Summary
Hypervirulent Klebsiella pneumoniae (hvKp) is a significant causative agent of invasive hepatic abscess syndrome in Asia, presenting substantial clinical challenges due to its intricate pathogenesis. This study revealed the crucial role of the gut microbiota in fortifying the host’s defense against hvKp infection by enhancing interleukin-22 (IL-22), probably through regulating downstream antimicrobial peptides such as Reg3β. In antibiotic-treated mice, we observed that gut microbiota disruption impaired the transformation of tryptophan to indole, a key ligand for the aryl hydrocarbon receptor (AhR), consequently affecting the regulatory functions of IL-22. Our experimental findings revealed that administering rIL-22 or indole propionic acid notably diminished the translocation of hvKp from the intestine to the liver. This research not only underscores the pivotal role of the gut microbiome in modulating tryptophan metabolism and the IL-22 pathway but also highlights its critical function in preventing hvKp migration from the colon to the liver.
Subject areas: Molecular biology, Microbiology, Medical Microbiology
Graphical abstract

Highlights
-
•
Gut microbiome turn tryptophan to IPA, boosting IL-22-Reg3β for intestinal defense
-
•
Antibiotics inhibit IL-22 and Reg3β function by disturbing tryptophan metabolism
-
•
Antibiotics aid gut hvKp translocation to liver via IL-22 and Reg3β down-regulation
-
•
IPA or rIL-22 therapies improve intestinal resistance in antibiotic treated mice
Molecular biology; Microbiology; Medical Microbiology
Introduction
In the mid-1980s, investigators in Taiwan first identified a distinctive syndrome of monomicrobial Klebsiella pneumoniae (K. pneumoniae, Kp) pyogenic liver abscess (KLA), predominantly in diabetic individuals.1 Subsequently, community-acquired K. pneumoniae liver abscess has become an important health problem in parts of Asia, accounting for 80% of all pyogenic liver abscess cases.2,3 This invasive infection often escalates to metastatic complications such as meningitis and endophthalmitis, significantly increasing mortality rates. Patients with KLA alone have a mortality rate of approximately 10%4; among those with metastatic meningitis, this rate increases to 30%–40%.5,6 Some studies have indicated high intestinal colonization rates of hypervirulent Klebsiella pneumoniae (hvKp), a subtype of Kp, in the healthy Asian population.7 This hypervirulent strain is characterized by its iron uptake capabilities, mediated by the aerobactin and salmochelin proteins encoded by the iuc and iro genes, respectively, and its hypermucoid phenotype caused by overexpressed capsule genes (rmpA and rmpA2). There is a notable genetic similarity between hvKp strains found in liver abscesses and those in the intestines.8,9,10 This connection was further supported by a mouse model-based study in which oral administration of hvKp led to liver abscesses,11 suggesting an intestinal origin for KLA.
Clinical research has shown that a history of prior antibiotic use is an independent risk factor for KLA,12 highlighting the possible connection between antibiotic-induced intestinal flora disruption and KLA development. These antibiotics disrupt the equilibrium of the gut microbiota, potentially affecting the expression of vital host immune factors, one of which is interleukin-22 (IL-22).13 IL-22 is a cytokine predominantly expressed in the gut by RORγt + type 3 innate lymphocytes (ILC3s). IL-22 expression relies on the activation of the aryl hydrocarbon receptor (AhR) by indole.14 Recent research has established a connection between intestinal dysbiosis and KLA.15 In our previous research, we noted elevated levels of IL-22 in patients with KLA. Given these findings, IL-22 is potentially a critical factor in KLA development.
In elderly mice, a decrease in intestinal macrophage numbers leads to reduced secretion of growth arrest-specific 6 (Gas6), facilitating the invasion of K. pneumoniae into the intestinal epithelium and its subsequent translocation to the liver. This mechanism may explain the higher incidence of K. pneumoniae infections in older individuals.16 However, the mechanism by which prior antibiotic use contributes to KLA remains incompletely understood. In our study, we utilized a mouse model of liver abscess induced by oral gavage of hvKp to explore the impact of antibiotic exposure on the intestinal immune response to Klebsiella pneumoniae and its impact on susceptibility to KLA. Our findings show that antibiotic-induced intestinal dysbiosis decreases indole derivative levels, reducing IL-22 levels. Reg3β, a C-type lectin antimicrobial protein (AMP) found predominantly in the small intestine and regulated by IL-22, is known for its bactericidal effects on gram-positive and gram-negative bacteria.17,18 The weakened bactericidal activity of Reg3β may results in the significant accumulation of hvKp on the intestinal epithelial surface, promoting its translocation through the epithelial defense barrier into the liver, thereby causing hvKp liver abscesses.
Results
Antibiotic use disrupts the intestinal microbiota composition in noninfected mice
We evaluated the impact of antibiotic treatment on the intestinal microbiota using 16S rRNA gene sequencing analysis of the intestinal microbiota in noninfected ABX (antibiotic-treated) and CON (control) mice (Figure 1A). The rarefaction curve demonstrated adequate sequencing depth (Figure 1B). The significant differences in the Chao1 and Shannon indices indicated that the community richness and community diversity in noninfected ABX mice were lower than those in noninfected CON mice (Figures 1C and 1D). The principal coordinate analysis (PCoA) results demonstrated significant differences between the two groups, indicating substantial variation in their microbial community composition (Figure 1E). Significant variations in the microbiota composition at both the phylum and genus levels are shown in Figures 1F and 1G, respectively. According to the aforementioned data, the dominant phyla of both the noninfected CON and ABX groups were Bacteroidetes (54.67% vs. 2.63%), Firmicutes (36.74% vs. 5.28%), and Proteobacteria (0.24% vs. 90.86%). Unlike in the noninfected CON group, in the noninfected ABX group, only Proteobacteria were enriched, and the other phyla were markedly reduced, with a significant reduction in the Bacteroidetes phylum. An abundance of the phylum Bacteroidetes has been reported to be associated with the production of commensal colonization factors, which are sufficient to cf. protection against colonization and infection by K. pneumoniae.19 At the genus level, the genera Acinetobacter, Pseudomonas, Stenotrophomonas, Klebsiella, and Staphylococcus were significantly enriched in the noninfected ABX group. The most abundant genera in the noninfected CON group were Muribaculaceae, Lactobacillus, Prevotellaceae_UCG-001, and Bacteroides. In conclusion, our findings demonstrate profound alterations in the intestinal microbiota composition following antibiotic treatment in noninfected mice, characterized by a substantial reduction in microbial diversity and a shift in microbial dominance. The aforementioned results indicate that the intestinal microecological balance of the mice was successfully disrupted by oral antibiotic treatment.
Figure 1.
Antibiotic use disrupts the intestinal microbiota composition in noninfected mice
(A) Experimental scheme for (B–G). The mice were randomly divided into antibiotic (ABX) and control (CON) groups (n = 10). Mice in the ABX group were administered antibiotic suspensions via gavage for 7 consecutive days, while those in the CON group received an equal volume of PBS via gavage for 7 consecutive days. Fecal samples were collected from the mice on the 9th day.
(B) The rarefaction curve illustrates adequate sequencing depth, with the x axis representing the number of reads sampled and the y axis showing the Sobs index at the amplicon sequence variant (ASV) level.
(C and D) Alpha diversity comparison using Chao (C) and Shannon (D) indices at the ASV level between ABX and CON mice, revealing significant differences (p = 0.00003653 and p = 0.0008743, Wilcoxon rank-sum test).
(E) Principal coordinate analysis (PCoA) based on Bray-Curtis dissimilarity at the ASV level (p = 0.001, ANOSIM).
(F) Distinct microbiota compositions between ABX and CON mice according to phylum-level community bar plot analysis.
(G) Heatmap of genera in ABX and CON mice. Heatmap of the genera in the top 30 in terms of total abundance at the classification level (n = 10). The color of the spots in the right panel represents the mean relative abundance in each sample (the colors blue, white, and red represent low abundance, intermediate abundance, and high abundance, respectively). The x axis and y axis are the sample and species clustering trees, respectively.
