Abstract
Pollen grains play a critical role in environmental, agricultural, and allergy research despite their tiny dimensions. The accurate classification of pollen grains remains a significant challenge, mainly attributable to their intricate structures and the extensive diversity of species. Traditional methods often lack accuracy and effectiveness, prompting the need for advanced solutions. This study introduces a novel deep learning framework, PollenNet, designed to tackle the intricate challenge of pollen grain image classification. The efficiency of PollenNet is thoroughly evaluated through stratified 5-fold cross-validation, comparing it with cutting-edge methods to demonstrate its superior performance. A comprehensive data preparation phase is conducted, including removing duplicates and low-quality images, applying Non-local Means Denoising for noise reduction, and Gamma correction to adjust image brightness. Furthermore, Explainable AI (XAI) is utilized to enhance the interpretability of the model, while Receiver Operating Characteristic (ROC) curve analysis serves as a quantitative method for evaluating the model's capabilities. PollenNet demonstrates superior performance when compared to existing models, with an accuracy of 98.45 %, precision of 98.20 %, specificity of 98.40 %, recall of 98.30 %, and f1-score of 98.25 %. The model also maintains low Mean Squared Error (0.03) and Mean Absolute Error (0.02) rates. The ROC curve analysis, the low False Positive Rate (0.016), and the False Negative Rate (0.017) highlight the reliability and dependability of the model. This study significantly improves the efficacy of classifying pollen grains, indicating an important advancement in the application of deep learning for ecological research.
Keywords: Pollen grain classification, Image processing, Deep learning, PollenNet model, Explainable AI
1. Introduction
Pollen grains are tiny fragments produced by plants as part of their reproductive process and have diverse shapes. Pollen morphology often exhibits distinguishing characteristics that may be used to identify the family it belongs to. However, in some cases, unique properties may be seen at the genus or species level. Pollen's extensive array of morphological changes makes it significant in several scientific disciplines since it facilitates the precise identification of the plants from which the pollen grains came [1]. Palynology is the scientific field that specifically studies and analyzes pollen grains, spores, and other minuscule biological particles. This area has many applications, including paleoclimate reconstruction [2], paleoecology [3], biodiversity [4], allergy research [5], studies of flora [6], plant reproductive biology [7], petroleum exploration [8], establishing plant taxonomic connections [9], forensic science [10], and more. The study of pollen grains, including their identification, monitoring, classification, and certification, has attracted considerable attention because of their many applications. These applications include archeology and honey certification [11], predicting allergies caused by airborne pollen [12], and food technology [13]. Traditional pollen tests include the analysis of microscopic images and the identification of pollen grains by comparing them to established species, a process performed by palynologists. However, the analysis of pollen is a laborious and challenging procedure that necessitates the physical preparation of samples, the microscopic identification and localization of pollen grains, and ultimately, the quantification of the abundance of various species within a specific sample. This procedure is distinguished by its lengthy duration, susceptibility to human mistakes, and need for substantial expertise in many tree species [14]. The categorization of pollen grains has developed into a costly analytical process that requires the identification and classification of features by a licensed palynologist. Undoubtedly, it continues to be the most exhaustive, precise, and dependable method. Nevertheless, it impedes scientific progress by wasting significant amounts of time and resources [15].
Computer science, including techniques in image processing and machine learning analysis, has lately seen significant advances in several fields of study. This achievement has facilitated a strong partnership between palynologists and computer scientists to tackle the aforementioned issues. The progress in artificial intelligence, specifically in deep learning, combined with the use of image processing, has enabled the execution of complex analysis with remarkable outcomes in several occupations [16]. Advanced Machine Learning approaches, including deep neural networks, have produced impressive outcomes in addressing many problems across several domains, all while maintaining cost-effectiveness [17]. A crucial determinant in attaining success in deep learning is the existence of a significant volume of annotated data. The efficacy of deep learning techniques is directly proportional to the amount of data, promoting the development of comprehensive datasets across many scientific fields [18]. Research on automated systems for identifying pollen grains will enable the development of tools that botanists and palynologists may use. These tools only need a normal bright-field microscope, an image-capturing camera for data collection, and a computer for data processing. This will obviate the need for specialist equipment. These gadgets possess the capacity to separate pollen grains from the images and assist researchers in classifying them according to species. The contributions of this research are as follows.
-
1.
Development of a novel deep learning model to precisely and effectively classify pollen grains.
-
2.
Implementation of advanced image processing techniques, such as Non-local Means denoising for noise reduction and Gamma correction for image improvement.
-
3.
Employing nine metrics for evaluation, including ROC curves, confusion matrices and stratified 5-fold cross-validation to validate the robustness and broad applicability of PollenNet.
-
4.
Explainable AI (XAI) techniques such as Grad-CAM and Saliency Maps have been applied to enhance the interpretability and transparency of PollenNet's decision-making process.
2. Literature review
Over the last several decades, researchers have made significant advancements in creating automated techniques for classifying pollen grains, a critical aspect of environmental and ecological research [30,31]. Traditional methods, although creative, generally relied heavily on explicit rule-based algorithms, which often encountered challenges when handling the intricate and nuanced aspects of pollen grain classification. The incorporation of machine learning and deep learning has profoundly revolutionized this domain. Through data-driven learning, these advanced algorithms have greatly enhanced the efficiency and accuracy of the classification process. Machine learning has shown exceptional adaptability and precision, particularly in pollen grain analysis. It has shown expertise in identifying and categorizing various types of pollen, which is crucial for understanding changes in the environment, monitoring allergies, and studying plant diversity. This technical innovation has not only accelerated research but also generated potential for deeper comprehension in the realm of ecological and environmental studies.
Rebiai et al. [19] have introduced a system for classifying and automatically recognizing multi-focal microscopic pollen images. The framework is meant to be adaptable and employs a strategy similar to brute force to achieve efficiency for different kinds and sizes of pollen images. First, the best focus is selected using the absolute gradient technique. In the end, a standardized method is used to extract and choose characteristics, and their classification is assessed using Hierarchical Cluster Analysis (HCA). Manikis et al. [20] acquired a classified dataset from the Hellenic Mediterranean University in another research. The collection included 564 microscopic pollen images. The researchers devised a fully automated analysis procedure with the objective of classifying the images into six distinct pollen categories. The task of image categorization was executed with the Random Forest (RF) classifier. The estimation of generalization performance and avoidance of overfitting was conducted utilizing a repeating nested cross-validation (nested-CV) template. The findings indicated an average accuracy rate of 88.24 %. The dataset supplied by Battiato et al. [21] contains more than 13,000 items. The identification of these items was achieved by a customized segmentation approach used to aerobiological samples to classify pollen. The data has undergone preprocessing to reduce significant background noise in the digital slide scans. The experiment is carried out using conventional Machine Learning techniques, such as Random Forest (RF), Support Vector Machines (SVM), AdaBoost, and Multilayer Perceptron (MLP). The item's texture was analyzed using the Local Binary Pattern (LBP) and Histogram of Oriented Gradient (HOG) methods. The research determines that the RBF-SVM model with HOG obtains the maximum performance among all the models, with an accuracy of 86.58 %.
Similarly, Grant-Jacob et al. [22] proposed detecting and capturing moving pollen grains by shining three laser beams on them. The inceptionV3 method was used to differentiate between two unique types of pollen grains and a category representing no pollen based on their dispersion patterns. The prediction accuracy of this model reached 86 %. In addition, Xu et al. [23] proposed a collaborative learning strategy to improve the precision of pollen segmentation and classification by using a weakly supervised method. The detection model is used to discern regions of pollen in the unprocessed images. To improve the precision of the classification, the pollen grains were partitioned into sections using a pre-trained U-Net model that leverages unsupervised pollen contour features. The segmented pollen regions were fed into a Convolutional Neural Network (CNN) model to get the activation maps. The categorization accuracy reaches a maximum of 86.6 %. Azari et al. [24] conducted research that employed the optical characteristics of pollen species to identify them. The authors evaluated several categorization algorithms in the Python scikit-learn module, such as Logistic Regression, SVM, Multi-layer Perception, and K-Nearest Neighbors. The authors opted for a Logistic Regression model among the available choices since it is easier to understand. The investigation yielded memory ratings ranging from 70 % to 85 % for the majority of classes, except for Corylus, which only attained a recall of 42 %.
In a comparable manner, Punyasena et al. [25] provide an automated methodology for examining pollen. This procedure involves the automated scanning of slides holding pollen samples, followed by the use of CNN models to detect and classify various kinds of pollen automatically. The researchers trained the data using the ResNet34 model. The average accuracy of the fine-tuned ResNet34 model, determined by computing the mean of the 25 separate per-class accuracies, is 89.5 %. Similarly, Menad et al. [26] showcased the use of deep Convolutional Neural Networks (CNN) in categorizing images of pollen grains for the purpose of identification. The neural network has 8 hidden layers. The first 5 layers comprise convolutional neurons responsible for processing image representations, while the following 3 levels are fully connected layers used for image classification. The model was subjected to a combined process of training and testing that spanned 4000 epochs. The model reached its highest level of accuracy, 85.1 %, during the 200th epoch and consistently maintained this level of performance throughout.
In addition, Romero et al. [27] explain a method for analyzing pollen that utilizes optical super-resolution microscopy and machine learning to create a more accurate and efficient approach. The scientists developed three convolutional neural network (CNN) classification models, namely the Maximum Projection Model (MPM), Multislice Model (MSM), and Fused Model (FM). These models were trained on a dataset including pollen samples from 16 distinct classes. The data was categorized, and the resulting average accuracies for all models ranged from 83 % to 90 %, with the FM model obtaining the greatest accuracy of 90.30 %. Daunys et al. [28] developed an approach to train many models, including as ConvNets, RepVGG, ResNet, EfficientNet, Vision transformer (ViT), ViT Compact Convolutional Transformer (ViT-CCT), and VOLO, for the purpose of classifying distinct kinds of pollen. The efficacy of these models was then assessed by comparing their outcomes on test sub-samples that were not used during the training phase. The fusion of RepVGG, ResNet, and VOLO models achieved a peak classification accuracy of 72.4 %.
