Abstract
Denosumab is a monoclonal anti-RANKL antibody that inhibits bone resorption, increases bone mass, and reduces fracture risk. Denosumab discontinuation causes an extensive wave of rebound resorption, but the cellular mechanisms remain poorly characterized. We utilized in situ hybridization (ISH) as a direct approach to identify the cells that activate osteoclastogenesis through the RANKL/OPG pathway. ISH was performed across species, skeletal sites, and following recombinant OPG (OPG:Fc) and parathyroid hormone 1–34 (PTH) treatment of mice. OPG:Fc treatment in mice induced an increased expression of RANKL mRNA mainly in trabecular, but not endocortical bone surface cells. Additionally, a decreased expression of OPG mRNA was detected in bone surface cells and osteocytes of both compartments. A similar but more pronounced effect on RANKL and OPG expression was seen one hour after PTH treatment. These findings suggest that bone surface cells and osteocytes conjointly regulate the activation of osteoclastogenesis, and that OPG:Fc treatment induces a local accumulation of osteoclastogenic activation sites, ready to recruit and activate osteoclasts upon treatment discontinuation. Analysis of publicly available single-cell RNA sequencing (scRNAseq) data from murine bone marrow stromal cells revealed that Tnfsf11+ cells expressed high levels of Mmp13, Limch1, and Wif1, confirming their osteoprogenitor status. ISH confirmed co-expression of Mmp13 and Tnfsf11 in bone surface cells of both vehicle- and OPG:Fc-treated mice. Under physiological conditions of human/mouse bone, RANKL is expressed mainly by osteoprogenitors proximate to the osteoclasts, while OPG is expressed mainly by osteocytes and bone-forming osteoblasts.
Subject terms: Osteoporosis, Bone, Osteoporosis
Introduction
Bone remodeling consists of a highly coordinated sequence of events including (i) activation, (ii) initial bone resorption by primary osteoclasts, (iii) reversal-resorption where secondary osteoclasts intermixed with osteoblastic reversal cells extend the bone resorption phase and prepare the bone surfaces for (iv) bone formation by osteoblasts.1–3 An uncoupling of this process, where bone resorption occurs with a delayed subsequent bone formation, causes a net bone loss, ultimately leading to bone diseases like osteoporosis.4,5 Today, anti-resorptive drugs like denosumab have become first-line of treatment for osteoporosis.6,7 Denosumab, a monoclonal antibody against RANKL, efficiently inhibits osteoclast development and resorption, increases bone mineral density (BMD), and reduces fracture risk in patients with postmenopausal osteoporosis.8–10 Paradoxically, denosumab discontinuation has been associated with extensive bone resorption, leading to a drop in BMD, returning to or even below baseline levels.8,11 This extensive bone resorption upon treatment discontinuation, commonly referred to as “rebound resorption”, increases the risk of multiple vertebral fractures.12–16
The mechanism leading to rebound resorption remains largely unresolved. Multiple studies have emerged, suggesting possible mechanisms responsible for this phenomenon. In humans, the suggested mechanisms include an increased pool of circulating pre-osteoclasts,17 and an increased proportion of empty osteocyte lacunae, reflecting an increased pool of dead osteocytes during denosumab treatment.18 In murine models, it has been shown that recombinant OPG:Fc treatment, like denosumab, inhibits osteoclast development resulting in fission of osteoclasts into osteomorphs.19 Osteomorphs may accumulate during treatment and re-fuse into functional osteoclasts upon treatment discontinuation, leading to extensive bone resorption.19 Recently, it has been suggested that long term denosumab treatment of mice expressing human RANKL leads to loss of the newly embedded osteocytes and their expression of OPG, possibly setting the stage for rebound bone loss upon treatment discontinuation.20 However, further studies are needed to directly pinpoint the cells that orchestrate the activation of osteoclastogenesis, both under physiological conditions and before the rebound resorption following denosumab treatment.
In order to gain a deeper insight into the cellular mechanisms that orchestrate the activation of osteoclastogenesis, and the subsequent rebound resorption observed after denosumab discontinuation, it is critical to identify the specific cell populations that provide the necessary signaling either stimulating or inhibiting osteoclastogenesis – and to understand their spatial organization within the bone microenvironment. Previous studies, with some dating back to the late 1990’s and early 2000’s have localized Tnfsf11 mRNA expression to chondrocytes, osteoblasts and bone lining cells in mice,21,22 while Tnfsf11 and Tnfrsf11b mRNA have been localized to chondrocytes, osteoblasts, lining cells, and early osteocytes in rats.23 In vitro studies exploring variations in RANKL and OPG expression during osteoblast differentiation suggest that RANKL is mostly expressed in early osteoblasts, while OPG is mostly expressed in mature osteoblasts.24,25 More recent functional studies using established Dmp1-cre and Sost-cre knockout models suggest that osteocytes are the main source of RANKL, while osteoblasts are the main source of OPG.26,27 However, uncertainty arises, as Dmp1 and Sost promoters are not exclusively activated in one specific cell population.20,22,27–29 Therefore, the precise spatial distribution of the most abundant sources of RANKL and OPG, and how they are affected during RANKL-inhibiting treatment remain largely unresolved. The present study aimed to investigate the hypothesis that osteoclastogenesis is regulated conjointly by bone surface osteoprogenitors and osteocytes through local regulation of the RANKL/OPG-pathway. Moreover, we hypothesized that accumulation of local osteoclastogenic activation sites, here defined as upregulated Tnfsf11 and/or downregulated Tnfrsf11b mRNA expression in osteoblastic cells localized near the bone surfaces, is a key driver of rebound resorption upon discontinuation of denosumab treatment. To address this, we utilized highly sensitive in situ hybridization (ISH) as a direct approach to spatially identify the cells that orchestrate the activation of osteoclastogenesis. This was performed by investigating the cellular expression of the murine Tnfsf11 and human TNFSF11 gene, encoding RANKL and the murine Tnfrsf11b and human TNFRSF11B gene, encoding OPG, as well as their prevalence after two weeks of OPG:Fc treatment. Moreover, previous studies have shown that acute PTH treatment is capable of rapidly increasing Tnfsf11 and decreasing Tnfrsf11b expression in osteoblastic cells in vitro.30,31 Based on these findings, we utilized a single injection of PTH as a biological probe to address which cells respond to systemic PTH stimuli by regulating their expression of Tnfsf11 and Tnfrsf11b in mice.
In brief, the cellular expression of RANKL and OPG was investigated in (1) tibiae from OPG:Fc-treated C57BL/6 J female mice, (2) publicly available scRNAseq data from enriched bone marrow stroma of C57BL/6 J male mice, (3) femora from C57BL/6 J female mice treated with a single injection of PTH (1–34), (4) vertebrae from male and female C57BL/6 J mice, (5) temporal bones of Sprague-Dawley rats, (6) human cortical bone specimens obtained from the proximal femur of healthy adolescents, and (7) human iliac crest bone biopsies from healthy adults.
Results
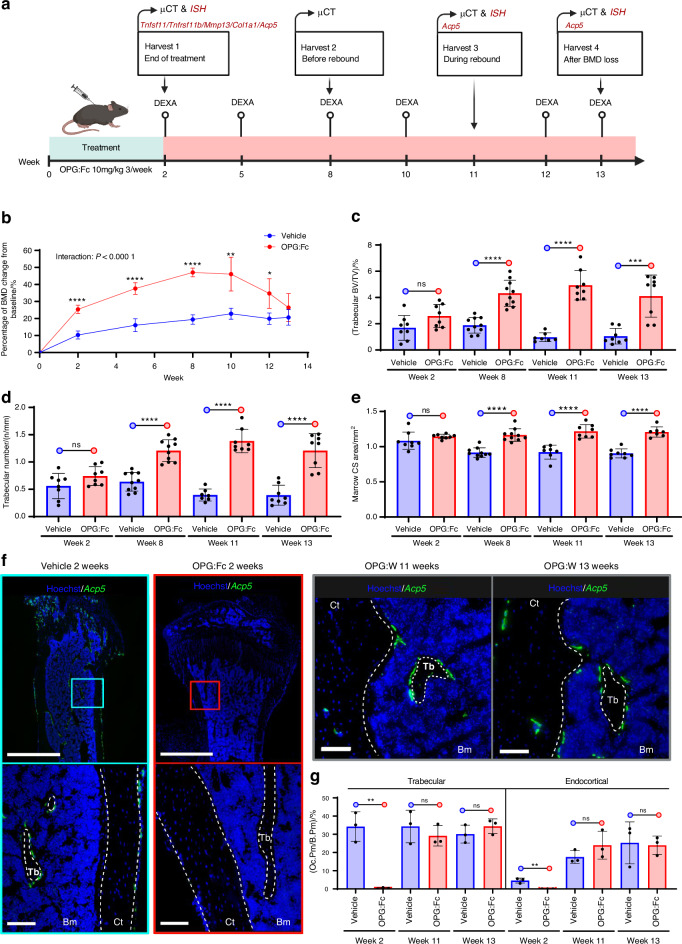
OPG:Fc treatment potently increases BMD and ablates osteoclasts from bone surfaces in mice, while treatment withdrawal leads to rebound resorption
To identify the cellular mechanisms that activate osteoclastogenesis and to address whether blocking RANKL may induce an increased abundance of local activation sites, we injected healthy 6–8-week-old female mice with OPG:Fc or vehicle 3 times a week for 2 weeks followed by treatment withdrawal, as illustrated in the study design (Fig. 1a). Longitudinal dual-energy X-ray absorptiometry (DEXA) revealed a significant interaction between time and treatment, with an initial increase in BMD, peaking at week 8 due to a prolonged half-life of OPG:Fc, followed by a decrease in BMD to vehicle levels by week 13 (Fig. 1b). Tibiae and femora were harvested following the 2 weeks of treatment, and at 8, 11, and 13 weeks after baseline (6, 9, and 11 weeks after withdrawal, respectively) (Fig. 1a). Micro-computed tomography (µCT) analysis of the femora revealed a notable increase in trabecular bone volume and trabecular thickness, accompanied by a decrease in trabecular separation at week 8, 11, and 13, in OPG:Fc-treated mice compared to vehicle, while trabecular thickness was increased at all time points compared to vehicle (Fig. 1c, d and Fig. S1a, b). Of note, at week 13, the distinction in trabecular architecture was less pronounced, suggesting a relative decrease in trabecular volume during the rebound bone loss. µCT-analysis further showed an increase in the diaphyseal marrow cross-sectional areas at week 8, 11, and 13, while total diaphyseal cross-sectional areas were increased at all timepoints compared to vehicle. This suggests that OPG:Fc treatment induced periosteal apposition and increased endosteal resorption during withdrawal (Fig. 1e and Fig. S1c). To confirm the absence of osteoclasts after OPG:Fc treatment and their reoccurrence after treatment withdrawal, we examined the presence of Acp5+ (encodes the osteoclast marker enzyme tartrate-resistant acid phosphatase [TRAcP]) osteoclasts on trabecular and endocortical bone surfaces using ISH. After 2 weeks, OPG:Fc treatment, like denosumab, successfully ablated Acp5+ osteoclasts from bone surfaces, while treatment withdrawal resulted in recruitment of Acp5+ osteoclasts to the bone surfaces at 11 and 13 weeks. Importantly, histological assessment revealed that Acp5+ osteoclasts predominantly occupied trabecular bone surfaces at 13 weeks during the rebound period, while endocortical bone surfaces were occupied by less osteoclasts in the OPG:Fc-treated mice (Fig. 1f, g and Fig. S2).
Fig. 1.
OPG:Fc treatment Increases BMD and ablates osteoclasts, while treatment withdrawal results in rebound resorption. a Study design illustrating 6–8-week-old, female mice that were injected with vehicle or OPG:Fc (10 mg/kg) 3 times a week for 2 weeks followed by a withdrawal period. b Longitudinal DEXA scans of the left hind limb were carried out every second or third week, and at the end of the study. c–e Ex vivo µCT femoral parameters at week 2, 8 11 or 13. f ISH of Acp5 using tissue sections of the tibia shows Acp5+ osteoclasts (green) present on trabecular and endocortical bone surfaces in vehicle-treated but not in OPG:Fc-treated mice, with return of Acp5+ cells following treatment withdrawal. g Oc.Pm/B.Pm determined using Acp5 ISH-stained sections after 2 weeks of treatment and at week 11 and 13 following withdrawal. f White dashed lines highlight trabecular, endocortical, and periosteal bone surfaces. c–g Data is expressed as mean ± SD. P values are calculated using a mixed-effects analysis in (b), and using an unpaired t test in (c–e), and (g). ****P values < 0.000 1, ***P values < 0.001, **P values < 0.01, *P values < 0.05. n = 3–8/group. Scale bars: (f) overview = 1 mm and high magnifications = 100 µm. Ct cortical, Tb trabecular, Bm bone marrow, OPG:W OPG:Fc withdrawal, Oc.Pm/B.Pm osteoclast perimeter/Bone perimeter, CS cross-sectional
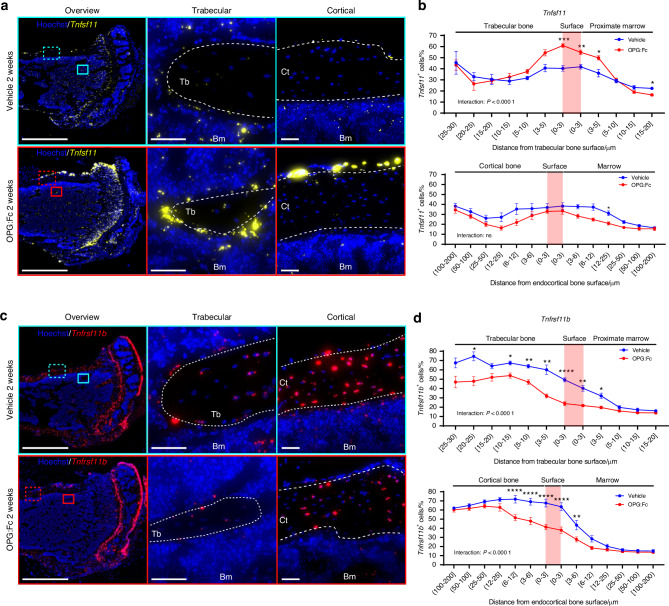
Tnfsf11 and Tnfrsf11b are expressed by distinct cell populations in mice
Next, we investigated the expression of Tnfsf11 and Tnfrsf11b to localize the cells that activate osteoclastogenesis. We found that Tnfsf11 and Tnfrsf11b expression is differently distributed when considering the zones beneath, above, and at the bone surfaces. The expression of Tnfsf11 was most prominent in bone surface cells located on periosteal, endocortical, and trabecular bone surfaces, which to our knowledge has not previously been directly illustrated (Fig. 2a). To determine the location of Tnfsf11+ cells we quantified the percentage of Tnfsf11+ cells and measured the distance of these cells to trabecular and endocortical bone surfaces. In vehicle-treated mice, we found that the percentage and staining intensities of Tnfsf11+ cells remained relatively stable across different distances from the bone surfaces, with a slight increase near the bone surface and deeper osteocytes (Fig. 2b and Fig. S3a). Likewise, we examined the spatial expression of Tnfrsf11b. Tnfrsf11b ISH revealed intense expression mainly amongst osteocytes, and to a lesser extent in the bone surface cells. Moreover, Tnfrsf11b+ cell populations residing in the marrow seemed less abundant (Fig. 2c). We further quantified the percentage of Tnfrsf11b+ cells and measured the distance of these cells from trabecular and endocortical bone surfaces. In vehicle-treated mice, osteocytes and cells near the bone surface were the most abundant population of Tnfrsf11b+ cells in both cortical and trabecular bone, while a lower proportion of Tnfrsf11b+ cells resided in the marrow (Fig. 2d). A similar pattern was seen in Tnfrsf11b staining intensities (Fig. S3b).
