Abstract
We determined that the highly pathogenic avian reovirus strain 176 (ARV-176) possesses an enhanced ability to establish productive infections in HD-11 avian macrophages compared to avian fibroblasts. Conversely, the weakly pathogenic strain ARV-138 shows no such macrophagotropic tendency. The macrophage infection capability of the two viruses did not reflect differences in the ability to either induce or inhibit nitric oxide production. Moderate increases in the ARV-138 multiplicity of infection resulted in a concomitant increase in macrophage infection, and under such conditions the kinetics and extent of the ARV-138 replication cycle were equivalent to those of the highly infectious ARV-176 strain. These results indicated that both viruses are apparently equally capable of replicating in an infected macrophage, but they differ in the ability to establish productive infections in these cells. Using a genetic reassortant approach, we determined that the macrophagotropic property of ARV-176 reflects a post-receptor-binding step in the virus replication cycle and that the ARV-176 M2 genome segment is required for efficient infection of HD-11 cells. The M2 genome segment encodes the major μ-class outer capsid protein (μB) of the virus, which is involved in virus entry and transcriptase activation, suggesting that a host-specific influence on ARV entry and/or uncoating may affect the likelihood of the virus establishing a productive infection in a macrophage cell.
The avian reoviruses (ARV) differ from the prototypical mammalian reoviruses (MRV) based on several biological properties other than just their distinct host ranges. Unlike MRV, ARV is naturally pathogenic in its avian host, lacks hemagglutinating ability, and is one of the few nonenveloped viruses capable of inducing syncytium formation in infected cell cultures and in vivo (14, 18, 24, 28). Although ARV pathogenesis has been extensively described (5, 6, 15, 34), the viral factors that influence ARV-host cell interactions and pathogenesis remain poorly understood.
We have been investigating two ARV strains that possess distinct pathogenic and syncytium-inducing potentials. Previous results demonstrated that ARV-176 is highly pathogenic relative to ARV-138 in an embryonated egg model of virus pathogenesis, an attribute that correlates with the relative fusogenic capability of the virus (8). Both viruses infect and replicate with equal efficiency in cultured fibroblast cells, they display 94 to 98% amino acid sequence identity in the three sequenced S-class genome segment-encoded proteins (7a), and all 10 of their individual genome segments can be resolved by electrophoretic analysis (8); these properties make these two ARV strains ideal parental virus candidates for genetic and molecular approaches to identify viral determinants of host interaction and pathogenicity. We previously used a genetic reassortant approach to reveal that the S1 genome segment of ARV-176 is solely responsible for the syncytium-inducing property of the virus (8). Subsequent molecular and biochemical studies confirmed the role of the S1 genome segment and its encoded 10-kDa protein in cell fusion (30). Genetic studies also revealed that while the S1 genome segment, and by inference syncytium formation, makes a significant contribution to the pathogenic potential of ARV, other genetically encoded viral properties also contribute to ARV-176 pathogenicity (8). Aside from the distinct pathogenic and syncytium-inducing characteristics of these two ARV strains, no other distinguishing viral attributes have been identified that could influence virus-host interactions.
Macrophages may be a preferred target cell population for ARV replication (38), which could conceivably contribute to the transient immunosuppression observed following ARV infection (25). Furthermore, there is evidence suggesting virus strain-specific differences in the ability of ARV to infect cultured macrophages (23). However, the relationship between macrophage infection and pathogenesis remains unclear (12, 38). These studies prompted us to evaluate ARV-176 and ARV-138 macrophage interactions using an avian macrophage cell line, HD-11. ARV-176 was approximately 25-fold more efficient at establishing productive infections in macrophage versus fibroblast cell cultures, while ARV-138 showed no such macrophagotropic property. Genetic studies indicated that the ARV-176 M2 genome segment, which encodes the major μ-class outer capsid protein of the virus, is necessary for enhanced macrophage infection. In view of the role of this outer capsid protein in virus entry and transcriptase activation, differences in the endosomal entry pathway between fibroblasts and macrophages could contribute to the observed differences in ARV macrophage infection.
MATERIALS AND METHODS
Virus strains and cells.
ARV-176 and ARV-138 have been previously described (8). Both strains were plaque purified and amplified to passage 4 using a multiplicity of infection of 0.01 in the continuous quail fibroblast cell line QM5. The QM5 cell line has been previously described (8). HD-11 is a continuous chicken macrophage cell line developed by transforming bone marrow-adherent cells with the replication defective avian retrovirus MC29 (1). The HD-11 cells were obtained from John Adams (University of California, Los Angeles) and were grown in RPMI 1640 medium supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin at 37°C.
Focus-forming assay.
The relative infectivities of ARV-176 and ARV-138 in QM5 fibroblasts and HD-11 macrophages were compared and quantified using a focus-forming assay. Concentrated virus stocks were obtained by differential centrifugation of infected QM5 cell lysates as previously described (9). Serial virus dilutions were used to infect QM5 and HD-11 cell monolayers, and the monolayers were fixed with methanol at 9 to 16 h postinfection (hpi). Fixed monolayers were stained using polyclonal rabbit antiserum raised against virus structural proteins and a secondary goat anti-rabbit immunoglobulin G conjugated with alkaline phosphatase (Life Technologies) as reported previously (9).
Quantification of infectious foci was achieved using the computer software Image-Pro Plus (version 4.0). Duplicate stained cell monolayers were observed by bright-field microscopy at a magnification of ×100 using a Nikon Diaphot-TMD inverted microscope and were photographed with a Sony DXC-950 color video camera. Five separate fields per duplicate dilution were quantified. The experiment was repeated at least three times for each virus. Captured images were subjected to background spatial filtering set to bright at 100 pixels to obtain an equal distribution of light intensity throughout each image capture. To avoid counting objects smaller than individual cells the minimum area filter range was set to 5 pixels. An 8-bit gray scale was selected for quantification. The intensity range selection (0 to 255) was set on manual and to the level which identified the areas in the cell monolayers stained with antireovirus antibodies. Twofold dilutions were used to ensure a linear dose response between foci counted and viral concentration. The relative infectivity was reported as the HD-11/QM5 focus-forming ratio.
The reassortant viruses were similarly analyzed to determine their relative infectivities. A one-way analysis of variance using Tukey's pairwise comparisons was used to analyze the relative infectivities of the parental and reassortant viruses and to assign the viruses to one of three phenotypic groups. The family error rate (the probability that there are not three distinct groups) had a P value of <0.05, and the individual error rate (the probability that any given virus is not in a specific group) had a P value of <0.0001. The M2 genome segment was identified as a predictor of macrophage infection efficiency using a generalized linear model to fit a regression model predicting the log of the relative infectivity (P < 0.001).