Depletion of the intestinal microbiota impairs host defenses against Klebsiella pneumoniae liver abscess
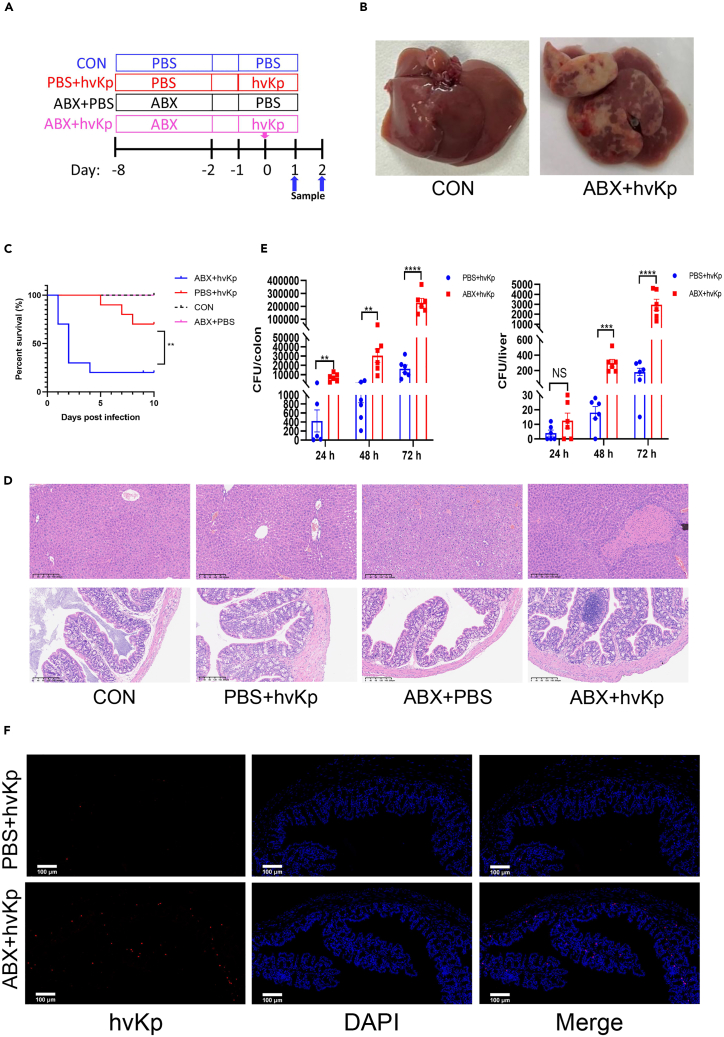
Broad-spectrum antibiotics were used to deplete the gut flora in mice. Subsequently, a liver abscess model was induced via hvKp gavage to clarify the effect of antibiotic use on liver abscesses in mice infected with hvKp (Figure 2A). Forty-eight hours after infection with hvKp, all ABX group mouse livers developed obvious abscesses visible to the naked eye (Figure 2B). The antibiotic-treated group (ABX + hvKp group) exhibited greater mortality rates than the PBS + hvKp group after infection with hvKp (Figure 2C). At 48 h postinfection, visually, the colon in the ABX + hvKp group was noticeably swollen and inflamed compared to that in the PBS + hvKp group (Figure S1). The ABX+ hvKp group displayed macroscopically distinct liver abscesses, and liver cell necrosis, microabscess formation, and altered liver cell structures were observed by H&E staining. Additional H&E staining revealed a thinner colonic wall, increased inflammatory cell infiltration, irregular villi, and a marked reduction or absence of crypts and goblet cells (Figure 2D). These results highlighted the enhanced colonic pathology in the ABX group after hvKp infection. Moreover, an increased bacterial burden within the colon and liver of the ABX group indicated a diminished capacity to eliminate pathogens (Figure 2E), and fluorescence in situ hybridization (FISH) staining results (Figure 2F) likewise showed more hvKp colonization in the colon of ABX mice than in the colon of PBS + hvKp group mice. The results indicate that antibiotic-induced depletion of the intestinal microbiota significantly impairs the host’s defense against bacterial invasion, leading to increased susceptibility and severity of Klebsiella pneumoniae liver abscesses in mice.
Figure 2.
Depletion of the intestinal microbiota impairs host defense against Klebsiella pneumoniae liver abscess
(A) Treatment scheme used to analyze differences in the susceptibility of hvKp to antibiotics. Six- to eight-week-old mice were randomly divided into the following four groups—the CON group (PBS + PBS), PBS + hvKp group, ABX + PBS group and ABX + hvKp group—and were gavaged with antibiotics or PBS for 7 days prior to the oral inoculation of hvKp or PBS. The mice were euthanized at 24, 48 and 72 h post-infection, and their colons and livers were harvested.
(B) The appearance of the liver in the CON (left) and ABX + hvkp (right) groups. Randomly selected images are shown for each group.
(C) Effect of antibiotics on the survival of mice infected with hvKp. Survival was monitored daily for 10 consecutive days (n = 10).
(D) H&E-stained images of liver tissue sections (top) and proximal colon cross-sections (bottom) from each group of mice. Randomly selected images are shown for each group. Scale bar: 200 μm.
(E) Bacterial counts in the colon and liver were determined at different points in time after hvKp infection in the PBS + hvKp and ABX + hvKp groups. Whole colon (left) and liver (right) tissues from each mouse were homogenized in 1 mL of PBS, and 20 μL of each homogenate was then plated onto blood agar plates and incubated at 37°C overnight. The number of colony forming units (CFUs) was determined the next day. Each dot represents the value from an individual mouse (n = 6).
(F) Colon sections were taken from PBS or ABX mice at 48 h after infection with hvKp and stained with a K. pneumoniae probe (red) and DAPI (blue). Scale bar: 100 μm.
Throughout, the data are presented as the mean ± SEM. NS: not significant, ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
Antibiotic use attenuated the protective effect of IL-22 on hvKp-induced liver abscesses
The underlying mechanism by which depletion of the gut microbiota by antibiotics significantly increases susceptibility to hvKp infection was subsequently investigated, and the role of IL-22, a cytokine primarily acting on epithelial cells in the intestines, skin, and respiratory tract, was explored. Both the qPCR and ELISA results confirmed that hvKp infection induced an increase in IL-22 in mice without antibiotic treatment, but this increase was suppressed in ABX mice (Figures 3A and 3B). Furthermore, we compared the serum IL-22 levels between liver abscess patients and healthy controls. Individuals with liver abscesses exhibited higher IL-22 levels than their healthy counterparts (Figure 3C). Interestingly, IL-22 levels did not significantly increase in patients with liver abscesses of other bacterial origins, which likely stemmed from primary hepatobiliary disorders rather than intestinal infections. Furthermore, a retrospective study of clinical specimens revealed that IL-22 levels were lower in patients with KLA who had a history of antibiotic use than in those without a history of antibiotic use (Figure 3D). This distinction underscores the protective role of IL-22 specifically in hvKp infection scenarios. We also investigated whether IL-10 and IL-17, which belong to the IL-10 family along with IL-22, play roles in antibiotic-induced bacterial invasion of the gut (Figures 3A and 3B). The results showed that IL-17 and IL-10 did not significantly affect hvKp infection in mice. Plasma IL-6 levels in ABX mice infected with hvKp were greater than those in control mice (Figure 3E). We also examined the mRNA expression levels of molecules downstream of IL-22, such as Cldn2 (claudin 2), Ocln (occludin), and Reg3β (regenerating islet-derived 3 beta), as well as ZO1 (zona occludens 1) and Tff3 (trefoil factor 3), which play protective roles in the intestine (Figure S2).20,21,22,23 Considering the trend of Reg3β expression in this study and its antimicrobial properties against gram-negative bacteria,17 which may play a protective role in hvKp infection, we conducted further protein analysis of Reg3β. Both the qPCR and western blotting results showed that Reg3β expression increases during hvKp infection. However, this increase was not detected in ABX mice (Figures 3F–3H). Based on the qPCR results of occludin and claudin-2, immunofluorescence staining was performed to examine the expression levels of them in the colon group (Figure S3). In conclusion, our results highlight the diminished protective effect of IL-22 in KLA due to antibiotic use. This effect of IL-22 may be mediated through the regulation of Reg3β.
Figure 3.
Antibiotic use attenuated the protective effect of IL-22 on hvKp-induced liver abscesses
(A) mRNA expression levels of Il-22 (n = 6), Il-10 (n = 4–5), and Il-17a (n = 6) in the colons of PBS and ABX mice at 24 h post infection with hvKp or control solvent (PBS).
(B) Plasma was obtained by blood sampling from the eyeballs of mice 48 h after infection. Plasma concentrations of IL-22 (n = 5), IL-10 (n = 4), and IL-17A (n = 6) in the four groups determined by ELISA.
(C) Serum IL-22 concentrations in clinical specimens from healthy individuals (n = 50), KLA patients (n = 41), and NKLA patients (non-K pneumoniae liver abscess, n = 20).
(D) Comparison of serum IL-22 levels in KLA patients with (n = 8) and without (n = 33) a history of antibiotic use.
(E) Plasma was obtained by blood sampling from the eyeballs of mice 48 h after infection. Plasma IL-6 concentrations in four groups of mice measured by ELISA (n = 6).
(F) mRNA expression levels of Reg3β in the colon of mice at 24 h post infection (n = 4).
(G) Western blot analysis showing Reg3β protein expression in the colons of different groups at 48 h post infection (n = 3).
(H) The relative densitometric intensities of Reg3β were determined and then normalized to those of β-actin (n = 3).
Throughout, the data are presented as the mean ± SEM. NS: not significant, ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
Fecal microbiota transplantation alleviates hvKp liver abscess in antibiotic-treated mice
To delineate the influence of the gut microbiota, we replenished the gut microbiota in ABX mice by administering fecal suspensions from healthy C57BL/6 mice (Figure 4A). Forty-eight hours after hvKp gavage, mice that underwent fecal microbiota transplantation (FMT) displayed significantly lower bacterial loads in both colonic and hepatic tissues than those observed in ABX-treated mice (Figure 4B). These results showed that hvKp infection in mice was attenuated after FMT compared to that in the ABX group. The H&E staining results of liver and colon sections from the mice showed the same trends (Figure 4C). Plasma IL-22 levels were elevated in FMT mice, albeit below control levels, and IL-6 levels were decreased (Figures 4D and 4E). Additionally, the upregulation of Reg3β expression in FMT mice was revealed by western blotting (Figures 4F and 4G). These results collectively suggest that FMT effectively reduces the severity of hvKp liver abscesses in mice treated with antibiotics, demonstrating its potential as a therapeutic intervention.
Figure 4.
Fecal microbiota transplantation alleviates hvKp liver abscess in antibiotic-treated mice
(A) Treatment scheme used to analyze fecal microbiota transplantation (FMT) differences in susceptibility to hvKp. Mice were randomly divided into three groups: CON, ABX, and FMT. Before fecal transplantation, mice in the ABX and FMT groups were gavaged with antibiotics for 7 days, while PBS was used as a control for mice in the CON group. Mice in the FMT group were gavaged with fecal suspensions, and mice in the ABX and CON groups were gavaged with PBS for 3 days. Finally, 3 groups of mice were gavaged with 1.5 × 108 CFU of hvKp. At 48 h post infection, the mice were euthanized, and the colon and liver were harvested.
