Abstract
Two types of endogenous ecotropic murine leukemia viruses (MuLVs), termed AKV- and Cas-E-type MuLVs, differ in nucleotide sequence and distribution in wild mouse subspecies. In contrast to AKV-type MuLV, Cas-E-type MuLV is not carried by common laboratory mice. Wild mice of Mus musculus (M. m.) castaneus carry multiple copies of Cas-E-type endogenous MuLV, including the Fv-4r gene that is a truncated form of integrated MuLV and functions as a host's resistance gene against ecotropic MuLV infection. Our genetic cross experiments showed that only the Fv-4r gene was associated with resistance to ecotropic F-MuLV infection. Because the spontaneous expression of infectious virus was not detected in M. m. castaneus, we generated mice that did not carry the Fv-4r gene but did carry a single or a few endogenous MuLV loci. In mice not carrying the Fv-4r gene, infectious MuLVs were isolated in association with three of six Cas-E-type endogenous MuLV loci. The isolated viruses showed a weak syncytium-forming activity for XC cells, an interfering property of ecotropic MuLV, and a slight antigenic variation. Two genomic DNAs containing endogenous Cas-E-type MuLV were cloned and partially sequenced. All of the Cas-E-type endogenous MuLVs were closely related, hybrid-type viruses with an ecotropic env gene and a xenotropic long terminal repeat. Duplications and a deletion were found in a restricted region of the hypervariable proline-rich region of Env glycoprotein.
Multiple DNA copies of the murine leukemia virus (MuLV) genome, called endogenous MuLV, are present in the chromosomal DNA of Mus musculus (M. m.) mice. MuLVs are largely classified into four groups (ecotropic, xenotropic, amphotropic, and polytropic [or dualtropic] viruses) on the bases of the host range and the interfering properties that are mainly determined by the receptor-binding specificity of the viral envelope (Env) glycoprotein. Ecotropic MuLVs infect mouse cells expressing the receptor for ecotropic MuLVs, mCAT-1 (1). There are two types of endogenous ecotropic MuLVs with a slight sequence divergence. AKV-type endogenous MuLV is carried by many laboratory strains of mice and has been well characterized. Castaneus ecotropic (Cas-E-type) MuLVs are not carried by common laboratory mice and therefore have not been characterized in detail. They are carried by geographically separated M. m. subspecies; AKV-type MuLVs were found in M. m. musculus mice populating the northern part of China, Korea, and Japan, and Cas-E-type MuLVs were found in M. m. castaneus mice populating southern Asia (from Pakistan to Japan) (23, 34). They appear to have diverged along with the subspeciation of M. musculus, probably separated by more than 105 to 106 years (23).
Infectious Cas-E-type MuLVs were isolated from mice from limited areas of California in the United States (6, 9, 15; for a review, see reference 8). Interestingly, unlike AKV-type MuLVs, infectious Cas-E-type MuLVs were reported to be transmitted through sex and milk but not in the germ line in this wild mouse population (10, 11). However, it is not clear by these studies whether endogenous Cas-E-type MuLVs can produce infectious viruses and whether the previously isolated infectious Cas-E-type MuLVs are directly related to endogenous MuLVs.
The Fv-4r gene is a truncated Cas-E-type endogenous MuLV (19, 22, 41), which is highly homologous in the env gene to infectious Cas-Br-E MuLV (46) isolated from a Californian wild mouse (15). The Fv-4r gene expresses an Env glycoprotein in various tissues (20, 30) and functions as a host resistance gene against infection by ecotropic MuLVs, probably via a receptor interference mechanism (21, 30, 56). Most M. m. castaneus mice carry both the Fv-4r gene and other Cas-E-type endogenous MuLVs (23, 34), but our attempts to isolate infectious ecotropic MuLVs from M. m. castaneus have failed. This could be accounted for either by the inability of the endogenous MuLVs to produce infectious viruses or by the presence of the dominant resistance gene Fv-4r, even if other endogenous viruses are competent to produce infectious viruses.
To understand the properties of Cas-E-type endogenous MuLVs, we crossed M. m. castaneus mice with laboratory mice free of endogenous ecotropic MuLV and established several mouse lines which did not carry the Fv-4r gene but did carry a single or a few endogenous Cas-E-type MuLVs. We could isolate and characterize infectious MuLVs from these mice. Furthermore, two genomic DNA clones containing endogenous Cas-E-type MuLVs were isolated and then partially sequenced. The nucleotide sequence data indicated that all of the three endogenous Cas-E-type MuLVs we cloned are similar in the env gene but distinct in the long terminal repeat (LTR) from the infectious Cas-E-type Cas-Br-E MuLV previously isolated from Californian wild mice.
MATERIALS AND METHODS
Mice.
Bgr and Ttg mice were originally trapped in Bogor, Indonesia, and Titung, People's Republic of China (K. Moriwaki, personal communication). Based on morphological and anatomical features, these mice were thought to belong to M. m. castaneus (K. Moriwaki and N. Sakaizumi, personal communication). Bgr mice had been bred as a closed colony in Moriwaki's laboratory, National Institute of Genetics, Japan, and given to our laboratory. (WHT × Ttg)F1 mice were developed for other experimental purposes and were a kind gift from N. Sakaizumi, Niigata University, Niigata, Japan. NFS/NJcl (NFS/N) and WHT (54) mice are the laboratory inbred mice susceptible to NB-tropic Friend leukemia virus complex (data not shown). NFS/N mice were purchased from Clea Japan, Inc., Tokyo, Japan, and have no AKV-type or Cas-E-type endogenous MuLV, while WHT mice carried one copy of AKV-type endogenous MuLV and no Cas-E-type endogenous MuLV.
Cells.
Most of the cell lines used in this experiment were grown in Dulbecco's modified Eagle medium (DMEM) with 50 μg of kanamycin (Meiji Seika, Ltd., Tokyo, Japan)/ml and 7% fetal calf serum (FCS) in 5% CO2 at 37°C. SC-1 (14), NIH 3T3, YH7, D8b5 (62), and D3h1g (62) were mouse cells susceptible to ecotropic MuLVs. SC-1 was a clonal line of a Californian wild mouse embryo (14). D8b5 and D3h1g were clonal cell lines derived from embryo of the inbred mouse strain DDD (62). C-182 cells were persistently infected with a defective murine sarcoma virus (MSV) but not a helper MuLV (2). XC cells were used for titration of ecotropic MuLV (50).
Viruses.
Infectious MuLVs were isolated from mice by cocultivation of SC-1 cells with spleen cells which had or had not been cultured for 2 days in RPMI 1640 medium containing 10% FCS, 5 × 10−5 M 2-mercaptoethanol, and 2 μg of concanavalin A (ConA) (Sigma)/ml. Titers of infectious ecotropic MuLVs were measured by UV-XC assay (50). Friend, Moloney, and AKV strains of ecotropic MuLVs were originally provided by A. Ishimoto (Kyoto University, Kyoto, Japan). Amphotropic 4070A, dualtropic AKR13, Cas-Br-M, and Cas-2S-M MuLVs were gifts of J. Hartley (National Institutes of Health, Bethesda, Md.). Cas-Br-M and Cas-2S-M were mouse-tropic (M, equal to ecotropic) viruses isolated from brain or spleen of Lake Casitas wild mice (15). MSV pseudotypes were prepared by infection of C-182 (2) by various MuLVs.
Membrane immunofluorescence.
MuLV antigens expressed on the cell surface were analyzed by flow cytometer (FACStar, Becton Dickinson, or Epics Profile II, Beckman Coulter). Briefly, cultured fibroblast cells were trypsinized to make a cell suspension and were washed three times with DMEM supplemented with 1% FCS and 0.05% NaN3. Cells (1 × 105 to 5 × 105) were incubated on ice for 30 min with biotinylated or nonbiotinylated monoclonal antibodies (MAbs) or antisera in DMEM containing 1% FCS and 0.05% NaN3 and then were washed three times. The cells treated with biotinylated MAbs were then stained with 0.5 μg of streptavidin-coupled phycoerythrin (Streptavidin-PE; PharMingen) per 100 μl on ice for 30 min. The cells treated with nonbiotinylated antisera were stained with fluorescent isothiocyanate-conjugated rabbit anti-mouse immunoglobulin G (IgG) (MBL, Nagoya, Japan) or anti-goat IgG [F(ab′)2] (Cappel Products, Durham, N.C.) on ice for 30 min. The cells were then washed twice and kept in phosphate-buffered saline containing 1% paraformaldehyde until used.
