Abstract
The DNA-binding with one finger (Dof) gene family is a class of plant-specific transcription factors involved in diverse biological processes, including response to biotic and abiotic stresses. Members of this family have been reported in the cultivated potato Solanum tuberosum, but clues to the roles of several Dof genes are still lacking. Potato wild relatives represent a genetic reservoir for breeding as they could provide useful alleles for adaptation to the environment and tolerance to biotic and abiotic stresses. We performed an in silico analysis to identify genes belonging to the Dof family in the wild potato S. commersonii, confirming that the identified Dof genes can be grouped in four classes (A, B, C, D), as reported for cultivated potato. A special focus was dedicated to Cycling Dof Factors (CDFs), which play a crucial role in plant responses to abiotic stresses. Analysis of available RNA-seq data confirmed CDF genes as regulated by stresses and often in a tissue specific manner. To ascertain their involvement in the stress response, S. tuberosum and S. commersonii plantlets growing in vitro were subjected to salt stress (80mM NaCl) for short (2 days) and prolonged (7 days) times. Analysis of phenotypic traits and qRT-PCR expression profiles of target CDF genes in aerial and root tissues showed differences between the two species. In addition, after saline treatment, changes in total phenols, proline, and malondialdehyde suggested a diverse perception of saline stress in S. commersonii vs. S. tuberosum. Overall, this study provided useful clues to the involvement of CDF genes in salt response and promoted the identification of potential candidate genes for further functional studies.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-75412-2.
Subject terms: Plant sciences, Plant stress responses, Transcriptomics
Introduction
Crops belonging to the Solanaceae family are often characterized by poor tolerance to abiotic stresses and their yield and quality are seriously compromised by ongoing climate change1. Among these, potato (Solanum tuberosum L.), the third most consumed crop in the world, faces various stress factors. As a result, many breeding programs have now reoriented their objectives and strategies towards improving stress tolerance and resilience. Importantly, the availability of data from an increasing number of RNA-seq experiments for different species allows the estimation of the expression profiles of several stress-related genes involved in crucial functions of plant growth and development; this creates novel and promising research opportunities.
In this context, comparative genomics studies between wild relatives and cultivated Solanaceae varieties have emerged as an effective strategy to explore the genetic diversity and inheritance patterns of beneficial alleles, with the aim of identifying useful genes for breeding purposes2. Numerous studies have outlined the crucial role of transcription factors (TFs) in controlling cellular and metabolic processes by regulating genes that trigger signaling pathways. Among them, TFs such as ERF, MYB, WRKY and AP2/EREBP have received extensive research attention3. By contrast, the Dof (DNA-binding with one finger) gene family has received less attention than others.
Dofs are plant-specific transcriptional regulators with multiple functions. They have been characterized in several Solanaceae: 34 in tomato4, 35 in potato5, 33 in pepper6, and 29 in eggplant7. Phylogenesis within the genus Solanum indicates that Dof genes are distributed into four major clusters (A, B, C and D), with clusters B, C, and D possibly further divided into subgroups4,6,8–10. The conservation of cluster D in the analyzed species is particularly interesting, since it includes Dof genes homologous to Cycling DOF Factor (CDF) genes in Arabidopsis, involved in responses to abiotic stress via the regulation of the C-repeat binding factor regulator (CBF)11–13. Furthermore, several members of this gene family are well known for their roles in growth and development processes such as seed germination, flowering, and leaf senescence14,15. Dof TFs are responsible for the regulation of secondary metabolic processes, such as the biosynthesis of glucosinolates and flavonoids and the cell cycle16–18. They also control various physiological processes, such as the formation of interfascicular cambium and vascular development19, axial leaf patterning20, photoperiodic flowering, circadian clock21–24, hormone signaling pathways15,25. Recent studies have highlighted that Dof TFs are key regulatory hubs of several phytohormone pathways and are responsible for the crosstalk between signaling pathways and the response to many abiotic stresses11,15,26–29. The expression of Dofs, especially those within cluster D, has been highly induced by salt, drought, cold, heat stress15 and reverse genetic approaches have demonstrated their role in mechanisms of tolerance30–34. In Arabidopsis, the AtCDF3 gene has been shown to regulate several stress-responsive TFs (e.g., CBF, DREB2A and ZAT10)11,35 as the cdf3-1 mutant was found to be sensitive to drought and cold stresses, whereas its overexpression increases osmotic stress resistance. In addition, overexpression of SlCDF1 or SlCDF3 increased tolerance and resistance to salt in tomato plants28.
Overexpression of the BnCDF1 gene in Brassica napus promoted cold tolerance through the regulation of several stress-responsive genes such as CBF1, CBF2, COR15A36. Furthermore, Dof TFs participate in modulating the expression of genes linked to the regulation of carbon (C) metabolism and nitrogen (N) assimilation37. In fact, a growing number of studies describe the possible role of Dof TFs in coordinating the carbon and nitrogen balance to ensure plant vital processes. Among others, Domínguez-Figueroa et al.37 reported that modulation of the expression of genes related to C and N assimilation by Arabidopsis AtCDF3 leads to improved root development. Similarly, overexpression of SlCDF3 increased tomato biomass and yield by enhancing sucrose and nitrogen assimilation35. Despite the potential role of Dof TF in abiotic stress responses, data on their function in potato under such conditions are limited. Notably, among the CDF members of Dof TF family, StCDF1 has been identified as playing a key role in regulating tuberization and potato life cycle38. Additionally, a recent study has highlighted StCDF1 involvement in regulate water loss by affecting stomatal development and diurnal opening38,39.
Taken together, these findings prompted us to study CDF genes within the Dof family in the wild potato Solanum commersonii to ascertain their involvement in salt stress tolerance compared to S. tuberosum. Wild species are known to be valuable sources of resistance traits, and S. commersonii, as a wild diploid potato, is particularly well-adapted to harsh environmental conditions compared to its cultivated counterparts40–43. Additionally, S. commersonii provides access to genomics resources that are lacking for other wild potato relatives43. For reliable comparative molecular and biochemical studies, we selected the doubled monoploid clone DM of S. tuberosum. This diploid genetic background facilitates a robust comparison with S. commersonii.
We performed a comparative in silico analysis to identify and characterize members of the Dof gene family in S. commersonii. To further investigate the role of CDF genes, we interrogated available RNA-seq data and explored CDF expression patterns in several organ and tissues of S. tuberosum varieties and in response to different abiotic stresses. Then, to highlight potential differences in stress response, in vitro grown plantlets of S. tuberosum and S. commersonii were subjected to salt stress for short (2 days) and longer (7 days) times and the transcriptional changes of CDF genes along with phenotypic and biochemical variations in stress markers were evaluated. Comparison of expression patterns of CDF genes revealed different transcriptional behavior between S. tuberosum and S. commersonii, both with respect to tissue- and species-specific responsiveness. In addition, phenotypic data indicated superior root plasticity of S. commersonii, suggesting a potential more effective root-to-shoot signaling. Finally, data collected on the content of total polyphenols, malondialdehyde and proline showed a marked metabolic variation over time of stress in S. tuberosum. This effect was negligible in S. commersonii.