Our study introduces an innovative approach that utilizes deep learning models to tackle the challenges associated with accurately classifying images of pollen grains. Traditional methods sometimes had difficulties in dealing with the intricate and irregular nature of pollen images, resulting in limited accuracy and adaptability. To address these issues, we focused primarily on creating and using a wide variety of sophisticated models, with a particular emphasis on our distinctive and customized development, PollenNet. Among the models considered, including AlexNet [33], MobileNetV2 [34], VGG16 [35], and ResNet50 [36], this model was selected for its exceptional capabilities in image processing and classification. These skills make it particularly suitable for the complex task of categorizing pollen. PollenNet exemplifies our dedication to advancing the field of pollen analysis. The integrated architecture of this system is purposefully designed to use the capabilities of its components, leading to outstanding accuracy and efficiency in classifying various types of pollen. This endeavor is not just focused on improving the precision of classification; it represents significant progress in the capabilities of pollen grain imaging and analysis. We want to set a new standard in the classification of pollen grains by using cutting-edge models. This will substantially contribute to environmental and ecological studies by providing crucial insights.
3. Research methodology and strategy
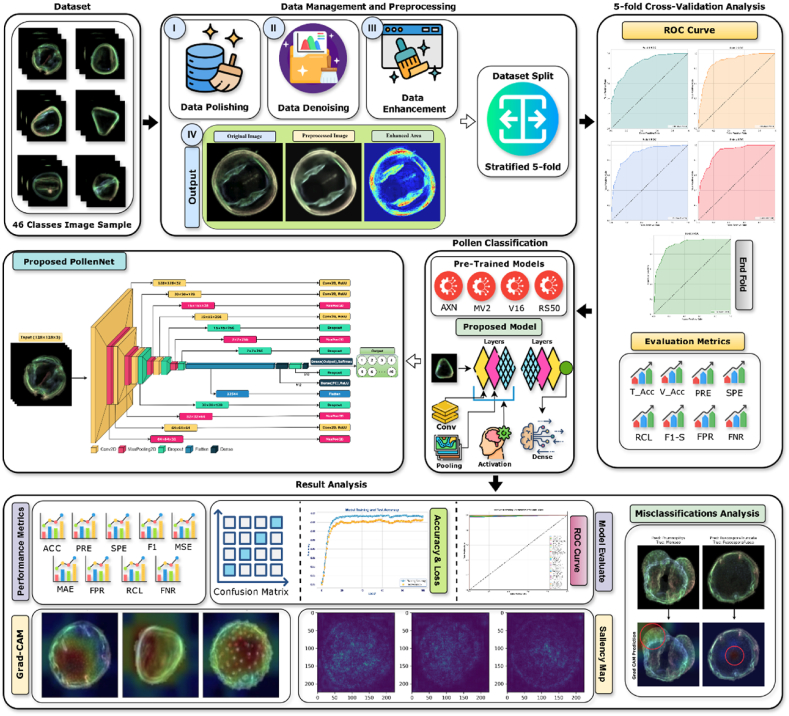
Our study's methodology involves a methodical approach to data preparation, which ensures the integrity and clarity of information fed into the deep learning framework. We begin by conducting thorough data preprocessing, utilizing techniques such as data extraction and refinement to remove any redundancies and flaws, resulting in a polished dataset. Subsequently, advanced image processing algorithms, such as non-local means denoising and gamma correction, are applied, which are crucial in reducing noise and improving image fidelity. After this extensive preprocessing, our proposed model, PollenNet, was implemented, and its subsequent responsibility was to classify pollen grains. Fig. 1 demonstrates the entire methodology of the proposed study.
Fig. 1.
The entire workflow of the proposed model for pollen classification.
3.1. Data managing and processing techniques
The efficacy of deep learning models heavily depends on the quality of the input data. Considering this, our data preprocessing phase was meticulously designed to improve the quality of the raw dataset. This study incorporates a sequence of cutting-edge techniques, commencing with the essential preprocessing steps of data extraction, refinement, denoising, and enhancement. The original dataset underwent a thorough process of data polishing, during which we systematically identified and eliminated duplicates, unclear images, and distorted images. This comprehensive refinement was crucial in building an adequate framework for the succeeding steps of feature extraction, classification, and examining the hidden variables in our data. Furthermore, we employed Non-local Means to reduce image noise while preserving essential structural details, thus refining the visual clarity of our data. Meanwhile, Gamma Correction was applied to adjust the intensity of the images, allowing our model to detect and learn from the nuanced elements required for precise classification. These preprocessing steps are crucial in transforming raw data into a clean and reliable format suitable for sophisticated analytical tasks.
3.1.1. Data extracting
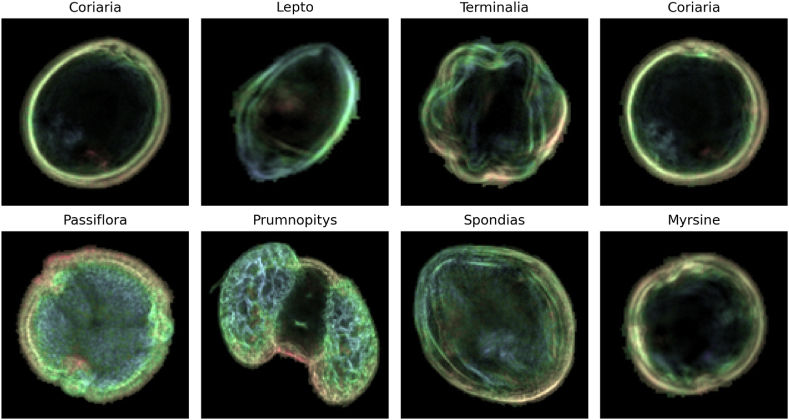
The study used the New Zealand pollen dataset [29], including 19,667 samples of pollen grains. The dataset comprises dark field microscopic images acquired using the Classifynder system, an autonomous system setup specifically tailored for automated pollen analysis. Designed to function independently, the system leverages fundamental shape descriptors to pinpoint pollen grains on standard microscope slides using a low-power objective lens. Upon identification, it engages a high-power objective lens to systematically capture high-resolution images of each pollen entity across multiple focal planes, creating a ’Z-stack’. The most sharply defined sections of the Z-stack are carefully selected and combined into a single, comprehensive composite image. The final image undergoes a segmentation process to separate the pollen grain from its background. Then, fake coloring techniques are applied to highlight the depth of information, which improves the level of detail for future classification tasks. Fig. 2 shows the sample images from the pollen dataset.
Fig. 2.
Samples of the 46 classes retrieved from the selected pollen dataset.
3.1.2. Duplicate image removal
This technique uses the MD5 hash function, a widely used cryptographic hash function that produces a 128-bit (16-byte) hash value. For any given image , the MD5 hash function maps it to a hash value , as shown in equation (1):
| (1) |
Since MD5 is a deterministic function, identical images will yield the same hash value, facilitating the identification of duplicates.
Algorithm 1
MD5-Based Duplicate Image Detection.
3.1.3. Repeating image removal
The Structural Similarity Index (SSIM) is employed for repeating image removal. For two images and , SSIM is defined as shown in equation (2):
| (2) |
Where , are the average pixel values, , are the variances and is the covariance of the images. and are constants to stabilize division with weak denominators.
Algorithm 2
Structural Similarity Index (SSIM) Based Repeating Image Removal Algorithm.
3.1.4. Unclear and distorted image removal
Blurriness is quantified using the variance of the Laplacian of an image. The Laplacian of an image , denoted as , measures its second spatial derivative. The variance of the Laplacian is given by equation (3):
| (3) |
Color variance is assessed by calculating the standard deviation of the pixel intensities across each color channel. For a color channel of an image , the standard deviation can be derived using equation (4):
| (4) |
A low mean of these standard deviations across all color channels indicates color uniformity, which is often a sign of distortion. These procedures can be observed in Algorithm 3.
Algorithm 3
Quantitative Assessment of Image Quality Through Blurriness and Color Variance Evaluation.
3.1.5. Non-local means denoising
This study employs the Non-local Means Denoising technique to reduce noise within the pollen image dataset, which is categorized into 46 groups. This method is chosen for its capability to diminish noise while preserving important image details effectively. The underlying principle of Non-local Means Denoising is that the value of a pixel can be more accurately estimated by a weighted average of all other pixel values in the image rather than only considering immediate neighbors. The denoised value of a pixel at position in a noisy image is determined by the relationship presented in equation (5):
| (5) |
Here, represents the weight determined by the similarity between small patches centered around the pixels at and , respectively. Furthermore, is the normalization factor. Algorithm 4 provides a straightforward overview of the procedural steps required to execute perform of Means Denoising.
Algorithm 4
Efficient Implementation of Non-local Means Denoising for Enhanced Image Clarity.
3.1.6. Gamma correction
Gamma correction is a crucial non-linear process in image processing used to encode and decode the brightness, also known as tristimulus values in images. The image brightness is adjusted by altering pixel intensity values, as described by the power-law formula in equation (6).
| (6) |
Here, ‘’ represents the original intensity of a pixel, denotes the gamma value, and is the gamma-corrected intensity of the pixel. The adjustment of the gamma value enables systematic supervision of image luminance. When the value of is less than 1, the image is intensified, thereby enhancing visibility in regions with lower brightness. On the contrary, when the value of surpasses 1, the image exhibits a decrease in brightness, hence enhancing the distinction in areas characterized by increased luminosity. Algorithm 5 represents the procedure of gamma correction.
Algorithm 5
Gamma Correction Procedure on pollen images.
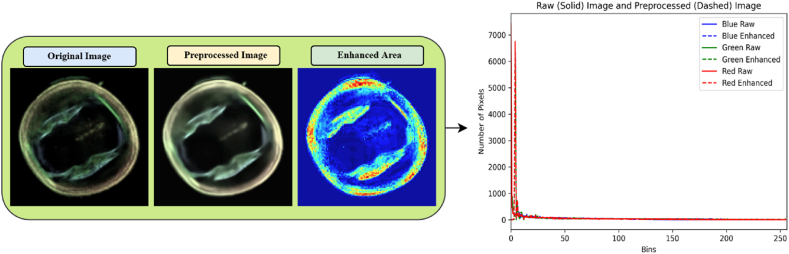
Fig. 3 displays the original image, the image after preprocessed, and a third image that emphasizes the enhanced areas. The ‘Enhanced Area’ image will augment the visibility of the differentiations, especially in regions where noise has been effectively reduced and enhanced the features. The histogram compares the distribution of pixel intensities across the channels before and after image processing, illustrating the effect of enhancement techniques on image contrast and clarity.
Fig. 3.
Demonstrates the original image, its preprocessed version, and the enhancement map alongside a histogram detailing the pixel intensity adjustments across the channels.
