Abstract
The molecular determinants responsible for flavivirus host cell binding and tissue tropism are largely unknown, although domain III of the envelope protein has been implicated in these functions. We examined the solution properties and antagonist activity of Langat virus domain III. Our results suggest that domain III adopts a stably folded structure that can mediate binding of tick-borne flaviviruses but not mosquito-borne flaviviruses to their target cells. Three clusters of phylogenetically conserved residues are identified that may be responsible for the vector-specific antagonist activity of domain III.
Flaviviruses (family Flaviviridae, genus Flavivirus) are organized into distinct vector-specific classes and serocomplex-specific subgroups (6), although little is known of the molecular determinants that dictate vector-specific pathogenesis, host cell specificity, and tissue tropism. Both mosquito-borne and tick-borne flaviviruses are responsible for epidemics throughout the developing world and pose serious public health threats in developed countries (7, 9, 10). Vaccines are only available to help control infection by yellow fever, Japanese encephalitis, Central European tick-borne encephalitis (TBE), and louping ill flaviviruses (1, 2, 16, 17). No antiviral therapy is available to treat any flavivirus infection.
Flavivirus infection requires attachment and entry into a target cell, mediated by binding of the viral envelope (E) proteins to cell surface receptors. The host cell receptor (or receptors) and the region (or regions) of the E protein responsible for flavivirus attachment are unknown, although heparan sulfate has been suggested as one factor mediating the interaction between dengue 2 virus and its target cells (3). It is unknown whether different cell surface receptors and/or envelope-receptor interactions are responsible for flavivirus pathogenesis, host range, and tissue tropism.
The structure of the E protein ectodomain from TBE virus has been determined (13). This ectodomain forms a homodimer, with each dimer subunit organized into three domains, designated I, II, and III. Comparisons of E proteins from wild-type viruses with those of attenuated or escape mutant viruses have identified a number of residues in domain III that may be responsible for receptor recognition (4, 8, 12–14). We examined the solution properties of domain III of the E protein from Langat virus (a tick-borne flavivirus) and the ability of this domain to function as an antagonist for virus infectivity. We demonstrate that recombinant domain III adopts a highly stable folded structure in solution and shows reduced infectivity in tick-borne flaviviruses but not mosquito-borne flaviviruses. This suggests that interactions between the envelope protein and its host cell receptor may be vector specific, although it is likely that other factors also influence flavivirus vector preference. Sequence and structure analyses identified a small number of residues that may be responsible for vector-specific receptor binding.
Cloning and expression of domain III of Langat virus E protein.
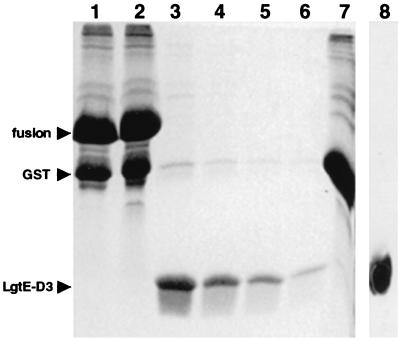
cDNA encoding domain III of the Langat virus E protein (E protein residues 300 to 395; LgtE-D3) was amplified by PCR. Template DNA for the PCR was plasmid pUC18 containing cDNA encoding Langat virus structural proteins. The PCR product was subcloned into pGEX-2T expression vector (Pharmacia), and the fidelity of the cloned sequence was confirmed by DNA sequencing (T. Woods, unpublished data). Escherichia coli DH5α cells were transformed with pGEX-2T expressing LgtE-D3 and grown at 37°C in 2xYT medium. The cultures were chilled to 16°C, and expression of a glutathione S-transferase (GST)–LgtE-D3 fusion protein was induced by the addition of 1 mM isopropyl-1-β-d-galactopyranoside. Cultures were maintained at 16°C for 6 to 12 h, pelleted, and frozen. Pellets were resuspended in ice-cold PBST buffer (10 mM sodium phosphate, 2 mM potassium phosphate [pH 7.4], 140 mM NaCl, 3 mM KCl, 0.1% [vol/vol] Tween 20) and lysed with mild sonication. Glutathione-agarose beads were added to the supernatant and mixed gently at 4°C for 2 to 4 h. The beads were washed with PBST buffer and high-salt PBST buffer (PBST with 500 mM NaCl) and the GST–LgtE-D3 fusion protein was eluted from the beads with Tris-glutathione buffer (50 mM Tris [pH 8.1], 20 mM glutathione). Eluted GST–LgtE-D3 protein was cleaved by treatment with thrombin at room temperature for 12 to 16 h. Alternatively, GST–LgtE-D3 protein could be cleaved on the glutathione-agarose beads and LgtE-D3 could be eluted (Fig. 1). The supernatant was concentrated and applied to a size exclusion column (Biosep SEC-S3000 column; Phenomenex). Recombinant LgtE-D3 was >98% pure, as determined by Coomassie blue-stained sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and immunogenic (data not shown).
