Abstract
Human immunodeficiency virus type 1 (HIV-1) subtype C is now the predominant subtype in the global epidemic. This subtype is encountered in southern Africa and parts of Asia, where the epidemic is rapidly spreading. One possible explanation for these epidemiological observations is that this subtype has genetic characteristics that may contribute to its spread and/or pathogenic potential. In this report, we describe the construction of MJ4, an infectious chimeric molecular clone of HIV-1 subtype C that replicates in donor peripheral blood mononuclear cells and macrophages. We also tested this clone for its ability to use the chemokine receptors CCR1, CCR2b, CCR3, CXCR4, and CCR5 and found that the clone utilizes only CCR5 as the coreceptor for cell entry. The MJ4 clone will be useful in further biological and virological characterization of HIV-1 subtype C and will be an important tool in the continuing efforts to understand what may constitute protective immunity in HIV-1. The clone may also be used in experimental design of vaccine candidates that may be directed against HIV-1 subtype C.
A significant challenge in the global effort to develop a vaccine against human immunodeficiency virus type 1 (HIV-1) is the extensive genetic variation observed among viral strains from different countries. Phylogenetic analysis has shown that HIV-1 sequences can be classified into three main groups designated M (for major), O (outlier), and N (non-M, non-O) (17, 29, 41, 43). Group M viruses are responsible for the majority of HIV-1 infections in the world (13, 21, 29) and can be subdivided into subtypes A through D, F, G, H, J, K, and circulating recombinant forms (CRFs). Genetic subtypes show differences of as much as 24% in amino acid sequence (15, 21), which raises the possibility that a vaccine candidate developed from one subtype may not be equally efficacious for other subtypes. Despite this concern, most immunological and virological characterization of HIV-1 has been carried out only with subtype B reagents, perhaps because of their ease of availability in Europe and North America. A successful global HIV-1 vaccine will have to be effective against non-B subtype viruses. Therefore, it is necessary to develop and characterize reagents that can be used in vaccine development and testing studies for non-B subtypes.
HIV-1 subtype C is the most prevalent subtype in southern Africa and in parts of Asia (20, 21, 25, 33, 36). The reasons for the predominance of subtype C in the HIV-1 pandemic (6, 12, 50) are not entirely clear, but biological differences from other subtypes cannot be ruled out. Most studies describing full-genome clones of subtype C viruses have been restricted to phylogenetic and other sequence analyses (15, 33, 42).
Subtype C viruses differ from other subtypes by having a premature truncation of the rev open reading frame and an enlarged Vpu protein (15). Studies have not yet been undertaken to address whether these genetic differences translate into biological differences. The long terminal repeat elements (LTRs) of different HIV-1 subtypes reveal differences that appear to have biological significance. Subtype C LTRs contain three and sometimes four copies of the NF-κB enhancer element and, in cotransfection studies with an expression vector for Rel p65, showed higher transcriptional activation of a reporter gene than subtype B LTRs (28). A differential response to the proinflammatory cytokine tumor necrosis factor alpha (TNF-α) has also been observed; the level of response appears to correlate with the number of NF-κB sites found in the LTR and is thus highest for subtype C (22, 27). In another transient transfection experiment that involved LTRs from 29 patients from a geographically diverse population, subtype C LTRs were found to more potently transactivate a reporter gene in a cell line than the LTRs of other subtypes (30).
It has been suggested that HIV-1 subtype C is unique in the evolution of its coreceptor utilization, which may affect its transmission and pathogenesis. A syncytium-inducing (SI) phenotype and CXCR4 chemokine receptor utilization were reported to be rare among subtype C isolates, even when the isolates were obtained from late-stage AIDS patients (4, 7, 49). Consistent with these findings, it has been observed that V3 sequence variability in HIV-1 subtype C is reduced and that the V3 sequence is characterized by a lack of basic amino acids, which among subtype B isolates is a feature of viruses that use CCR5 for cell entry (36). These observations may indicate that infection with subtype C may have an outcome different from that of infection with other subtypes in view of studies in animal models that suggest different pathogenic sequelae for infection with CCR5- or CXCR4-utilizing strains (2, 3, 18).
In this report, we describe the construction and the replication kinetics of MJ4, a chimeric infectious molecular clone of HIV-1 subtype C from Botswana. The MJ4 molecular clone will facilitate studies of molecular determinants of biological activity for HIV-1 subtype C by the introduction of mutations and other genomic alterations and will also be an important reagent for immunological characterization of HIV-1 subtype C in the continuing effort toward vaccine development.
MATERIALS AND METHODS
Patient samples, DNA extraction, and plasmid constructs.
Blood samples were obtained from anonymous infected donors from Gaborone (96BW06) and Molepolole (96MOLE1), Botswana. The serostatus of each patient was established by enzyme-linked immunosorbent assay (ELISA) and Western blot analysis. Patients' clinical information, amplification, cloning, and initial characterization of noninfectious molecular clones are reported elsewhere (31). In brief, patients' blood samples were cocultured for 14 days with donor peripheral blood mononuclear cells (PBMCs). High-molecular-weight genomic DNA was extracted from the PBMCs using the Qiagen Genomic-tip 100/G kit. (Chatsworth, Calif.) A full-length HIV-1 clone designated C.96BW06.J4 (J4) was constructed from sample 96BW06. This clone failed to replicate in permissive cultures in vitro, and Western blot analysis suggested a defect in envelope glycoprotein processing (31). Several HIV-1 subtype C envelopes were amplified, cloned, and tested for ability to trans-complement and mediate cell entry of an HIV-1 construct with a defective envelope gene. The envelope function analysis assay is a modification of the method described by Helseth et al. (19). Our procedure includes two plasmids; one encodes subtype C envelope glycoproteins and the other is a full-length HIV-1 proviral clone that has a deletion in the env gene and also has the bacterial chloramphenicol acetyltransferase (CAT) gene in place of the nef gene (pHXBΔenvCAT). The two plasmids were cotransfected into COS-1 cells, and the resulting supernatants were used to infect target cells. The amount of CAT in the target cells reflected the ability of the test envelope clone to mediate cell entry of the HIV-1 provirus. The pSVIII/MOLE1 envelope expressor plasmid (from patient 96MOLE1) consistently gave higher CAT levels in target U87.CD4.CCR5 cells than the dualtropic envelope expressor plasmid pSVIII/89.6 (referred to below as 89.6) (44), which was used as the positive control. In subsequent experiments, pSVIII/MOLE1 was seen to complement in trans the C.96BW06.J4 clone, which resulted in the production of infectious virions (31).
Infectious chimeric HIV-1 subtype C clone.