3.1.7. Dataset split
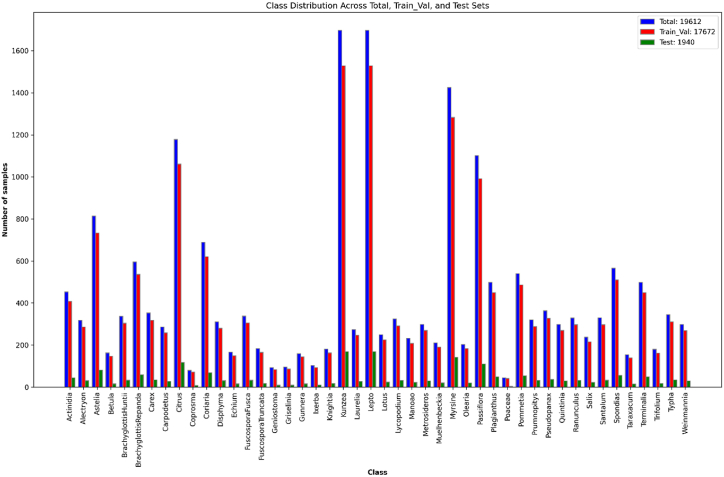
This study initially divided the dataset into a training and validation set (90 %) and a testing set (10 %) to ensure robust model evaluation. The total dataset consisted of 19,612 images after applying various preprocessing steps, including removing unclear and distorted images, duplicate images, and repeating image techniques. This resulted in a final dataset distribution of 17,672 images for training and validation and 1940 for testing.
To further ensure the robustness of the model evaluation, stratified 5-fold cross-validation [42] was applied to the training and validation set. This method involves dividing the dataset into five equally sized folds while maintaining the class distribution in each fold. Each iteration uses four folds for training and one for validation. This process is repeated five times, with each fold being used once as the validation set. This approach helps rigorously evaluate and tune the model, ensuring that the class distribution remains consistent across all folds and reducing the risk of overfitting. The stratified 5-fold cross-validation ensures the proposed model is tested on all data points while preserving the proportion of classes in each fold. This method comprehensively evaluates the model's performance, allowing for fine-tuning of hyperparameters and improving generalization. Fig. 4 depicts the distribution of the final datasets, illustrating the balanced allocation of images across the training, validation and test sets.
Fig. 4.
Detailed overview of the final pollen dataset.
3.2. Proposed model (PollenNet) structure and design
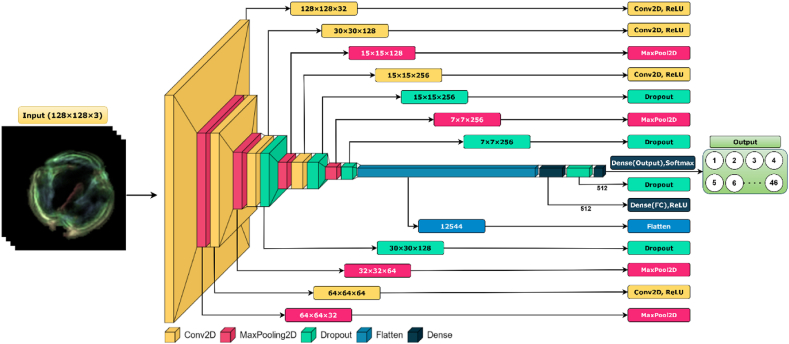
The PollenNet model is a customized Convolutional Neural Network (CNN) particularly developed for classifying pollen grains based on microscopic images. The system architecture is designed to precisely capture the intricate characteristics of pollen grains, which are crucial for proper classification.
The network accepts input images of shape 128 × 128 × 3. The model includes four convolutional blocks, each comprising a convolutional layer, a max pooling layer, and batch normalization. The first convolutional block, applies 32 filters , each of size 3 × 3, to the input image using the convolutional operation, as given in equation (7):
| (7) |
Where denotes the convolutional operation, is the bias vector, and ReLU is the activation function. Furthermore, reduces the spatial dimensions by applying a 2 × 2 max pooling operation. Then, normalizes the output of the pooling layer.
Subsequent convolutional Blocks (2nd, 3rd and 4th), follow the similar pattern with the increasing number of filters , , and include dropout layers with a rate of 0.5 after the 2nd and 4th blocks. The output and final convolutional block is flattened into a single vector , transforming the 2D feature maps into 1D feature vector.
Then, the flattened vector is passed through dense layers. The first dense layer transforms the vector, as represented in equation (8):
| (8) |
Where and are the weights and biases of the dense layer, respectively.
The final dense layer with the SoftMax activation function classifies the image into 46 categories. To convert the linear output (logits) into a probability distribution, the logits are computed by applying a linear transformation to , as indicated in equation (9):
| (9) |
Where and are the weights and biases of the output layer.
The SoftMax function is then applied to these logits to obtain the probability distribution, as illustrated in equation (10):
| (10) |
The SoftMax function itself is mathematically defined by equation (11):
| (11) |
Where is the ith logit corresponding to the ith class, N is the total number of classes, and the index j iterates over each class in the output layer, ensuring that the dominator is the sum of the exponential values of all class logits. Algorithm 6 illustrates the detailed architecture of the proposed model.
Algorithm 6
PollenNet for Pollen Grain Classification.
In the Pollen Net model, each convolutional block is characterized by an increasing number of filters, denoted by , where represents the block index. The quantity of filters doubles with each subsequent convolutional block to capture more complex features. The output of each block is represented by , with the subscript indicating the block's sequential position in the network. The convolutional layers are together with the ReLU activation function, max-pooling operations for spatial dimension reduction, and batch normalization to standardize the inputs to each layer. Additionally, dropout is strategically applied after 2nd and 4th blocks to prevent overfitting by randomly zeroing a subset of features.
Following the progression through the convolutional blocks, the final output is flattened into a one-dimensional vector , providing a format suitable for dense layer processing. The first dense layer, characterized by the weights and biases , transforms the vector with a ReLU activation function to create , a representation that encapsulates the learned high-level features. The model then computes the logits, a set of raw prediction values that are not yet normalized, using the weights and biases of the output layer.
The final step in classification is the application of the softmax function, which converts the logits into a normalized probability distribution over the possible pollen categories, ensuring that the sum of probabilities for all classes equals one. This is achieved by exponentiating each logit and dividing by the sum of all exponentiated logits, as encapsulated by the softmax operation in the network. Table 1 illustrates the detailed specifications of the proposed model PollenNet.
Fig. 5 portrays the detailed architecture of the proposed model. The model comprised four convolutional blocks, each comprising a Conv2D layer with a ReLU activation function. Following the convolutional operations, max-pooling layers are employed to reduce spatial dimensions and parameter counts, thus increasing the computational efficiency. Furthermore, Dropout layers are strategically placed after specific convolutional layers to reduce overfitting by randomly deactivating a fraction of neurons during training. Then, flattening the multidimensional convolutional features into a one-dimensional vector is a crucial step before the classification process. The flattened vector is then fed into a dense layer with ReLU activation followed by a dropout layer to enforce the regularization further. The final dense layer uses the softmax activation function to distribute the probabilities across 46 distinct pollen categories where each neuron corresponds to one category.
Table 1.
Detailed layer-by-layer architecture and parameter specification of the PollenNet convolutional neural network model for pollen grain image classification.
| Layer (Name) | Stride & Padding | Output Shape | Karnel | Activation Function | Dropout | Parameters |
|---|---|---|---|---|---|---|
| Input Layer | – | 128 × 128 × 3 | – | – | – | 0 |
| Conv 1 (Conv2D) | Stride: 1, Padding: Same | 128 × 128 × 32 | 3 × 3 | ReLU | – | 896 |
| Max Pooling 1 | Stride: 2, Padding: Valid | 64 × 64 × 32 | 2 × 2 | – | – | 0 |
| Conv 2 (Conv2D) | Stride: 1, Padding: Same | 64 × 64 × 64 | 3 × 3 | ReLU | – | 18,496 |
| Max Pooling 2 | Stride: 2, Padding: Valid | 32 × 32 × 64 | 2 × 2 | – | – | 0 |
| Conv 3 (Conv2D) | Stride: 1, Padding: Valid | 30 × 30 × 128 | 3 × 3 | ReLU | 0.5 | 73,856 |
| Max Pooling 3 | Stride: 2, Padding: Valid | 15 × 15 × 128 | 2 × 2 | – | – | 0 |
| Conv 4 (Conv2D) | Stride: 1, Padding: Same | 15 × 15 × 256 | 3 × 3 | ReLU | 0.5 | 295,168 |
| Max Pooling 4 | Stride: 2, Padding: Valid | 7 × 7 × 256 | 2 × 2 | – | 0.5 | 0 |
| Flatten Layer | – | 12544 | – | – | – | 0 |
| Dense (Fully Connected) | – | 512 | – | ReLU | 0.5 | 6423040 |
| Dense (Output) | – | 46 | – | softmax | – | 23598 |
Fig. 5.
Schematic representation of the PollenNet architecture models for grain classification, detailing the convolutional, pooling, dropout and dense layer.
4. Result and analysis
This section presents a comprehensive overview of the study's findings. It begins with a performance analysis of the applied models, highlighting their effectiveness and comparative advantages. The complexity of the model's predictive abilities is further explored through confusion matrices and ROC curves, serving as illustrative measures of classifier performance. The assessment approach is supported by a robust stratified 5-fold cross-validation process, ensuring the results' reliability and generalizability. Explainable AI (XAI) is employed to provide clarity on the interpretative aspects of model predictions, offering a clear perspective on the underlying decision-making processes. We deliberately selected four pre-trained distinct models in our analysis: AlexNet, MobileNetV2, VGG16, and ResNet50. The models were chosen based on their distinct abilities and proficiencies in the domains of image processing and classification. The proposed PollenNet model, specifically developed for this work, was intended to tackle the unique challenges of categorizing pollen grain images. The selection of AlexNet, an early deep learning model, was based on its well-documented effectiveness in image recognition tasks, which serves as a dependable benchmark for comparison. The MobileNetV2 model, known for its effectiveness and speed, was chosen to assess performance in a resource-constrained setting. The decision to include VGG16 was based on its renowned characteristics of simplicity and depth and its remarkable capacity to record visual textures and patterns. Ultimately, ResNet50, which incorporates a residual learning framework, was selected to evaluate the effectiveness of more complex network architectures in this particular domain. The integration of these models provided a comprehensive analysis of several architectural techniques, extending from traditional to innovative, in the field of pollen grain image classification [32]. By implementing a varied and comprehensive method, we successfully evaluated and compared many techniques, augmenting the scope and comprehensiveness of our study's findings and conclusions.