The antibodies used were BALB/c anti-BALB/c-Fv-4r (C4W) alloantiserum (20), goat anti-Rauscher leukemia virus gp70 (provided by the Division of Cancer Cause and Prevention, National Cancer Institute, Bethesda, Md.), MAb4D2 (26), MAb282 (18), and MAb36. These MAbs were derived from hybridoma cells made by fusion of the mouse myeloma cell lines and spleen cells from a BALB/c mouse immunized with C4W spleen and thymus cells. MAb282 and MAb4D2 were biotinylated and MAb36 was not biotinylated. BALB/c anti-C4W, MAb4D2, MAb282, and MAb36 are reactive with Fv-4r MuLV SU (18, 20, 29).
Southern blot analysis.
Chromosomal DNAs were extracted from the liver or tail by the phenol extraction method (52) with modifications. Ten micrograms of DNA digested with restriction enzymes was fractionated by electrophoresis through 0.7% agarose gel, was transferred to NitroPlus 2000 (Micron Separations, Inc., Westborough, Mass.), and was hybridized to a 32P-labeled Fv4renv probe derived from a 690-bp BamHI-BamHI fragment of the Fv-4renv region (19). Hybridization was carried out at 51°C for 18 to 30 h in 50% formamide, 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 50 mM sodium phosphate (pH 6.5), 1× Denhardt's solution, and 0.1% sodium dodecyl sulfate (SDS). Washing was carried out at 51°C for 20 min in 2× SSC and 0.1% SDS, for 30 min in 0.1× SSC and 0.1% SDS, and then for 20 min in 2× SSC. Hybridization and washing were done in rotation in a hybridization oven. Hybridization signals were detected by exposing the membrane to Kodak AR film or the imaging plate of a Bio Imaging Analyzer (Fuji Bas 2000; Fuji PhotoFilm Co., LTD, Tokyo, Japan).
Cloning of endogenous MuLVs.
A genomic DNA library was constructed by ligation of Bgr liver DNA, which was partially digested with Sau3AI and was selected by size (>10 kb), and BamHI-cleaved arms of lambda phage EMBL3 (Stratagene, La Jolla, Calif.). Recombinant phages containing endogenous MuLV were selected by a hybridization signal with the Fv4renv probe (19). Inserts of the recombinant phages were subcloned into the SalI site of pBluescript II SK(+) phagemid (Stratagene).
Nucleotide sequence accession numbers.
The sequences of the endogenous viruses reported in this study have been deposited with DDBJ under accession numbers AB050720 (Frg1) and AB050721 (Frg3).
RESULTS
Genetic analysis of the relationship between Cas-E-type endogenous MuLV loci and the host's resistance to infection by ecotropic Friend leukemia virus.
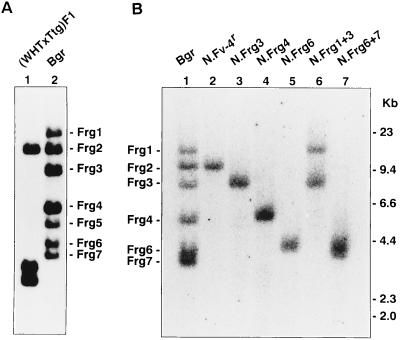
Previous studies demonstrated that M. m. castaneus mice carry multiple copies of Cas-E-type endogenous MuLVs but no AKV-type endogenous MuLV (23, 34). This study examined M. m. castaneus mice trapped in two distinct areas, termed Ttg and Bgr. Southern blot analysis for HindIII-digested DNAs using the Fv4renv probe specifically hybridizing to Cas-E-type MuLVs (19) indicated that a (WHT × Ttg)F1 mouse carried three fragments, and three Bgr mice which were kept in a closed breeding colony carried six or seven fragments (Fig. 1). WHT mice had no Cas-E-type endogenous MuLV, and (WHT × Ttg)F1 mice had only one fragment of AKV-type endogenous MuLV, which was derived from the parental WHT mice (data not shown), indicating that Bgr and WHT mice had no AKV-type but had multiple Cas-E-type endogenous MuLVs. These endogenous virus patterns were typical of M. m. castaneus.
FIG. 1.
Southern blot analysis of Cas-E-type endogenous MuLVs. HindIII-digested mouse DNAs were hybridized with the Fv-4renv probe (19). (A) Lane 1, (WHT × Tgt)F1; lane 2, Bgr. (B) Lane 1, Bgr; lane 2, N.Fv-4r (= Frg2); lane 3, N.Frg3; lane 4, N.Frg4; lane 5, N.Frg6; lane 6, N.Frg1 and -3; lane 7, N.Frg6 and -7. Mice of lanes 2 to 7, which were produced by backcrossing a Bgr mouse to NFS/N mice, carried the indicated endogenous MuLV(s) derived from the Bgr mouse. The Frg5 band was detectable in two Bgr mice (panel A and data not shown) but not in another Bgr mouse (panel B). The Frg5 band did not show a Mendelian inheritance (see text).
The 9.6-kb HindIII fragment was common to these mice (Fig. 1A). This fragment represents the Fv-4r gene because it also hybridized with a probe derived from the cellular flanking sequence 3′ to the Fv-4r MuLV (19) (data not shown). Southern blot analysis of Ttg and Bgr DNAs with several other restriction enzymes and the probes of the env and cellular flanking regions indicated that the restriction enzyme fragment length polymorphism of the Fv-4r gene of these wild mice was the same as that of Fv-4 congenic C4W mice whose Fv-4r alleles were derived from M. m. molossinus (19) (data not shown).
Bgr, (NFS/N × Bgr)F1, and (WHT × Ttg)F1 mice were tested for susceptibility to infection by Friend leukemia virus complex (FLV) that contains both the replication-competent ecotropic virus, F-MuLV, and the replication-defective spleen focus-forming virus, SFFV. These mice were strongly resistant to both FLV-induced splenomegaly and replication of F-MuLV (data not shown). To test the possible genetic association of endogenous Cas-E-type MuLV loci with the resistance to FLV, (NFS/N × Bgr)F1 and (WHT × Ttg)F1 mice were mated with NFS/N laboratory mice which are susceptible to FLV and have neither the Cas-E-type nor the AKV-type endogenous MuLV. To avoid milk-transmitted viral or unknown factors from wild mice as suggested for Californian wild mice (10), male wild-derived mice were mated with female NFS/N mice.
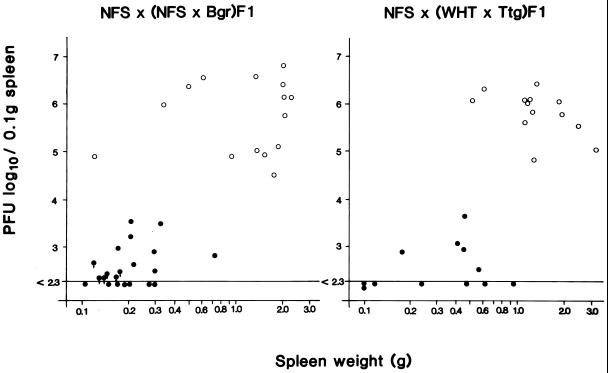
The FLV-induced splenomegaly was segregated in the NFS/N × (NFS/N × Bgr)F1 backcross mice (Fig. 2, left panel). Liver or tail DNAs of each backcross mouse were analyzed for the presence or absence of each endogenous MuLV by Southern blot analysis. The endogenous virus loci of Bgr mice were named Frg1 to Frg7 according to the order of HindIII fragment sizes (Fig. 1). Based on the segregation pattern of the virus loci in the backcross mice, the parental Bgr mouse was considered to carry two copies of each endogenous viral locus (homozygous at each locus), except for Frg5, which did not show a Mendelian inheritance. The reason was unknown. Frg2, which represents the Fv-4r gene as indicated above, was well associated with the suppression of F-MuLV replication (less than 5 × 103 PFU/0.1 g of spleen) but was not completely associated with the suppression of splenomegaly (Fig. 2, left panel). We usually judge a spleen of more than 0.4 g as showing splenomegaly. In 36 backcross mice, there were 3 unexpected mice; one Fv-4rs mouse was resistant to replication of F-MuLV but did develop splenomegaly (0.75 g), and two Fv-4ss mice were permissive to F-MuLV but did not develop splenomegaly (0.1 and 0.3 g).
FIG. 2.
