Abstract
Background: Inflammation is the major contributor to the pathophysiology of ischemic stroke (IS). Long non-coding ribonucleic acids (lncRNAs) metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) and tumor necrosis factor and heterogeneous nuclear ribonucleoprotein L-related immunoregulatory (THRIL) have been demonstrated to be up-regulated in inflammation and atherosclerosis. Therefore, we aimed to study the expression profile of these lncRNAs after IS.
Methods: This observational case-control study was conducted in Namazi Hospital, Shiraz, Iran. The real-time polymerase chain reaction (RT-PCR) measured the sequential changes in circulating levels of MALAT1 and THRIL on days 1, 3, and 5 after IS. The receiver operating characteristic (ROC) curve analysis was used to estimate the diagnostic and prognostic potential of lncRNAs with the area under the curve (AUC).
Results: In patients with IS, the relative MALAT1 and THRIL expressions were significantly higher than the controls (P < 0.001 and P < 0.01, respectively), on days 1, 3, and 5 after stroke.
We showed a significantly increase in lncRNAs expression on day five compared to days 1 and 3 after stroke. Moreover, a positive correlation was detected between MALAT1 expression and time within the first 24 hours after stroke (r = 0.27, P = 0.03). Logistic regression analysis showed a significant positive association between MALAT1 and THRIL and the risk of stroke evolution. We found a potential diagnostic marker for MALAT1 with an AUC of 0.78.
Conclusion: We demonstrated the significant sequential upregulation in MALAT1 and THRIL expression on days 1, 3, and 5 after IS with a significant positive association with the risk of stroke. MALAT1 also significantly correlated with time within the first 24 hours after stroke.
Key Words: Long Noncoding RNA, MALAT1 Long Noncoding RNA, THRIL Long Noncoding RNA, Ischemic Stroke
Introduction
Ischemic stroke (IS) is the first cause of morbidity and one of the most important causes of mortality worldwide.1 There is no accurate, rapid, cost-effective laboratory biomarker for the diagnosis of IS.2
Long non-coding ribonucleic acids (lncRNAs) are ribonucleic acids (RNAs) with more than 200 nucleotides without contributing to protein synthesis.3 In the past decade, authors have identified the potential of different lncRNAs as diagnostic and prognostic markers in IS.2,4-6 Several studies have demonstrated the significant relations of lncRNAs metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) and tumor necrosis factor and heterogeneous nuclear ribonucleoprotein L-related immunoregulatory (THRIL) in the pathogenesis of inflammation, atherosclerosis, and endothelial cell dysfunction.7-14
A significant increase in MALAT1 and THRIL has also been reported in the peripheral blood of patients with coronary artery disease.15-18 The cholesterol-filled foam cell accumulation is accelerated in blood vessels by MALAT1, enhancing lipid uptake in macrophages by CD36 expression and promoting atherosclerosis.19 Changes in the expression pattern of MALAT1 and THRIL in IS, especially in atherosclerotic stroke, are still unclear. Ren et al. showed the downregulation of MALAT1 in patients with IS within 24 hours from stroke onset.20 Another study has reported MALAT1 upregulation in the G variant of rs619586, which was associated with an increased risk of IS.21
Therefore, in this study, we evaluated the sequential changes in expression levels of two key lncRNAs associated with atherosclerosis and inflammation (MALAT1 and THRIL) in peripheral blood of patients with confirmed large-artery atherosclerosis (LAA) and small-vessel disease (SVD) IS, to determine their expression levels at different time points on days 1, 3, and 5 after the event to identify the association between these lncRNAs with the clinical and laboratory parameters as well as their potential diagnostic biomarker in IS.
Materials and Methods
Participants: This case-control study was conducted in the neurology ward of Namazi Hospital at Shiraz University of Medical Sciences, Shiraz, south of Iran, between August 2019 and August 2020. All consecutive patients with IS were recruited.
The control group was composed of a representative sample of Shiraz population which were sex and age-matched with cases.
According to the Recognition of Stroke in the Emergency Room (ROSIER) scale, IS was screened as a focal neurological deficit of sudden onset that persisted beyond 24 hours. Radiological confirmation was conducted by a brain computed tomography (CT) or magnetic resonance imaging (MRI). A definite diagnosis was made according to the guidelines for the early management of patients with acute IS (AIS).13 The inclusion criteria for stroke included patients older than 18 years, filling the informed consent form with confirmed SVD or LAA according to TOAST classification,22 and within 0-24 hours after symptom onset. Patients with a transient ischemic attack (TIA) were excluded. Cases with intra-parenchymal hemorrhage (IPH), dissection, vasculitis, reversible vasoconstriction syndrome, cerebral venous thrombosis (CVT), Moyamoya disease, hypoperfusion syndromes, iatrogenic stroke, and post-infectious stroke were excluded as well. This study defined patients with diabetes and hypertension (HTN) according to the guidelines.23,24 The stroke severity was evaluated by the National Institutes of Health Stroke Scale (NIHSS) score on admission.25 The outcomes were obtained three months after admission according to the modified Rankin scale (mRS) blinded to the level of lncRNA.26 Ethics approval for this study was obtained by the local ethics committee of Shiraz University of Medical Sciences (IR.SUMS.REC.1398.17988). Written informed consent was provided from all subjects (or their proxy respondents). Blood sampling from patients was performed in 3 times after onset of stroke [day 1 (n = 59), day 3 (n = 43), and day 5 (n = 25) after stroke].