Fig. 2.
OPG:Fc treatment induces a changed Tnfsf11 and Tnfrsf11b expression pattern. Tibiae from 8-week-old female mice were harvested after being injected thrice weekly for two weeks with either vehicle or OPG:Fc (10 mg/kg). Following the final dose of OPG:Fc at the end of the two-week period, Tnfsf11 and Tnfrsf11b ISH was performed on tissue sections. a Tnfsf11 (yellow) illustrated in bone sections from vehicle and OPG:Fc-treated mice. b Percentage of Tnfsf11+ cells against the distance from the trabecular or endocortical bone surface. c Tnfrsf11b (red) illustrated in bone sections from vehicle and OPG:Fc-treated mice. d Percentage of Tnfrsf11b+ cells against the distance from the trabecular or endocortical bone surface. a, c White dashed lines highlight trabecular, endocortical, and periosteal bone surfaces. Data is expressed as mean ± SEM. P values are calculated using a two-way ANOVA. ****P values < 0.000 1, ***P values < 0.001, **P values < 0.01, *P values < 0.05. n = 8/group. Scale bars: (a, b) overview = 1 mm and high magnifications (a) = 100 µm, (b) = 50 µm. Ct cortical, Tb trabecular, Bm bone marrow, OPG:W OPG:Fc withdrawal
OPG:Fc treatment induces a change in the Tnfsf11 and Tnfrsf11b expression pattern in mice
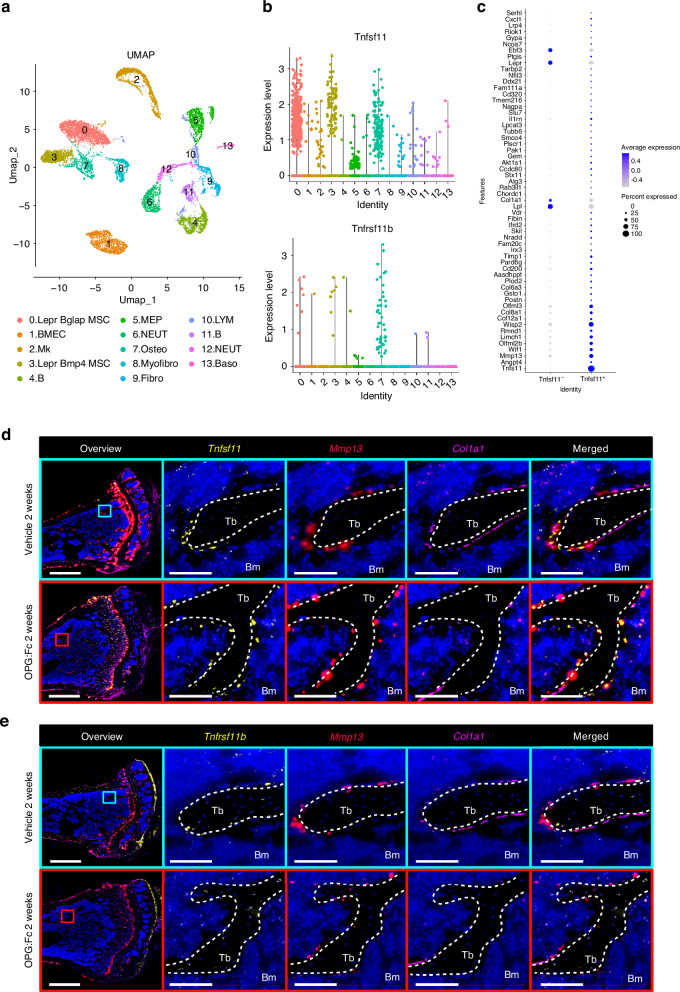
Since denosumab treatment is associated with rebound bone loss upon treatment discontinuation, we hypothesized that denosumab treatment may alter the expression of Tnfsf11 and Tnfrsf11b. Therefore, we examined the expression of Tnfrsf11 and Tnfrsf11b at the end of 2 weeks OPG:Fc treatment. ISH revealed a noticeable increase in the expression of Tnfsf11 especially on trabecular bone surfaces and the periosteum of the cortex, while endocortical surfaces seemed less affected (Fig. 2a). We further quantified the percentage of Tnfsf11+ cells and the Tnfsf11 staining intensity and found a significant interaction between regions and treatment (Fig. 2b and Fig. S3a). Moreover, the percentage of Tnfsf11+ cells and the Tnfsf11 staining intensity increased significantly on the trabecular bone surface in OPG:Fc-treated mice compared to vehicle-treated mice (Fig. 2b and Fig. S3a), suggesting that Tnfsf11 expression is mainly upregulated in trabecular bone surface cells upon OPG:Fc treatment. Examining the abundance of Tnfsf11 expressing cells in the cortical compartment, no interaction between region and treatment was detected. Moreover, the abundance of Tnfsf11+ cells was not increased in OPG:Fc-treated mice (Fig. 2b). However, interaction between treatment and region was detected when comparing staining intensities between vehicle- and OPG:Fc-treated mice across the regions (Fig. S3a). In contrast to the expression of Tnfsf11, the expression of Tnfrsf11b was strongly reduced upon OPG:Fc treatment in both the cortical and trabecular compartment (Fig. 2c). We further quantified the percentage of Tnfrsf11b+ cells and the Tnrfsf11b staining intensity and found a significant interaction between regions and treatment (Fig. 2d and Fig. S3b). Finally, the abundance of Tnfrsf11b+ cells was persistently lower on the trabecular and endocortical bone surfaces and among osteocytes residing close to the surface in OPG:Fc-treated mice compared to vehicle (Fig. 2d), while Tnrfsf11b staining intensity was lower in almost all cortical and trabecular osteocytes in OPG:Fc-treated mice compared to vehicle (Fig. S3b).
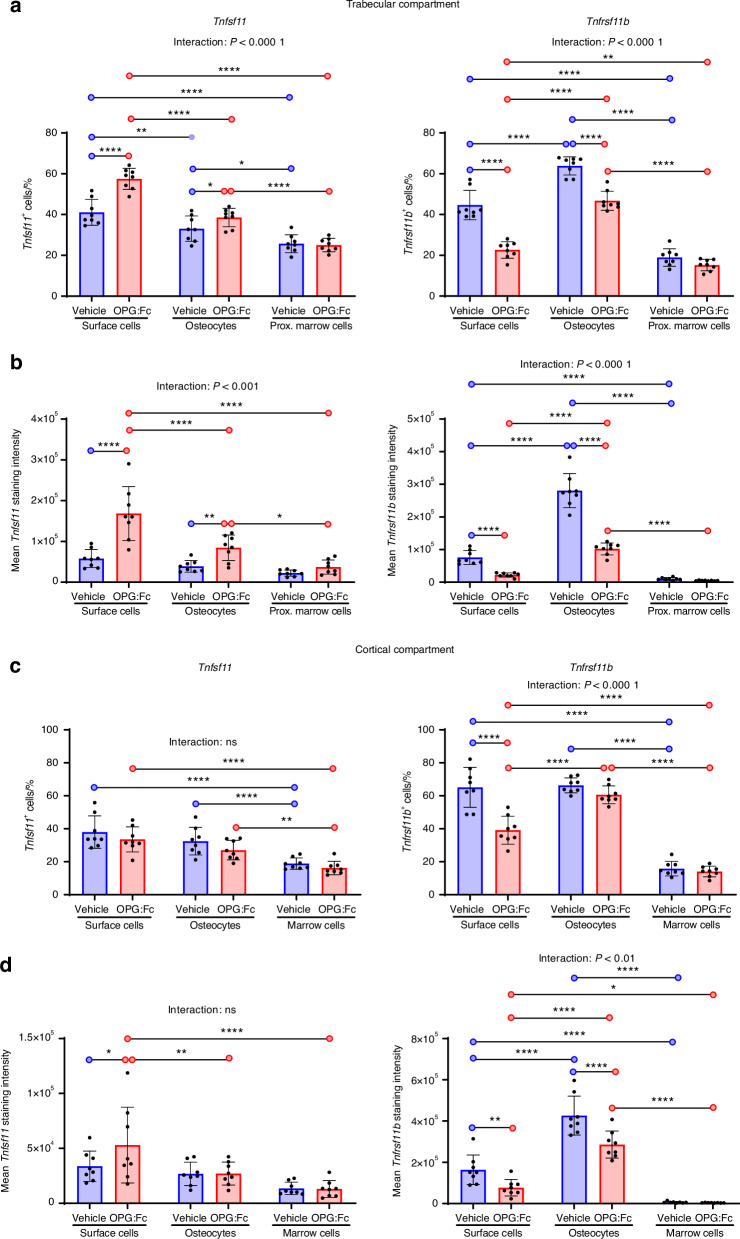
OPG:Fc treatment in mice induces an increase in Tnfsf11 and a decrease in Tnfrsf11b expression in the trabecular bone
Since denosumab discontinuation is associated with multiple spontaneous vertebral fractures, we hypothesized that the rebound mainly affects trabecular and not cortical bone.9 Therefore, we aimed to assess the expression of Tnfsf11 and Tnfrsf11b across cell types and in the two bone compartments. In each compartment, we defined three distinct cell types based on their spatial position. Cells residing on the bone surface, including bone lining cells, osteoblasts, and reversal cells were collectively defined as bone surface cells. Moreover, bone matrix embedded cells were defined as osteocytes, while cells residing in the marrow were defined as marrow cells. Then, we examined the impact of 2 weeks of OPG:Fc treatment on these cell types.
In the trabecular compartment, we found that the vehicle-treated mice had a significantly higher proportion of trabecular bone surface cells expressing Tnfsf11+ compared to osteocytes and proximate marrow cells. Next, we compared the percentage of Tnfsf11+ in vehicle and OPG:Fc-treated mice and found that Tnfsf11+ was strongly upregulated among bone surface cells compared to vehicle. Notably, the percentage of Tnfsf11+ bone surface cells were significantly higher compared to osteocytes, and proximate marrow cells in OPG:Fc treated mice (Fig. 3a). Although less prominent, the percentage of Tnfsf11+ osteocytes were also significantly increased in OPG:Fc-treated mice compared to vehicle-treated mice (Fig. 3a). No difference was detected in proximate marrow cells between vehicle and OPG:Fc treated mice (Fig. 3a). We further quantified the percentage of Tnfrsf11b+ cells and found a higher proportion of osteocytes expressing Tnfrsf11b+ compared to bone surface cells and proximate marrow cells in vehicle-treated mice. In addition, surface cells also presented with a higher percentage of Tnfrsf11b+ compared to proximate marrow cells (Fig. 3a). Similarly, we examined the effect of OPG:Fc treatment on Tnfrsf11b expression. Here, we found that OPG:Fc treatment resulted in a significant decrease in the percentage of Tnfrsf11b+ bone surface cells and osteocytes (Fig. 3a). No difference was detected between proximate marrow cells upon OPG:Fc treatment (Fig. 3a). The increase in Tnfsf11+ and decrease in Tnfrsf11b+ cells resulted in a significantly increased Tnfsf11+/Tnfrsf11b+ cell ratio among all cell types upon OPG:Fc treatment, with the most prominent increase observed in bone surface cells (Fig. S3c). Examining the changes in Tnfsf11 and Tnfrsf11b on histological sections, the Tnfsf11 and Tnfrsf11b staining intensity appeared highly affected by OPG:Fc treatment (Fig. 2a, c). Therefore, we quantified the mean staining intensity and found that the Tnfsf11 staining intensity was increased almost 3-fold in bone surface cells, and by 2-fold in osteocytes upon OPG:Fc treatment (Fig. 3b). Notably, bone surface cells exhibited a significantly higher staining intensity compared to osteocytes and proximate marrow cells in the OPG:Fc-treated mice (Fig. 3b). Tnfrsf11b staining intensity was decreased almost 3-fold in bone surface cells, more than 3-fold in osteocytes, and were unaltered in marrow cells (Fig. 3b). The changes in Tnfsf11 and Tnfrsf11b staining intensities, resulted in an 11-fold increase in the Tnfsf11/Tnfrsf11b staining intensity ratios amongst bone surface cells and approximately a 3-fold increase in the Tnfsf11/Tnfrsf11b staining intensity ratios in marrow cells proximate to the bone surface (Fig. S3c).
Fig. 3.
OPG:Fc treatment has differential effects on Tnfsf11 and Tnfrsf11b expression in cortical and trabecular bone. Tnfsf11+ and Tnfrsf11b+ cell percentage and cell staining intensities were quantified in bone surface cells, osteocytes, and marrow cells and compared across cell types and between vehicle and OPG:Fc-treated mice. All data is collected from ISH stained sections of the tibia. a Percentage of Tnfsf11+ and Tnfrsf11b+ trabecular surface cells, trabecular osteocytes, and proximate marrow cells. b Mean Tnfsf11 and Tnfrsf11b cell staining intensities in trabecular surface cells, trabecular osteocytes and proximate marrow cells c Percentage of Tnfsf11+ and Tnfrsf11b+ endocortical surface cells, cortical osteocytes and marrow cells. d Mean Tnfsf11 and Tnfrsf11b cell staining intensities in endocortical surface cells, cortical osteocytes, and marrow cells. Data are shown as mean ± SD. P values are calculated using a two-way ANOVA. ****P values < 0.000 1, ***P values < 0.001, **P values < 0.01, *P values < 0.05. n = 8/group
OPG:Fc treatment does not result in an increased expression of Tnfsf11 in the cortical compartment
In the cortical compartment, the percentage of Tnfsf11+ cells did not differ between the vehicle and the OPG:Fc-treated mice. However, a significantly higher proportion of endocortical surface cells and cortical osteocytes were Tnfsf11+ compared to marrow cells. Interestingly, we found a 40% reduction in Tnfrsf11b+ bone surface cells, while the abundance of osteocytes and marrow cells were unaffected by OPG:Fc treatment (Fig. 3c). Accordingly, Tnfsf11+/Tnfrsf11b+ cell ratios were significantly higher among bone surface cells in the OPG:Fc-treated mice compared to vehicle, but were unchanged in osteocytes and marrow cells (Fig. S3d). Like in the trabecular bone, we examined the effect of OPG:Fc treatment on Tnfsf11 and Tnfrsf11b staining intensity on cortical osteocytes, endocortical surface cells, and marrow cells (Fig. 3d). In the OPG:Fc-treated mice, Tnfsf11 staining intensity in bone surface cells was increased by approximately 1.5-fold compared to vehicle. Moreover, Tnfsf11 staining intensity was higher in bone surface cells than in osteocytes and marrow cells, in the OPG:Fc-treated mice (Fig. 3d). Tnfrsf11b staining intensity was higher in cortical osteocytes than in endocortical surface cells and marrow cells, and was significantly reduced in cortical osteocytes and endocortical surface cells upon OPG:Fc treatment (Fig. 3d). Tnfsf11/Tnfrsf11b staining intensity ratios seemed unaffected by OPG:Fc treatment, but were generally higher in marrow cells (Fig. S3d).