Segregation of splenomegaly and F-MuLV replication in NFS × (NFS × Bgr)F1 (left panel) and NFS × (WHT × Ttg)F1 (right panel) backcross mice after injection of Friend leukemia virus complex. The closed circles and the open circles indicate mice carrying Fv-4rs or not carrying the Fv-4r gene (Fv-4ss), respectively. Ten days after virus injection, the spleen weight of an individual mouse was measured, and the spleen homogenates were used for titration of F-MuLV by the UV-XC test on C-182 cells. Liver or tail DNA of each mouse was used for Southern blot analysis, as shown in Fig. 1. An arrow attached to the circle means that the virus was undetectable and, thereby, was below the indicated titer.
In 25 NFS/N × (WHT × Ttg)F1 backcross mice, the Fv-4r gene was clearly associated with the suppression of F-MuLV replication but less clearly with the suppression of splenomegaly; 7 of 12 Fv-4rs mice developed moderate splenomegaly (0.4 to 0.9 g) (Fig. 2, right panel). The two Cas-E-type endogenous virus loci derived from the parental Ttg mouse (Fig. 1A) were not associated with the moderate splenomegaly (data not shown).
These cross experiments indicated that only the Fv-4r gene but not the other Cas-E-type endogenous viral loci could clearly suppress the replication of F-MuLV in both Bgr and Ttg mice. However, in our general experience with laboratory mice, we did not observe FLV-induced moderate splenomegaly without replication of the helper F-MuLV in either homozygous Fv-4rr or heterozygous Fv-4rs mice. The reason for the unexpected results is not known.
Expression of infectious MuLVs by endogenous Cas-E-type MuLV loci.
We were unsuccessful in isolating infectious virus from M. m. castaneus mice. To test whether the endogenous MuLV loci can produce infectious viruses, we tried to generate mice carrying a single or a few proviral loci by serial backcrossing of Bgr progenies to NFS/N. In the second or third backcross generation, mice carrying a single endogenous virus locus of Frg3, Frg4, or Frg6 were found (Fig. 1B) and were further mated with NFS/N mice. We then attempted to recover infectious viruses from spleen cells of these backcross mice. Spleen cells stimulated or unstimulated with mitogen ConA for 2 days in vitro were further cocultured with MuLV-susceptible SC-1 cells for 2 to 4 weeks. Virus antigen expressed in SC-1 cells was detected by membrane immunofluorescence assay using polyclonal BALB/c anti-C4W serum (20). Viruses were readily detected in mice carrying the Frg3 locus (three of three) and the Frg4 locus (four of five), aged 3 to 5 months (Table 1). Only one of four Frg6-positive mice produced viruses. This virus-producing mouse was 11 months old, while the nonproducing mice were 3 or 4 months old. The low frequency of virus recovery from the Frg6-positive mice might be due to the property of the viral genome that could produce infectious viruses with a low titer or at a low efficiency. Thus, at least three of six provirus loci of Bgr mice appeared to have the potential to produce infectious viruses.
TABLE 1.
Recovery of infectious viruses from hybrid mice of NFS/N × M. m. castaneusa
| Endogenous virus locus (i) | Age (mo) assayed | No. of virus-positive mice/total no. of mice |
|---|---|---|
| Frg3 | 3–4 | 3/3 |
| Frg4 | 3–5 | 4/5 |
| Frg6 | 3–11 | 1/4 |
| Frg1, -3 | 2–4 | 4/4 |
| Frg6, -7 | 3 | 2/3 |
| Frg1, -2, -3, -4, -6, -7 | 2 | 0/1 |
| Frg1, -2, -3, -4, -7 | 11 | 0/2 |
| Frg1, -2, -3, -6, -7 | 2–11 | 0/2 |
| Frg1, -2, -3, -7 | 11 | 0/1 |
Mice carrying the indicated endogenous MuLV locus (i) were tested for the production of infectious MuLVs. Spleen cells stimulated or unstimulated with ConA in vitro were cocultured with SC-1 cells for 2 to 4 weeks. Virus production in the cocultured SC-1 cells was examined by the UV-XC test and flow cytometric analysis by using the BALB/c anti-C4W serum, which reacts with the Fv-4r env protein (20; see Table 2). All of the recovered viruses induced XC syncytia and expressed viral antigens recognized by the BALB/c anti-C4W serum.
We also tested mice carrying the multiple endogenous virus loci derived from Bgr mice (Fig. 1B). Infectious viruses were isolated from all of the four mice carrying Frg1 and Frg3 and from two of three mice carrying Frg6 and Frg7 (Table 1). In contrast, mice carrying Frg2 (= Fv-4r) and an additional three to five proviral loci did not express a detectable amount of the virus (Table 1), suggesting that the Fv-4r gene suppressed the expression or spread of Cas-E-type endogenous MuLVs in mice.
All of the isolated viruses induced XC syncytia, which are a characteristic of most ecotropic MuLVs, but the syncytia were much smaller and less clear than those induced by other ecotropic MuLV strains (see below). The mitogenic stimulation of spleen cells with ConA seemed to have no enhancing effect on the recovery of infectious viruses, because in all of the virus-positive mice, viruses were detected from both stimulated and unstimulated spleen cells (data not shown).
The virus isolated from mice carrying either the Frg3, Frg4, or Frg6 locus was termed Frg3, Frg4, or Frg6 virus, respectively. Antigenic properties of these viruses were examined with virus-infected SC-1 cells by the membrane immunofluorescence assay, using several antibodies. Polyclonal BALB/c anti-C4W serum reacted to SC-1 cells infected by the Frg3, Frg4, Frg6, Cas-Br-M, or Cas-2S-M viruses (Table 2) and to cells transfected with Fv-4r DNA (22) but not to cells infected by any other ecotropic (Friend, Moloney, and AKV strains) (Table 2), amphotropic (4070A strain), and dualtropic (AKR13 strain) MuLVs (data not shown). Thus, all of the Cas-E-type MuLVs tested were recognized by the BALB/c anti-C4W serum. We tested three MAbs which were produced by immunization of BALB/c with C4W cells. MAb4D2 was reactive to SC-1 cells infected with the Frg3, Frg4, or Frg6 viruses, while MAb282 and MAb36 were reactive only to Frg3 virus-infected SC-1 cells (Table 2). These antigenic properties indicated that the three isolated viruses were closely related to Fv-4r MuLV, Cas-Br-M, and Cas-2S-M, all of which show ecotropic interfering properties (15, 22) but also exhibited slight variations in their viral antigens.
TABLE 2.
Reactivities of antibodies to virus-infected SC-1 cells in membrane immunofluorescence assay
| SC-1 cells infected or transfected witha | Mean fluorescent intensityb
|
||||
|---|---|---|---|---|---|
| MAb
|
Polyclonal Ab
|
||||
| 4D2 | 282 | 36 | BALB/c anti-C4W | Goat anti-gp70 | |
| None | 0.9 | 0.7 | 0.5 | 1.0 | 0.7 |
| FL2.3-Fv4r (tf) | 21.3 | 20.4 | 2.4 | 6.5 | 1.1 |
| Friend | 0.7 | 0.7 | 0.4 | 1.4 | 2.6 |
| AKV | 0.8 | 0.7 | 0.5 | 1.5 | 4.2 |
| Moloney | 0.9 | 0.6 | 0.4 | 0.8 | 0.8 |
| Frg3 V | 51.7 | 146.1 | 27.7 | 43.2 | 1.9 |
| Frg4 V | 36.5 | 1.1 | 0.5 | 9.2 | 3.3 |
| Frg6 V | 26.1 | 0.8 | 0.8 | 27.5 | 0.6 |
| Cas-Br-M | 0.7 | 1.5 | 0.5 | 17.5 | 0.8 |
| Cas-2S-M | 0.7 | 0.7 | 0.5 | 14.7 | 0.8 |
SC-1 cells were persistently infected with the indicated viruses or transfected (tf) with FL2.3-Fv4r DNA derived from the Fv-4r gene (37).
Reactivities of the antibodies (Ab) to the cell membrane of infected or transfected cells are indicated as the mean fluorescent intensity of the flow cytometric analysis. MAb4D2 and MAb282 were biotinylated and the other antibodies were not biotinylated. Cells treated with the biotinylated antibodies were then treated with Streptavidin-PE, and the cells treated with the unbiotinylated antibodies were treated with fluorescent isothiocyanate-conjugated rabbit anti-mouse IgG or anti-goat IgG [F(ab′)2].
Host range of the isolated MuLVs.