Results
Identification of CDF proteins in S. commersonii and chromosomal distribution
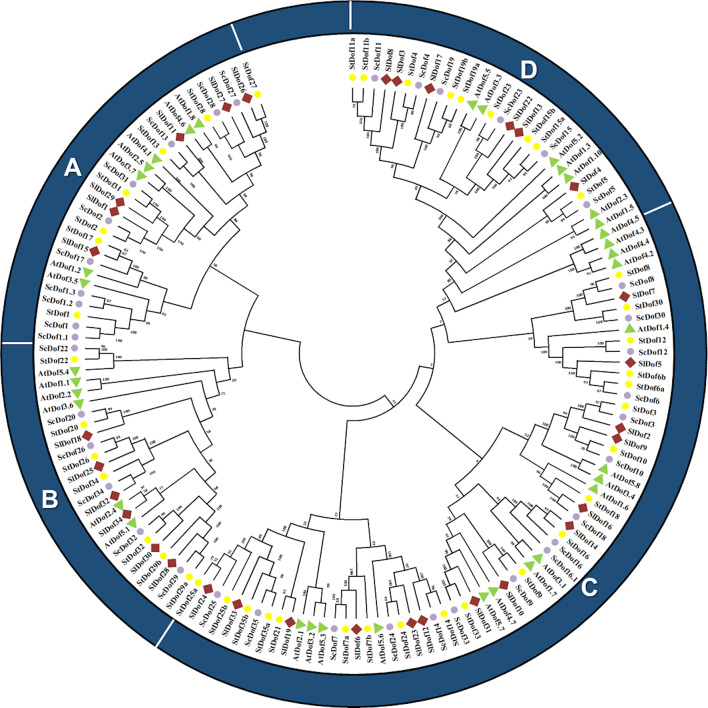
Thirty-eight (38) Dof genes were identified in Solanum commersonii and were named following the nomenclature of S. tuberosum Dof genes5. To gain some functional clues about the phylogenetic relationships between Dof genes in wild and cultivated potato, tomato, and Arabidopsis, we constructed an unrooted cladogram tree from the multiple sequence alignment of 151 full-length Dof proteins (Supplementary Table 1). Within the tree, the four known major clusters (A, B, C and D) could be distinguished (Fig. 1), consistently with what was reported for the cultivated potato5.
Fig. 1.
Cladogram tree including Dof proteins of S. commersonii (violet), S. tuberosum (yellow), S. lycopersicum (red) and A. thaliana (green).
In S. commersonii, group D included genes known as CDF. This group displayed six genes (ScDof4, ScDof5, ScDof11, ScDof15, ScDof19 and ScDof23), namely ScDof4 and ScDof5 are homologues of CDF2, ScDof11 ortholog of the Arabidopsis CDF3 gene, ScDof15 ortholog of CDF5, ScDof19 ortholog of CDF1, and ScDof 23 of CDF4. Unlike S. tuberosum, no transcript variants were detected for ScDof11, ScDof15 and ScDof19 within this group. Overall, S. commersonii showed similar Dof clustering to S. tuberosum and the other species under investigation.
ScDof genes were distributed across all chromosomes except 7 and 12.Two genes, ScDof1.3 and ScDof1, were located on scaffold 275 of the genome assembly ASM123980v143 (Supplementary Fig. 1). ScDof genes showed a non-uniform distribution along the chromosomes, with a single ScDof gene found on chromosome 9 and ten genes on chromosome 2, which exhibited the largest number of ScDof. This distribution aligns with previous results for pepper and tomato5,9.
Expression profiles of CDF genes in different tissues upon stresses
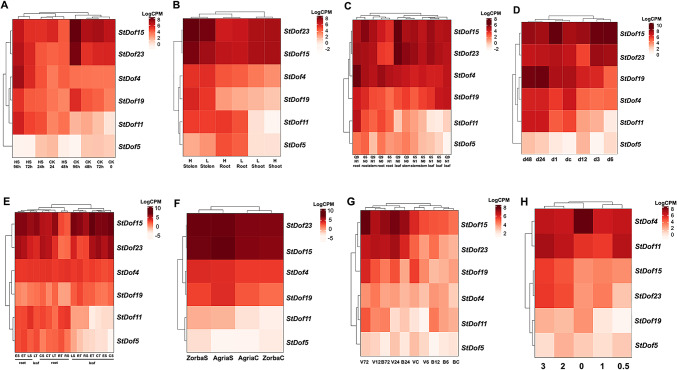
Due to the lack of information on CDF genes in wild potato, we utilized publicly available S. tuberosum RNA-sequencing data (Supplementary Table 2) to analyze gene expression profiles of CDF genes across various tissues and organs subjected to salt stress, drought, nitrogen deprivation, as well as biotic infections and wounds (Fig. 2; Supplementary File 1). The transcriptional response of Dof genes in cultivated potato Longshu n. 5 subjected to salt stress44 revealed the progressive induction of Dof11 and Dof4 genes over the time of stress application (Fig. 2A). Interestingly, the expression of Dof23 and Dof15 increased during saline treatment but not as much as in the absence of stress; on the contrary, StDof5 showed the lowest expression level compared to the other Dof genes analyzed under stressed and non-stressed conditions. RNA-seq data analysis from leaves, stolons, and roots of the Indian potato variety “Kufri Jyoti” grown with varied nitrogen supplies showed that the expression of target Dof genes was not affected by nitrogen45.
Fig. 2.
Heatmaps and dendrograms showing clustering of samples and CDF genes with similar expression patterns. RNA-sequencing data were used to derive the expression profiles (rendered in counts per million, CPM) of StDof4 StDof5, StDof11, StDof15, StDof19, StDof23 genes (A) Potato variety “Longshu No. 5” grown in medium containing 500 mM NaCl (High salt group, HS) or 0 mM NaCl (control group, CK) for 24, 48, 72, and 96 h. (B) Potato plants (shoots, roots and stolons) grown in aeroponic culture with low N (0.2 mM) and high (4 mM, control) nitrogen (N) supply. (C) Two potato varieties subjected to nitrogen deficiency (N0) or not (N1). (D) Aboveground tissues of variety Desiree at 0, 1, 3, 6, 12, 24 and 48 h after drought stress. (E) Leaves and roots of two S. tuberosum subsp. andigena varieties with contrasting drought tolerance (S = susceptible, T = tolerant) E = early response 40 min after drought induction; L = late response 120 min after drought induction; R = recovery phase. (F) Leaves of “Agria” and “Zorba” varieties exposed to water deprivation for 25 days (S) and normally irrigated (C). (G) Susceptible (“Valor)” and tolerant (“BP1”) varieties at 0, 6, 12, 24, and 72 h post-inoculation with P. c. brasiliense. (H) Potato tuber tissue collected at 0, 0.5, 1, 2 and 3 days post-wounding.
