Abstract
Iron is an essential mineral that supports numerous biological functions. Studies have reported associations between iron dysregulation and certain cardiovascular and neurodegenerative diseases, but the direction of influence is not clear. Our goal was to use computational approaches to better understand the role of genetically predicted iron levels on disease risk. We meta-analyzed genome-wide association study summary statistics for serum iron levels from two cohorts and two previous meta-analyses. We then obtained summary statistics from 11 neurodegenerative, cerebrovascular, cardiovascular or lipid traits to assess global and regional genetic correlation between iron levels and these traits. We used two-sample Mendelian randomization (MR) to estimate causal effects. Sex-stratified analyses were also carried out to identify effects potentially differing by sex. Overall, we identified three significant global correlations between iron levels and (i) coronary heart disease, (ii) triglycerides, and (iii) high-density lipoprotein (HDL) cholesterol levels. A total of 194 genomic regions had significant (after correction for multiple testing) local correlations between iron levels and the 11 tested traits. MR analysis revealed two potential causal relationships, between genetically predicted iron levels and (i) total cholesterol or (ii) non-HDL cholesterol. Sex-stratified analyses suggested a potential protective effect of iron levels on Parkinson’s disease risk in females, but not in males. Our results will contribute to a better understanding of the genetic basis underlying iron in cardiovascular and neurological health in aging, and to the eventual identification of new preventive interventions or therapeutic avenues for diseases which affect women and men worldwide.
Keywords: Serum iron, Genetic correlation, Mendelian randomization (MR), Sex-stratified, Cardiovascular, Neurodegeneration
Subject terms: Genetic association study, Computational biology and bioinformatics
Introduction
Iron is an essential mineral that facilitates many biological functions throughout the human body. For instance, it is key for essential functions at the systemic and cellular levels, such as oxygen transfer and storage, production and function of neurotransmitters, redox balance, inflammation regulation and cellular growth1,2.
Iron is necessary for the formation of hemoglobin, a vital oxygen-transporting protein present in the red blood cells. The delivery of oxygen from the lungs to tissues is important for the generation of adenosine triphosphate (ATP), the currency of cellular energy. Iron is also necessary for numerous enzymes involved in energy metabolism. Altogether, these functions highlight iron’s critical role in sustaining energy-intensive cellular processes, including those related to cardiovascular or neurological health3.
The ability of cells to produce energy, especially cells of the brain or heart with high energy demands, could be compromised without an adequate supply of iron (Supplementary Fig. 1). Such cells include neurons, responsible for maintaining the brain’s energy-demanding cognitive functions, and cardiomyocytes, which require substantial energy to support essential cardiac functions. In contrast, excessive iron levels can trigger the generation of free radicals, leading to oxidative stress, cellular damage, and inflammatory responses. Furthermore, these free radicals can induce the accumulation of iron in specific organs, which can result in tissue damage and impaired organ function4.
Given the importance of iron for sustaining heart and brain health, it is unsurprising that disturbances in iron metabolism have been implicated in the pathogenesis of a range of chronic diseases, including cardiovascular (CVD) and neurodegenerative diseases (NDD). CVD and NDD impose a significant burden on global health, contributing to considerable morbidity, mortality, and socio-economic implications. Understanding the underlying mechanisms driving the development and progression of these diseases is therefore essential for developing effective preventive and therapeutic strategies to improve health.
The association between iron levels (or body iron stores, measured by ferritin levels) and several CVD and NDD is reported with inconsistent or contradictory results by numerous observational and prospective studies5–10. For example, with regard to iron and CVD, a systematic review meta-analysis of prospective studies found that higher serum iron status may confer a protective effect against coronary heart disease or myocardial infarction11. Conversely, conflicting results are presented in other studies, such as Sempos et al., which reported no evidence for an association between serum ferritin levels and the development of cardiovascular endpoints in a prospective cohort study12. Additionally, Knuiman et al. reported that there was insufficient evidence regarding the role of serum ferritin levels levels as a risk factor for cardiovascular disease in a longitudinal prospective cohort study in Western Australia13.
As observational studies can be impacted by confounding factors and reverse causality, contributing to the inconsistency of reported results, alternative approaches are required to better assess how and whether iron levels are linked to CVD and NDD risk, and to determine the most plausible direction of effect.
Genetic factors contribute to inter-individual in iron levels, and the overall heritability for iron has been reported as ranging from 14 to 32%14–17. Additionally, many CVD and NDD have a significant genetic component18–20. Exploring the impact of iron levels on CVD and NDD from a genetic perspective presents an opportunity to identify the mechanisms and biological pathways relating iron levels and complex disease risk. Such knowledge can be actionable given that iron levels can be regulated, up to a certain extent, through dietary interventions and supplements.
Iron metabolism varies by sex, with differing absorption rates and overall levels in the body21. Additionally, hormonal fluctuations throughout the menstrual cycle can impact iron absorption and regulation in females22. These variations in iron metabolism could play crucial roles in determining disease outcomes23.
We aimed to use large-scale genetic variant-trait association summary statistics and computational genetics methods to better understand the effect of genetically predicted serum iron levels on cardiovascular or neurodegenerative diseases, as well as the direction of these relationships. Given the established sex differences for iron levels and the diseases of interest, we also carried out complementary sex-stratified analyses.
Methods
Summary statistics for meta-analysis of serum iron levels
Genome-wide association study (GWAS) summary statistics for serum iron levels were obtained from the following two cohorts for meta-analysis: Baltimore Longitudinal Study of Aging (BLSA) study24, the Montreal Heart Institute (MHI) Biobank as well as from two previously published meta-analyses: Benyamin et al. and Moksnes et al. (Table 1)16,25. For the former meta-analysis, we used the summary statistics from their discovery analysis, which combined 11 cohorts that did not overlap with other cohorts used for our meta-analysis. The latter meta-analyzed three cohorts: the Trøndelag Health Study—HUNT, the Michigan Genomics Initiative (MGI), and SardiNIA26–28.
Table 1.
Summary of cohorts of individuals of European-like genetic ancestry for meta-analysis or serum iron levels.
| Cohort | N | % Female | Distribution of iron measurements (average ± SD) µmol/L |
|---|---|---|---|
| Benyamin et al.25 meta-analysis | 23,986 | 44 | 18.63 ± 6.35 |
| Baltimore Longitudinal Study of Aging (BLSA) | 480 | 49 | 15.65 ± 5.32 |
| Montreal Heart Institute Biobank (MHI Biobank) | 2089 | 38 | 13.28 ± 6.91 |
| (Median ± IQR) | |||
|---|---|---|---|
| Moksnes et al.16 meta-analysis | |||
| HUNT | 56,667 | 47 | 15 ± 8 |
| Michigan Genomics Initiative (MGI) | 10,504 | 42 | 12.2 ± 8.9 |
| SardiNIA | 5930 | 43 | 15 ± 7.6 |
| Interval | 40,197 | 50 | 17 ± 9 |
| deCODE | 123,314 | 37 | 15 ± 8 |
| Total sample size | 263,167 | ||
IQR interquartile range, SD standard deviation.
Additionally, we obtained sex-stratified summary statistics for serum iron from the following three cohorts for which such data were available: Montreal Heart Institute (MHI) Biobank, MGI Biobank and SardiNIA26,27.
All summary statistics for analysis had assessed individuals inferred to be genetically similar to a European genetic ancestry reference population, which we refer to as individuals with European-like genetic ancestry. Further details of the cohorts included can be found in Supplementary Methods.
Meta-analysis of serum iron levels summary statistics
We meta-analyzed GWAS summary statistics for serum iron to increase statistical power for our subsequent analyses. We used the sample-size weighting scheme in METAL to perform a fixed-effects meta-analysis29. The sample-size weighting scheme allows for the analysis of studies with different scales of measurements. All summary statistics included were derived from individuals of European-like genetic ancestry29. Autosomal SNPs and short indels were included, and chromosome X variants were included when available. We used the LiftOver tool to ensure that all summary statistics were on human genome build GRCh3730. A meta-analysis using sex-combined summary statistics was conducted by combining summary statistics from two previous meta-analyses (Benyamin et al.25 and Moksnes et al.16) and two other cohorts: the Baltimore Longitudinal Study of Aging (BLSA)24 and the Montreal Heart Institute (MHI) Biobank. We used FUMA (version 1.5.2) to annotate our iron meta-analysis results31.