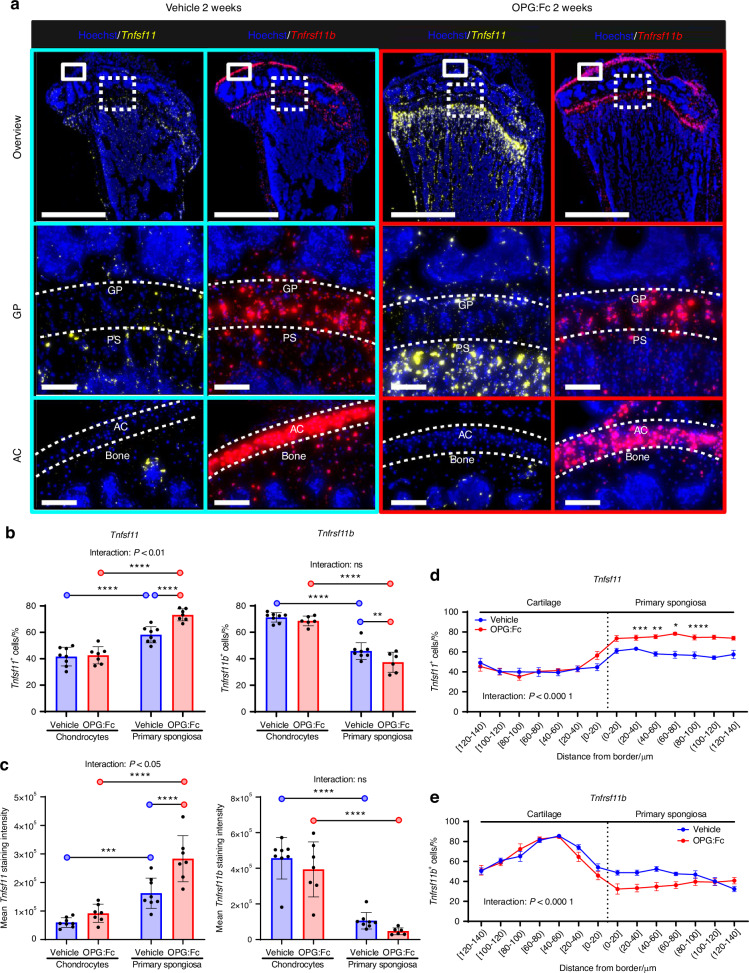
Tnfrsf11b expression by chondrocytes in the growth plate and articular cartilage is high compared to the primary spongiosa, where it becomes even lower upon OPG:Fc treatment
We explored other local sources of RANKL and OPG. ISH for Tnfsf11 and Tnfrsf11b revealed a prominent expression of Tnfrsf11b in chondrocytes of the epiphyseal growth plate and the articular cartilage, while Tnfsf11 expression seemed higher in the primary spongiosa below the growth plate (Fig. 4a). Moreover, Tnfsf11 expression seemed to be upregulated upon OPG:Fc treatment in the primary spongiosa (Fig. 4a). Quantification of the Tnfsf11 and Tnfrsf11b expression in the chondrocytes of the growth plate and primary spongiosa revealed that approximately 40% of chondrocytes expressed Tnfsf11, while the proportion of Tnfsf11+ cells increased to almost 60% in the primary spongiosa immediately below the growth plate (Fig. 4b). The proportion of Tnfsf11+ cells increased in the primary spongiosa, but not in chondrocytes, upon OPG:Fc treatment. In contrast, the proportion of Tnfrsf11b+ cells were higher in chondrocytes than in the primary spongiosa (Fig. 4b). OPG:Fc treatment significantly reduced the proportion of Tnfrsf11b+ cells in the primary spongiosa (Fig. 4b). Moreover, OPG:Fc treatment increased the Tnfsf11+/Tnfrsf11b+ cell ratio significantly in the cells in the primary spongiosa, but not in the chondrocytes (Fig. S4a). To further examine the expression of Tnfsf11 and Tnfrsf11b, we quantified staining intensities and found a higher Tnfsf11 staining intensity in the primary spongiosa than in chondrocytes, while Tnfrsf11b staining intensity was higher in chondrocytes (Fig. 4c). Moreover, Tnfsf11 staining intensity was enhanced by OPG:Fc treatment, while Tnfrsf11b staining intensities were generally not affected (Fig. 4c). The Tnfsf11/Tnfrsf11b staining intensity ratio increased more than 4-fold in the primary spongiosa upon OPG:Fc treatment (Fig. S4a). To visualize the expression pattern of Tnfsf11 and Tnfrsf11b, we quantified the percentage of positive cells, and related their distance to an interface line separating the chondrocytes from the primary spongiosa (Fig. 4d, e and Fig. S4b, c). For both Tnfsf11 and Tnfrsf11b, a significant interaction between regions and treatment was detected (Fig. 4d, e and Fig. S4b, c). Moreover, The percentage of Tnfsf11+ cells and Tnfsf11 staining intensities remained low in the cartilage, and increased as the tissue transitioned to primary spongiosa, with OPG:Fc treatment increasing the Tnfsf11+ cells and Tnfsf11 staining intensity in the primary spongiosa (Fig. 4d and Fig. S4b). Conversely, Tnfrsf11b+ cells and Tnfrsf11b staining intensity increased gradually in chondrocytes of the growth plate, while it decreased again in the primary spongiosa (Fig. 4e and Fig. S4c). OPG:Fc treatment induced a decrease in Tnfrsf11b staining intensity in the primary spongiosa (Fig. S4c).
Fig. 4.
Chondrocytes of the epiphysial growth plate of the tibia express high levels of Tnfrsf11b, while cells residing in the primary spongiosa express high levels of Tnfsf11. All results on this figure are female mice treated with vehicle or OPG:Fc for two weeks. a ISH of Tnfrsf11b (red) present at high levels in the growth plate of the tibia and articular cartilage and Tnfsf11 (yellow), present at the primary spongiosa. b Percentage of Tnfsf11+ and Tnfrsf11b+ cells, and Tnfsf11+/Tnfrsf11b+ cell ratios in chondrocytes and cells within the primary spongiosa. c Mean staining intensities of Tnfsf11 and Tnfrsf11b, and Tnfsf11/Tnfrsf11b staining intensity ratios in chondrocytes and cells within the primary spongiosa. d, e Percentage of Tnfsf11+ and Tnfrsf11b+ cells against the distance from the border between the primary spongiosa and chondrocytes. b, c Data are shown as mean ± SD. d, e Data are shown as mean ± SEM. P values are calculated using a two-way ANOVA or a mixed-effects analysis. b–e ****P values < 0.000 1, ***P values < 0.001, **P values < 0.01, *P values < 0.05. n = 8/group. Scale bars: Overview = 1 mm, high magnification images = 100 µm. GP growth plate, AC articular cartilage, PS primary spongiosa
Bone surface osteoprogenitors expressing the gene encoding matrix metallopeptidase 13 (Mmp13) also express Tnfsf11
To identify the main cell populations expressing Tnfsf11 and Tnfrsf11b we utilized the previously published and publicly available scRNAseq datasets.32 Enriched bone marrow stromal cells were classified into 13 different cell clusters (Fig. 5a). In these 13 identified cell clusters, Tnfsf11+ and Tnfrsf11b+ cells were abundant in cluster 0, 3, and 7 (Fig. 5b). Key gene markers defining the identity of bone marrow cell clusters included Lepr, Cxcl12, Adipoq, and Bglap for cluster 0, lepr, Cxcl12, Adipoq, and Bmp4 for cluster 3, and Col1a1, Alpl, Bglap, Bglap2, and Bglap3 for cluster 7. Moreover, Mmp13 was expressed in all three cell clusters (Fig. S5a). Since our ISH assessment showed that Tnfrsf11b was mainly expressed by osteocytes, and the scRNAseq data only includes bone marrow stromal cells, we decided to continue with Tnfrsf11 for further characterization of the cell populations that express this gene. We identified 58 differentially expressed genes in Tnfsf11+ cells compared to Tnfsf11− cells in cluster 0, 3, and 7 (Fig. 5c). Of these genes, a high proportion of Tnfsf11+ cells had a high expression of genes such as Mmp13, Wisp2, Limch1, and Wif1 and a low expression of genes such as Col1a1, Lepr, and Lpl (Fig. 5c). Conversely, a high proportion of Tnfsf11- cells had a high expression of Col1a1, Lepr. and Lpl, and a low expression of Mmp13, Wisp2, Limch1 (Fig. 5c). Since we have previously established that osteoprogenitor cells including reversal cells, canopy cells and bone lining cells express MMP13 in humans,33,34 we hypothesized that these cells also express Tnfsf11. Multiplex ISH of Tnfsf11 or Tnfrsf11b together with Mmp13, and Col1a1, revealed that the majority of Tnfsf11+ bone surface cells were Mmp13+, and Col1a1-, and that these Tnfsf11/Mmp13+ cells remained on the bone surfaces after OPG:Fc treatment (Fig. 5d). Tnfrsf11b/Mmp13/Coll1a1 multiplex ISH confirmed that Tnfrsf11b was mainly expressed by Mmp13-/Col1a1- osteocytes, and showed that only a few bone surface cells were Tnfrsf11b+/Mmp13+/Col1a1+(Fig. 5e) and found some Tnfrsf11b+/ Col1a1high osteoblasts (data not shown).
Fig. 5.
Single cell RNA-seq data of enriched bone marrow stroma of male mice and multiplex ISH of tissue sections from OPG:Fc-treated female mice. a Bone marrow cells were classified into 13 cell clusters by UMAP: (0) Lepr+ Bglap+ mesenchymal stem cell (MSC), (1) bone marrow endothelial cell (BMEC), (2) megakaryocyte (Mk), (3) Lepr+ Bmp4hi MSC, (4) B cell, (5) myeloerythroid progenitor (MEP), (6) neutrophil (NEUT), (7) osteolineage cell (Osteo), (8) myofibroblast (Myofibro), (9) fibroblast (Fibro), (10) lymphocyte (LYM), (11) B cell, (12) neutrophil (NEUT), (13) basophil (Baso). b Expression of Tnfrsf11b and Tnfsf11 within each bone marrow cell cluster. c All differentially expressed genes identified in Tnfsf11+ cell compared to Tnfsf11− cells within clusters 0, 3, 7. d Tnfsf11/Mmp13/Col1a1, or (e) Tnfrsf11b/Mmp13/Col1a1 multiplex ISH conducted on adjacent sections of tibiae collected from mice treated with either vehicle or OPG:Fc for two weeks. d, e White dashed lines outline trabecular bone surfaces. Tb Trabecular, Bm bone marrow
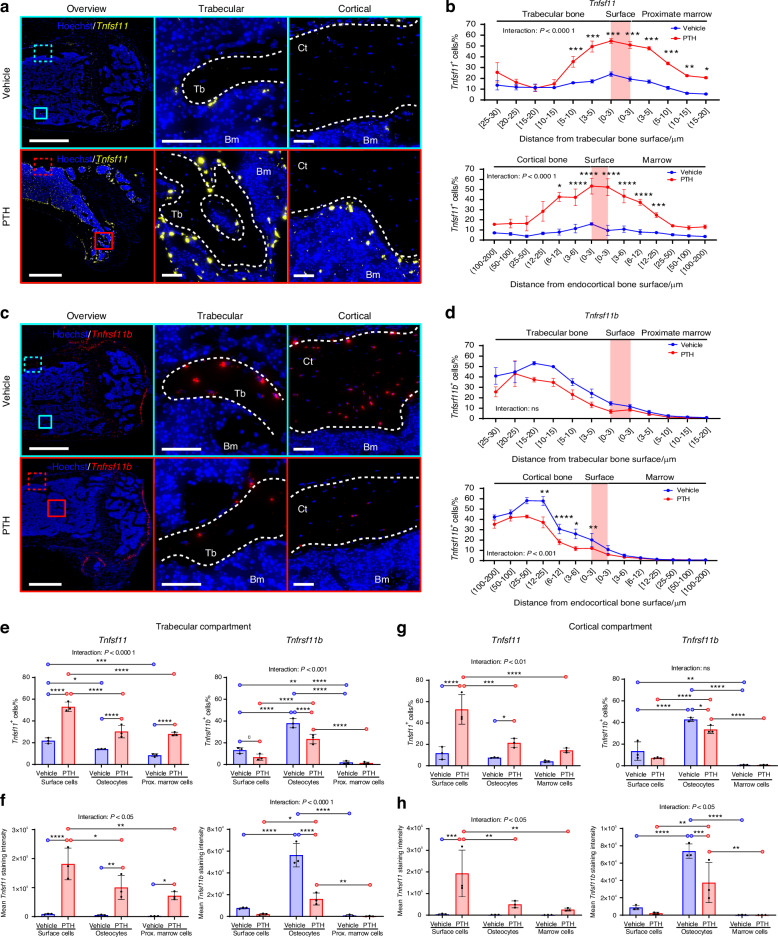
Bone surface cells and osteocytes near the bone surface respond to systemic PTH stimulus
Previous studies suggest that Tnfsf11 and Tnfrsf11b expression is regulated upon PTH treatment with the most notable effect observed one hour after treatment.30,31 Therefore, we utilized a single PTH treatment dose as a biological probe to induce Tnfsf11 expression in mice, in order to (1) identify the spatial location of cells that respond to systemic PTH stimuli by regulating the expression of Tnfsf11 and Tnfrsf11b, and (2) to validate that the main activator of osteoclastogenesis is cells residing near or on the bone surfaces. Histological assessment of Tnfsf11 mRNA expression in vehicle-treated mice revealed subtle expression of Tnfsf11 in bone surface cells, with limited expression in osteocytes. Upon PTH treatment, mRNA expression was highly increased in cells near the trabecular and endocortical bone surfaces (Fig. 6a). Next, we quantified cells and related their distances to the trabecular and cortical bone surfaces. A significant interaction between treatment and region was detected in both the trabecular and cortical compartment. Specifically, we found that PTH treatment highly increased Tnfsf11 expression in bone surface cells, and osteocytes and marrow cells near the bone surface compared to vehicle—a finding similar to what we observed in the mice treated with OPG:Fc (Fig. 6b and Fig. S6a). Similarly, we assessed the expression of Tnfrsf11b using ISH and found that mainly osteocytes near the bone surfaces expressed Tnfrsf11b compared to marrow cells and surface cells—this osteocytic Tnfrsf11b expression was reduced by PTH(1–34) treatment (Fig. 6c). Quantification of Tnfrsf11b expressing cells, and their distances to the trabecular or endocortical bone surfaces revealed a significant interaction between treatment and region in all cases, but not when relating the abundance of Tnfrsf11b+ cells to the distance to the trabecular bone surfaces (Fig. 6d and Fig. S6b). PTH treatment reduced Tnfrsf11b expression in bone surface cells, and osteocytes near the bone surface compared to vehicle—this was reflected by a decreased staining intensity in trabecular osteocytes and a decrease in both the abundance and staining intensity of Tnfrsf11b+ cells in cortical osteocytes near the bone surfaces (Fig. 6d and Fig. S6b).