MuLVs are classified into four classes based on their host-range properties and interference properties that are determined mainly by the receptor binding specificity of the virus. Ecotropic MuLVs infect only mouse and rat cells, xenotropic MuLVs infect nonmouse cells, and polytropic (or dualtropic) and amphotropic MuLVs infect both mouse and nonmouse cells. To examine the host-range properties, we repeatedly tried to obtain high titer virus stocks of each virus isolate. However, none of the isolates achieved a titer of more than 104 PFU/ml when measured on SC-1 cells by the XC plaque assay.
Cultured cells of various non-M. m. origins were examined for susceptibilities to infection by Frg3, Frg4, Frg6, and Cas-Br-M viruses by both the XC plaque assay and the membrane immunofluorescence assay. Neither of the assay systems detected the successful infection of Mus dunni cells, rat NRK cells, rabbit SIRC cells, and mink lung cells by all of the viruses (data not shown).
Limited mouse cells were susceptible to the isolated viruses when examined with the XC plaque assay (Table 3). Various mouse genes control the susceptibility of cells to MuLVs. Fv-1 is one of the well-characterized host genes determining the susceptibility. Fv-1n-type mouse cells carrying two Fv-1n alleles at the Fv-1 locus are permissive to N-tropic MuLVs, and Fv-1b type mouse cells carrying two Fv-1b alleles are permissive to B-tropic MuLVs (47). SC-1 cells of wild mouse origin exceptionally have no Fv-1 restriction, termed Fv-1o type, and are highly susceptible to many ecotropic MuLV strains (14). The Frg3, Frg4, and Frg6 viruses infected SC-1 cells but not NIH 3T3 (Fv-1n-type) or YH7 (Fv-1b-type) cells, while in the same experiment, various MuLVs of the common laboratory strains showed the expected Fv-1 tropism (Table 3).
TABLE 3.
Infectivity of the isolated MuLVs
| MuLVa | Fv-1 tropismb | Infectivity (PFU/ml) on cells:
|
||
|---|---|---|---|---|
| SC-1 (Fv-1o type) | NIH 3T3 (Fv-1n type) | YH7 (FV-1b type) | ||
| Frg3 | 2,100 | <10 | <10 | |
| Frg4 | 650 | <10 | <10 | |
| Frg6 | 6,300 | <10 | <10 | |
| Moloney | NB | 7,000,000 | 85,000 | 290,000 |
| AKV623 | N | 560,000 | 8,300 | 820 |
| Friend | N | 1,500,000 | 78,000 | 1,300 |
| WN1802B | B | 53,000 | <10 | 8,500 |
| Cas-Br-M | N | 550,000 | 42,000 | 1,000 |
| Cas-2S-M | N | 210,000 | 15,000 | 640 |
The cells were infected by 10-fold dilutions of the indicated viruses and were analyzed by the UV-XC assay at 5 days after infection.
N, N-tropic; B, B-tropic; NB, NB-tropic.
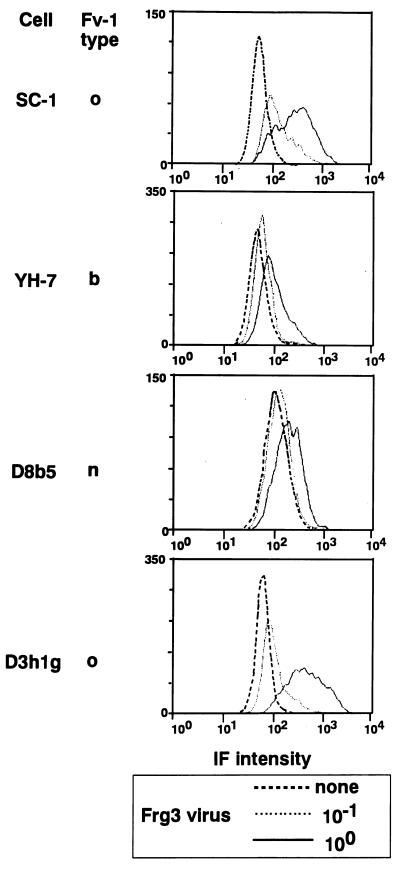
Flow cytometric analysis was employed to detect the infection of four mouse cells by the Frg3 virus. At 5 days after infection, SC-1 cells expressed the virus antigens with a higher proportion and at a higher amount than YH-7 cells (Fig. 3), findings which were compatible with the results obtained with the XC assay (Table 3). D8b5 and D3h1g cell clones were derived from an embryo of the DDD mouse strain (62). DDD mice are of the Fv-1n type, but repeated cell cloning generated cell clones that lost the Fv-1n restriction, termed Fv-1o. D3h1g was one of the Fv-1o-type cells, and D8b5 cells retain the Fv-1n restriction (62). Infection of D3h1g (Fv-1o) cells by the Frg3 virus induced a higher amount of viral antigens than D8b5 (Fv-1n) cells (Fig. 3). Thus, the two Fv-1o-type cells tested were relatively susceptible to infection by the Frg3 virus.
FIG. 3.
Membrane immunofluorescence analysis of cells infected by Frg3 virus. SC-1, YH-7, D8b5, and D3h1g cells were infected by an undiluted or a 10-fold diluted solution of an Frg3 virus stock. Five days after the infection, the cells were treated with BALB/c anti-C4W serum followed by fluorescent isothiocyanate-conjugated anti-mouse IgG. Immunofluorescence (IF) intensities were measured by a flow cytometer, and the results are shown in histograms.
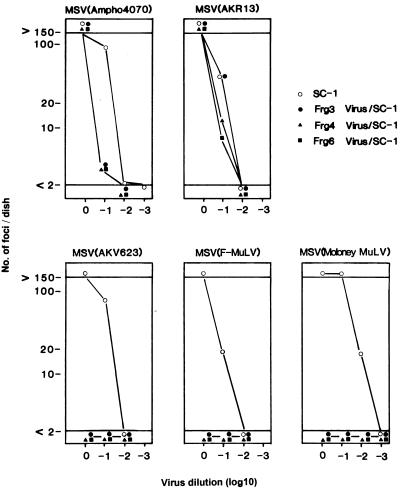
The interference properties of the isolated viruses were tested on SC-1 cells. If these viruses are actually ecotropic MuLVs, then cells infected with the viruses should interfere with superinfection only by ecotropic MuLVs. SC-1 cells were persistently infected with Frg3, Frg4, or Frg6 virus and then were superinfected by MSV pseudotyped with helper ecotropic (AKV623, Friend, and Moloney strains), amphotropic (ampho4070 strain) or dualtropic (AKR13) MuLVs. The infectivities of the pseudotype viruses were scored by MSV-induced foci. Only the MSVs pseudotyped with the three ecotropic MuLV strains were restricted in infecting these persistently infected cells (Fig. 4), suggesting that all of the isolated viruses belong to the ecotropic interference group.
FIG. 4.
Interfering activity of the Frg3, Frg4, and Frg6 viruses toward superinfection. SC-1 cells persistently infected with the viruses were superinfected with MSV pseudotyped with amphotropic (4070A), dualtropic (AKR13), or ecotropic (AKV623, Friend, and Moloney) MuLVs. MSV-induced foci were counted 5 to 7 days after infection.
Nucleotide sequences of two endogenous MuLVs.
We cloned two genomic DNAs containing endogenous Cas-E-type MuLV from a lambda phage library of Bgr mouse DNA. The clones were found to contain the Frg1 or Frg3 endogenous MuLV. The identification of the particular endogenous virus was evidenced by the results that probes derived from the cellular sequences 5′ or 3′ to each endogenous MuLV specifically hybridized to the Frg1 or Frg3 fragment of Bgr DNA (data not shown).