GWAS summary statistics for cardiovascular- and neurodegenerative-related traits
We selected 11 large GWAS (> 20,000 cases for binary traits) conducted in individuals of European-like genetic ancestry (so to match the genetic ancestry of our iron meta-analysis) for neurodegenerative (Alzheimer’s disease32, Parkinson’s disease33, amyotrophic lateral sclerosis34), cardiovascular (coronary artery disease35, heart failure36), cerebrovascular (stroke37) and lipid (triglycerides, and the following cholesterol: total, low-density lipoprotein, high-density lipoprotein, non-high-density lipoprotein38) traits (Supplementary Fig. 2).
Global genetic correlations between iron and cardiovascular or neurodegenerative traits
We used cross-trait linkage disequilibrium score regression (LDSC; v1.0.1) to assess genome-wide genetic correlations (rg) between serum iron and each of the 11 traits of interest, based on common autosomal single nucleotide polymorphisms (SNPs)39. In brief, LDSC regresses the product of the marginal z-scores in the GWAS summary statistics for the two traits on the LD scores of each SNP. We processed the summary statistics for each trait through LDSC’s munge-sumstats.py script, which retained 1,217,312 common autosomal SNPs from the HapMap phase 3 project. Data from the European super-population of the 1000 Genomes phase 3 project was used for the LD reference panel (available at https://github.com/bulik/ldsc). Due to the long-range and complex linkage disequilibrium within the major histocompatibility complex region on chromosome 6, SNPs within this region are excluded.
Local genetic correlations between iron and cardiovascular or neurodegenerative traits
We used LAVA (Local Analysis of [co]Variant Association; v0.0.6) to estimate regional genetic correlations between iron and the 11 traits of interest. LAVA divides the autosomes into 2459 non-overlapping loci based on the European super-population of the 1000 Genomes Project as the linkage disequilibrium reference40. LAVA estimates local genetic correlation for each iron-trait pair at loci with genetic signal in both traits. Specifically, first, a univariate test was performed to identify which loci have genetic signal by testing for per-trait local heritability. Then, for loci where both traits had a p-value in the univariate test that passed Bonferroni correction (p < 0.05/2459), we then performed the bivariate (genetic correlation) analysis. We also applied a Bonferroni correction at the bivariate stage, reporting loci passing a p-value threshold for each iron-trait pair tested of 0.05/number of comparisons. Summary statistics for Alzheimer’s and Parkinson’s included proxy cases (individuals who have a close relative with the disease of interest, but who do not have the disease themselves). For comparison, we also carried out a sensitivity analysis using summary statistics that excluded proxy cases for Alzheimer’s41 and Parkinson’s33, which have lower sample sizes (N cases = 21,982 and N controls = 41,944 for Alzheimer’s, and N cases = 17,996 and N controls = 16,502 for Parkinson’s).
Causal inference approaches (Mendelian randomization) to elucidate links between iron and cardiovascular or neurodegenerative traits
Mendelian randomization (MR) is an epidemiological instrumental variable approach that uses genetic variant-trait associations to assess causal relationships between genetically predicted values of an exposure and an outcome, given certain assumptions. We carried out a series of two-sample (MR) analyses with iron as the exposure and one of the 11 selected traits as the outcome. There are three core assumptions that must be met for the causal inference to be valid. First, the genetic variants selected as genetic instruments are robustly associated with the exposure. Second, the genetic instruments are not associated with confounding factors, measured or unmeasured. Finally, the relationship between the genetic instruments and the outcome is only through the exposure. The first assumption can be quantitatively evaluated by calculating the F statistic for the selected genetic instrument and the exposure, where an F statistic greater than 10 is standardly considered a robust association. There is no quantitative test per se to ensure the validity of the second and third assumptions, but sensitivity analyses, including those that assess the presence of horizontal pleiotropy, such as MR-Egger, are commonly conducted to ensure the consistency of results.
We verified that the selected genetic variants do not exhibit evidence of association with confounding traits (see “Results” section for details) or with the tested outcomes by searching in the GWAS Catalog and in PhenoScanner (v2)42,43. Variants that did exhibit evidence were excluded.
We used the R package TwoSampleMR (version 0.5.7) to carry out two-sample MR analyses44. For our primary analysis, we used the inverse variance weighted approach with random effects, followed by MR Egger, weighted median and weighted mode as secondary analyses to assess the consistency of the results. We report the Cochran’s Q test for heterogeneity in causal estimates for the inverse variance weighted method across the genetic variants used as instruments. We performed the MR Egger intercept test to assess the presence of pleiotropic variants45. We also performed MR-PRESSO (version 1.0) using the R package MR-PRESSO to identify and remove horizontal pleiotropic outliers from our MR analyses46, as well as a leave-one-out analysis to evaluate whether the results were influenced by a specific genetic instrument. Finally, for each iron-outcome pair tested, a directionality test was performed using TwoSampleMR to assess the validity of the assumption that the exposure (genetically predicted iron levels) causes the outcome rather than the other way around.
Sex-stratified analyses
We also performed a sex-stratified meta-analysis following a similar protocol as the sex-combined meta-analysis, using data from the three cohorts for which we had access to sex-stratified summary statistics (SardiNIA, Montreal Heart Institute (MHI) Biobank, and Michigan Genomics Initiative (MGI) Biobank). The total sample size tested at each genetic variant was up to 11,547 females and 9090 males. Then, we estimated global and local genetic correlations, and finally, we performed MR using the same workflow as previously described with the sex-stratified iron meta-analysis summary statistics and sex-specific summary statistics from the following 3 conditions with large-scale sex-stratified summary statistics available from European-like study samples: Parkinson’s disease33, coronary artery disease35 and amyotrophic lateral sclerosis47 (summary statistics provided by co-authors RPB and RLM). As in the sex-combined summary statistics, the summary statistics for all studies used here were derived from individuals of European-like genetic ancestry.
Results
Meta-analysis of iron summary statistics
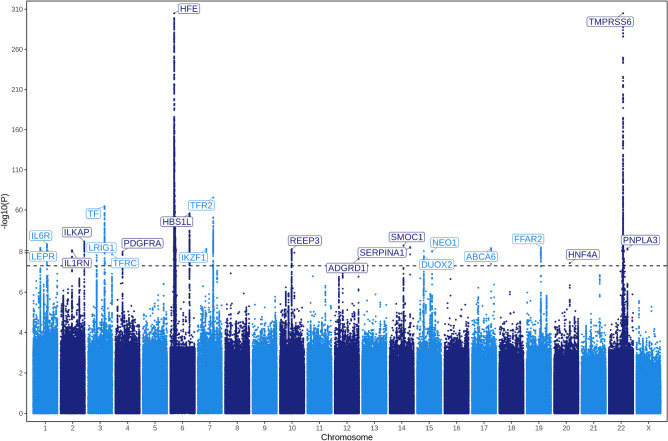
In our meta-analysis of serum iron GWAS studies, we identified a total of 23 genome-wide significant (p-value < 5 × 10–8) loci (Fig. 1, Table 2). No significant associations were identified on the X chromosome, but sample sizes available for testing variants on this chromosome were limited as the Benyamin et al. and BLSA studies included in our meta-analysis did not provide association statistics for chromosome X variants. The most significant associations were observed on chromosome 22 at TMPRSS6 (p-value = 2.17 × 10–375), and on chromosome 6 at HFE (p-value = 2.23 × 10–375). 20 of the 23 loci had been previously identified at genome-wide significance in at least one of the GWAS studies included in our meta-analysis. The three loci that had not been previously identified (corresponding to closest genes, TFRC, PDGFRA and NEO1) were all suggestive signals in the Moksnes et al.16 serum iron meta-analysis (p < 2 × 10–6). As expected for a complex trait, the most significant variants fell into intergenic or intronic regions (Supplementary Fig. 3).
