Abstract
In recent years, CD9 has been extensively studied as a potential biomarker for cancer. However, the biological role of CD9 in gliomas remains unclear. This study investigates the function of CD9 in gliomas and its molecular mechanisms. Utilizing pan-cancer analysis with TCGA, CGGA, and GEO databases, differential expression of CD9 was observed in 11 tumor types within the TCGA cohort, and it was associated with patient survival rates. Analysis of the CGGA glioma database revealed that patients with high CD9 expression had lower survival rates. The area under the ROC curve (AUC) for GSE16011 was greater than 0.7, indicating a high discriminative ability. Through gene set enrichment analysis (GSEA), immune-related analysis, and CD9 mutation detection, CD9 was found to have the strongest correlation with neutrophil involvement (cor = 0.30, P < 0.05), and the high CD9 expression group exhibited higher rejection responses and TIDE scores, suggesting a lower likelihood of successful immunotherapy. The high CD9 expression group was more sensitive to 81 drugs, indicating potential therapeutic effects for gliomas. Furthermore, overexpression of CD9 in gliomas may be associated with gene mutations. Down-regulation or up-regulation of CD9 expression in the glioblastoma cell line LN229 showed that CD9 could positively regulate the migratory ability of LN229 cells. Further, several marker genes, such as VEGFR-2, TGF-β1, CASP1 and PI3K, were down regulated in CD9 knockdown cell lines and up regulated in CD9 overexpression cell lines, compared with control cell line. This study preliminarily explores the role of CD9 in gliomas and its prognostic value, providing new insights for personalized treatment strategies in glioma therapy.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-74109-w.
Keywords: Glioma, CD9, Survival, Prognostic value, Bioinformatics
Subject terms: Cancer, Neuroscience, Medical research, Oncology
Introduction
Gliomas are the most common primary tumors of the central nervous system (CNS), originating from neuroglial cells, as a prevalent form of CNS tumor, and are notably aggressive and malignant1. Approximately 80–85% of adult malignant brain tumors are gliomas, which are characterized by diffuse infiltration into the brain parenchyma. The fifth edition of the World Health Organization (WHO) classification of CNS tumors categorizes these tumors into grades 1–4, with about 49% of malignant brain tumors being glioblastomas and 30% being diffuse infiltrative low-grade gliomas2. Lower grade gliomas (LGGs) encompass grades 1 and 2, while high-grade gliomas (HGGs) include grades 3 and 43. Currently, the classification of gliomas has entered the era of molecular profiling. Based on molecular genetic characteristics, gliomas can be classified into different molecular subtypes. The most commonly identified subtypes include IDH-mutant gliomas, 1p/19q codeleted gliomas, and MGMT promoter methylated gliomas4. IDH mutations are present in the majority of lower-grade gliomas (LGGs), with mutations in IDH1 and IDH2 being the most prevalent. These mutations alter cellular epigenetics by inhibiting histone and DNA methylation, thereby impeding the normal differentiation process of cells and creating a cellular state conducive to malignant transformation5,6. Moreover, compared to their IDH wild-type counterparts, IDH-mutant gliomas are associated with a better prognosis and treatment response7. Gliomas with 1p/19q codeletion, characterized by the concurrent loss of chromosomal arms 1p and 19q, are typically associated with oligodendrogliomas and are linked to a favorable chemotherapy response and longer survival. Gliomas with MGMT promoter methylation result in reduced expression of the MGMT protein, which is associated with increased sensitivity to chemotherapeutic agents, thereby correlating with a better treatment response and prognosis4. The consensus treatment strategy for gliomas remains a combination of surgical resection, chemotherapy, and radiotherapy, yet the overall 5-year survival rate for patients remains low, and the rate of postoperative recurrence is high, significantly impacting the quality of life of patients8. Therefore, it is urgent to identify potential therapeutic targets to advance precision and individualized treatment.
CD9, also known as Motility-related protein-1 (MRP-1), is a member of the Transmembrane 4 superfamily (TM4SF)9. CD9 is presented on a variety of human cell membranes and extracellular vesicle membranes, playing important roles in intercellular contact, cell-extracellular matrix interactions, integrin-dependent cell migration, signal transduction, membrane fusion, apoptosis, inflammation, proliferation, and differentiation. These processes contribute to the mediation of various diseases such as infections, tumors, and inflammatory conditions, influencing their onset and progression10–12. On one hand, CD9 acts as a tumor suppressor plays an inhibitory role in tumor growth and progression. In prostate cancer patients, the progression of the disease is accompanied by a decrease in CD9 levels due to the absence or mutation of its transcripts, with significant differences observed between clinically localized and advanced stages of the disease. This suggests that CD9 inactivation may play a crucial role in the progression of prostate cancer, as confirmed in clinical settings13. On the other hand, CD9 may also affect tumor recurrence, progression, and colonization by promoting the formation of tumor blood and lymphatic vessels14,15, and has been extensively studied in recent years as a tumor marker. The expression levels of CD9 are closely related to the prognosis of various tumors. In pancreatic cancer, high expression of CD9 has been correlated with poorer prognosis16, and in cutaneous melanoma, high CD9 expression is associated with adverse outcomes and a higher rate of lymph node metastasis17. Blake et al.18 observed that in the non-small cell lung cancer (NSCLC) model A549 cell line, CD9 enhanced cell migration towards chemotactic agents, including IL-16. Podergajs et al.19 performed transcriptomic analysis on GBM tissues and compared them with normal brain tissues, discovering a significant upregulation of the CD9 gene. An over twofold increase in CD9 expression was associated with a shorter survival period in patients. GBM stem cells with silenced CD9 exhibited altered activation patterns in protein kinase B (Akt), mitogen-activated protein kinase (MapK), and signal transducer and activator of transcription protein 3 (Stat3) signaling pathways. In vivo orthotopic xenografts in nude mice with CD9-silenced GBM stem cells resulted in prolonged survival times. However, this has limited implications for treatment guidance, and has not clarified effective therapeutic targets and the biological role of CD9 in gliomas. This study primarily employs bioinformatics analysis methods to investigate the association between CD9 and the prognosis of glioma patients. It further examines the biological pathways involved, the relationship with the immune microenvironment, and drug sensitivity. This research aims to provide new theoretical support and reference for guiding the rational treatment of glioma patients and prolonging their survival.
Materials and methods
Data source
To begin with, transcriptomic data and survival information for 33 cancers were downloaded from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/). Among them, glioblastoma multiforme (GBM) and brain lower grade glioma (LGG) were combined to form the glioma samples, which contained 687 tumor samples and was referred to as the TCGA-glioma (GBMLGG) dataset. Additionally, expression data, survival information, and clinical information for the CGGA_693 cohort containing 693 glioma samples were downloaded from the Chinese Glioma Genome Atlas (CGGA) database (http://www.cgga.org.cn/), called the CGGA-glioma dataset. Furthermore, GSE16011 and validation set CSE68848 were retrieved from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/), where GSE16011 included 276 glioma samples, 8 control samples and CSE68848 included 443 glioma samples, 28 control samples.
CD9 pan-cancer analysis
To examine CD9 expression in different types of cancer, CD9 gene expression were analyzed and compared in 32 cancer tumor and control groups from the TCGA cohort using Wilcoxon test. Then, to evaluate the prognostic value of CD9 in various cancers, all cancer samples in TCGA cohort were divided into high and low expression groups depending on median value of CD9 expression, and Kaplan–Meier (K–M) survival analysis was performed by survival package (v 3.5-3) to obtain the survival differences in overall survival (OS), disease specific survival (DSS), and progression free interval (PFI) between 2 expression groups in pan-cancers using Log-rank test (P < 0.05).