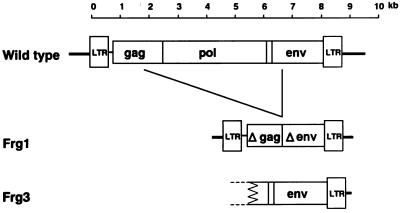
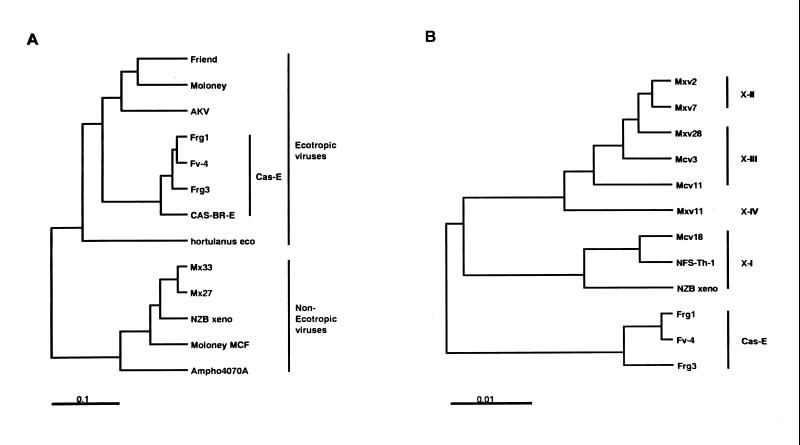
Partial sequencing of the endogenous virus clones indicated that the Frg3 MuLV had a complete env and 3′ LTR, while the Frg1 MuLV had a large internal deletion from the gag to env genes (Fig. 5). Compared to the Frg3 MuLV, the Frg1 MuLV was missing the amino terminal 64 amino acids (aa) of the Env protein. The 5′ end of the Frg1 env region joined to a nucleotide equivalent to the 1,467th nucleotide of the AKV p30 (CA) region of the gag gene (17). The env to 3′ LTR regions of the Frg1 and Frg3 MuLVs were highly homologous and best colinearly aligned to the Fv-4r MuLV when compared with other MuLVs. Another Cas-E-type infectious MuLV, Cas-Br-E, was closely related in the env region to, but distinct in the LTR region from, the three Cas-E-type endogenous MuLVs (see below). The env regions of the four Cas-E-type MuLVs had 88 to 98% nucleotide homologies and 90 to 98% amino acid homologies. Amino acid homologies were lower with other ecotropic MuLVs (AKV, Moloney, and Friend strains) (76 to 79%), polytropic MuLVs (71 to 73%), amphotropic MuLV (69%), and xenotropic MuLVs (64 to 71%). A phylogenetic analysis of various MuLV Env proteins showed that the env genes were largely classified into two groups, the ecotropic MuLV group and the nonecotropic (amphotropic, xenotropic, and polytropic) MuLV group (Fig. 6A). The ecotropic MuLV group had three clusters. The first cluster included the four Cas-E-type MuLVs; the second cluster included the AKV, Friend, and Moloney strains; and the third cluster had only the ecotropic MuLV isolated from Mus hortulanus (59). It should be noted that the AKV-type endogenous MuLVs are carried by about three-fourths of laboratory mouse strains (24), and both the Friend and Moloney strains were isolated from laboratory mice.
FIG. 5.
Structure of Frg1 and Frg3 endogenous MuLVs. Compared to a wild-type endogenous MuLV, Frg1 MuLV had a deletion from gag to env genes and Frg3 had complete env and 3′ LTR regions. The nucleotide sequences have been deposited in DDBJ under accession no. AB050720 (Frg1) and no. AB050721 (Frg3).
FIG. 6.
Phylogenetic analyses of env and LTR U3 genes of Cas-E-type MuLVs. (A) Deduced amino acid sequences of the env genes were analyzed by the unweighted pair-group method with arithmetic means (UPGMA). Sequence sources were as follows: Friend (31), Moloney (53), AKV (17), Fv-4 (19, 41), Cas-Br-E (46), hortulanus eco (59), modified polytropic Mx33 (55), polytropic Mx27 (55), NZB xeno (43), Moloney MCF (4), and amphotropic 4070A (45). (B) The U3 sequences were aligned according to the characteristic sequences (28, 58) (see Fig. 8), and then the phylogenetic analysis was done by the UPGMA method in combination with Kimura's two-parameter method. Sequence sources were as follows: Mxv2 (58), Mxv7 (58), Mxv28 (58), Mcv3 (58), Mcv11 (58), Mxv11 (58), Mcv18 (58), NFS-Th-1 (28), NZB xeno (43), and Fv-4 (19).
When the env regions of the four Cas-E-type MuLVs were compared in more detail, the most prominent differences were seen in two regions: the proline-rich hypervariable region located at the middle portion of the SU domain and the 3′ end of the TM domain (Fig. 7). In the latter region, Cas-Br-E MuLV was different from the other three but was related to amphotropic 4070 MuLV, so that this region and the adjacent 3′ LTR were possibly derived from amphotropic MuLV, a probable recombination counterpart.
FIG. 7.
Alignment of the amino acid sequences of the four Cas-E-type MuLV env genes. Regions I, II, and III represent the disulfide-bonded structural elements conserved in ecotropic MuLVs (38). The hypervariable proline-rich region is indicated. The 8-aa direct repeats found in the proline-rich region of Frg1 and Fv-4r are underlined with a dotted line and a double line, respectively. A 5-aa deletion of Cas-Br-E, compared with Frg3, is boxed. Asterisks or dots shown under the sequences indicate the position in which all of the four MuLVs have the same amino acid or in which three of the four MuLVs have the same amino acid, respectively.
It was previously noted that the Fv-4r env gene has a 24-bp tandem repeat in the proline-rich region (41). The Frg1 MuLV also had a tandem repeat of the same size at the same position as the Fv-4r MuLV (Fig. 7). However, the duplicated Frg1 sequences differed from those of the Fv-4r MuLV by 2 of 24 bp, resulting in one nonsynonymous substitution. The Frg3 env gene appeared to be normal. Compared to the Frg3 env gene, the Cas-Br-E env gene had a 15-bp deletion at a position probably only 3 or 7 bp upstream of where the duplications occurred in the Frg1 and Fv-4r MuLVs.
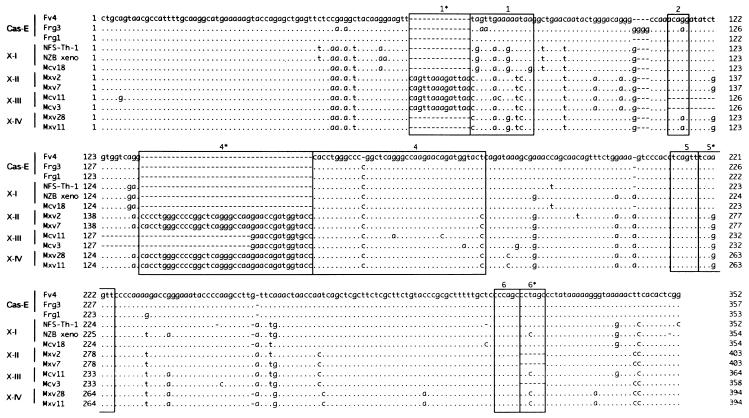
In the LTR region, the Frg1, Frg3, and Fv-4r MuLVs were most colinearly aligned with xenotropic MuLVs, with 92 to 93% nucleotide homologies, while the infectious Cas-Br-E of Cas-E-type MuLV was closely related to amphotropic MuLV (45). The U3 region of LTR is more polymorphic than the R and U5 regions of LTR. A number of sequences of xenotropic U3 were isolated and proposed to classify into four types (X-I to -IV) based on the unique structural features (28, 58). According to their alignment and classification, the three Cas-E U3 sequences shared a few unique characters with the X-I type U3, such as the presence of only one copy of region 1 and region 4 (Fig. 8). The Cas-E U3s also had several single-base changes distinguishable from the X-I type and the other types, as shown in Fig. 8 at nucleotide positions 53, 58, 76, 92, 130, 257, and 343 of the Fv-4r U3. A phylogenetic analysis of the LTR U3 region, estimated after the alignment (58), showed that the Frg1, Frg3, and Fv-4r MuLVs formed a closely related gene cluster which was slightly distinct from the xenotropic U3 types (Fig. 6B).
FIG. 8.
Alignment of LTRU3 sequences of Cas-E-type ecotropic MuLVs and xenotropic MuLVs. Based on the characteristic sequences (1, 1∗, 2, 4, 4∗, 5, 5∗, 6, and 6∗) (28, 58), the U3 sequences of Cas-E-type MuLV (Fv-4, Frg3, and Frg1) were aligned, along with the xenotropic U3 sequences. Dots indicate the nucleotide identity and dashes indicate the absence of a nucleotide. Xenotropic U3 sequences are grouped into X-1 (NFS-Th-1, NZB xeno, and Mcv18), X-II (Mxv2 and Mxv7), X-III (Mcv11 and Mcv3), and X-IV (Mxv28 and Mxv11) (58).