The research encompassed five algorithms, including the proposed PollenNet model and four existing transfer learning models. The algorithms were implemented to carry out the crucial task of detecting and classifying images of pollen grains. The classification of pollen grain images is essential in several scientific and environmental contexts, as it significantly influences the understanding of pollen-related allergies, the monitoring of ecological changes, and the progress of agriculture and climatology. Accurately identifying and categorizing pollen grains is essential for monitoring plant diversity and may aid in predicting allergic reactions, significantly impacting public health and environmental studies. To fully evaluate these models, each one underwent intensive training for 100 epochs, and their performance was meticulously documented at every stage. The rigorous training procedure was essential for accurately evaluating the ability of each model to analyze and classify complex pollen grain images, which display diverse morphological characteristics. The effectiveness and efficiency of these models in classifying pollen grains were evaluated using certain performance measures, calculated using equations (12), (13), (14), (15), (16), (17), (18), (19), (20). This approach provides a comprehensive comprehension of the performance of each algorithm when used for the complex task of analyzing pollen grain images. This work is crucial for advancing our understanding of environmental and ecological processes.
4.1. Environmental setup
The deep learning models used in our study were principally developed using the TensorFlow (version 2.0) frameworks and the Keras library (version 2.10.0), both of which were coded in Python version 3.7. We used the Seaborn and Matplotlib tools to enhance data visualization, which is renowned for creating detailed and intuitive graphical representations of our results. The computational evaluation of these models was conducted utilizing a system equipped with high-performance computing capabilities. The machine used an AMD Ryzen 7 CPU operating at a clock speed of 3.90 GHz. In addition, it had 32 GB of RAM, providing enough capacity for processing large datasets and complex calculations. We used an AMD Radeon RX 580 series GPU for graphic processing, which confers significant benefits for deep learning tasks. To ensure the stability and compatibility of all software components used in our investigation, we conducted the whole operation inside a Windows 10 environment.
4.2. Evaluation metrics
We used a range of statistical measures, including Accuracy, Specificity, Recall, Precision, False Positive Rate (FPR), F1-score, Mean Squared Error (MSE), and Mean Absolute Error (MAE), to thoroughly evaluate our model's performance in classifying pollen images [37].
The model's accuracy is defined as the ratio of accurate predictions to the total number of predictions produced.
| (12) |
Here, = True Positives, = True Negatives, = False Positives and = False Negatives.
Denotes the proportion of pertinent instances out of the total instances that were predicted as positive by the model. It offers valuable insight regarding the accuracy of positive predictions.
| (13) |
The parameter denotes the percentage of true positive instances that the model successfully predicted, thereby showcasing its capability to identify positive cases.
| (14) |
This metric quantifies the true negative rate, which serves as an indicator of the model's efficacy in precisely classifying the negative instance among all other instances.
| (15) |
The percentage of negative instances that are erroneously categorized as positive.
| (16) |
Achieves harmony between Precision and Recall through their integration into a single metric. Additionally, this metric evaluates the capability of a model to distinguish between positive and negative data.
| (17) |
The FNR is a metric that measures the proportion of actual positive instances that were incorrectly classified as negative by the model.
| (18) |
The difference between the estimator and what is estimated is calculated as the average of the squares of the errors or deviations.
| (19) |
Where is the actual value, is the predicted value and is the number of observations.
Evaluates the average magnitude of the errors in a set of predictions without considering their direction. It's the mean over the test sample of the absolute differences between prediction and actual observation, where all individual differences have equal weight.
| (20) |
4.3. Performance evaluation of all applied models
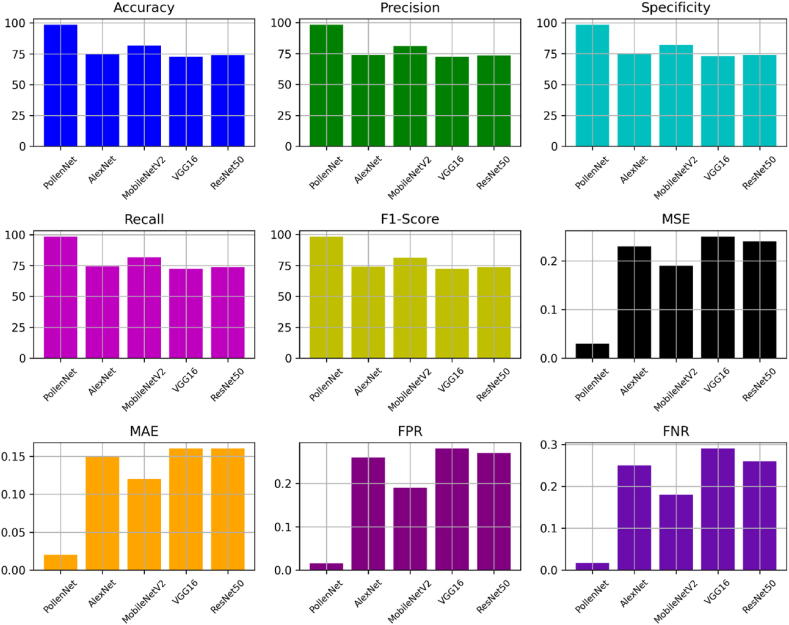
This study thoroughly evaluated five distinct models to determine their efficacy in precisely classifying pollen images with high dependability. Fig. 6 displays the performance of all the models. After analyzing the models 'PollenNet,' 'AlexNet,' 'MobileNetV2,' 'VGG16,' and 'ResNet50,' we have identified significant attributes and benefits in their performance metrics. PollenNet demonstrates outstanding accuracy, earning an impressive test accuracy of 98.45 %, validation accuracy of 98.60 % and a precision of 98.20 %. PollenNet's exceptional precision is shown in its ability to classify positive instances when accurately categorizing pollen grains. Furthermore, the impressive specificity of 98.40 % showcases its strong ability to precisely identify negative samples, which is essential for reducing the occurrence of false positives in critical classification tasks.
Fig. 6.
Performance comparison among all applied models for pollen grain classification.
In comparison, the other models, such as AlexNet, MobileNetV2, VGG6 and ResNet50, demonstrated varying but typically lower performance levels. AlexNet, despite showing a decent recall of 74.30 %, lacked test accuracy at 74.80 %, indicating problems in consistently detecting true positive instances. Furthermore, MobileNetV2 and VGG6, with test accuracies of 81.60 % and 72.68 %, respectively and moderate F-scores, exhibited a balance between precision and recall but failed to match PollenNet's balanced and high-performance results. ResNet50, achieving test accuracy of 73.85 % and specificity of 74.10 %, demonstrated the lack of capacity during pollen grain classification. All models have F1 scores, demonstrating an appropriate compromise between accuracy and recall. Among them, PollenNet has the highest score of 98.25 %. The models demonstrate different performance levels when assessed using Mean Squared Error (MSE) and Mean Absolute Error (MAE) as measures of error rates. PollenNet has a Mean Squared Error (MSE) of 0.03 and a Mean Absolute Error (MAE) of 0.02, indicating a significant degree of precision in its predictive capabilities. PollenNet excels in the crucial aspect of the False Positive Rate, with a very low FPR of 0.016 and False Negative Rate (FNR) of 0.017. This demonstrates its effectiveness in decreasing incorrect positive classifications.
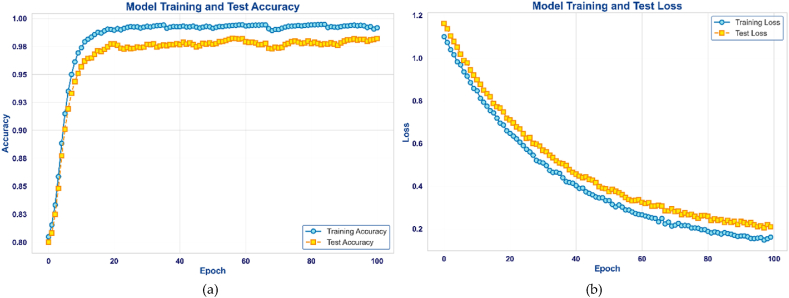
PollenNet stands out because of its remarkable accuracy, precision, and low false positive rate. These specific features make it ideal for the complex and precise task of classifying pollen grains. While the different models have distinct advantages, they also have areas that might be improved, such as reaching an acceptable balance between accuracy, recall, and reducing error rates. Fig. 7 illustrates the performance metrics of the proposed PollenNet model during the training and testing phase. Subfigure (a) depicts the model's training and test accuracy over 100 epochs, displaying an upward trend in accuracy that eventually levels off at a high level, indicating the model's effective learning from the data. Subfigure (b) illustrates the training and test loss. The training loss decreases rapidly and maintains a low value as the epochs of number increase, indicating effective learning by the model. The test loss also decreases initially but begins to fluctuate as the epoch progresses, a typical occurrence during performance on new, unseen data.
Fig. 7.
Demonstrates the (a) accuracy and (b) loss of the proposed PollenNet model.
These results provide valuable insights into each model's pros and cons in identifying pollen grain images. By analyzing these patterns, we may get a more thorough knowledge of the model's ability to distinguish distinct types of pollen. This component is crucial in environmental monitoring, allergy prediction, and plant biodiversity studies. Furthermore, Table 2 summarizes the performance metrics of the five models used in the investigation. Here, the training accuracy is denoted as ‘T_Acc’, validation accuracy is denoted as ‘V_Acc’ and test accuracy is denoted as ‘T_Ac’.
Table 2.