CD9 expression analysis in glioma
To further understand expression trend of CD9 in glioma, the expression of CD9 in tumors and controls was analyzed and compared by Wilcoxon test in GSE16011, external validation of CD9 expression in different groups was also performed using the validation set GES68848. Moreover, CD9 expression differences in various clinical characteristics (age, gender, IDH mutation, and radiation) of TCGA-glioma and in different clinical characteristics (grade, IDH mutation, and X1p19q codeletion status) in CGGA-glioma were compared (P < 0.05). In CGGA-glioma, glioma samples were also sorted into high/low expression groups based on CD9 expression median value. K–M survival curves were plotted to assess OS difference in 2 expression groups through survival package with log-rank test. Notably, no DSS or PFI information in CGGA-glioma. In addition, to assess the diagnostic capability of CD9 for glioma, Receiver Operating Characteristic (ROC) curve in GSE16011 was drawn using pROC package (v 1.18.0)20.
Gene set enrichment analysis (GSEA) for CD9
The clusterProfiler package was utilized to perform GSEA on the CD9 gene with adj. P value < 0.05. In TCGA-glioma, Spearman’s correlation coefficients were calculated between CD9 and all other genes. The genes were then ranked in ascending order based on their correlation coefficients to obtain a list of related genes. The KEGG gene set (c2.cp.kegg.v2023.1.Hs.symbols.gmt) from MsigDB database was selected as the background gene set. On the other hand, differential analysis was performed between 2 expression groups (high vs. low expression groups), log2FC was calculated for all genes, and then all genes were sorted by log2FC in ascending order to obtain the list of related genes corresponding to CD9. The background gene set was HALLMARK gene set from the MsigDB database, which was h.all.v2023.1.Hs.symbols.gmt (adj. P < 0.05).
Immune cell infiltration analysis
In order to assess immune levels of CD9 high/low expression groups in glioma patients, the TCGA-glioma cohort was applied to analyze infiltration of 22 immune cells through CIBERSORT algorithm in IOBR package (v 0.99.9)21, and scores were computed with each immune cell. Next, the immune cells with low expressing levels were filtered, and the differences in remaining immune cells between 2 expression groups were compared by Wilcoxon test (P < 0.05). Further, correlation analysis was performed between immune cells as well as CD9 and immune cells via Spearman’s method.
Immune checkpoints and prediction of immunotherapy effects
Immune checkpoints were crucial in preventing autoimmune effects. In TCGA-gliomas, the expression levels of 48 immune checkpoints were compared between CD9 high/low expression groups using Wilcoxon test (P < 0.05). Then, correlation of differential immune checkpoints and CD9 was analyzed by Spearman’s method. The TIDE score was utilized to assess the likelihood of tumor immune escape. The TIDE, dysfunction, and exclusion scores were obtained from the TIDE database for TCGA-glioma patients.
Drug sensitivity analysis
In chemotherapy, the ability of the drug to inhibit cell growth was stronger when the semi-inhibitory concentration (IC50) of drug was lower, indicating that the tumor was more sensitive to the drug, resulting in more effective cancer treatment. The R pRRophetic package (v 0.5)22 was utilized to evaluate the IC50 values of drugs in TCGA-glioma samples. Then, correlation analysis of differential drugs and CD9 expression in TCGA-glioma was performed using Spearman’s correlation analysis.
Mutation analysis of CD9
To understand the mutation profile of CD9, TCGA-glioma data were selected to analyze the mutation profile of CD9 through cBioPortal (http://www.cbioportal.org/).
Cell lines and cell culture
The glioblastoma cell lines LN229 (right frontal parieto-occipital glioblastoma of human brain) was purchased from American Type Culture Collection (ATCC). The cells were maintained in an incubator at 37 °C with 5% CO2 and cultured in high glucose DMEM medium, supplemented with 10% fetal bovine serum, 100 mg/mL streptomycin and 100 U/mL penicillin.
Cell transfection
The full length of CD9 was synthesized and cloned into YOE-LV001 to construct the overexpression vector. The overexpression and shRNA vectors were transfected into LN229 using Lipofectamine 2000 (Invitrogen, USA) following the manufacturer’s instructions. The LN229 cells were also transfected with YOE-LV001and shRNA of non-targeting control sequence as negative controls.
RNA isolation and quantitative real-time polymerase chain reaction
Total RNA was extracted using TRIZOL reagent (Solarbio, China) and quantified by Nanodrop2000 spectrophotometer (Thermo Fisher Scientific, USA). Then, the total RNA was synthesized into first strand cDNA with PrimeScript RT Reagent Kit (Takara, China). The cDNA was amplified with 2x M5 Ultra SYBR Mixture (Mei5bio, China) on a Bio-Rad CFX96 system (Bio-Rad, USA). β-actin was used as the reference gene. The expression data were calculated with2−ΔΔCT. The primers sequences are detailed in Supplementary Table S1.
Western blotting
GBM cells were collected and lysed in RIPA lysis buffer (Beyotime, China). The protein concentrations were determined using a BCA Protein Assay Kit (Beyotime, China). Total cell lysates containing 26 µg of protein were separated using SDS-PAGE and transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, USA). After blocking with 5% non-fat dry milk in TBST for 30 min, membranes were incubated with primary antibodies (anti-CD9 antibody diluted at 1:1000) at 4 °C overnight. Then, washed membranes were hybridized with HRP-conjugated secondary antibody (diluted at 1:5000) at room temperature for two hours. Protein bands were visualized by ECL (Santa Cruz Biotechnology, USA) and detected using the ECL Detection System.
Transwell assay
For transwell assays, 1 × 106 cells were plated in 200 µL of serum-free medium on upper chambers. After incubation for 24 h, migrated cells in lower chambers were fixed with methanol, stained with crystal violet, and photographed using a microscope. Cell counting was conducted using ImageJ.
Statistical analysis
R software (v 4.2.2) was applied to execute statistical analysis, with P < 0.05 as statistical significance.