RNA extraction and real-time polymerase chain reaction (RT-PCR): At the first, total RNAs were extracted from blood specimens by using TRIzol Reagent (GeneAll, Seoul, South Korea) according to the acid guanidinium thiocyanate-phenol-chloroform (AGPC) method. Then complementary deoxyribonucleic acid (cDNA) synthesis was performed by the OneStep RT-PCR Kit (AddBio, Seoul, South Korea) according to the manufacturer’s instructions. Regular expressions (REs) of MALAT1 and THRIL were measured in all specimens using the RealQ Plus 2x Master Mix Green High ROX™ (AddBio, Seoul, South Korea). All reactions were conducted in StepOnePlus™ RealTime PCR equipment (Applied Biosystems, Foster City, CA, USA) in duplicate with the thermal-cycling settings of 10 minutes at 95 °C (1 repeat) accompanied by 40 cycles for 15 seconds at 95 °C, 25 seconds at 60 °C, and 25 seconds at 72 °C. Thermal cycling in the melting phase was as follows: 15 seconds at 95 °C, 25 seconds at 60 °C, and 15 seconds at 72 °C. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene was used as a normalizer. The results were calculated with the 2−ΔCt method.27 The primer sequences are used as follows:
MALAT1:
Forward: 5′-TCAGTGTTGGGGCAATCTT-3′
Reverse: 5′-CGTTCTTCCGCTCAAATCC-3′
THRIL:
Forward: 5′-TGTGATCCATACTCCTCGG-3′
Reverse: 5′-AGGCAAGGGAGTTTCAGAA-3′
GAPDH:
Forward: 5′-GCATCTTCTTTTGCGTCG-3′
Reverse: 5′-TGTAAACCATGTAGTTGAGGT-3′ (27).
The chi-square test and independent two-sample t-test were used to compare two categorical and two numeric variables. We used logistic regression analyses between cases and controls to evaluate the association of lncRNAs expression levels and clinical parameters with the risk of IS. The significant time interaction for MALAT1 and THRIL expression was estimated by linear mixed model analysis after adjusting the group. The lncRNAs expression level was shown as mean ± standard error (SE). The relationship between lncRNAs levels with clinical parameters was analyzed using linear regression. We used the Spearman correlation to identify the correlation between NIHSS and lncRNA expression. The receiver operating characteristic (ROC) curve analysis was used to estimate the diagnostic and prognostic potential of MALAT1 and THRIL with the area under the curve (AUC). The analyses were done using the SPSS software (version 19, SPSS Inc., Chicago, IL, USA) and GraphPad Prism 5.0. The P < 0.05 was considered a significant value.
Results
Demographic and clinical parameters of patients with IS and controls: A total of 59 patients with IS and 63 controls were included in the study. Vascular risk factors such as diabetes and HTN were more prevalent in cases compared to controls (P < 0.05) (Table 1). In laboratory results, there were no significant differences in the levels of total cholesterol, triglyceride (TG), and low-density lipoprotein (LDL) between IS cases and controls; however, high-density lipoprotein (HDL) level was significantly lower in IS cases compared to controls.
Table 1.
Demographic and clinical characteristics of the patients and controls
| Characteristics | Cases (n = 59) | Controls (n = 65) | P |
|---|---|---|---|
| Gender [n (%)] | |||
| Men | 42 (71.20) | 43 (68.25) | 0.900* |
| Women | 17 (28.80) | 20 (31.75) | |
| Age (year) (mean ± SD) | 63.90 ± 1.80 | 64.66 ± 1.70 | 0.900** |
| BMI (kg/m2) (mean ± SD) | 26.15 ± 0.60 | 25.81 ± 3.97 | 0.900** |
| NIHSS at admission [n (%)] | |||
| ≤ 6 | 22 (37.30) | ||
| ≥ 7 | 37 (62.70) | ||
| Aspect [n (%)] | |||
| < 6 | 14 (23.70) | ||
| ≥ 6 | 45 (76.30) | ||
| mRS at admission [n (%)] | |||
| 0-2 | 12 (20.33) | ||
| 3-6 | 47 (79.66) | ||
| mRS at 3 months [n (%)] | |||
| 0-2 | 24 (40.67) | ||