Progeny virus yield and viral protein synthesis.
To compare the replication abilities per infected cell of ARV-176 and ARV-138 in QM5 and HD-11 cells, monolayer cultures were infected with the minimum dose of virus required for a >90% infection as determined by immunostaining. After attachment for 1 h at 37°C, the inoculum was removed and the monolayers were washed three times with warm phosphate-buffered saline followed by the overlay of 1% fresh medium. Duplicate wells containing infected monolayers were harvested at 24 or 48 hpi by scraping the cells into the culture medium, cells were disrupted by three freeze-thaw cycles, and the total infectious progeny virus titer was determined by plaque assay on QM5 cell monolayers as previously described (9).
The kinetics and extent of viral protein synthesis in monolayers of HD-11 and QM5 cells, infected as described above, were analyzed by [35S]methionine pulse-labeling and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as previously described (9).
Greiss assay.
A standard assay was used to quantify the levels of nitrite present in the culture medium as an indicator of the relative inducible nitric oxide synthase (iNOS) activity (11). To measure iNOS activity in infected or uninfected HD-11 cells, monolayers were incubated in the absence or presence of 500 ng of lipolysaccharide (LPS) per ml for 6 h. Cells were then either mock infected or infected with equivalent QM5 focus-forming units of ARV-176 or ARV-138 under conditions that gave a >90% infection by ARV-176. The monolayers were incubated with 500 μl of phenol red-free RPMI 1640 at 37°C for 12 to 16 h. Following incubation, 100 μl of Greiss reagent, composed of a 1:1 mixture of 1% sulfanilamide in 2.5% phosphoric acid and 0.1% naphthylenediamine dihydrochloride in 2.5% phosphoric acid, was added to 100 μl of sample supernatants and the absorbance at 550 nm was measured. To determine the concentration of nitrite production from HD-11 cells, a sodium nitrite standard curve with a range of 500 to 0.5 μM was created using a 5 mM sodium nitrite stock solution diluted in phenol red-free RPMI 1640.
Isolation of ARV reassortants.
ARV reassortants were generated by slight modifications of standard techniques used to make MRV reassortants (13). Briefly, subconfluent monolayers of QM5 cells in 24-well tissue culture plates were infected with a mixture of ARV-138 and ARV-176, at multiplicities of infection of 5:5, 12:3, and 3:12 PFU per cell per clone. Infected cells were incubated at 37°C for 24 h (approximately one round of replication), freeze-thawed twice, and disrupted with an ultrasonic sonicator to dissociate clumps. Serial dilutions of viral lysates were prepared and plated under medium 199–1% agar as described previously (9). Individual plaques representing putative reassortants separated by at least 1 cm were picked and amplified through two passages.
RNA analysis.
Each of the twice-passaged putative reassortant stock clones was used to infect subconfluent QM5 monolayers in P100 dishes. Cytoplasmic extracts were prepared from each infection as described previously (13). Briefly, cells were harvested when they showed a >50% cytopathic effect, nuclei were removed, and double-stranded RNA was phenol-chloroform extracted from the cytoplasmic fractions. RNA was precipitated with ethanol, dried, and resuspended in electrophoresis sample buffer (0.24 M Tris [pH 6.8], 1.5% dithiothreitol, 1% SDS). Samples were heated to 65°C and resolved by SDS-PAGE (16.0 by 16.0 by 0.1 cm gel) under standard Laemmli conditions (typical conditions were 12.5% acrylamide gels electrophoresed at 12 mA for 68 h). All reassortants were analyzed on multiple gels and under different electrophoretic conditions (7, 10, and 12.5% acrylamide gels, altered times of electrophoresis) to obtain maximal resolution of the individual L-, M-, and S-class genome segments. Gels were then stained with ethidium bromide, and RNA was visualized on a UV light box and photographed with Polaroid film. Alternatively, images were captured with a Bio-Rad Gel Doc 2000 system and manipulated with Adobe Photoshop.
RESULTS
Strain-specific differences in ARV infection of HD-11 macrophage cultures.
The HD-11 myelomonocytic cell line is a chicken bone marrow-derived cell line obtained by transformation with the replication-defective avian retrovirus MC29 (1). This cell line displays many macrophage-like properties, including high phagocytic capability and expression of Fc receptors, secretion of interleukin-1B, macrophage inflammatory protein 1α, interleukin-8, and tumor necrosis factor alpha (TNF-α), and iNOS activity following stimulation with TNF-α and/or LPS (1, 31, 33, 39, 40). We used the HD-11 cell line as a model system to assess ARV-176 and ARV-138 macrophage interactions using a focus-forming assay.
Both viruses were first standardized using the focus-forming assay in the permissive QM5 quail fibroblast cell line. QM5 monolayers were infected with serial dilutions of ARV-176 or ARV-138 virus stocks, and monolayers were fixed and immunostained at 16 hpi using ARV-specific antiserum (Fig. 1). Previous results indicated that this time point is prior to the release of progeny virus particles (9); therefore, the antigen-positive foci represent the primary foci of infection. As previously reported (8) and as shown in Fig. 1a and b, ARV-176 is more fusogenic than ARV-138, resulting in larger, though in equivalent numbers, syncytial foci of infection in the infected QM5 monolayers.
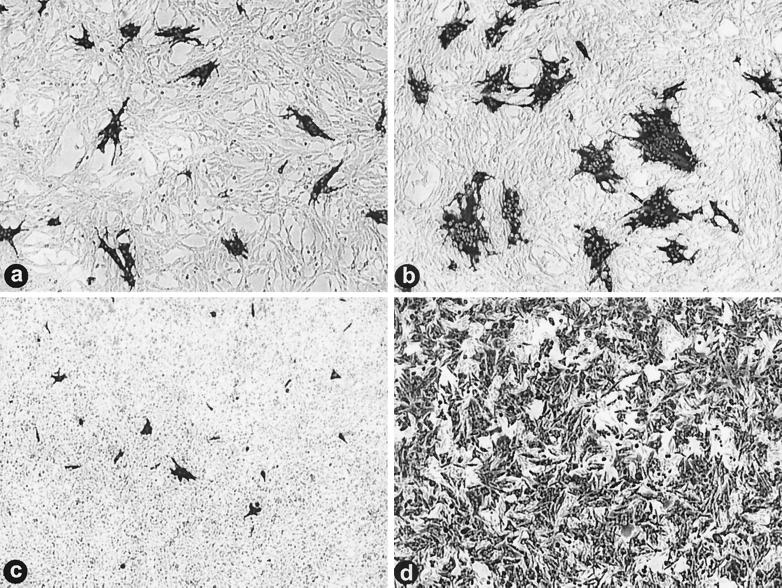
FIG. 1.