Fig. 1.
Manhattan plot of the results from the GWAS meta-analysis of serum iron using a sample-size weighted z-score method resulting in a total sample size of 263,167 individuals of European-like genetic ancestry. The dashed line presents the threshold to denote genome-wide significance (5 × 10–8). We replaced extremely significant values in our analysis with a minimum non-zero value that plotting libraries can handle. Note rescaling on the y-axis represented by two horizontal lines (“=”).
Table 2.
Summary of top hits in meta-analysis of serum iron GWAS.
| SNP | Nearest protein coding gene | Position (GRCh37) | Effect allele | Direction | Z score | Meta-analysis p value | MAF | Beta | SE |
|---|---|---|---|---|---|---|---|---|---|
| rs35945185 | LEPR | chr1:66,137,239 | A | ???+ | 7.384 | 1.54e−13 | 0.38 | 0.022 | 0.003 |
| rs4129267 | IL6R | chr1:154,426,264 | T | ++++ | 8.719 | 2.802e−18 | 0.36 | 0.025 | 0.003 |
| rs7580634 | IL1RN | chr2:113,866,854 | T | ?−−− | − 6.39 | 1.663e−10 | 0.42 | − 0.019 | 0.003 |
| rs6726348 | ILKAP | chr2:239,084,119 | T | ++−+ | 9.495 | 2.193e−21 | 0.45 | 0.026 | 0.003 |
| rs12633819 | LRIG1 | chr3:66,448,307 | A | −−+− | − 8.277 | 1.259e−16 | 0.36 | − 0.024 | 0.003 |
| rs8177252 | TF | chr3:133,480,174 | A | ++++ | 17.07 | 2.494e−65 | 0.35 | 0.049 | 0.003 |
| rs6583288 | TFRC | chr3:195,809,564 | A | −+−− | − 5.68 | 1.349e−08 | 0.26 | − 0.018 | 0.003 |
| rs7655524 | PDGFRA | chr4:55,354,337 | A | −−−− | − 5.746 | 9.131e−09 | 0.39 | − 0.016 | 0.003 |
| rs1800562 | HFE | chr6: 26,093,141 | A | ++++ | 41.442 | 2.23e−375 | 0.04 | 0.29 | 0.007 |
| rs7776054 | HBS1L | chr6:135,418,916 | A | −−−− | − 15.848 | 1.451e−56 | 0.26 | − 0.05 | 0.003 |
| rs6976036 | IKZF1 | chr7:50,429,442 | T | ?+−− | − 6.939 | 3.96e−12 | 0.5 | − 0.02 | 0.003 |
| rs2075672 | TFR2 | chr7:100,240,296 | A | −−−− | − 18.482 | 2.861e−76 | 0.38 | − 0.052 | 0.003 |
| rs7897379 | REEP3 | chr10:65,301,725 | T | −+−− | − 6.88 | 5.982e−12 | 0.49 | − 0.019 | 0.003 |
| rs4759827 | ADGRD1 | chr12:131,492,288 | T | ?+−− | − 5.59 | 2.276e−08 | 0.29 | − 0.018 | 0.003 |
| rs1958078 | SMOC1 | chr14:70,354,858 | A | −+−− | − 8.208 | 2.252e−16 | 0.17 | − 0.03 | 0.004 |
| rs28929474 | SERPINA1 | chr14:94,844,947 | T | ?−?+ | 7.635 | 2.256e−14 | 0.02 | 0.079 | 0.01 |
| rs1961660 | DUOX2 | chr15:45,401,636 | T | ?++− | − 5.644 | 1.658e−08 | 0.07 | − 0.032 | 0.006 |
| rs531019 | NEO1 | chr15:73,598,607 | A | −−+− | − 6.045 | 1.496e−09 | 0.47 | − 0.017 | 0.003 |
| rs77542162 | ABCA6 | chr17:67,081,278 | A | ?−+− | − 7.286 | 3.2e−13 | 0.01 | − 0.106 | 0.015 |
| rs34536858 | KRTDAP, FFAR2 | chr19:35,950,174 | A | ?+++ | 7.573 | 3.651e−14 | 0.29 | 0.024 | 0.003 |
| rs1800961 | HNF4A | chr20:43,042,364 | T | ?−−+ | 5.508 | 3.629e−08 | 0.04 | 0.041 | 0.007 |
| rs855791 | TMPRSS6 | chr22:37,462,936 | A | −−−− | − 41.443 | 2.17e−375 | 0.39 | − 0.116 | 0.003 |
| rs3747207 | PNPLA3 | chr22:44,324,855 | A | ?−++ | 7.129 | 1.012e−12 | 0.22 | 0.025 | 0.003 |
MAF minor allele frequency, from the 1000 Genomes European super-population, SE standard error of the Beta.
Direction of effect of the four input summary statistics corresponding to the following order: Benyamin et al. meta-analysis, Baltimore Longitudinal Study of Aging, Montreal Heart Institute Biobank, Moksnes et al. meta-analysis. The symbols −, + and ? in this column indicate that relative to the effect allele, there was a negative effect, a positive effect or the variant was not assessed, respectively in that particular dataset.
The estimated heritability of serum iron levels on the observed scale using LD score regression based on the 1000 Genomes European genetic ancestry super-population as the LD reference was estimated to be 0.0362 ± 0.008.
Global genetic correlations between iron and cardiovascular or neurodegenerative traits
We identified four significant correlations (p < 0.05) between serum iron and a tested trait (Supplementary Fig. 4). Specifically, we observed a significant positive genetic correlation between iron levels and HDL cholesterol. On the other hand, negative correlations were observed for iron and cardiovascular diseases (coronary artery disease and heart failure), as well as with triglycerides.