Results
The DSS was significantly different between CD9 high and low expression groups for GBM LGG
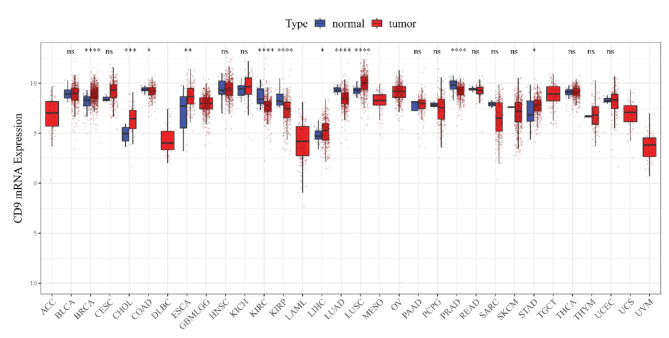
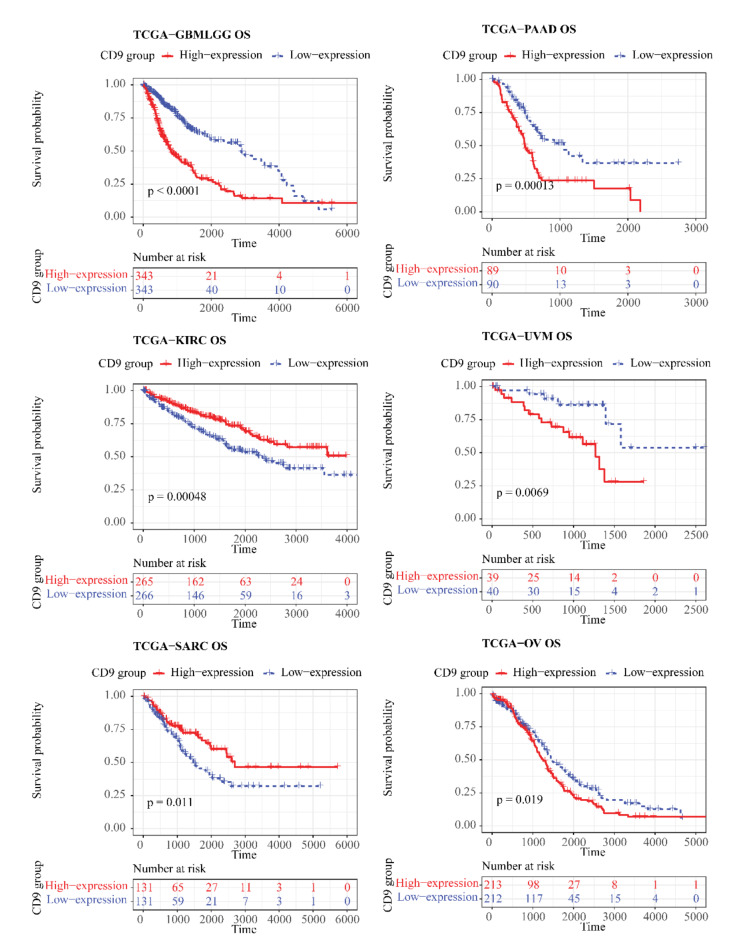
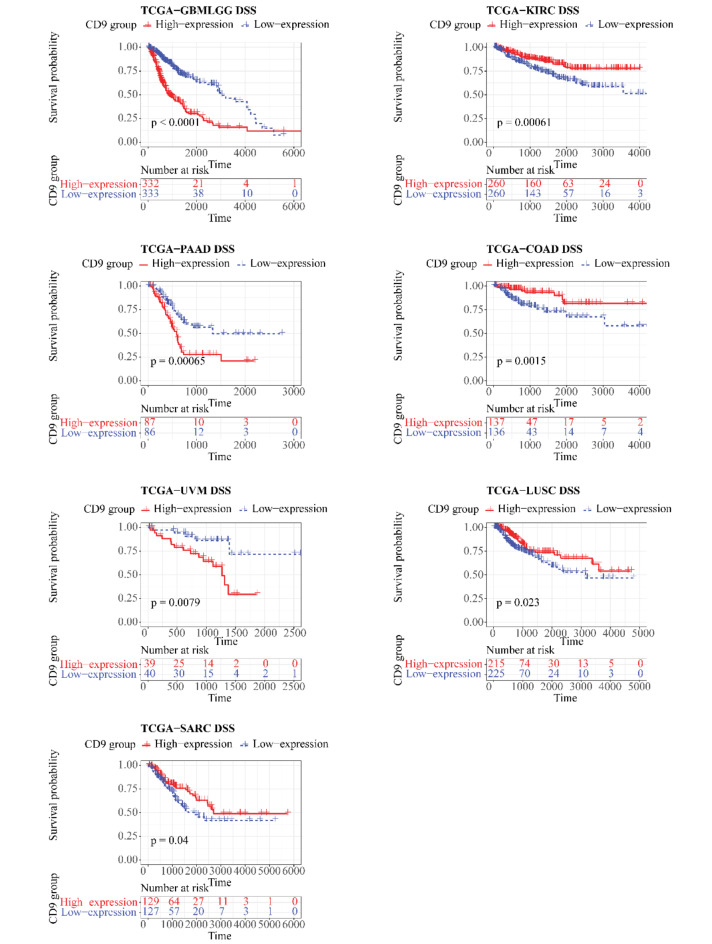
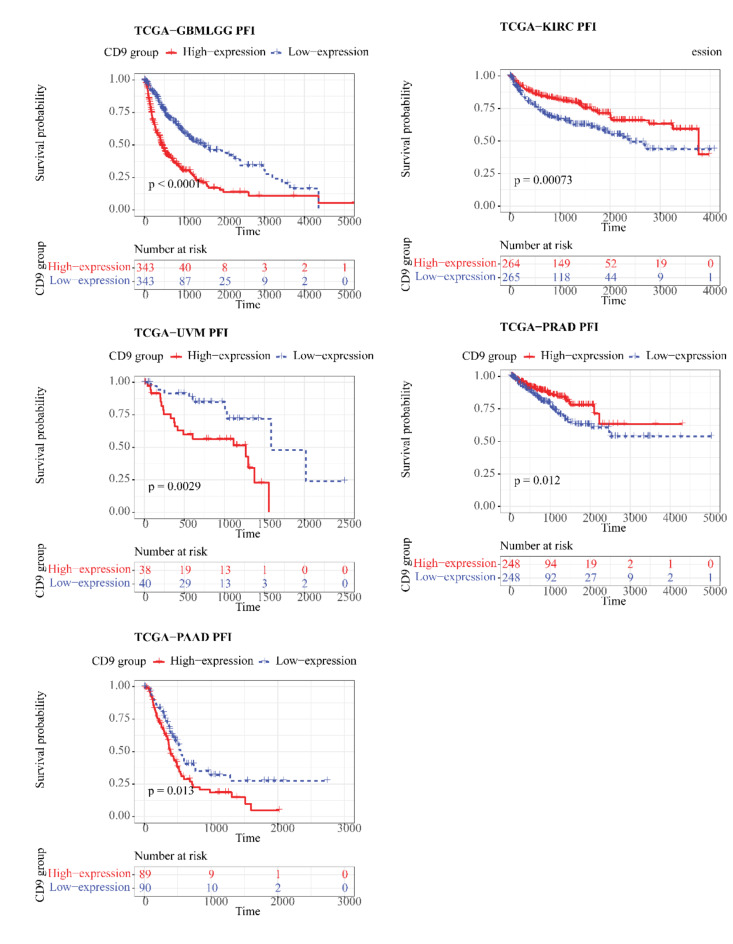
In the TCGA cohort, CD9 was differentially expressed between tumor and control samples in 11 kinds of cancers (P < 0.05), (Fig. 1) for instance, breast cancer (BRCA), cholangiocarcinoma (CHOL), colorectal cancer (COAD), etc. Notable differences (P < 0.05) were existed between CD9 high/low expression groups for 6 out of 32 cancers in OS, namely GBMLGG, pancreatic adenocarcinoma (PAAD), KIRC, uveal melanoma (UVM), sarcomatoid carcinoma (SARC), and ovarian cancer (OV) (Fig. 2). For DSS, remarkable differences (P < 0.05) were seen between 2 expression groups for GBMLGG, kidney renal clear cell carcinoma (KIRC), PAAD, COAD, UVM, lung squamous cell carcinoma (LUSC), and SARC of 31 cancers (no DSS data for acute myeloid leukemia (LAML)) (Fig. 3). In regard to PFI, there were significant differences (P < 0.05) between both expression groups for 5 (GBMLGG, KIRC, UVM, prostate adenocarcinoma (PRAD), PAAD) of the 31 cancers (no LAML) (Fig. 4).
Fig. 1.
CD9 expression levels in different types of cancers from the TCGA database (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
Fig. 2.
Kaplan–Meier OS curve of human tumors with high and low CD9 expression analyzed by the TCGA database.
Fig. 3.
Kaplan–Meier DSS curve of human tumors with high and low CD9 expression analyzed by the TCGA database.
Fig. 4.
Kaplan–Meier PFI curve of human tumors with high and low CD9 expression analyzed by the TCGA database.