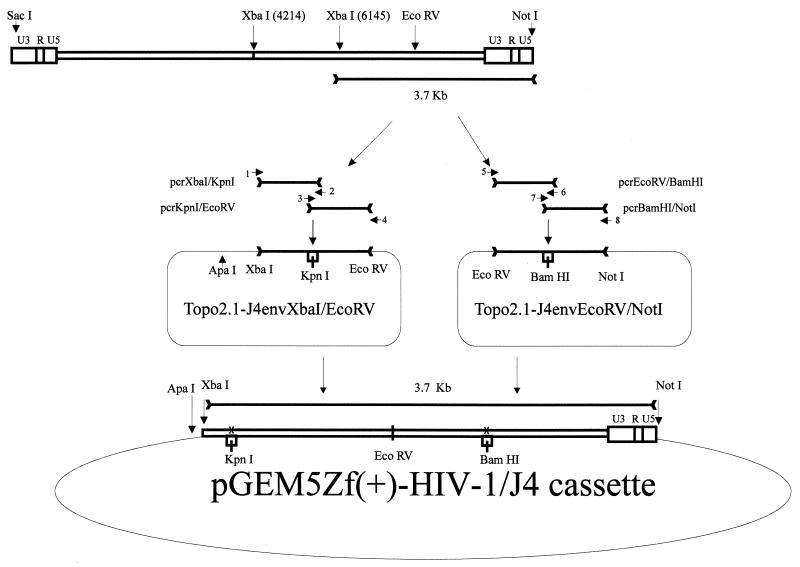
To facilitate the subcloning of the functional envelope from the pSVIII/MOLE1 clone, convenient restriction enzyme cloning sites were introduced into the full-length clone C.96BW06.J4 (Fig. 1). PCR was performed on clone C.96BW06.J4 to amplify the fragment located between the XbaI site at nucleotide 6145 and the KpnI site (engineered) at nucleotide 6345, using primers Xba-MUTJ4 (primer 1 in Fig. 1; 5′-GTATATCTAGAATATAGGAAACTTGTAAGACAAAGAAAG-3′) and J4-Kpn1(R) (primer 2; 5′-CCACACAGGTACCCCATAATAGACTGTG-3′). A second reaction to amplify a fragment located between the KpnI site and an EcoRV site (nucleotide 8057) was performed using primers J4-KpnI (primer 3; 5′-CACAGTCTATTATGGGGTACCTGTGTGG-3′) and EcoRV-J4(R) (primer 4; 5′-CATGTTGTCCCAGATATCTCCTAGAGATTTATTACTCC-3′). A third reaction was then performed to amplify the fragment located between the EcoRV site and the BamHI site (engineered, at nucleotide 8445), using primers EcoRV-J4(F) (primer 5; 5′-GGAGTAATAAATCTCTAGGAGATATCTGGGACAGACATG-3′) and J4-BamHI(R) (primer 6; 5′-CAGGCAAGTGCTAAGGATCCGTTCACTAATCG-3′). A fourth amplification from the BamHI site to the NotI site located at nucleotide 9880 was also run using primers J4-BamHI (primer 7; 5′-CGATTAGTGAACGGATCCTTAGCACTTGCCTG-3′) and J4-NotI(R) (primer 8; 5′-CGGATCCGCGGCGGCCGCGCACCCATCTCTCTCCTTC-3′). Relevant restriction sites are underlined in the primer sequences. The conditions for all four PCRs were as follows: a 2-min denaturation step at 94°C followed by 30 cycles of 94°C for 15 s, 55°C for 30 s, and 72°C for 2 min. A final extension step at 72°C for 7 min was included. Fifty nanograms of plasmid DNA was used as the template, and each primer was added at a concentration of 10 pmol per reaction. Amplification was carried out using the high-fidelity pfu polymerase (Promega Corp., Madison, Wis.). The PCR products were agarose gel purified and resuspended in 20 μl of 10 mM Tris buffer. Two microliters each of the products from reactions 1 and 2 were then combined, and the same was done for the products from reactions 3 and 4. With these PCR products serving as both primers and templates, another PCR was conducted for 20 cycles, which were run using the conditions outlined above. Sense and antisense primers were then added: Xba-MUTJ4 and EcoRV-J4(R) for reactions 1 and 2 and EcoRV-J4(F) and J4-NotI(R) for reactions 3 and 4. An additional 20 cycles were then run. These PCR samples thus yielded two products: an XbaI-to-EcoRV fragment (1.9 kb) with an engineered KpnI site and an EcoRV-to-NotI fragment (1.8 kb) that contained an engineered BamHI site. Both fragments were then cloned into the relevant restriction sites in the pCR2.1-Topo vector after digestion. The XbaI-EcoRV fragment was then digested with ApaI (in the vector) and EcoRV and cloned into the corresponding sites in pGEM-5Zf(+) (Promega Corp.). The EcoRV-NotI fragment was subsequently introduced into the EcoRV-NotI cloning site in pGEM-5Zf(+). To exchange the nonfunctional C.96BW06.J4 envelope for the functional MOLE1 envelope, we digested the pGEM-5Zf(+) subclone with the restriction enzymes KpnI and BamHI. The subclone containing the functional envelope was then transferred back into the HIV-1 C.96BW06.J4 backbone by digestion with NotI and partial digestion with XbaI (C.96BW06.J4 has an additional XbaI site at nucleotide 4214). Nucleotide sequencing of the XbaI-NotI fragment was carried out to ensure that the only changes were the engineered KpnI and BamHI sites.
FIG. 1.
Construction of the HIV-1 subtype C infectious clone. Four PCRs of the parental clone, C.96BW06.J4, were performed: reaction 1, from the XbaI to the KpnI site; reaction 2, from the KpnI to the EcoRV site; reaction 3, from the EcoRV to the BamHI site; and reaction 4, from the BamHI to the NotI site. Reactions 1 and 2, as well as reactions 3 and 4, were then combined for further amplification, resulting in an XbaI-to-EcoRV fragment that contained a new KpnI site and an EcoRV-to-NotI fragment with a BamHI site. Both the XbaI-EcoRV and EcoRV-NotI fragments were separately cloned into the pCR2.1-Topo vector. The XbaI-EcoRV fragment was then cloned into the ApaI-EcoRV sites in the pGEM-5Zf(+) vector, followed by the EcoRV-NotI piece. The unique KpnI and BamHI sites in this subclone then allowed the exchange of corresponding C.96BW06.J4 and pSVIII/MOLE1 envelopes. The XbaI-NotI subclone that contained the functional envelope was then inserted back into the C.96BW06.J4 backbone.
Cells and cell lines.
COS-1 and 293T cells were propagated in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS). U87.CD4 cells were obtained from the National Institutes of Health (NIH) AIDS Research and Reference Reagent Program (Rockville, Md.). U87.CD4 cells expressing chemokine receptors CCR1, CCR2b, CCR3, CXCR4, and CCR5 were maintained in DMEM supplemented with 15% FBS, 1 μg of puromycin/ml, and 300 μg of G418/ml. U87.CD4 cells were cultured in the same medium but without puromycin. All the U87 cells were cultured in 12-well flat-bottom plates in 1,000 μl (total volume) of culture medium.
PBMCs and macrophages were obtained from anonymous HIV-1-negative donors and were separated by density gradient centrifugation on lymphocyte separation medium (Organon Teknika, Corp., Durham, N.C.). PBMCs were cultured in RPMI 1640 supplemented with 10% FBS, 5 μg of phytohemagglutinin (Sigma, St. Louis, Mo.)/ml, and 20 U of interleukin-2 (Becton Dickinson Labware, Bedford, Mass.)/ml for 72 h prior to infection. Macrophages were prepared from PBMCs according to the protocol of Gartner et al. (16). In brief, density-separated PBMCs were propagated in RPMI 1640 culture medium supplemented with antibiotics and 10% fetal calf serum. Nonadherent cells were removed 7 days after seeding by rinsing the cultures three times with growth medium. Adherent cells were then infected and grown in RPMI medium. Both PBMCs and macrophages were seeded at 2 × 106/well in 24- or 48-well plates and were propagated in 1,000 μl of medium.
Transfection and infection.