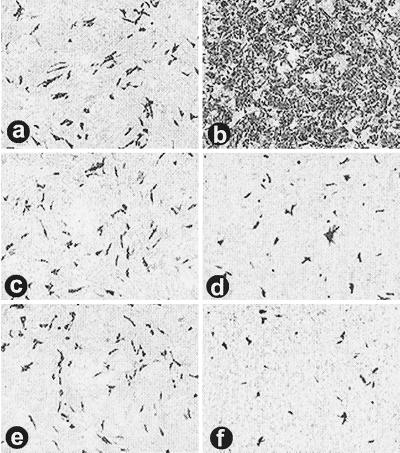
ARV-176 shows a predilection for macrophages. Diluted stocks of ARV-138 and ARV-176 were used to infect QM5 fibroblasts (a and b, respectively). Infected cell monolayers were incubated at 37°C and fixed at 16 hpi before being immunostained using a reovirus-specific rabbit polyclonal antiserum and goat anti-rabbit immunoglobulin G conjugated with alkaline phosphatase to detect viral foci of infection. Virus dilutions that gave equivalent numbers of foci in QM5 fibroblasts were then used to infect HD-11 macrophages (c and d) that were similarly fixed and immunostained at 9 hpi.
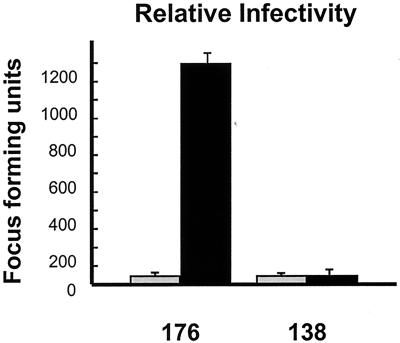
Infection of HD-11 macrophages using virus stocks diluted to give equivalent QM5 focus-forming units/ml gave very different results. While equivalent aliquots of ARV-138 gave approximately equal numbers of infectious foci in both QM5 and HD-11 cells (Fig. 1a and c), ARV-176 displayed a dramatic increase in the number of foci established in the HD-11 cells (Fig. 1d). For presentation purposes, the HD-11 cells were fixed and stained at 9 hpi to minimize the overlap of individual foci due to syncytium formation. Extended incubation to 16 hpi before fixation did not contribute to an increase in the numbers of foci observed with ARV-138 (data not shown). The relative infectivities of the two viruses in QM5 and HD-11 cells were quantified using the focus-forming assay and serial virus dilutions, as described in Materials and Methods (Fig. 2). In repeated experiments, the relative infectivity of ARV-138 in HD-11 versus QM5 cells ranged from 0.8 to 1.2. Conversely, ARV-176 displayed a consistent and reproducible HD-11/QM5 relative infectivity ratio of 25 to 30:1. These results clearly indicated that ARV-176 preferentially establishes productive infections in macrophage cell cultures.
FIG. 2.
Relative infectivities of ARV-176 and ARV-138 in macrophages versus fibroblasts. Both viruses were serially diluted and used to infect monolayers of QM5 fibroblasts (gray bars). Virus dilutions that gave equivalent numbers of foci of infection in QM5 cells were similarly used to infect HD-11 macrophages (black bars). Infected monolayers were immunostained to detect viral foci of infection as described for Fig. 1. The average number of foci of infection per field at a magnification of ×100 was determined after correcting for the relative virus dilution, as described in Materials and Methods. Results are presented as means and standard errors from a representative experiment.
ARV-176 and ARV-138 do not differentially stimulate or inhibit NO production.
Nitric oxide (NO) is a reactive nitrogen species produced by iNOS in macrophages activated by TNF-α and/or LPS. NO is a potential potent inhibitor of the replication of numerous viruses, including ARV replication in HD-11 cells (25, 26). Studies have also shown strain-specific differences in the ability of avian influenza viruses to inhibit NO production in HD-11 cells, a property that correlated with virus virulence and viral replication (21). It therefore seemed plausible that the observed difference in the ability of ARV-176 and ARV-138 to establish productive infections in HD-11 macrophages might reflect a strain-specific difference in the activation or inhibition of iNOS activity.
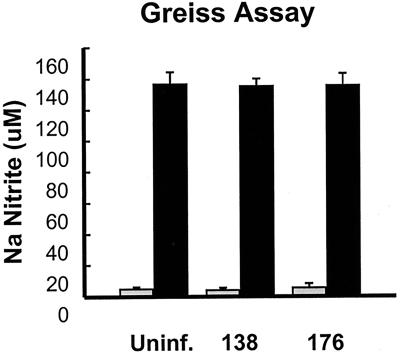
To test this hypothesis, we examined iNOS activity in ARV-infected HD-11 cells by measuring the accumulation of sodium nitrite, the stable end product of NO reduction, in the culture medium. Uninfected or ARV-infected HD-11 cell monolayers all showed equivalent low basal levels of sodium nitrite in the culture medium, indicating that neither virus stimulates NO production (Fig. 3). We also examined whether either virus could inhibit NO production in activated HD-11 cells. Incubation of HD-11 cells in the presence of LPS stimulated iNOS activity, as indicated by the high levels of sodium nitrite detected in culture supernatants at 12 h posttreatment (Fig. 3) and as previously reported (33). Neither virus was capable of inhibiting NO production by activated macrophages (Fig. 3), and the replication of both viruses was equally impaired by LPS treatment (data not shown), as previously reported for ARV-176 (26). Therefore, the differential ability of these two ARV strains to establish productive infections in HD-11 macrophages is not related to NO production and/or inhibition.
FIG. 3.
Neither ARV-176 nor ARV-138 differentially affects NO production by infected macrophages. HD-11 cell monolayers were incubated for 6 h in the presence (black bars) or absence (gray bars) of LPS at 500 ng/ml followed by infection with equivalent concentrations of QM5 focus-forming units of ARV-176 or ARV-138 per milliliter. At 12 hpi the supernatants were harvested and assayed for the presence of nitrite using standard protocols. Nitrite concentrations were determined relative to a standard sodium nitrite curve. Uninf., uninfected cells.