CD9 highly expressed in glioma group of GSE16011
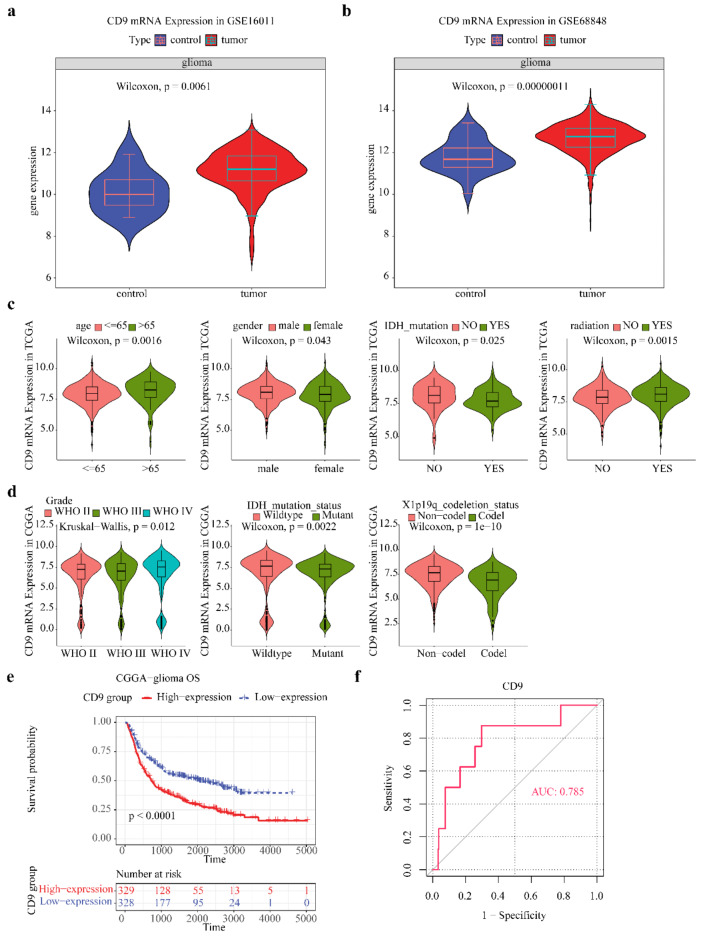
Differences in expression showed that CD9 highly expressed in tumor group of GSE16011 (Fig. 5a). This is consistent with the external validation results (Fig. 5b). In TCGA-glioma, CD9 expression was higher in age > 65, male, no IDH mutation, and radiation (Fig. 5c). CD9 expression difference in grade, IDH mutation, and X1p19q codeletion status of CGGA-glioma were also remarkable (Fig. 5d). The CD9 pan-cancer prognostic analysis indicated that CD9 was linked to OS, DSS, and PFI survival in glioma. Further, we analyzed the difference in OS between high/low expression groups of CGGA-glioma, and found that the survival rate of patients was lower in CD9 high expression group (Fig. 5e). ROC curve has a good ability to distinguish samples in GSE16011, with area under curve (AUC) = 0.785 (Fig. 5f).
Fig. 5.
CD9 is highly expressed in tumor groups. (a) GSE16011 database showed that CD9 mRNA expression in tumor tissues was higher than that in normal tissues; (b) Validation set GES68848 showed that CD9 mRNA expression in tumor tissues was higher than that in normal tissues. (c) TCGA database indicated that CD9 mRNA expression was high in patients older than 65 years-old, male, IDH wild type, and receiving radiotherapy; (d) CGGA database indicated that the expression level of CD9 was WHO4 > WHO2 > WHO3, and CD9 mRNA was highly expressed in patients with IDH wild type and X1p19q without co-deletion; (e) CGGA database indicated that patients with glioma with high expression of CD9 had lower OS; (f) Area under ROC curve (AUC) = 0.785, indicating that CD9 has a high differentiation ability for GSE16011 samples.
KEGG and Hallmark pathways involved in CD9
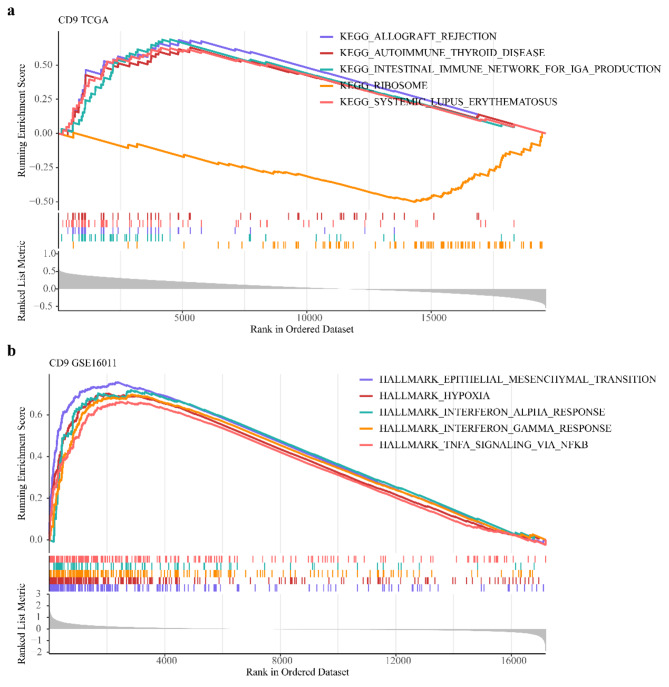
By GSEA in TCGA-glioma, CD9 was enriched in ‘allograft rejection’, ‘intestinal immune network for IGA production’, ‘ribosome’ etc. pathways in KEGG (Fig. 6a). In GSE16011, CD9 was enriched in ‘epithelial mesenchymal transition’, ‘hypoxia’, ‘interferon alpha response’, etc. Hallmark pathways (Fig. 6b).
Fig. 6.
KEGG and Hallmark pathways involved in CD9. (a) KEGG enrichment pathway of GSEA (top5); (b) HALLMARK enrichment pathway of GSEA (top5).
CD9 was associated with neutrophils
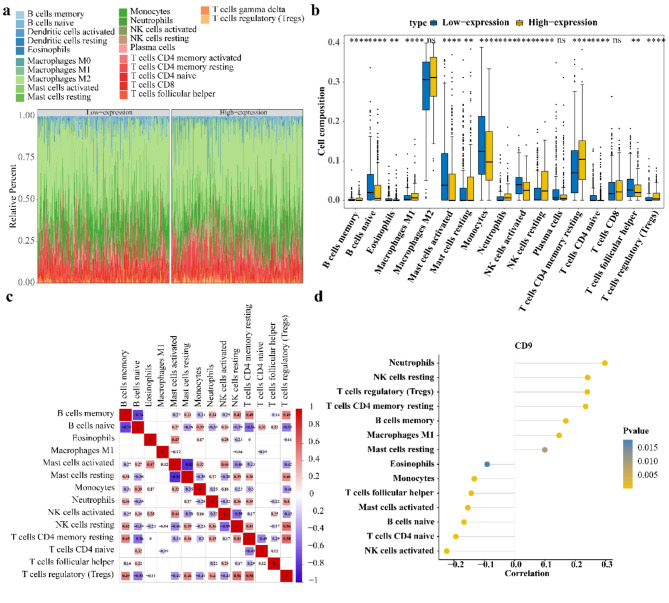
The stacked plot illustrated the abundance of the 22 immune cells between CD9 high/low expression groups samples in TCGA-glioma (Fig. 7a). Then, of 17 remaining immune cells, abundance of 14 immune cells differed markedly in 2 expression groups (Fig. 7b). For the 14 differential immune cells, regulatory T cells (Tregs) had the highest positive correlation with resting memory CD4 T cells (cor = 0.58, P < 0.05) and resting mast cells had the highest negative correlation with activated mast cells (cor = − 0.82, P < 0.05) (Fig. 7c). Additionally, CD9 had the highest correlation with neutrophils (cor = 0.30, P < 0.05) (Fig. 7).
Fig. 7.
Association analysis between CD9 and immune cells. (a) The abundance of immune cells in the samples of high and low CD9 expression groups in the TCGA database; (b) The immune cells of the high and low CD9 expression groups were infiltrated differentially; Yellow represents high CD9 expression group, blue represents low CD9 expression group. (c) Differential immune cell correlation heat map; Red represents positive correlation, blue represents negative correlation, the darker the color, the higher the correlation; Blank indicates no significant (d) CD9 is associated with differential immune cells.