(B) Bacterial counts in the colon and liver were determined 48 h after hvKp infection. Whole colon (left) and liver (right) tissues from each mouse were homogenized in 1 mL of PBS, and 20 μL of each homogenate was plated onto blood agar plates and incubated at 37°C overnight. The numbers of CFUs were determined the next day. Each dot represents the value from an individual mouse (n = 6).
(C) H&E-stained images of liver tissue sections (top) and proximal colon cross-sections (bottom) from control (left), ABX (middle), and FMT (right) mice at 48 h post infection. Randomly selected images are shown for each group. Scale bar: 200 μm.
(D) Plasma was obtained by blood sampling from the eyeballs of mice 48 h after infection. Plasma concentrations of IL-22 in the CON (n = 5), ABX (n = 5), and FMT (n = 10) groups were measured by ELISA.
(E) Plasma concentrations of IL-6 in each group at 48 h post infection were measured by ELISA (n = 6).
(F) Detection of Reg3β by western blotting. Colon tissues were collected, homogenized, and analyzed using an anti-Reg3β antibody (n = 3).
(G) The relative densitometric intensities of Reg3β were determined and then normalized to those of β-actin (n = 3).
Throughout, the data are presented as the mean ± SEM. NS: not significant, ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
Recombinant IL-22 administration provided protection against hvKp in ABX mice
To counteract the reduction in intestinal IL-22 expression in ABX mice, recombinant IL-22 (rIL-22, 0.5 μg) was administered 4 h after hvKp infection (Figure 5A). Compared with untreated ABX mice, mice receiving rIL-22 treatment exhibited improved 10-day survival rates and significantly lower hvKp bacterial loads in both liver and colon tissues (Figures 5B and 5C). Compared with those in ABX mice, histological analyses revealed decreased mucosal erosion, crypt destruction, and inflammatory cell infiltration in rIL-22 mice (Figure 5D). IL-6 was not restored after rIL-22 supplementation, and this increase might be related to immune activation in response to hvKp infection (Figure 5E). Importantly, Reg3β mRNA and protein levels in the colon of rIL-22-treated mice were notably lower than those in the ABX group (Figures 5F–5H). These results confirm the pivotal role of rIL-22 supplementation in alleviating hvKp infection in antibiotic-treated mice, suggesting its therapeutic potential in such infection scenarios.
Figure 5.
Recombinant IL-22 administration provided protection against hvKp in ABX mice
(A) Treatment scheme used to analyze the effect of rIL-22 on susceptibility to hvKp. Six- to eight-week-old mice were gavaged with antibiotics for 7 days prior to oral inoculation of hvKp. Four hours after infection, the mice were injected intraperitoneally with 0.5 μg of rIL-22 (rIL-22 group) or an equal amount of PBS (CON group). Mice were euthanized 24 and 48 h after infection, and the colon and liver were harvested.
(B) Effect of rIL-22 on the survival of mice infected with hvKp. Survival rates were monitored daily for 10 consecutive days (n = 10).
(C) Bacterial counts in the colons and livers were determined 48 h after hvKp infection. Whole colon (left) and liver (right) tissues from each mouse were homogenized in 1 mL of PBS, and 20 μL of each homogenate was plated onto blood agar plates and incubated at 37°C overnight. The numbers of CFUs were determined the next day. Each dot represents the value from an individual mouse (n = 6).
(D) H&E-stained images of liver tissue sections (top) and proximal colon cross-sections (bottom) from CON (left) and ABX + rIL-22 group (right) mice at 48 h post infection. Randomly selected images are shown for each group. Scale bar: 200 μm.
(E) Plasma was obtained by blood sampling from the eyeballs of mice 48 h after infection. Plasma concentrations of IL-6 at 48 h post infection were measured by ELISA (n = 8–9).
(F) mRNA expression levels of Reg3β in the colons of the indicated mice at 24 h after infection (n = 4–5).
(G) Western blot analysis showing Reg3β protein expression in the colons of different groups at 48 h post infection (n = 3).
(H) The relative densitometric intensities of Reg3β were determined and then normalized to those of β-actin (n = 3).
Throughout, the data are presented as the mean ± SEM. NS: not significant, ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
Disruption of tryptophan metabolism in antibiotic-treated mice affects susceptibility to KLA
Our results suggest that the gut microbiota in ABX mice influences susceptibility to hvKp-induced liver abscess by altering IL-22 production. This effect may stem from the influence of the microbiota on tryptophan metabolism within the gastrointestinal tract. Tryptophan is metabolized into indole derivatives such as indole-3-acetic acid (IAA) by gut bacteria or into kynurenine (Kyn) by host cells via indoleamine 2,3-dioxygenase. Indole derivatives are AhR ligands known to promote local IL-22 production. We performed tryptophan metabolism analyses on mouse (noninfected ABX [antibiotic-treated] and CON [control] mice) and clinical case specimens (liver abscess patients [KLA] and healthy controls [healthy]). Tryptophan metabolism assays revealed that IAA and indole propionic acid (IPA) constitute significant components of tryptophan metabolism (Figures 6A and 6B). ABX mice exhibited impaired IAA production and low IPA levels, likely due to an underdeveloped gut immune system (Figure 6C), similar to clinical patient specimens (Figure 6D). These results collectively indicate disrupted tryptophan (Trp) metabolism as a key factor in the increased susceptibility to KLA in antibiotic-treated mice and patients.
Figure 6.
Tryptophan metabolism in ABX mice significantly deviated from that in control mice
(A) Cluster analysis plot: comparative expression patterns of intestinal tryptophan metabolites in the ABX group versus the CON group (n = 10).
(B) Cluster analysis plot: expression patterns of tryptophan metabolites in clinical specimens. Comparison between KLA (n = 14) and healthy individuals (n = 10), showing divergent metabolite profiles.
(C) Histogram of the relative abundance of mouse intestinal tryptophan metabolites in the ABX and CON groups (n = 10).
(D) Histogram of relative abundance: Analysis of tryptophan metabolites in clinical specimens from KLA (n = 14) and healthy individuals (n = 10).
Indole propionic acid therapy reduces the KLA burden in antibiotic-treated mice
Significant differences in IPA compared to those in the control group were observed in both our clinical specimens and mouse models (Figure 7A). Despite the minimal IAA differences observed in clinical specimens between the disease and control groups (Figure 7A), considering the therapeutic effects of IAA in other disease models, we supplemented our ABX mouse KLA model with either IAA or IPA at 20 mg/kg/day (Figure 7C). IPA administration markedly improved IL-22 expression deficits in ABX mice (Figure 7D). Administration of IPA to ABX mice significantly reduced hvKp bacterial loads in the colon and liver tissues (Figure 7E). However, IAA plays no role in this process (Figures 7D and 7E). These findings suggest that antibiotics disrupt the intestinal flora, leading to decreased colonic IL-22 expression by altering tryptophan metabolism. This disruption contributes to increased susceptibility to KLA in ABX mice by impairing IL-22 signaling pathways.
Figure 7.
Indole propionic acid therapy reduces KLA burden in antibiotic-treated mice
(A) IAA Concentrations: Comparative analysis of IAA levels in feces from CON and ABX mice (left, n = 10) and clinical samples from healthy controls (n = 10) and KLA patients (right, n = 14).
(B) IPA concentrations: Comparative analysis of IPA levels in feces from CON and ABX mice (left, n = 10) and clinical samples from healthy controls and KLA patients (right, n = 14).
(C) Experimental scheme for (C–E). Mice were randomly divided into four groups (n = 6 in each group): CON, ABX, and indole groups. The ABX mice were treated with a 7-day continuous gavage of antibiotic suspension. Four hours after daily antibiotic gavage, the indole groups began receiving IPA/IAA treatment at 20 mg/kg/day through gavage. The control group was gavaged with an equal volume of solvent. After 7 days, all groups were gavaged with 1.5 × 108 CFU of hvKp. At 48 h post-infection, the mice were euthanized, and blood, liver tissue, and colon tissue samples were collected for analysis.
(D) Plasma was obtained by blood sampling from the eyeballs of mice 48 h after infection. Plasma concentrations of IL-22 in the four groups were measured by ELISA (n = 5).
(E) Bacterial counts in the colons (left) and livers (right) were determined 48 h after infection. Colon and liver tissues were homogenized in PBS, and the homogenates were plated onto blood agar plates and incubated overnight. The numbers of CFUs were determined the next day. Each dot represents the value from an individual mouse (n = 6).
Throughout, the data are presented as the mean ± SEM. NS: not significant, ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
Discussion
Clinical research has shown that a history of antibiotic exposure is an independent risk factor for KLA,12 with further evidence connecting gut dysbiosis to this disease.15 However, the exact mechanisms by which antibiotics promote the development of KLA are not fully understood. Our research reveals the pathogenesis of KLA and highlights the role of metabolic pathways, particularly the reduction in IPA following antibiotic use, in affecting host immunity. This reduction leads to lower levels of IL-22, which may impair the bactericidal function of Reg3β and compromises the intestinal barrier. This process facilitates the migration of hvKp from the intestinal tract to the liver and promotes abscess formation. Our study sheds light on the host immune response mechanisms underlying KLA, providing a basis for innovative treatments, such as probiotics expressing recombinant IL-22, and advancing immunotherapy approaches.