| 3-6 | 35 (59.32) | ||
| Vascular risk factors [n (%)] | |||
| HTN | |||
| Yes | 31 (52.50) | 19 (30.15) | 0.010* |
| No | 28 (47.50) | 44 (69.84) | |
| Diabetes | |||
| Yes | 21 (35.60) | 10 (15.88) | 0.010* |
| No | 38 (64.40) | 53 (84.12) | |
| Smoking | |||
| Yes | 10 (15.90) | 8 (12.69) | 0.300* |
| No | 49 (83.10) | 55 (87.30) | |
| Drinking | |||
| Yes | 2 (3.40) | 2 (3.18) | 0.600* |
| No | 57 (96.60) | 61 (96.82) | |
| HLP | |||
| Yes | 19 (32.20) | 19 (30.15) | 0.400* |
| No | 40 (67.80) | 44 (69.84) | |
| Laboratory findings (mean ± SD) | |||
| TG (mg/dl) | 126.70 ± 7.08 | 132.81 ± 9.80 | 0.600** |
| Total cholesterol (mg/dl) | 161.20 ± 5.60 | 162.19 ± 4.10 | 0.900** |
| LDL (mg/dl) | 97.60 ± 4.50 | 90.60 ± 3.70 | 0.200** |
| HDL (mg/dl) | 33.70 ± 0.90 | 44.21 ± 1.50 | 0.003** |
| Types of stroke [n (%)] | |||
| LAA | 29 (49.20) | - | |
| SVD | 30 (50.80) | - |
HTN: Hypertension; LDL: Low-density lipoprotein; HDL: High-density lipoprotein; HLP: Hyperlipidemia; TG: Triglyceride; LAA: Large-artery atherosclerosis; SVD: Small-vessel disease; BMI: Body mass index; NIHSS: National Institutes of Health Stroke Scale; mRS: Modified Rankin scale; SD: Standard deviation
Chi-square test;
Independent two-sample t-test
The levels of MALAT1 and THRIL lncRNAs in peripheral blood of patients with IS at different time points: In the present study, we evaluated MALAT1 and THRIL expression on days 1, 3, and 5 after stroke, in 59, 43, and 25 cases, respectively, and 63 controls. Blood sampling was repeated from 25 cases on days 1, 3, and 5 after stroke. All 59 patients were not hospitalized for five days, and 34 were discharged earlier. We had only 25 fixed cases hospitalized for a total of five days. Thus, we used linear mixed model analysis. After adjusting the group, a significant time interaction was detected for MALAT1 and THRIL expression. The level of THRIL on days 1 and 3 was significantly lower than the expression on day five, respectively, and the MALAT1 level on day 1 was significantly lower than the expression on day 5 (Table 2). By logistic regression analysis, increasing MALAT1 and THRIL levels showed a significant association with the risk of IS. After adjusting for relevant clinical and laboratory variables [body mass index (BMI), HTN, hyperlipidemia (HLP), diabetes mellitus (DM), smoking], elevated MALAT1 and THRIL levels remained significant [P = 0.001, odds ratio (OR) = 1.52, 95% confidence interval (CI): 1.18-1.96 and P = 0.03, OR = 1.26, 95% CI: 1.01-1.57, respectively).
Table 2.
Time interaction for MALAT1 and THRIL expression estimated by mixed model analysis after adjusting the group
| Parameter |
Estimate
THRIL |
P |
95% CI
|
Estimate MALAT1 | P |
95% CI
|
||
|---|---|---|---|---|---|---|---|---|
| Lower | Upper | Lower | Upper | |||||
| Intercept | 9.64 | < 0.001 | 7.08 | 12.20 | 6.80 | < 0.001 | 4.91 | 8.69 |
| (Day 1) | -6.47 | < 0.001 | -9.36 | -3.59 | -2.69 | 0.011 | -4.77 | -0.62 |
| (Day 3) | -5.67 | < 0.001 | -8.68 | -2.66 | -0.08 | 0.934 | -2.24 | 2.06 |
| (Day 5) Ref | 0 | 0 | . | - | - | |||
After adjusting the group, the significant time interaction was detected for MALAT1 and THRIL expression. The level of THRIL on days 1 and 3 significantly was lower than the expression on day 5, respectively, and the MALAT1 level on day 1 significantly was lower than the expression on day 5.
CI: Confidence interval
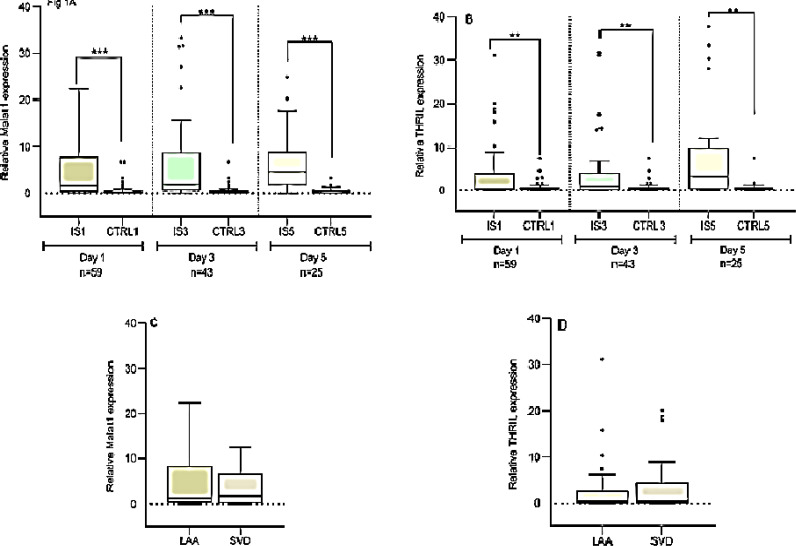
MALAT1 expression in patients with IS was significantly higher than the controls at different time points on day 1 (4.10 ± 0.65 vs. 0.63 ± 0.16), day 3 (6.70 ± 1.40 vs. 0.60 ± 0.20), and day 5 (6.50 ± 1.30 vs. 1.10 ± 0.36, respectively) (All P < 0.001) (Figure 1A).