Glioma patients with CD9 high expression might have a higher likelihood of immune escape and a lower success rate of immunotherapy
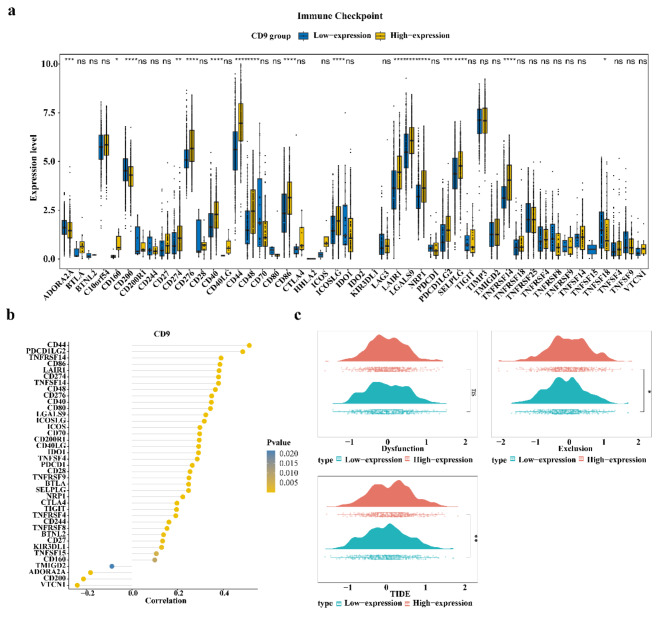
Regarding 48 immune checkpoints, 40 immune checkpoints were differentially expressed in the CD9 high and low expression groups of TCGA-glioma (P < 0.05) (Fig. 8a). The correlation analysis revealed that 39 immune checkpoints were correlated with CD9 (Fig. 8b). Among them, CD44 had the highest positive correlation with CD9 (cor = 0.51, P < 0.05). Moreover, exclusion and TIDE scores were significantly higher in the CD9 high expression group, implying a higher likelihood of immune escape and less successful immunotherapy (Fig. 8c).
Fig. 8.
Glioma patients with high CD9 expression may have a lower success rate of immunotherapy. (a) Immune checkpoints with different expression of CD9 in high and low expression groups (P value < 0.05); (b) Correlation between differential immune checkpoint and CD9; (c) TIDE fractions in high and low CD9 expression groups; Red indicates the CD9 high expression group, green indicates the CD9 low expression group.
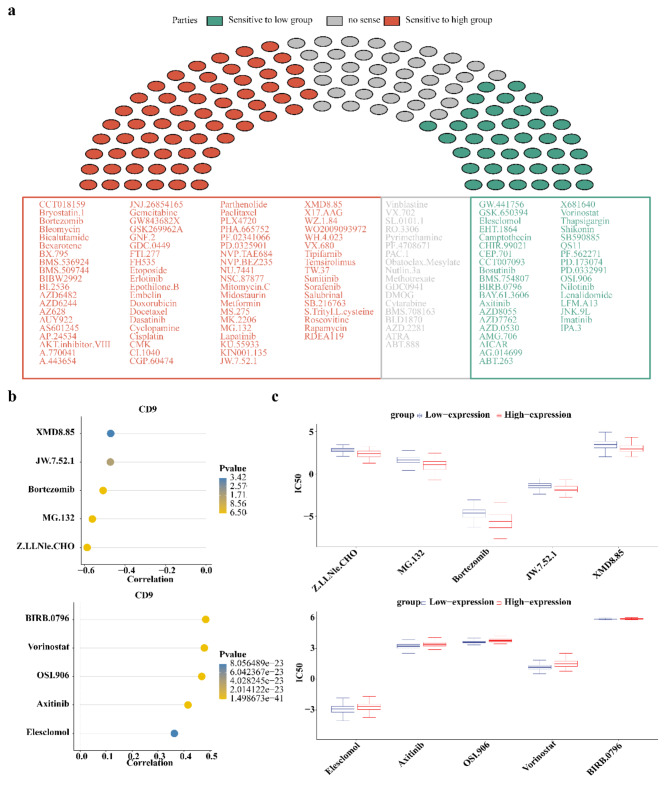
BIRB.0796 and Z.LLNle.CHO could be potential drugs for gliomas
In 138 anticancer drugs, the IC50 of a total of 119 drugs had notable differences between CD9 high/low expression groups (P < 0.05), of which 81 drugs had higher sensitivities in CD9 high expression group and 38 drugs had higher sensitivities in CD9 low expression group (Fig. 9a). The top 5 drugs most positively and negatively correlated with CD9 were presented. BIRB.0796 was highest positively correlated with CD9 (cor = 0.48, P < 0.05) and Z.LLNle.CHO had the highest negative correlation with CD9 (cor = − 0.59, P < 0.05) (Fig. 9b). The IC50 of these 10 drugs in 2 expression groups were also shown (Fig. 9c).
Fig. 9.
Drug sensitivity analysis of patients with high or low CD9 expression level. (a) Drug sensitivity of patients in high and low CD9 expression groups. Red indicates drugs with high sensitivity in patients with high CD9 expression, green indicates drugs with high sensitivity in patients with low CD9 expression, and gray indicates drugs with no sensitivity difference between high and low CD9 expression groups; (b) Correlation between drug IC50 and CD9. The top five drugs most negatively associated with CD9 are shown above, and the top five drugs most positively associated with CD9 are shown below. (c) IC50 values of the top5 drugs most positively and negatively correlated with CD9. The top 5 drugs most negatively correlated with CD9 are shown above, and the top 5 drugs most positively correlated with CD9 are shown below.
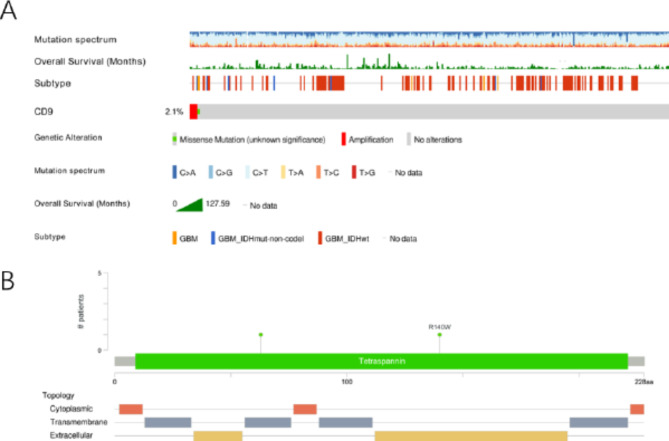
High CD9 expression in gliomas might be related to gene mutation
The mutation frequency of CD9 was 2.1% in TCGA-glioma. Most of the mutations were amplification (Fig. 10a). Figure 10b showed the distribution of mutations in the structural domains of the CD9 protein.
Fig. 10.
The expression level of CD9 in gliomas might be related to gene mutation; (a) Mutation analysis of CD9; (b) CD9 protein domain mutation distribution.
