Abstract
The pursuit of translational biomarkers is complex due to the heterogeneous human pathophysiology, but critical for disease diagnosis, prognosis, monitoring therapeutic efficacy, and for patient stratification. In HIV-associated neurocognitive impairment (NCI), biomarkers that delineate the trajectory of neuropathogenesis and neurocognitive sequelae are critical, particularly considering confounders such as substance use, including Methamphetamine (METH). METH use is a significant health concern among persons living with HIV (PWH), aggravating cognitive deficits and neuroinflammation despite of antiretrovirals, introducing elements in the microenvironment that are fundamentally differerent in relation to non-METH users, such as high levels of dopamine (DA) affecting HIV-innate immune targets. Yet, current biomarkers do not detect these differences. We hypothesized that predefined DA-induced signatures detectable in peripheral blood leukocytes, can distinguish HIV+ METH users compared to HIV-negative or PWH that are non METH users. The elevated expression of CD8A, CREBBP, CCL5, and combinations of dopaminergic pathway transcripts clustered METH users with detectable CSF viral load and major depressive disorder (MDD), indicating neuroimmune-mechanistic links. Cathecol-o-methyltransferase (COMT) gene polymorphisms affecting DA metabolism improved the identification of PWH using METH with biomarkers. The results indicate that underlying immunedopaminergic mechanisms provide signatures and genotypes that can identify PWH that are METH users and their attributes.
Keywords: Methamphetamine, Dopamine, Biomarkers, HIV, Cognition
1. Introduction
The pursuit of translational biomarkers is complex, arising from the heterogeneity inherent in human pathophysiology. Biomarkers are indispensable tools for disease diagnosis and prognostication and harbingers of therapeutic efficacy. Furthermore, they facilitate patient stratification in clinical research and enable the monitoring of treatment response. A critical yet underexplored domain within biomarker research is the elucidation of neuropathogenesis, particularly in the context of HIV-associated neurocognitive impairment (NCI). The identification of biomarkers that can delineate the trajectory of HIV pathogenesis and its neurocognitive sequelae is paramount, especially when considering the confounding influence of comorbid substance use disorders.
Among the substances commonly misused, methamphetamine (Meth) is notably prevalent within cohorts of people with HIV (PWH) compared to people without HIV (PWoH) (Atkinson et al., 2009; Halkitis et al., 2001; Semple et al., 2004), acting as a catalyst for risky HIV transmission behaviors and exacerbating neurocognitive impairment. Meth exerts its influence on the central nervous system by dysregulating dopaminergic neurotransmission, a pathway already compromised by HIV and its proteins, as shown in human specimens and in various models of neuroHIV (Baek et al., 2020; Kesby et al., 2017, 2019; Basova et al., 2021; Fitting et al., 2006; Moran et al., 2012, 2013; Nath et al., 2000). This dysregulation provides a plausible link to the elevated addiction rates observed among PWH. Meth and other stimulants, act on the reward system by elevating the neurotransmitter dopamine (DA), produced by dopaminergic neurons in the brain. On the other hand, HIV and its proteins, suppress components of the dopaminergic system (Baek et al., 2020; Kesby et al., 2017, 2019; Zauli et al., 2000), in part explaining the high prevalence of addiction in this population.
The dysregulation in the dopaminergic system affects directly immune response components, due to the expression of DA receptors on the surface of immune cells, particularly in cells that are HIV targets and contributors to the inflammatory process in the brain and elsewhere (Basova et al., 2018; Calderon et al., 2017; Coley et al., 2015; Gaskill et al., 2009, 2012), and resulting in important differences in inflammatory markers (Basova et al., 2023a). The brain microenvironment of PWH that are Meth users is fundamentally differerent in relation to non-METH users, including due to high levels of dopamine (DA) affecting HIV-innate immune targets. Immune cells in the periphery may express signs of response to this hyperdopaminergic environment. Yet, current biomarkers do not detect these differences. DA-associated immune signatures may be important tools to identify inflammatory processes due to substance use, but also to fully capture the impact of comorbidities in the population of PWH. DA-dependent inflammatory signatures may be critically related to uncontrolled viral load in substance users, particularly in populations at risk of psychosocial stress (Li et al., 2022; Cherenack et al., 2023). A recent review of 28 study cohorts has indicated that in 23 of them stimulant substance use is independently associated with detectable viral loads despite HIV treatments (Ross et al., 2023). We have recently discussed the importance of syndemic substance use and other psychosocial and mental health stressors to address the challenges of achieving viral suppression (Grelotti DJM et al., 2024). Interestingly, an evidence-based behavioral intervention trial in PWH that are Meth users has been shown to be effective at decreasing the drug use, but also at modifying pathways and epigenetic signatures activated by DA in leukocytes (Carrico et al., 2024). These observations further highlight the importance of the neuro-immune-dopaminergic cross-communication in the context HIV infection, and further suggest that DA signatures in leukocytes are important tools to assess immune processes in Meth users.