Characterization of ARV infection and replication in HD-11 macrophages.
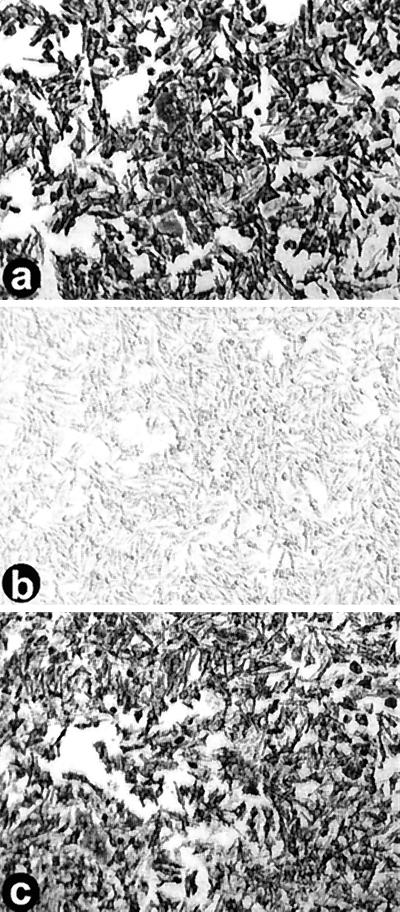
One mechanism that might account for the differences in virus infectivity observed in the two cell types is that the HD-11 macrophage line consists of a mixed population of cells, some of which are exclusively susceptible to infection by ARV-176. Such a situation has been reported in other virus systems where, for example, the stage of macrophage differentiation or the expression of cell surface receptors on a subpopulation of macrophages dictates the outcome of virus infection (4, 20, 35). To test this hypothesis, HD-11 cells were infected with various amounts of ARV-138. Stepwise twofold increases in the QM5-standardized ARV-138 inoculum resulted in concomitant stepwise increases in the number of foci of infection in both the QM5 and HD-11 cells. Essentially complete infection by ARV-138 of the HD-11 cell monolayer was achieved by a 16- to 32-fold increase in the concentration of the standardized inoculum (Fig. 4). Clearly, both ARV-138 and ARV-176 are capable of establishing productive infections in all of the cells present in an HD-11 cell monolayer, suggesting that there is no distinct subpopulation of HD-11 cells that is specifically permissive for ARV-176 infection. Rather, the balance between host cell defenses and virus infection appears to be tipped more in favor of the virus in the case of ARV-176 such that this strain of ARV is more efficient at establishing a productive infection in any given macrophage cell.
FIG. 4.
ARV-138 is capable of complete infection of HD-11 macrophage monolayers. HD-11 macrophages were left uninfected (b) or were infected with increasing concentrations of standardized QM5 focus-forming units of ARV-176 (a) or ARV-138 (c) per milliliter. Monolayers were fixed and immunostained as described for Fig. 1 to reveal viral foci of infection. The ARV-138 inoculum in panel c corresponds to a 32-fold increase in the QM5 focus-forming units compared to the ARV-176 inoculum in panel a.
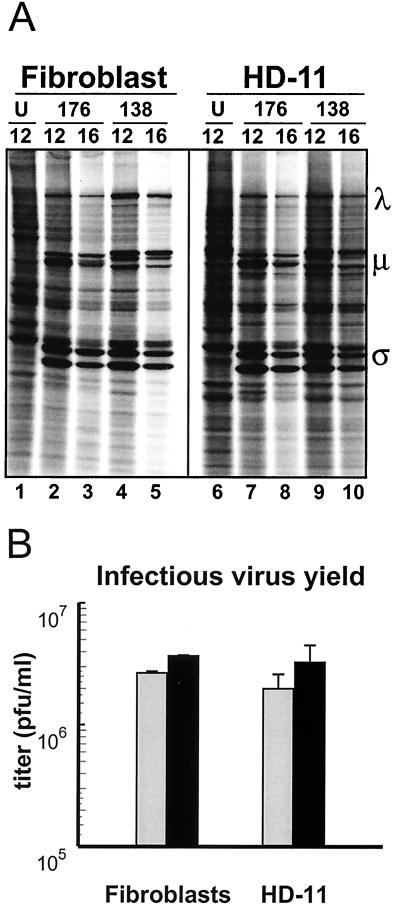
The immunostaining protocol used in the focus-forming assay indicated that both viruses could establish equivalent productive infections in macrophage cells, at least to the point where robust virus translation occurs. It was not clear, however, whether both viruses were equally capable of completing their replication cycle within a particular infected macrophage. To more clearly assess the kinetics and extent of the virus replication cycle within an infected cell, virus translation and progeny virus production were evaluated in QM5 and HD-11 cell cultures infected with the minimum virus inoculum required for complete infection of the monolayer, as indicated in Fig. 4. The results indicated that under these infection conditions, there were no obvious differences in either the rate or extent of virus translation or progeny virus production when comparing the various virus-cell combinations (Fig. 5). The kinetics and extent of virus translation were equivalent when comparing either one virus to the other or one particular virus in either cell line with predominant viral proteins evident at 12 hpi and diminishing by 16 hpi (Fig. 5A). The same situation applied when cells were examined at 4 hpi (data not shown). Similar to the results observed with virus translation, neither virus showed preferential progeny virus production in a particular cell type under infection conditions sufficient to infect the majority of cells in the monolayer (Fig. 5B). The previously reported enhanced replicative ability of ARV-138 versus ARV-176 in quail fibroblasts (8) was conserved in HD-11 cells; ARV-138 progeny titers were approximately threefold higher than those of ARV-176 (Fig. 5B). These results indicated that ARV-176 possesses an enhanced ability to establish a productive infection in any given macrophage cell but not an enhanced ability to replicate within a productively infected cell.
FIG. 5.
ARV-176 and ARV-138 protein synthesis and replication are similar in fibroblasts and macrophages. (A) Cell monolayers of quail QM5 fibroblasts or HD-11 macrophages were left uninfected (U) or infected with ARV-176 (176) or ARV-138 (138) using the minimum concentrations of virus inocula required for complete infection of the monolayer (see Fig. 4). Cultures were pulse-labeled for 1 h with [35S]methionine at 12 and 16 hpi, and the radiolabeled cell lysates were fractioned by SDS-PAGE and detected by autoradiography. The locations of the major λ, μ, and ς classes of viral proteins are indicated on the right. (B) Monolayers of quail QM5 fibroblasts or HD-11 macrophage cells were infected with ARV-176 (gray bars) or ARV-138 (black bars) as described for panel A. Infected cultures were harvested at 48 hpi, and the yield of infectious progeny virions was determined by a plaque assay on QM5 cell monolayers. Results are presented as means and standard errors from a representative experiment.