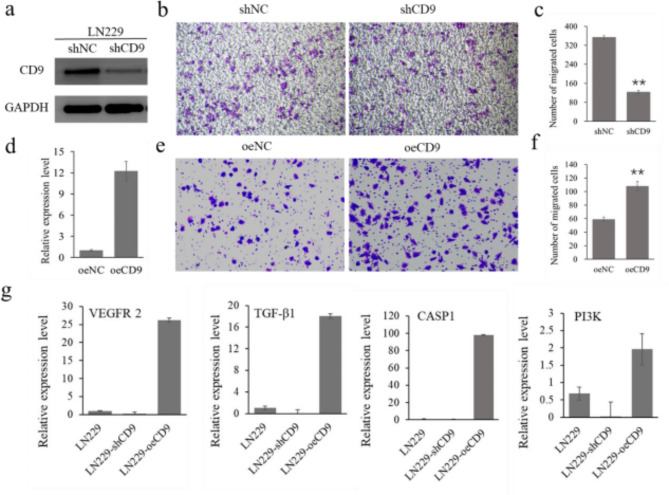
CD9 could regulate the migration of glioblastoma cell lines
To investigate the role of CD9 on migration of glioblastoma cell lines, short hairpin RNA was designed to knock down CD9 expression in LN229 cell line. The knockdown efficiency was evaluated using western blot analysis (Fig. S1) and shCD9-1 was selected for further study (Fig. 11a). Subsequently, the trans well assays was carried out to evaluate cell proliferation and the results showed that CD9 knockdown significantly decreased the migratory ability of LN229 cells (Fig. 11b, c). Additionally, LN229 cells were transfected with CD9 overexpression vector and negative control respectively. qRT-PCR analysis showed that the expression level of CD9 was notably upregulated in CD9 overexpression lines (Fig. 11d). Transwell assay demonstrated that the migratory ability of LN229 cells with overexpression of CD9 was significantly increased compared to the negative control (Fig. 11e, f). Then, vascular endothelial growth factor receptor-2 (VEGFR-2), transforming growth factor beta1 (TGF-β1), caspase 1(CASP1) and phosphoinositide 3-kinase (PI3K), which were differentially expressed in Hallmark pathways, were selected to check if the gene expression level was associated with CD9. The results showed that all these genes were down regulated in CD9 knockdown cell lines and up regulated in CD9 overexpression cell lines, compared with control LN229 cell lines (Fig. 11g). All these results demonstrated that CD9 could increase the migration of glioblastoma cells through mediating several essential pathways.
Fig. 11.
CD9 increased cell migration of glioblastoma cell lines. (a) western blot analysis of CD9 abundance in vector-control cells (shNC) and CD9 short hairpin cells (shCD9); (b,c) trans-well assay results showed that knockdown of CD9 gene significantly inhibited the proliferative ability of LN229 cell lines; (d) qRT-PCR of CD9 mRNA expression in vector-control cells (oeNC) and CD9-overexpressing cells (oeCD9); (e,f) trans-well assay showed that overexpression of CD9 gene significantly enhanced the proliferative ability of LN229 cell lines; (g) qRT-PCR of VEGFR2, TGF-β1, CASP1 and PI3K mRNA expression in LN229, shCD9 and oeCD9 cell lines. Asterisks indicate significant differences with respect to values of vector control (Student’s t-test): **P < 0.01.
Discussion
Glioma is the most common primary tumor of the central nervous system, and based on histological characteristics, gliomas can be classified into astrocytomas, oligodendrogliomas, and ependymomas, among others. Studies have revealed that the expression of CD9 in astrocytic tumors correlates with their degree of malignancy23. Conversely, in non-astrocytic gliomas, including oligodendrogliomas, anaplastic oligodendrogliomas, and ependymomas, immunohistochemical techniques have detected no immunoreactivity for CD9, indicating that the CD9 molecule is no longer expressed in these types of brain tumor counterparts24.Its overexpression is directly correlated with a lower survival rate and is considered a potential marker of glioma stem cells (GSC)19. This study employs bioinformatics methods to retrieve relevant information from multiple databases, exploring the association between CD9 and tumorigenesis, its prognostic value in glioma, and the potential molecular mechanisms involved.
In our investigation of transcriptomic data and survival information for pan-cancer and gliomas across TCGA, CGGA, and GEO databases, we identified differential expression of CD9 in tumor versus control samples across 11 cancer types, which correlated with patient survival outcomes. Specifically, in diseases such as GBMLGG, PAAD, UVM, and OV, higher expression levels of CD9 were associated with poorer prognoses. Conversely, in KIRC, SARC, COAD, LUSC, and PRAD, patients with elevated CD9 expression demonstrated more favorable survival outcomes. This suggests that there may be similar developmental patterns among these cancers, which could inform subsequent research on gliomas by referencing the molecular mechanisms involved in the tumorigenesis and progression of other tumors. This could not only guide more in-depth studies but also provide a theoretical foundation for the development of broad-spectrum anticancer therapies.
Compared to its expression in healthy brain tissue, CD9 is overexpressed on glioblastoma stem cells, and may be associated with the CD9 mutation. It has been found that during the progression of prostate cancer, the expression of CD9 is significantly reduced or even lost, which may be associated with the absence and mutation of CD9 mRNA25. The silencing of CD9 in glioblastoma stem cells followed by xenotransplantation into nude mice has been shown to prolong survival19. The pro-apoptotic and anti-proliferative properties of anti-CD9 monoclonal antibodies have been confirmed in vitro in human gastric cancer cell lines and in vivo in SCID mice engrafted with these malignant cell lines19. Our study demonstrates that in the CGGA glioma cohort, patients with high CD9 expression have a lower survival rate. Differences in CD9 expression persist across ages, genders, IDH mutation statuses, and radiotherapy conditions.
The expression of CD9 increased after neoadjuvant chemoradiotherapy in colorectal adenocarcinoma patients26, Jennrich S27 reported that CD9 and CD81-positive extracellular vesicles (EVs) were detected in the supernatant of all glioblastoma cells, and the release of CD9 and CD81-positive EVs increased after radiotherapy-induced cell apoptosis. Although EVs secreted by non-irradiated cells did not predict radiosensitivity, increased EV release after irradiation was associated with a cytotoxic response to radiation therapy 72 h after irradiation, suggesting a novel application of EVs in the monitoring of anticancer therapy. Our study suggests that the expression of CD9 in glioma patients after radiotherapy is higher than that in patients without radiotherapy, which is consistent with the literature report, suggesting that there may be a certain correlation between radiotherapy and CD9 expression. It is hypothesized that radiotherapy induces epigenetic remodeling, such as DNA methylation and histone modifications28, which in turn affects cellular signaling pathways. These alterations can influence the expression of CD9. Additionally, during radiotherapy, the DNA damage response activates repair mechanisms, conferring radioresistance to the tumor. This process involves pathways related to damage detection, signal transduction, and activation of cell cycle checkpoints. CD9 is implicated in various physiological activities of the cell, including signal transduction and cell proliferation. Therefore, the occurrence of a DNA damage response may stimulate the expression of CD9. At this juncture, if CD9 function is compromised, it could lead to impaired apoptotic mechanisms and obstructed signal transduction, disrupting the normal function of signaling pathways. This disruption may affect the DNA damage response and repair pathways, potentially resulting in genetic mutations. Further studies are needed to verify the effect of radiotherapy on CD9. Studies have identified that mutations in isocitrate dehydrogenase (IDH) lead to the accumulation of D-2-Hydroxyglutarate (D2HG) within cells, surpassing physiological levels. This accumulation acts to inhibit normal cellular differentiation by inducing epigenetic dysfunctions, thereby promoting tumor progression29. Epigenetic mechanisms, such as DNA methylation and histone modifications, may regulate the expression of the CD9 gene by affecting its chromatin structure, playing a pivotal role particularly in the downregulation of CD930. Our research has revealed that, compared to the wild-type IDH, the expression of CD9 is significantly downregulated in the IDH mutant group. Consequently, it can be inferred that the epigenetic alterations caused by IDH mutations may regulate the expression of CD9. However, to elucidate the specific reasons for this differential expression, further studies involving functional experiments such as overexpression or knockdown of epigenetic modifying enzymes, as well as the analysis of clinical data, are required.
In the GSE16011 tumor group, high expression of CD9 was observed, with an area under the ROC curve (AUC) greater than 0.7, indicating a high discriminatory capacity. Based on these findings, we conducted an in-depth investigation into the impact of CD9 on the survival of patients with gliomas, as well as potential signaling pathways, immunological relevance, and drug sensitivity. Based on the diversity of cell types and associated molecules, CD9 exhibits a multitude of biological activities, such as cell adhesion, migration, signal transduction, inflammation, and processes related to tumorigenesis14,31–34. STAT3 is a signaling pathway associated with inflammation, and Shi et al.35 discovered that CD9 stabilizes GP130 by inhibiting ubiquitin-dependent lysosomal degradation, thereby promoting the activation of STAT3 in SC). CD9 is essential for the self-renewal and proliferation of GSC. Our results are consistent with literature studies, suggesting that CD9 is an adverse prognostic factor for glioma.