Figure 1.
The expression levels of MALAT1 and THRIL in patients, controls on days 1, 3, and 5 after stroke A and B: Independent Student’s t-test revealed that MALAT1 and THRIL expressions were significantly higher in patients with ischemic stroke (IS) relative to controls on days 1, 3, and 5 after stroke; C and D: The comparison of blood levels of MALAT1 and THRIL in large-artery atherosclerosis (LAA) and small-vessel disease (SVD) Results were expressed as mean ± standard error of the mean (SEM)
**P < 0.010; ***P < 0.001
On the first day after stroke, the MALAT1 lncRNA level was higher than the control and this upregulation remained at the high level on days 3 and 5 after stroke onset.
THRIL expression in patients with IS significantly was higher than the controls on day 1 (3.16 ± 0.79 vs. 0.70 ± 0.14), day 3 (3.90 ± 1.10 vs. 0.71 ± 0.17), and day 5 (9.30 ± 2.70 vs. 2.60 ± 1.80, respectively) (All P < 0.01) (Figure 1B). There were no significant differences in the expression of two lncRNAs between patients with LAA and patients with SVD (Figures 1C and 1D).
Association of MALAT1 and THRIL expression with clinical parameters : We used linear regression analysis to detect the association between the lncRNAs levels with clinical parameters and types of stroke in 59 patients (Table 3).
Table 3.
Linear regression analysis for the association between clinical parameters with MALAT1 and THRIL lncRNA levels in patients with ischemic stroke (IS)
| Variables | Beta MALAT1 | P | Lower | Upper | Beta THRIL | P | Lower | Upper |
|---|---|---|---|---|---|---|---|---|
| Constant | 0.197 | -4.953 | 23.332 | 0.878 | -16.469 | 19.201 | ||
| Age | -0.102 | 0.572 | -0.162 | 0.091 | 0.122 | 0.511 | -0.107 | 0.212 |
| Sex | 0.150 | 0.341 | -1.795 | 5.080 | 0.056 | 0.728 | -3.581 | 5.089 |
| mRS | 0.224 | 0.201 | -0.372 | 1.724 | 0.024 | 0.893 | -1.233 | 1.411 |
| NIHSS | -0.262 | 0.143 | -0.458 | 0.068 | 0.158 | 0.390 | -0.189 | 0.475 |
| BMI | 0.078 | 0.605 | -0.212 | 0.361 | 0.048 | 0.759 | -0.306 | 0.417 |
| DM | 0.137 | 0.452 | -2.342 | 5.176 | 0.114 | 0.542 | -3.293 | 6.187 |
| HTN | 0.161 | 0.350 | -1.812 | 5.017 | 0.114 | 0.522 | -2.924 | 5.688 |
| HLP | 0.040 | 0.819 | -3.289 | 4.140 | 0.067 | 0.712 | -3.818 | 5.550 |
| Smoker | 0.055 | 0.714 | -3.267 | 4.732 | -0.045 | 0.772 | -5.775 | 4.313 |
| SVD | -0.223 | 0.195 | -5.599 | 1.176 | -0.114 | 0.520 | -5.648 | 2.896 |
| IHD | 0.035 | 0.831 | -3.135 | 3.882 | 0.008 | 0.963 | -4.323 | 4.526 |
NIHSS: National Institutes of Health Stroke Scale; mRS: Modified Rankin scale; BMI: Body mass index; DM: Diabetes mellitus; IHD: Ischemic heart disease; HTN: Hypertension; HLP: Hyperlipidemia; SVD: Small-vessel disease
There were no significant correlations between MALAT1 and THRIL expression with clinical parameters and type of stroke (LAA and SVD) in patients with IS. We found a non-significant negative correlation between MALAT1 expression and NIHSS (Beta = -0.333, P = 0.120).
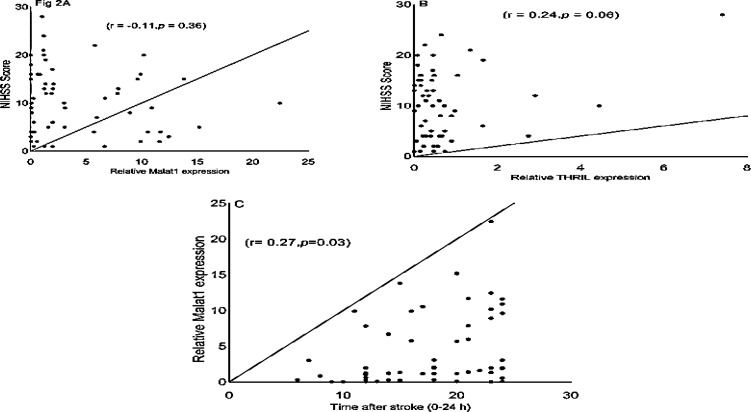
Spearman correlation of MALAT1 and THRIL lncRNAs with NIHSS and sampling time after stroke: Moreover, the Spearman correlation was also detected between the expression levels of MALAT1 and THRIL with NIHSS score in the patients with IS. As shown in figure 2A, the level of MALAT1 expression in all of the patients (n = 59) was not correlated with NIHSS scores (r = -0.11, 95% CI = -0.36 to 0.14, P = 0.360). We also found a non-significant positive correlation between THRIL expression and stroke severity (r = 0.24, 95% CI = -0.01 to 0.46, P = 0.060) (Figure 2B).