DISCUSSION
Four Cas-E-type MuLVs, Fv-4r, Cas-Br-E, Frg1, and Frg3, have been molecularly cloned and sequenced so far, and two are reported here. All of them were highly homologous in the env region, while the three endogenous viruses, Frg1, Frg3, and Fv-4r, were distinct in the LTR region from the infectious Cas-Br-E virus isolated from Californian wild mice. The LTRs of the three endogenous viruses were closely related to xenotropic MuLV LTRs, while the Cas-Br-E LTR was related to the amphotropic MuLV LTR (46). Xenotropic MuLVs appear older than Cas-E-type MuLVs because the xenotropic env and LTR sequences were present in all M. m. subspecies (34, 57, 58), while the env sequences of Cas-E-type MuLVs were found in only M. m. castaneus subspecies and its derivative M. m. molossinus (23, 34). Therefore, endogenous Cas-E-type MuLVs are likely to be recombinants between an ecotropic env gene of unknown origin and the LTR of the preexisting xenotropic MuLV.
The extensive molecular analyses of nonecotropic endogenous MuLVs showed that wild mice carry many recombinant-type viruses compared to laboratory mice (57, 58). An M. m. castaneus mouse had 26 fragments of the xenotropic env sequences, the majority of which were considered to be associated with the polytropic LTR. The same mouse also carried 35 fragments of xenotropic LTR sequences. Endogenous MuLV genomes with a combination of xenotropic env and xenotropic LTR were also detected by the PCR amplification method, but they seemed to be less frequent (57). It was not known from the studies what env sequence is associated with most of the xenotropic LTRs in M. m. castaneus. Our data suggest that the Cas-E-type endogenous ecotropic MuLV is one of the probable candidates.
Cas-E-type ecotropic MuLVs were first isolated from wild mice from several areas of California in the United States, and most of the virological studies of this type of virus have been performed with these viruses (6, 9, 15). Wild mice from these areas appear to be a mixture of two subspecies; one is M. m. castaneus, which migrated from southern Asia, and the other is M. m. domesticus, which migrated from western Europe (8). About 85% of the mice of the areas were viremic, with a mixture of amphotropic MuLV and Cas-E-type ecotropic MuLV, and this mouse population carried the Akvr-1R gene, identical to the Fv-4r gene, with a gene frequency of 47% (12). In contrast, we were not able to isolate infectious ecotropic MuLVs from M. m. castaneus mice of southern Asia, in which the gene frequency of the Fv-4r gene was more than 85% (23). The present study indicated that some of the endogenous Cas-E-type MuLVs have a potential to express infectious viruses and that the Fv-4r endogenous MuLV suppresses the expression or spread of the infectious viruses from the potent viral loci. Thus, the low gene frequency of the Fv-4r resistance gene in Californian wild mice may have increased the chances for replication-competent proviruses to spread in mice and to generate new recombinant viruses such as Cas-Br-E.
Restriction maps of a number of infectious ecotropic MuLVs isolated from Californian wild mice were reported (6). There were slight variations in their LTRs. Some appear to be like amphotropic LTRs, and the others appear to be like xenotropic LTRs. We suspect that these infectious viruses included both viruses expressed directly from the endogenous viruses and recombinant viruses consisting of the env gene from the endogenous viruses and the LTR gene from amphotropic MuLV, like the Cas-Br-E strain. Amphotropic MuLVs have been isolated only from particular areas of California (15, 48) and are not present as an endogenous virus in any M. m. subspecies (44).
Infectious Cas-E-type MuLVs were successfully isolated from mice of the NFS-Bgr crosses if these mice did not carry the Fv-4r endogenous MuLV but did carry some endogenous MuLVs (Table 1). Three (Frg3, Frg4, and Frg6) of the six endogenous virus loci of Bgr mice were associated with the expression of infectious viruses, implying that the proviruses encoded replication-competent MuLV. Of the rest of the three loci, two were apparently defective in structure; the Fv-4r (= Frg2) gene is a truncated provirus (22) and the Frg1 provirus had a large internal deletion in the gag-pol-env region (Fig. 5). The presence of both defective and nondefective endogenous viruses in Cas-E-type MuLVs resembles the status of AKV-type MuLVs in laboratory mice (24). These are in contrast to the endogenous polytropic MuLVs, all of which are considered to be defective but are recoverable only as recombinant viruses with the polytropic env gene of endogenous virus origin after infection with exogenous viruses (7).
All of the three infectious viruses isolated from the Bgr-NFS crosses showed the ecotropic interference property (Fig. 4), although they exhibited antigenic variations in the Env proteins (Table 2). These viruses produced XC syncytia, which, however, were less clear than those of the other ecotropic MuLVs tested (Table 3). Detailed virological analyses of these viruses were limited because we could not obtain high titer stocks of these viruses. SC-1 cells were relatively susceptible to these viruses and are considered to lack the Fv-1 restriction (14). Another mouse cell line, D3h1g, which phenotypically lost the Fv-1 restriction (62), was likewise slightly susceptible, although its parental type cells with the Fv-1n-type restriction were resistant (Fig. 3). The reason is unknown why these viruses can replicate only in the two cell lines which phenotypically lost the Fv-1 restriction. Cells of wild mouse origin generally showed weaker Fv-1 restriction patterns than those of laboratory mice (32, 33). Thus, further studies are needed to clarify the interaction between host genes and the Cas-E-type MuLVs.
All of the sequenced Cas-E-type MuLVs had the closely related env sequences. However, slight variations were seen in the proline-rich region which is involved in the SU-SU interaction (39), the SU-TM interaction (13, 35), and the viral fusion activity (35). The sequence comparison of this region suggests a possible descent relationship among the four viruses (Fig. 7). The Frg3 provirus seems to be a prototypic virus. A 15-bp deletion from the Frg3 env would generate a Cas-Br-E-like env, and two independent events of 24-bp duplication in the Frg3 env would produce the Frg1- and Fv-4r-like MuLVs. These rearrangements occurred only within three to seven nucleotides which lie in the C-terminal portion of the proline-rich region. The proline-rich region of MuLV consists of about 40 aa and is shown to be tolerant of artificial modifications; deletions of up to 32 aa and insertions of a 252-aa unrelated protein domain did not disrupt the Env function as infectious virus (27, 60). The C-terminal portion of the proline-rich region was more tolerant than the N-terminal portion (27). Actually, the proline-rich region of the Fv-4r env with the 8-aa tandem repeat was competent for viral replication (18, 41). Thus, the Fv-4r and Frg1 MuLVs are examples of the naturally occurring rearrangement in the proline-rich region, and such rearrangements might be present at a high frequency in nature.
The truncated endogenous Cas-E-type MuLV, Fv-4r, confers resistance to exogenous infection by various laboratory strains of ecotropic MuLV (22, 25, 56). This provirus also restricts the in vivo spread of endogenous ecotropic viruses of both the Cas-E type (Table 1) and the AKV type (42). None of the other endogenous MuLVs of Bgr and Ttg mice have such a resistance function against FLV infection (Fig. 2). There are several other examples indicating that endogenous retroviral genomes are involved in the susceptibilities of the host to retroviral diseases. Avian endogenous virus loci, ev-3, ev-6, and ev-9, confer resistance to exogenous infection by subgroup E avian sarcoma/leukosis virus, via an env-mediated resistance mechanism (49). High expressions of endogenous Env proteins are associated with the mouse Rmcf gene (5, 16, 51) and with a gene carried by M. m. castaneus (40), both of which control resistance to dualtropic MuLV infection, although the responsible endogenous viruses have not been identified. In addition, a gag gene product of the endogenous MuERV-L associated with the Fv-1 gene is proposed to interfere with the replication cycle of exogenous MuLVs (3). However, in any of these cases, it is unclear why these particular endogenous viruses exert the resistance functions, despite the presence of multiple related endogenous viruses in the vertebrate genome. For the Fv-4r gene, critical regions for the resistance function, for example, the env gene or its putative promoter region, are not known. Indeed, we do not find any unusual sequence characteristics in the env and LTR of the Fv-4r MuLV compared with those of the other Cas-E-type MuLVs (Fig. 7 and 8).
Unusual FLV-induced splenomegaly was observed in the genetic cross experiments for Bgr and Ttg mice in which the Fv-4r gene was clearly associated with the suppression of F-MuLV replication but not with the strong suppression of splenomegaly (Fig. 2). In general, the FLV-induced splenomegaly needs the replication of F-MuLV helper virus because SFFV, the acute splenomegaly-inducing virus, is defective. SFFV encodes a deleted recombinant Env protein that stimulates the erythropoietin receptor and leads to a constitutive growth signal for the infected erythroid cells (36). One possibility is that, since the FLV complex usually contains polytropic viruses, the splenomegaly would have been induced by SFFV and a polytropic helper virus. Another possibility is that SFFV with a polytropic envelope could have induced the splenomegaly in the absence of an ecotropic helper virus, as previously demonstrated (61). However, these possibilities cannot explain the complete resistance of (NFS/N × Bgr)F1 mice and Bgr mice to the same preparation of FLV complex.