FIG. 1.
Coomassie blue-stained composite sodium dodecyl sulfate-polyacrylamide gel showing expression and purification of recombinant LgtE-D3. Lane 1, GST–LgtE-D3 fusion protein bound to glutathione-conjugated agarose beads. Lane 2, total protein at the initiation of thrombin cleavage of bound GST–LgtE-D3. Lane 3, soluble protein after thrombin cleavage of bound GST–LgtE-D3. Lanes 4 to 6, soluble protein recovered after sequential washes of glutathione-agarose beads with low-salt buffer. Lane 7, protein bound to glutathione-agarose beads after thrombin cleavage. Lane 8, Western immunoblot of purified LgtE-D3 probed with rabbit polyclonal antibody generated against recombinant LgtE-D3 (Alpha Diagnostics, Inc.). The GST–LgtE-D3 fusion protein migrates as a single band at ∼37 kDa. Recombinant LgtE-D3 migrates as a single band at ∼10.7 kDa.
Solution properties of LgtE-D3.
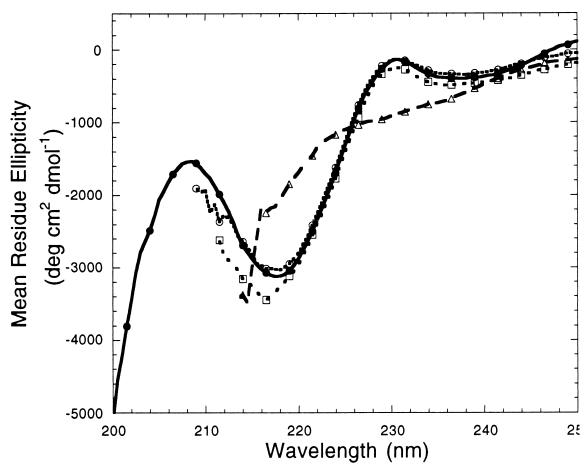
LgtE-D3 was highly soluble and could be concentrated to >40 mg/ml in Tris buffer. Its circular dichroism (CD) spectra showed a local minimum at around 215 to 220 nm (Fig. 2), characteristic of proteins composed primarily of a β-sheet secondary structure (19). Deconvolution of the CD spectra into component secondary-structure basis spectra indicated that LgtE-D3 contained ∼40% β-sheet and ∼60% random coil elements. This composition was consistent with the secondary structural elements observed in the crystal structure of TBE virus envelope protein, where ∼55% of domain III formed β-sheets (13). The CD spectra were unchanged in 2 and 4 M urea (Fig. 2), indicating that domain III is highly stable and resistant to denaturation even in the presence of high urea concentrations. However, in 8 M urea, the CD spectra were similar to those obtained for proteins composed entirely of a random coil structure (19), demonstrating that domain III was largely unfolded in 8 M urea.
FIG. 2.