Reassortant analysis implicates the ARV-176 M2 genome segment in HD-11 infectivity.
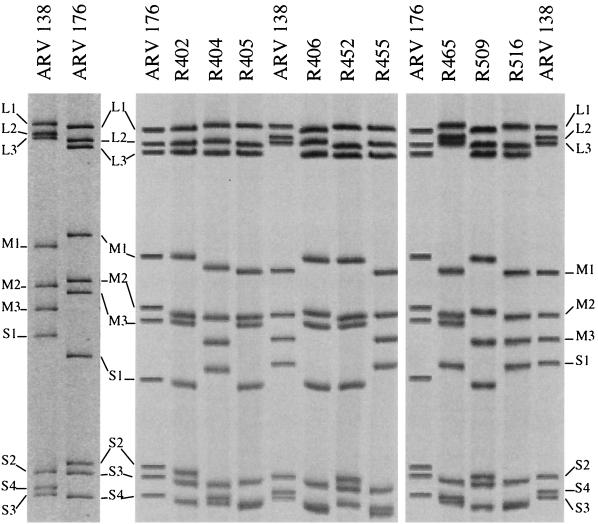
We undertook a genetic approach to identify the viral factors involved in the distinct macrophagotropic properties of ARV-138 and ARV-176. A panel of 29 reassortant viruses was isolated following coinfection of QM5 cells by the two virus strains. A select panel of the reassortant genomes is shown in Fig. 6. The assignment of genome segments to a particular parental virus was accomplished by repeated analysis under different electrophoretic conditions designed to specifically resolve individual L-, M-, and S-class genome segments (see Materials and Methods). Viral stocks were prepared from each of the reassortants, and their infectivities were standardized using the QM5 focus-forming assay as described above. Serial dilutions of the standardized inocula were then used to infect HD-11 cell monolayers, and the relative focus-forming propensity of each clone in both cell types was determined (Fig. 7).
FIG. 6.
Genome segment profiles of parental and reassortant ARV virions. Viral double-stranded RNA was isolated from concentrated virus stocks of the parental and reassortant viruses, and individual genome segments were resolved on SDS–12.5% PAGE gels. Gels were stained with ethidium bromide and visualized under UV illumination. Images were captured and enhanced with Adobe Photoshop, and negative images were printed. The parental ARV-176 and ARV-138 genome segment profiles are shown by themselves in the left-hand panel and included as markers on the gels that contain the reassortant genomes. Only a selected number of reassortants used in this study are shown.
FIG. 7.
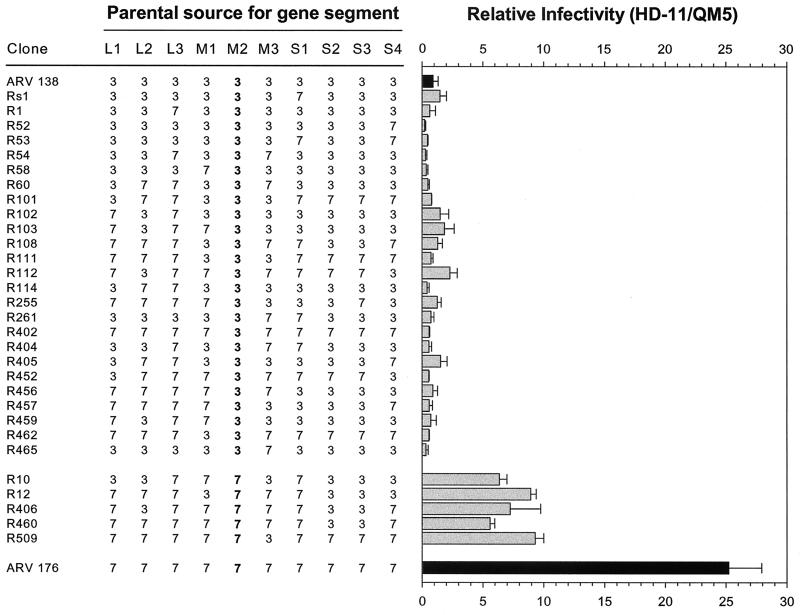
Genome segment assignments and relative infectivities of ARV reassortants. Parental (black bars) and reassortant (gray bars) clones are identified at the extreme left. For each reassortant, the parental identity of each genome segment, as determined by its relative mobility by SDS-PAGE, is indicated (3 for ARV-138 and 7 for ARV-176). Reassortant virus stocks were standardized to give equal and countable numbers of focus-forming units in quail QM5 fibroblasts. The standardized inocula were then used to infect QM5 or HD-11 cell monolayers. The infected QM5 and HD-11 cell monolayers were fixed and immunostained, and the average number of foci per field was determined as described for Fig. 2. The relative infectivity was calculated as the ratio of HD-11/QM5 foci, and values were normalized to the relative infectivity of ARV-138 arbitrarily set to a value of 1. Results are presented as means and standard errors from three or more separate experiments.
With the two parental viruses included with the reassortants, the viruses visually and statistically (P < 0.05) segregated into three separate groups based on their macrophage infection propensities. The group with the highest relative infectivity contains ARV-176 alone, with an HD-11/QM5 average relative infectivity of 25. The second group contains five reassortants whose average relative infectivities ranged from approximately 6 to 9. This group contains all of the viruses that possess the ARV-176 M2 genome segment. The third group contains the remaining 24 reassortants, none of which contain the ARV-176 M2 genome segment and all of which segregate with ARV-138. The average relative infectivities of this last group range from approximately 0.3 to 2. Most of the variability in the relative infectivities of the last group reflected minor inherent variations in the assay, as evidenced by the relative infectivities obtained for two separate reassortants with identical genome segment profiles (R459 and R103 have relative infectivities of 0.7 and 1.9, respectively). The most notable reassortant in the ARV-138 group was R402, a monoreassortant containing the ARV-138 M2 genome segment in an otherwise ARV-176 genetic background (Fig. 6). R402 had a relative infectivity of approximately 0.6 and was indistinguishable from ARV-138 when examined by the focus-forming assay (Fig. 8). These genetic studies clearly indicated that the ARV-176 M2 genome segment is necessary for efficient macrophage infection (P < 0.001).