Fig. 6.
PTH treatment induces a changed Tnfsf11 and Tnfrsf11b expression pattern. Tnfsf11 and Tnfrsf11b ISH performed on sections of femora obtained from 12-week-old female mice, 1 h after they were injected with a single dose of PTH (80 µg/kg). a Tnfsf11 (yellow) ISH illustrated in sections of the femur from vehicle and PTH treated mice. b Percentage of Tnfsf11+ cells against the distance from the trabecular or endocortical bone surface. c Tnfrsf11b (red) ISH illustrated in bone sections from vehicle and PTH treated mice. d Percentage of Tnfrsf11b+ cells against the distance from the trabecular or endocortical bone surface. e Percentage of Tnfsf11+ and Tnfrsf11b+ trabecular surface cells, trabecular osteocytes, and proximate marrow cells. f Mean Tnfsf11 and Tnfrsf11b cell staining intensities in trabecular surface cells, trabecular osteocytes and proximate marrow cells. g Percentage of Tnfsf11+ and Tnfrsf11b+ endocortical surface cells, cortical osteocytes and marrow cells. h Mean Tnfsf11 and Tnfrsf11b cell staining intensities in endocortical surface cells, cortical osteocytes, and marrow cells. a–c White dashed lines highlight trabecular, endocortical, and periosteal bone surfaces. In (b, d) data is expressed as mean ± SEM. In (e–h) data is expressed as mean ± SD. P values are calculated using a two-way ANOVA or mixed-effects analysis. ****P values < 0.000 1, ***P values < 0.001, **P values < 0.01, *P values < 0.05. n = 3/group. Scale bars: (a, b) Overview = 1 mm and high magnifications (a) = 100 µm or (b) = 50 µm. Ct cortical, Tb trabecular, Bm bone marrow
We subdivided the cell populations into bone surface cells, osteocytes and marrow cells to compare Tnfsf11 and Tnfrsf11b expression between cell types and to determine the impact of PTH treatment on the mRNA expression. In the trabecular compartment PTH induced a 2–3-fold increase in the proportion of surface cells, osteocytes and marrow cells that expressed Tnfsf11 (Fig. 6e). Notably, the proportion of bone surface cells expressing Tnfsf11 was 2-fold higher compared to marrow cells and osteocytes in the PTH-treated mice (Fig. 6e). Moreover, the Tnfsf11 staining intensities in bone surface cells, osteocytes and marrow cells were 18–35-fold higher in the PTH-treated mice compared to vehicle. Notably, bone surface cells had approximately a 2-fold higher Tnfsf11 staining intensity than osteocytes and marrow cells after PTH-treatment (Fig. 6e). Assessing the expression of Tnfrsf11b across cell types in the vehicle group, we found nearly a 3-fold higher proportion of osteocytes expressing Tnfsf11b compared to bone surface cells, while a 20-fold higher proportion was observed in osteocytes compared to marrow cells (Fig. 6e). The proportion of osteocytes expressing Tnfrsf11b was significantly reduced by nearly 2-fold in osteocytes and bone surface cells after PTH treatment, while marrow cells were not affected (Fig. 6e). Similar findings in osteocytes were found for staining intensities, while the Tnfrsf11b staining intensities were not changed in surface cells and marrow cells in the PTH-treated mice compared to vehicle (Fig. 6f).
In the cortical compartment, a 4.5-fold higher proportion of bone surface cells expressed Tnfsf11 in the PTH-treated compared to vehicle-treated mice (Fig. 6g). Moreover, nearly a 3-fold higher proportion of osteocytes expressed Tnfsf11 in PTH-treated mice compared to vehicle-treated mice. No significant differences were detected in marrow cells between PTH- and vehicle-treated mice (Fig. 6g). Comparing the proportion of bone surface cells to osteocytes and marrow cells within the PTH-treated mice, the proportion of Tnfsf11+ bone surface cells was 2.5-fold higher compared to osteocytes and nearly 4-fold higher compared to marrow cells (Fig. 6g). Similar findings were found for Tnfsf11 staining intensities (Fig. 6h). Furthermore, we assessed the expression of Tnfrsf11b across cell types in the vehicle group and found approximately a 3-fold higher proportion of osteocytes expressing Tnfsf11b compared to bone surface cells, while the proportion of Tnfrsf11b+ osteocytes was nearly 58-fold than that of marrow cells (Fig. 6g). Moreover, the proportion of osteocytes expressing Tnfrsf11b, and the Tnfrsf11b staining intensities were 1–2-fold lower in the PTH-treated mice compared to vehicle-treated mice (Fig. 6g, h).
When comparing PTH- to vehicle-treated mice, the ratio of Tnfsf11/Tnfsf11b positive cells and ratio of staining intensity trended toward an increase in the PTH treated group but was only significantly different for marrow cells (Fig. S6c, d).
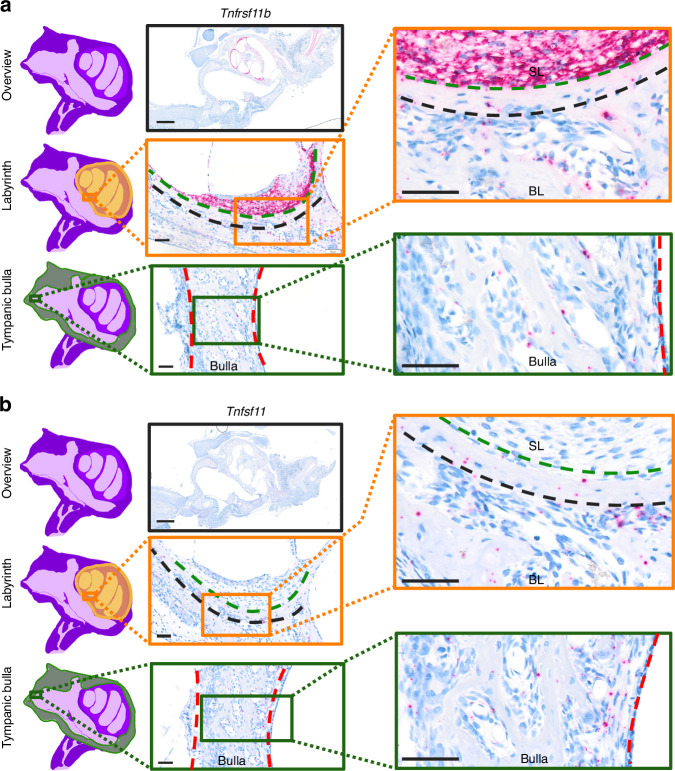
The inner ear of rats is protected from bone remodeling by high Tnfrsf11b and low Tnfsf11 expression
To better understand the cellular mechanisms that influence the activation of osteoclastogenesis, we utilized the inner ear as a unique anatomic location. Here, bone remodeling is highly inhibited in the bone immediately surrounding the inner ear, while the bulla of the middle ear undergo bone modeling with resorption on one side and formation on the other side.35–38 ISH of Tnfsf11 and Tnfrsf11b expression in the inner ear, revealed an extremely high local expression of Tnfrsf11b in the fibrocytes of the spiral ligament of the cochlea, while a minority of the osteocytes expressed Tnfrsf11b (Fig. 7a). In the bone immediately adjacent to the spiral ligament, bone remodeling was clearly inhibited, as bone remodeling took place at a distance from the Tnfrsf11b+ fibrocytes (Fig. 7a). We examined Tnfsf11 expression in adjacent to Tnfrsf11b-stained sections, and found little or no Tnfsf11+ cells in the spiral ligament and the bone immediately outside the spiral ligament, while Tnfsf11+ cells were increased in the bone remodeling spaces more distant from the spiral ligament, including in the bony labyrinth and in the tympanic bulla of the middle ear (Fig. 7b).
Fig. 7.
Fibrocytes of the spiral ligament express high levels of Tnfrsf11b. All data are obtained from a 10-day-old Sprague-Dawley rat. ISH on sections through the inner and middle ear, with representative overview image and high magnification images of the spiral ligament lining the bony labyrinth of the inner ear and the tympanic bulla of the middle ear for (a)Tnfrsf11b and (b) Tnfsf11 ISH of an adjacent section. The green dashed line separates the fibrocytes of the spiral ligament from the bony labyrinth. The black dashed line separates bone protected from remodeling from bone undergoing remodeling. The red dashed line outlines a region of the tympanic bulla of the middle ear. Scale bars: overview = 1 mm, high magnification images = 50 µm. BL bony labyrinth, SL spiral ligament
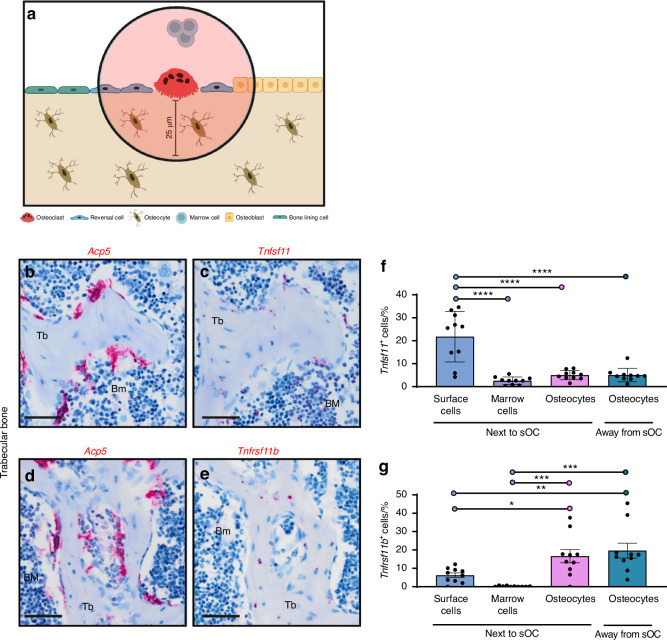
Bone surface cells proximate to osteoclasts are the most abundant source of Tnfsf11 in mice
Previously, it has been illustrated that reversal cells form cell extensions that extend beneath the bone resorbing osteoclast, demonstrating direct cell-cell contact between reversal cells and bone resorbing osteoclasts.33,39 Therefore, we investigated Tnfsf11 and Tnfrsf11b expression in the local environment surrounding the osteoclasts in female and male mouse vertebral bone serial sections hybridized to Tnfsf11, Tnfrsf11b, or Acp5 probes in order to characterize the environment close to the osteoclasts and to address the possibility of sexual dimorphism in Tnfsf11 and Tnfrsf11b expression. We quantified the expression in cells that were up to 25 µm away from the osteoclasts residing on trabecular and endocortical bone surfaces, including mononuclear osteoblastic cells on the bone surface, marrow cells above the bone surface, and osteocytes below the bone surface. Collectively, these cells were defined as cells located next to the osteoclasts. Then, we quantified the expression of Tnfsf11 and Tnfrsf11b in osteocytes located more than 25 µm away from an osteoclast (Fig. 8a). Histological images illustrate Acp5+ osteoclasts on the bone surface (Fig. 8b, d). Since we found no obvious differences in Tnfsf11 and Tnfrsf11b expression between female and male mice (Fig. S7a, b), the two groups were combined to increase statistical power. Tnfsf11+ cells were mainly detected on the bone surface and only to some extent in osteocytes (Fig. 8c), while Tnfrsf11b+ cells were mainly osteocytes (Fig. 8e). Next, we quantified the proportion of Tnfsf11+ and Tnfrsf11b+ cells. A higher proportion of bone surface cells expressed Tnfsf11, compared to osteocytes and marrow, while no difference was detected between marrow cells and osteocytes (Fig. 8f). In contrast, the proportion of Tnfrsf11b+ cells was highest in osteocytes, but did not differ between marrow cells and surface cells. Moreover, the osteocytic expression of Tnfrsf11b was not affected by the proximity to osteoclasts (Fig. 8g).
Fig. 8.