Far-UV CD spectra of recombinant LgtE-D3 incubated with increasing amounts of denaturant. Spectra from LgtE-D3 in 0 M urea (solid curve, solid circles), 2 M urea (dotted curve, open circles), 4 M urea (dotted curve, open squares), and 8 M urea (dashed curve, open triangles) are shown. The LgtE-D3 concentration was 0.3 mg/ml in Tris buffer (pH 7.4). Spectra were collected on an Aviv 62 DS circular dichrometer operating with a 0.5-nm step increment and a 1-s interval. Cylindrical quartz cuvettes with a 0.1-cm path length were used for all measurements. All sample spectra were recorded five times, averaged, and corrected for buffer contributions. Measurements were considered unreliable when the instrument dynode voltage exceeded 410 V and were not included in subsequent analyses.
Sedimentation velocity experiments performed at 23°C with a Beckman Optima XL-A analytical ultracentrifuge showed that LgtE-D3 was a single low-molecular-weight species with a sedimentation coefficient (15) of 1.3S in physiological buffer at room temperature (data not shown). Analysis of equilibrium velocity measurements in physiological buffer gave a mean molecular mass of 10.7 kDa for LgtE-D3. This molecular mass was within 0.5% of the theoretical molecular mass calculated from the LgtE-D3 amino acid sequence. The model of LgtE-D3 as a noninteracting monomer in solution provided a very good fit to the experimental equilibrium velocity data (data not shown).
Antagonist activity of LgtE-D3.
The ability of LgtE-D3 to protect cells from infection with tick-borne and mosquito-borne flaviviruses was tested. LgtE-D3 significantly reduced the amount of observed cytopathic effect (CPE) in Vero cells infected with the two tick-borne viruses but not with the mosquito-borne viruses (Table 1). In addition, there was a significant delay in the appearance of CPE in cells challenged with tick-borne viruses and protected with LgtE-D3 relative to unprotected cells (data not shown). The protection afforded Vero cells by LgtE-D3 was quantitated by examining virus titers produced in cells challenged with tick-borne viruses. Approximately 10-fold less tick-borne virus was produced from cells protected with LgtE-D3 than from unprotected cells (Table 2). These observations suggest that domain III functioned as a vector-specific antagonist to interfere with flavivirus binding to host cell receptors. This is the first direct experimental evidence that domain III is involved in host cell receptor binding. Recent immunofluorescence studies support the conclusion that LgtE-D3 binds to a membrane-associated host cell receptor (M. Holbrook, personal communication). The observed vector-specific differences could result from the tick-borne and mosquito-borne flaviviruses' interacting with an identical receptor binding site via different intermolecular contacts, with the mosquito-borne flaviviruses binding to this site with greater affinity than the tick-borne flaviviruses. Alternatively, vector specificity could result from flaviviruses' recognizing different cell surface receptors or different binding sites on the same cell surface receptor.
TABLE 1.
Qualitative examination of CPE in Vero cellsa
| Flavivirus | Vector | CPE of LgtE-D3b |
|---|---|---|
| Dengue virus type 4 | Mosquito | + |
| Japanese encephalitis virus | Mosquito | + |
| Yellow fever virus | Mosquito | + |
| Langat virus | Tick | − |
| Powassan virus | Tick | − |
Cells were grown until 60 to 70% confluent, washed with cold PBS, and incubated on ice with 20 μM LgtE-D3. Cells were challenged with virus at a multiplicity of infection of 0.5 and incubated again for 30 min on ice. The cells were then supplemented with 2 ml of Dulbecco's modified Eagle's medium containing 2% (vol/vol) fetal bovine serum (viral maintenance medium) and incubated at 37°C in 5% carbon dioxide.
Presence (+) or absence (−) of CPE compared to control cells.
TABLE 2.