In details, high expression of Dof 23 and Dof 15 was detected in all tissues and conditions, while Dof5 and Dof11 were mainly expressed in roots regardless of nitrogen concentration (Fig. 2B). In roots, shoots and leaves of two S. tuberosum varieties grown under low- and high-nitrogen fertilization conditions46, a similar trend was observed for Dof23, Dof4 and Dof15. These genes were unevenly regulated by stress but highly expressed. By contrast, Dof5 and Dof11 also showed a root-specific expression (Fig. 2C).
Under drought stress conditions monitored at different time points (0, 1, 3, 6, 12, 24 and 48 h)47 (Fig. 2D), aboveground tissues from “Dèsirèe” showed early activation of Dof15 and Dof23, with subsequent upregulation detected for Dof19, Dof11, whereas Dof4, which was already activated in the control condition (dc), reduced its expression until 12 h but restored initial induction after 24–48 h. Noteworthy, Dof5 had low expression at all time points. Dof15, Dof23 and Dof4 showed high expression in both leaves and roots of two varieties of S. tuberosum subsp. andigena, one susceptible and one tolerant to drought stress48. Dof19 was mainly expressed in leaves, while Dof5 and Dof11 were expressed in roots and almost downregulated in leaves; this suggests tissue-specific activation of these genes (Fig. 2E). A high level of expression for Dof23, Dof15, Dof4 and Dof19, a slight induction for Dof11and a downregulation of Dof5 were detected in the leaves of potato varieties “Agria” and “Zorba” under water deprivation for 25 days49 (Fig. 2F). Dof23, Dof15 and Dof19 were induced and Dof5 was downregulated in stems of two potato varieties with different susceptibility to soft rot enterobacterium50, where the expression profiles of TFs did not appear to be variety-specific (Fig. 2G). In potato tubers during the early stages of the wound-healing process51, Dof4 and Dof11 showed the highest expression level at time point 0 while maintaining a high transcriptional level even during the subsequent three days of wound stress. The expression of Dof15 and Dof23 gradually increased over time, while a low level of expression was observed for Dof5 and Dof19 over time (Fig. 2H).
Overall, analysis of available RNA-seq data indicated that Dof expression, particularly for Dof5 and Dof11, appears to be tissue-specific. In several dataset these genes were expressed in roots and at a very low level in leaves under all stress conditions. By contrast, Dof23, Dof15, Dof4 and Dof19 were constitutively highly expressed and even more transcriptionally active following stress treatments.
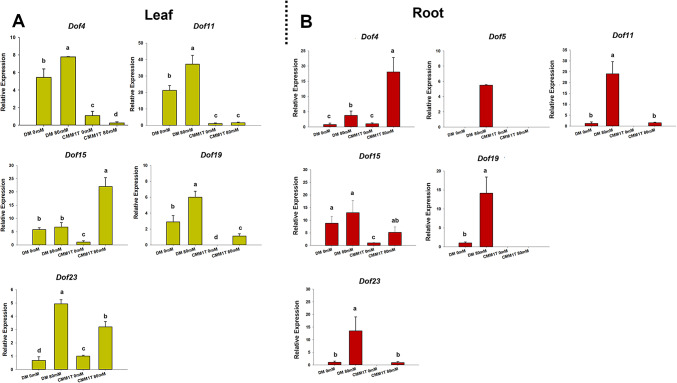
Transcriptional behavior of StCDF and ScCDF after salt stress treatment
Quantitative RT-PCR experiments were performed to explore CDF expression under salt stress treatment (Fig. 3A-B). In leaves, after 2 days of salt treatment, Dof4 showed significant induction only in clone DM of S. tuberosum group Phureja, whereas in clone CMM1T of the wild species S. commersonii the gene was almost inactive (Fig. 3A). No transcription for the Dof5 gene was detected under any condition. Dof11 showed constitutive upregulation in DM compared to CMM1T, with induction in DM leaves following stress. Transcription of Dof15 was highly induced only in CMM1T leaves. Both clones exhibited an increase in Dof19 transcriptional activity after stress, with a greater extent observed in DM leaves. Substantial induction of Dof23 was observed in clones of both species, particularly in DM. Overall, in DM leaves, all target genes (except Dof15) were induced by stress, whereas in CMM1T only Dof15, Dof19 and Dof23 showed stress-related upregulation (Fig. 3A). In roots (Fig. 3B), Dof4 showed significant induction in CMM1T roots compared to DM, while Dof5 was not constitutively expressed in either DM or CMM1T, although transcriptional induction was observed in DM under stress. Similarly, Dof11 was induced in DM under stress and to a lesser extent in CMM1T. In addition, Dof15 was significantly induced only in CMM1T under stress. The remaining Dofs, Dof19 and Dof 23, were upregulated after 48 h of salt stress in DM roots. In terms of quantitative aspect, in CMM1T the expression of Dof4 was 10-fold greater under stress conditions compared to the control. Similarly, the transcriptional activity of Dof15 in roots was 5-fold greater in CMM1T while it was not significantly different in stressed DM roots. In contrast, Dof 5, Dof11, Dof19, Dof 23 were more stress-induced in DM. After seven days of salt stress, we were unable to detect Dof TFs expression, likely underlying the involvement of these genes in early transcriptional responses to stress.
Fig. 3.
Quantitative RT-PCR expression profiles of target CDF genes. (A) leaves and (B) roots of S. tuberosum DM and S. commersonii CMM1T after 2 days of treatment with 0 mM and 80mM NaCl. The y-axes represent the relative expression of each target gene. All data are expressed as means ± standard error of three independent pools analyzed in three technical replicates. Different letters indicate statistically significant differences among samples (Tukey’s HSD test p ≤ 0.05).
Phenotypic response to salt stress
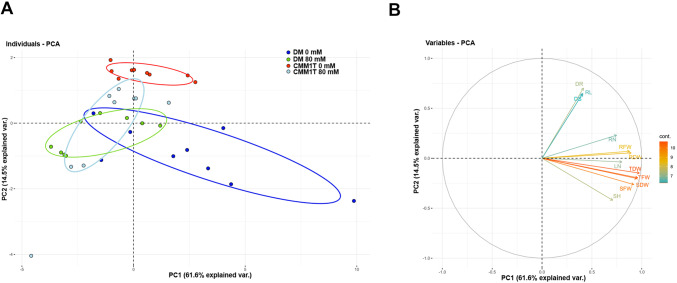
The PCA in Fig. 4 provides an overview of all phenotypic data during stress in DM and CMM1T. The first two principal components (PC) accounted for 76.1% of the total variance (Fig. 4A). Notably, CMM1T showed consistent behavior before and after stress. DM exhibited greater variability, although it showed a more homogeneous response after salt stress. Particularly, in DM salt stress significantly reduced the number of leaves while the number of roots was reduced marginally. Shoot height and root length also decreased significantly, highlighting impaired growth. In contrast, CMM1T showed stability in the number of leaves and shoot height, but a significant reduction in the number of roots and root length under salt stress, emphasizing clone-specific responses, particularly in root growth (Fig. 4B and Supplementary Tables 4 and 5).
Fig. 4.