Surface cells proximate to osteoclasts mostly express Tnfsf11 while osteocytes express Tnfrsf11b. Representative images in this figure are of adjacent vertebral bone sections from a 12-week-old female mouse, while quantifications are from male and female mice. a Methodological approach. b Acp5 ISH and c Tnfsf11 ISH on adjacent section. d Acp5 ISH and (e) Tnfrsf11b ISH on adjacent section. Histograms illustrate the mean percentage of (f) Tnfsf11+ and (g) Tnfrsf11b+ cells. f, g Data are shown as mean ± SD and P values are calculated using one-way ANOVA. ****P values < 0.000 1, ***P values < 0.001, **P values < 0.01, *P values < 0.05. n = 10/group. Scale bars: = 50 µm. Tb Trabecular, Bm bone marrow. Illustrations were created using BioRender.com
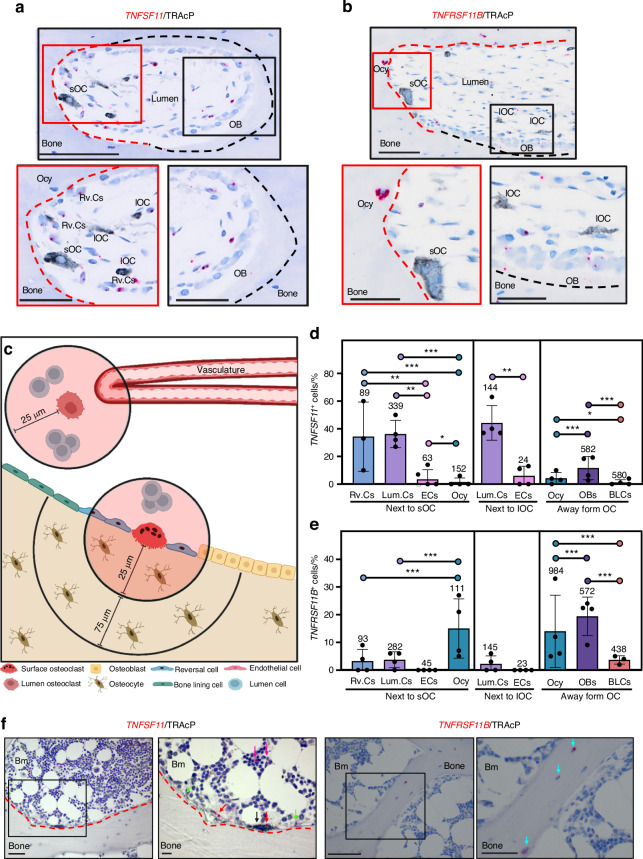
Osteoprogenitor cells proximate to the osteoclasts are the most abundant source of TNFSF11 in humans
To determine whether a similar expression pattern exists in humans, we investigated femoral cortical bone sections from adolescents and Jamshidi iliac crest bone sections from healthy adults. Chromogenic ISH for TNFSF11 or TNFRSF11B expression was combined with an immunohistochemical staining for TRAcP. Histological images illustrated the expression of TNFSF11 in reversal cells and lumen cells residing close to osteoclasts (Fig. 9a), which previously has been demonstrated to be osteoprogenitor cells.1,3,5,34,40 TNFRSF11B was found predominantly in osteocytes close to intracortical canals and in osteoblasts on the bone surface (Fig. 9b). The cellular expression of TNFSF11 and TNFRSF11B was quantified within 25 µm from surface osteoclasts or lumen osteoclasts, including mononuclear reversal cells on the bone surface, mononuclear lumen cells, and endothelial cells (Fig. 9c). Moreover, the cellular expression of TNFSF11 and TNFRSF11B was quantified in osteocytes 0–100 µm away from surface osteoclasts, in osteoblasts on osteoid surfaces, and in bone lining cells on quiescent bone surfaces (Fig. 9c). Then, we calculated the likelihood of detecting TNFSF11 and TNFRSF11B across different cell types. We found that reversal cells next to osteoclasts on eroded surfaces and lumen cells next to surface or lumen osteoclasts were more likely to express TNFSF11 compared to endothelial cells of the vascular structures, osteocytes (Fig. 9d and Table S1), osteoblasts on osteoid surfaces, and bone lining cells on quiescent surfaces (Table S1). Contrary to the TNFSF11 expression pattern, osteocytes and osteoblasts were more likely to express TNFRSF11B compared to reversal cells, lumen cells and bone lining cells (Fig. 9e and Table S1) Similar to the cortical compartment, histological assessment of TNFSF11 and TNFRSF11B in trabecular compartment revealed that TNFSF11 was mainly expressed by reversal cells and canopy cells close to osteoclasts, while TNFRSF11B was mainly expressed by trabecular osteocytes (Fig. 9f).
Fig. 9.
TNFSF11 and TNFRSF11B expression in human cortical and trabecular bone. a–e Data is obtained from cortical femoral bone specimens acquired from 4 adolescent females, and (f) from iliac crest bone sections from healthy humans. a Representative image of a selected cutting cone from TNFSF11 ISH (red), combined with a TRAcP (black) immunostained section. b Representative images of a selected cutting cone from TNFRSF11B (red) ISH, combined with a TRAcP (black) immunostained section. Red and black dashed lines in (a, b), respectively indicate eroded and formative surfaces, as determined by Masson’s Trichrome stained adjacent sections. c Methodological approach. d Histograms illustrate the mean percentage of TNFSF11+ cells located either next to surface osteoclasts, away from surface osteoclasts, or next to lumen osteoclasts. e Histogram illustrating the mean percentage of TNFRSF11B+ located either next to surface osteoclasts, away from surface osteoclasts, or next to lumen osteoclasts. The number above each bar is the number of cells quantified. f Representative images of human iliac crest bone sections stained for TNFSF11 or TNFRSF11B ISH (red), combined with a TRAcP immunostaining (black). TRAcP+ Osteoclasts (black arrow), TNFSF11+ Canopy cells (red arrows), reversal cells (green arrows), marrow cells (purple arrows), and TNFRSF11B+ osteocytes (blue arrows). d, e Data are shown as mean ± SD. Clustered logistic regression was utilized to determine the likelihood of TNFSF11 or TNFRSF11B being expressed in one cell population compared with another. ***P values < 0.001, **P values < 0.01, *P values < 0.05. P values, odds ratios, and corresponding 95% confidence intervals are shown in Table S1. Scale bars: overview = 100 µm, high magnification images = 50 µm. sOC surface osteoclast, lOC lumen osteoclast, Ocy osteocyte, OB osteoblast, Rv.C reversal cell, Lum.C lumen cell, EC vascular endothelial cell, BLC bone lining cell, Illustrations were created using BioRender.com
Discussion
The identification of cells orchestrating the activation of osteoclastogenesis through the RANKL/OPG-pathway have been debated since the discovery of the RANKL/OPG system in the 1990s,26,27,41–46 but the exact and most abundant cellular sources of RANKL and OPG has remained unresolved until now. In the present study, we investigated the cellular in situ mRNA expression of TNFSF11 (encoding RANKL) and TNFRSF11B (encoding OPG) across mouse, rat, and human bone, across different skeletal sites and compartments, and upon OPG:Fc treatment in mice, reflecting the events leading to rebound resorption upon denosumab discontinuation. Moreover, we utilized publicly available scRNAseq data from enriched bone marrow stoma of mice to identify and characterize cell populations that express RANKL. Finally, we utilized a single injection of PTH to map the landscape of the cells that orchestrate the activation of osteoclastogenesis through the RANKL/OPG-pathway.
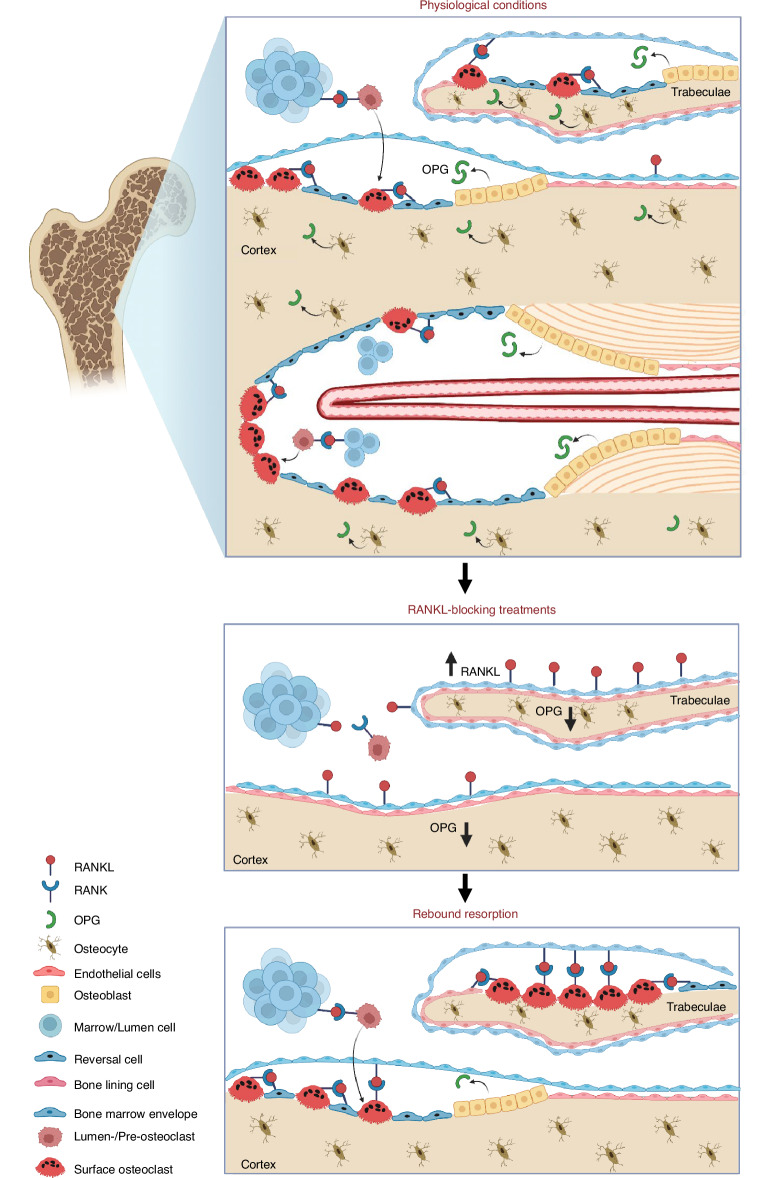
The study demonstrates that osteoprogenitors close to the bone surface (bone lining cells, reversal cells, and lumen/canopy cells) are the most abundant source of RANKL, whereas osteocytes, bone-forming osteoblasts, fibrocytes of the inner ear, and chondrocytes of the growth plate and articular cartilage are the most abundant source of OPG. Two weeks of OPG:Fc treatment resulted in accumulation of osteoclastic activation sites on trabecular bone surfaces, characterized by increased expression of RANKL mostly by bone surface cells and reduced local expression of OPG by bone surface cells and osteocytes. However, this was not the case on endocortical bone surfaces, where bone surface cells and osteocytes exhibited a reduced OPG expression, while RANKL expression were less affected. Accordingly, the osteoclastic rebound upon OPG:Fc withdrawal likely occurs mainly on trabecular surfaces, and not on endocortical surfaces, which probably explain why denosumab discontinuation may increase the risk of multiple vertebral fractures, as vertebrae predominately consist of trabecular bone.47 Taken together, these findings suggest that treatments that inhibit the function of RANKL induce an accumulation of local osteoclastogenic activation sites that are ready to induce osteoclastogenesis upon treatment withdrawal, as summarized in Fig. 10 and discussed below.
Fig. 10.
Concluding figure illustrating key findings. Under physiological conditions surfaces are activated by e.g. PTH stimulus and occupied by osteoclasts that resorb bone. Osteoprogenitors are recruited to the bone surface, where they continue providing the necessary RANKL for continuous recruitment of osteoclasts until bone formation is initiated where osteoblasts, together with osteocytes provide inhibitory OPG. During inhibition of RANKL (by e.g. OPG:Fc or denosumab treatment), osteoclasts are abolished, also leading to the abolishment of osteoblast lineage cells due to the coupling of bone resorption and formation. Moreover, trabecular bone surface cells increase the production of RANKL, while OPG production is decreased in trabecular surface cells and osteocytes. RANKL production remains unchanged in endocortical surface cells and osteocytes, while OPG is reduced in bone surface cells and osteocytes near the surface. Taken together, this reflects an accumulation of local activation sites with an increase in RANKL production in trabecular, but not endocortical bone surfaces, and a decrease in OPG in bone surface cells, and osteocytes close to the trabecular and endocortical surfaces. This shift in local RANKL/OPG production eventually sets stage for rebound bone resorption upon treatment discontinuation. Illustrations were created using BioRender.com
Several studies have presented novel insights into the mechanisms contributing to rebound resorption upon denosumab discontinuation, potentially leading to bone loss and multiple vertebral fractures in patients.15,18–20,48–51 One possible mechanism is that osteoclasts have been shown to undergo fission, forming mononucleated osteomorphs. This fission is induced by denosumab treatment, as recently shown in an OPG:Fc-treated murine model.19 Here, the notion is that these accumulating osteomorphs re-fuse upon treatment withdrawal, thereby contributing to the rebound resorption.19 In line with this, another study has reported an increased pool of circulating pre-osteoclasts during denosumab treatment.17 Nonetheless, the accumulation of pre-osteoclasts and osteomorphs, whether locally or in the circulation, only partially explains the rebound resorption associated with denosumab discontinuation. Pre-osteoclasts need to be recruited and stimulated to fuse and initiate bone resorption on multiple bone surfaces before excess bone resorption occurs. The present study demonstrates that OPG:Fc treatment in mice results in a local shift in the expression of RANKL/OPG mostly on the trabecular bone surfaces, resulting in extensive recruitment of osteoclasts on bone surfaces upon treatment withdrawal, in line with previous research where OPG:Fc-treated mice showed extensive resorption following treatment withdrawal.19,52
This spatial link between the increased RANKL/OPG expression ratio and the wave of osteoclasts strongly supports the notion that an accumulation of local activation sites, reflected by increased RANKL expression and a decrease in OPG expression near the bone surfaces, is a key driver of rebound resorption upon treatment discontinuation. The elevated RANKL/OPG ratios in serum of denosumab-treated patients suggests that a similar phenomenon probably occur in humans.49 The present study shows that expression of RANKL was preferentially increased in trabecular bone surface cells upon OPG:Fc, while RANKL expression in cortical bone surface cells, as well as osteocytes and marrow cells appeared less affected by the OPG:Fc treatment. Since bone resorption and formation are coupled processes,3,49 treatments like denosumab in humans and OPG:Fc in mice inhibit both bone resorption and formation.19,53,54 Therefore, the observed higher prevalence of RANKL-expressing bone surface cells in the OPG:Fc-treated mice are likely bone lining cells or reversal cells. Under normal physiological conditions, this accumulation would have resulted in recruitment and activation of an osteoclast on the bone surface, resetting these activation sites. Previous studies have emphasized the indirect, yet an important role bone lining cells play in controlling bone resorption, as they cover the quiescent bone surfaces together with the neighboring bone marrow envelope.1,34,55 Moreover, bone lining cells have been proposed to function as “gate-keepers” that either prevent or allow initiation of bone resorption.56 Firstly, bone lining cells may release collagenases such as MMP13, which digest the unmineralized lamina limitans covering the quiescent surface, exposing bone minerals to osteoclastic contacts.57 Secondly, bone lining cells retract from the bone surface, making the surface accessible for recruited osteoclasts to attach, fuse and resorb bone.33,39,58–60 Thirdly, bone lining cells re-colonize the resorbed bone surfaces as osteoblastic reversal cells (osteoprogenitors), maintaining direct contact with secondary osteoclasts deepening the erosion.3,33,55 Collectively, the exclusive position of bone lining cells directly on the quiescent bone surface, together with their expression of RANKL and their role in preparing the surface for osteoclastic bone resorption, supports the pivotal role of bone lining cells and reversal cells in the activation of osteoclastogenesis and rebound resorption upon denosumab discontinuation.