Previous research has shown that antibiotics disrupt the balance of the gut microbiota and thereby interfere with the normal expression of host immune factors, particularly IL-22, which plays a critical role in the antimicrobial defense of the intestinal epithelium.24,25 Our prior clinical observations also revealed an increase in IL-22 levels among KLA patients. Building on these insights, our study explored the role of IL-22 in the pathogenesis of antibiotic-associated KLA and aimed to elucidate its critical impact on disease progression. We found that ABX mice had a reduced ability to eliminate K. pneumoniae, and hvKp infection induced an increase in IL-22, which was inhibited in ABX mice. Supplementation with IL-22 alleviated hvKp infection in ABX mice. The specific alteration in IL-22 levels, as opposed to IL-17A and IL-10, suggested that the gut microbiota predominantly modulates the immune response to hvKp through IL-22. This is possibly because K. pneumoniae can bypass IL-17A defenses by encapsulation for effective colonization.19 However, rIL-22 supplementation did not reduce IL-6 levels, probably because of the strong systemic response to hvKp infection. This response might overshadow the effects of IL-22, or there could be a synergistic interaction between IL-22 and IL-6.26 Intriguingly, the serum IL-22 levels in patients with liver abscesses caused by pathogens other than hvKp did not significantly deviate from those in healthy controls, likely reflecting the varied etiologies of liver abscesses. Liver abscesses caused by other pathogens are usually secondary to the breakdown of the hepatobiliary system barrier caused by primary hepatobiliary diseases27,28 because there is less damage to the intestinal barrier involved, which may not trigger the same IL-22-mediated immune response. Moreover, our study also revealed a notable difference in IL-22 levels between KLA patients with and without a history of antibiotic use (within the last 30 days), as shown in Figure 3D. This finding implies that the history of antibiotic use plays a crucial role in modulating the host’s defense mechanisms against KLA by influencing IL-22 levels.
The microbiome is a key regulator of IL-22 activity via the synthesis of L-Trp-derived AhR ligands.24,29,30,31,32 Our results revealed a marked reduction in L-tryptophan (L-Trp)-derived metabolites, especially IPA, in the feces of both KLA patients and ABX mice. Moreover, supplementing IPA in the ABX mouse model substantiated its therapeutic potential. However, the role of IAA in hvKp liver abscess remains ambiguous, potentially because we did not find a suitable experimental concentration or duration of drug administration. According to our microbiome sequencing results, the number of Bacteroides and the abundance of Lactobacillus were significantly reduced in ABX mice. Studies have shown that Lactobacillus species metabolize Trp into indole-3-aldehydes, promoting AhR-dependent IL-22 expression.24 Furthermore, Bacteroidetes play a crucial role in defending against K. pneumoniae colonization and transmission.19 In addition, an increased presence of Acinetobacter and Pseudomonas was noted in ABX mice. The phenomenon of cross-protection and cross-feeding between Klebsiella pneumoniae and Acinetobacter baumannii is instrumental in facilitating their coexistence.33 Moreover, the biomass of Klebsiella pneumoniae within biofilms cocultured with Pseudomonas was significantly greater than that in mono-culture biofilms, suggesting the possibility of metabolic cooperation within mixed-species biofilms.34 Different bacterial enzymes are responsible for the production of certain indole derivatives in the process of tryptophan catabolism.35 These previous studies imply a necessity for an in-depth investigation into the precise modalities through which the intestinal microbiota engages in dialogue with the host immune apparatus. In particular, future research should identify which bacterial species produce specific AhR ligands, such as IPA, providing molecular insights and identifying new therapeutic targets for KLA.
The gut is a primary site of antimicrobial defense, with intestinal epithelial cells secreting various AMPs, including β-defensins, regenerating proteins, and S100 proteins.17,36 In human anatomy, the primary producers of AMPs are specialized Paneth cells, which are predominantly located in the small intestine and are less abundant in the proximal colon; IL-22 plays a pivotal role in the development of Paneth cells and expression of AMPs.37,38 Studies suggest that Reg3β binds to lipopolysaccharide (LPS), killing gram-negative bacteria.39,40 The human gut is a reservoir for hypervirulent K. pneumoniae,7,41,42,43 particularly in the Chinese population,44 with hvKp carriage being a risk factor for invasive infections and liver abscesses.10,45,46 However, most research on K. pneumoniae pathogenic mechanisms has focused on virulence,47,48,49,50 overlooking the defense of intestinal AMPs against hvKp. Our study revealed the potential protective role of Reg3β, which is regulated by IL-22, in KLA pathogenesis. Claudin-2, a prototypic pore-forming claudin, forms actively gated channels that are selective for water and small cations.51,52 IL-22 stimulation leads to the upregulation of epithelial claudin-2 expression and can thereby elicit water efflux, diarrhea, and pathogen clearance in the intestine.53 Additionally, occludin, which is crucial for tight junction integrity in epithelial and endothelial cells, plays a pivotal role in maintaining barrier functions.54 In our KLA model, an unexpected increase in claudin-2 expression was observed in the intestines of ABX mice despite a decrease in IL-22, suggesting that severe infections in ABX mice may drive this increase, boosting intestinal permeability and facilitating pathogen elimination. Furthermore, research indicates that IL-1, IL-6, IL-13, and tumor necrosis factor (TNF) also potentially elevate claudin-2 expression.53,55,56,57,58 Notably, occludin expression was reduced in ABX mice, indicating enhanced vulnerability of the colonic epithelium to infection, with a more significant decrease in the ABX group than that observed in the control group, highlighting the impact of infection severity on tight junction proteins.
In conclusion, our study underscores the pivotal influence of antibiotic-induced intestinal dysbiosis on the host immune response toward hvKp using a mouse model of liver abscess induced by oral gavage of hvKp. Our findings revealed that such dysbiosis curtails the production of indole derivatives, specifically IPA, and diminishes IL-22 levels. In addition, the down-regulation of Reg3β was also involved. The cascade effect of IPA-IL-22 may significantly reduce the expression of Reg3β, an essential antimicrobial peptide, thereby potentially facilitating the proliferation of hvKp. The ensuing disruption of the intestinal barrier permits hvKp to translocate to the liver, consequently increasing susceptibility to KLA. This finding underscores the importance of understanding the interplay between antibiotic use, intestinal microbiota, and immune responses in the pathogenesis of KLA.
Limitations of the study
Our investigation, while contributing valuable insights, has certain limitations, particularly in microbiota transplantation experiments, where the transplanted group showed better therapeutic effects than healthy mice. Such observations suggest a complex interplay of factors, potentially involving competitive exclusion or a fortified immune response post FMT in resisting hvKp infection. Further exploration is needed to pinpoint the specific cellular entities involved in indole-mediated modulation of IL-22 and the regulatory dynamics between IL-22 and Reg3β within the context of KLA. Several literatures have confirmed the role of Reg3β as a downstream molecule of IL-22 in combating Gram-negative bacteria. Based on our experimental findings, the down-regulation of Reg3β can be observed during hvKp infection in ABX mice. Therefore, we suppose the downstream effects of IL-22 is due to Reg3β. However, more evidences from Reg3β depletion and supplementation model are needed to define the protective role of Reg3β dependent on IL-22 in hvKp-induced liver abscess.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Xiuyu Xu (xuxiuyu85@126.com).
Materials availability
There are restrictions to the availability of the hypervirulent Klebsiella pneumoniae (hvKp) strain due to our use of clinical specimens isolated from patients.
Data and code availability
-
•
Data have been deposited at Figshare and are publicly available as of the date of publication.
-
•
The strain genomic data, metabolomic datasets and mouse gut microbiota 16S rRNA datasets generated during the current study will be available in the public repository. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Acknowledgments
This study was supported by a grant from the Chongqing Municipal Science and Technology Commission Natural Science Foundation General Program (cstc2020jcyj-msxmX0199). We thank The First Affiliated Hospital of Chongqing Medical University, the School of Laboratory Medicine of Chongqing Medical University, and all the authors involved in this study.
Author contributions
Conceptualization, X.X, X.Y., and L.W.; methodology, X.Y., L.W., H.W., and Y.X.; investigation, X.Y., L.W., H.W., Y.X, Y.C., X.M., X.J., and H.X.; writing – original draft, X.Y., L.W., H.W; writing – review & editing, X.Y., L.W., and X.X.; funding acquisition, X.X.; resources, H.W. and X.X.; supervision, H.W. and X.X.