Figure 2.
The Spearman correlation between expression levels of MALAT1 and THRIL with National Institutes of Health Stroke Scale (NIHSS) and the time after stroke in patients with ischemic stroke (IS) A: The Spearman correlation between expression of MALAT1 and NIHSS in patients; B: Between THRIL expression and NIHSS; C: Between MALAT1 expression and time after stroke
We found a significant positive correlation between MALAT1 expression and time of blood sampling within the first 24 hours after stroke (r = 0.27, 95% CI = 0.01 to 0.49, P = 0.030) (Figure 2C).
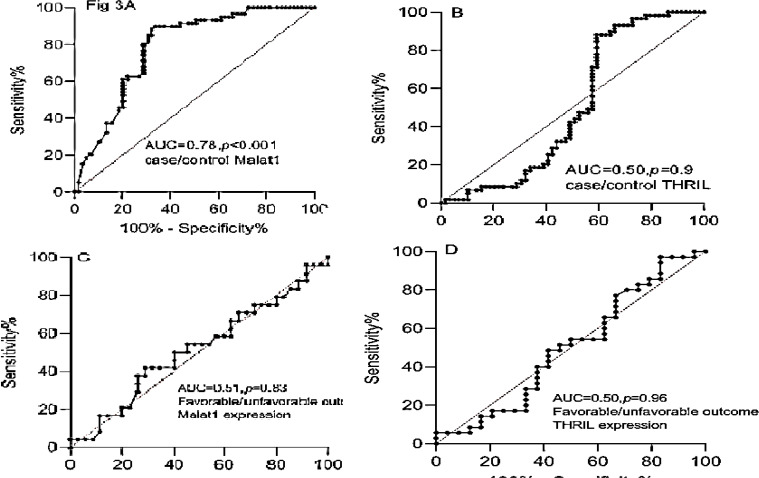
Diagnostic value and prediction of functional outcome of MALAT1 and THRIL expression levels in peripheral blood of patients with IS: ROC curve analyses were used to evaluate the diagnostic value of MALAT1 and THRIL lncRNAs for discriminating patients with IS from the controls which revealed that MALAT1 could be used as a diagnostic marker with an AUC of 0.78 (P < 0.001) (Figure 3A). The sensitivity and specificity were 79.6% and 71.2%, respectively.
Figure 3.
Receiver operating characteristic (ROC) curve analyses used to evaluate the diagnostic and prognostic values of MALAT1 and THRIL long non-coding ribonucleic acids (lncRNAs)
A and B: ROC curve analyses of MALAT1 and THRIL for discriminating patients with ischemic stroke (IS) from the controls; C and D: ROC curve analyses of MALAT1 and THRIL for predicting functional outcome
AUC: Area under the curve
ROC curve analysis of THRIL expression between cases and controls revealed that THRIL had a potential diagnostic marker for discriminating IS with an AUC of 0.50 (P = 0.900), and the sensitivity and specificity of 47.5% and 45.8%, respectively (Figure 3B).
In our study, mRS score of 3-6 in 3 months after stroke was considered an unfavorable functional outcome. Using a ROC curve analysis, MALAT1 and THRIL levels showed no significant predictive prognosis for a 3-month unfavorable outcome relative to a favorable outcome with an AUC of 0.51 (95% CI: 0.36-0.66, P = 0.800) and AUC of 0.50 (95% CI: 0.34-0.65, P = 0.900), respectively (Figures 3C and 3D). The sensitivity and specificity were 54.2%-54.3% and 51.4%-54.1%, respectively.