Under physiological conditions in mice, the RANKL-expressing cells close to osteoclasts are more likely to be bone surface cells than osteocytes or marrow cells. In humans, these RANKL-expressing cells are more likely to be osteoprogenitors, including reversal cells and lumen/canopy cells located near osteoclasts – rather than osteocytes, marrow cells, or the more distant bone-forming osteoblasts and bone lining cells. Taken together, this suggests that osteoprogenitors, not osteocytes, are the most abundant source of RANKL. This is in contradiction with previous studies, suggesting that osteocytes are the most abundant sources of RANKL based on conditional knockout models, such as the Dmp1-cre;Tnfsf11fl/fl and Sost-cre;Tnfsf11fl/fl models.27,61 However, Dmp1 and Sost promoters are not exclusively activated in osteocytes, as several lineage-tracing studies have shown that these cre models also result in staining of non-osteocytic cell populations, including cells on the bone surface and in the marrow.20,22,27–29 Our data suggests that these often-overlooked bone surface osteoprogenitor cells are likely important in the activation of osteoclastogenesis. Previously, we have established that bone lining cells, reversal cells, intracortical lumen cells, and bone marrow envelope/canopy cells express markers such as Runt-related transcription factor 2 (RUNX2), CD56, CD271, and collagen type 3 in human bone, markers characteristic to osteoprogenitors,3,34,40,62 as well as receptors for osteoclastic coupling factors.63 This supports the concept that osteoprogenitors on or above the bone surface, including bone lining cells, reversal cells, lumen cells, and bone marrow envelope cells are capable of producing RANKL and recruiting osteoclasts to the bone surface. In addition, osteoprogenitors may also respond to osteoclastic coupling factors, suggesting bidirectional communication between osteoprogenitors and osteoclasts.63 To characterize the cell populations expressing RANKL and OPG, we retrieved publicly available scRNAseq data of enriched bone marrow stroma isolated from mice. Interestingly, RANKL and OPG was expressed mostly by three cell clusters – in these clusters, a high proportion of RANKL+ cells were identified to express high levels of Mmp13. This was again validated in vehicle and OPG:Fc-treated mice by confirming that RANKL expressing cells on the bone surface also express RANKL. This is in line with our previous findings, where Mmp13 has been detected at high levels in osteoprogenitor cells (i.e. reversal cells, bone lining cells and canopy cells) on the bone surfaces,33,34 further supporting the notion that osteoprogenitor cells on the bone surface activate osteoclastogenesis and recruit osteoclasts to the bone surfaces under physiological conditions and following withdrawal of RANKL blocking treatments.
In a recently published study, which aimed to refine the identity of mesenchymal cell types associated with murine periosteal and endosteal bone, 11 different cell clusters were identified—two of these were termed Osteo-CAR (defined by Limch1, Wif1, Angpt4 and Kcnk2 expression) and pre-osteoblasts (defined by Spp1, Mmp13, Slit2, and Vdr expression), respectively.64 Nookaew et al. confirmed the spatial organization of these cells using ISH for markers Limch1 and Spp1, locating them closely to endosteal bone surfaces.64 In the present study, we interestingly identified three of these genes (Limch1, Wif1, Mmp13) to be highly expressed by cells that also express RANKL—further highlighting that these are osteoprogenitors. Interestingly, Nookaew et al. found that denosumab treatment of humanized RANKL mice did not have any effect on the abundance of Limch1+ Osteo-CAR cells, while it only slightly reduced the abundance of Spp1+ pre-ostebolasts.64 Combined with data of the present study, these findings indicate that osteoprogenitors near the bone surface play a pivotal role in the rebound resorption observed following denosumab discontinuation, which should be investigated further.
Since the time-window from the first expression of RANKL in e.g. bone lining cells, to osteoclasts are recruited to the bone surface is very narrow, it is difficult to capture the cells that first express RANKL in vivo. The recruitment of osteoclasts likely occurs within hours, as suggested by a highly increased RANKL expression in osteoblastic cells only a few hours after PTH treatment, which returns to baseline within 24 h post PTH treatment, suggesting a short-term and transient increase in RANKL upon PTH treatment.30,31 This aligns well with the present findings, where an upregulation of RANKL expression and downregulation of OPG expression was seen in mostly in bone surface cells and osteocytes near the bone surface—further supporting our findings in the OPG:Fc-treated mice.
The expression of OPG was observed mainly in bone surface cells (predominantly bone-forming osteoblasts), osteocytes, and fibrocytes of the rat inner ear. Moreover, the OPG expression by bone surface cells and osteocytes residing near the cortical and trabecular bone surfaces was significantly reduced upon OPG:Fc treatment in mice. This is consistent with a recent study that examined the effect of denosumab treatment in humanized RANKL mice, and reported a reduced OPG expression by cortical and trabecular osteocytes, and that sclerostin-negative osteocytes close to the bone surface are the predominant source of OPG.20 However, some of the OPG-expressing cells appear as bone surface cells, which may be OPG-expressing osteoblasts that are lost upon denosumab treatment, as these osteoblasts are also a source of OPG,26 consistent with the finding of the present study where osteoblasts were found to be an abundant source of OPG. Likewise, denosumab treatment reduces the expression of both osteoclast and bone-forming osteoblast-related genes in postmenopausal women following a single dose of denosumab,51,65 and bone formation parameters in osteoporotic patients treated with denosumab,53 as osteoblastic bone formation is normally tightly coupled to osteoclastic bone resorption during the bone remodeling process.3,49 These findings further support the view that bone-forming osteoblasts on the bone surface are a source of OPG, which during denosumab treatment is abolished, leading to loss of newly embedded osteocytes. Nonetheless, it is likely that loss of osteoblasts and newly formed osteocytes both – but not necessarily to a similar extent—contribute to the reduced expression of OPG near the bone surface upon blocking of RANKL.
In addition, a high OPG-expression was also observed in chondrocytes in the tibial epiphyseal growth plate and in articular cartilage, while RANKL was much less expressed in the chondrocytes and highly expressed in the primary spongiosa beneath the epiphyseal growth plate. The expression patterns of OPG in chondrocytes are in line with the findings of an older isotope-based ISH study on rats,66 as well as those of the recent study on a humanized RANKL murine model.20 On the other hand, RANKL expression is high in the primary spongiosa where, where it promotes osteoclast recruitment for resorption of the calcified cartilage and its subsequent remodeling into trabecular bone. The expression of OPG by chondrocytes in the growth plate and articular cartilage, as well as by fibrocytes lining the otic capsule of the inner ear, may serve as a protective mechanism,20,38,66 as abnormal bone resorption at these sites is seen in OPG deficient mice.37,67 Interestingly, the endogenic OPG-mediated inhibition of osteoclastic bone resorption by fibrocytes lining the inner ear does not lead to an increased abundance of RANKL-expressing cells. This implies that not all RANKL-blocking conditions lead to an accumulation of osteoclastogenic activation sites.
One limitation of this study is that we utilized ISH to detect the cells that express RANKL and OPG, which may not necessarily reflect expression at the translational level. However, previous detections of RANKL and OPG using immunohistochemical techniques, where OPG immunoreactivity has been reported in osteoblasts, pre-osteoblasts, and osteocytes68—whereas RANKL immunoreactivity has been reported in canopy and bone lining cells,69,70 support translation of mRNA at the positions corresponding with the present ISH. Secondly, we used young mice to study the cellular mechanisms leading to rebound following OPG:Fc withdrawal. Future studies should preferably investigate these mechanisms in older, and ovariectomized mice, as it has previously been shown that estrogen deficiency specifically increases RANKL expression in bone lining cells in mice.69 Similarly, while we utilized bone specimens from adolescents, allowing for spatiotemporal analysis of cellular events involved in the activation of osteoclastogenesis, it is essential to recognize that our findings may not generalize to other age groups. Finally, the present study utilized AI-based technology to quantify cells, which could potentially influence the accuracy of cell counts due to various factors, including variability in cell morphology and high cell densities, requiring a large training data to optimize.
In summary, we utilized multiple models to assess the local sources of RANKL and OPG. In each model, OPG appears as a protective factor acting over a broad matrix zone, whereas RANKL is responsible for a localized activation of osteoclastogenesis near the bone surfaces. We demonstrate that under physiological conditions, osteoprogenitors, including bone lining cells, reversal cells, lumen cells, and canopy cells are the main source of RANKL, while osteocytes, osteoblasts, fibrocytes lining the inner ear, and chondrocytes of the growth plate and articular cartilage are the major sources of OPG. PTH treatment in mice illustrated that bone surface cells, osteocytes and marrow cells close to the bone surface are capable responding to systemic PTH by increasing RANKL expression and decreasing OPG expression, with bone surface cells being the primary responder to this stimulus. Furthermore, we showed that OPG:Fc treatment in murine models mimics denosumab treatment in humans, and results in accumulation of osteoclastogenic activation sites with increased RANKL-expressing bone surface cells on trabecular surfaces, but not on endocortical bone surfaces. Moreover, OPG:Fc treatment results in a decreased expression of OPG in bone surface cells and osteocytes close to the bone surface, resulting in a local shift in the RANKL/OPG ratio, setting the stage for rebound resorption upon discontinuation. Further characterization of cell populations using scRNAseq of murine enriched bone marrow stroma revealed that Tnfsf11+ cells had a high expression of genes such as Mmp13, Wisp2, Limch1, and Wif1, and a low expression of genes such as Col1a1, Lepr, and Lpl. Multiplex ISH further confirmed the co-expression of Mmp13 and Tnfsf11 in bone surface cells of both vehicle and OPG:Fc-treated mice. Taken together, these findings suggest that bone surface osteoprogenitors and osteocytes conjointly regulate the activation of osteoclastogenesis in a physiological state and following denosumab discontinuation.
Methods
Animal experiments
OPG:Fc treatment of C57BL/6 J female mice
Three groups of 6–8-week-old female C57BL/6 J mice (n = 8/group) were obtained from Australian BioRsources and subjected to a treatment regimen with 10 mg/kg OPG:Fc (Amgen inc.), or vehicle (saline) administered i.p. thrice weekly for 2 weeks. One cohort of mice was sacrificed at the end of the 2 weeks of treatment, while three other cohorts were treated for 2 weeks followed by treatment withdrawal for 6, 9, and 11 weeks, i.e., sacrificed at week 8, 11, and 13, respectively (Fig. 1a). Bones were harvested immediately after sacrifice. Animal experiments were performed in accordance with approved protocols from the Garvan Institute/St Vincent’s Hospital Animal Ethics committee (ARA 21/17 and 18/03) and conducted in accordance with the Australian Code of Practice and Use of Animals for Scientific purposes. All animals were acclimatized upon arrival and housed under a constant temperature of 21.4 °C in ventilated cages with a 12-h light/dark cycle. Mice were provided with standard chow and water ad libitum.
Dual-energy X-ray absortiometry (DEXA)
To assess bone mineral density (BMD) in OPG:Fc- or vehicle-treated mice, DEXA was performed every two or three weeks and at the end of the study. Prior to scanning, mice were anesthetized with 3%–5% inhaled isoflurane using the Faxitron Ultrafocus DEXA system (Hologic, USA). BMD was assessed using the Vision DEXA system (Hologic, USA), with a manually delineated region of interest covering the left hind limb.
Micro-computed tomography (µCT)
Right femora from OPG:Fc- and vehicle-treated mice were scanned using a SkyScan 1772 µCT scanner (Bruker, Belgium). The scans were performed using a 0.5 mm aluminum filter, isotropic voxel size of 5 µm, and an X-ray voltage of 50 kV and current of 200 µA. The bone micro-architecture was examined using a 0.5-mm-high volume of interest (VOI) starting 3 mm proximal to the distal femoral growth plate and quantified using the CTAn software (Bruker, Belgium). Scans were reconstructed using SkyScan nRecon (Bruker, Belgium) and the range of attenuation coefficients used was between 0.0 and 0.12. The µCT images were segmented using a global threshold value of 792 mg HA/cm3. The trabecular analysis included: trabecular thickness, trabecular separation, and trabecular bone volume/tissue volume (BV/TV). Additional analyses included cortical bone, and marrow space cross-sectional areas.
PTH treatment of C57BL/6 J female mice
12-week-old female C57BL/6 J mice (Jackson Laboratories, strain 00664) were subjected to a single dose of 80 µg/kg PTH (1–34) (Bachem), or vehicle (saline) administered i.p. (n = 3/group). Mice were sacrificed 1 h following PTH injection and femora were harvested. Animal experiments were performed in accordance with approved protocols from the institutional Animal Care and Use Committee of Harvard Medical School (2016N000303) and conducted in accordance with relevant guidelines ad laws. All animals were acclimatized upon arrival and housed under a constant temperature of 21.4 °C in ventilated cages with a 12-h light/dark cycle. Mice were provided with standard chow and water ad libitum.
C57B6 female and male mice
12-week-old female and male C57BL/6 J mice (n = 5/gender) were obtained from Janvier Labs. Animals were sacrificed, and vertebrae were harvested. Animal experiments were performed in accordance with approved protocols from the Danish Ethical Committee for Animal Studies (2018-15-0201-01436). All animals were acclimatized upon arrival and housed under constant temperature of 21 °C in ventilated cages with a 12-h light/dark cycle. Mice were provided with standard chow and water ad libitum.
Sprague-Dawley rats
10-day-old male Sprague-Dawley rats (n = 4) were obtained from Charles River. Animals were first acclimatized and then sacrificed, followed by harvest of the crania, harboring the inner ear. Animal experiments were approved by the Danish Ethical Committee for Animal Studies (P-19-315). All animals were housed under a constant temperature of 22 °C in ventilated cages with a 12-h light/dark cycle. Rats were provided with standard chow and water ad libitum.
Human bone specimen
Human cortical bone specimens were obtained from the proximal femur of 4 healthy adolescent females (aged 14–16 years) undergoing corrective surgery for Coxa Valga, a deformity of the hip. The study was approved by the Danish National Committee on Biomedical Research Ethics (Project ID: S-2012- 0193). 3-mm Jamshidi Iliac crest bone biopsies was obtained from 2 healthy females (aged 33 and 49) and 2 males (aged 27 and 41). The study was approved by the Danish National Committee of Biomedical Research Ethics (Project ID S-20110112). Written informed consent was received prior to participation in this study.
Tissue preparation
All tissue samples were fixed immediately in 4% paraformaldehyde for 24–48 h, decalcified in 0.5 mol·L−1 EDTA, paraffin-embedded, serially sectioned into 3.5-µm-thick sections. All sections were stored at 4 °C before the subsequent experiments.
Masson trichrome stain
Every 5th human bone section was Masson trichrome stained as previously described3,62 to select sections with active intracortical bone remodeling. Singleplex ISH was performed on an adjacent section from each individual, as described below.