Local genetic correlations between iron and cardiovascular or neurodegenerative traits
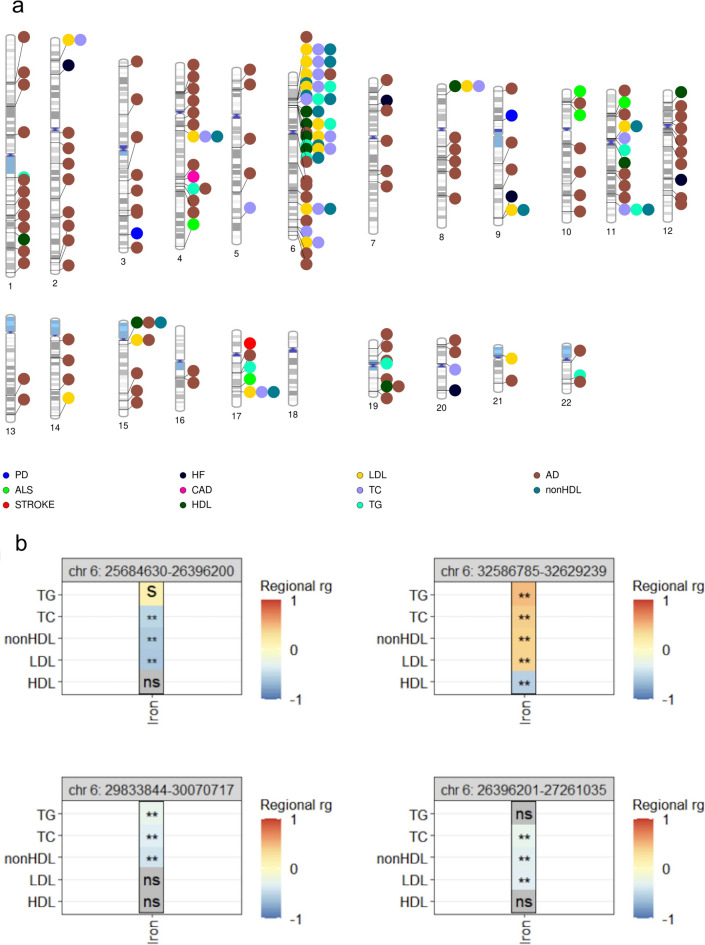
Local genetic correlation can differ from global genetic correlation, i.e. from the genome-wide average correlation. For a pair of traits, if certain genomic loci are positively correlated, but other loci are negatively correlated, these effects could be canceled out in a global genetic correlation analysis. A published example of such a scenario is between HDL and LDL. Globally, these two traits are not strongly significantly correlated with each other, but locally, multiple regions are significantly correlated48. Additionally, global genetic correlation analysis can miss findings if strong genetic correlations are restricted to specific genomic regions49. We therefore assessed local/regional genetic correlations between iron and the eleven traits of interest. We identified a total of 194 significantly correlated loci (presented in Supplementary Table 1) between iron and the tested traits (when testing Alzheimer’s and Parkinson’s disease data that included proxies). The number of significant loci for each iron-trait pair tested ranged from one locus (between iron and either stroke or coronary artery disease) to 106 loci (between iron and Alzheimer’s disease) (Fig. 2a). To gain insight from the 106 loci significantly correlated between iron and Alzheimer’s disease, we carried out a gene-set enrichment analysis. Specifically, for each of the 106 loci, we obtained the list of protein coding genes falling into the genomic coordinates and used the Gene2Func function of FUMA (version 1.5.2) to identify enriched gene-sets using all protein coding genes as the background gene-set31. We applied a Bonferroni correction and required a minimum of 5 genes overlapping with a gene-set. Of the significantly enriched gene-sets, those related to metals homeostasis and cellular response, including for copper and zinc showed evidence of enrichment, as well as gene-sets related to fatty acids, oxidation by cytochrome P450 and eicosanoids (Supplementary Fig. 5). The enriched gene-set of eicosanoids (lipid-based signaling molecules involved in innate immune response)50 may suggest that the innate immune system could be a potential link between genetically predicted iron levels and Alzheimer’s disease. Indeed, multiple lines of evidence have highlighted the role of the immune system in Alzheimer’s disease, as previously reviewed51,52, and iron homeostasis is known to be tied to inflammatory response, which has also been previously reviewed53,54. When using the Alzheimer’s and Parkinson’s summary statistics excluding proxy cases, one locus remained significantly correlated between serum iron levels and Alzheimer’s and one locus remained significantly correlated between serum iron levels and Parkinson’s, likely due to limited power. Both those loci were also significant in the primary analysis including proxy cases, and there was agreement in the direction of effects.
Fig. 2.
Local genetic correlations between iron and NDD or CVD. Chromosomal visualization (created using PhenoGram78) of significantly correlated regions between iron and the 11 neurodegenerative or cardiovascular traits tested as listed in Supplementary Table 1 (a). Illustration of selected significant shared regional correlations on chromosome 6 (either just upstream or within the major histocompatibility complex) between serum iron and lipid traits tested (b). **Significant local correlation after Bonferroni correction; s: p-value < 0.05 but not significant at the Bonferroni correction threshold; ns non-significant (p-value > 0.05), AD Alzheimer’s disease, PD Parkinson’s disease, ALS amyotrophic lateral sclerosis, CAD coronary artery disease, HF heart failure, TC total cholesterol, TG triglycerides, HDL high-density lipoprotein cholesterol, LDL low-density lipoprotein cholesterol, nonHDL non-high-density lipoprotein cholesterol.
We noted that 58 (30%) of all significant loci were found between iron and a tested lipid trait (specifically, HDL cholesterol, LDL cholesterol, total cholesterol or triglycerides, corresponding to 36 unique genomic regions, 19 of which are situated within or nearby the major histocompatibility complex region on chromosome 6 (Fig. 2b). The major histocompatibility region is known for its complex long-range linkage disequilibrium patterns and for encoding a set of highly polymorphic genes that are key for immune response and immune system function55. This observation warrants further work on immunity and inflammation to assess the relationship between genetically predicted iron levels and lipid traits in downstream studies.
The locus (chr22:37,364,005–38,718,589, GRCh37) encompassing the genes most significantly associated with serum iron in our meta-analysis, TMPRSS6, was significantly correlated with triglycerides (rg = 0.370075, p-value = 9.63e-06). The local genetic correlation analyses did not show any correlation with any of the traits tested for locus on chromosome 6 with the second most significant signal (HFE).
Causal inference approaches (Mendelian randomization) to elucidate links between iron and cardiovascular or neurodegenerative traits
The completed MR-STROBE checklist is found in Supplementary Table 2.
To assess the validity of the three core MR assumptions, we selected independent genetic variants at genome-wide significant loci (p < 5 × 10–8). Through searches through the GWAS Catalog and PhenoScanner database, we also excluded variants with evidence of association with various traits that could be confounders acting on the exposure and the various outcomes tested. Such traits included cardiovascular diseases (including coronary artery disease, hypertension, and cardiomegaly), metabolic traits (including body mass index, body fat percentage), neurological disorders (including multiple sclerosis), inflammatory diseases (such as Crohn’s disease and rheumatoid arthritis), respiratory diseases (including asthma), and specific causes of mortality (such as malignant neoplasms and intestinal perforation). Additionally, variants identified as potential outliers via MR-PRESSO were removed from analyses. Finally, variants associated with the outcomes (cardiovascular and neurodegenerative traits) were not selected as genetic instruments.
Two-sample MR also assumes that the participants contributing to the summary statistics of the exposure are different from the participants contributing to the summary statistics of the outcome. To our knowledge, there should be minimal (if any) overlap for our analyses assessing iron levels as the exposure for each of the 11 outcomes.
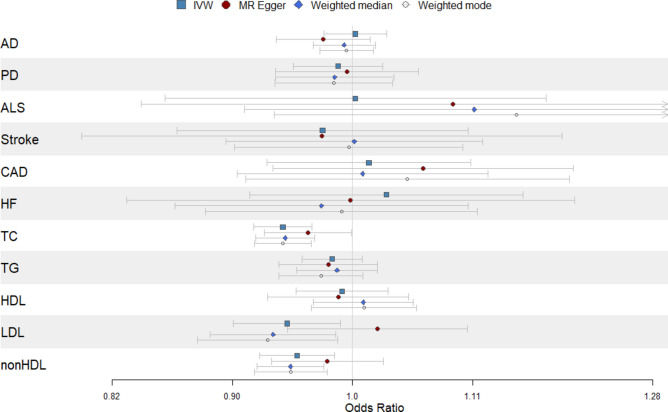
Our primary analysis, using the inverse variance weighted approach, yielded evidence of a significant (p < 0.0045, accounting for eleven exposure-outcome comparisons) causal effect between genetically predicted iron levels and total cholesterol (OR [95% CI] = 0.94 [0.92–0.97], p = 3.3 × 10–6) and non-high-density lipoprotein cholesterol (OR [95% CI] = 0.95 [0.92–0.98], p = 3.9 × 10–3) (Fig. 3), suggesting a protective effect of iron. That is to say, modestly higher genetically predicted iron levels result in either modestly lower total cholesterol or non-high-density lipoprotein cholesterol. Secondary analyses using either the weighted median or weighted mode approach offered support for the results of the inverse variance weighted approach (p < 0.0045 and consistent direction of effect). The genetic instruments used in each analysis, including their corresponding F statistic (all greater than the accepted threshold of 10), and their corresponding effect sizes and standard errors in the exposure and outcome GWAS are presented in Supplementary Table 3. Results from the primary and secondary analyses are presented in Supplementary Table 4. Although MR Egger results did not reach statistical significance, the direction of effect (i.e. direction of the beta) was consistent, reinforcing the observed results.
Fig. 3.