PCA based on phenotypic variables. Classification of S. tuberosum DM and S. commersonii CMM1T clones based on their response to salt stress. (A) PCA plot showing DM and CMM1T after the application of 0 mM and 80 mM NaCl. (B) Bi-plot reporting the relationships among all the traits under investigation shoot height (SH), root length (RL), leaf number (LN), root number (RN), shoot fresh weight (SFW) and root fresh weight (RFW), root dry weight (RDW) and shoot dry weight(SDW), total dry weight (TDW), total fresh weight (TFW), days of emission of roots (DR) and days of emission of shoots (DS).
Total phenols, proline and MDA content
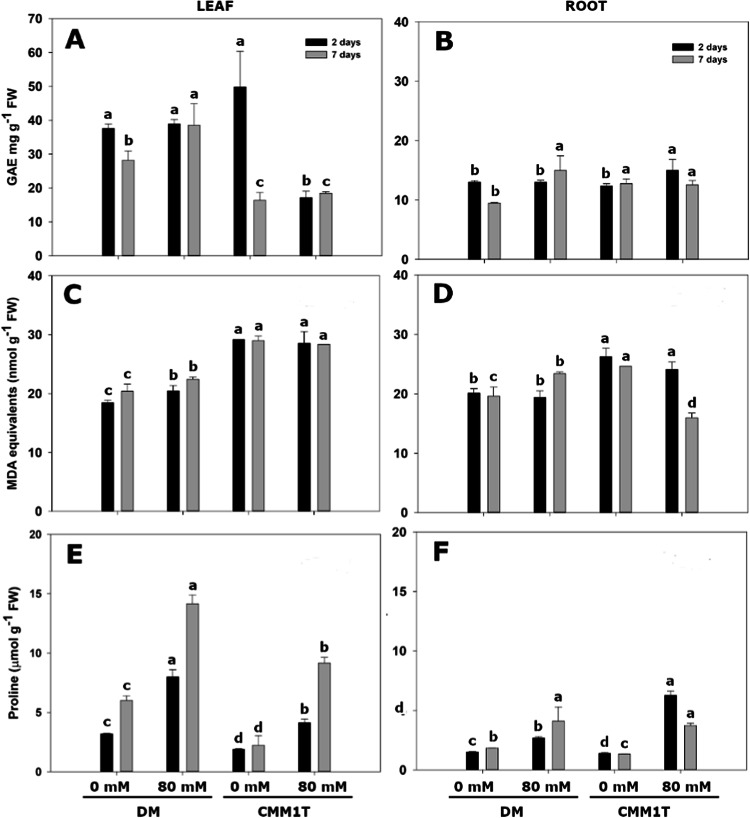
We measured changes in the total phenol content (TPC) in DM and CMM1T leaves and roots in response to salt stress, aiming to understand their antioxidant contribution to plant cells (Fig. 5A-F). No significant difference in TPC was observed in DM leaves two days after stress application (Fig. 5A); after seven days a significant increase was evident. Conversely, CMM1T leaves exhibited a significant decrease in total phenols under short-term stress (~ 30 mg g−1 FW of GAE, to approximately 10 mg), with no change during prolonged stress. In roots (Fig. 5B), although TPC under unstressed condition for both DM and CMM1T was comparable and almost fourfold lower than in leaves (~ 40 mg versus ~ 10 mg), salt stress in DM roots had a response similar to that observed in leaves. By contrast, in CMM1T a slight increase in TPC was detected 2 days after stress application.
Fig. 5.
Total phenolics, malondialdehyde and proline contents. Total phenolic content expressed as gallic acid equivalents (GAE) in leaves (A) and roots (B) of S. tuberosum DM and S. commersonii CMM1T clones. Malondialdehyde (MDA) content, as a lipid peroxidation marker, in leaves (C) and roots (D). Proline content in leaves (E) and roots (F). Measurements were taken after 2 and 7 days with NaCl at 0 mM (control) and 80 mM (stressed). FW = fresh weight. Bars represent means ± standard error of three independent pools analysed in three technical replicates. Within each time point different letters indicate statistically significant differences among samples (Tukey’s HSD test p ≤ 0.05).
Regarding malondialdehyde (MDA), DM leaves showed a significant increase after 2 and 7 days of stress (from 18 to 20 nmol g−1), whereas no significant changes were observed in CMM1T leaves at both time points (Fig. 5C). In DM roots, an increase in MDA content was statistically significant only after prolonged stress (Fig. 5D), while CMM1T roots showed a slight reduction after two days, with a marked decrease of approximately 1.5-fold after 7 days.
Salt stress increased the amount of proline in both species and tissues (Fig. 5E-F). The proline content detected in leaves and roots ranged from 1 to 14 µmol g−1 FW (Fig. 5E). In DM leaves, the amount of proline increased almost four- and three-fold after 2 and 7 days, respectively, while in CMM1T leaves it almost doubled and quadrupled after the same periods. In DM roots, the increase in proline was less pronounced than in CMM1T (Fig. 5F). After 2 days of stress, it increased from 1 to 2 µmol g−1, reaching the amount of 4 µmol g−1 at 7 days. Regarding CMM1T roots, the increase in proline content was of approximately 6 µmol g−1 after two days of stress compared to the control. However, when stress was prolonged up to 7 days, this amount was reduced to 2 µmol g −1 FW.
Discussion
The growing interest in Dof plant-specific TFs is based on their multiple roles in plants. Literature extensively covers the identification, functional characterization and evolutionary changes of Dofs in both monocotyledonous or dicotyledonous species13,25,52. This is particularly true for Dof TFs involved in plant response to abiotic stress, identified as crucial regulators of genes contributing to environmental adaptation. In our study, we identified a total of 38 Dof factors in the wild potato S. commersonii. Typically, Dof family comprises 20 to 50 member genes, and the number of ScDofs identified mirrored that of cultivated potato and was similar to those found in other Solanaceae9,28,53. Consistent with previous studies, alignment of ScDof protein sequences revealed a highly conserved Dof domain, suggesting that these TFs have been evolutionarily conserved among plants. The ScDof cladogram tree allows us to distinguish four main groups. Clade D includes six CDF genes. In S. tuberosum, three genes of Clade D, the StDof11, StDof15 and StDof19, have transcript variants53. Unfortunately, the latter variants have not yet been identified and annotated in S. commersonii. The presence of transcript variants is generally associated with phenotypic plasticity, particularly in immunity, circadian rhythm, and flowering period54–56. However, as reported by Smith et al.57, several phenotypic changes associated with transcript variants may not be universally beneficial, possibly including a mix of beneficial, neutral, and deleterious effects.
Transcriptional data available for S. tuberosum CDF genes collected from different organs and after exposure to drought, saline treatment and ABA indicated that transcript variants “a” and “b” did not show distinctive behavior from each other, nor were they related to tissue-specific induction and reactivity to stress53. Notably, expression of the StDof15 b isoform under stress was negligible compared to the counterpart53. To better understand the stress responsiveness of selected Dof TFs, we interrogated eight publicly available transcriptome datasets44–51 and derived the expression profiles of CDF genes in S. tuberosum stressed tissues. Under different stress conditions, the expression profiles of target CDF genes were comparable. So far, StDof5 and StDof11 were found to be constitutively upregulated in roots, also showing tissue-specific regulation, whereas transcription of Dof15, Dof23, Dof4 and Dof19 was induced in almost all analyzed tissues and stress conditions.