FIG. 8.
The ARV-176 M2 genome segment is required for enhanced macrophage infection. Virus stocks of ARV-176 (a and b), ARV-138 (c and d), and the ARV-138 M2 monoreassortant R402 (e and f) were standardized to give equivalent concentrations of focus-forming units in quail QM5 fibroblasts (a, c, and e). The same virus dilutions were used to infect HD-11 macrophage monolayers (b, d, and f). Cells were immunostained as described for Fig. 1 to reveal viral foci of infection.
It was also apparent that additional virus genes or gene constellations exert an auxiliary influence on macrophage infection, since none of the ARV-176 M2-containing reassortants displayed the full infectivity of the parental ARV-176 virus (Fig. 7). The asymmetry in the distribution of the ARV-176 M2 genome segment among the panel of reassortants precluded any meaningful statistical analysis to identify potential second gene effects. However, the R509 monoreassortant (ARV-176 with an ARV-138 M3 genome segment) suggests that the M3 genome segment likely makes at least a minor (approximately twofold) contribution to the enhanced ability of ARV-176 to infect macrophages. The remaining ARV-176 M2 reassortants all contain the ARV-138 S2 and S3 genome segments, suggesting the possibility that either or both of these genome segments might also impact ARV infection of macrophages (approximately two- to threefold). Therefore, while it is clear from the present analysis that the ARV-176 M2 genome segment is required for efficient macrophage infection, the M2 genome segment alone is not sufficient for conferring the complete macrophagotropic property of ARV-176, indicating that second gene effects modulate ARV macrophage infection.
DISCUSSION
Our current results revealed that there are clear virus strain-specific differences in the tropism of ARV for HD-11 macrophages. We also demonstrated that this differential tropism does not reflect differences in the iNOS pathway, that the two viruses differ in the ability to establish a productive infection in macrophages but not in the ability to replicate within an infected macrophage, and that the M2 genome segment of ARV-176 is required for the efficient establishment of a productive infection in HD-11 macrophages.
The distinct macrophagotropic properties of ARV-176 and ARV-138 represent the third defined difference in the attributes of these viruses, the others being the extent of syncytium formation and embryo pathogenesis. We have previously shown that the S1 genome segment, which encodes the p10 fusion protein of ARV, contributes approximately 60% of the embryo pathogenic potential of ARV-176 (8). In view of the ability of various strains of ARV to infect macrophages in vivo and under culture conditions (12, 19, 23, 38), the transient immunosuppression that accompanies ARV-176 infection (25), and the central role of the macrophage in orchestrating the early immune response to virus infection, it is conceivable that the preferential infection of macrophages by ARV-176 in vivo could partially contribute to the S1 genome segment-independent events that influence ARV pathogenesis. However, this would need to be directly evaluated in vivo, since the macrophage infection propensity of ARV-176 has never been evaluated in infected animals and the relationship between ARV macrophage infection and virus pathogenicity remains unclear (12, 38). Similar confusion regarding the correlation between virus pathogenicity and macrophage infection exists with other viruses, such as herpes simplex virus (10, 17). A panel of ARV reassortants derived from two viruses with defined quantitative differences in their pathogenicity and macrophage infection independent of their relative replicative ability affords an opportunity to more precisely define the role, if any, of macrophage infection in ARV pathogenesis using animal or embryo models.
Aside from the possible implications for ARV pathogenesis, our present results have also revealed several interesting features of ARV-macrophage interactions and identified a virus gene that influences these interactions. Macrophage activation and NO production in response to virus infection can profoundly influence the outcome of virus-macrophage interactions in other virus systems (27). However, such events do not appear to contribute to the ability of different ARV strains to productively infect HD-11 macrophages. Neither ARV-176 nor ARV-138 stimulated iNOS activity following HD-11 infection, and both strains were equally susceptible to the antiviral effects of NO produced by LPS-activated HD-11 cells (Fig. 3). We have not as yet examined other cellular factors that may contribute to the differences in the relative infectivities of these two ARV strains, although our reassortant analysis suggests some potential candidates (see below).
The differential macrophage infection property of these two ARV strains is a dose-dependent phenomenon and is easily overcome by modest increases in the ARV-138 inoculum (Fig. 4). This suggests that there is not a subpopulation of HD-11 cells exclusively susceptible to ARV-176 infection, as occurs, for example, with herpesviruses or Theiler's virus, where the efficiency of macrophage infection reflects the state of macrophage activation or differentiation (4, 16, 35). Moreover, the analysis of virus translation and replication under infection conditions that result in the productive infection of the majority of the cells in a monolayer indicated that both viruses replicate equally well within an infected fibroblast or macrophage. In fact, the less infectious ARV-138 strain actually replicates to slightly higher titers in either cell line once the barrier to productive infection is overcome by moderate increases in the inoculum concentration (Fig. 5). Apparently, the viruses differ only in the ability to establish a productive infection in macrophage cells and not in the ability to replicate within any given infected cell. It seems reasonable to speculate that one or more of the steps in the ARV-176 replication cycle are enhanced in HD-11 cells relative to QM5 fibroblasts such that the likelihood of the virus establishing a productive infection in a macrophage cell is increased.
Our genetic studies provided evidence that a postattachment step of the ARV-176 replication cycle contributes to enhanced macrophage infection, since the S1 genome segment did not correlate with the propensity for macrophage infection. In addition to encoding the p10 fusion protein (30), the S1 genome segment also encodes the ςC cell attachment protein of ARV (22, 29). Since the parental source of the S1 genome segment was randomly distributed amongst the reassortant viruses (Fig. 7), it is unlikely that the differential macrophage infection property of these two strains of ARV reflects a receptor-binding phenomenon. We have recently confirmed that both viruses attach to HD-11 cells with equal efficiencies (D. O'Hara and R. Duncan, unpublished data). Consequently, there is a host-specific influence on some postattachment stage of the ARV replication cycle.
The genetic implication of the M2 genome segment in the enhanced infection of HD-11 cells by ARV-176 suggests a possible enhanced entry or uncoating step in the virus replication cycle. The M2 genome segment encodes the μB major outer capsid protein of ARV (37), the homolog of the MRV μ1 capsid protein. Cleavage and subsequent removal of the μ-class outer capsid protein is associated with the endosomal membrane interactions and conformational changes in the capsid required for delivery of the transcriptionally active core particle to the cytoplasm (2, 32, 36). As with MRV, ARV entry is also a low-pH-dependent event that is accompanied by specific cleavage of the major μ-class outer capsid protein (7). Consequently, differences between ARV-138 and ARV-176 in the rate of cleavage of the M2-encoded μB protein, in its membrane interaction properties, or in the subsequent capsid rearrangements required for activation of the core-associated transcriptase activity (3) could lead to altered delivery of the transcriptionally active core particle to the cytoplasm of macrophages.