ACKNOWLEDGMENTS
We are grateful to H. Yoshikura and A. Iwamoto of the University of Tokyo for providing D58b5 and D3h1g cells and to K. Moriwaki and N. Miyashita, National Institute of Genetics, for providing Bgr and (WHT × Ttg)F1 mice. We also thank A. Ishimoto, University of Kyoto, and J. W. Hartley, National Institutes of Health, for providing many virus strains.
This study was supported in part by grants from the Science and Technology Agencies of Japan and the Ministry of Agriculture, Forestry and Fisheries of Japan.
REFERENCES
- 1.Albritton L M, Tseng L, Scadden D, Cunningham J M. A putative murine ecotropic retrovirus receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell. 1989;57:659–666. doi: 10.1016/0092-8674(89)90134-7. [DOI] [PubMed] [Google Scholar]
- 2.Bassin R H, Tuttle N, Fischinger P J. Rapid cell culture assay technic for murine leukaemia viruses. Nature. 1971;229:564–566. doi: 10.1038/229564b0. [DOI] [PubMed] [Google Scholar]
- 3.Best S, Le Tissier P, Towers G, Stoye J P. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature. 1996;382:826–829. doi: 10.1038/382826a0. [DOI] [PubMed] [Google Scholar]
- 4.Bosselman R A, van Straaten F, Van Beveren C, Verma I M, Vogt M. Analysis of the env gene of a molecularly cloned and biologically active Moloney mink cell focus-forming proviral DNA. J Virol. 1982;44:19–31. doi: 10.1128/jvi.44.1.19-31.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buller R S, Sitbon M, Portis J L. The endogenous mink cell focus-forming (MCF) gp70 linked to the Rmcf gene restricts MCF virus replication in vivo and provides partial resistance to erythroleukemia induced by Friend murine leukemia virus. J Exp Med. 1988;167:1535–1546. doi: 10.1084/jem.167.5.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chattopadhyay S K, Oliff A I, Linemeyer D L, Lander M R, Lowy D R. Genomes of murine leukemia viruses isolated from wild mice. J Virol. 1981;39:777–791. doi: 10.1128/jvi.39.3.777-791.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cloyd M W, Hartley J W, Rowe W P. Lymphomagenicity of recombinant mink cell focus-inducing murine leukemia viruses. J Exp Med. 1980;151:542–552. doi: 10.1084/jem.151.3.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gardner M B. Retroviruses and wild mice: an historical and personal perspective. Adv Cancer Res. 1994;65:169–201. doi: 10.1016/s0065-230x(08)60068-8. [DOI] [PubMed] [Google Scholar]
- 9.Gardner M B. Type C viruses of wild mice: characterization and natural history of amphotropic, ecotropic, and xenotropic MuLv. Curr Top Microbiol Immunol. 1978;79:215–259. doi: 10.1007/978-3-642-66853-1_5. [DOI] [PubMed] [Google Scholar]
- 10.Gardner M B, Chiri A, Dougherty M F, Casagrande J, Estes J D. Congenital transmission of murine leukemia virus from wild mice prone to the development of lymphoma and paralysis. JNCI. 1979;62:63–70. [PubMed] [Google Scholar]
- 11.Gardner M B, Klement V, Rongey R R, McConahey P, Estes J D, Huebner R J. Type C virus expression in lymphoma-paralysis-prone wild mice. J Natl Cancer Inst. 1976;57:585–590. doi: 10.1093/jnci/57.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gardner M B, Rasheed S, Pal B K, Estes J D, O'Brien S J. Akvr-1, a dominant murine leukemia virus restriction gene, is polymorphic in leukemia-prone wild mice. Proc Natl Acad Sci USA. 1980;77:531–535. doi: 10.1073/pnas.77.1.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gray K D, Roth M J. Mutational analysis of the envelope gene of Moloney murine leukemia virus. J Virol. 1993;67:3489–3496. doi: 10.1128/jvi.67.6.3489-3496.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hartley J W, Rowe W P. Clonal cells lines from a feral mouse embryo which lack host-range restrictions for murine leukemia viruses. Virology. 1975;65:128–134. doi: 10.1016/0042-6822(75)90013-6. [DOI] [PubMed] [Google Scholar]
- 15.Hartley J W, Rowe W P. Naturally occurring murine leukemia viruses in wild mice; characterization of a new “amphotropic” class. J Virol. 1976;19:19–25. doi: 10.1128/jvi.19.1.19-25.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartley J W, Yetter R A, Morse H C. A mouse gene on chromosome 5 that restricts infectivity of mink cell focus-forming recombinant murine leukemia viruses. J Exp Med. 1983;158:16–24. doi: 10.1084/jem.158.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herr W. Nucleotide sequence of AKV murine leukemia virus. J Virol. 1984;49:471–478. doi: 10.1128/jvi.49.2.471-478.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ikeda H, Kato K, Suzuki T, Kitani H, Matsubara Y, Takase-Yoden S, Watanabe R, Kitagawa M, Aizawa S. Properties of the naturally occurring soluble surface glycoprotein of ecotropic murine leukemia virus: binding specificity and possible conformational change after binding to receptor. J Virol. 2000;74:1815–1826. doi: 10.1128/jvi.74.4.1815-1826.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ikeda H, Laigret F, Martin M A, Repaske R. Characterization of a molecularly cloned retroviral sequence associated with Fv-4 resistance. J Virol. 1985;55:768–777. doi: 10.1128/jvi.55.3.768-777.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ikeda H, Odaka T. A cell membrane “gp70” associated with Fv-4 gene: immunological characterization, and tissue and strain distribution. Virology. 1984;133:65–76. doi: 10.1016/0042-6822(84)90426-4. [DOI] [PubMed] [Google Scholar]
- 21.Ikeda H, Odaka T. Cellular expression of murine leukemia virus gp70-related antigen on thymocytes of uninfected mice correlates with Fv-4 gene-controlled resistance to Friend leukemia virus infection. Virology. 1983;128:127–139. doi: 10.1016/0042-6822(83)90324-0. [DOI] [PubMed] [Google Scholar]
- 22.Ikeda H, Sugimura H. Fv-4 resistance gene: a truncated endogenous murine leukemia virus with ecotropic interference properties. J Virol. 1989;63:5405–5412. doi: 10.1128/jvi.63.12.5405-5412.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inaguma Y, Miyashita N, Moriwaki K, Huai W C, Jin M L, He X Q, Ikeda H. Acquisition of two endogenous ecotropic murine leukemia viruses in distinct Asian wild mouse populations. J Virol. 1991;65:1796–1802. doi: 10.1128/jvi.65.4.1796-1802.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jenkins N A, Copeland N G, Taylor B A, Lee B K. Organization, distribution, and stability of endogenous ecotropic murine leukemia virus DNA sequences in chromosomes of Mus musculus. J Virol. 1982;43:26–36. doi: 10.1128/jvi.43.1.26-36.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kai K, Ikeda H, Yuasa Y, Suzuki S, Odaka T. Mouse strain resistant to N-, B-, and NB-tropic murine leukemia viruses. J Virol. 1976;20:436–440. doi: 10.1128/jvi.20.2.436-440.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kai K, Sato H, Odaka T. Relationship between the cellular resistance to Friend murine leukemia virus infection and the expression of murine leukemia virus-gp70-related glycoprotein on cell surface of BALB/c-Fv-4wr mice. Virology. 1986;150:509–512. doi: 10.1016/0042-6822(86)90315-6. [DOI] [PubMed] [Google Scholar]
- 27.Kayman S C, Park H, Saxon M, Pinter A. The hypervariable domain of the murine leukemia virus surface protein tolerates large insertions and deletions, enabling development of a retroviral particle display system. J Virol. 1999;73:1802–1808. doi: 10.1128/jvi.73.3.1802-1808.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khan A S, Martin M A. Endogenous murine leukemia proviral long terminal repeats contain a unique 190-base-pair insert. Proc Natl Acad Sci USA. 1983;80:2699–2703. doi: 10.1073/pnas.80.9.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kitagawa M, Aizawa S, Kamisaku H, Ikeda H, Hirokawa K, Sado T. Cell-free transmission of Fv-4 resistance gene product controlling Friend leukemia virus-induced leukemogenesis: a unique mechanism for interference with viral infection. Blood. 1995;15:1557–1563. [PubMed] [Google Scholar]
