigal J.A., Saudemont A. B cell regulation in cancer and anti-tumor immunity. Cell. Mol. Immunol. 2017;14:662–674. doi: 10.1038/cmi.2017.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Molfetta R., Paolini R. The controversial role of intestinal mast cells in colon cancer. Cells. 2023;12:459. doi: 10.3390/cells12030459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang W., Zhao Z., Li F. Natural killer cell dysfunction in cancer and new strategies to utilize NK cell potential for cancer immunotherapy. Mol. Immunol. 2022;144:58–70. doi: 10.1016/j.molimm.2022.02.015. [DOI] [PubMed] [Google Scholar]
- 48.Zhang Q., Li H., Mao Y., Wang X., Zhang X., Yu X., Tian J., Lei Z., Li C., Han Q. Apoptotic SKOV3 cells stimulate M0 macrophages to differentiate into M2 macrophages and promote the proliferation and migration of ovarian cancer cells by activating the ERK signaling pathway. Int. J. Mol. Med. 2020;45:10–22. doi: 10.3892/ijmm.2019.4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen Z., Wu J., Wang L., Zhao H., He J. Tumor-associated macrophages of the M1/M2 phenotype are involved in the regulation of malignant biological behavior of breast cancer cells through the EMT pathway. Med. Oncol. 2022;39:83. doi: 10.1007/s12032-022-01670-7. [DOI] [PubMed] [Google Scholar]
- 50.Weber J. Immune checkpoint proteins: a new therapeutic paradigm for cancer—preclinical background: CTLA-4 and PD-1 blockade, Seminars in oncology. Elsevier. 2010:430–439. doi: 10.1053/j.seminoncol.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 51.Aponte P.M., Caicedo A. Stemness in cancer: stem cells, cancer stem cells, and their microenvironment. Stem Cell. Int. 2017;2017 doi: 10.1155/2017/5619472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Monteiro J., Fodde R. Cancer stemness and metastasis: therapeutic consequences and perspectives. Eur. J. Cancer. 2010;46:1198–1203. doi: 10.1016/j.ejca.2010.02.030. [DOI] [PubMed] [Google Scholar]
- 53.Grosse-Wilde A., Fouquier d'Herouel A., McIntosh E., Ertaylan G., Skupin A., Kuestner R.E., del Sol A., Walters K.A., Huang S. Stemness of the hybrid epithelial/mesenchymal state in breast cancer and its association with poor survival. PLoS One. 2015;10 doi: 10.1371/journal.pone.0126522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Y., Wang Z., Ajani J.A., Song S. Drug resistance and Cancer stem cells. Cell Commun. Signal. 2021;19:19. doi: 10.1186/s12964-020-00627-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fusco M.J., West H.J., Walko C.M. Tumor mutation burden and cancer treatment. JAMA Oncol. 2021;7 doi: 10.1001/jamaoncol.2020.6371. 316-316. [DOI] [PubMed] [Google Scholar]
- 56.Liu Z., Jiang Z., Gao Y., Wang L., Chen C., Wang X. TP53 mutations promote immunogenic activity in breast cancer. Journal of oncology. 2019;2019 doi: 10.1155/2019/5952836. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Clinical data for this research were obtained from the GEO (https://www.ncbi.nlm.nih.gov/geo/) and TCGA (https://portal.gdc.cancer.gov/) databases and can be downloaded from the provided websites. Datasets and custom scripts are available upon request.