Chromogenic ISH combined with immunostaining
Singleplex chromogenic ISH was performed using a modified version of the RNAScope 2.5 HD procedure (310035, Advanced Cell Diagnostics [ACD]). Sections were deparaffinized in xylene, dehydrated in 99% ethanol, and blocked with 3% hydrogen peroxide for 30 min at room temperature. Target retrieval and nuclei permeabilization was performed by incubating samples in ACD’s Target Retrieval Reagent (Catalog 322000) in an 80–90 °C water bath, followed by treatment with 10%–20% pepsin in diethylpyrocarbonate- (DEPC-) treated water. Subsequently, sections were probe hybridized overnight at 40 °C using 20 double-Z probe pairs with complementary RNA to the target mRNA. Tissue sections were incubated with following probes diluted 1:1 in probe diluent: mouse Acp5 (Mm-Acp5, catalog 465001), mouse Tnfsf11 (Mm-Tnfsf11, catalog 410921), mouse Tnfrsf11b (Mm-Tnfrsf11b, catalog 488961), rat Tnfsf11 (Rn-Tnfsf11, catalog 813111), rat Tnfrsf11b (Rn-Tnfrsf11b, catalog 813091), human TNFSF11 (Hs-TNFSF11, catalog 412271), and human TNFRSF11B (Hs-TNFRSF11B, catalog 412291). Following probe hybridization, a 6-step branch amplification was carried out according to the manufacturer’s instructions. The signal was further amplified using digoxygenin (DIG)-labeled tyramide from a tyramide signal amplification (TSA) plus DIG Reagent kit (NEL748001KT, PerkinElmer). Sections were washed in TBS/Tween and incubated with Anti-DIG FAB fragments conjugated to alkaline phosphatase (11093274910, Roche). The mRNA signal was developed using Liquid Permanent Red (K064030-2, Agilent).
To detect mature osteoclasts in human bone, sections were further stained for TRAcP using immunohistochemistry. Tissue sections were first washed in demineralized water, blocked in casein diluted in TBS, followed by incubation with a primary mouse IgG2b anti-tartrate-resistant acid phosphatase (TRAcP) antibody (clone 9C5, MABF96, Merck Millipore). Subsequently, sections were washed in TBS, blocked in 3% hydrogen peroxide in TBS, and incubated with horseradish peroxidase (HRP)-conjugated secondary anti-mouse antibody (BrightVision, Immunologic). The sections were washed in TBS and the signal was developed using Deep Space Black (BC-BRI4015H, HISTOLAB). All sections were counterstained with Mayer’s hematoxylin, and mounted in Aquatex. Slides were scanned using an Olympus VS200 Slidescanner or a Hamamatsu slide scanner.
Singleplex and multiplex fluorescence ISH
Singleplex fluorescent ISH was performed using a modified version of the RNAscope 2.5 high-definition procedure (322310, ACD), on sectioned tibiae harvested from vehicle and OPG:Fc-treated mice. The staining was performed similarly to the chromogenic ISH with minor modifications. In short, sections were deparaffinized in xylene, blocked with 3% hydrogen peroxide, followed by target retrieval and tissue permeabilization using a custom pretreatment reagent (catalog 300040). Sections were hybridized using 20 double-Z probe pairs targeting mouse Acp5 (Mm-Acp5, catalog 465001), mouse Tnfsf11 (Mm-Tnfsf11, catalog 410921), mouse Tnfrsf11b (Mm-Tnfrsf11b, catalog 488961). Following hybridization, branch amplification was carried out according to the manufacturer’s instructions and the signal was developed with HRP followed by incubation with the fluorochrome Opal 570.
Multiplex fluorescent ISH was performed using a modified version of the RNAscope Multiplex Flourecent Assay (323110, ACD). Tissue sections were deparaffinized and pretreated as in the singleplex ISH. Following, sections were hybridized using multiplexing probes simultaneously. The probes consisted of 20 double-Z probe pairs with three distinct spectral channels (C), targeting mouse Tnfsf11 (C1-Mm-Tnfsf11, catalog 410921) or mouse Tnfrsf11b (C1-Mm-Tnfrsf11b, catalog 488961) in combination with mouse Mmp13 (C3-Mm-Mmp13, catalog 427601, ACD) and mouse Col1a1 (C4-Mm-Col1a1, catalog, 319371ACD). Following probe hybridization, a 3-step branch amplification was carried out according to the manufacturer’s instructions. The signal was developed using sequential TSA-enhancement with fluorochrome Opal 570 for channel 1, Opal 620 for channel 3 and Opal 690 for channel 4. Tissue sections were blocked using a hydrogen peroxidase blocker followed by incubation with a new peroxidase-conjugated probe for the subsequent channel. In singleplex- and multiplex-stained sections, nuclei in were stained using 2.5 µg/mL Hoechst (H3569, Invitrogen) and mounted in Prolong gold antifade reagent (P36939, Invitrogen). Slides were scanned using an Olympus VS200 slide scanner.
Quantification and spatial analysis using HALO AI
AI-based quantification and spatial analysis was performed using HALO and HALO AI (Indica Labs) on tissue sections of the tibia collected from OPG:Fc- and vehicle-treated mice. Three regions of interest (ROIs) were defined on all scanned sections. The first ROI was defined as a 3-mm-long region, and included the marrow and the trabecular bone compartment, while the cortical bone, growth plate, and primary spongiosa were excluded. In the vehicle-treated mice, measurements began 400 µm distal to the proximal epiphyseal growth plate, while in the OPG:Fc-treated mice, measurements began 600 µm distal to the proximal epiphyseal growth plate, due to the increased length of the primary spongiosa in OPG:Fc-treated mice (Fig. S8a). The nuclei were quantified on a single cell level using a pre-trained deep-learning-based neuronal network (HALO AI, Indica Labs), which was improved on these sections by marking approximately 3 000 additional nuclei before the analysis. Each cell’s mRNA staining intensity of either Tnfsf11 or Tnfrsf11b mRNA was quantified using HALO’s thresholding parameters, which were determined using the real-time tuning function. All parameters were held constant in-between samples. Following nuclei quantification, a spatial analysis was conducted by first drawing an interface line on all trabecular bone surfaces. The distance of every single cell from the interface line was measured up to 20 µm into the marrow 50 µm into the trabecular bone. Surface cells were defined as cells that were less than 3 µm above or below the interface line, while marrow cells were defined as cells more than 3 µm above the interface line and osteocytes as cells more than 3 µm below the interface line. A second 2.5-mm-long ROI was defined, encompassing both cortical compartments, and the marrow compartment. The ROI started 1 mm distal to the proximal epiphyseal growth plate and extended 2.5 mm in both groups. The primary spongiosa, trabecular bone, periosteum, and transcortical canals were excluded from the analysis, to focus on the endocortical surface cells and cortical osteocytes (Fig. S8b). The number of nuclei and Tnfsf11 and Tnfrsf11b mRNA staining intensities were quantified and a spatial analysis was performed by first drawing an interface line, outlining the endocortical bone surfaces of both cortices, and measuring the distance of each cell from the interface line and up to 500 µm into the marrow and 300 µm into the cortical bone. Finally, a third ROI encompassing the growth plate and the primary spongiosa was drawn and the nuclei and their Tnfsf11 and Tnfrsf11b mRNA intensity was quantified. Following, a spatial analysis was performed, where the single cell distances up to 200 µm above or below an interface line, separating the primary spongiosa from the growth plate were measured (Fig. S8c).
The same spatial analysis pipeline was employed to analyze the Tnfsf11 and Tnfrsf11b expression in PTH-treated mice, as in OPG:Fc-treated mice. In short, AI-based quantification and spatial analysis was performed using HALO and HALO AI (Indica Labs) on tissue sections of the femur collected from PTH- and vehicle-treated mice. For first ROI encompassing the trabecular and marrow compartment, the ROI started 400 µm proximal to the distal femoral growth plate and extended 4 mm (Fig. S9a). For the ROI encompassing both cortical bone compartments and the marrow compartment, the analysis started 1 mm proximal to the distal femoral and extended 5 mm (Fig. S9a).
Manual quantification and spatial analysis of chromogenic/Fluorescent ISH
Vehicle and OPG:Fc-treated mice
Osteoclast perimeter and total bone perimeters were manually measured in HALO (HALO AI, Indica Labs) based on Acp5 ISH to determine trabecular and endocortical Oc.Pm/B.pm in vehicle and OPG:Fc-treated mice at week 2, 11, and 13. All perimeters were measured in the previously defined ROIs employed in the Tnfsf11/Tnfrsf11 spatial analysis.
Human bone specimens
The percentage of TNFSF11+ and TNFRSF11B+ cells in adjacent human femoral bone sections was calculated by manually counting TNFSF11+, TNFRSF11B+, and negative cells within a distance of 25 µm from TRAcP+ surface osteoclasts and lumen osteoclasts. The quantified cells were included matrix-embedded osteocytes, mononuclear reversal cells on the eroded bone surface, mononuclear lumen cells, and endothelial cells of vascular structures. Collectively, these cells were defined as peri-osteoclastic cell populations, residing next to osteoclasts. We further quantified the number of positive and negative osteocytes residing 25–100 µm away from surface osteoclasts. Finally, we quantified the number of positive and negative osteoblasts on osteoid surfaces and bone lining cells on quiescent bone surfaces – these cells were defined as cell populations residing away from osteoclasts. The different types of surfaces, including eroded, osteoid and quiescent surfaces, were validated using an adjacent Masson trichrome stained section.
Mouse vertebral bone
Paraffin-embedded lumbar vertebrae (L3–L4) were sectioned in the coronal plane. Subsequently, adjacent sections were stained for Tnfsf11, Acp5, or Tnfrsf11b mRNA using singleplex chromogenic ISH as described above. A ROI was defined by measuring 350 µm from both growth plates of L3 or L4, depending on the tissue quality, encompassing the trabecular and endocortical bone compartment (Fig. S9b). The percentage of Tnfsf11+ and Tnfrsf11+ cells was quantified by first identifying Acp5+ osteoclasts on the bone surfaces of one section, followed by localization of the osteoclast on the adjacent section stained for Tnfsf11 or Tnfrsf11. Following, Tnfsf11+, Tnfrsf11b+, and negative cells that were up to 25 µm away from osteoclasts residing on trabecular or endocortical bone surfaces were manually quantified; collectively, these were defined as cells next to osteoclasts. Osteocytes further away from osteoclasts, were defined as cells located more than 25 µm away from an osteoclast.
Bone marrow scRNA-seq bioinformatic analysis
Mouse bone marrow scRNAseq datasets (accession number GSM3674243, GSM3674244, GSM3674245, and GSM3674246) were downloaded from the NCBI GEO database.32 Seurat (v.5.0.0) was used to analyze the raw count matrix.71 Following a standard quality analysis and prefiltering step, gene expression for each cell was normalized by the total expression, multiplied by 10 000, and log transformed. The top 2 000 variable genes were selected by the FindVariableGenes function. A linear transformation was applied using ScaleData function followed by dimensional reduction analysis. Principal Component Analysis (PCA) was performed using the RunPCA function, followed by Uniform Manifold Approximation and Projection (UMAP) algorithm using the RunUMAP function. Cell cluster biomarkers were identified using the FindMakers function. Cell cluster identity was assigned based on published literature32 and the Cell Taxonomy Database (https://ngdc.cncb.ac.cn/celltaxonomy/). Genes of interest were visualized as violin plots using the VlnPlot function. FindMakers function was also applied to identify differentially expressed genes (DEGs) between Tnfsf11+ and Tnfsf11- cells using Wilcoxon Rank Sum test. P value was adjusted by Bonferroni correction. DEGs meeting the criteria of adjusted P value less than 0.05 and log2 fold change (log2FC) higher than 0.5 were considered significant and visualized by dot plots using the DotPlot function.
Statistics
One-way ANOVAs, two-way ANOVAs, multiple comparisons test, unpaired, and paired t-tests were conducted in GraphPad Prism V8 (GraphPad Software, Inc., La Jolla, CA, USA). One-way ANOVA and comparison tests were conducted using Holm-Sidak’s multiple comparisons test. Two-way ANOVA and comparison tests were conducted using either Sidak’s multiple comparisons test or Tukey’s multiple comparisons test. Mixed-effects analysis was conducted using Sidak’s multiple comparisons test. In the figures, data are expressed as either means with error bars representing standard deviation or means with error bars representing standard error of the mean. P values < 0.05 were considered statistically significant. Clustered logistic regressions were conducted using STATA (STATA 18, StataCorp, College Station, TX, USA) and were utilized to calculate odds ratios (ORs), i.e., the likelihood of a greater or smaller prevalence of a given factor in one cell population compared with another cell population. An OR greater than one indicates a higher prevalence of a given factor in one cell population compared to another, while an OR smaller than one indicates a lower prevalence. Human data included observations from 4 biopsies treating each donor as a cluster. ORs, confidence intervals, and statistical significance were calculated by using the upstream cell population as a reference. The prevalence of a given factor was visualized in histograms illustrating, the percentage of cells positive for selected genes.
Supplementary information
Acknowledgements
We thank Søren Harving for providing the investigated human bone specimens, and Birgit MacDonald, Signe Marie Andersen, and Ariel Castro-Martinez for their technical contributions. The study was supported by Odense University Hospital’s Fund for Free Research (A5111), the Region of Southern Denmark’s Fund for Free Research (A955), and the VELUX Foundations (VELUX25723). The study was supported by the Novo Nordisk Foundation. Amgen Inc provided the OPG:Fc under a material transfer agreement.
Author contributions
Contributions T.L.A., C.M.A., J.M.D., and J.F.C., conceptualized and designed the study. B.M.E. contributed to study design. T.L.A., J.F.C., M.M.M., J.S.T., A.B., M.S.S., and L.J.H. designed the animal studies. X.G.D., J.F.C. handled and coordinated the PTH treatment of murine models. A.S.K. and M.M.M. handled and coordinated OPG:Fc treatment of murine models. A.S.K., V.T., C.M. produced µCT and DEXA data. B.M.E., K.S.L., M.H.N. optimized and conducted ISH experiments, B.M.E., K.S.L., M.H.N., and C.M.A. performed cell quantifications, B.M.E., and V.B.K. acquired histological images. B.M.E. performed the data and statistical analyses. B.M.E. and T.L.A. interpreted the results and wrote the first draft of the manuscript. All authors approved the final version of the manuscript.
Data availability
The data sets generated in the present study are available from the corresponding author upon reasonable request.
Competing interests
M.M.M. and X.G.D. have received honoraria from Amgen. J.F.C. has participated in Alexion, Kyowa Kirin and Amgen advisory boards. J.F.C. has received funding from Novartis. M.B. and J.S.T. have received a research grant from Keros Therapeutics for an unrelated study. T.L.A. has collaborated with Amgen and is the custodian of the biopsies from the FREEDOM study, and in part received free reagents from ACD bioscience (now part of the Bio-Techne Corporation) for the specific study. Amgen Inc provided the OPG:Fc under a material transfer agreement.
Supplementary information
The online version contains supplementary material available at 10.1038/s41413-024-00362-4.