- 30.Kitagawa M, Aizawa S, Kamisaku H, Sado T, Ikeda H, Hirokawa K. Distribution of Fv-4 resistant gene product in Friend leukemia virus-resistant Fv-4r mouse strain. Exp Hematol. 1996;24:1423–1431. [PubMed] [Google Scholar]
- 31.Koch W, Hunsmann G, Friedrich R. Nucleotide sequence of the envelope gene of Friend murine leukemia virus. J Virol. 1983;45:1–9. doi: 10.1128/jvi.45.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kozak C A. Analysis of wild-derived mice for Fv-1 and Fv-2 murine leukemia virus restriction loci: a novel wild mouse Fv-1 allele responsible for lack of host range restriction. J Virol. 1985;55:281–285. doi: 10.1128/jvi.55.2.281-285.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kozak C A, Chakraborti A. Single amino acid changes in the murine leukemia virus capsid protein gene define the target of Fv1 resistance. Virology. 1996;225:300–305. doi: 10.1006/viro.1996.0604. [DOI] [PubMed] [Google Scholar]
- 34.Kozak C A, O'Neill R R. Diverse wild mouse origins of xenotropic, mink cell focus-forming, and two types of ecotropic proviral genes. J Virol. 1987;61:3082–3088. doi: 10.1128/jvi.61.10.3082-3088.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lavillette D, Maurice M, Roche C, Russell S J, Sitbon M, Cosset F L. A proline-rich motif downstream of the receptor binding domain modulates conformation and fusogenicity of murine retroviral envelopes. J Virol. 1998;72:9955–9965. doi: 10.1128/jvi.72.12.9955-9965.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li J P, D'Andrea A D, Lodish H F, Baltimore D. Activation of cell growth by binding of Friend spleen focus-forming virus gp55 glycoprotein to the erythropoietin receptor. Nature. 1990;343:762–764. doi: 10.1038/343762a0. [DOI] [PubMed] [Google Scholar]
- 37.Limjoco T I, Dickie P, Ikeda H, Silver J. Transgenic Fv-4 mice resistant to Friend virus. J Virol. 1993;67:4163–4168. doi: 10.1128/jvi.67.7.4163-4168.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Linder M, Linder D, Hahnen J, Schott H H, Stirm S. Localization of the intrachain disulfide bonds of the envelope glycoprotein 71 from Friend murine leukemia virus. Eur J Biochem. 1992;203:65–73. doi: 10.1111/j.1432-1033.1992.tb19828.x. [DOI] [PubMed] [Google Scholar]
- 39.Linder M, Wenzel V, Linder D, Stirm S. Structural elements in glycoprotein 70 from polytropic Friend mink cell focus-inducing virus and glycoprotein 71 from ecotropic Friend murine leukemia virus, as defined by disulfide-bonding pattern and limited proteolysis. J Virol. 1994;68:5133–5141. doi: 10.1128/jvi.68.8.5133-5141.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lyu M S, Nihrane A, Kozak C A. Receptor-mediated interference mechanism responsible for resistance to polytropic leukemia viruses in Mus castaneus. J Virol. 1999;73:3733–3736. doi: 10.1128/jvi.73.5.3733-3736.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Masuda M, Yoshikura H. Construction and characterization of the recombinant Moloney murine leukemia viruses bearing the mouse Fv-4 env gene. J Virol. 1990;64:1033–1043. doi: 10.1128/jvi.64.3.1033-1043.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Odaka T, Ikeda H, Akatsuka T. Restricted expression of endogenous N-tropic XC-positive leukemia virus in hybrids between G and AKR mice: an effect of the Fv-4r gene. Int J Cancer. 1980;25:757–762. doi: 10.1002/ijc.2910250611. [DOI] [PubMed] [Google Scholar]
- 43.O'Neill R R, Buckler C E, Theodore T S, Martin M A, Repaske R. Envelope and long terminal repeat sequences of a cloned infectious NZB xenotropic murine leukemia virus. J Virol. 1985;53:100–106. doi: 10.1128/jvi.53.1.100-106.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O'Neill R R, Hartley J W, Repaske R, Kozak C A. Amphotropic proviral envelope sequences are absent from the Mus germ line. J Virol. 1987;61:2225–2231. doi: 10.1128/jvi.61.7.2225-2231.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ott D, Friedrich R, Rein A. Sequence analysis of amphotropic and 10A1 murine leukemia viruses: close relationship to mink cell focus-inducing viruses. J Virol. 1990;64:757–766. doi: 10.1128/jvi.64.2.757-766.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perryman S M, McAtee F J, Portis J L. Complete nucleotide sequence of the neurotropic murine retrovirus CAS-BR-E. Nucleic Acids Res. 1991;19:1707. doi: 10.1093/nar/19.7.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pincus T, Rowe W P, Lilly F. A major genetic locus affecting resistance to infection with murine leukemia viruses. II. Apparent identity to a major locus described for resistance to friend murine leukemia virus. J Exp Med. 1971;133:1234–1241. doi: 10.1084/jem.133.6.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rasheed S, Gardner M B, Chan E. Amphotropic host range of naturally occurring wild mouse leukemia viruses. J Virol. 1976;19:13–18. doi: 10.1128/jvi.19.1.13-18.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robinson H L, Astrin S M, Senior A M, Salazar F H. Host susceptibility to endogenous viruses: defective, glycoprotein-expressing proviruses interfere with infections. J Virol. 1981;40:745–751. doi: 10.1128/jvi.40.3.745-751.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rowe W P, Pugh W E, Hartley J W. Plaque assay techniques for murine leukemia viruses. Virology. 1970;42:1136–1139. doi: 10.1016/0042-6822(70)90362-4. [DOI] [PubMed] [Google Scholar]
- 51.Ruscetti S, Matthai R, Potter M. Susceptibility of BALB/c mice carrying various DBA/2 genes to development of Friend murine leukemia virus-induced erythroleukemia. J Exp Med. 1985;162:1579–1587. doi: 10.1084/jem.162.5.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 53.Shinnick T M, Lerner R A, Sutcliffe J G. Nucleotide sequence of Moloney murine leukaemia virus. Nature. 1981;293:543–548. doi: 10.1038/293543a0. [DOI] [PubMed] [Google Scholar]
- 54.Staats J. Standardized nomenclature for inbred strains of mice: eighth listing. Cancer Res. 1985;45:945–977. [PubMed] [Google Scholar]
- 55.Stoye J P, Coffin J M. The four classes of endogenous murine leukemia virus: structural relationships and potential for recombination. J Virol. 1987;61:2659–2669. doi: 10.1128/jvi.61.9.2659-2669.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Suzuki S. FV-4: a new gene affecting the splenomegaly induction by Friend leukemia virus. Jpn J Exp Med. 1975;45:473–478. [PubMed] [Google Scholar]
- 57.Tomonaga K, Coffin J M. Structure and distribution of endogenous nonecotropic murine leukemia viruses in wild mice. J Virol. 1998;72:8289–8300. doi: 10.1128/jvi.72.10.8289-8300.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tomonaga K, Coffin J M. Structures of endogenous nonecotropic murine leukemia virus (MLV) long terminal repeats in wild mice: implication for evolution of MLVs. J Virol. 1999;73:4327–4340. doi: 10.1128/jvi.73.5.4327-4340.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Voytek P, Kozak C A. Nucleotide sequence and mode of transmission of the wild mouse ecotropic virus, HoMuLV. Virology. 1989;173:58–67. doi: 10.1016/0042-6822(89)90221-3. [DOI] [PubMed] [Google Scholar]
- 60.Weimin Wu B, Cannon P M, Gordon E M, Hall F L, Anderson W F. Characterization of the proline-rich region of murine leukemia virus envelope protein. J Virol. 1998;72:5383–5391. doi: 10.1128/jvi.72.7.5383-5391.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wolff L, Ruscetti S. Malignant transformation of erythroid cells in vivo by introduction of a nonreplicating retrovirus vector. Science. 1985;228:1549–1552. doi: 10.1126/science.2990034. [DOI] [PubMed] [Google Scholar]
- 62.Yoshikura H, Tejima S, Kuchino T, Segawa K, Odaka T. Characterization of N-type and dually permissive cells segregated from mouse fibroblasts whose Fv-1 phenotype could be modified by another independently segregating gene(s) J Virol. 1982;41:145–152. doi: 10.1128/jvi.41.1.145-152.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]