Decreased tick-borne flavivirus production in protected Vero cellsa
| Challenge virus | Virus titer (log PFU)
|
Protection factorb | |
|---|---|---|---|
| Without LgtE-D3 | With LgtE-D3 | ||
| Langat virus | 6.6 | 5.7 | 7.9 |
| Powassan virus | 6.0 | 5.1 | 7.9 |
Cells were challenged with virus as described in the text. Aliquots of the supernatant were harvested at selected time points and frozen. Virus titers were determined by plaque assay using confluent Vero cells overlaid with 5 ml of 1% agar overlay (2% agar with an equal volume of viral maintenance medium). Plates were incubated at 37°C in 5% CO2 for 5 to 10 days and plaques were visualized with 1% agar overlay supplemented with 2% (vol/vol) neutral red (Sigma).
Protection factor = (PFU without LgtE-D3)/(PFU with LgtE-D3).
Molecular basis of vector-specific antagonist activity.
Since LgtE-D3 displays vector-specific antagonist activity, an analysis of vector-based invariant residues was performed to identify domain III regions that mediate host cell binding (Fig. 3). Analogous sequence comparisons in the hemagglutinin receptor-binding site of influenza A viruses have identified phylogenetically conserved residues that determine receptor specificity (11, 18). Domain III amino acid sequences showed ∼80 to 95% sequence identity among tick-borne flaviviruses, ∼50 to 90% identity among mosquito-borne flaviviruses, and ∼50% identity among two non-vector-borne flaviviruses. Twelve residues were invariant, and nine residues were highly conserved (>93%) in the sequences examined. Six amino acids, at positions 324, 355, 381, 382, 384, and 394 (TBE virus numbering), were completely conserved within a flavivirus vector class but differed between the tick-borne and mosquito-borne viruses. In addition, vector-specific insertions and deletions occurred at positions 309, 369, 376, 378, and 386, either at loops between β-sheets or in extended coils (Fig. 4).
FIG. 3.
Amino acid sequence alignment of domain III of flavivirus E proteins. Residues that are conserved in at least 15 of the 16 sequences are indicated by a star above the alignment. Shaded boxes indicate positions of conserved vector-specific residues and insertions. Amino acid numbering refers to TBE virus residue positions. The virus and vector classes are indicated along the left-hand margin. Protein sequences were obtained from Swiss-Protein database entries and correspond to Langat virus (LGT; strain V, accession number P29837), Powassan virus (POW; accession number Q04538), Central European TBE virus (accession number Q01299), louping ill virus (LI; accession number P22338), yellow fever virus (YF; strain 1899/81, accession number P29165), Murray Valley encephalitis virus (MV; accession number P05769), Japanese encephalitis virus (JE; strain Nakayama, accession number P27395), St. Louis encephalitis virus (SLE; accession number P09732, strain MS1-7), Kunjin virus (KUN; strain MRM61C, accession number P14335), West Nile virus (WN; strain RO97-50, accession number Q9WHD1), dengue virus type 1 (DEN1; strain AHF82-80, accession number P27912), dengue virus type 2 (DEN2; isolate Malaysia M2, accession number P14338), dengue virus type 3 (DEN3; accession number P27915), dengue virus type 4 (DEN4; accession number P09866), Rio Bravo virus (RIO; accession number AF144692), and Apoi virus (accession number AF160193).
FIG. 4.
Structure of TBE virus envelope protein. (A) Top view of the E protein homodimer. (B) Side view of the E protein homodimer. Orientation nomenclature is adapted from that of Rey et al. (13), where top suggests a view towards and normal to the virus surface, and side suggests a view tangent to the virus surface, with the viral membrane below the protein. Each monomer of the homodimer is shaded differently, and the dashed lines delineate the domain III boundaries. (C) Isolated domain III viewed approximately perpendicular to the twofold axis of the E protein homodimer, corresponding to the solvent-exposed lateral surface of domain III. Positions of conserved vector-specific residues and insertions in domain III are indicated with spheres. Amino acid numbering refers to the TBE virus residue positions. Domain III structures were displayed using Swiss-Pdb Viewer (5; http://www.expasy.ch/spdbv).