In addition to identifying a virus strain-specific difference in a postattachment step of the ARV replication cycle in macrophage cells, there is also a clear host-dependent effect on this same postattachment step. It is conceivable that differences in the environment of the endosome-lysosome compartment of macrophages versus fibroblasts could contribute to altered processing of the ARV-176 outer capsid and virus entry or uncoating. We are currently pursuing a molecular analysis of the ARV μB protein and a biochemical analysis of the early stages of the virus replication cycle in HD-11 cells in order to specifically identify the nature of the host-specific effect on ARV macrophage infection. Entry studies monitoring the rate and extent of parental and reassortant virus μB cleavage in conjunction with sequence analysis of the M2 genome segments should serve to clearly determine whether the μB protein influences steps in the virus entry pathway that contribute to virus-and cell-specific differences in ARV tissue tropism.
ACKNOWLEDGMENTS
We thank Jingyun Shou for technical assistance, Leonard MacLean for assistance with statistical analysis, and Maya Shmulevitz for insightful discussions.
This research was supported by grants from the Medical Research Council of Canada to R.D. and to K.M.C. Studentship support from the Natural Sciences and Engineering Research Council of Canada (to D.O.) and from the Manitoba Health Research Council (to M.P.) is gratefully acknowledged.
REFERENCES
- 1.Beug H, von Kirchbach A, Doderlein G, Conscience J F, Graf T. Chicken hematopoietic cells transformed by seven strains of defective avian leukemia viruses display three distinct phenotypes of differentiation. Cell. 1979;18:375–390. doi: 10.1016/0092-8674(79)90057-6. [DOI] [PubMed] [Google Scholar]
- 2.Borsa J, Sargent M D, Kay C M, Oikawa K. Circular dichroism of intermediate particles of reovirus: elucidation of the mechanism underlying the specific monovalent cation effects on uncoating. Biochim Biophys Acta. 1976;451:619–627. doi: 10.1016/0304-4165(76)90157-4. [DOI] [PubMed] [Google Scholar]
- 3.Borsa J, Sargent M D, Lievaart P A, Copps T P. Reovirus: evidence for a second step in the intracellular uncoating and transcriptase activation process. Virology. 1981;111:191–200. doi: 10.1016/0042-6822(81)90664-4. [DOI] [PubMed] [Google Scholar]
- 4.Bruun T, Kristoffersen A K, Rollag H, Gegre M. Interaction of herpes simplex virus with mononuclear phagocytes is dependent on the differentiation stage of the cells. APMIS. 1998;106:305–314. doi: 10.1111/j.1699-0463.1998.tb01351.x. [DOI] [PubMed] [Google Scholar]
- 5.Clark F D, Ni Y, Collisson E W, Kemp M C. Characterization of avian reovirus strain-specific polymorphisms. Avian Dis. 1990;34:304–314. [PubMed] [Google Scholar]
- 6.Drastini Y, McKenna P K, Kibenge F S B, Lopez A. Chymotrypsin and trypsin sensitivities of avian reoviruses. Can J Vet Res. 1994;58:75–78. [PMC free article] [PubMed] [Google Scholar]
- 7.Duncan R. The low pH-dependent entry of avian reovirus is accompanied by two specific cleavages of the major outer capsid protein μ2C. Virology. 1996;219:179–189. doi: 10.1006/viro.1996.0235. [DOI] [PubMed] [Google Scholar]
- 7a.Duncan R. Extensive sequence divergence and phylogenetic relationships between the fusogenic and nonfusogenic orthoreoviruses: a species proposal. Virology. 1999;260:316–328. doi: 10.1006/viro.1999.9832. [DOI] [PubMed] [Google Scholar]
- 8.Duncan R, Sullivan K. Characterization of two avian reoviruses that exhibit strain-specific quantitative differences in their syncytium-inducing and pathogenic capabilities. Virology. 1998;250:263–272. doi: 10.1006/viro.1998.9371. [DOI] [PubMed] [Google Scholar]
- 9.Duncan R, Chen Z, Walsh S, Wu S. Avian reovirus-induced syncytium formation is independent of infectious progeny virus production and enhances the rate, but is not essential, for virus-induced cytopathology and virus egress. Virology. 1996;224:453–464. doi: 10.1006/viro.1996.0552. [DOI] [PubMed] [Google Scholar]
- 10.Gortz J, Brake B, Harle-Grupp V, Falke D. Replication of HSV-1 in murine peritoneal macrophages: comparison of various virus strains with different properties. Arch Virol. 1991;79:173–187. doi: 10.1007/BF01310810. [DOI] [PubMed] [Google Scholar]
- 11.Green L, Wagner D, Glogowski J, Skipper P, Wishnok J, Tannenbaum S. Analysis of nitrate, nitrite, and [15N]nitrate in biological fluids. Anal Biochem. 1982;126:131–138. doi: 10.1016/0003-2697(82)90118-x. [DOI] [PubMed] [Google Scholar]
- 12.Haffer K. In vitro and in vivo studies with an avian reovirus derived from a temperature-sensitive mutant clone. Avian Dis. 1984;28:669–676. [PubMed] [Google Scholar]
- 13.Hazelton P R, Coombs K M. The reovirus mutant tsA279 has temperature-sensitive lesions in the M2 and L2 genes: the M2 gene is associated with decreased viral protein production and blockade in transmembrane transport. Virology. 1995;207:46–58. doi: 10.1006/viro.1995.1050. [DOI] [PubMed] [Google Scholar]
- 14.Hieronymus D R K, Villegas P, Kleven S H. Identification and serological differentiation of several reovirus strains isolated from chickens with suspected malabsorption syndrome. Avian Dis. 1983;27:246–254. [PubMed] [Google Scholar]
- 15.Hieronymus D R K, Villegas P, Kleven S H. Characteristics and pathogenicity of two avian reoviruses isolated from chickens with leg problems. Avian Dis. 1983;27:255–259. [PubMed] [Google Scholar]