Declaration of interests
The authors declare no competing interests.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| REG3B Polyclonal Antibody | Thermofisher | Cat #PA5-47700 |
| HRP∗Polyclonal Rabbit Anti Sheep IgG (h+l) | Immunoway | Cat #RS030231 |
| Goat Anti-Rabbit IgG H&L (HRP) | Abcam | Cat #Ab6721; RRID: AB_955447 |
| Rabbit Anti-beta-Actin Monoclonal Antibody, HRP Conjugated, Clone13E5 | Cell Signaling Technology | Cat # 5125; RRID: AB_1903890 |
| Anti-Claudin 2 Antibody | Abcam | Cat #ab53032; RRID: AB_869174 |
| Occludin (E6B4R) Rabbit mAb | Cell Signaling Technology | Cat #91131; RRID: AB_2934013 |
| Bacterial and virus strains | ||
| Hypervirulent Klebsiella pneumoniae | The first affiliated hospital of Chongqing Medical University | 2203184066 |
| Chemicals, peptides, and recombinant proteins | ||
| IL-22-Protein-Mouse-CHO | MedChemExpress | Cat # HY-P72793 |
| Vancomycin | Macklin | Cat # V871983-5g |
| Metronidazole | Macklin | Cat # M813526-5g |
| Neomycin sulfate | Macklin | Cat # N814740-5g |
| Ampicillin | Macklin | Cat # A830931-5g |
| Indole-3-acetic acid | Macklin | Cat # I6311-5g |
| 3-Indolepropionic acid | Macklin | Cat # I811687-5g |
| PageRuler Prestained Protein Ladder | Thermofisher | Cat # 26616 |
| LB Broth, powder | Solarbio | Cat # L1010 |
| RNAiso Plus reagent | TaKaRa | Cat # 9108 |
| Critical commercial assays | ||
| Color PAGE Gel Rapid Preparation Kit | Epizyme | Cat # PG113 |
| SYBR Green Pro Taq HS Premix Ⅳ | Agbio | Cat # AG11746 |
| Evo M-MLV RT Kit with gDNA Clean for qPCR | Agbio | Cat # AG11705 |
| ELISA MAX™ Deluxe Set Mouse IL-6 | Biolegend | Cat # 431304 |
| ELISA MAX™ Deluxe Set Mouse IL-22 | Biolegend | Cat # 436304 |
| ELISA MAX™ Deluxe Set Mouse IL-10 | Biolegend | Cat # 431414 |
| ELISA MAX™ Deluxe Set Mouse IL-17A | Biolegend | Cat # 432504 |
| Deposited data | ||
| Raw and analyzed data | This paper | Figshare: https://doi.org/10.6084/m9.figshare.25971472 |
| Mouse gut microbiota 16S rRNA | This paper | https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1120022 |
| Strain genomic data | This paper | NCBI database: JBDOIS000000000 |
| Tryptophan Metabolite Analysis | This paper | MetaboLights: MTBLS10364 |
| Experimental models: Organisms/strains | ||
| Mouse:C57BL/6J | Beijing Spfbiotech | RRID: IMSR_JAX:000664 |
| Oligonucleotides | ||
| Primers for qPCR, see Table S1 | This paper | N/A |
| Software and algorithms | ||
| GraphPad Prism 8.0 | GraphPad Software | https://www.graphpad-prism.cn/ |
| ImageJ | ImageJ Software | https://imagej.nih.gov/ij/ |
| BioRender | BioRender Software | BioRender:GE271UBGFG |
Experimental model and study participant details
Bacterial strain
The hypervirulent Klebsiella pneumoniae (hvKp) strain used in this study was isolated from liver drainage fluid from a liver abscess patient at the First Affiliated Hospital of Chongqing Medical University. The capsular serotype of the strain was first identified by amplifying the wzi gene using the primers wzi_for2 (GTGCCGCGAGCGCTTTCTATCTTGGTATTCC) and wzi_rev (GAGAGCCACTGGTTCCAGAATTTGACCGC) and subsequent sequencing. Two microliters of the extracted DNA was used as the template for PCR. The 50-μL PCR mixture contained 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 1.5 mM MgCl, 0.2 μM of each primer, 100 μM of each deoxynucleoside triphosphate, and 0.17 μL of Taq polymerase. After denaturation at 94°C for 2 minutes, 30 cycles were performed (94°C for 30 seconds, 55°C for 40 seconds, and 72°C for 30 seconds), followed by a final elongation step at 72°C for 5 minutes. The amplification products were then sequenced, and the sequences were compared against the capsular typing database (https://bigsdb.Pasteur.fr/klebsiella/), verifying that the strain was serotype K1 (Figure S4 and Data S1). Whole-genome sequencing was conducted to further characterize the strain, revealing that the capsular type of the strain is K1, which is typically associated with hypervirulent K. pneumoniae (Kp). The genome sequence analysis confirmed the hypervirulent nature of the strain (iroB+, rmpA+, iutA+, iucA+, iucB+, iucC+, iucD+).59 The presence of the aerobactin coding genes iucA+, iucB+, iucC+, iucD+, and iutA+ is typically associated with high-virulence plasmids. The strain information, including genomic data, has been deposited in the NCBI database: JBDOIS000000000, ensuring its availability and accessibility for further research and verification within the scientific community.
Animals
The experimental animals used in this study were healthy male WT C57BL/6J mice, aged 6–8 weeks, purchased from Beijing Spfbiotech Co., Ltd. The mice were housed in a standard SPF-grade animal facility at Chongqing Medical University. All animal experiments described in this study were reviewed and approved by the Animal Ethics Committee of Chongqing Medical University.
Antibiotics disrupt the gut microbiota in mice
Six- to eight-week-old male C57BL/6 mice weighing 18–20 grams were randomly divided into groups to ensure similar body weights for various treatments. For the antibiotic-induced gut microbiota dysbiosis procedure, the mice were divided into CON (control: PBS gavage) and ABX (antibiotics: broad-spectrum antibiotic gavage) groups. The antibiotic mixture included vancomycin (10 mg/mL, t1/2: 4-11 hours), neomycin sulfate (20 mg/mL, t1/2: 2 to 3 hours), metronidazole (20 mg/mL, t1/2: 8 hours), and ampicillin (20 mg/mL, t1/2: 1 hour), each dissolved in PBS. Approximately 200 μL of the mixture was administered via gavage daily for 7 days. Forty-eight hours after the final administration of antibiotics via oral gavage, antibiotics were administered via subsequent procedures. Given the half-life of the antibiotics, a period of 48 hours is sufficient to for complete metabolization, preventing interference with subsequent experiments. Mouse feces were collected 48 hours after the last antibiotic gavage.
Procedure for generating hypervirulent Kp-infected mice
Mice were fasted overnight prior to the day of infection. Subgroups received 200 μL of 1.5 × 108 CFUs of hvKp; 3.0 × 108 CFUs of hvKp were used in the survival experiments. Gavage was performed using a 12-gauge gavage needle, with the control groups receiving sterile 1 × PBS. Post infection, the mice were fasted for an additional 6 hours before they resumed feeding. At 24 or 48 hours post hvKp infection, the mice were weighed and anesthetized with sodium pentobarbital, and blood was collected via eyeball removal into 1.5 mL EP tubes with anticoagulant. Then, the mice were euthanized by cervical dislocation. The livers and colons were quickly removed. For bacterial load experiments, tissues were rinsed with PBS and kept in 1.5 mL EP tubes on ice. For protein and RNA experiments, tissues were frozen in liquid nitrogen for 30 minutes and stored at -80°C. For HE/FISH staining, tissues were stored in 4% paraformaldehyde or FISH preservation solution.
Fecal microbiota transplantation experiment
For the fecal preparation and transplantation procedure, experimental mice were randomly divided into three groups: CON (control), ABX (antibiotic-treated), and FMT (fecal microbiota transplantation). Mice in the ABX and FMT groups received antibiotics via oral gavage for 7 days, while those in the CON group received an equivalent volume of PBS. Forty-eight hours after the last antibiotic administration, the mice in the FMT group underwent a three-day FMT, whereas those in the ABX and CON groups received PBS. Fecal pellets were collected from ten healthy male mice, homogenized in sterile PBS (50 mg feces/mL), and centrifuged at 100 × g for 5 minutes at 4°C to obtain the supernatant for transplantation. Each mouse in the FMT group was inoculated with 200 μL of fecal suspension daily for three days via gavage, while mice in the CON and ABX groups received an equivalent volume of PBS. Forty-eight hours after the final FMT, the three groups underwent the same infection procedure as before.
rIL-22 treatment
For rIL-22 treatment, the mice were divided into ABX + rIL-22 and CON groups. Both groups underwent antibiotic-induced gut microbiota dysbiosis and hvKp infection. Six hours postinfection, the ABX + rIL-22 group received intraperitoneal injections of 0.5 μg of rIL-22 dissolved in PBS, while the control group received PBS injections. It is critical to avoid puncturing any organ during hvKp injection.
Indole treatment
For indole treatment, the mice were divided into ABX (antibiotics + control solvent), IAA (antibiotics + 3-indole acetic acid), IPA (antibiotics + 3-indole acetic acid), and CON (PBS + solvent) groups. Indoles (3-indole acetic acid, 3-indole propionic acid) were dissolved in NaOH and adjusted to pH 7 with HCl, and the concentration was set to 2 mg/mL. Four hours after daily antibiotic gavage, 200 μL of 3-indole acetic acid or 3-indole propionic acid solution was administered via gavage to the IAA or IPA group, respectively, following the same gavage procedure: the control group received an equivalent volume of solvent. Forty-eight hours after the final antibiotic gavage, the four groups underwent the same hvKp infection procedure as before.
Human samples
Healthy control participants (n = 50) and patients with Klebsiella pneumoniae liver abscess (n = 41) and liver abscess caused by other pathogens (n = 20) were enrolled. The inclusion criteria were as follows: patients newly diagnosed with pyogenic liver abscess at the First Affiliated Hospital of Chongqing Medical University between November 2021 and July 2023 who had undergone liver puncture and had a positive microbial culture result after admission. All patients with malignant liver tumors, cholangitis, bile duct stones, or hepatocellular carcinoma initially presenting as pyogenic liver abscess were excluded. Additionally, patients with a history of pyogenic liver abscess and those with negative culture results were excluded. The Ethics Committee of the First Affiliated Hospital of Chongqing Medical University approved all study participants. Fecal samples were collected from all participants using sterile fecal collectors, immediately labeled, homogenized with a sterile stick, and then stored at −80°C. Patient serum samples were collected from the Department of Laboratory Medicine at the First Affiliated Hospital of Chongqing Medical University.