Mendelian randomization results with genetically predicted iron levels as the exposure and each of the 11 neurodegenerative, cardiovascular or cerebrovascular traits tested as the outcome with inverse-variance weighted (IVW) approach as the primary analysis and MR Egger, weighted median and weighted mode as secondary analyses. AD Alzheimer’s disease, PD Parkinson’s disease, ALS amyotrophic lateral sclerosis, CAD coronary artery disease, HF heart failure, TC total cholesterol, TG triglycerides, HDL high-density lipoprotein cholesterol, LDL low-density lipoprotein cholesterol, nonHDL non-high-density lipoprotein cholesterol.
We constructed leave-one-out plots, scatterplots of effect sizes in the exposure versus the outcome and funnel plots for each of the exposure-outcome pairs tested (Supplementary Figs. 6–8).
The directionality tests suggested that for all iron-outcome pairs tested, the direction of iron as the exposure was most probable. Results of the directionality test are presented in Supplementary Table 5. Heterogeneity tests for the inverse variance weighted method suggest some heterogeneity for certain iron-outcome pairs tested, including for iron with non-HDL (Q = 25.7, p = 0.04), which may bias the causal estimates (Supplementary Table 6). None of the analyses, apart from low-density lipoprotein cholesterol (MR Egger intercept = -0.003, p = 0.04) showed evidence of horizontal pleiotropy as per the MR Egger intercept test (Supplementary Table 7).
Sex-stratified analyses
Our sex-stratified meta-analyses of serum iron using sex-stratified summary statistics from three cohorts for which sex-stratified results were available (Montreal Heart Institute (MHI) Biobank, Michigan Genomics Initiative (MGI) Biobank, and SardiNIA) yielded the same two strongest genome-wide significant signals as the overall meta-analysis, corresponding to the genes HFE and TMPRSS6 on chromosomes 6 and 22 respectively (Supplementary Fig. 9). Similar to the overall meta-analysis, there were no significant associations on the X chromosome in the sex-stratified analyses, but sample size was lower on this chromosome compared to the autosomes, as the SardiNIA cohort did not test X chromosome variants for association.
Global genetic correlations did not reveal statistically significant associations between iron levels and Parkinson’s disease, coronary artery disease, or amyotrophic lateral sclerosis for the sex-stratified analyses (Supplementary Fig. 10). However, our analysis of local genetic correlations identified significantly correlated loci for iron levels and these three diseases (Supplementary Figs. 11–13, Supplementary Table 8). A locus on chromosome 12 (chr12:23,923,799–25,058,714, GRCh37) was significantly correlated between iron and Parkinson’s disease in females, whereas two loci were correlated in the corresponding male analysis (chr8: 98,132,520–99,388,944 and chr19:55,714,086–56,411,086). There were two loci significantly correlated between iron and coronary artery disease in the female analysis (chr19: 54,602,371–55,182,973 and chr20:42,185,672–43,008,890) and another two in the male analysis (chr6:110,647,843–111,363,443 and chr9:136,770,927–137,593,527). One locus on chromosome 9 (chr9:132,193,799–132,999,452) showed significant correlation in the male analysis between iron and amyotrophic lateral sclerosis and there were no significantly correlated loci in the corresponding female analysis. These sex-stratified local correlations also need to be replicated in larger sample sizes.
Finally, through sex-specific MR, we observed a potential near-significant causal effect of iron levels on Parkinson’s disease among females in our primary analysis using the inverse variance weighted approach (OR [95% CI] = 0.79 [0.64–0.98], p = 0.03). Direction of effect was consistent for the secondary analyses. Three genetic instruments were used in the stratified MR analyses (Supplementary Table 9). No significant causal effects were detected for coronary artery disease or amyotrophic lateral sclerosis (Supplementary Figs. 14–17, Supplementary Table 10). Directionality test results suggested that iron as the exposure (i.e. not the outcome) was most likely direction (Supplementary Table 11). We observed neither evidence of heterogeneity through heterogeneity test results from the inverse variance weighted approach (Supplementary Table 12) nor the presence of horizontal pleiotropy through the MR Egger intercept test (Supplementary Table 13).
Discussion
Serum iron is important for various bodily functions, including those involved in the cardiovascular and neurological systems. We aimed to better understand the contribution of genetically predicted serum iron levels on cardiovascular and neurodegenerative diseases by applying computational approaches to large-scale GWAS summary statistics. To accomplish this aim, we meta-analyzed iron studies to increase statistical power and used eleven large cardiovascular- or neurodegenerative-related GWAS results. We supported our primary analyses with sex-stratified analyses, data permitting, given the established sex differences in iron levels and the various traits tested.
In summary, our meta-analysis of serum iron studies identified 20 genome-wide significant loci previously reported and unveiled 3 novel loci (which were subthreshold significant in a previous meta-analysis, thus highlighting the importance of increasing sample size). Further investigation into these loci may reveal novel biological mechanisms underlying iron homeostasis.
The results of our global genetic correlations analyses underscore a complex interplay between serum iron levels and lipid profiles, with a significant positive genetic correlation observed between iron levels and HDL cholesterol. This finding suggests that higher, but within the normal range, genetically predicted iron levels is correlated with a possibly beneficial lipid profile, since HDL cholesterol is generally associated with reduced risk of cardiovascular disease56,57. Negative genetic correlations were observed between iron and the following traits: coronary artery disease, heart failure and triglyceride levels, which could suggest that iron plays a protective role that contributes to improved cardiovascular health.
Local genetic correlations analysis further elucidated the genetic architecture underlying the relationship between iron and neurodegenerative and cardiovascular traits. Differences in the number of significant regions/loci identified could also reflect differences in study sample size, statistical power, or the prevalence of these conditions in the populations studied. The largest number of associated loci (n = 106) between iron and Alzheimer’s disease could suggest a genetic connection between iron levels and the risk of this neurodegenerative condition. However, correlation does not imply causation, nor does it inform on direction of effect. Causal inference analysis, through Mendelian randomization (MR), did not yield significant findings between iron levels and Alzheimer’s disease, but the directionality test pointed to iron levels to Alzheimer’s as the most likely direction.
MR analyses revealed a potential positive causal effect of iron levels on lipid profile traits, notably total, LDL and non-HDL cholesterol, suggesting a possible beneficial effect of higher iron levels on lipid profiles. However, the presence of heterogeneity for certain iron-outcome pairs, such as in the analysis of iron with non-HDL cholesterol, may have biased the causal estimates.
Experimental and clinical findings concerning sex-related differences in iron metabolism for certain cardiovascular58,59 and neurodegenerative21 traits are receiving increased attention. Females, particularly those of childbearing age, generally have a higher prevalence of iron deficiency and anemia than males23,60. In contrast, while menstruating females may have lower iron stores, males are more likely to be iron overloaded61,62.
In addition, sex hormones play a crucial role in regulating iron homeostasis. Estrogen and progesterone influence iron regulation through hepcidin activity, thus contributing to variations in iron levels throughout the menstrual cycle22. Clinical and experimental evidence suggests that high estrogen levels correlate with increased systemic iron availability63. Conversely, very low testosterone levels can be observed in cases of severe iron overload64, while increased testosterone levels have been found to increase dietary iron absorption and body iron stores by inhibiting hepcidin synthesis65.
These differences in iron metabolism underline the importance of carrying out sex-stratified analyses to allow for the discovery of tailored interventions and treatments. Our sex-specific MR analysis suggested a potential causal effect of iron levels on Parkinson’s disease in females, indicating that higher genetically predicted iron levels within the normal range could play a protective role against the development of Parkinson’s in females. Results in males were insignificant, but further analyses in larger sample sizes should be performed to assess whether differences in statistical power can explain the differing results in the sex-stratified analyses.