Final overview of findings of all applied models for pollen image classification.
| Model | T_Acc | V_Acc | T_Ac | Precision | SPE | Recall | F1-S | MSE | MAE | FPR | FNR |
|---|---|---|---|---|---|---|---|---|---|---|---|
| PollenNet | 99.50 % | 98.60 % | 98.45 % | 98.20 % | 98.40 % | 98.30 % | 98.25 % | 0.03 | 0.02 | 0.016 | 0.017 |
| AlexNet | 80.47 % | 75.20 % | 74.80 % | 73.90 % | 74.80 % | 74.30 % | 74.10 % | 0.23 | 0.15 | 0.26 | 0.25 |
| MobileNetV2 | 84.91 % | 82.10 % | 81.60 % | 81.10 % | 82.00 % | 81.60 % | 81.35 % | 0.19 | 0.12 | 0.19 | 0.18 |
| VGG16 | 77.35 % | 73.00 % | 72.68 % | 72.25 % | 73.10 % | 72.50 % | 72.37 % | 0.25 | 0.16 | 0.28 | 0.29 |
| ResNet50 | 79.50 % | 74.00 % | 73.85 % | 73.50 % | 74.10 % | 73.70 % | 73.60 % | 0.24 | 0.16 | 0.27 | 0.26 |
The results of our study demonstrate that the PollenNet model showed significant promise, as it successfully categorized images of pollen grains. PollenNet had an impressive test accuracy rate of 98.45 %, combined with a precision of 98.20 %, indicating its strong ability to categorize various types of pollen precisely. The model demonstrated a remarkable degree of specificity, achieving a rate of 98.40 %, and a recall rate of 98.30 %, indicating its ability to reliably classify positive instances while minimizing the incidence of false negatives. The F1-score, a metric that captures the compromise between accuracy and recall, was calculated to be 98.25 %, showing the model's overall robustness. When analyzing error measurements, PollenNet demonstrated outstanding accuracy with a low Mean Squared Error (MSE) of 0.03 and Mean Absolute Error (MAE) of 0.02. The model exhibited an impressively low False Positive Rate (FPR) of 0.016 and a False Negative Rate (FNR) of 0.017, indicating its exceptional accuracy in accurately classifying and differentiating various pollen types. Throughout the 5-fold cross-validation process, PollenNet excelled consistently, showcasing its robustness and reliability. Eventually, the model outperformed all applied pre-trained models in the comparison and demonstrated its superiority in the given classification task.
4.4. Confusion matrix and ROC curve
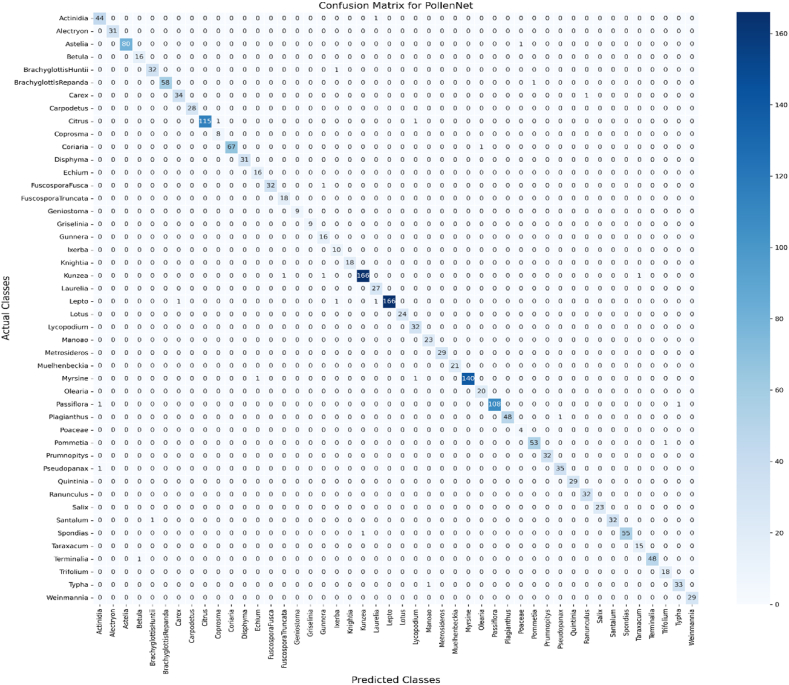
During the testing phase of our research, we evaluated the effectiveness of the models by utilizing confusion matrices to analyze the patterns of accurate categorization and inaccurate categorization for images of pollen grains. The models were assessed using a comprehensive test dataset comprising 1940 samples across 46 distinct classes of pollen. Fig. 8 presents the confusion matrix of the PollenNet model, which provides a detailed summary of its classification performance for the various types of pollen grains.
Fig. 8.
Confusion Matrix for PollenNet model Classification. The matrix illustrates the prediction distribution across actual classes with a pronounced diagonal indicating a high rate of correct classifications and minimal off-diagonal entries signifying few misclassifications.
The axes of the confusion matrix correspond to the actual and predicted class labels. Diagonal entries in the matrix represent correctly classified samples, highlighted by darker shades, while off-diagonal cells represent instances of misclassification, with lighter shades indicating a lower frequency of occurrences. A prominent dark diagonal line is observed in the matrix, reflecting PollenNet's high accuracy in correctly classifying the majority of samples.
For instance, the class “Kunzea” had 169 test samples, of which 166 were correctly classified, reflecting the model's high precision for this class. Similarly, “Lepto” had 169 test samples, with 166 correctly classified. The “Citrus” class had 117 test samples, and the proposed model accurately identified 115. Classes such as “Alectryon”, with 31 samples, and “Astelia”, with 81 samples, also show high correct classification rates, further emphasizing the model's precision and recall capabilities. These high numbers and the diagonal for most classes signify PollenNet's robustness in correctly identifying pollen grains. The lighter shades in off-diagonal cells indicate fewer misclassifications, such as in the “FuscosporaFusca” and “Pseudopanax” classes, where the misclassification was negligible. Overall, the confusion matrix shows that PollenNet achieved an impressive accuracy rate of 98.45 %. This high accuracy is crucial for applications requiring precise identification of pollen grains, underscoring PollenNet's effectiveness and reliability.
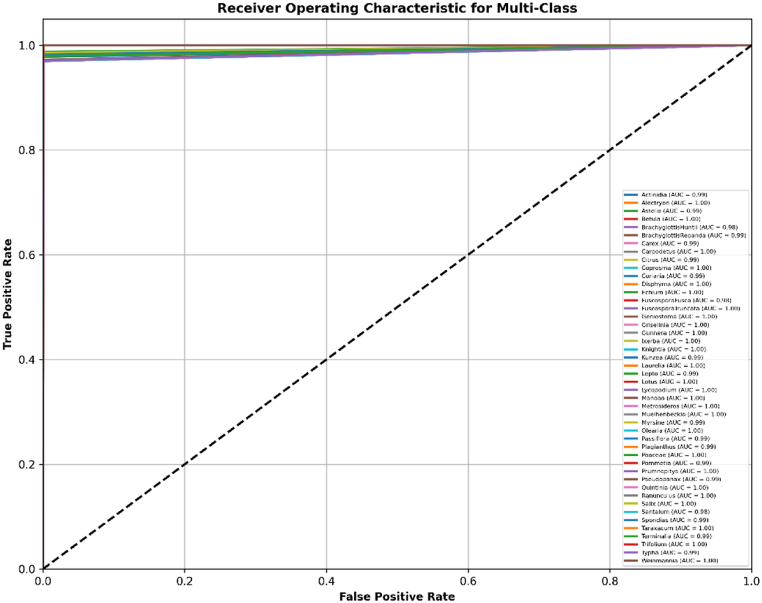
Fig. 9 shows the comprehensive ROC curves for the PollenNet model, evaluating its classification performance across 46 different pollen grain classes. Each curve represents the trade-off between the True Positive Rate (Sensitivity) and False Positive Rate (1-specificity) for a specific class. Particularly, classes such as Betula, Manoao, and Metrosideros exhibit exceptionally high AUC values of 1.00, indicating perfect or near-perfect discrimination between positive and negative instances. Other classes, including Actinidia and Citrus, display slightly lower AUC values, such as 0.99, reflecting strong but slightly less than perfect classification performance.
Fig. 9.
Analysis of ROC curve and AUC scores for multi-class classification of the proposed model where each curve represents one of the classes.
The AUC values are remarkably consistent, with most classes having AUCs close to 1.00, showcasing the model's robustness. The lower AUC values, such as 0.98 for FuscosporaFusca and Santalum, were observed, still indicating a strong performance. The diagonal dashed line in the plot represents the performance of a random classifier, with a 50 % chance of correct classification. The distance of the PollenNet curves from this line underscores its superior performance. The ROC analysis in this figure further confirms PollenNet's ability to accurately distinguish between various pollen grains accurately, maintaining high precision and recall across a diverse set of classes. This detailed evaluation of class-specific AUC values highlights the model's effectiveness in multi-class classification tasks, ensuring reliable identification across all categories.
4.5. Stratified 5-fold cross-validation
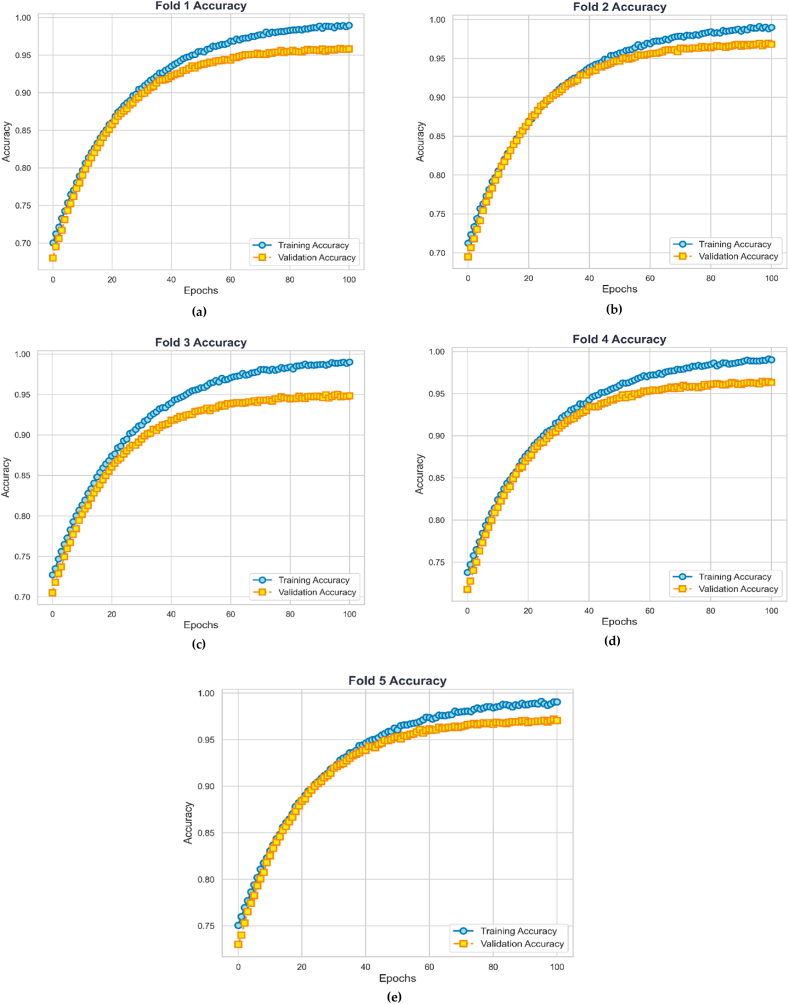
Fig. 10 represents the Stratified 5-fold cross-validation accuracy of PollenNet. The figure illustrates the training and validation accuracy of PollenNet across five distinct cross-validation folds. Recent studies have employed the method of monitoring each fold during k-fold cross-validations by tracking training and validation accuracy and losses across epochs [43,44]. This approach is widely used in deep learning research to gain deeper insights into the model's learning dynamics and ensure robust performance evaluation across different data splits. Each subplot (a-e) corresponds to a separate fold, depicting the model's performance over 100 training epochs. The blue lines represent the training accuracy, while the orange line denotes validation accuracy for each fold. The plots collectively indicate a consistent achievement of high accuracy, with training and validation accuracies maintaining above 98 % and 95 %, respectively, across all folds. This performance pattern underscores the robustness and generalizability of PollenNet, demonstrating its capacity to maintain high predictive accuracy even with varying subsets of data.