In leaf samples from plantlets treated with 80mM NaCl, quantitative PCR pointed out the upregulation of Dof15, Dof23 and Dof19, and the downregulation of Dof4 in CMM1T. Salt stress induced all the target genes in DM roots; whereas in CMM1T, Dof5 and Dof19 gene expression was undetectable and Dof11, Dof15, and Dof23 were induced by stress but to a lower extent compared to DM stressed roots. Transcription of ScDof4 was markedly activated compared to DM. It has been proposed that CDF1-5 may have a redundant function in regulating photoperiod, thereby controlling flowering time in Arabidopsis11,22,28. However, whether they maintain a similar function regarding stress responses is still under investigation. Indeed, it has been functionally confirmed that CDF1 and CDF3, corresponding to Dof19 and Dof11, play an important role in regulating the photoperiodic flowering response in tomato as well as in Arabidopsis11,24,26,35,37. In addition to regulating the flowering period, Arabidopsis CDF2, orthologous to StDof4, has been recently shown to have a key role in growth promotion58. Although the function of CDF2 in belowground tissues is not well established, in aboveground tissues CDF2 physically interacts with the bHLH PIF4 to activate target genes and induce hypocotyl elongation, enhancing auxin biosynthesis. We hypothesized that upregulation of Dof4 in stressed roots of CMM1T might be linked to a specific role in root sensing and response, but additional studies are needed to confirm this hypothesis. Recently Xu et al.59 revealed that the A. thaliana CDF4, orthologous to the potato Dof23 gene, is a senescence-related TF. They reported that its overexpression promotes leaf senescence by upregulating endogenous ABA levels, while its downregulation increases tolerance to stress by improving detoxification of reactive oxygen species. Further, the senescence-related role of StCDF1, corresponding to StDof19, has just been elucidated by Shi et al.60, who found that constitutive overexpression of StCDF1 not only caused a delay in the onset of potato senescence, but also promoted a faster progression of senescence and shortened the life cycle. The expression profiles obtained from qPCR experiments showed intriguing transcriptional activity for both ScDof23 and ScDof19, whose expression was significantly reduced in stressed leaves and roots of CMM1T compared to DM. This result may be consistent with increased stress tolerance of CMM1T to slowed senescence processes.
A recent in vitro screening for salt tolerance in different accessions of wild and cultivated potato demonstrated that root architecture plays a crucial role in mediating stress response to salt treatment in wild potato accessions41. The phenotypic data we gathered confirm that S. commersonii shows better phenotypic plasticity than S. tuberosum in response to salt stress; indeed, unlike CMM1T, the growth of DM plants was impaired after undergoing salt stress. Furthermore, data collected on the main metabolic markers of oxidative stress suggest that in the short term most of the leaf phenolics of wild potato are rapidly consumed, perhaps to cope with the propagation of the state of oxidative stress, while in the leaves of S. tuberosum the phenolic content increases over the time frame of the experiment. Proline accumulation was greater in the leaves and roots of cultivated potato grown under stress, although the pattern of accumulation was similar between the two species. Nevertheless, the accumulation of proline could be indicative of a lower requirement for osmotic balance in the wild species compared to the cultivated potato. In line with this view, in the roots of CMM1T the reduction of MDA content after seven days of stress indicates the absence of oxidative stress, which was instead detected in the leaves and roots of DM.
Based on current findings and previous results, we believe that in S. commersonii roots play a decisive role as the first line of defense against salt stress, most likely by limiting stress propagation to aboveground tissues.
The transcriptional and phenotypic analyses provide valuable insights for future research directions. The data suggest that ScDof4 may reflect an evolutionary adaptation in signaling mechanism between wild and cultivated potatoes under salinity stress. The up-regulation of ScDof4 in S. commersonii could indicate the wild species’ enhanced ability to tolerate stress by promoting root growth. Similarly, the limited expression of ScDof19 and ScDof23 during salinity stress makes them promising candidates for further investigations. These molecular traits may highlight in the wild species a superior capacity to regulate growth and senescence under stress. Further studies should explore the network linking these Dofs to upstream and downstream signaling pathways. In particular, functional characterization of ScDof4 and ScDof23 could shed light on their roles in supporting the greater abiotic stress tolerance observed in S. commersonii.
Materials and methods
Identification of Dofgenes in S. commersonii
A BLASTp search was conducted using an E-value threshold of 10⁻³ against the S. commersonii protein complement using the Dof genes already annotated in S. tuberosum, S. lycopersicum and A. thaliana as queries. The protein sequences of S. commersonii were retrieved from the Dryad database43. The hidden Markov model (HMM) profile PF02701 was downloaded from the InterPro database61 and was searched against the S. commersonii proteins using HMMER62 by setting an e-value of 10−3.
Phylogenetic analysis of Dof genes
To systematically classify ScDof genes and determine phylogenetic relationships with Dof members of other plant species, a multiple-sequence alignment was conducted using the ClustalW program with default parameters63 for all available Dof protein sequences from A. thaliana (N.=36), S. lycopersicum (N.=34), S. tuberosum (N.=43) and S. commersonii (N.=38) (Supplementary Table 1). Phylogenetic analysis of full-length Dof protein sequences of all the above-mentioned species was performed using MEGA X64 with the neighbor-joining (NJ) method, setting the following parameters: poisson correction, random seed and pairwise deletion. Bootstrapping (1,000 replicates) was used to infer confidence values on the cladogram tree. Only clades with confidence values greater than 55 were selected for the consensus tree, which was visualized with Figtree version 1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/).
Chromosomal location of the ScDof genes
The chromosomal location of the ScDof genes was retrieved from the S. commersonii genome assembly ASM1825827v165. ScDof genes were mapped to the corresponding chromosomes using the online toolMG2C_v2.1 (http://mg2c.iask.in/mg2c_v2.1/). ScDof1.3 and ScDof1 were located on scaffold 275 of the GCA_001239805.1 (ASM123980v1)43 genome assembly.
Analysis of gene expression profiles of target genes based on available RNA-seq data
Raw RNA-sequencing data in FASTQ format associated with publicly accessible works listed in Supplementary Table 2 were downloaded from the Sequence Read Archive using the fastq-dump utility. The samtools-faidx utility66 was used to extract the sequence of the following genes: PGSC0003DMG400025129, PGSC0003DMG400037029, PGSC0003DMG400001330, PGSC0003DMG400019528, PGSC0003DMG400018408, PGSC0003DMG400033046 from the indexed genome reference sequence (SolTub_3.0) retrieved from Ensembl Plants. The coordinates of the genes were obtained from the file Solanum_tuberosum.SolTub_3.0.56.chr.gff3 downloaded from the Ensembl Plants database. Bowtie267 was used to perform end-to-end read alignments in sensitive mode. The samtools-idxstats utility66 was used to obtain the number of reads mapped to each target gene. Expression values were then rendered in CPM (Counts Per Million) according to the following formula: CPM = readsMappedToGene * 1/totalNumReads * 106.