Subconfluent COS-1 or 293T cells were transfected with 7 μg of plasmid DNA using the Fugene 6 transfection reagent (Boehringer Mannheim, Indianapolis, Ind.). As a positive control, we used the subtype B proviral clone HXB2RU3, which is a derivative of HXB2 that has intact vpu, vpr, and nef genes (37, 52). Another positive control was HXB2RU3CI, which is equivalent to HXB2RU3 except for the V3 loop, which has been exchanged with a CCR5-tropic V3 loop (47, 51). Seventy-two hours posttransfection, culture supernatants were filtered through 0.45-μm-pore-size filter units (Nalgene, Rochester, N.Y.) and HIV-1 virions were quantified by p24 antigen ELISA (NEN Life Science Products, Boston, Mass.). Equivalent amounts of virus (as determined by the amount of p24 antigen in the culture supernatant) were used to infect 2 × 106 PBMCs, macrophages, or U87.CD4 glioma cells with or without chemokine receptors. Twenty-four hours postinfection, cells were washed three times with phosphate-buffered saline (PBS), and fresh medium was then added. Infection of the target cells was monitored for as long as 21 days by p24 ELISA. All infected cultures were sampled and fed with 50% replacement of the total culture volume at days 7 and 14. U87 glioma cells were split 1:3 at days 7 and 14.
Viral pellets.
Culture supernatants from transfected cells were centrifuged at 800 × g for 15 min. Supernatants were then filtered through 0.45-μm-pore-size filter units, overlaid on 4 ml of a 20% sucrose cushion, and centrifuged at 20,000 rpm (Beckman SW28 rotor) for 2 h. The supernatant was then discarded, and any remaining fluid was dried using cotton swabs. The viral pellet was then resuspended in 100 μl of lysis buffer (0.15 M NaCl, 0.05 M Tris-HCl [pH 7.2], 1% Triton X-100, 1% sodium deoxycholate, and 0.1% sodium dodecyl sulfate). The total amount of protein recovered was quantified with the Bio-Rad (Hercules, Calif.) protein assay kit. Then 12 μg of total protein for each sample was mixed with reducing buffer (0.08 M Tris-HCl [pH 6.8], 0.1 M dithiothreitol, 2% sodium dodecyl sulfate, 10% glycerol, and 0.2% bromophenol blue), boiled for 3 min, and resolved on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels with a linear gradient of 4 to 15% polyacrylamide.
Immunoblotting.
Resolved proteins were transferred passively by placing the gel between two nitrocellulose membranes (Schleicher & Schuell, Keene, N.H.) and placing a weight on top of the cassette for 48 h. Viral proteins were visualized by immunoblotting with pooled sera from HIV-1-seropositive individuals infected with subtype C from Botswana and subtype B from the United States.
Sequencing and sequence analysis.
To sequence the genome, a primer-walking strategy was used on purified plasmid DNA, and overlapping contiguous sequences were obtained throughout the genome to ensure accuracy of sequence output. The primers were approximately 300 nucleotides apart on each strand of the genome. More than 100 different sequencing primers were used, and their sequences are available upon request. Automatic sequencing was performed using a model 373A automated DNA Sequenator (Applied Biosystems, Inc., Foster City, Calif.). Individual contiguous sequences of proviral DNA were assembled using the Sequencher program (Gene Codes Corp., Ann Arbor, Mich.). Multiple sequence alignment was carried out using the Clustal W program, version 1.7. Phylogenetic analysis was performed by the neighbor-joining method, correcting for multiple substitutions, and the reliability of the branching pattern was estimated by 100 bootstrap resampling. The Njplot (35) program was used to view sequence relatedness.
Nucleotide sequence accession number.
The GenBank accession number for the nucleotide sequence of the MJ4 clone is AF321523.
RESULTS
Our previous characterization of eight subtype C full-length clones from Botswana showed that none of them were replication competent in vitro despite their lacking any obvious inactivating mutations such as translational stop codons, frameshifts, deletions, or defective packaging or processing signals. Two of the eight clones could be trans-complemented by a functional envelope clone to yield infectious virions (31). In this study, we used standard cloning techniques to construct a chimeric HIV-1 subtype C infectious molecular clone (Fig. 1). Several steps were necessary to construct this clone, but subsequent resequencing of the XbaI-NotI fragment revealed that all reading frames in the genetically manipulated segment were preserved. The new chimeric clone contains a 2.1-kb KpnI-BamHI envelope fragment from the sample MOLE1. The first 119 and the last 341 nucleotides of the env gene are derived from the original noninfectious clone C.96BW06.J4. The new recombinant clone, termed MJ4, has the entire second exon of tat and a portion of the rev gene as well as most of the env coding region—all derived from the MOLE1 sample.
Viral protein expression profile.
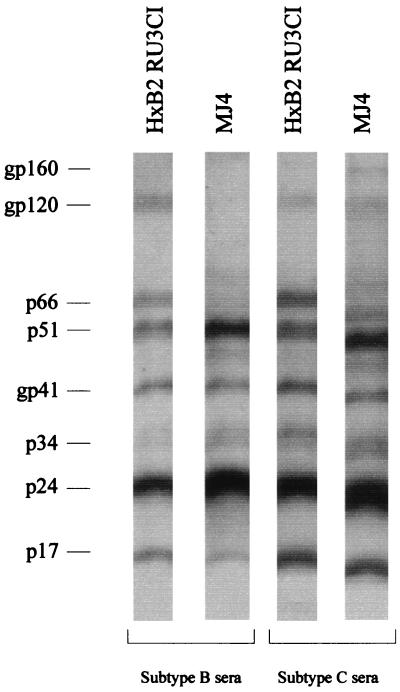
HIV-1 protein expression from transfected cells was assessed by Western blotting using pooled sera from HIV-1-infected individuals from Botswana (subtype C) and from individuals infected in the United States (subtype B). The results from the viral lysate are shown in Fig. 2. When compared with molecular clone HXB2RU3CI, which was used as the positive control, clone MJ4 expressed all the proteins required for HIV-1 infectivity, namely, the gag gene-encoded proteins p24 and p17, the pol gene-encoded proteins p66, p51, and p34, and the env gene-encoded proteins gp120 and gp41.
FIG. 2.
Analysis of viral proteins by Western blotting. 293T cells were transfected with plasmid DNAs of the HXB2RU3CI and MJ4 molecular clones. Cell culture supernatants were collected at 72 h posttransfection and overlaid on a 20% sucrose cushion. After centrifugation, viral pellets were resuspended in lysis buffer and viral proteins were separated by 4 to 15% linear SDS-PAGE. Proteins were transferred to nitrocellulose membranes and analyzed by immunoblotting with pooled sera from individuals from Botswana known to be infected with HIV-1 subtype C; a separate blot, retained from the same SDS-PAGE gel, was analyzed using sera of individuals from the United States who are infected with subtype B.
Analysis of replication competence of clone MJ4 in PBMCs.
To generate viral progeny to test for the ability to replicate in target cells in vitro, 7 μg of plasmid DNA was transfected into either COS-1 or 293T cells. Production of viral progeny was determined 72 h posttransfection. In both cell types, viral yield was always over 50 ng/ml, as measured by the concentration of the p24 core protein in the culture supernatant. The subtype B clone, HXB2RU3CI, was used as the positive control. This clone is identical to the T-cell-tropic, CXCR4-utilizing clone HXB2RU3 except for the V3 loop, which has been swapped with a CCR5-tropic loop. This clone was used as the positive control because most subtype C isolates have been shown to use CCR5 as a coreceptor for cell entry, and its parental clone (HXB2RU3) is a well-characterized molecular clone. An amount of virus corresponding to 500 pg of p24 was used to infect PBMCs from six different donors. Infection was assessed by measurement of the p24 antigen concentration in tissue culture supernatant at days 1, 7, 14, and 21 postinfection. All donor PBMCs were readily infected, as indicated by the p24 levels at the highest peak compared to the p24 levels at day 1 (Table 1). In five of six donor PBMCs tested, the subtype B recombinant molecular clone HXB2RU3CI replicated to a peak p24 antigen level that was higher than that of clone MJ4. Despite the fact that HXB2RU3CI and MJ4 reached peak p24 levels at different time points, the peak p24 level for HXB2RU3CI was 1.4 to 3.3 times higher than that of MJ4 in those five donors. One donor PBMC culture (donor 4) was relatively resistant to infection, with both the subtype B and C clones reaching nearly equal (2 ng/ml) peak p24 concentrations.