Fig. 10.
Each graph (a–e) represents one of the 5-folds, showing training and validation accuracy over epochs.
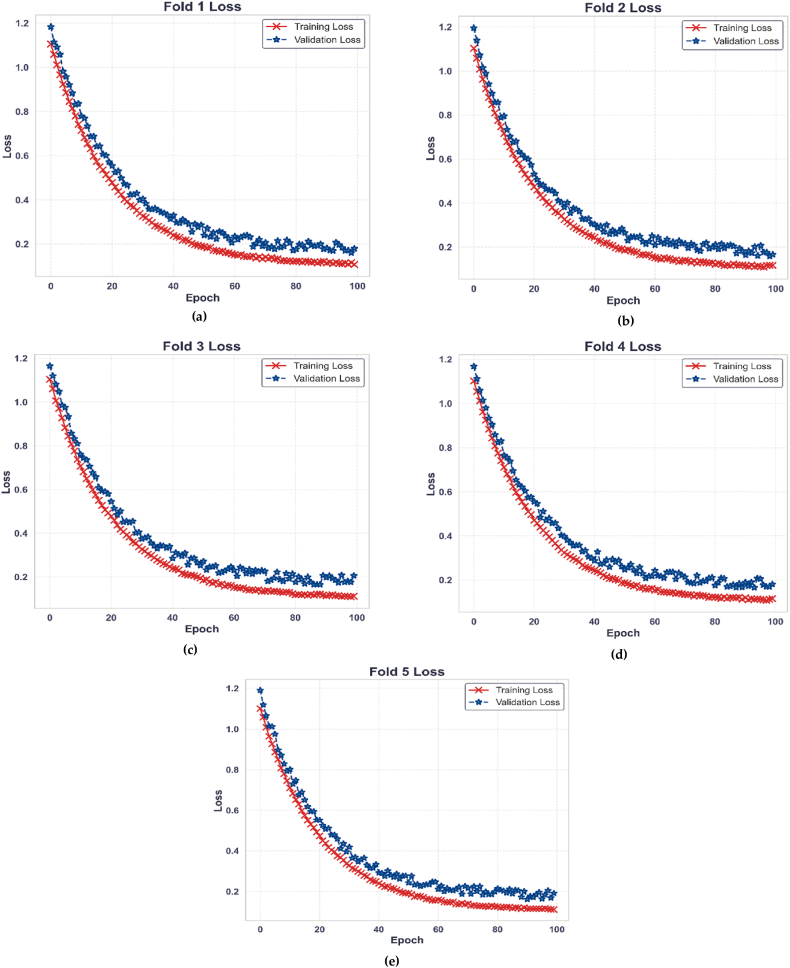
Fig. 11 depicts loss metrics during the stratified 5-fold cross-validation of PollenNet. Subfigures (a-e) illustrate the training and validation loss as a function of epochs for each fold. The red line represents the training loss, decreasing steadily as the model learns from the data. The blue line depicting the validation loss shows an overall drop with some fluctuation, indicating that the model is encountering and adjusting to new patterns within the validation set. The consistency of this pattern across all 5-folds demonstrates PollenNet's effective learning and generalization capability across folds under the cross-validation framework.
Fig. 11.
Training and validation loss across 5-fold (a–e) in PollenNet's cross-validation.
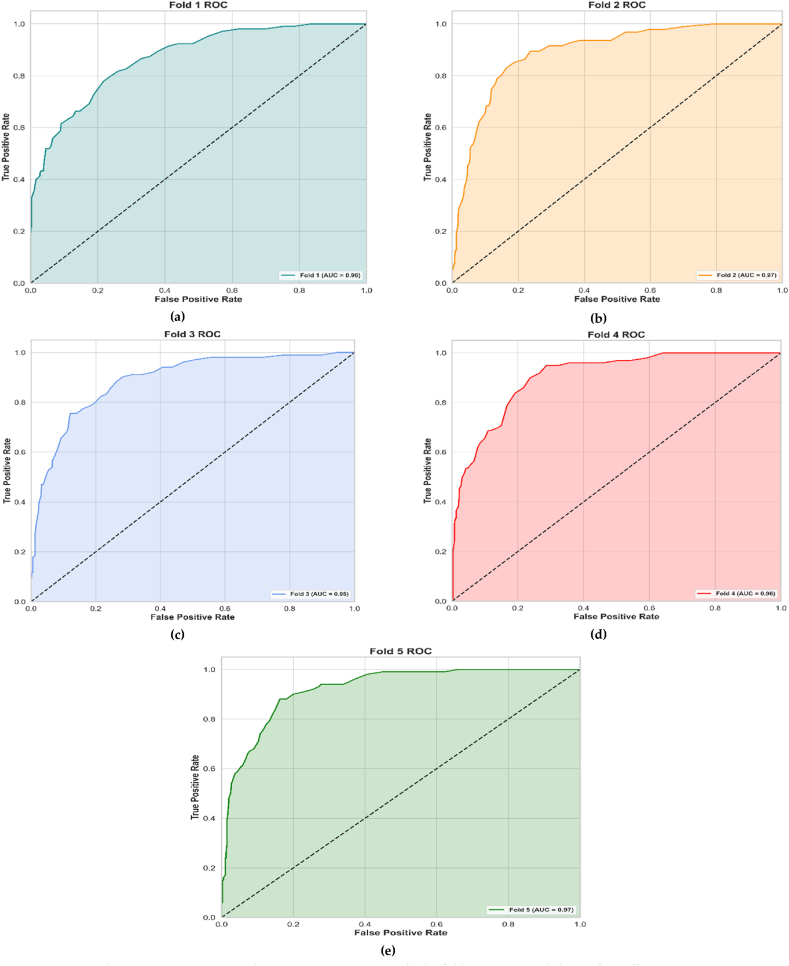
The Receiver Operating Characteristic (ROC) curves and Area Under the Curve (AUC) scores serve as essential indicators in the attempt to gain a deep comprehension of a model's performance across different classifications. Fig. 12(a–e) exhibits the ROC curves and AUC scores for each fold in the 5-fold cross-validation of PollenNet. The ROC curves for Fold 1 through Fold 5 are displayed, with AUC values of 0.96, 0.97, 0.95, 0.96, and 0.97, respectively. Each curve, accompanied by its shaded area, indicates the model's ability to classify different classes across all folds correctly. The shaded areas under each curve visually represent the AUC, providing an intuitive understanding of the model's performance. The curves trending towards the upper left corner confirms the higher true positive rates and low false positive rates, signifying the model's reliable performance. The consistency in the AUC scores across all folds demonstrates PollenNet's robustness and effectiveness in accurately handling the diverse and complex nature of the dataset despite any class imbalances or morphological variations. These results affirm the model's generalizability and ability to maintain high predictive accuracy across varying validation sets.
Fig. 12.
ROC curves and AUC scores across stratified 5-fold (a–e) cross-validation for PollenNet.
Furthermore, Table 3 provides a comprehensive overview of PollenNet's performance across five validation folds, illustrating the model's consistency and robustness. Each fold demonstrates high training accuracy (T_Acc), ranging from 98.00 % to 98.99 %, indicating the model's effectiveness in learning from the training data. The validation accuracy (V_Acc) range of 95.00 %–97.20 % demonstrates the model's robust generalization capabilities on unseen data. Precision values, which measure the proportion of relevant instances correctly identified, remain consistently high between 94.10 % and 96.70 %, reflecting the model's accuracy in retrieving relevant data. Specificity (SPE), which evaluates the model's ability to identify negative results correctly, is uniformly high across all folds, underscoring the model's efficacy in minimizing false positives. Recall values ranging from 94.20 % to 96.75 % further indicate the model's strong capability to identify relevant instances. The F1-Score (F1-S), which balances precision and recall, is consistently high across all folds. Additionally, the False Positive Rates (FPR) and False Negative Rates (FNR) are low, reinforcing the model's effectiveness in minimizing errors. Overall, the results demonstrate that PollenNet performs reliably across different folds, maintaining high accuracy, precision, and low error rates, which underscores the model's robustness in classification tasks.
Table 3.
Performance metrics for proposed PollenNet across five validation folds.
| Model | T_Acc | V_Acc | Precision | SPE | Recall | F1-S | FPR | FNR |
|---|---|---|---|---|---|---|---|---|
| Fold 1 | 98.68 % | 96.00 % | 95.50 % | 95.70 % | 95.60 % | 95.55 % | 0.05 | 0.04 |
| Fold 2 | 98.90 % | 97.00 % | 96.50 % | 96.60 % | 96.60 % | 96.55 % | 0.04 | 0.03 |
| Fold 3 | 98.00 % | 95.00 % | 94.10 % | 94.30 % | 94.20 % | 94.15 % | 0.06 | 0.05 |
| Fold 4 | 98.75 % | 96.50 % | 95.85 % | 96.00 % | 95.90 % | 95.88 % | 0.04 | 0.04 |
| Fold 5 | 98.99 % | 97.20 % | 96.70 % | 96.80 % | 96.75 % | 96.72 % | 0.03 | 0.03 |
5. Explainable AI (XAI) for model prediction analysis
In the field of Explainable AI (XAI) [39], visualization techniques such as Gradient-weighted Class Activation (Grad-CAM), saliency maps, and misclassification analysis play a pivotal role in demonstrating the internal mechanisms of neural networks such as PollenNet. Grad-CAM generates heatmaps that reveal the region's most influence on the model's predictions, while saliency maps detail the specific pixels that capture the model's attention. In addition, carefully examining misclassified images may indicate subtle anomalies in the dataset or identify areas where the model may benefit from further training. Together, these methods enhance our understanding of PollenNet's decision-making process.