Our study revealed that CD9 exhibits the highest correlation with neutrophils. Neutrophils, as integral components of the tumor microenvironment (TME), are abundantly present in circulation and can exert carcinogenic effects by promoting angiogenesis and immune suppression36. Glioblastoma releases a variety of chemokines and cytokines that enhance the recruitment of neutrophils within the TME37. This increased recruitment of neutrophils is associated with the progression of glioblastoma and may also contribute to the development of therapeutic resistance38. CD9 is expressed in multiple cell types, including immune and tumor cells, and is involved in intercellular interactions, signal transduction, and cell migration. Studies have found that CD9 can regulate the stability of the gp130 protein, which mediates the activation of key pro-survival pathways such as angiogenesis39. Therefore, it can be hypothesized that CD9 may indirectly promote the formation of the GBM vascular network by affecting the angiogenic function of neutrophils. Given the relevance of CD9 to neutrophils, immunotherapeutic strategies might need to consider targeting CD9 as a potential immunomodulatory target. By inhibiting the expression or function of CD9, it may be possible to reduce the pro-tumorigenic activities of neutrophils, thereby enhancing anti-tumor immune responses.
Our study identifies BIRB.0796 and Z.LLNle.CHO as potential pharmacological agents for the treatment of glioma. BIRB.0796 has been identified as an inhibitor of MMP-1 and MMP-10 mRNA expression. MMP-1 and MMP-10 are pivotal members of the matrix metalloproteinase (MMP) family, which play a crucial role in the degradation of the extracellular matrix, tissue remodeling, and the invasion and metastasis of tumors. These enzymes facilitate the alteration of the cellular microenvironment by degrading components of the extracellular matrix, thereby influencing biological behaviors such as cell proliferation, migration, and apoptosis. Clinically, the expression levels of MMP-1 and MMP-10 have become important markers for assessing cancer progression and prognosis40–42. Z-LLNle-CHO, a γ-secretase inhibitor, impedes the activity of γ-secretase and the Akt-mediated pro-survival pathway, thus inhibiting cell proliferation and survival43. Studies have found that the combination of γ-secretase inhibitors (GSIs) with radiotherapy is a rational strategy for treating patients with non-small cell lung cancer (NSCLC). Treatment with GSIs following radiotherapy can significantly enhance radiation-mediated cytotoxicity in tumor cells44. Given its ability to induce apoptosis in tumor cells and affect cell survival pathways, it may contribute to overcoming resistance in glioblastoma multiforme (GBM), although this requires specific research on GBM cell lines and temozolomide (TMZ) resistance to validate.
When CD9 is used as a potential tumor-associated antigen molecular target, the widespread expression of CD9 in various tissues and organs must be considered, and the effect of using anti-CD9 antibodies, especially when combined with toxins or cytotoxic drugs, can be complicated.
In our cellular experiments, we discovered that CD9 enhances the migratory capacity of glioblastoma cells. We conducted qRT-PCR analyses on several pathway genes identified through bioinformatics analysis and observed that key genes such as VEGFR-2 and PI3K exhibited corresponding expression changes in cell lines with upregulated or downregulated CD9. These findings suggest that CD9’s influence on the migratory ability of glioblastoma cell lines may be associated with these pathways. Our results indicate that CD9 could be a potential oncogenic driver and therapeutic target in glioblastoma.
Our study delves into the role of CD9 in cancer progression and patient clinical outcomes, providing novel scientific support for personalized treatment strategies in gliomas. However, there are limitations to our research. Although we have conducted preliminary investigations into the function of CD9 in gliomas at the cellular and molecular levels, our findings lack direct in vivo evidence. Future research should include animal model experiments to validate the role of CD9 in the onset and progression of gliomas and to assess its potential value in clinical therapy. Additionally, our study did not encompass all possible glioma subtypes. Comparisons between IDH mutant and wild-type variants were not addressed. More comprehensive clinical samples and data are needed to conduct a deeper analysis of CD9 expression across different glioma subtypes and its relationship with clinical characteristics.
Conclusion
In this study, we primarily employed bioinformatics methods to analyze the differential expression of CD9 across various tumor types and its prognostic value in gliomas. Our findings indicate that high expression of CD9 is associated with poor prognosis in glioma patients. We further investigated the association between CD9 expression and patient prognosis in glioma, delving into the underlying biological pathways, the connection with the immune microenvironment, and drug sensitivity. Our findings offer new theoretical support and reference for the treatment of patients with glioma, potentially guiding more effective therapeutic strategies.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by the National and Provincial Clinical Key Specialty Capacity Building Project of 2020 - Li Wenbin and the Beijing Tiantan Hospital Talent Introduction Program - Li Wenbin (RCYJ-2020-2025-LWB).
Abbreviations
- GBM
Glioblastoma multiforme
- GBMLGG
Glioblastoma multiforme and lower-grade glioma
- PAAD
Pancreatic adenocarcinoma
- UVM
Uveal melanoma
- OV
Ovarian cancer
- KIRC
Renal cell carcinoma
- SARC
Sarcoma
- COAD
Colorectal adenocarcinoma
- LUSC
Lung squamous cell carcinoma
- PRAD
Prostate adenocarcinoma
- HNSCC
Head and neck squamous cell carcinoma
- AUC
Area under curve
- DCs
Human dendritic cells
- EVs
Extracellular vesicles
- GSC
Glioma stem cells
Author contributions
J.J. Bioinformatics analysis, validation experiment operation, data management and article writing; W.B.L. Experimental design, data management, article revision; B.J. Guidance of bioinformatics analysis and cell experiments.
Data availability
The TCGA-glioma (GBMLGG) dataset utilized in this study was sourced from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/), while the CGGA-glioma dataset was acquired from the Chinese Glioma Genome Atlas (CGGA) database (http://www.cgga.org.cn/). Additionally, GSE16011 and validation set CSE68848 were obtained from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/).
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Bo Jiang, Email: jiangboprof@163.com.
Wen-bin Li, Email: liwenbin@ccmu.edu.cn.