TABLE 1.
Analysis of MJ4 clone replication kinetics in PBMCs from six donorsa
| Donor | Viral clone | Concn of p24 antigen in culture supernatant (pg/ml)
|
|||
|---|---|---|---|---|---|
| Day 1 | Day 7 | Day 14 | Day 21 | ||
| 1 | MJ4 | 126 | 6,003 | 37,968 | 18,227 |
| HXB2RU3CI | 411 | 18,452 | 80,000 | 63,577 | |
| C.96BW06.J4 | 159 | 460 | 171 | 172 | |
| 2 | MJ4 | 16 | 30 | 6,487 | 12,398 |
| HXB2RU3CI | 10 | 81 | 294 | 16,998 | |
| C.96BW06.J4 | 14 | 9 | 8 | 57 | |
| 3 | MJ4 | 10 | 98 | 25,855 | 36,394 |
| HXB2RU3CI | 8 | 341 | 7,266 | 49,199 | |
| C.96BW06.J4 | 9 | 9 | 7 | 52 | |
| 4 | MJ4 | 204 | 172 | 1,612 | 2,087 |
| HXB2RU3CI | 331 | 889 | 2,004 | 1,824 | |
| C.96BW06.J4 | 181 | 162 | 100 | 6 | |
| 5 | MJ4 | 140 | 185 | 12,794 | 14,817 |
| HXB2RU3CI | 146 | 3,019 | 48,336 | 34,386 | |
| C.96BW06.J4 | 130 | 88 | 146 | 150 | |
| 6 | MJ4 | 191 | 696 | 20,472 | 15,263 |
| HXB2RU3CI | 52 | 7,260 | 49,600 | 32,194 | |
| C.96BW06.J4 | 208 | 87 | 267 | 325 | |
The parental clone C.96BW06.J4 and the subtype B, CCR5-utilizing clone HXB2RU3CI were used as controls. 293T or COS-1 cells were cotransfected with 7 μg of plasmid DNA. Seventy-two hours later, p24 antigen was quantified by ELISA. Culture supernatant corresponding to 500 pg of p24 was then used to infect 2 × 106 cells. The following day, the target cells were washed three times with PBS, and then new medium was added. Infection was monitored by p24 ELISA.
Culture supernatant from one of the primary PBMC cultures was also used to secondarily infect other PBMC cultures after filtration through 0.45-μm-pore-size filters and standardization of the virus titer by p24 antigen concentration. These PBMCs were infected, as determined by the rise in p24 antigen levels in culture supernatant over a 21-day period, and PCR amplification confirmed the presence of the MJ4 envelope fragment from the genomic DNA of these secondary infection cells (data not shown).
Replication in macrophages.
It has been reported previously that HIV-1 subtype C primary isolates from Ethiopia, unlike subtype B viruses, failed to replicate in primary monocyte-derived macrophage cultures until these cultures were cocultivated with Jurkattat cells (4). We investigated the abilities of virions from cells transfected with MJ4 to infect macrophages from five different HIV-1-negative donors. MJ4 was able to infect all the macrophage cultures (Table 2), but its growth pattern in these cells, compared with that of the subtype B control, was somewhat different from that seen in PBMCs. MJ4 and HXB2RU3CI readily infected macrophages, but the latter reached peak p24 antigen titers that were higher than those of MJ4 by a factor of 1.6 to 18.6.
TABLE 2.
Replication analysis of the molecular clones MJ4, HXB2RU3CI, and C.96BW06.J4 in donor macrophages from five individualsa
| Donor | Viral clone | Concn of p24 antigen in culture supernatant (pg/ml)
|
|||
|---|---|---|---|---|---|
| Day 1 | Day 7 | Day 14 | Day 21 | ||
| A | MJ4 | 17 | 1,828 | 3,284 | 10,197 |
| HXB2RU3CI | 9 | 5,562 | 6,621 | 42,867 | |
| C.96BW06.J4 | 5 | 18 | 15 | 16 | |
| B | MJ4 | 6 | 2,973 | 2,384 | 984 |
| HXB2RU3CI | 5 | 47,123 | 55,329 | 12,111 | |
| C.96BW06.J4 | 5 | 47 | 61 | 56 | |
| C | MJ4 | 5 | 3,588 | 9,074 | 6,325 |
| HXB2RU3CI | 5 | 20,991 | 28,409 | 15,244 | |
| C.96BW06.J4 | 5 | 24 | 18 | 87 | |
| D | MJ4 | 8 | 2,145 | 3,924 | 1,683 |
| HXB2RU3CI | 7 | 3,426 | 4,387 | 6,296 | |
| C.96BW06.J4 | 10 | 77 | 172 | 378 | |
| E | MJ4 | 25 | 836 | 5,451 | 2,794 |
| HXB2RU3CI | 5 | 1,350 | 10,254 | 4,630 | |
| C.96BW06.J4 | 6 | 36 | 18 | 17 | |
Replication was followed by quantification of p24 core protein concentration in tissue culture.
Statistical analysis of kinetics of replication in PBMCs and macrophages.
We investigated whether, based on the observed data, there was a significant difference in the p24 antigen peak between MJ4 and HXB2RU3CI. Because the peak p24 antigen values for the viruses were not normally distributed and because there was no mathematical transformation that would normalize the data, we used a nonparametric test, the Wilcoxon matched-pair signed-rank test. The data are considered to be matched pairs because in each case the two viruses were grown in the same environment. We found that the peak reached by HXB2RU3CI was significantly higher than that reached by MJ4 (P = 0.0044). The difference was significant even when PBMCs and macrophages were considered separately (P = 0.0464 and P = 0.0431, respectively). We also tested whether one virus reached peak titers sooner than the other. Since the day the peak was reached was also not normally distributed, we again used the Wilcoxon matched-pair signed-rank test. The difference was not statistically significant.
Characterization of coreceptor utilization by the MJ4 clone.
Several recent reports have suggested that HIV-1 subtype C viruses may be unique in that variants that use coreceptors other than CCR5 may be very rare. CCR5 and CXCR4 are the main chemokine receptors known to be utilized by most primary isolates of HIV-1 as coreceptors for cell entry, but some variants may use others. We therefore decided to characterize clone MJ4 with regard to the use of common chemokine receptors. The results of replication as measured by p24 antigen in culture supernatants of U87.CD4 glioma cells expressing CCR5 and CXCR4 are shown in Table 3. The clone was able to use only the chemokine receptor CCR5 as a coreceptor for cell entry. Replication in CCR5-expressing U87 glioma cell lines was somewhat different from that seen in PBMCs and macrophages in that both HXB2RU3CI and MJ4 replicated to higher titers in the U87.CD4.CCR5 cells than in PBMCs or macrophages, reaching peak p24 antigen titers of more than 100 ng/ml. Neither MJ4 nor HXB2RU3CI virus replicated in U87.CD4.CXCR4 cells, but HXB2RU3, the CXCR4-utilizing virus used as a positive control, replicated to high titers as expected. MJ4 also failed to replicate in cells expressing chemokine receptor CCR1, CCR2(b), or CCR3, and in U87.CD4 cells that did not express any chemokine receptor (data not shown).