Discussion
Our results indicated that the relative level of MALAT1 was significantly elevated in peripheral blood of patients with IS compared to controls. The protective, anti-apoptotic, and anti-inflammatory roles of MALAT1 in brain microvasculature and animal model were reported by Liu et al.28 Ren et al. reported the downregulation and protective role of MALAT1 in patients with IS.20 The protective roles of lncRNA MALAT1 in neurological or cerebrovascular diseases were also demonstrated through activating phosphatidylinositol 3-kinase (PI3K)29 via inhibition of pro-apoptotic or pro-inflammatory factors.28,30 Fathy et al. found an association between the polymorphism and MALAT1 expression in patients with IS. In Fathy et al.'s study, patients with the G variant of MALAT1 rs619586 showed a higher serum MALAT1 expression with a negative association with NIHSS.21
We could show the high expression level of MALAT1 on days 1, 3, and 5 in 59 patients after stroke without significant difference between diabetic and nondiabetic patients and without significant correlation with NIHSS. These results were inconsistent with Ren et al.'s study.20 This controversy could be attributed to the types of stroke, time of blood sampling, specific polymorphism of MALAT1 gene in the Iranian population, or sample size. We had 59 IS cases with LAA and SVD, while in Ren et al.'s study, the MALAT1 level was assessed in 120 patients with IS with all types of stroke in the first 24 hours after stroke. We found a significant positive correlation between MALAT1 expression level with sampling time within 24 hours after stroke. Additionally, mix model analysis has also shown a significant increase of MALAT1 expression on day five relative to day one after stroke. Therefore, it seems that the MALAT1 expression increased over time. According to Fathy et al.'s study,21 the population's specific polymorphism (G variant of MALAT1 rs619586) may be considered the leading cause of MALAT1 upregulation in our patients. Therefore, more studies based on MALAT1 lncRNA polymorphisms are warranted to confirm these findings.
Ren et al.'s study detected a significant negative correlation between MALAT1 expression and NIHSS.20 However, we found non-significant negative Spearman correlations between MALAT1 expression and NIHSS score, and also linear regression showed a non-significant negative relationship between MALAT1 expression with NIHSS. This represents the possible protective role of MALAT1 following IS. Therefore, evaluating the MALAT1 expression in a larger sample size and in the optimal time window might show its significant correlation with NIHSS. The first hours after the onset of stroke have been shown as the optimal time window in previous studies.31,32 In our study, most patients were admitted while more than 12 hours had elapsed from their stroke symptoms, and blood samplings were repeated on days 3 and 5 after stroke. It seems that the MALAT1 levels in the first hours (< 12 hours) after IS could show a significant correlation with NIHSS.
For the first time, we evaluated the THRIL expression in peripheral blood of patients with IS. In our study, THRIL was significantly up-regulated on the first day after IS, and its expression level was higher on day five compared to THRIL on days 1 and 3. We could find only a non-significant positive correlation between THRIL expression and stroke severity.
Previous studies showed the THRIL upregulation following in vitro and in vivo models of cerebral ischemia-reperfusion injury.33,34 Hypoxia-induced increase of THRIL expression and the protective role of THRIL inhibition against injuries has been demonstrated by in-vitro studies.34,35 THRIL inhibition increased cell viability, migration, and invasion, and it decreased cell apoptosis by up-regulating microRNA (miR)-99a expression in H9C2 cells.34 THRIL could aggravate the cerebral damage following hypoxia by binding to miR-24-3p and activating the nuclear factor kappa B (NF-κB) p65 signaling pathway.33 Moreover, Lin and Bao reported the alleviation of myocardial injury after THRIL inhibition in the coronary heart disease (CHD) model.36 In this study, we could not find a significant correlation between THRIL expression and stroke severity and a significant diagnostic value for THRIL expression within 24 hours after stroke.
ROC curve analysis showed that the MALAT1 expression within 24 hours after stroke (more than 12 hours) in patients with LAA and SVD stroke had a significant diagnostic value with an AUC of 0.78 (P < 0.001) relative to control. This result is consistent with Ren et al.’s study. They reported a diagnostic value with an AUC of 0.79 for MALAT1 level in patients with IS relative to control on the first day after stroke.20
Conclusion
In concordance with ongoing studies regarding potential role of lncRNAs as diagnostic and prognostic markers, we showed a significant sequential upregulation in MALAT1 and THRIL expression after IS. We also found a potential diagnostic accuracy for MALAT1 expression with an AUC of 0.78. To confirm our results, studies with a larger sample size are warranted.
Acknowledgments
The authors would like to thank the Office of Vice Chancellor for Research in Shiraz University of Medical Sciences for financial support (grant No. #17988) and also stroke ward of Nemazi Hospital.
Notes:
How to cite this article: Bayat M, Hooshmandi E, Karimi N, Rahimi M, Tabrizi R, Asadabadi T, et al. Sequential in expression of long non-coding RNAs THRIL and MALAT1 after ischemic stroke. Curr J Neurol 2024; 23(1): 74-82.
Conflict of Interests
The authors declare no conflict of interest in this study.