In vitro salt stress treatment
Plant material included in vitro-grown plants of a doubled monoploid clone of S. tuberosum group Phureja (DM1-3-516-R44, referred to as DM) and of a clone of the wild species S. commersonii derived from the accession PI243503 (referred to as CMM1T). To obtain a sufficient number of plants, several in vitro propagation cycles were carried out on a substrate containing Murashige and Skoog68 salts with the addition of 10 g L−1 of sucrose and 8.0 g L−1 of agar (pH 5.7–5.8). In vitro plantlets were placed in growth chambers at 24 °C, with an irradiance of 200 µmol 96 m−2 s−1 under a 16/8 h (light/dark) photoperiod and maintained for two months before being used in the test. For each clone, five stem cuttings 1.0–1.5 cm long, with one or two auxiliary buds were placed in plastic tubes with approximately 20 ml of semisolid MS medium. The salt stress experiment was performed by imposing a concentration of 80 mM NaCl (treated plants) or 0 mM NaCl (control plants) for either 2 or 7 days. For each species, 90 plantlets were used. Aboveground and belowground tissue samples from 30 DM and CMM1T plantlets were collected after 7 days at 0 and 80mM NaCl for phenotyping, while the remaining 60 plantlets were used for molecular and biochemical analyses during short (2 days) and prolonged (7 days) stress treatment. That is, for each species and timing, 30 plantlets in a complete randomized design were used to form 3 pools consisting of 10 plantlets each. Plant material, divided into leaves and roots from each pool, was frozen at -80 °C and stored for molecular and biochemical analyses.
Phenotypic responses to salinity
In vitro plantlets were washed in distilled water, dried on filter paper, and separated into shoots and roots. The length of shoot and roots was measured after 7 days of treatment. As reported by Garramone et al.41, several traits were measured to observe stress-induced effects: shoot height (SH, in mm), root length (RL, in mm), number of leaves (LN), number of roots (RN), shoot (SFW) and root (RFW) fresh weight (in mg per plantlet), root (RDW) and shoot (SDW) dry weight (in mg per plantlet), total dry weight (TDW, in mg per plantlet), total fresh weight (TFW, in mg per plantlet), days of root (DR) and shoot (DS) emergence.
Evaluation of proline, total phenols and malondialdehyde content
Fresh leaf and root samples (50 mg) were homogenized in liquid nitrogen and extracted with 70:30 ethanol: water mixture (v/v). Then, 50 µL of extracts were mixed with 100 µL of acidic ninhydrin reagent [1% ninhydrin (w/v) in 60% acetic acid (v/v) and 20% ethanol (v/v)] following a previously described procedure69. Proline concentration was determined from a standard curve and calculated on a fresh weight basis (µmol proline per gram of FW) using three biological and three technical replicates. The total polyphenol content (TPC) of potato leaves and roots was determined using the Folin–Ciocalteu colorimetric method according to70. Briefly, 20 µL of diluted leaf extract and 5 µL of Folin–Ciocalteu reagent were added to 145 µL of ultrapure water in a 96-well microplate. Then, 30 µL of Na2CO3 (20%, w/v) was added to each well and the reaction mixtures were incubated at 25 °C for 45 min. Absorbances were read at 725 nm with a Multiskan Go microplate spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Gallic acid (GA) was used as the reference standard and TPC was estimated from the GA calibration curve (range 5–200 µg/mL, 7 concentration levels; R2 = 0.999). The results were expressed as GA equivalents per 100 g of dried leaf (mg GAE 100 g − 1 FW, mean ± standard deviation of three biological replicates). Lipid peroxidation was estimated by determining the malondialdehyde (MDA) content in potato leaves and roots. One hundred (100) mg of dried samples were homogenized in 2 mL of 0.1% TCA. The homogenate was centrifuged at 15,000×g for 10 min at 4 °C. A 0.5 mL aliquot of the supernatant was mixed with 1.5 mL of 0.5% TBA prepared in TCA 20% and incubated at 90 °C for 20 min. After stopping the reaction in an ice bath, the samples were centrifuged at 10,000× g for 5 min. The absorbance of the supernatant was then measured at 532 nm. After subtracting the non-specific absorbance at 600 nm, the MDA concentration (three replicates per treatment) was determined using the extinction coefficient 155 mM−1 cm−1.
RNA isolation and quantitative real-time PCR (qPCR) assays
One hundred (100) mg of leaf and root pools (three biological replicates) taken from DM and CMM1T plantlets subjected to 0 and 80 mM NaCl for 2 and 7 days, were ground under nitrogen and subsequently used to extract RNA using TRIzol™ (Invitrogen) according to the manufacturer’s protocol. RNA samples were treated with DNase I Amp Grade (Invitrogen) to remove genomic DNA contamination. The purity and concentration of total RNA were determined with a spectrophotometer (NanoDrop ND-1000; Cellbio, Italy). One µg of RNA was reverse transcribed into cDNA using SuperScript™ II First-Strand cDNA Synthesis Reverse Transcriptase. The Reverse Transcriptase (RT) mastermix contained: RT primers (50µM RT primer mix or 10µM oligo (dT)15), 10mM dNTPs, RTase-specific buffer, 0.1 M DTT (SuperScript II only), and 40U RNaseOUT. For CDF expression profiles, qRT primers were designed on nucleotide sequences regions with high level of similarity between S. tuberosum and S. commersonii. Quantitative RT-PCR analysis was performed using SYBR Green dye on a 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). Each 20 µL qPCR reaction contained 0.8 µL of each primer pair, 2 µL of cDNA diluted 1:10 and 10 µL of SYBR Green Mix (Applied Biosystems, Foster City, CA, USA) and 6 µL of H2O. SDS 2.3 and RQ Manager 1.2 software (both Applied Biosystems, Foster City, CA, USA) were used for data processing. Melting curves of PCR products were analyzed for the presence of a single peak. All reactions were performed on one biological pool and three technical replicates, and fold change measures were calculated with the 2−ΔΔCT method. For genes whose expression was not detectable in each sample (therefore it was not possible to compare their expression between the control sample and the treated sample) the analysis was performed using the 2−ΔCT method. Relative expression was normalized to CT values of the S. tuberosum Elongation Factor (EF) gene (GenBank accession number: XM_006343390) used as a housekeeping gene. For each gene the analysis was carried out using as internal calibrator the genotype which in control conditions presented the lowest level of gene expression, to which the value of 1 was therefore attributed. The list of primers is shown in Supplementary Table 3.
Statistical analysis
Quantitative PCR and biochemical data underwent analysis of variance (ANOVA) using SigmaPlot 12 software (Systat Software, Inc., San Jose, California), and multiple pairwise comparisons were assessed using Tukey’s test (p value < 0.05, n = 9). Principal components analysis (PCA) was performed using the factoextra package, and the PCA plot was generated using the ggbiplot package in R. Statistical differences in the phenotypic traits were evaluated by Student’s t-test.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This study was carried out within the Agritech National Research Center and received funding from the European Union Next-GenerationEU (PIANO NAZIONALE DI RIPRESA E RESILIENZA (PNRR) – MISSIONE 4 COMPONENTE 2, INVESTIMENTO 1.4 – D.D. 1032 17/06/2022, CN00000022). The author would like to thank Dr. Elisa Cappetta for her helpful advice on technical issues.