TABLE 3.
Replication in U87.CD4 cells expressing CCR5 or CXCR4a
| Coreceptor | Viral clone | Concn of p24 antigen in culture supernatant (pg/ml)
|
|||
|---|---|---|---|---|---|
| Day 1 | Day 7 | Day 14 | Day 21 | ||
| CCR5 | MJ4 | 45 | 1,524 | 15,925 | 137,196 |
| HXB2RU3CI | 52 | 350 | 70,000 | 150,000 | |
| C.96BW06.J4 | 112 | 75 | 54 | 164 | |
| HXB2RU3 | 7 | 11 | 11 | 7 | |
| CXCR4 | MJ4 | 3 | 14 | 17 | 10 |
| HXB2RU3CI | 4 | 48 | 37 | 155 | |
| C.96BW06.J4 | 3 | 11 | 13 | 8 | |
| HXB2RU3 | 4 | 183,540 | 899,680 | 844,240 | |
Five hundred picograms of p24 viral supernatants from transfected 293T cells were used to infect U87.CD4 cells. Cells were washed three times with PBS after 24 h, and fresh growth medium was added. Infection was monitored by quantification of the p24 core protein by ELISA. The cells were split 1:3, and fresh medium was added at days 7 and 14.
Sequence analysis.
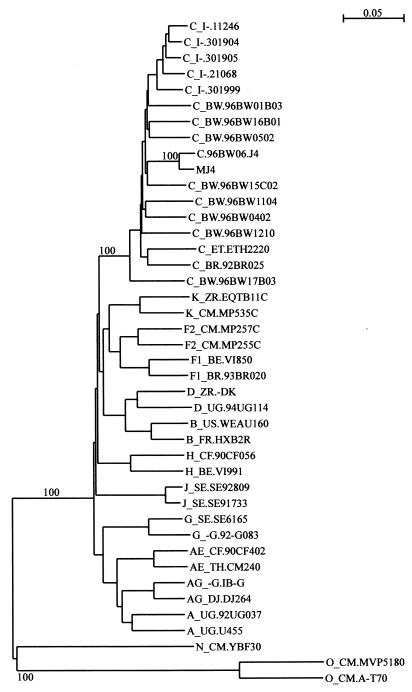
Previous studies have identified mainly subtype C but also a recombinant genome from Botswana (33, 34). The southern African region is, however, dominated by HIV-1 subtype C, and this genotype is at the center of various vaccine and pathogenesis study efforts. We therefore analyzed the genome of the new infectious clone to ensure that it belonged to subtype C. The phylogenetic tree resulting from this analysis is shown in Fig. 3. The MJ4 sequence is tightly clustered with subtype C sequences, which confirms that the clone is of this subtype. Bootscanning analysis, which utilizes reference sequences from each of the group M genetic subtypes to assign a subtype to a query sequence throughout its genome, confirmed the clone to be of HIV-1 subtype C in every locus (data not shown).
FIG. 3.
Phylogenetic tree analysis of clones MJ4 and C.96BW06.J4. The tree was generated by the neighbor-joining method. Corrections were made for multiple substitutions, and reliability of the branching pattern was estimated by 100 bootstrap resampling. The reference sequences for different HIV-1 groups and subtypes were obtained from the HIV database at http://hiv-web.lanl.gov. As expected, MJ4 and its parental clone, C.96BW06.J4, were tightly clustered, both together and with subtype C sequences.
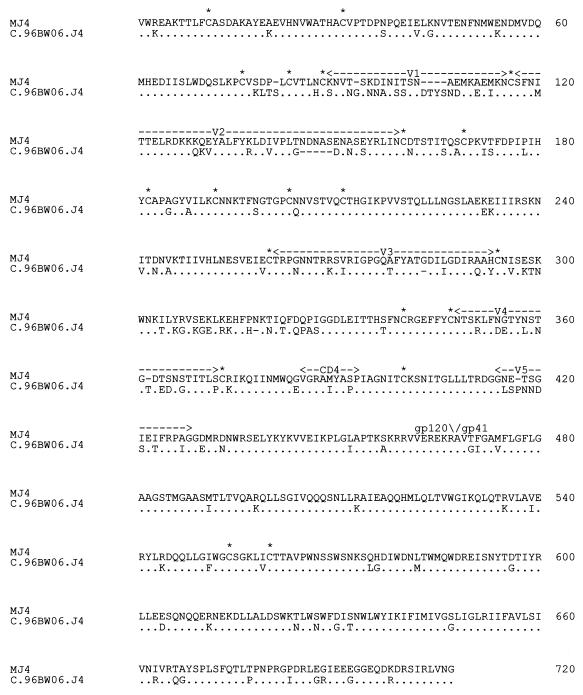
The infectious clone MJ4 differs from its noninfectious parental clone C.96BW06.J4 only in the 2.1-kb KpnI-BamHI fragment in the envelope. There are stretches of 700 and 699 amino acids for MJ4 and C.96BW06.J4, respectively, in this fragment. Alignment of the two clones in this region revealed 124 amino acid differences between the clones (Fig. 4). MJ4 had a 5-amino-acid deletion in the V1 region, and C.96BW06.J4 had a deletion of similar size in the V2 region. The envelope glycoprotein V3 loop has great biological significance in HIV-1, including determination of coreceptor tropism. There were eight amino acid differences in the V3 loop between the two clones, but none of these involved Arg-298, Pro-299, Thr-303, or Ala-328, which have been identified by mutagenesis as important for CCR5 utilization (51). Clone C.96BW06.J4 had a deletion of Gly-220 in the V3 loop. A number of other amino acid residues throughout the envelope glycoprotein have been identified by mutagenesis of the subtype B macrophage-tropic clone YU-2 as important for CCR5 binding (40). All of the residues whose mutagenesis resulted in more than a 90% reduction in CCR5 binding are conserved in both clones MJ4 and C.96BW06.J4, except for a threonine-123 mutation to aspartic acid in MJ4. Interestingly, the same mutation in YU-2 resulted in a 94% reduction in CCR5 binding. Various residues identified by mutagenesis as important for CD4 binding (24) are conserved in both clones, except for the MYAPP motif, which is located immediately downstream of the V4 loop. The fourth phenylalanine has been replaced by serine in MJ4, while in C.96BW06.J4 the first methionine has been mutated to isoleucine.
FIG. 4.
Amino acid alignment of the KpnI-BamHI fragments of the infectious clone MJ4 and the noninfectious clone C.96BW06.J4. The only difference between the two clones is in the aligned region. Dots under the MJ4 sequences indicate C.96BW06.J4 residues that are conserved. Cysteine residues are indicated by asterisks. Hypervariable regions V1 to V5, as well as the CD4 binding and gp120-gp41 binding regions, are indicated above the sequences.
We previously reported that the C.96BW06.J4 clone may be defective in envelope glycoprotein processing and/or packaging because of a high gp160/gp120 ratio in Western blot analysis of its viral lysate (31). However, it was not possible to tell what amino acid residues may be responsible for this defect, as the proteolytic cleavage site REKR at the junction of gp120 and gp41 was conserved. Mutation of cysteine residues within the envelope glycoprotein sequence is also known to affect envelope processing and viral infectivity (9, 45, 48). There are 20 cysteine residues in the KpnI-BamHI fragment of MJ4, and all are conserved in C.96BW06.J4.