References
- 1.Tabrizi R, Lankarani KB, Kardeh B, Akbari H, Azarpazhooh MR, Borhani-Haghighi A. A Comprehensive systematic review and meta-analysis on the risk factors of stroke in iranian population. Arch Iran Med. 2021;24(1):64–77. doi: 10.34172/aim.2021.10. [DOI] [PubMed] [Google Scholar]
- 2.Jickling GC, Sharp FR. Blood biomarkers of ischemic stroke. Neurotherapeutics. 2011;8(3):349–60. doi: 10.1007/s13311-011-0050-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2021;22(2):96–118. doi: 10.1038/s41580-020-00315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bao MH, Szeto V, Yang BB, Zhu SZ, Sun HS, Feng ZP. Long non-coding RNAs in ischemic stroke. Cell Death Dis. 2018;9(3):281. doi: 10.1038/s41419-018-0282-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu M, Li N, Luo P, Jing W, Wen X, Liang C, et al. Peripheral blood leukocyte expression of lncRNA MIAT and its diagnostic and prognostic value in ischemic stroke. J Stroke Cerebrovasc Dis. 2018;27(2):326–37. doi: 10.1016/j.jstrokecerebrovasdis.2017.09.009. [DOI] [PubMed] [Google Scholar]
- 6.Rezaei M, Mokhtari MJ, Bayat M, Safari A, Dianatpuor M, Tabrizi R, et al. Long non-coding RNA H19 expression and functional polymorphism rs217727 are linked to increased ischemic stroke risk. BMC Neurol. 2021;21(1):54. doi: 10.1186/s12883-021-02081-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu W, Tian L, Yue X, Liu J, Fu Y, Yan Y. LncRNA expression profiling of ischemic stroke during the transition from the acute to subacute stage. Front Neurol. 2019;10:36. doi: 10.3389/fneur.2019.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao H, Wang X, Lin C, An Z, Yu J, Cao H, et al. Exosomal MALAT1 derived from ox-LDL-treated endothelial cells induce neutrophil extracellular traps to aggravate atherosclerosis. Biol Chem. 2020;401(3):367–76. doi: 10.1515/hsz-2019-0219. [DOI] [PubMed] [Google Scholar]
- 9.Cai Q, Gao ML, Huang LS, Chen HS, Pan LH. MALAT1/miRNA-203/Wnt5a: A potential mechanism for regulating coronary artery disease. Int J Cardiol. 2021;329:48. doi: 10.1016/j.ijcard.2020.12.046. [DOI] [PubMed] [Google Scholar]
- 10.Liu JY, Yao J, Li XM, Song YC, Wang XQ, Li YJ, et al. Pathogenic role of lncRNA-MALAT1 in endothelial cell dysfunction in diabetes mellitus. Cell Death Dis. 2014;5(10):e1506. doi: 10.1038/cddis.2014.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiao J, Lu Y, Yang X. THRIL mediates endothelial progenitor cells autophagy via AKT pathway and FUS. Mol Med. 2020;26(1):86. doi: 10.1186/s10020-020-00201-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han Y, Qiu H, Pei X, Fan Y, Tian H, Geng J. Low-dose sinapic acid abates the pyroptosis of macrophages by downregulation of lncrna-malat1 in rats with diabetic atherosclerosis. J Cardiovasc Pharmacol. 2018;71(2):104–12. doi: 10.1097/FJC.0000000000000550. [DOI] [PubMed] [Google Scholar]
- 13.Powers WJ, Rabinstein AA, Ackerson T, Adeoye OM, Bambakidis NC, Becker K, et al. Guidelines for the early management of patients with acute ischemic stroke: 2019 update to the 2018 guidelines for the early management of acute ischemic stroke: A guideline for healthcare professionals From the American Heart Association/American Stroke Association. Stroke. 2019;50(12):e344–e418. doi: 10.1161/STR.0000000000000211. [DOI] [PubMed] [Google Scholar]
- 14.Li H, Zhu X, Hu L, Li Q, Ma J, Yan J. Loss of exosomal MALAT1 from ox-LDL-treated vascular endothelial cells induces maturation of dendritic cells in atherosclerosis development. Cell Cycle. 2019;18(18):2255–67. doi: 10.1080/15384101.2019.1642068. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Lv F, Liu L, Feng Q, Yang X. Long non-coding RNA MALAT1 and its target microRNA-125b associate with disease risk, severity, and major adverse cardiovascular event of coronary heart disease. J Clin Lab Anal. 2021;35(4):e23593. doi: 10.1002/jcla.23593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toraih EA, El-Wazir A, Alghamdi SA, Alhazmi AS, El-Wazir M, Abdel-Daim MM, et al. Association of long non-coding RNA MIAT and MALAT1 expression profiles in peripheral blood of coronary artery disease patients with previous cardiac events. Genet Mol Biol. 2019;42(3):509–18. doi: 10.1590/1678-4685-GMB-2018-0185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiu S, Sun J. lncRNA-MALAT1 expression in patients with coronary atherosclerosis and its predictive value for in-stent restenosis. Exp Ther Med. 2020;20(6):129. doi: 10.3892/etm.2020.9258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qi H, Shen J, Zhou W. Up-regulation of long non-coding RNA THRIL in coronary heart disease: Prediction for disease risk, correlation with inflammation, coronary artery stenosis, and major adverse cardiovascular events. J Clin Lab Anal. 2020;34(5):e23196. doi: 10.1002/jcla.23196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huangfu N, Xu Z, Zheng W, Wang Y, Cheng J, Chen X. LncRNA MALAT1 regulates oxLDL-induced CD36 expression via activating beta-catenin. Biochem Biophys Res Commun. 2018;495(3):2111–7. doi: 10.1016/j.bbrc.2017.12.086. [DOI] [PubMed] [Google Scholar]