Ethics approval and consent to participate
All animal experiments complied with the guidelines provided by the Institutional Animal Care and Use Committee of Chongqing Medical University, Chongqing, China. The Committee on the Ethics of Animal Experiments at Chongqing Medical University approved this study (permit number: IACUC-CQMU-2024-0053). All participants in the study received approval from the Ethics Committee of the First Affiliated Hospital of Chongqing Medical University (permit number: K-2023-299).
Method details
Sample DNA extraction, Illumina MiSeq sequencing and microbial diversity and statistical analysis
Mouse feces were collected 48 hours after the last antibiotic gavage. The genomic DNA of the mouse fecal microbial community was extracted using the PF Mag-Bind Stool DNA Kit (Omega Biotek, Georgia, USA) according to the manufacturer's instructions. The integrity of the extracted DNA was assessed via 1% agarose gel electrophoresis, and the DNA concentration and purity were measured with a NanoDrop 2000 spectrophotometer (Thermo Scientific, USA).
The V3-V4 region of the 16S rRNA gene was amplified using the forward primer 338F (5’-ACTCCTACGGGAGGCAGCAG-3’) and reverse primer 806R (5’-GGACTACHVGGGTWTCTAAT-3’), both of which contain barcode sequences. The PCR conditions were as follows: initial denaturation at 95°C for 3 minutes, followed by 27 cycles of 95°C for 30 seconds, 55°C for 30 seconds, and 72°C for 30 seconds, with a final extension at 72°C for 10 minutes. Each sample was amplified in triplicate using a 20 μL PCR system, which included 4 μL of 5 × TransStart FastPfu buffer, 2 μL of 2.5 mM dNTPs, 0.8 μL of each primer (5 μM), 0.4 μL of TransStart FastPfu DNA Polymerase, and 10 ng of template DNA. PCR products were extracted from 2% agarose gels, purified, and quantified using a Quantus™ Fluorometer (Promega, USA).
The purified amplicons were pooled in equimolar amounts and sequenced on an Illumina PE300/PE250 platform (Illumina, San Diego, USA) at Majorbio Bio-Pharm Technology Co., Ltd. (Shanghai, China) following standard protocols. The sequences were demultiplexed, quality filtered using fastp (0.19.6), and merged with FLASH (v1.2.11). High-quality sequences were denoised using the DADA2 plugin in QIIME 2 (version 2020.2) to generate amplicon sequence variants (ASVs). Sequences were rarefied to 20,000 per sample to minimize sequencing depth effects, ensuring an average Good’s coverage of 97.90%. Taxonomic assignment of ASVs was performed using the naive Bayes classifier in QIIME 2 with the SILVA 16S rRNA database (v138).
Alpha diversity indices, including the Chao and Shannon indices, were calculated using mothur (version 1.30.2), and differences between groups were analyzed using the Wilcoxon rank-sum test. Beta diversity and community structure were examined via principal coordinate analysis (PCoA) based on the Bray‒Curtis distance, and statistical significance was assessed using PERMANOVA. Differential abundance analysis was conducted using LEfSe to identify significantly different bacterial taxa between groups (LDA > 2, p < 0.05).
Determination of liver and colon bacterial load
Liver and colon tissues were obtained from mice 48 hours after hvKp infection. Whole tissues were rinsed twice with sterile PBS, placed into a sterilized homogenizer with 1 mL of PBS, ground until no residue remained, and transferred to a 1.5 mL EP tube kept on ice. Three 1.5 mL EP tubes were prepared, labeled (1–3), and filled with 0.9 mL of PBS each. First, 0.1 mL of the original tissue homogenate was added to the first tube (tube 1) and mixed thoroughly, followed by serial 10-fold dilutions into the second (tube 2) and third (tube 3) tubes. From these dilutions, 20 μL samples of the 10-fold, 100-fold, and 1000-fold dilutions were inoculated onto blood agar plates. The plates were incubated upright for 30 minutes, inverted and incubated at 37°C for 16 hours for colony counting. Accurate identification and quantification of hvKp were ensured through colony morphology observation, string tests and MALDI–TOF MS. The dilution factors for liver bacterial load determination included the original solution and 10-fold and 100-fold diluted solutions, while colon bacterial load determination included 10-fold, 100-fold, and 1000-fold diluted solutions, according to the preliminary experimental findings.
Quantitative PCR analysis
Colon tissues were obtained from mice 24 hours after hvKp infection. Total RNA was extracted from mouse colon tissue using RNAiso Plus following the manufacturer’s protocol. Total RNA was reverse transcribed into cDNA according to the instructions of the reverse transcription kit. Target gene expression levels were quantified using a Bio-Rad CFX96 Real-Time system with SYBR Premix Ex Taq. The primer pairs used in the study are listed in Table S1. PCR primers were synthesized by Sangon (Shanghai, China).
Cytokine analysis
Following anesthesia, the eyeballs were removed from the mice, and blood was collected into 1.5 mL Eppendorf tubes pretreated with anticoagulant. Subsequently, the blood was centrifuged to separate the plasma. Plasma IL-22, IL17A, IL-10, and IL-6 levels were measured using commercial kits (ELISA MAX Deluxe, Biolegend). All steps were performed strictly following the manufacturer’s protocol.
H&E staining and immunofluorescence
Liver and colon tissues were obtained from mice 48 hours after hvKp infection. The tissues were perfused with paraformaldehyde, fixed overnight by immersion in paraformaldehyde, embedded in paraffin, and cut into 5 μm sections. For H&E staining, the sections were first deparaffinized, rehydrated, and stained with hematoxylin and eosin. The tissue section slides were subsequently redehydrated and cleared in xylene. The sections were then mounted using neutral gum and viewed under a light microscope. The tissue section slides were also stained with Anti-Claudin 2 Antibody and Occludin (E6B4R) Rabbit mAb, followed by incubation with a secondary antibody, and incubated with DAPl to stain nuclear. From each experimental group, three mice were randomly selected, and one tissue section per mouse was processed. Randomly selected images are shown for each group.
FISH of mouse colon tissue
Colon tissues were obtained from mice 48 hours after hvKp infection. Mouse colon tissues were fixed in FISH fixative for more than 12 hours, followed by dehydration through an ethanol gradient and paraffin embedding. Sections were deparaffinized and treated with proteinase K (20 μg/mL) at 37°C for 5 minutes. After rinsing with distilled water and PBS, the sections were prehybridized before applying a hybridization solution containing a Klebsiella pneumoniae probe (5'-CCT ACA CAC CAG CGT GCC-3') at a concentration of 1.5 μM. Hybridization was performed overnight at 40°C. Post hybridization washes were followed by blocking with BSA. Nuclear counterstaining was performed with DAPI. The sections were observed and imaged using a Nikon upright fluorescence microscope with the following settings: a UV excitation wavelength of 330-380 nm with emission at 420 nm for blue fluorescence and a Cy3 excitation wavelength of 510-560 nm with emission at 590 nm for red fluorescence. Scale bar = 100 μm. From each experimental group, three mice were randomly selected, and one tissue section per mouse was processed.
Western blot analysis
Colon tissues were obtained from mice 48 hours after hvKp infection. The tissues were generated using RIPA buffer containing PMSF, and the proteins were separated on 12.5% SDS‒PAGE gels, electrotransferred to PVDF membranes and subjected to western blotting. The primary antibodies used for western blotting were Rabbit Anti-beta-Actin Monoclonal Antibody (Cell Signaling Technology, Cat # 5125) and REG3B Polyclonal Antibody (Thermofisher, Cat #PA5-47700). Protein bands were visualized utilizing HRP-conjugated secondary antibodies coupled with enhanced chemiluminescence (ECL) detection. The band intensities were analyzed using ImageJ software.
Tryptophan metabolite analysis
Mouse feces were collected 48 hours after the last antibiotic gavage. The 33 standards in the tryptophan pathway were accurately weighed and dissolved in 50% methanol to prepare standard solutions, which were then combined and diluted to create mixed standard solutions of varying concentrations. For metabolite extraction, 25 mg of mouse fecal sample was homogenized in 10 μL of an internal standard (Trp-D5, 4000 ng/mL) and 190 μL of extraction solution (methanol: water = 4:1) at –10°C using a high-throughput tissue crusher, followed by sonication at 5°C. After centrifugation, the supernatant was analyzed using a Nexera Series LC-40 system coupled with a QTRAP® 6500+ mass spectrometer. Metabolites were separated on an ACQUITY UPLC® HSS T3 column (2.1 × 150 mm, 1.8 μm) at 40°C with a gradient mobile phase of water containing 0.1% formic acid (solvent A) and acetonitrile containing 0.1% formic acid (solvent B) over 18 minutes. Quality control samples were injected every 10 samples to ensure analytical stability. The data were processed with Sciex OS software.
Quantification and statistical analysis
Two-tailed unpaired Student’s t test with or without Welch’s correction was used to assess the differences between two groups depending on the homogeneity of variance. One-way ANOVA or Welch’s ANOVA was used to assess the differences in means between groups when the variances were homogeneous or not. The data are presented as the means ± SEMs of 3 independent experiments. Survival analysis over time following infection was conducted using the Kaplan‒Meier method supplemented by log-rank tests. All the statistical procedures were performed using GraphPad Prism software version 8.0. The significance levels were defined as: ∗ p < 0.05, ∗∗ p < 0.01, ∗∗∗ p < 0.001, and ∗∗∗∗ p < 0.0001.
Published: August 30, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.110849.