Author contributions
TD, NDA, DC and RA designed the research. AP and RG performed most of the experiments. TD and NDA analyzed the data. TD and VDA drafted the manuscript. NDA, VDA and RA reviewed the manuscript. DC helped with general supervision and critically edited the article. All authors read and approved the final manuscript.
Data availability
Data is provided within the manuscript or supplementary information files.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Teresa Docimo, Email: teresa.docimo@ibbr.cnr.it.
Nunzio D’Agostino, Email: nunzio.dagostino@unina.it.
References
- 1.Fahad, S. et al. Crop production under drought and heat stress: Plant responses and management options. Front. Plant. Sci.8, (2017). [DOI] [PMC free article] [PubMed]
- 2.Iorizzo, M., Mengist, M. F. & D’Agostino, N. Perspectives of advanced genetics and genomics approaches to exploit solanum wild crop relatives for breeding. (2021). 10.1007/978-3-030-30343-3_13
- 3.Baillo, E. H., Kimotho, R. N., Zhang, Z. & Xu, P. Transcription factors associated with abiotic and biotic stress tolerance and their potential for crops improvement. Genes10, 771 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai, X. et al. Genome-wide analysis of plant-specific Dof transcription factor family in tomato. J. Integr. Plant. Biol.55, 552–566 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Venkatesh, J. & Park, S. W. Genome-wide analysis and expression profiling of DNA-binding with one zinc finger (Dof) transcription factor family in potato. Plant. Physiol. Biochem.94, 73–85 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Kang, W. H., Kim, S., Lee, H. A., Choi, D. & Yeom, S. I. Genome-wide analysis of Dof transcription factors reveals functional characteristics during development and response to biotic stresses in pepper. Sci. Rep.6, 33332 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei, Q. et al. Genome-wide identification and characterization of Dof transcription factors in eggplant (Solanum melongena L.). PeerJ6, e4481 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lijavetzky, D., Carbonero, P. & Vicente-Carbajosa, J. Genome-wide comparative phylogenetic analysis of the rice and Arabidopsis Dof gene families. BMC Evol. Biol.3, 17 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu, Z. et al. Genome-wide identification and expression profile of Dof transcription factor gene family in pepper (Capsicum annuum L.). Front. Plant. Sci.7, (2016). [DOI] [PMC free article] [PubMed]
- 10.Liu, Y. et al. Genome-wide analysis of wheat DNA-binding with one finger (Dof) transcription factor genes: evolutionary characteristics and diverse abiotic stress responses. BMC Genom.21, 276 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corrales, A. R. et al. Multifaceted role of cycling DOF factor 3 (CDF3) in the regulation of flowering time and abiotic stress responses in Arabidopsis. Plant. Cell. Environ.40, 748–764 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Shi, Y., Ding, Y. & Yang, S. Molecular regulation of CBF signaling in cold acclimation. Trends Plant. Sci.23, 623–637 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Waschburger, E. L. Turchetto-zolet, A. C. DOF gene family expansion and diversification. Genet. Mol. Biol.3, 1–9 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yin, L. et al. Transcription Factor dynamics in cross-regulation of plant hormone signaling pathways. bioRxiv: The preprint server for biology 2023.03.07.531630 (2023).
- 15.Wang, Z. et al. Emerging roles of plant DNA-binding with one finger transcription factors in various hormone and stress signaling pathways. Front. Plant. Sci.13, (2022). [DOI] [PMC free article] [PubMed]
- 16.Skirycz, A. et al. The DOF transcription factor OBP1 is involved in cell cycle regulation in Arabidopsis thaliana. Plant. J.56, 779–792 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Skirycz, A. et al. Transcription factor AtDOF4;2 affects phenylpropanoid metabolism in Arabidopsis thaliana. New. Phytol175, 425–438 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Skirycz, A. et al. DOF transcription factor AtDof1.1 (OBP2) is part of a regulatory network controlling glucosinolate biosynthesis in Arabidopsis. Plant. J.47, 10–24 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Le Hir, R. & Bellini, C. The plant-specific dof transcription factors family: New players involved in vascular system development and functioning in Arabidopsis. Front. Plant. Sci.4, (2013). [DOI] [PMC free article] [PubMed]
- 20.Kim, H. S. et al. The DOF transcription factor Dof5.1 influences leaf axial patterning by promoting Revoluta transcription in Arabidopsis. Plant. J.64, 524–535 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Wu, Q. et al. Constitutive expression of OsDof4, encoding a C2-C2 zinc finger transcription factor, confesses its distinct flowering effects under long- and short-day photoperiods in rice (Oryza sativa L.). BMC Plant. Biol.17, 166 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fornara, F. et al. Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev. Cell.17, 75–86 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Blair, E. J., Goralogia, G. S., Lincoln, M. J., Imaizumi, T. & Nagel, D. H. Clock-controlled and Cold-Induced CYCLING DOF FACTOR6 alters growth and development in Arabidopsis. Front. Plant. Sci.13, (2022). [DOI] [PMC free article] [PubMed]
- 24.Goralogia, G. S. et al. CYCLING DOF FACTOR 1 represses transcription through the TOPLESS co-repressor to control photoperiodic flowering in Arabidopsis. Plant. J.92, 244–262 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zou, X. & Sun, H. DOF transcription factors: specific regulators of plant biological processes. Front. Plant. Sci. 14, (2023). [DOI] [PMC free article] [PubMed]
- 26.Carrillo, L. et al. Ectopic expression of the AtCDF1 transcription factor in potato enhances tuber starch and amino acid contents and yield under open field conditions. Front. Plant. Sci.14, (2023). [DOI] [PMC free article] [PubMed]
- 27.Renau-Morata, B. et al. CDF transcription factors: Plant regulators to deal with extreme environmental conditions. J. Exp. Bot.71, 3803–3815 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Corrales, A. R. et al. Characterization of tomato Cycling Dof factors reveals conserved and new functions in the control of flowering time and abiotic stress responses. J. Exp. Bot.65, 995–1012 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Waqas, M. et al. Role of DNA-binding with one finger (Dof) transcription factors for abiotic stress tolerance in plants. Trans. Factors Abiotic Stress Tolerance Plants (2020). 10.1016/B978-0-12-819334-1.00001-0
- 30.Ravindran, P., Yong, S. Y., Mohanty, B. & Kumar, P. P. An LRR-only protein regulates abscisic acid-mediated abiotic stress responses during Arabidopsis seed germination. Plant. Cell. Rep.39, 909–920 (2020). [DOI] [PubMed] [Google Scholar]
- 31.Zhang, C. et al. Genome design of hybrid potato. Cell184, 3873-3883e12 (2021). [DOI] [PubMed] [Google Scholar]
- 32.Li, G., Xu, W., Jing, P., Hou, X. & Fan, X. Overexpression of VyDOF8, a Chinese wild grapevine transcription factor gene, enhances drought tolerance in transgenic tobacco. Environ. Exp. Bot.190, 104592 (2021). [Google Scholar]
- 33.Ramírez Gonzales, L. et al. Potato CYCLING DOF FACTOR 1 and its lncRNA counterpart StFLORE link tuber development and drought response. Plant. J.105, 855–869 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Zhu, X. et al. Calcium-dependent protein kinase 32 gene maintains photosynthesis and tolerance of potato in response to salt stress. Sci. Hortic.285, 110179 (2021). [Google Scholar]
- 35.Renau-Morata, B. et al. Ectopic expression of CDF3 genes in tomato enhances biomass production and yield under salinity stress conditions. Front. Plant. Sci.8, (2017). [DOI] [PMC free article] [PubMed]
- 36.Xu, J. & Dai, H. Brassica napus Cycling Dof Factor1 (BnCDF1) is involved in flowering time and freezing tolerance. Plant. Growth Regul.80, 315–322 (2016). [Google Scholar]
- 37.Domínguez-Figueroa, J. et al. The Arabidopsis transcription factor CDF3 is involved in Nitrogen responses and improves nitrogen use efficiency in tomato. Front. Plant. Sci.11, (2020). [DOI] [PMC free article] [PubMed]
- 38.Kloosterman, B. et al. Naturally occurring allele diversity allows potato cultivation in northern latitudes. Nature495, (2013). [DOI] [PubMed]
- 39.Yu, B. et al. Relationship between Potato Canopy-air temperature difference and drought tolerance. Acta Agron. Sin.44, (2018).