References
- 1.Yang, K. et al. Glioma targeted therapy: insight into future of molecular approaches. Mol. Cancer. 21. 10.1186/s12943-022-01513-z (2022). [DOI] [PMC free article] [PubMed]
- 2.Schaff, L. R. & Mellinghoff, I. K. Glioblastoma and other primary brain malignancies in adults: a review. JAMA. 329, 574–587 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang, Y. et al. Pretreatment geriatric assessments of elderly patients with glioma: development and implications. Aging Dis. 11, 448–461 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glioma subclassifications and their clinical significance. Neurotherapeutics. 14 (2), 284–297 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waitkus, M. S., Diplas, B. H. & Yan, H. Isocitrate dehydrogenase mutations in gliomas. Neuro Oncol. 18 (1), 16–26 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu, C. et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 483 (7390), 474–478 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hai, Y. et al. IDH1 and IDH2 mutations in gliomas. N Engl. J. Med. 360 (8), 765–773 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu, S., Tan, L., Li, X., Fan, F. & Liu, Z. Immunotherapy for glioma: current management and future application. Cancer Lett. 476, 1–12 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Dharan, R. et al. Transmembrane proteins tetraspanin 4 and CD9 sense membrane curvature. Proc. Natl. Acad. Sci. U S A. 119, e2208993119. 10.1073/pnas.2208993119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fabre, T. et al. Identification of a broadly fibrogenic macrophage subset induced by type 3 inflammation. Sci. Immunol. 8, eadd8945. 10.1126/sciimmunol.add8945 (2023). [DOI] [PubMed] [Google Scholar]
- 11.Li, Y. et al. CD9 exacerbates pathological cardiac hypertrophy through regulating GP130/STAT3 signaling pathway. iScience. 26, 108070. 10.1016/j.isci.2023.108070 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nicolai, L. et al. Vascular neutrophilic inflammation and immunothrombosis distinguish severe COVID-19 from influenza pneumonia. J. Thromb. Haemost. 19, 574–581 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang, J. C. et al. Down-regulation of CD9 expression during prostate carcinoma progression is associated with CD9 mRNA modifications. Clin. Cancer Res. 13, 2354–2356 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Lorico, A., Lorico-Rappa, M., Karbanová, J., Corbeil, D. & Pizzorno, G. CD9, a tetraspanin target for cancer therapy? Exp. Biol. Med. (Maywood). 246, 1121–1138 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ondruššek, R. et al. Prognostic value and multifaceted roles of tetraspanin CD9 in cancer. Front. Oncol. 13, 1140738. 10.3389/fonc.2023.1140738 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han, X. et al. Neoadjuvant chemotherapy endows CD9 with prognostic value that differs between tumor and stromal areas in patients with pancreatic cancer. J. Clin. Lab. Anal. 36, e24517. 10.1002/jcla.24517 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lucarini, G. et al. Tetraspanin CD9 expression predicts sentinel node status in patients with cutaneous melanoma. Int. J. Mol. Sci. 23, 4775. 10.3390/ijms23094775 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blake, D. J., Martiszus, J. D., Lone, T. H. & Fenster, S. D. Ablation of the CD9 receptor in human lung cancer cells using CRISPR/cas alters migration to chemoattractants including IL-16. Cytokine. 111, 567–570 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Podergajs, N. et al. Transmembrane protein CD9 is glioblastoma biomarker, relevant for maintenance of glioblastoma stem cells. Oncotarget. 7, 593–609 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robin, X. et al. pROC: an open-source package for R and S + to analyze and compare ROC curves. BMC Bioinform. 12, 77. 10.1186/1471-2105-12-77 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeng, D. et al. IOBR: multi-omics immuno-oncology biological research to decode tumor microenvironment and signatures. Front. Immunol. 12, 87975. 10.3389/fimmu.2021.687975 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geeleher, P., Cox, N. & S Huang, R. pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS One. 9, e107468. 10.1371/journal.pone.0107468 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haydo, A. et al. Combining organotypic tissue culture with light-sheet microscopy (OTCxLSFM) to study glioma invasion. EMBO Rep. 24 (12), e56964. 10.15252/embr.202356964 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawashima, M., Doh-ura, K., Mekada, E., Fukui, M. & Iwaki, T. CD9 expression in solid non-neuroepithelial tumors and infiltrative astrocytic tumors. J. Histochem. Cytochem. 50 (9), 1195–1203 (2002). [DOI] [PubMed] [Google Scholar]
- 25.Wang, J-C. et al. Down-regulation of CD9 expression during prostate carcinoma progression is associated with CD9 mRNA modifications. Clin. Cancer Res. 13 (8), 2354–2361 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Khushman, M. et al. The impact of neoadjuvant concurrent chemoradiation on exosomal markers (CD63 and CD9) expression and their prognostic significance in patients with rectal adenocarcinoma. Oncotarget. 15, 1490–1498 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jennrich, S. et al. CD9- and CD81-positive extracellular vesicles provide a marker to monitor glioblastoma cell response to photon-based and Proton-based radiotherapy. Front. Oncol. 12, 947439. 10.3389/fonc.2022.947439 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peng, Q. et al. A perspective of epigenetic regulation in radiotherapy. Front. Cell. Dev. Biol. 9, 624312. 10.3389/fcell.2021.624312 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waitkus, M. S., Diplas, B. H. & Yan, H. Biological role and therapeutic potential of IDH mutations in cancer. Cancer Cell. 34 (2), 186–195 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Bruyne, E. et al. Epigenetic silencing of the tetraspanin CD9 during disease progression in multiple myeloma cells and correlation with survival. Clin. Cancer Res. 14 (10), 2918–2926 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Umeda, R. et al. Structural insights into tetraspanin CD9 function. Nat. Commun. 11 10.1038/s41467-020-15459-7 (2020). [DOI] [PMC free article] [PubMed]
- 32.Reyes, R., Cardeñes, B., Machado-Pineda, Y. & Cabañas, C. Tetraspanin CD9: a key regulator of cell adhesion in the immune system. Front. Immunol. 9, 863. 10.3389/fimmu.2018.00863 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Powner, D., Kopp, P. M., Monkley, S. J., Critchley, D. R. & Berditchevski, F. Tetraspanin CD9 in cell migration. Biochem. Soc. Trans. 39, 563–567 (2011). [DOI] [PubMed] [Google Scholar]
- 34.Brosseau, C., Colas, L., Magnan, A. & Brouard, S. CD9 tetraspanin: a new pathway for the regulation of inflammation? Front. Immunol. 9, 2316. 10.3389/fimmu.2018.02316 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi, Y. et al. Tetraspanin CD9 stabilizes gp130 by preventing its ubiquitin-dependent lysosomal degradation to promote STAT3 activation in glioma stem cells. Cell. Death Differ. 24, 167–180 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiong, S., Dong, L. & Cheng, L. Neutrophils in cancer carcinogenesis and metastasis. J. Hematol. Oncol. 14 (1), 173 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun, C. et al. Neutrophils in glioma microenvironment: from immune function to immunotherapy. Front. Immunol. 15 (1393173). 10.3389/fimmu.2024.1393173 (2024). [DOI] [PMC free article] [PubMed]
- 38.Liang, J. et al. Neutrophils promote the malignant glioma phenotype through S100A4. Clin. Cancer Res. 20 (1), 187–198 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi, Y. et al. Tetraspanin CD9 stabilizes gp130 by preventing its ubiquitin-dependent lysosomal degradation to promote STAT3 activation in glioma stem cells. Cell. Death Differ. 24 (1), 167–180 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Turner, N. A., Warburton, P., O’Regan, D. J., Ball, S. G. & Karen, E. Porter.Modulatory effect of interleukin-1α on expression of structural matrix proteins, MMPs and TIMPs in human cardiac myofibroblasts: role of p38 MAP kinase. Matrix Biol. 29 (7), 613–620 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scheau, C. et al. The role of matrix metalloproteinases in the epithelial-mesenchymal transition of hepatocellular carcinoma. Anal Cell Pathol. 9423907. 10.1155/2019/9423907 (2019). [DOI] [PMC free article] [PubMed]
- 42.Cui, N. & Khalil, M. H. R. A. Biochemical and biological attributes of matrix metalloproteinases. Prog Mol. Biol. Transl. Sci. 147, 1–73 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fukuzaki, Y., Sugawara, H. & Yamanoha, B. 532 nm low-power laser irradiation recovers γ-secretase inhibitor-mediated cell growth suppression and promotes cell proliferation via akt signaling. PLoS One. 8 (8), e70737. 10.1371/journal.pone.0070737 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mizugaki, H. et al. M Nishimura.γ-Secretase inhibitor enhances antitumour effect of radiation in notch-expressing lung cancer. Br. J. Cancer. 106 (12), 1953–1959 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jong, D. AT1412, a patient-derived antibody in development for the treatment of CD9 positive precursor B-acute lymphoblastic leukemia. Blood. 134, 4461. 10.1182/blood-2019-127894 (2019). [Google Scholar]
- 46.Neviani, V. et al. Site-specific functionality and tryptophan mimicry of lipidation in tetraspanin CD9. FEBS J. 287, 5323–5344. 10.1111/febs.15295 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The TCGA-glioma (GBMLGG) dataset utilized in this study was sourced from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/), while the CGGA-glioma dataset was acquired from the Chinese Glioma Genome Atlas (CGGA) database (http://www.cgga.org.cn/). Additionally, GSE16011 and validation set CSE68848 were obtained from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/).