Supplemental information
References
- 1.Liu Y.C., Cheng D.L., Lin C.L. Klebsiella pneumoniae liver abscess associated with septic endophthalmitis. Arch. Intern. Med. 1986;146:1913–1916. [PubMed] [Google Scholar]
- 2.Chung D.R., Lee S.S., Lee H.R., Kim H.B., Choi H.J., Eom J.S., Kim J.S., Choi Y.H., Lee J.S., Chung M.H., et al. Emerging invasive liver abscess caused by K1 serotype Klebsiella pneumoniae in Korea. J. Infect. 2007;54:578–583. doi: 10.1016/j.jinf.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 3.Siu L.K., Yeh K.M., Lin J.C., Fung C.P., Chang F.Y. Klebsiella pneumoniae liver abscess: a new invasive syndrome. Lancet Infect. Dis. 2012;12:881–887. doi: 10.1016/S1473-3099(12)70205-0. [DOI] [PubMed] [Google Scholar]
- 4.Wang J.H., Liu Y.C., Lee S.S., Yen M.Y., Chen Y.S., Wang J.H., Wann S.R., Lin H.H. Primary liver abscess due to Klebsiella pneumoniae in Taiwan. Clin. Infect. Dis. 1998;26:1434–1438. doi: 10.1086/516369. [DOI] [PubMed] [Google Scholar]
- 5.Fang C.T., Chen Y.C., Chang S.C., Sau W.Y., Luh K.T. Klebsiella pneumoniae meningitis: timing of antimicrobial therapy and prognosis. QJM. 2000;93:45–53. doi: 10.1093/qjmed/93.1.45. [DOI] [PubMed] [Google Scholar]
- 6.Tang L.M., Chen S.T. Klebsiella pneumoniae meningitis: prognostic factors. Scand. J. Infect. Dis. 1994;26:95–102. doi: 10.3109/00365549409008596. [DOI] [PubMed] [Google Scholar]
- 7.Lin Y.T., Siu L.K., Lin J.C., Chen T.L., Tseng C.P., Yeh K.M., Chang F.Y., Fung C.P. Seroepidemiology of Klebsiella pneumoniae colonizing the intestinal tract of healthy Chinese and overseas Chinese adults in Asian countries. BMC Microbiol. 2012;12:13. doi: 10.1186/1471-2180-12-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung D.R., Lee H., Park M.H., Jung S.I., Chang H.H., Kim Y.S., Son J.S., Moon C., Kwon K.T., Ryu S.Y., et al. Fecal carriage of serotype K1 Klebsiella pneumoniae ST23 strains closely related to liver abscess isolates in Koreans living in Korea. Eur. J. Clin. Microbiol. Infect. Dis. 2012;31:481–486. doi: 10.1007/s10096-011-1334-7. [DOI] [PubMed] [Google Scholar]
- 9.Siu L.K., Fung C.P., Chang F.Y., Lee N., Yeh K.M., Koh T.H., Ip M. Molecular typing and virulence analysis of serotype K1 Klebsiella pneumoniae strains isolated from liver abscess patients and stool samples from noninfectious subjects in Hong Kong, Singapore, and Taiwan. J. Clin. Microbiol. 2011;49:3761–3765. doi: 10.1128/JCM.00977-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fung C.P., Lin Y.T., Lin J.C., Chen T.L., Yeh K.M., Chang F.Y., Chuang H.C., Wu H.S., Tseng C.P., Siu L.K. Klebsiella pneumoniae in gastrointestinal tract and pyogenic liver abscess. Emerg. Infect. Dis. 2012;18:1322–1325. doi: 10.3201/eid1808.111053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tu Y.C., Lu M.C., Chiang M.K., Huang S.P., Peng H.L., Chang H.Y., Jan M.S., Lai Y.C. Genetic requirements for Klebsiella pneumoniae-induced liver abscess in an oral infection model. Infect. Immun. 2009;77:2657–2671. doi: 10.1128/IAI.01523-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin Y.T., Liu C.J., Yeh Y.C., Chen T.J., Fung C.P. Ampicillin and amoxicillin use and the risk of Klebsiella pneumoniae liver abscess in Taiwan. J. Infect. Dis. 2013;208:211–217. doi: 10.1093/infdis/jit157. [DOI] [PubMed] [Google Scholar]
- 13.Dudakov J.A., Hanash A.M., van den Brink M.R.M. Interleukin-22: immunobiology and pathology. Annu. Rev. Immunol. 2015;33:747–785. doi: 10.1146/annurev-immunol-032414-112123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hendrikx T., Duan Y., Wang Y., Oh J.-H., Alexander L.M., Huang W., Stärkel P., Ho S.B., Gao B., Fiehn O., et al. Bacteria engineered to produce IL-22 in intestine induce expression of REG3G to reduce ethanol-induced liver disease in mice. Gut. 2019;68:1504–1515. doi: 10.1136/gutjnl-2018-317232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng Y., Ding Y., Xu M., Chen H., Zhang H., Liu Y., Shen W., Li J. Gut Microbiota Contributes to Host Defense Against Klebsiella pneumoniae-Induced Liver Abscess. J. Inflamm. Res. 2021;14:5215–5225. doi: 10.2147/JIR.S334581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsugawa H., Ohki T., Tsubaki S., Tanaka R., Matsuzaki J., Suzuki H., Hozumi K. Gas6 ameliorates intestinal mucosal immunosenescence to prevent the translocation of a gut pathobiont, Klebsiella pneumoniae, to the liver. PLoS Pathog. 2023;19 doi: 10.1371/journal.ppat.1011139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mukherjee S., Hooper L.V. Antimicrobial defense of the intestine. Immunity. 2015;42:28–39. doi: 10.1016/j.immuni.2014.12.028. [DOI] [PubMed] [Google Scholar]
- 18.Sanos S.L., Vonarbourg C., Mortha A., Diefenbach A. Control of epithelial cell function by interleukin-22-producing RORgammat+ innate lymphoid cells. Immunology. 2011;132:453–465. doi: 10.1111/j.1365-2567.2011.03410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sequeira R.P., McDonald J.A.K., Marchesi J.R., Clarke T.B. Commensal Bacteroidetes protect against Klebsiella pneumoniae colonization and transmission through IL-36 signalling. Nat. Microbiol. 2020;5:304–313. doi: 10.1038/s41564-019-0640-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y., Mumm J.B., Herbst R., Kolbeck R., Wang Y. IL-22 Increases Permeability of Intestinal Epithelial Tight Junctions by Enhancing Claudin-2 Expression. J. Immunol. 2017;199:3316–3325. doi: 10.4049/jimmunol.1700152. [DOI] [PubMed] [Google Scholar]
- 21.Nong H., Yuan H., Lin Y., Chen S., Li Y., Luo Z., Yang W., Zhang T., Chen Y. IL-22 promotes occludin expression by activating autophagy and treats ulcerative colitis. Mol. Cell. Biochem. 2024;479:1443–1450. doi: 10.1007/s11010-023-04806-z. [DOI] [PubMed] [Google Scholar]
- 22.Kuo W.T., Odenwald M.A., Turner J.R., Zuo L. Tight junction proteins occludin and ZO-1 as regulators of epithelial proliferation and survival. Ann. N. Y. Acad. Sci. 2022;1514:21–33. doi: 10.1111/nyas.14798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Podolsky D.K., Gerken G., Eyking A., Cario E. Colitis-associated variant of TLR2 causes impaired mucosal repair because of TFF3 deficiency. Gastroenterology. 2009;137:209–220. doi: 10.1053/j.gastro.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zelante T., Iannitti R.G., Cunha C., De Luca A., Giovannini G., Pieraccini G., Zecchi R., D'Angelo C., Massi-Benedetti C., Fallarino F., et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39:372–385. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 25.Keir M., Yi T., Lu T., Ghilardi N. The role of IL-22 in intestinal health and disease. J. Exp. Med. 2020;217 doi: 10.1084/jem.20192195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee D., Jo H., Go C., Jang Y., Chu N., Bae S., Kang D., Kim Y., Kang J.S. The Roles of IL-22 and Its Receptor in the Regulation of Inflammatory Responses in the Brain. Int. J. Mol. Sci. 2022;23 doi: 10.3390/ijms23020757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mavilia M.G., Molina M., Wu G.Y. The Evolving Nature of Hepatic Abscess: A Review. J. Clin. Transl. Hepatol. 2016;4:158–168. doi: 10.14218/JCTH.2016.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peng Y.C., Lin C.L., Sung F.C. Risk of pyogenic liver abscess and endoscopic sphincterotomy: a population-based cohort study. BMJ Open. 2018;8 doi: 10.1136/bmjopen-2017-018818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lamas B., Richard M.L., Leducq V., Pham H.P., Michel M.L., Da Costa G., Bridonneau C., Jegou S., Hoffmann T.W., Natividad J.M., et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat. Med. 2016;22:598–605. doi: 10.1038/nm.4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monteleone I., Rizzo A., Sarra M., Sica G., Sileri P., Biancone L., MacDonald T.T., Pallone F., Monteleone G. Aryl hydrocarbon receptor-induced signals up-regulate IL-22 production and inhibit inflammation in the gastrointestinal tract. Gastroenterology. 2011;141:237–248.e1. doi: 10.1053/j.gastro.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 31.Lamas B., Hernandez-Galan L., Galipeau H.J., Constante M., Clarizio A., Jury J., Breyner N.M., Caminero A., Rueda G., Hayes C.L., et al. Aryl hydrocarbon receptor ligand production by the gut microbiota is decreased in celiac disease leading to intestinal inflammation. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.aba0624. [DOI] [PubMed] [Google Scholar]