5.1. Grad-CAM analysis for visualizing decision-making patterns
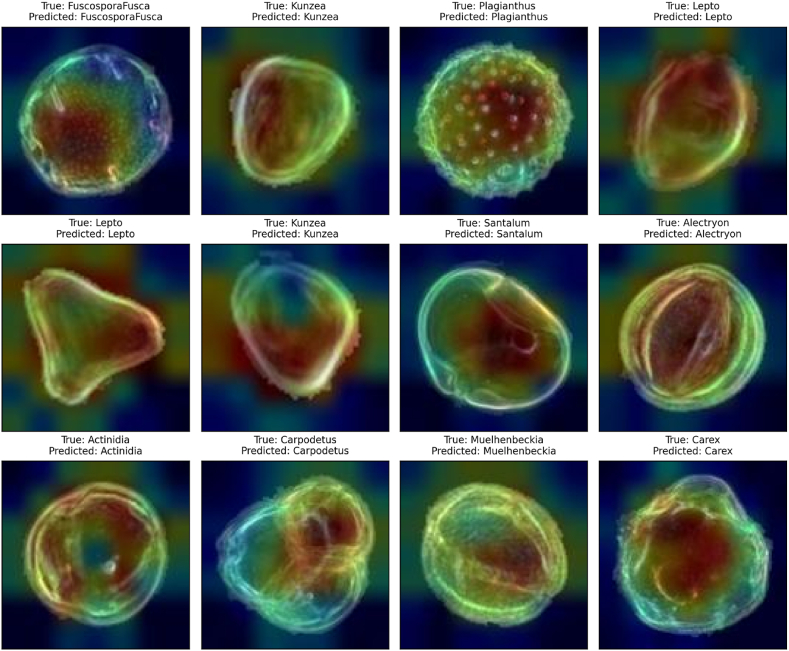
Fig. 13 depicts Grad-CAM [[38], [40]] visualization of PollenNet's classifications. Displayed heatmaps for correctly classified pollen grains by PollenNet, indicating the image's most influential areas for the classification. The levels from blue to red on the heatmaps represent the model's focus from least to most influential, with red areas being key to the correct class identification. These visualizations confirm PollenNet's capability to successfully recognize and leverage distinct features for each class, such as the central pollen structure in ‘FucosporaFusca’ and the defining edges in ‘Kunzea’. The consistency in PollenNet's attention to relevant features across various classes underlines its exceptional performance in comprehensive feature extraction for accurate pollen image classification.
Fig. 13.
Grad-CAM heatmaps demonstrate PollenNet's accurate feature detection across various correctly classified pollen grains.
5.2. Saliency map analysis for feature visualization
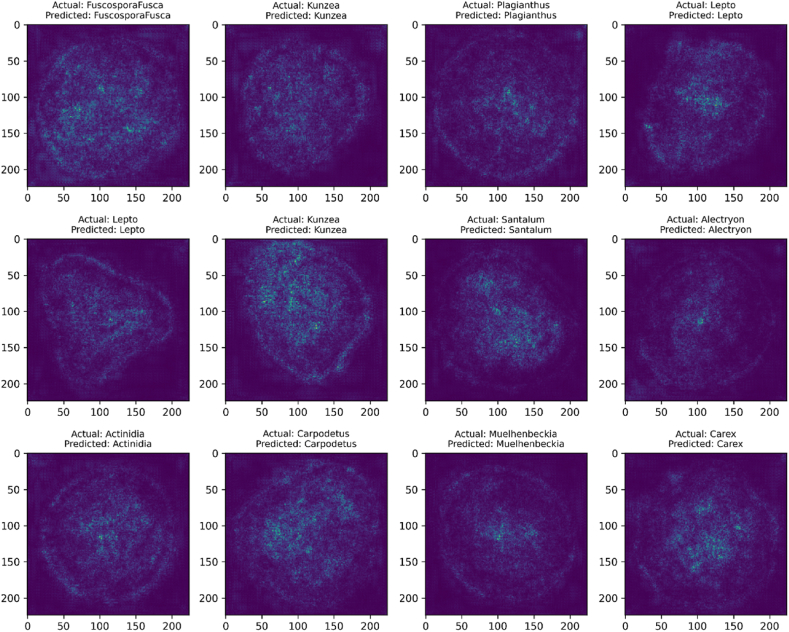
Fig. 14 displays the saliency maps [41] generated by PollenNet for the purpose of classifying pollen grain images. The presented saliency maps highlight the precise areas inside each image that were identified as the most significant by PollenNet during the categorization procedure. Saliency maps are a form of visual explanation that illuminates the gradient of the model's attention, where brighter regions indicate a higher contribution to the model's predictive conclusion. These particular maps correspond to various classes of pollen grains where the model's focus is clearly on identifying specific textural and structural features that are crucial for recognizing each category. The saliency maps produced by PollenNet effectively highlight important regions, demonstrating its advanced ability to discover distinctive features and accurately distinguish across intricate categories in the dataset.
Fig. 14.
Saliency maps highlight the focus area utilized by PollenNet to successfully classify different pollen grain classes.
6. Study limitations (interpreting model misclassifications with Grad-CAM)
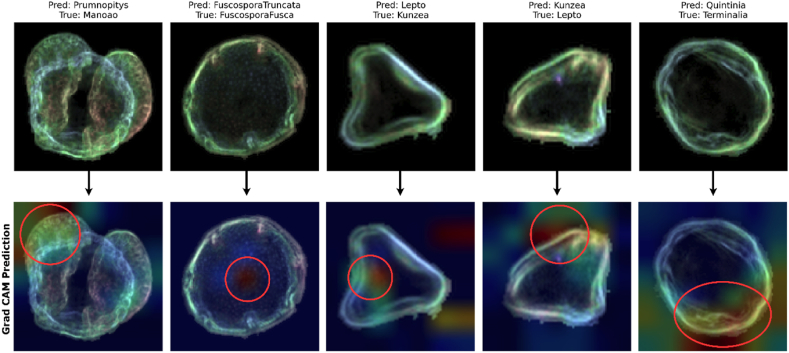
Fig. 15 illustrates the use of Grad-CAM to identify PollenNet's areas of focus that resulted in misclassification. Despite achieving an impressive accuracy in training, testing and validation the PollenNet occasionally misinterprets images. Each subfigure, with a red circle, depicts an input image superimposed with a heatmap that outlines the regions deemed critical by PollenNet during the prediction phase. The warmer colors highlight segments that significantly influenced PollenNet's decision-making process. The model's significant accuracy across a diverse dataset with 46 classes and more than 17000 images indicates that the misclassification shown may be attributable to nuances within the dataset. Such nuances could intrinsically class ambiguities, subtle variations within class instances or imbalances in representative features. These visual examinations imply that, while PollenNet demonstrates exceptional performance, there remains an opportunity for further fine-tuning and dataset augmentation to address these edge cases, thereby enhancing the boundaries of its classification capabilities.
Fig. 15.
Grad-CAM heat maps on specific misclassified images by PollenNet showing regions impacting its predictions.
7. Conclusions
The study presents 'PollenNet' as a novel approach for categorizing images of pollen grains. It exhibits exceptional accuracy (98.45 %) and effectiveness. The instrument has substantial promise in ecological research, particularly in environmental studies and agricultural applications, as shown by its accuracy, specificity, recall, and F1 score. The minimal error statistics provide further validation of its reliability. PollenNet's future possibilities include augmenting the dataset to encompass a broader spectrum of pollen varieties and exploring novel image processing techniques. These efforts have significant potential to enhance PollenNet's dependability and usefulness. Additional investigation might also focus on the use of PollenNet in real-world scenarios to aid with agricultural planning and allergy forecasting. Furthermore, the methodologies used in this study have the potential to be adapted for similar challenges in several scientific disciplines, hence promoting interdisciplinary progress. PollenNet is a significant advance in the classification of pollen, facilitating broader scientific inquiries and practical applications. This underscores the pivotal significance of machine learning in advancing environmental and botanical studies.
Data availability
All data generated or analyzed during this study are included in this published article. It also available in- https://doi.org/10.6084/m9.figshare.12370307.v1.
CRediT authorship contribution statement
F M Javed Mehedi Shamrat: Writing – review & editing, Writing – original draft, Visualization, Software, Resources, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Mohd Yamani Idna Idris: Writing – review & editing, Supervision, Project administration. Xujuan Zhou: Writing – review & editing, Project administration, Funding acquisition. Majdi Khalid: Writing – review & editing, Project administration. Sharmin Sharmin: Writing – original draft, Visualization, Software, Resources, Methodology, Investigation, Formal analysis, Data curation. Zeseya Sharmin: Writing – review & editing, Validation, Project administration. Kawsar Ahmed: Writing – review & editing, Validation, Supervision, Project administration, Conceptualization. Mohammad Ali Moni: Writing – review & editing, Validation, Supervision, Software, Project administration.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Contributor Information
F M Javed Mehedi Shamrat, Email: javedmehedicom@gmail.com.
Mohd Yamani Idna Idris, Email: yamani@um.edu.my.
Xujuan Zhou, Email: xujuan.zhou@usq.edu.au.
Majdi Khalid, Email: mknfiai@uqu.edu.sa.
Sharmin Sharmin, Email: sharmin9889@gmail.com.
Zeseya Sharmin, Email: zeseya15@gmail.com.
Kawsar Ahmed, Email: kawsar.ict@mbstu.ac.bd, k.ahmed@usask.ca.
Mohammad Ali Moni, Email: mmoni@csu.edu.au.