DISCUSSION
In this paper we have described the construction and biological analysis of the first infectious proviral DNA molecular clone of HIV-1 subtype C from the African continent. This clone is a chimeric molecule, because the majority of env was derived from sample 96MOLE1 while the rest of the genome was from sample 96BW06. This clone is derived from primary viral isolates, because the viruses were not propagated in immortalized cell lines prior to DNA amplification. MJ4 utilizes CCR5 as the coreceptor for entry into susceptible cells, consistent with observations from other studies that field isolates of HIV-1 subtype C viruses mostly use this coreceptor for cell entry. In addition, most viruses that are transmitted from individual to individual are known to be macrophage-tropic and thus CCR5 using (10, 11, 53, 54). This clone may be important in elucidating genetic determinants that underlie HIV-1 subtype C disease pathogenesis, such as its apparent failure to evolve to use other chemokine receptors to mediate cell entry during disease progression. Most studies of HIV-1 coreceptor utilization have relied on subtype B molecular clones, while a few have been conducted with primary isolates from other subtypes. Given the high prevalence of HIV-1 subtype C in the global epidemic and its predominance in the most heavily affected region of southern Africa, there is an obvious need to address whether viral genetic factors and/or a complex interaction between viral and host factors may be playing a yet unrecognized role in the spread of the virus.
It is worthy of note that the clone MJ4 replicated to significantly lower peak titers than the subtype B positive control in both PBMCs and macrophages. The reasons for this are not clear and may be just clone specific, especially because the positive control used in our experiments is a derivative of the laboratory-adapted molecular clone HXB2RU3, which is known to replicate to high titers. Further studies are needed to determine what factors may contribute to the differences observed in this study. Possible clone differences in entry mechanisms, transcription activity, packing capacity, induction of cytopathic effects, and other characteristics may individually or collectively play a role. However, it has been observed previously that HIV-1 subtype C primary isolates failed to replicate for as long as 28 days in macrophages until the macrophages were cocultured with Jurkattat cells (4). The apparent failure of HIV-1 subtype C viruses to replicate to high titers may not be subtype specific but may simply imply that these viruses have the typical slow/low, non-syncytium-inducing (NSI) phenotype, like that found in some primary subtype B isolates (1, 14, 46). The statistically significant differences observed in this study suggest that viral genetic factors, independent of cell donor differences, are responsible for the replication kinetics observed. We did not quantify the ability of MJ4 to induce cytopathic effects in PBMCs and macrophages, which may impact the ability of the virus to replicate to high titers in vitro. Recently, Chen et al. (8) reported the construction of chimeric simian/human immunodeficiency viruses (SHIV) that bear HIV-1 subtype C envelope glycoproteins. Although these envelope glycoproteins were functional, some of the constructs failed to replicate in vitro but were able to infect experimental primates. The authors suggested the possibility that the target cell in vivo for subtype C envelope-bearing SHIV may be missing in PBMC culture. Taken together, these studies warrant further investigation into HIV-1 subtype C biology.
The presence of extra NF-κB enhancer element sites within subtype C LTRs has led to the speculation that these viruses may have a replicative advantage in target cells (4, 22, 27, 28, 30). This argument is bolstered by transfection experiments involving reporter gene assays that show higher transcriptional activation of HIV-1 subtype C LTRs compared to that for other subtypes. However, few studies have compared the replication or cytopathic effects of viral isolates or clones of different subtypes in diverse primary target cells. It is noteworthy that these studies have not exhaustively explored the contribution of biologically relevant cofactors, such as proinflammatory cytokines, to the replication of HIV-1 subtype C in laboratory experiments. In this study, we compared the replication kinetics of two different clones belonging to subtypes B and C. We have shown that HXB2RU3CI (subtype B) replicates to higher titers than MJ4 (subtype C) in macrophages and PBMCs. Future studies will include the investigation of how factors such as cytokines may influence HIV-1 replication in a subtype-dependent manner, if at all.
The epidemic spread and pathogenesis of HIV-1 subtype C may differ from those of other subtypes. Studies in Tanzania show that HIV-1 subtype C has a higher odds ratio of perinatal transmission than the other two subtypes (38). In circulating intersubtype recombinants, certain genetic loci of subtype C appear to have been selected in association with transmission (5, 39). In Kenya, a cross-sectional study showed that plasma HIV RNA levels were highest, and CD4 lymphocyte counts were lowest, among women infected with HIV-1 subtype C compared to those infected with subtypes A and D (32). In a west African cohort of female commercial sex workers, individuals infected with subtype C were reported to be more likely to develop AIDS than those infected with subtype A (23).
This is the second infectious molecular clone of HIV-1 subtype C reported, the first having been cloned from an Indian isolate after propagation in immortalized cell lines (26). HIV-1 vaccine development, targeted mainly at rapidly spreading strains, is a priority. This clone and others like it are more representative of these genotypes and will be important tools for identifying correlates of protection, epitopes that may confer resistance to infection or disease, as well as for facilitating the testing of potential vaccines.
ACKNOWLEDGMENTS
T. Ndung'u and B. Renjifo contributed equally to this work.
We thank Tun-Hou Lee and Jean-Louis Sankale for comments on the manuscript and Chanc E VanWinkle for editorial assistance. Mary Fran McLane and Victor Pena-Cruz provided expertise on tissue culture. We thank Geoffrey Eisen and Peter Gilbert for help with statistical analysis.
This study was supported in part by NIH grants R35 CA39805 and R01 HD37783 and by grant D43 TW00004 from the Fogarty International Center.