Parkinson’s is a progressive neurodegenerative disease, manifesting as motor and non-motor symptoms marked by neuronal loss in the pars compacta of the substantia nigra66. Despite extensive research, the primary disease mechanisms remain unclear, although several theories have been proposed, including mitochondrial dysfunction, inflammation, abnormalities in protein management and oxidative stress67. Various factors contributing to oxidative stress in Parkinson’s have been investigated, including aging, neuroinflammation and iron overload68. Animal studies using neurotoxin models of Parkinson’s disease have demonstrated that excess iron levels in the brain can contribute to loss of dopaminergic neurons, a hallmark of the disease69. Sex-differing factors in Parkinson’s disease epidemiology and clinical progression have been previously reported70–72. There may be neuroprotective effects of estrogen in females, delaying Parkinson’s disease age of onset73, and other work has reported that estrogen treatment can improve motor and cognitive function in females with Parkinson’s74.
We acknowledge limitations of our work. First, all cohorts assessed were of European-like genetic ancestry, which does not represent the world’s diversity. It is imperative to include diverse sets of participants, as data become available to allow for the generalizability of findings.
Second, we acknowledge the importance of sex-stratified analyses given sex-biases in iron levels as well as the neurodegenerative and cardiovascular traits of interest. Sex, as a biological variable, exerts a wide range of effects on genome function and resulting phenotypes, and these effects can be mediated by a variety of considerations including sex-differing gene expression or differing hormonal levels. However, sample sizes in our sex-stratified analyses were limited due to lack of availability of sex-stratified results. Increased sample sizes in these analyses may reveal sex-biased effects for which we did not have sufficient power to detect in the current analyses. On a similar note, chromosome X was not assessed in all studies included in our analyses, leading to less power to detect findings on that chromosome compared to the autosomes.
Third, research has indicated systemic iron status may not correlate well with either brain or cardiac iron levels75–77. Further investigations are needed to understand the role of iron in cardiovascular and neurological health, accounting for both systemic and organ-specific levels.
Another limitation pertains to the variation in iron status across different age groups and life stages. For instance, iron levels are influenced by the characteristics of female life stages, such as prepuberty, post-puberty, pregnancy, breastfeeding and menopause. Failure to control for these factors is a limitation, and incorporating information on life stage in future work when such data are available at-scale will enhance our understanding of the impact of genetically predicted serum iron levels on disease risk.
What is more, we note that iron levels of participants in the cohorts included in our meta-analysis tended to be lower than or within the normal range for serum iron levels (see Table 1). Future work assessing individuals with iron levels above the normal range may increase power.
In future investigations, our understanding can be broadened by examining additional iron biomarkers, such as transferrin and ferritin, alongside serum iron, to gain additional insight of the role of genetically predicted iron distribution in the body on cardiovascular and neurological health. Moreover, as data become available, longitudinal analyses to uncover the temporal relationship between iron dysregulation and the onset and progression of cardiovascular and neurodegenerative diseases should be pursued.
We also encourage exploration into other substances, like copper and magnesium, which share metabolic pathways with iron, for their interactions and associations with cardiovascular and neurodegenerative disease risk, potentially informing dietary or supplemental interventions.
In conclusion, our findings underscore potential links between genetically predicted serum iron levels in cardiovascular and neurodegenerative diseases, warranting further research to elucidate causal mechanisms and explore preventive and therapeutic strategies, including the consideration of sex-differing differences. While our findings presented here provide initial insights, future studies incorporating larger sample sizes, other types of genetic variation in addition to SNPs, are needed to replicate and validate these findings. Additionally, integrating data on hormonal status and increasing genetic diversity of the analysis cohorts will be key to provide insights into effects that may differ by sex or genetic ancestry.
Supplementary Information
Acknowledgements
This research was enabled in part by support provided by Calcul Quebec https://www.calculquebec.ca, the Digital Research Alliance of Canada (alliancecan.ca). SAGT was funded by a Junior 1 award and is now funded by a Junior 2 award from the Fonds de recherche du Québec—Santé (FRQS; https://frq.gouv.qc.ca) and acknowledges support from two Canadian Institutes of Health Research (CIHR) Project Grants (PJT 183817 and PJT 191687), a CIHR Catalyst grant (AD6 192920), and the Alzheimer Society Research Program by the Alzheimer Society of Canada with co-funding from the CIHR—Institute of Aging (Funding Reference Number 189919). FLD was funded by an FRQS postdoctoral training award. This research was undertaken, in part, thanks to funding from the Canada Research Chairs Program to MPD. JCT holds the Canada Research Chair in personalized and translational medicine and the Université de Montréal endowed research chair in atherosclerosis. This work was supported in part by the Intramural Research Program of the National Institute on Aging. RPB is supported by funding from the MND Association (Byrne/Oct22/979-799). The authors acknowledge the Michigan Genomics Initiative participants, Precision Health at the University of Michigan Medical School Central Biorepository, and the University of Michigan Advanced Genomics Core for providing data and specimen storage, management, processing, and distribution services, and the Center for Statistical Genetics in the Department of Biostatistics at the School of Public Health for genotype data curation, imputation, and management in support of the research reported in this publication.
Author contributions
WB, FLD and SAGT wrote the initial draft of the manuscript. All authors reviewed the manuscript. WB performed the analyses and prepared figures and tables. FLD assisted in analysis and results interpretation. SAGT conceptualized hypothesis, devised the analytical approach and assisted in results interpretation. CB, EF, AM, MS, FC were responsible for analyses in the SardiNIA cohort. DB, MCC, SP, MPD, JCT were responsible for analyses in the Montreal Heart Institute cohort. TT and LF were responsible for analyses in the Baltimore Longitudinal Study of Aging cohort. BV, JW and MZ were responsible for analyses in the Michigan Genomics Initiative cohort. RPB and RLM were responsible for the sex-stratified genome-wide association data for amyotrophic lateral sclerosis.
Data availability
Information on how to access the iron summary statistics for Benyamin et al.25 and Moksnes et al.16 and the 11 cardiovascular and neurodegenerative summary statistics used here are publicly available with access information provided in the respective publications. Baltimore Longitudinal Study of Aging (BLSA): Data for BLSA can be accessed through submission of a project proposal at https://www.blsa.nih.gov/. Michigan Genomics Initiative (MGI): The individual level genetic and clinical MGI data reported in this study cannot be deposited in a public repository because of patient confidentiality. Summary statistics can be requested for download by emailing a completed Data Use Agreement form available at https://precisionhealth.umich.edu/our-research/documents-for-researchers/ to phdatahelp@umich.edu. Montreal Heart Institute Biobank: The individual level genetic and clinical data reported in this study cannot be deposited in a public repository because of patient confidentiality. Summary statistics can be requested through the corresponding author. SardiNIA: Serum iron summary statistics from the SardiNIA cohort are available upon request. Summary statistics from the iron meta-analysis performed in the context of this work are publicly available at https://my.locuszoom.org/gwas/111911/?token=a254c600e06a417a9d1e8527cb620b31, https://my.locuszoom.org/gwas/904129/?token=ff44329da5de47fb81fc657d967e10b7 (male-only) and https://my.locuszoom.org/gwas/835579/?token=d16e2ce5101a44fd8078df5f18a08e45 (female-only).
Code availability
Code is available in the following GitHub repository: https://github.com/GaglianoTaliun-Lab/iron_cardio-neuro.
Declarations
Competing interests
MPD has minor equity interest in Dalcor Pharmaceuticals. MPD has a patent Methods for Treating or Preventing Cardiovascular Disorders and Lowering Risk of Cardiovascular Events issued to Dalcor Pharmaceuticals, no royalties received; a patent Genetic Markers for Predicting Responsiveness to Therapy with HDL-Raising or HDL Mimicking Agent issued to Dalcor Pharmaceuticals, no royalties received; and a patent Methods for using low dose colchicine after myocardial infarction, assigned to the Montreal Heart Institute. JCT holds research grants from Amarin, Ceapro, DalCor Pharmaceuticals, Esperion, Ionis, Merck, Novartis and Pfizer; fees from DalCor Pharmacuticals, HLS Pharmaceuticals, Pendopharm and Pfizer; and minor equity interest from DalCor Pharmaceuticals. The other authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-76245-9.