- 16.Jelachich M L, Bandyopadhyay P, Blum K, Lipton H L. Theiler's virus growth in murine macrophage cell lines depends on the state of differentiation. Virology. 1995;209:437–444. doi: 10.1006/viro.1995.1276. [DOI] [PubMed] [Google Scholar]
- 17.Jiang Y M, Daikoku T, Yamamoto M, Morishima T, Nishiyama Y. Growth and cytopathogenicity of herpes simplex virus in a macrophage cell line, RAW264: a good indicator of intraperitoneal pathogenicity. Microbiol Immunol. 1995;39:905–909. doi: 10.1111/j.1348-0421.1995.tb03276.x. [DOI] [PubMed] [Google Scholar]
- 18.Kibenge F S B, Jones R C, Savage C E. Effects of experimental immunosuppression on reovirus-induced tenosynovitis in light-hybrid chickens. Avian Pathol. 1985;16:73–92. doi: 10.1080/03079458708436354. [DOI] [PubMed] [Google Scholar]
- 19.Kibenge F S B, Gwaze G E, Jones R C, Chapman A F, Savage C E. Experimental reovirus infection in chickens: observations on early viraemia and virus distribution in bone marrow, liver, and enteric tissues. Avian Pathol. 1985;14:87–98. doi: 10.1080/03079458508436210. [DOI] [PubMed] [Google Scholar]
- 20.Kowalchyk K, Plagemann P G. Cell surface receptors for lactate dehydrogenase-elevating virus on subpopulation of macrophages. Virus Res. 1985;2:211–229. doi: 10.1016/0168-1702(85)90010-3. [DOI] [PubMed] [Google Scholar]
- 21.Lyon J A, Hinshaw V S. Inhibition of nitric oxide induction from avian macrophage cell lines by influenza virus. Avian Dis. 1993;37:868–873. [PubMed] [Google Scholar]
- 22.Martinez-Costas J, Grande A, Varela R, García-Martínez C, Benavente J. Protein architecture of avian reovirus S1133 and identification of the cell attachment protein. J Virol. 1997;71:59–64. doi: 10.1128/jvi.71.1.59-64.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mills J N, Wilcox G E. Replication of four antigenic types of avian reovirus in subpopulations of chicken leukocytes. Avian Pathol. 1993;22:353–361. doi: 10.1080/03079459308418926. [DOI] [PubMed] [Google Scholar]
- 24.Nibert M L, Schiff L A, Fields B N. Reoviruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Press; 1996. pp. 1557–1623. [Google Scholar]
- 25.Pertile T L, Sharma J M, Walser M M. Reovirus infection in chickens primes splenic adherent macrophages to produce nitric oxide in response to T cell-produced factors. Cell Immunol. 1995;164:207–216. doi: 10.1006/cimm.1995.1163. [DOI] [PubMed] [Google Scholar]
- 26.Pertile T L, Karaca K, Sharma J M, Walser M M. An antiviral effect of nitric oxide: inhibition of reovirus replication. Avian Dis. 1996;40:342–348. [PubMed] [Google Scholar]
- 27.Reiss C S, Komatsu T. Does nitric oxide play a critical role in viral infections? J Virol. 1998;72:4547–4551. doi: 10.1128/jvi.72.6.4547-4551.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robertson M D, Wilcox G E. Avian reovirus. Vet Bull. 1986;56:726–733. [Google Scholar]
- 29.Shapouri M R S, Arella M, Silim A. Evidence for the multimeric nature and cell binding ability of avian reovirus ς3 protein. J Gen Virol. 1996;77:1203–1210. doi: 10.1099/0022-1317-77-6-1203. [DOI] [PubMed] [Google Scholar]
- 30.Shmulevitz M, Duncan R. A new class of fusion-associated small transmembrane (FAST) proteins encoded by the non-enveloped fusogenic reoviruses. EMBO J. 2000;19:902–912. doi: 10.1093/emboj/19.5.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sick C, Schneider K, Staeheli P, Weining K C. Novel chicken CXC and CC chemokines. Cytokine. 2000;12:181–186. doi: 10.1006/cyto.1999.0543. [DOI] [PubMed] [Google Scholar]
- 32.Sturzenbecker L J, Nibert M L, Furlong D, Fields B N. Intracellular digestion of reovirus particles requires a low pH and is an essential step in the viral infectious cycle. J Virol. 1987;61:2351–2361. doi: 10.1128/jvi.61.8.2351-2361.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sung Y J, Hotchkiss J H, Austic R E, Dietert R R. l-Arginine-dependent production of a reactive nitrogen intermediate by macrophages of a uricotelic species. J Leukoc Biol. 1991;50:49–56. doi: 10.1002/jlb.50.1.49. [DOI] [PubMed] [Google Scholar]
- 34.Tang K N, Fletcher O J, Villegas P. The effect on newborn chicks of oral inoculation of reovirus isolated from chickens with tenosynovitis. Avian Dis. 1987;31:584–590. [PubMed] [Google Scholar]
- 35.Tenney D J, Morahan P S. Differentiation of the U937 macrophage cell line removes an early block of HSV-1 infection. Viral Immunol. 1991;4:91–102. doi: 10.1089/vim.1991.4.91. [DOI] [PubMed] [Google Scholar]
- 36.Tosteson M T, Nibert M L, Fields B N. Ion channels induced in lipid bilayers by subvirion particles of the nonenveloped mammalian reoviruses. Proc Natl Acad Sci USA. 1993;90:10549–10552. doi: 10.1073/pnas.90.22.10549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Varela R, Benavente J. Protein coding assignment of avian reovirus strain S1133. J Virol. 1994;68:6775–6777. doi: 10.1128/jvi.68.10.6775-6777.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.von Bulow V, Klasen A. Effects of avian reoviruses on cultured chicken bone-marrow-derived macrophages. Avian Pathol. 1983;12:179–198. doi: 10.1080/03079458308436162. [DOI] [PubMed] [Google Scholar]
- 39.Weining K C, Sick C, Kaspers B, Staeheli P. A chicken homolog of mammalian interleukin-1 beta: cDNA cloning and purification of active recombinant protein. Eur J Biochem. 1998;258:994–1000. doi: 10.1046/j.1432-1327.1998.2580994.x. [DOI] [PubMed] [Google Scholar]
- 40.Zhang S, Lillehoj H S, Ruff M D. Chicken tumor necrosis-like factor. I. In vitro production by macrophages stimulated with Eimeria tenella of bacterial lipopolysaccharide. Poult Sci. 1995;74:1304–1310. doi: 10.3382/ps.0741304. [DOI] [PubMed] [Google Scholar]