- 20.Ren H, Wu F, Liu B, Song Z, Qu D. Association of circulating long non-coding RNA MALAT1 in diagnosis, disease surveillance, and prognosis of acute ischemic stroke. Braz J Med Biol Res. 2020;53(12):e9174. doi: 10.1590/1414-431X20209174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fathy N, Kortam MA, Shaker OG, Sayed NH. Long noncoding RNAs MALAT1 and ANRIL gene variants and the risk of cerebral ischemic stroke: An association study. ACS Chem Neurosci. 2021;12(8):1351–62. doi: 10.1021/acschemneuro.0c00822. [DOI] [PubMed] [Google Scholar]
- 22.Adams HP Jr, Bendixen BH, Kappelle LJ, Biller J, Love BB, Gordon DL, et al. Classification of subtype of acute ischemic stroke Definitions for use in a multicenter clinical trial. TOAST Trial of Org 10172 in Acute Stroke Treatment. Stroke. 1993;24(1):35–41. doi: 10.1161/01.str.24.1.35. [DOI] [PubMed] [Google Scholar]
- 23.Unger T, Borghi C, Charchar F, Khan NA, Poulter NR, Prabhakaran D, et al. 2020 International Society of Hypertension Global Hypertension Practice Guidelines. Hypertension. 2020;75(6):1334–57. doi: 10.1161/HYPERTENSIONAHA.120.15026. [DOI] [PubMed] [Google Scholar]
- 24.American Diabetes Association. Diagnosis and classification of diabetes mellitus. Diabetes Care. 2004;27(Suppl 1):S5–S10. doi: 10.2337/diacare.27.2007.s5. [DOI] [PubMed] [Google Scholar]
- 25.Williams LS, Yilmaz EY, Lopez-Yunez AM. Retrospective assessment of initial stroke severity with the NIH Stroke Scale. Stroke. 2000;31(4):858–62. doi: 10.1161/01.str.31.4.858. [DOI] [PubMed] [Google Scholar]
- 26.Nunn A, Bath PM, Gray LJ. Analysis of the modified rankin scale in randomised controlled trials of acute ischaemic stroke: A systematic review. Stroke Res Treat. 2016;2016:9482876. doi: 10.1155/2016/9482876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3(6):1101–8. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 28.Liu C, Zhang C, Yang J, Geng X, Du H, Ji X, et al. Screening circular RNA expression patterns following focal cerebral ischemia in mice. Oncotarget. 2017;8(49):86535–47. doi: 10.18632/oncotarget.21238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xin JW, Jiang YG. Long noncoding RNA MALAT1 inhibits apoptosis induced by oxygen-glucose deprivation and reoxygenation in human brain microvascular endothelial cells. Exp Ther Med. 2017;13(4):1225–34. doi: 10.3892/etm.2017.4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Masoumi F, Ghorbani S, Talebi F, Branton WG, Rajaei S, Power C, et al. Malat1 long noncoding RNA regulates inflammation and leukocyte differentiation in experimental autoimmune encephalomyelitis. J Neuroimmunol. 2019;328:50–9. doi: 10.1016/j.jneuroim.2018.11.013. [DOI] [PubMed] [Google Scholar]
- 31.Wang J, Zhao H, Fan Z, Li G, Ma Q, Tao Z, et al. Long noncoding RNA H19 promotes neuroinflammation in ischemic stroke by driving histone deacetylase 1-dependent M1 microglial polarization. Stroke. 2017;48(8):2211–21. doi: 10.1161/STROKEAHA.117.017387. [DOI] [PubMed] [Google Scholar]
- 32.Xiao Z, Qiu Y, Lin Y, Medina R, Zhuang S, Rosenblum JS, et al. Blocking lncRNA H19-miR-19a-Id2 axis attenuates hypoxia/ischemia induced neuronal injury. Aging (Albany NY) 2019;11(11):3585–600. doi: 10.18632/aging.101999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuai F, Zhou L, Zhou J, Sun X, Dong W. Long non-coding RNA THRIL inhibits miRNA-24-3p to upregulate neuropilin-1 to aggravate cerebral ischemia-reperfusion injury through regulating the nuclear factor kappaB p65 signaling. Aging (Albany NY) 2021;13(6):9071–84. doi: 10.18632/aging.202762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xia J, Jiang N, Li Y, Wei Y, Zhang X. The long noncoding RNA THRIL knockdown protects hypoxia-induced injuries of H9C2 cells through regulating miR-99a. Cardiol J. 2019;26(5):564–74. doi: 10.5603/CJ.a2018.0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sheng C, Hu F, Wu L. Retraction notice to "Geniposide alleviates hypoxia-induced injury by down-regulation of lncRNA THRIL in rat cardiomyocytes derived H9c2 cells" [Eur. J. Pharmacol. 854 (2019) 28-38] Eur J Pharmacol. 2022;929:175125. doi: 10.1016/j.ejphar.2022.175125. [DOI] [PubMed] [Google Scholar]
- 36.Lin L, Bao J. Long non-coding RNA THRIL is upregulated in coronary heart disease and binds to microRNA-424 to upregulate TXNIP in mice. Microvasc Res. 2021;138:104215. doi: 10.1016/j.mvr.2021.104215. [DOI] [PubMed] [Google Scholar]