References
- 1.Halbritter H., Ulrich S., Grímsson F., Weber M., Zetter R., Hesse M., Buchner R., Svojtka M., Frosch-Radivo A. Springer; 2018. Illustrated Pollen Terminology. [Google Scholar]
- 2.Salonen J.S., Korpela M., Williams J.W., Luoto M. Machine-learning based reconstructions of primary and secondary climate variables from North American and European fossil pollen data. Sci. Rep. 2019;9(1) doi: 10.1038/s41598-019-52293-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnes C.M., Power A.L., Barber D.G., Tennant R.K., Jones R.T., Lee G.R.…Love J. Deductive automated pollen classification in environmental samples via exploratory deep learning and imaging flow cytometry. New Phytol. 2023;240(3):1305–1326. doi: 10.1111/nph.19186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balmaki B., Rostami M.A., Christensen T., Leger E.A., Allen J.M., Feldman C.R.…Dyer L.A. Modern approaches for leveraging biodiversity collections to understand change in plant-insect interactions. Frontiers in Ecology and Evolution. 2022;10 [Google Scholar]
- 5.Khanzhina N., Putin E. Digital Transformation and Global Society: First International Conference, DTGS 2016, St. Petersburg, Russia, June 22-24, 2016, Revised Selected Papers 1. Springer International Publishing; 2016. Pollen recognition for allergy and asthma management using gist features; pp. 515–525. [Google Scholar]
- 6.Tan Z., Yang J., Li Q., Su F., Yang T., Wang W.…Min L. PollenDetect: an open-source pollen viability status recognition system based on deep learning neural networks. Int. J. Mol. Sci. 2022;23(21) doi: 10.3390/ijms232113469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunker S., Motivans E., Rakosy D., Boho D., Maeder P., Hornick T., Knight T.M. Pollen analysis using multispectral imaging flow cytometry and deep learning. New Phytol. 2021;229(1):593–606. doi: 10.1111/nph.16882. [DOI] [PubMed] [Google Scholar]
- 8.Daood A., Ribeiro E., Bush M. International Symposium on Visual Computing. Springer International Publishing; Cham: 2016, December. Pollen grain recognition using deep learning; pp. 321–330. [Google Scholar]
- 9.Sobol M.K., Finkelstein S.A. Predictive pollen-based biome modeling using machine learning. PLoS One. 2018;13(8) doi: 10.1371/journal.pone.0202214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Riley K.C., Woodard J.P., Hwang G.M., Punyasena S.W. Progress towards establishing collection standards for semi-automated pollen classification in forensic geo-historical location applications. Rev. Palaeobot. Palynol. 2015;221:117–127. [Google Scholar]
- 11.Ngo T.N., Rustia D.J.A., Yang E.C., Lin T.T. Automated monitoring and analyses of honey bee pollen foraging behavior using a deep learning-based imaging system. Comput. Electron. Agric. 2021;187 [Google Scholar]
- 12.Schiele J., Rabe F., Schmitt M., Glaser M., Häring F., Brunner J.O.…Damialis A. 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC) IEEE; 2019, July. Automated classification of airborne pollen using neural networks; pp. 4474–4478. [DOI] [PubMed] [Google Scholar]
- 13.Sevillano V., Aznarte J.L. Improving classification of pollen grain images of the POLEN23E dataset through three different applications of deep learning convolutional neural networks. PLoS One. 2018;13(9) doi: 10.1371/journal.pone.0201807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vega G.L. Université de Bourgogne; 2015. Image-based Detection and Classification of Allergenic Pollen (Doctoral Dissertation. [Google Scholar]
- 15.del Pozo-Baños M., Ticay-Rivas J.R., Cabrera-Falcón J., Arroyo J., Travieso-González C.M., Sánchez-Chavez L., Pérez S.T., Alonso J.B., Ramírez-Bogantes M. 2012. Image Processing for Pollen Classification. [Google Scholar]
- 16.Sonka M., Hlavac V., Boyle R. Springer; 2013. Image Processing, Analysis and Machine Vision. [Google Scholar]
- 17.Shakil R., Akter B., Shamrat F.J.M., Noori S.R.H. A novel automated feature selection based approach to recognize cauliflower disease. Bulletin of Electrical Engineering and Informatics. 2023;12(6):3541–3551. [Google Scholar]
- 18.Shamrat F.J.M., Azam S., Karim A., Ahmed K., Bui F.M., De Boer F. High-precision multiclass classification of lung disease through customized MobileNetV2 from chest X-ray images. Comput. Biol. Med. 2023;155 doi: 10.1016/j.compbiomed.2023.106646. [DOI] [PubMed] [Google Scholar]
- 19.Rebiai A., Seghir B.B., Hemmami H., Zeghoud S., Siham T., Kouadri I., Terea H., Brahmia F. Clustering and Discernment of bee pollen using an image analysis system. Algerian Journal of Chemical Engineering AJCE. 2021;1(2):41–48. [Google Scholar]
- 20.Manikis G.C., Marias K., Alissandrakis E., Perrotto L., Savvidaki E., Vidakis N. 2019 IEEE International Conference on Imaging Systems and Techniques (IST) IEEE; 2019, December. Pollen grain classification using geometrical and textural features; pp. 1–6. [Google Scholar]
- 21.Battiato S., Ortis A., Trenta F., Ascari L., Politi M., Siniscalco C. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops. 2020. Detection and classification of pollen grain microscope images; pp. 980–981. [Google Scholar]
- 22.Grant-Jacob J.A., Praeger M., Eason R.W., Mills B. In-flight sensing of pollen grains via laser scattering and deep learning. Engineering Research Express. 2021;3(2) [Google Scholar]
- 23.Li J., Xu Q., Cheng W., Zhao L., Liu S., Gao Z.…You H. Weakly supervised collaborative learning for airborne pollen segmentation and classification from SEM images. Life. 2023;13(1):247. doi: 10.3390/life13010247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.El Azari H., Renard J.B., Lauthier J., Dudok de Wit T. A laboratory evaluation of the new automated pollen sensor beenose: pollen discrimination using machine learning techniques. Sensors. 2023;23(6):2964. doi: 10.3390/s23062964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Punyasena S.W., Haselhorst D.S., Kong S., Fowlkes C.C., Moreno J.E. Automated identification of diverse Neotropical pollen samples using convolutional neural networks. Methods Ecol. Evol. 2022;13(9):2049–2064. [Google Scholar]
- 26.Menad H., Ben-Naoum F., Amine A. JERI. 2019, April. Deep convolutional neural network for pollen grains classification. [Google Scholar]
- 27.Romero I.C., Kong S., Fowlkes C.C., Jaramillo C., Urban M.A., Oboh-Ikuenobe F.…Punyasena S.W. Improving the taxonomy of fossil pollen using convolutional neural networks and superresolution microscopy. Proc. Natl. Acad. Sci. USA. 2020;117(45):28496–28505. doi: 10.1073/pnas.2007324117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Daunys G., Šukienė L., Vaitkevičius L., Valiulis G., Sofiev M., Šaulienė I. Comparison of computer vision models in application to pollen classification using light scattering. Aerobiologia. 2022:1–13. [Google Scholar]
- 29.Holt Katherine. New Zealand pollen. figshare. Dataset. 2020 doi: 10.6084/m9.figshare.12370307.v1. [DOI] [Google Scholar]
- 30.Battiato S., Ortis A., Trenta F., Ascari L., Politi M., Siniscalco C. 2020 IEEE International Conference on Image Processing (ICIP) IEEE; 2020, October. Pollen13k: a large scale microscope pollen grain image dataset; pp. 2456–2460. [Google Scholar]
- 31.Battiato S., Ortis A., Trenta F., Ascari L., Politi M., Siniscalco C. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops. 2020. Detection and classification of pollen grain microscope images; pp. 980–981. [Google Scholar]
- 32.Mondol C., Shamrat F.J.M., Hasan M.R., Alam S., Ghosh P., Tasnim Z., Ahmed K., Bui F.M., Ibrahim S.M. Early prediction of chronic kidney disease: a comprehensive performance analysis of deep learning models. Algorithms. 2022;15(9):308. [Google Scholar]
- 33.Krizhevsky A., Sutskever I., Hinton G.E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012;25 [Google Scholar]
- 34.Sandler M., Howard A., Zhu M., Zhmoginov A., Chen L.C. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2018. Mobilenetv2: inverted residuals and linear bottlenecks; pp. 4510–4520. [Google Scholar]
- 35.Simonyan K., Zisserman A. Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556. 2014 [Google Scholar]
- 36.He K., Zhang X., Ren S., Sun J. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016. Deep residual learning for image recognition; pp. 770–778. [Google Scholar]
- 37.Sutradhar A., Al Rafi M., Ghosh P., Shamrat F.J.M., Moniruzzaman M., Ahmed K.…Moni M.A. An intelligent thyroid diagnosis system utilising multiple ensemble and explainable algorithms with medical supported attributes. IEEE Transactions on Artificial Intelligence. 2023 [Google Scholar]
- 38.Zhou B., Khosla A., Lapedriza A., Oliva A., Torralba A. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016. Learning deep features for discriminative localization; pp. 2921–2929. [Google Scholar]
- 39.Arrieta A.B., Díaz-Rodríguez N., Del Ser J., Bennetot A., Tabik S., Barbado A.…Herrera F. Explainable Artificial Intelligence (XAI): concepts, taxonomies, opportunities and challenges toward responsible AI. Inf. Fusion. 2020;58:82–115. [Google Scholar]
- 40.Selvaraju R.R., Cogswell M., Das A., Vedantam R., Parikh D., Batra D. Proceedings of the IEEE International Conference on Computer Vision. 2017. Grad-cam: visual explanations from deep networks via gradient-based localization; pp. 618–626. [Google Scholar]
- 41.Simonyan K., Vedaldi A., Zisserman A. Deep inside convolutional networks: visualising image classification models and saliency maps. arXiv preprint arXiv:1312.6034. 2013 [Google Scholar]
- 42.Szeghalmy S., Fazekas A. A comparative study of the use of stratified cross-validation and distribution-balanced stratified cross-validation in imbalanced learning. Sensors. 2023;23(4):2333. doi: 10.3390/s23042333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rastogi D., Johri P., Tiwari V., Elngar A.A. Multi-class classification of brain tumour magnetic resonance images using multi-branch network with inception block and five-fold cross validation deep learning framework. Biomed. Signal Process Control. 2024;88 [Google Scholar]
- 44.Bashivan P., Rish I., Yeasin M., Codella N. Learning representations from EEG with deep recurrent-convolutional neural networks. arXiv preprint arXiv:1511.06448. 2015 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article. It also available in- https://doi.org/10.6084/m9.figshare.12370307.v1.