References
- 1.Abbaspour, N., Hurrell, R. & Kelishadi, R. Review on iron and its importance for human health. J. Res. Med. Sci. 19, 1 (2014). [PMC free article] [PubMed] [Google Scholar]
- 2.Nairz, M. & Weiss, G. Iron in infection and immunity. Mol. Asp. Med. 75, 864. 10.1016/j.mam.2020.100864 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Paul, B. T., Manz, D. H., Torti, F. M. & Torti, S. V. Mitochondria and iron: Current questions. Expert Rev. Hematol. 10, 47. 10.1080/17474086.2016.1268047 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Emerit, J., Beaumont, C. & Trivin, F. Iron metabolism, free radicals, and oxidative injury. Biomed. Pharmacother. 55, 3. 10.1016/S0753-3322(01)00068-3 (2001). [DOI] [PubMed] [Google Scholar]
- 5.Basuli, D., Stevens, R. G., Torti, F. M. & Torti, S. V. Epidemiological associations between iron and cardiovascular disease and diabetes. Front. Pharmacol. 5, 117. 10.3389/fphar.2014.00117 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Medeiros, M. S. et al. Iron and oxidative stress in Parkinson’s disease: An observational study of injury biomarkers. PLoS ONE 11, e0146129 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moliner, P. et al. Association between norepinephrine levels and abnormal iron status in patients with chronic heart failure: Is iron deficiency more than a comorbidity? J. Am. Heart Assoc. 8, 887 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang, L., Li, C., Chen, X., Li, S. & Shang, H. Abnormal serum iron-status indicator changes in amyotrophic lateral sclerosis (ALS) patients: A meta-analysis. Front. Neurol. 11, 380. 10.3389/fneur.2020.00380 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo, S., Mao, X., Li, X. & Ouyang, H. Association between iron status and incident coronary artery disease: a population based-cohort study. Sci. Rep. 12, 1 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eftekhari, M. H., Mozaffari-Khosravi, H., Shidfar, F. & Zamani, A. Relation between body iron status and cardiovascular risk factors in patients with cardiovascular disease. Int. J. Prev. Med. 4, 1 (2013). [PMC free article] [PubMed] [Google Scholar]
- 11.De Das, S., Krishna, S. & Jethwa, A. Iron status and its association with coronary heart disease: Systematic review and meta-analysis of prospective studies. Atherosclerosis 238, 296–303 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Sempos, C. T. et al. Serum ferritin and death from all causes and cardiovascular disease: The NHANES II Mortality Study. National Health and Nutrition Examination Study. Ann. Epidemiol. 10, 441–448 (2000). [DOI] [PubMed] [Google Scholar]
- 13.Knuiman, M. W., Divitini, M. L., Olynyk, J. K., Cullen, D. J. & Bartholomew, H. C. Serum ferritin and cardiovascular disease: A 17-year follow-up study in Busselton, Western Australia. Am. J. Epidemiol. 158, 144–149 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Njajou, O. T. et al. Heritability of serum iron, ferritin and transferrin saturation in a genetically isolated population, the Erasmus Rucphen Family (ERF) study. Hum. Hered. 61, 1 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Sayed-Tabatabaei, F. A. et al. Heritability of the function and structure of the arterial wall: Findings of the Erasmus Rucphen family (ERF) study. Stroke 36, 1 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Moksnes, M. R. et al. Genome-wide meta-analysis of iron status biomarkers and the effect of iron on all-cause mortality in HUNT. Commun. Biol. 5, 591 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bell, S. et al. A genome-wide meta-analysis yields 46 new loci associating with biomarkers of iron homeostasis. Commun. Biol. 4, 1 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tada, H., Fujino, N., Hayashi, K., Kawashiri, M. & Takamura, M. Human genetics and its impact on cardiovascular disease. J. Cardiol. 79, 5. 10.1016/j.jjcc.2021.09.005 (2022). [DOI] [PubMed] [Google Scholar]
- 19.Bellou, E., Stevenson-Hoare, J. & Escott-Price, V. Polygenic risk and pleiotropy in neurodegenerative diseases. Neurobiol. Dis. 142, 953. 10.1016/j.nbd.2020.104953 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baker, E. et al. What does heritability of Alzheimer’s disease represent? PLoS ONE 18, e0281440 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kezele, T. G. & Ćurko-Cofek, B. Age-related changes and sex-related differences in brain iron metabolism. Nutrients 12, 601. 10.3390/nu12092601 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Badenhorst, C. E., Forsyth, A. K. & Govus, A. D. A contemporary understanding of iron metabolism in active premenopausal females. Front. Sports Act. Living 4, 937. 10.3389/fspor.2022.903937 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ryan, B. J., Charkoudian, N. & McClung, J. P. Consider iron status when making sex comparisons in human physiology. J. Appl. Physiol. 132, 1 (2022). [DOI] [PubMed] [Google Scholar]
- 24.Shock, N., Greulick, R. & Andres, R. Normal Human Aging: The Baltimore Study of Aging 84 (NIH Publication, 1984). [Google Scholar]
- 25.Benyamin, B. et al. Novel loci affecting iron homeostasis and their effects in individuals at risk for hemochromatosis. Nat. Commun. 5, 4926 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zawistowski, M. et al. The Michigan genomics initiative: A biobank linking genotypes and electronic clinical records in Michigan Medicine patients. Cell Genom. 3, 100257 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pilia, G. et al. Heritability of cardiovascular and personality traits in 6,148 Sardinians. PLoS Genet. 2, 1 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krokstad, S. et al. Cohort profile: The HUNT study, Norway. Int. J. Epidemiol. 42, 968–977 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Willer, C. J., Li, Y. & Abecasis, G. R. METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hinrichs, A. S. et al. The UCSC genome browser database: Update 2006. Nucleic Acids Res. 34, D590–D598 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watanabe, K., Taskesen, E., van Bochoven, A. & Posthuma, D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 8, 1826 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wightman, D. P. et al. A genome-wide association study with 1,126,563 individuals identifies new risk loci for Alzheimer’s disease. Nat. Genet. 53, 1276–1282 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blauwendraat, C. et al. Investigation of autosomal genetic sex differences in Parkinson’s disease. Ann. Neurol. 90, 35–42 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Rheenen, W. et al. Common and rare variant association analyses in amyotrophic lateral sclerosis identify 15 risk loci with distinct genetic architectures and neuron-specific biology. Nat. Genet. 53, 1636–1648 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aragam, K. G. et al. Discovery and systematic characterization of risk variants and genes for coronary artery disease in over a million participants. Nat. Genet. 54, 1803–1815 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shah, S. et al. Genome-wide association and Mendelian randomisation analysis provide insights into the pathogenesis of heart failure. Nat. Commun. 11, 163 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mishra, A. et al. Stroke genetics informs drug discovery and risk prediction across ancestries. Nature 611, 115–123 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Graham, S. E. et al. The power of genetic diversity in genome-wide association studies of lipids. Nature 600, 675–679 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bulik-Sullivan, B. et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 47, 1236 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Werme, J., van der Sluis, S., Posthuma, D. & de Leeuw, C. A. An integrated framework for local genetic correlation analysis. Nat. Genet. 54, 274–282 (2022). [DOI] [PubMed] [Google Scholar]