The 11 vector-specific positions were clustered in three spatially distinct regions in domain III (Fig. 4), which may be responsible for the vector-specific antagonist activity of domain III. Region 1, formed by residues 309, 384, and 386, was located on the solvent-exposed lateral face of domain III. These residues were flanked by invariant residues, and at residue 386 three homologous residues were inserted in mosquito-borne flaviviruses relative to tick-borne flaviviruses. Thus, the structural and chemical properties of region 1 may be similar among vector-specific flaviviruses, allowing this region to form vector-specific interactions with cell surface receptors. Recent studies showed that mutations in region 1 were associated with virus attenuation (8, 13, 14). Region 2, formed by residues 324, 355, 376, 378, 381, 382, and 394, made extensive contacts between the β-sheet termini of domain III (Fig. 4). The majority of these residues were partially buried within the domain III structure and thus likely shielded from intermolecular interactions. Region 3, containing residue 369, was part of a solvent-exposed loop (Fig. 4). Between residues 369 and 370 of the mosquito-borne flaviviruses, one additional residue was inserted in the dengue and yellow fever viruses and three additional residues were inserted in the non-dengue and non-yellow fever viruses relative to the tick-borne flaviviruses. Invariant residues were not adjacent to residue 369, implying that the tertiary structure and chemical properties of region 3 are different for different mosquito-borne flaviviruses.
Vector-specific residues in the non-vector-borne flaviviruses had similarities to both the tick- and mosquito-borne flaviviruses. The non-vector-borne and mosquito-borne flaviviruses had the same residues at positions 324, 355, and 381 and had a conserved deletion at position 309. The non-vector-borne and tick-borne flaviviruses had similar residues at positions 376 and 394 and conserved deletions at positions 378 and 386. The lysine residue at position 384 in the non-vector-borne flaviviruses was distinct from the conserved residue found in the mosquito- and tick-borne flaviviruses.
Domain III mutants can be used to delineate E protein receptor interaction surfaces and test if different E protein interaction surfaces are responsible for tissue tropism. Domain III and its derivatives may be useful first-generation vector-specific antagonists and as targets against which to generate antiviral agents that prevent binding of flavivirus to its host cell receptor. In addition, recombinant domain III may be valuable in the development of flavivirus vaccines and diagnostic reagents.
Acknowledgments
This work was supported by grant N65236-97-1-5811 (R.E.S.) from the Defense Advanced Research Projects Agency, a fellowship from the McLaughlin Foundation (M.H.), and the Sealy Center for Structural Biology.
We thank J. Lee and C. Chan for assistance with the analytical ultracentrifugation experiments, L. Lee for assistance with the CD experiments, and D. Konkel for critically reading the manuscript.