- 32.Natividad J.M., Agus A., Planchais J., Lamas B., Jarry A.C., Martin R., Michel M.L., Chong-Nguyen C., Roussel R., Straube M., et al. Impaired Aryl Hydrocarbon Receptor Ligand Production by the Gut Microbiota Is a Key Factor in Metabolic Syndrome. Cell Metabol. 2018;28:737–749.e4. doi: 10.1016/j.cmet.2018.07.001. [DOI] [PubMed] [Google Scholar]
- 33.Semenec L., Cain A.K., Dawson C.J., Liu Q., Dinh H., Lott H., Penesyan A., Maharjan R., Short F.L., Hassan K.A., Paulsen I.T. Cross-protection and cross-feeding between Klebsiella pneumoniae and Acinetobacter baumannii promotes their co-existence. Nat. Commun. 2023;14:702. doi: 10.1038/s41467-023-36252-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee K.W.K., Periasamy S., Mukherjee M., Xie C., Kjelleberg S., Rice S.A. Biofilm development and enhanced stress resistance of a model, mixed-species community biofilm. ISME J. 2014;8:894–907. doi: 10.1038/ismej.2013.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hyland N.P., Cavanaugh C.R., Hornby P.J. Emerging effects of tryptophan pathway metabolites and intestinal microbiota on metabolism and intestinal function. Amino Acids. 2022;54:57–70. doi: 10.1007/s00726-022-03123-x. [DOI] [PubMed] [Google Scholar]
- 36.Loonen L.M.P., Stolte E.H., Jaklofsky M.T.J., Meijerink M., Dekker J., van Baarlen P., Wells J.M. REG3gamma-deficient mice have altered mucus distribution and increased mucosal inflammatory responses to the microbiota and enteric pathogens in the ileum. Mucosal Immunol. 2014;7:939–947. doi: 10.1038/mi.2013.109. [DOI] [PubMed] [Google Scholar]
- 37.He G.W., Lin L., DeMartino J., Zheng X., Staliarova N., Dayton T., Begthel H., van de Wetering W.J., Bodewes E., van Zon J., et al. Optimized human intestinal organoid model reveals interleukin-22-dependency of paneth cell formation. Cell Stem Cell. 2022;29:1718–1720. doi: 10.1016/j.stem.2022.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gaudino S.J., Beaupre M., Lin X., Joshi P., Rathi S., McLaughlin P.A., Kempen C., Mehta N., Eskiocak O., Yueh B., et al. IL-22 receptor signaling in Paneth cells is critical for their maturation, microbiota colonization, Th17-related immune responses, and anti-Salmonella immunity. Mucosal Immunol. 2021;14:389–401. doi: 10.1038/s41385-020-00348-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miki T., Holst O., Hardt W.-D. The bactericidal activity of the C-type lectin RegIIIβ against Gram-negative bacteria involves binding to lipid A. J. Biol. Chem. 2012;287:34844–34855. doi: 10.1074/jbc.M112.399998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stelter C., Käppeli R., König C., Krah A., Hardt W.-D., Stecher B., Bumann D. Salmonella-induced mucosal lectin RegIIIβ kills competing gut microbiota. PLoS One. 2011;6 doi: 10.1371/journal.pone.0020749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin R.M., Cao J., Brisse S., Passet V., Wu W., Zhao L., Malani P.N., Rao K., Bachman M.A. Molecular Epidemiology of Colonizing and Infecting Isolates of Klebsiella pneumoniae. mSphere. 2016;1 doi: 10.1128/mSphere.00261-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gorrie C.L., Mirceta M., Wick R.R., Edwards D.J., Thomson N.R., Strugnell R.A., Pratt N.F., Garlick J.S., Watson K.M., Pilcher D.V., et al. Gastrointestinal Carriage Is a Major Reservoir of Klebsiella pneumoniae Infection in Intensive Care Patients. Clin. Infect. Dis. 2017;65:208–215. doi: 10.1093/cid/cix270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huynh B.T., Passet V., Rakotondrasoa A., Diallo T., Kerleguer A., Hennart M., Lauzanne A.D., Herindrainy P., Seck A., Bercion R., et al. Klebsiella pneumoniae carriage in low-income countries: antimicrobial resistance, genomic diversity and risk factors. Gut Microb. 2020;11:1287–1299. doi: 10.1080/19490976.2020.1748257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang J., Li Y., Tang N., Li J., Zhou J., Lu S., Zhang G., Song Y., Wang C., Zhong J., et al. The human gut serves as a reservoir of hypervirulent Klebsiella pneumoniae. Gut Microb. 2022;14 doi: 10.1080/19490976.2022.2114739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lam M.M.C., Wyres K.L., Duchêne S., Wick R.R., Judd L.M., Gan Y.H., Hoh C.H., Archuleta S., Molton J.S., Kalimuddin S., et al. Population genomics of hypervirulent Klebsiella pneumoniae clonal-group 23 reveals early emergence and rapid global dissemination. Nat. Commun. 2018;9:2703. doi: 10.1038/s41467-018-05114-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Russo T.A., Marr C.M. Hypervirulent Klebsiella pneumoniae. Clin. Microbiol. Rev. 2019;32 doi: 10.1128/CMR.00001-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Namikawa H., Oinuma K.-I., Yamada K., Kaneko Y., Kakeya H., Shuto T. Differences in severity of bacteraemia caused by hypermucoviscous and non-hypermucoviscous Klebsiella pneumoniae. Int. J. Antimicrob. Agents. 2023;61 doi: 10.1016/j.ijantimicag.2023.106767. [DOI] [PubMed] [Google Scholar]
- 48.Wang H., Guo Y., Liu Z., Chang Z. The Type VI Secretion System Contributes to the Invasiveness of Liver Abscess Caused by Klebsiella pneumoniae. J. Infect. Dis. 2023;228:1127–1136. doi: 10.1093/infdis/jiad166. [DOI] [PubMed] [Google Scholar]
- 49.Hu D., Chen W., Wang W., Tian D., Fu P., Ren P., Mu Q., Li G., Jiang X. Hypercapsule is the cornerstone of Klebsiella pneumoniae in inducing pyogenic liver abscess. Front. Cell. Infect. Microbiol. 2023;13 doi: 10.3389/fcimb.2023.1147855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Choby J.E., Howard-Anderson J., Weiss D.S. Hypervirulent Klebsiella pneumoniae - clinical and molecular perspectives. J. Intern. Med. 2020;287:283–300. doi: 10.1111/joim.13007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rosenthal R., Günzel D., Krug S.M., Schulzke J.D., Fromm M., Yu A.S.L. Claudin-2-mediated cation and water transport share a common pore. Acta Physiol. 2017;219:521–536. doi: 10.1111/apha.12742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Amasheh S., Meiri N., Gitter A.H., Schöneberg T., Mankertz J., Schulzke J.D., Fromm M. Claudin-2 expression induces cation-selective channels in tight junctions of epithelial cells. J. Cell Sci. 2002;115:4969–4976. doi: 10.1242/jcs.00165. [DOI] [PubMed] [Google Scholar]
- 53.Tsai P.-Y., Zhang B., He W.-Q., Zha J.-M., Odenwald M.A., Singh G., Tamura A., Shen L., Sailer A., Yeruva S., et al. IL-22 Upregulates Epithelial Claudin-2 to Drive Diarrhea and Enteric Pathogen Clearance. Cell Host Microbe. 2017;21:671–681.e4. doi: 10.1016/j.chom.2017.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Furuse M., Hirase T., Itoh M., Nagafuchi A., Yonemura S., Tsukita S., Tsukita S. Occludin: a novel integral membrane protein localizing at tight junctions. J. Cell Biol. 1993;123:1777–1788. doi: 10.1083/jcb.123.6.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heller F., Florian P., Bojarski C., Richter J., Christ M., Hillenbrand B., Mankertz J., Gitter A.H., Bürgel N., Fromm M., et al. Interleukin-13 is the key effector Th2 cytokine in ulcerative colitis that affects epithelial tight junctions, apoptosis, and cell restitution. Gastroenterology. 2005;129:550–564. doi: 10.1016/j.gastro.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 56.Suzuki T., Yoshinaga N., Tanabe S. Interleukin-6 (IL-6) regulates claudin-2 expression and tight junction permeability in intestinal epithelium. J. Biol. Chem. 2011;286:31263–31271. doi: 10.1074/jbc.M111.238147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamamoto T., Kojima T., Murata M., Takano K.-I., Go M., Chiba H., Sawada N. IL-1beta regulates expression of Cx32, occludin, and claudin-2 of rat hepatocytes via distinct signal transduction pathways. Exp. Cell Res. 2004;299:427–441. doi: 10.1016/j.yexcr.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 58.Amasheh M., Andres S., Amasheh S., Fromm M., Schulzke J.-D. Barrier effects of nutritional factors. Ann. N. Y. Acad. Sci. 2009;1165:267–273. doi: 10.1111/j.1749-6632.2009.04063.x. [DOI] [PubMed] [Google Scholar]
- 59.Russo T.A., Olson R., Fang C.T., Stoesser N., Miller M., MacDonald U., Hutson A., Barker J.H., La Hoz R.M., Johnson J.R. Identification of Biomarkers for Differentiation of Hypervirulent Klebsiella pneumoniae from Classical K. pneumoniae. J. Clin. Microbiol. 2018;56 doi: 10.1128/jcm.00776-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Data have been deposited at Figshare and are publicly available as of the date of publication.
-
•
The strain genomic data, metabolomic datasets and mouse gut microbiota 16S rRNA datasets generated during the current study will be available in the public repository. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.