References
- 1.Delaisse, J. M. et al. Re-thinking the bone remodeling cycle mechanism and the origin of bone loss. Bone141, 115628 (2020). [DOI] [PubMed]
- 2.FROST, H. M. Dynamics of bone remodeling. Bone Biodyn. 315-334 (1964).
- 3.Lassen, N. E. et al. Coupling of bone resorption and formation in real time: New knowledge gained from human Haversian BMUs. J. Bone Min. Res.32, 1395–1405 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Parfitt, A. M. The coupling of bone-formation to bone-resorption - a critical analysis of the concept and of its relevance to the pathogenesis of osteoporosis. Metab. Bone Dis. Relat.4, 1–6 (1982). [DOI] [PubMed] [Google Scholar]
- 5.Andersen, T. L. et al. Understanding coupling between bone resorption and formation: Are reversal cells the missing link? Am. J. Pathol.183, 235–246 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Reginster, J. Y. Antifracture efficacy of currently available therapies for postmenopausal osteoporosis. Drugs71, 65–78 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Compston, J. E., McClung, M. R. & Leslie, W. D. Osteoporosis. Lancet393, 364–376 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Tsourdi, E. et al. Fracture risk and management of discontinuation of denosumab therapy: A systematic review and position statement by ECTS. J. Clin. Endocr. Metab.106, 264–281 (2021). [DOI] [PubMed] [Google Scholar]
- 9.Tsourdi, E. et al. Discontinuation of denosumab therapy for osteoporosis: A systematic review and position statement by ECTS. Bone105, 11–17 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Ferrari, S. et al. Further nonvertebral fracture reduction beyond 3 years for up to 10 years of denosumab treatment. J. Clin. Endocr. Metab.104, 3450–3461 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Brown, J. P. et al. Bone remodeling in postmenopausal women who discontinued denosumab treatment: Off-treatment biopsy study. J. Bone Miner. Res.26, 2737–2744 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Cosman, F., Huang, S., McDermott, M. & Cummings, S. R. Multiple vertebral fractures after denosumab discontinuation: FREEDOM and FREEDOM extension trials additional post hoc analyses. J. Bone Min. Res. 37, 2112–2120 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anastasilakis, A. D. et al. Clinical features of 24 patients with rebound-associated vertebral fractures after denosumab discontinuation: Systematic review and additional cases. J. Bone Min. Res. 32, 1291–1296 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Cummings, S. R. et al. Vertebral fractures after discontinuation of denosumab: A post hoc analysis of the randomized placebo-controlled FREEDOM trial and its extension. J. Bone Min. Res.33, 190–198 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Ferrari, S. & Langdahl, B. Mechanisms underlying the long-term and withdrawal effects of denosumab therapy on bone. Nat. Rev. Rheumatol.19, 307–317 (2023). [DOI] [PubMed] [Google Scholar]
- 16.Fassio, A. et al. Changes in Dkk-1, sclerostin, and RANKL serum levels following discontinuation of long-term denosumab treatment in postmenopausal women. Bone123, 191–195 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Fontalis, A., Gossiel, F., Schini, M., Walsh, J. & Eastell, R. The effect of denosumab treatment on osteoclast precursor cells in postmenopausal osteoporosis. Bone Rep.13, 100457 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Jähn-Rickert, K. et al. Elevated bone hardness under denosumab treatment, with persisting lower osteocyte viability during discontinuation. Front. Endocrinol. 11, 250 (2020). [DOI] [PMC free article] [PubMed]
- 19.McDonald, M. M. et al. Osteoclasts recycle via osteomorphs during RANKL-stimulated bone resorption. Cell184, 1330 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu, Q. et al. Reduced osteoprotegerin expression by osteocytes may contribute to rebound resorption after denosumab discontinuation. JCI Insight8, e167790 (2023). [DOI] [PMC free article] [PubMed]
- 21.Kartsogiannis, V. et al. Localization of RANKL (receptor activator of NF kappa B ligand) mRNA and protein in skeletal and extraskeletal tissues. Bone25, 525–534 (1999). [DOI] [PubMed] [Google Scholar]
- 22.Matic, I. et al. Quiescent bone lining cells are a major source of osteoblasts during adulthood. Stem Cells34, 2930–2942 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Silvestrini, G. et al. Detection of osteoprotegerin (OPG) and its ligand (RANKL) mRNA and protein in femur and tibia of the rat. J. Mol. Histol.36, 59–67 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Gori, F. et al. The expression of osteoprotegerin and RANK ligand and the support of osteoclast formation by stromal-osteoblast lineage cells is developmentally regulated. Endocrinology141, 4768–4776 (2000). [DOI] [PubMed] [Google Scholar]
- 25.Thomas, G. P., Baker, S. U., Eisman, J. A. & Gardiner, E. M. Changing RANKL/OPG mRNA expression in differentiating murine primary osteoblasts. J. Endocrinol.170, 451–460 (2001). [DOI] [PubMed] [Google Scholar]
- 26.Cawley, K. M. et al. Local production of osteoprotegerin by osteoblasts suppresses bone resorption. Cell Rep.32, 108052 (2020). [DOI] [PMC free article] [PubMed]
- 27.Xiong, J. H. et al. Osteocytes, not osteoblasts or lining cells, are the main source of the RANKL required for osteoclast formation in remodeling bone. Plos One10, e0138189 (2015). [DOI] [PMC free article] [PubMed]
- 28.Root, S. H. et al. Perivascular osteoprogenitors are associated with transcortical channels of long bones. Stem Cells38, 769–781 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lim, J., Burclaff, J., He, G., Mills, J. C. & Long, F. Unintended targeting of Dmp1-Cre reveals a critical role for Bmpr1a signaling in the gastrointestinal mesenchyme of adult mice. Bone Res.5, 16049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walker, E. C. et al. Sustained RANKL response to parathyroid hormone in oncostatin M receptor-deficient osteoblasts converts anabolic treatment to a catabolic effect in vivo. J. Bone Miner. Res.27, 902–912 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Huang, J. C. et al. PTH differentially regulates expression of RANKL and OPG. J. Bone Min. Res.19, 235–244 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Baryawno, N. et al. A cellular taxonomy of the bone marrow stroma in homeostasis and leukemia. Cell177, 1915–1932.e1916 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abdelgawad, M. E. et al. Early reversal cells in adult human bone remodeling: osteoblastic nature, catabolic functions and interactions with osteoclasts. Histochemistry Cell Biol.145, 603–615 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Andersen, T. L. et al. A critical role of the bone marrow envelope in human bone remodeling. J. Bone Min. Res.38, 918–928 (2023). [DOI] [PubMed] [Google Scholar]
- 35.Frisch, T., Sorensen, M. S., Overgaard, S. & Bretlau, P. Estimation of volume referent bone turnover in the otic capsule after sequential point labeling. Ann. Otol. Rhinol. Laryngol.109, 33–39 (2000). [DOI] [PubMed] [Google Scholar]
- 36.Sorensen, M. S., Jorgensen, M. B. & Bretlau, P. Drift barriers in the postcartilaginous development of the mammalian otic capsule. Eur. Arch. Otorhinolaryngol.249, 56–61 (1992). [DOI] [PubMed] [Google Scholar]
- 37.Zehnder, A. F. et al. Osteoprotegrin knockout mice demonstrate abnormal remodeling of the otic capsule and progressive hearing loss. Laryngoscope116, 201–206 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zehnder, A. F., Kristiansen, A. G., Adams, J. C., Merchant, S. N. & McKenna, M. J. Osteoprotegerin in the inner ear may inhibit bone remodeling in the otic capsule. Laryngoscope115, 172–177 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Everts, V. et al. The bone lining cell:: Its role in cleaning Howship’s lacunae and initiating bone formation. J. Bone Miner. Res.17, 77–90 (2002). [DOI] [PubMed] [Google Scholar]
- 40.van Dijk Christiansen, P. et al. Osteoprogenitor recruitment and differentiation during intracortical bone remodeling of adolescent humans. Bone177, 116896 (2023). [DOI] [PubMed] [Google Scholar]
- 41.Eriksen, E. F., Eghbali-Fatourechi, G. Z. & Khosla, S. Remodeling and vascular spaces in bone. J. Bone Min. Res22, 1–6 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Boyce, B. F. & Xing, L. Functions of RANKL/RANK/OPG in bone modeling and remodeling. Arch. Biochem Biophys.473, 139–146 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yasuda, H. et al. Osteoclast differentiation factor is a ligand for osteoprotegerin/osteoclastogenesis-inhibitory factor and is identical to TRANCE/RANKL. Proc. Natl Acad. Sci. USA95, 3597–3602 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim, H. N., et al. Osteocyte RANKL is required for cortical bone loss with age and is induced by senescence. Jci Insight5, e138815 (2020). [DOI] [PMC free article] [PubMed]
- 45.Lacey, D. L. et al. Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell93, 165–176 (1998). [DOI] [PubMed] [Google Scholar]
- 46.Tatsumi, S. et al. Targeted ablation of osteocytes induces osteoporosis with defective mechanotransduction. Cell Metab.5, 464–475 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Clarke, B. Normal bone anatomy and physiology. Clin. J. Am. Soc. Nephrol.3, S131–S139 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Solling, A. S., Harslof, T. & Langdahl, B. Treatment with zoledronate subsequent to denosumab in osteoporosis: A 2-year randomized study. J. Bone Miner. Res.36, 1245–1254 (2021). [DOI] [PubMed] [Google Scholar]
- 49.Solling, A. S., Harslof, T., Jorgensen, N. R. & Langdahl, B. Changes in RANKL and TRAcP 5b after discontinuation of denosumab suggest RANKL mediated formation of osteoclasts results in the increased bone resorption. Osteoporos. Int. 34, 599–605 (2023). [DOI] [PubMed] [Google Scholar]
- 50.Tsai, J. et al. Denosumab reduces the number of circulating osteoclast precursor cells in postmenopausal women with osteoporosis. J. Bone Miner. Res.38, 188 (2023). [Google Scholar]
- 51.Drejer, L. A. et al. Trabecular bone deterioration in a postmenopausal female suffering multiple spontaneous vertebral fractures due to a delayed denosumab injection - A post-treatment re-initiation bone biopsy-based case study. Bone Rep.19, 101703 (2023). [DOI] [PMC free article] [PubMed]
- 52.Kim, A. S. et al. Temporal patterns of osteoclast formation and activity following withdrawal of RANKL inhibition. J. Bone Miner. Res.39, 484–497 (2024). [DOI] [PMC free article] [PubMed]
- 53.Reid, I. R. et al. Effects of denosumab on bone histomorphometry: the FREEDOM and STAND studies. J. Bone Min. Res.25, 2256–2265 (2010). [DOI] [PubMed] [Google Scholar]
- 54.Eastell, R. et al. Effects of denosumab on bone turnover markers in postmenopausal osteoporosis. J. Bone Miner. Res.26, 530–537 (2011). [DOI] [PubMed] [Google Scholar]
- 55.Kristensen, H. B., Andersen, T. L., Marcussen, N., Rolighed, L. & Delaisse, J. M. Osteoblast recruitment routes in human cancellous bone remodeling. Am. J. Pathol.184, 778–789 (2014). [DOI] [PubMed] [Google Scholar]
- 56.Miller, S. C. & Jee, W. S. S. The bone lining cell: a distinct phenotype? Calcif. Tissue Int.41, 1–5 (1987). [DOI] [PubMed] [Google Scholar]
- 57.Chambers, T. J., Thomson, B. M. & Fuller, K. Effect of substrate composition on bone resorption by rabbit osteoclasts. J. Cell Sci.70, 61–71 (1984). [DOI] [PubMed] [Google Scholar]
- 58.Karsdal, M. A., Fjording, M. S., Foged, N. T., Delaissé, J. M. & Lochter, A. Transforming growth factor-β-induced osteoblast elongation regulates osteoclastic bone resorption through a p38 mitogen-activated protein kinase- and matrix metalloproteinase-dependent pathway. J. Biol. Chem.276, 39350–39358 (2001). [DOI] [PubMed] [Google Scholar]
- 59.Perez-Amodio, S., Beertsen, W. & Everts, V. (Pre-)Osteoclasts induceretraction of osteoblasts before their fusion to osteoclasts. J. Bone Min. Res.19, 1722–1731 (2004). [DOI] [PubMed] [Google Scholar]
- 60.Ferrier, J., Xia, S. L., Lagan, E., Aubin, J. E. & Heersche, J. N. Displacement and translocation of osteoblast-like cells by osteoclasts. J. Bone Min. Res.9, 1397–1405 (1994). [DOI] [PubMed] [Google Scholar]
- 61.Nakashima, T. et al. Evidence for osteocyte regulation of bone homeostasis through RANKL expression. Nat. Med.17, 1231–1234 (2011). [DOI] [PubMed] [Google Scholar]
- 62.Andersen, T. L. et al. A physical mechanism for coupling bone resorption and formation in adult human bone. Am. J. Pathol.174, 239–247 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Borggaard, X. G., Nielsen, M. H., Delaisse, J. M., Andreasen, C. M. & Andersen, T. L. Spatial organization of osteoclastic coupling factors and their receptors at human bone remodeling sites. Front Mol. Biosci.9, 896841 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nookaew, I. et al. Refining the identity of mesenchymal cell types associated with murine periosteal and endosteal bone. J. Biol. Chem.300, 107158 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weivoda, M. M. et al. Identification of osteoclast-osteoblast coupling factors in humans reveals links between bone and energy metabolism. Nat. Commun. 11, 87 (2020). [DOI] [PMC free article] [PubMed]
- 66.Ikeda, T., Utsuyama, M. & Hirokawa, K. Expression profiles of receptor activator of nuclear factor kappaB ligand, receptor activator of nuclear factor kappaB, and osteoprotegerin messenger RNA in aged and ovariectomized rat bones. J. Bone Min. Res.16, 1416–1425 (2001). [DOI] [PubMed] [Google Scholar]
- 67.Mizuno, A. et al. Severe osteoporosis in mice lacking osteoclastogenesis inhibitory factor osteoprotegerin. Biochem. Biophys. Res. Commun.247, 610–615 (1998). [DOI] [PubMed] [Google Scholar]
- 68.Onyia, J. E. et al. In vivo demonstration that human parathyroid hormone 1-38 inhibits the expression of osteoprotegerin in bone with the kinetics of an immediate early gene. J. Bone Min. Res.15, 863–871 (2000). [DOI] [PubMed] [Google Scholar]
- 69.Streicher, C. et al. Estrogen regulates bone turnover by targeting RANKL expression in bone lining cells. Sci. Rep.7, 6460 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pereira, R. C. et al. Bone canopies in pediatric renal osteodystrophy. PLoS One11, e0152871 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hao, Y. et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat. Biotechnol.42, 293–304 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data sets generated in the present study are available from the corresponding author upon reasonable request.