- 41.Kunkle, B. W. et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat. Genet. 51, 414–430 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sollis, E. et al. The NHGRI-EBI GWAS Catalog: Knowledgebase and deposition resource. Nucleic Acids Res. 51, D977–D985 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kamat, M. A. et al. PhenoScanner V2: An expanded tool for searching human genotype-phenotype associations. Bioinformatics 35, 4851–4853 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hemani, G. et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife 7, 1 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burgess, S. & Thompson, S. G. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur. J. Epidemiol. 32, 377–389 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Verbanck, M., Chen, C.-Y., Neale, B. & Do, R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 50, 693–698 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Byrne, R. P. et al. Sex-specific risk loci and modified MEF2C expression in ALS. MedRxiv. 10.1101/2024.05.25.24307829 (2024).39006447 [Google Scholar]
- 48.Shi, H., Mancuso, N., Spendlove, S. & Pasaniuc, B. Local genetic correlation gives insights into the shared genetic architecture of complex traits. Am. J. Hum. Genet. 101, 1 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Rheenen, W., Peyrot, W. J., Schork, A. J., Lee, S. H. & Wray, N. R. Genetic correlations of polygenic disease traits: From theory to practice. Nat. Rev. Genet. 20, 1. 10.1038/s41576-019-0137-z (2019). [DOI] [PubMed] [Google Scholar]
- 50.Sheppe, A. E. F. & Edelmann, M. J. Roles of eicosanoids in regulating inflammation and neutrophil migration as an innate host response to bacterial infections. Infect. Immunity 89, 21. 10.1128/IAI.00095-21 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen, X. & Holtzman, D. M. Emerging roles of innate and adaptive immunity in Alzheimer’s disease. Immunity 55, 16. 10.1016/j.immuni.2022.10.016 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ennerfelt, H. E. & Lukens, J. R. The role of innate immunity in Alzheimer’s disease. Immunol. Rev. 297, 896. 10.1111/imr.12896 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wessling-Resnick, M. Iron homeostasis and the inflammatory response. Annu. Rev. Nutr. 30, 804. 10.1146/annurev.nutr.012809.104804 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mu, Q. et al. The role of iron homeostasis in remodeling immune function and regulating inflammatory disease. Sci. Bull. 66, 10. 10.1016/j.scib.2021.02.010 (2021). [DOI] [PubMed] [Google Scholar]
- 55.Horton, R. et al. Gene map of the extended human MHC. Nat. Rev. Genet. 5, 1489. 10.1038/nrg1489 (2004). [DOI] [PubMed] [Google Scholar]
- 56.Cho, Y. K. & Hee Jung, C. Hdl-c and cardiovascular risk: You don’t need to worry about extremely high HDL-C levels. J. Lipid Atheroscler. 10, 57. 10.12997/jla.2021.10.1.57 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ali, K. M., Wonnerth, A., Huber, K. & Wojta, J. Cardiovascular disease risk reduction by raising HDL cholesterol—Current therapies and future opportunities. Br. J. Pharmacol. 167, 1. 10.1111/j.1476-5381.2012.02081.x (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Das, S. K. et al. Females are protected from iron-overload cardiomyopathy independent of iron metabolism: Key role of oxidative stress. J. Am. Heart Assoc. 6, 1 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shapiro, J. S., Chang, H. C. & Ardehali, H. Iron and sex cross paths in the heart. J. Am. Heart Assoc. 6, 1 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li, X. et al. Iron deficiency and overload in men and woman of reproductive age, and pregnant women. Reprod. Toxicol. 118, 1 (2023). [DOI] [PubMed] [Google Scholar]
- 61.Harrison-Findik, D. D. Gender-related variations in iron metabolism and liver diseases. World J. Hepatol. 2, 1 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Anderson, G. J. & Bardou-Jacquet, E. Revisiting hemochromatosis: Genetic vs phenotypic manifestations. Ann. Transl. Med. 9, 1 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hamad, M., Bajbouj, K. & Taneera, J. The case for an estrogen-iron axis in health and disease. Exp. Clin. Endocrinol. Diabetes 128, 1677. 10.1055/a-0885-1677 (2020). [DOI] [PubMed] [Google Scholar]
- 64.Gabrielsen, J. S., Lamb, D. J. & Lipshultz, L. I. Iron and a man’s reproductive health: The good, the bad, and the ugly. Curr. Urol. Rep. 19, 1. 10.1007/s11934-018-0808-x (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bachman, E. et al. Testosterone suppresses hepcidin in men: A potential mechanism for testosterone-induced erythrocytosis. J. Clin. Endocrinol. Metab. 95, 1 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hayes, M. T. Parkinson’s disease and Parkinsonism. Am. J. Med. 132, 802–807 (2019). [DOI] [PubMed] [Google Scholar]
- 67.Olanow, C. W. & Tatton, W. G. Etiology and pathogenesis of Parkinson’s disease. Annu. Rev. Neurosci. 22, 123–144 (1999). [DOI] [PubMed] [Google Scholar]
- 68.Grubić Kezele, T. & Ćurko-Cofek, B. Age-related changes and sex-related differences in brain iron metabolism. Nutrients 12, 1 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shi, L. et al. The association of iron and the pathologies of Parkinson’s diseases in MPTP/MPP+-induced neuronal degeneration in non-human primates and in cell culture. Front. Aging Neurosci. 11, 215 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.de Souza Ferreira, L. P. et al. Sex differences in Parkinson’s disease: An emerging health question. Clinics 77, 121. 10.1016/j.clinsp.2022.100121 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Russillo, M. C. et al. Sex differences in Parkinson’s disease: From bench to bedside. Brain Sci. 12, 917. 10.3390/brainsci12070917 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cerri, S., Mus, L. & Blandini, F. Parkinson’s disease in women and men: What’s the difference? J. Parkinsons Dis. 9, 501–515 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jurado-Coronel, J. C. et al. Sex differences in Parkinson’s disease: Features on clinical symptoms, treatment outcome, sexual hormones and genetics. Front. Neuroendocrinol. 50, 18–30 (2018). [DOI] [PubMed] [Google Scholar]
- 74.Song, Y.-J. et al. The effect of estrogen replacement therapy on Alzheimer’s disease and Parkinson’s disease in postmenopausal women: A meta-analysis. Front. Neurosci. 14, 157 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Paterek, A. et al. Systemic iron deficiency does not affect the cardiac iron content and progression of heart failure. J. Mol. Cell Cardiol. 159, 16–27 (2021). [DOI] [PubMed] [Google Scholar]
- 76.Hirsch, V. G. et al. Cardiac iron concentration in relation to systemic iron status and disease severity in non-ischaemic heart failure with reduced ejection fraction. Eur. J. Heart Fail. 22, 2038–2046 (2020). [DOI] [PubMed] [Google Scholar]
- 77.Schipper, H. M. Neurodegeneration with brain iron accumulation—Clinical syndromes and neuroimaging. Biochim. Biophys. Acta Mol. Basis Dis. 1822, 16. 10.1016/j.bbadis.2011.06.016 (2012). [DOI] [PubMed] [Google Scholar]
- 78.Wolfe, D., Dudek, S., Ritchie, M. D. & Pendergrass, S. A. Visualizing genomic information across chromosomes with PhenoGram. BioData Min. 6, 1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Information on how to access the iron summary statistics for Benyamin et al.25 and Moksnes et al.16 and the 11 cardiovascular and neurodegenerative summary statistics used here are publicly available with access information provided in the respective publications. Baltimore Longitudinal Study of Aging (BLSA): Data for BLSA can be accessed through submission of a project proposal at https://www.blsa.nih.gov/. Michigan Genomics Initiative (MGI): The individual level genetic and clinical MGI data reported in this study cannot be deposited in a public repository because of patient confidentiality. Summary statistics can be requested for download by emailing a completed Data Use Agreement form available at https://precisionhealth.umich.edu/our-research/documents-for-researchers/ to phdatahelp@umich.edu. Montreal Heart Institute Biobank: The individual level genetic and clinical data reported in this study cannot be deposited in a public repository because of patient confidentiality. Summary statistics can be requested through the corresponding author. SardiNIA: Serum iron summary statistics from the SardiNIA cohort are available upon request. Summary statistics from the iron meta-analysis performed in the context of this work are publicly available at https://my.locuszoom.org/gwas/111911/?token=a254c600e06a417a9d1e8527cb620b31, https://my.locuszoom.org/gwas/904129/?token=ff44329da5de47fb81fc657d967e10b7 (male-only) and https://my.locuszoom.org/gwas/835579/?token=d16e2ce5101a44fd8078df5f18a08e45 (female-only).
Code is available in the following GitHub repository: https://github.com/GaglianoTaliun-Lab/iron_cardio-neuro.