- 40.Aversano, R. et al. Stochastic changes affect Solanum wild species following autopolyploidization. J. Exp. Bot.10.1093/jxb/ers357 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garramone, R. et al. In vitro assessment of salt stress tolerance in wild potato species. Agronomy13, 1784 (2023). [Google Scholar]
- 42.Fasano, C. et al. Transcriptome and metabolome of synthetic Solanum autotetraploids reveal key genomic stress events following polyploidization. New. Phytol210, 1382–1394 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Aversano, R. et al. The Solanum commersonii genome sequence provides insights into adaptation to stress conditions and genome evolution of wild potato relatives. Plant. Cell.27, 954–968 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li, Q. et al. Transcriptome analysis uncovers the gene expression profile of salt-stressed potato (Solanum tuberosum L.). Sci. Rep.10, 1–19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tiwari, J. K. et al. Transcriptome analysis of potato shoots, roots and stolons under nitrogen stress. Sci. Rep.10, 1–18 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo, H. et al. Transcriptome analysis reveals multiple effects of nitrogen accumulation and metabolism in the roots, shoots, and leaves of potato (Solanum tuberosum L.). BMC Plant. Biol.22, 1–12 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jian, H. et al. Construction of drought stress regulation networks in potato based on SMRT and RNA sequencing data. BMC Plant. Biol.22, 1–17 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ponce, O. P. et al. Transcriptome profiling shows a rapid variety-specific response in two Andigenum potato varieties under drought stress. Front. Plant. Sci.13, 1–20 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alvarez-Morezuelas, A., Barandalla, L. & Ritter, E. Transcriptome analysis of two tetraploid potato varieties under water-stress conditions. Int. J. Mol. Sci.23, 13905 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kwenda, S., Motlolometsi, T. V., Birch, P. R. J. & Moleleki, L. N. RNA-seq profiling reveals defense responses in a tolerant potato cultivar to stem infection by Pectobacterium carotovorum ssp. brasiliense. Front. Plant. Sci.7, (2016). [DOI] [PMC free article] [PubMed]
- 51.Woolfson, K. N. et al. Transcriptomic analysis of wound-healing in Solanum tuberosum (potato) tubers: evidence for a stepwise induction of suberin-associated genes. Phytochemistry206, 113529 (2023). [DOI] [PubMed] [Google Scholar]
- 52.Yanagisawa, S. Dof domain proteins: Plant-Specific transcription factors associated with diverse phenomena unique to plants. Plant. Cell. Physiol.45, 386–391 (2004). [DOI] [PubMed] [Google Scholar]
- 53.Venkatesh, J., Yu, J. W. & Park, S. W. Genome-wide analysis and expression profiling of the Solanum tuberosum aquaporins. Plant. Physiol. Biochem. PPB73, 392–404 (2013). [DOI] [PubMed] [Google Scholar]
- 54.Costanzo, S. & Jia, Y. Alternatively spliced transcripts of Pi-ta blast resistance gene in Oryza sativa. Plant. Sci.177, 468–478 (2009). [Google Scholar]
- 55.Macknight, R. et al. Functional significance of the alternative transcript processing of the Arabidopsis floral promoter FCA. Plant. Cell.14, 877–888 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Filichkin, S. A. & Mockler, T. C. Unproductive alternative splicing and nonsense mRNAs: A widespread phenomenon among plant circadian clock genes. Biol. Direct7, 20 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smith, C. C. R. et al. Genetics of alternative splicing evolution during sunflower domestication. Proc. Natl. Acad. Sci. USA 115, 6768–6773 (2018). [DOI] [PMC free article] [PubMed]
- 58.Gao, H. et al. PIF4 enhances DNA binding of CDF2 to co-regulate target gene expression and promote Arabidopsis hypocotyl cell elongation. Nat. Plants8, 1082–1093 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu, P., Chen, H. & Cai, W. Transcription factor CDF4 promotes leaf senescence and floral organ abscission by regulating abscisic acid and reactive oxygen species pathways in Arabidopsis. EMBO Rep.21, e48967 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shi, L. et al. Aging later but faster: How < scp > StCDF1 regulates senescence in Solanum tuberosum. New. Phytol242, 2541–2554 (2024). [DOI] [PubMed] [Google Scholar]
- 61.Paysan-Lafosse, T. et al. InterPro in 2022. Nucleic Acids Res.51, D418–D427 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Potter, S. C. et al. HMMER web server: 2018 update. Nucleic Acids Res.46, W200–W204 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Larkin, M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics23, 2947–2948 (2007). [DOI] [PubMed] [Google Scholar]
- 64.Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol.35, 1547–1549 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cho, K. S. et al. The complete chloroplast genome sequences of potato wild relative species, Solanum commersonii. Mitochondrial DNA Part. B1, 241–242 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Danecek, P. et al. Twelve years of SAMtools and BCFtools. Gigascience 10, (2021). [DOI] [PMC free article] [PubMed]
- 67.Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Murashige, T. & Skoog, F. A. Revised medium for rapid growth and bio assays with Tobacco tissue cultures. Physiol. Plant.15, 473–497 (1962). [Google Scholar]
- 69.Velikova, V., Yordanov, I. & Edreva, A. Oxidative stress and some antioxidant systems in acid rain-treated bean plants. Plant. Sci.151, 59–66 (2000). [Google Scholar]
- 70.Singleton, V. L., Orthofer, R. & Lamuela-Ravents, R. M. Analysis of total phenols and other oxidation substrates and antioxidants by means of folin-ciocalteu reagent. Methods Enzymol.299, 152–178 (1998). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is provided within the manuscript or supplementary information files.