REFERENCES:
- 1.Asjo B, Morfeldt-Manson L, Albert J, Biberfeld G, Karlsson A, Lidman K, Fenyo E M. Replicative capacity of human immunodeficiency virus from patients with varying severity of HIV infection. Lancet. 1986;ii:660–662. [PubMed] [Google Scholar]
- 2.Berkowitz R D, Alexander S, Bare C, Linquist-Stepps V, Bogan M, Moreno M E, Gibson L, Wieder E D, Kosek J, Stoddart C A, McCune J M. CCR5- and CXCR4-utilizing strains of human immunodeficiency virus type 1 exhibit differential tropism and pathogenesis in vivo. J Virol. 1998;72:10108–10117. doi: 10.1128/jvi.72.12.10108-10117.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berkowitz R D, Van't Wout A B, Kootstra N A, Moreno M E, Linquist-Stepps V D, Bare C, Stoddart C A, Schuitemaker H, McCune J M. R5 strains of human immunodeficiency virus type 1 from rapid progressors lacking X4 strains do not possess X4-type pathogenicity in human thymus. J Virol. 1999;73:7817–7822. doi: 10.1128/jvi.73.9.7817-7822.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bjorndal A, Sonnerborg A, Tscherning C, Albert J, Fenyo E. Phenotypic characteristics of human immunodeficiency virus type 1 subtype C isolates of Ethiopian AIDS patients. AIDS Res Hum Retrovir. 1999;15:647–653. doi: 10.1089/088922299310944. [DOI] [PubMed] [Google Scholar]
- 5.Blackard J T, Renjifo B R, Mwakagile D, Montano M A, Fawzi W W, Essex M. Transmission of human immunodeficiency type 1 viruses with intersubtype recombinant long terminal repeat sequences. Virology. 1999;254:220–225. doi: 10.1006/viro.1998.9504. [DOI] [PubMed] [Google Scholar]
- 6.Brookmeyer R, Mehendale S M, Pelz R K, Shepherd M E, Quinn T, Rodrigues J J, Bollinger R C. Estimating the rate of occurrence of new HIV infections using serial prevalence surveys: the epidemic in India. AIDS. 1996;10:924–925. [PubMed] [Google Scholar]
- 7.Cecilia D, Kulkarni S S, Tripathy S P, Gangakhedkar R R, Paranjape R S, Gadkari D A. Absence of coreceptor switch with disease progression in human immunodeficiency virus infections in India. Virology. 2000;271:253–258. doi: 10.1006/viro.2000.0297. [DOI] [PubMed] [Google Scholar]
- 8.Chen Z, Huang Y, Zhao X, Skulsky E, Lin D, Ip J, Gettie A, Ho D D. Enhanced infectivity of an R5-tropic simian/human immunodeficiency virus carrying human immunodeficiency virus type 1 subtype C envelope after serial passages in pig-tailed macaques (Macaca nemestrina) J Virol. 2000;74:6501–6510. doi: 10.1128/jvi.74.14.6501-6510.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dedera D, Gu R, Ratner L. Conserved cysteine residues in the human immunodeficiency virus type 1 transmembrane envelope protein are essential for precursor envelope cleavage. J Virol. 1992;66:1207–1209. doi: 10.1128/jvi.66.2.1207-1209.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 11.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 12.Essex M. Human immunodeficiency viruses of the developing world. Adv Virus Res. 1999;53:71–88. doi: 10.1016/s0065-3527(08)60343-7. [DOI] [PubMed] [Google Scholar]
- 13.European Commission (DG XII, INCO-DC) and the Joint United Nations Programme on HIV/AIDS. HIV-1 subtypes: implications for epidemiology, pathogenicity, vaccines, and diagnostics. Workshop report. AIDS. 1997;11:17–36. [PubMed] [Google Scholar]
- 14.Fenyo E M, Morfeldt-Manson L, Chiodi F, Lind B, von Gegerfelt A, Albert J, Olausson E, Asjo B. Distinct replicative and cytopathic characteristics of human immunodeficiency virus isolates. J Virol. 1988;62:4414–4419. doi: 10.1128/jvi.62.11.4414-4419.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao F, Robertson D, Carruthers C, Morrison S, Jian B, Chen Y, Barre-Sinoussi F, Girard M, Srinivasan A, Abimiku A, Shaw G, Sharp P, Hahn B. A comprehensive panel of near-full-length clones and reference sequences for non-subtype B isolates of human immunodeficiency virus type 1. J Virol. 1998;72:5680–5698. doi: 10.1128/jvi.72.7.5680-5698.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gartner S, Markovits P, Markovitz D M, Kaplan M H, Gallo R C, Popovic M. The role of mononuclear phagocytes in HTLV-III/LAV infection. Science. 1986;233:215–219. doi: 10.1126/science.3014648. [DOI] [PubMed] [Google Scholar]
- 17.Gurtler L, Hauser P, Erberle J, von Brunn A, Knapp S, Zekeng L, Tsague J, Kaptue L. A new subtype of human immunodeficiency virus type 1 (MVP5180) from Cameroon. J Virol. 1994;68:1581–1585. doi: 10.1128/jvi.68.3.1581-1585.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harouse J M, Gettie A, Tan R C H, Blanchard J, Cheng-Mayer C. Distinct pathogenic sequela in rhesus macaques infected with CCR5 or CXCR4 utilizing SHIVs. Science. 1999;284:816–819. doi: 10.1126/science.284.5415.816. [DOI] [PubMed] [Google Scholar]
- 19.Helseth E, Kowalski M, Gabuzda D, Olshevsky U, Haseltine W, Sodroski J. Rapid complementation assays measuring replicative potential of human immunodeficiency virus type 1 envelope glycoprotein mutants. J Virol. 1990;64:2416–2420. doi: 10.1128/jvi.64.5.2416-2420.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu D, Dondero T, Rayfield M, George J, Schochetman G, Jaffe H, Luo C, Kalish M, Weniger B, Pau C, Schable C, Curran J. The emerging genetic diversity of HIV. The importance of global surveillance for diagnostics, research, and prevention. JAMA. 1996;275:210–216. [PubMed] [Google Scholar]
- 21.Janssens W, Buve A, Nkengasong J N. The puzzle of HIV-1 subtypes in Africa. AIDS. 1997;11:705–712. doi: 10.1097/00002030-199706000-00002. [DOI] [PubMed] [Google Scholar]
- 22.Jeeninga R, Hoogenkamp M, Armand-Ugon M, De Baar M, Verhoef K, Berkhout B. Functional differences between the long terminal repeat transcriptional promoters of human immunodeficiency virus type 1 subtypes A through G. J Virol. 2000;74:3740–3751. doi: 10.1128/jvi.74.8.3740-3751.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanki P, Hamel D, Sankale J, Hsieh C, Thior I, Barin F, Woodcock S, Gueye-Ndiaye A, Zhang E, Montano M, Siby T, Marlink R, NDoye I, Essex M, MBoup S. Human immunodeficiency virus type 1 subtypes differ in disease progression. J Infect Dis. 1999;179:68–73. doi: 10.1086/314557. [DOI] [PubMed] [Google Scholar]
- 24.Kuiken C, Foley B, Hahn B, Marx P, McCutchan F, Mellors J, Mullins J, Wolinsky S, Korber B E. Human retroviruses and AIDS: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.Mex: Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; 1999. [Google Scholar]
- 25.Lole K, Bollinger R, Paranjape R, Gadkari D, Kulkarni S, Novak N, Ingersoll R, Sheppard H, Ray S. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mochizuki N, Otsuka N, Matsuo K, Shiino T, Kojima A, Kurata T, Sakai K, Yamamoto N, Isomura S, Dhole T, Takebe Y, Matsuda M, Tatsumi M. An infectious DNA clone of HIV type 1 subtype C. AIDS Res Hum Retrovir. 1999;15:1321–1324. doi: 10.1089/088922299310223. [DOI] [PubMed] [Google Scholar]
- 27.Montano M, Nixon C, Ndung'u T, Bussmann H, Novitsky V, Dickman D, Essex M. Elevated tumor necrosis factor-α activation of human immunodeficiency virus type 1 subtype C in southern Africa is associated with an NF-κB enhancer gain-of-function. J Infect Dis. 2000;181:76–81. doi: 10.1086/315185. [DOI] [PubMed] [Google Scholar]