REFERENCES
- 1.Barrett A D T. Japanese encephalitis and dengue vaccines. Biologicals. 1997;25:27–34. doi: 10.1006/biol.1997.0057. [DOI] [PubMed] [Google Scholar]
- 2.Barrett A D T. Yellow fever vaccines. Biologicals. 1997;25:17–25. doi: 10.1006/biol.1997.0056. [DOI] [PubMed] [Google Scholar]
- 3.Chen Y, Maquire T, Hileman R E, Fromm J R, Esko J D, Linhardt R J, Marks R M. Dengue virus infectivity depends on envelope protein binding to target cell heparan sulfate. Nat Med. 1997;3:866–871. doi: 10.1038/nm0897-866. [DOI] [PubMed] [Google Scholar]
- 4.Dunster L M, Wang H, Ryman K D, Miller B R, Watowich S J, Minor P D, Barrett A D T. Molecular and biological changes associated with HeLa cell attenuation of wild-type yellow fever virus. Virology. 1999;261:309–318. doi: 10.1006/viro.1999.9873. [DOI] [PubMed] [Google Scholar]
- 5.Guex N, Peitsch M C. SWISS-MODEL and the Swiss-Pdb Viewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 6.Kuno G, Chang G-J J, Tsuchiya K R, Karabatsos N, Cropp C B. Phylogeny of the genus Flavivirus. J Virol. 1998;72:73–83. doi: 10.1128/jvi.72.1.73-83.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lanciotti R S, Roehrig J T, Deubel V, Smith J, Parker M, Steele K, Crise B, Volpe K E, Crabtree M B, Scherret J H, Hall R A, MacKenzie J S, Cropp C B, Panigrahy B, Ostlund E, Schmitt B, Malkinson M, Banet C, Weissman J, Komar N, Savage H M, Stone W, McNamara T, Gubler D J. Origin of the West Nile virus responsible for an outbreak of encephalitis in the northeastern United States. Science. 1999;286:2333–2337. doi: 10.1126/science.286.5448.2333. [DOI] [PubMed] [Google Scholar]
- 8.Mandl C W, Allison S L, Holzmann H, Meixner T, Heinz F X. Attenuation of tick-borne encephalitis virus by structure-based site-specific mutagenesis of a putative flavivirus receptor binding site. J Virol. 2000;74:9601–9609. doi: 10.1128/jvi.74.20.9601-9609.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Monath T P. Dengue: the risk to developed and developing countries. Proc Natl Acad Sci USA. 1994;91:2395–2400. doi: 10.1073/pnas.91.7.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Monath T P, Heinz F X. Flaviviruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 961–1034. [Google Scholar]
- 11.Naeve C W, Hinshaw V S, Webster R G. Mutations in the hemagglutinin receptor-binding site can change the biological properties of an influenza virus. J Virol. 1984;51:567–569. doi: 10.1128/jvi.51.2.567-569.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ni H, Ryman K D, Wang H, Saeed M F, Hull R, Wood D, Minor P D, Watowich S J, Barrett A D T. Interaction of yellow fever virus French neurotropic vaccine strain with monkey brain: characterization of monkey brain membrane receptor escape variants. J Virol. 2000;74:2903–2906. doi: 10.1128/jvi.74.6.2903-2906.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rey F A, Heinz F X, Mandl C, Kunz C, Harrison S C. The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature. 1995;375:291–298. doi: 10.1038/375291a0. [DOI] [PubMed] [Google Scholar]
- 14.Ryman K D, Ledger T N, Campbell G A, Watowich S J, Barrett A D T. Mutation in a 17D-204 vaccine substrain-specific envelope protein epitope alters the pathogenesis of yellow fever virus in mice. Virology. 1998;244:59–65. doi: 10.1006/viro.1998.9057. [DOI] [PubMed] [Google Scholar]
- 15.Stafford W F I. Boundary analysis in sedimentation transport experiments: a procedure for obtaining sedimentation coefficient distributions using the time derivative of the concentration profile. Anal Biochem. 1992;203:295–301. doi: 10.1016/0003-2697(92)90316-y. [DOI] [PubMed] [Google Scholar]
- 16.Stephenson J R. Flavivirus vaccines. Vaccine. 1988;6:471–480. doi: 10.1016/0264-410x(88)90095-3. [DOI] [PubMed] [Google Scholar]
- 17.Venugopal K, Gould E A. Towards a new generation of flavivirus vaccines. Vaccine. 1994;12:966–975. doi: 10.1016/0264-410x(94)90329-8. [DOI] [PubMed] [Google Scholar]
- 18.Weis W, Brown J H, Cusack S, Paulson J C, Skehel J J, Wiley D C. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature. 1988;333:426–431. doi: 10.1038/333426a0. [DOI] [PubMed] [Google Scholar]
- 19.Yang J T, Wu C-S C, Martinez H M. Calculation of protein conformation from circular dichroism. Methods Enzymol. 1986;130:208–269. doi: 10.1016/0076-6879(86)30013-2. [DOI] [PubMed] [Google Scholar]