- 28.Montano M, Novitsky V, Blackard J, Cho N, Katzenstein D, Essex M. Divergent transcriptional regulation among expanding human immunodeficiency virus type 1 subtypes. J Virol. 1997;71:8657–8665. doi: 10.1128/jvi.71.11.8657-8665.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Myers G, Korber B, Berzofsky J A, Smith R F, Pavlakis G N. Human retroviruses and AIDS. Los Alamos, N.Mex: Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; 1997. [Google Scholar]
- 30.Naghavi M, Schwartz S, Sonnerborg A, Vahlne A. Long terminal repeat promoter/enhancer activity of different subtypes of HIV type 1. AIDS Res Hum Retrovir. 1999;15:1293–1303. doi: 10.1089/088922299310197. [DOI] [PubMed] [Google Scholar]
- 31.Ndung'u T, Renjifo B, Novitsky V A, McLane M F, Gaolekwe S, Essex M. Molecular cloning and biological characterization of full-length HIV-1 subtype C from Botswana. Virology. 2000;278:390–399. doi: 10.1006/viro.2000.0583. [DOI] [PubMed] [Google Scholar]
- 32.Neilson J, John G, Carr J, Lewis P, Kreiss J, Jackson S, Nduati R, Mbori-Ngacha D, Panteleeff D, Bodrug S, Giachetti C, Bott M, Richardson B, Bwayo J, Ndinya-Achola J, Overbaugh J. Subtypes of human immunodeficiency virus type 1 and disease stage among women in Nairobi, Kenya. J Virol. 1999;73:4393–4403. doi: 10.1128/jvi.73.5.4393-4403.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Novitsky V, Montano M, McLane M, Renjifo B, Vannberg F, Foley B, Ndung'u T, Rahman M, Makhema M, Marlink R, Essex M. Molecular cloning and phylogenetic analysis of human immunodeficiency virus type 1 subtype C: a set of 23 full-length clones from Botswana. J Virol. 1999;73:4427–4432. doi: 10.1128/jvi.73.5.4427-4432.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Novitsky V A, Gaolekwe S, McLane M F, Ndung'u T P, Foley B T, Vannberg F, Marlink R, Essex M. HIV type 1 A/J recombinant with a pronounced pol gene mosaicism. AIDS Res Hum Retrovir. 2000;16:1015–1020. doi: 10.1089/08892220050058434. [DOI] [PubMed] [Google Scholar]
- 35.Perriere G, Gouy M. WWW-query: an on-line retrieval system for biological sequence banks. Biochimie. 1996;78:364–369. doi: 10.1016/0300-9084(96)84768-7. [DOI] [PubMed] [Google Scholar]
- 36.Ping L, Nelson J, Hoffman I, Schock J, Lamers S, Goodman M, Vernazza P, Kazembe P, Maida M, Zimba D, Goodenow M, Eron J, Jr, Fiscus S, Cohen M, Swanstrom R. Characterization of V3 sequence heterogeneity in subtype C human immunodeficiency virus type 1 isolates from Malawi: underrepresentation of X4 variants. J Virol. 1999;73:6271–6281. doi: 10.1128/jvi.73.8.6271-6281.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ratner L, Fisher A, Jagodzinski L L, Mitsuya H, Liou R S, Gallo R C, Wong-Staal F. Complete nucleotide sequences of functional clones of the AIDS virus. AIDS Res Hum Retrovir. 1987;3:57–69. doi: 10.1089/aid.1987.3.57. [DOI] [PubMed] [Google Scholar]
- 38.Renjifo B, Fawzi W, Mwakagile D, Hunter D, Msamanga G, Spiegelman M, Garland C, Kagoma C, Kim A, Chaplin B, Hertzmark E, Essex M. Differences in perinatal transmission between HIV-1 genotypes. J Hum Virol. 2001;4:1–11. [PubMed] [Google Scholar]
- 39.Renjifo B, Gilbert P, Chaplin B, Vannberg F, Mwakagile D, Msamanga G, Hunter D, Fawzi W, Essex M. Emerging recombinant human immunodeficiency viruses: uneven representation of the envelope V3 region. AIDS. 1999;13:1613–1621. doi: 10.1097/00002030-199909100-00003. [DOI] [PubMed] [Google Scholar]
- 40.Rizzuto C D, Wyatt R, Hernandez-Ramos N, Sun Y, Kwong P D, Hendrickson W A, Sodroski J. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science. 1998;280:1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 41.Robertson D L, Anderson J P, Bradac J A, Carr J K, Foley B, Funkhouser R K, Gao F, Hahn B H, Kalish M L, Kuiken C, Learn G H, Leitner T, McCutchan F, Osmanov S, Peeters M, Pieniazek D, Salminen M, Sharp P M, Wolinsky S, Korber B. HIV-1 nomenclature proposal. Science. 2000;288:55–56. doi: 10.1126/science.288.5463.55d. [DOI] [PubMed] [Google Scholar]
- 42.Salminen M O, Johansson B, Sonnerborg A, Ayehunie S, Gotte D, Leinikki P, Burke D S, McCutchan F E. Full-length sequence of an Ethiopian human immunodeficiency virus type 1 (HIV-1) isolate of genetic subtype C. AIDS Res Hum Retrovir. 1996;12:1329–1339. doi: 10.1089/aid.1996.12.1329. [DOI] [PubMed] [Google Scholar]
- 43.Simon F, Mauclere P, Roques P, Loussert-Ajaka I, Muller-Trutwin M, Saragosti S, Georges-Courbot M, Barre-Sinoussi F, Brun-Vezinet F. Identification of a new human immunodeficiency virus type 1 distinct from group M and O. Nat Med. 1998;4:1032–1037. doi: 10.1038/2017. [DOI] [PubMed] [Google Scholar]
- 44.Sullivan N, Sun Y, Li J, Hofmann W, Sodroski J. Replicative function and neutralization sensitivity of envelope glycoproteins from primary and T-cell line-passaged human immunodeficiency virus type 1 isolates. J Virol. 1995;69:4413–4422. doi: 10.1128/jvi.69.7.4413-4422.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Syu W, Lee W, Du B, Yu Q, Essex M, Lee T. Role of conserved gp41 cysteine residues in the processing of human immunodeficiency virus envelope precursor and viral infectivity. J Virol. 1991;65:6349–6352. doi: 10.1128/jvi.65.11.6349-6352.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tersmette M, de Goede R E, Al B J, Winkel I N, Gruters R A, Cuypers H T, Huisman H G, Miedema F. Differential syncytium-inducing capacity of human immunodeficiency virus isolates: frequent detection of syncytium-inducing isolates in patients with acquired immunodeficiency syndrome (AIDS) and AIDS-related complex. J Virol. 1988;62:2026–2032. doi: 10.1128/jvi.62.6.2026-2032.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trujillo J, Wang W, Lee T, Essex M. Identification of the envelope V3 loop as a determinant of a CD4-negative neuronal cell tropism for HIV-1. Virology. 1996;217:613–617. doi: 10.1006/viro.1996.0158. [DOI] [PubMed] [Google Scholar]
- 48.Tschachler E, Buchow H, Gallo R C, Reitz M S., Jr Functional contribution of cysteine residues to the human immunodeficiency virus type 1 envelope. J Virol. 1990;64:2250–2259. doi: 10.1128/jvi.64.5.2250-2259.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tscherning C, Alaeus A, Fredriksson R, Bjorndal A, Deng H, Littman D, Fenyo E, Albert J. Differences in chemokine receptor usage between genetic subtypes of HIV-1. Virology. 1998;241:181–188. doi: 10.1006/viro.1997.8980. [DOI] [PubMed] [Google Scholar]
- 50.UNAIDS. The global source of HIV/AIDS information. AIDS epidemic update. Geneva, Switzerland: UNAIDS; 1996. –1999. [Google Scholar]
- 51.Wang W, Dudek T, Zhao Y, Brumblay H, Essex M, Lee T. CCR5 coreceptor utilization involves a highly conserved arginine residue of HIV type 1 gp120. Proc Natl Acad Sci USA. 1998;95:5740–5745. doi: 10.1073/pnas.95.10.5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu X, Yuan X, Matsuda Z, Lee T H, Essex M. The matrix protein of human immunodeficiency virus type 1 is required for incorporation of viral envelope protein into mature virions. J Virol. 1992;66:4966–4971. doi: 10.1128/jvi.66.8.4966-4971.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang L Q, MacKenzie P, Cleland A, Holmes E C, Brown A J, Simmonds P. Selection for specific sequences in the external envelope protein of human immunodeficiency virus type 1 upon primary infection. J Virol. 1993;67:3345–3356. doi: 10.1128/jvi.67.6.3345-3356.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu T, Mo H, Wang N, Nam D S, Cao Y, Koup R A, Ho D D. Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science. 1993;261:1179–1181. doi: 10.1126/science.8356453. [DOI] [PubMed] [Google Scholar]