Building on our previous work, which identified distinct transcriptional signatures with biomarker potential in HIV-infected monocytic cell lines—such as S100A8 and A9, components of the pro-inflammatory MRP8/14 complex (Basova et al., 2023b)—this study ventures into a broader analysis. We employ a systems biology framework to interrogate an extensive gene expression dataset from leukocytes of 202 individuals, encompassing both PWH and uninfected controls, with and without a history of Meth use, dependence and abuse. Our objective is to refine the specificity of biomarkers for HIV-associated NCI by discerning the transcriptional changes uniquely attributable to Meth use within the HIV-positive population.
By leveraging gene network analysis, we aim to dissect the composite immune and dopaminergic signaling alterations induced by Meth in the milieu of HIV infection. This methodology promises to transcend the limitations of single-gene assessments, which are often subject to confounding variables, and instead provide a holistic view of the perturbed biological networks. The insights gleaned from this analysis are anticipated to not only define biomarkers and refine diagnostic criteria for HIV-associated NCI but also to pinpoint novel therapeutic targets for intervention in the co-morbid context of Meth abuse and HIV.
We have identified individual transcriptional dopaminergic signatures in HIV-infected monocytic cell lines with biomarker value in humans, such as S100A8 and A9, which encode the pro-inflammatory complex MRP8/14, and can identify Meth users with detectable virus in the CSF (Basova et al., 2023a). We analyzed a comprehensive array of gene expression patterns that integrate immune and dopaminergic signaling using a systems biology methodology in total leukocytes from blood specimens derived from PWH and people without HIV (PWoH), chronic Meth users (i.e., meeting DSM-IV diagnostic criteria for lifetime methamphetamine dependence + METH abuse or dependence in the last 18 months) (METH+) or not (METH-) to search for additional biomarkers of HIV-associated NCI tailored to distinguish processes occurring in the brain of Meth users. The approach involving gene network analysis, increased the power of discovery, by expanding the focus on disease, biological processes, and pathways affected by Meth in the context of HIV, narrowed to consequences to viral host interactions, regulation of HIV replication, inflammation in the brain and effects on cognitive function, offering mechanistic insights and biomarker opportunities tailored to identify NCI in the context of substance use and its complexity. The approach is a development from single genes susceptible to confounders, towards the identification of sets of genes involved in processes of interest in the HIV-1 pathogenesis, discriminating the damaging effects of Meth.
2. Material and methods
Human cohorts, specimens and study design – Specimens were peripheral blood leukocytes isolated from participants that included 202 adults enrolled by NIH-funded studies at the University of California San Diego’s HIV Neurobehavioral Research Program (HNRP) and Translational Methamphetamine Research Center (TMARC) under informed consent and approved protocols. The subset of PWH and PWoH selected for this study were by design males, between 35 and 44 years old, due to cohort characteristics and to increase statistical power. The participants were divided based on HIV serostatus (HIV+/−) and Meth use (METH+/−). METH+ was defined as meeting lifetime DSM-IV criteria for methamphetamine use or dependence, and METH dependence or abuse within 18 months (LT Methamphetamine Dx), with 8.2% urine toxicology positive/current METH users. A cross-sectional design assembled the following groups: HIV-METH- (n = 47), HIV+METH- (n = 62), HIV-METH+ (n = 39), and HIV+METH+ (n = 54). Exclusion criteria were a history of non-HIV-related neurological, medical, or psychiatric disorders that affect brain function (e.g., schizophrenia, traumatic brain injury, epilepsy), learning disabilities, or dementia. Inclusion of major depressive disorder (MDD) and polysubstance use occurred in the cohort due to the high prevalence among Meth users and accounted for in the analysis models. Life time (LT) and current MDD and substance use diagnosis were established using computer-assisted Composite International Diagnostic Interview (CIDI, version 2.1. World Health Organization), with Lifetime MDD according to DSM-IV (Cattie et al., 2012; Kessler and Ustun, 2004). LT and current polysubstance use was further examined as described (Saloner et al., 2019) and by urine toxicology (alcohol, cannabis, cocaine, hallucinogen, inhalant, methamphetamine, opioids, PCP, sedatives or any other) respectively, performed as standard of care by the HNRP and TMARC at the time of blood collection, with positivity for one or more substances in addition to Meth. Supplementary Table 1 shows characteristics such as age, health and virological parameters, and psychiatric scores. The cohort was 61.9% White, 21.8% Hispanic, 12.9% Black, 2.0% Asian and 1.5% other ethnicity. At visits, neurocognitive scores were collected using a comprehensive neurocognitive test battery designed to optimize sensitivity to HIV and METH-associated deficits across seven cognitive domains (verbal fluency, speed of information processing, executive functions, learning, memory, motor) (Carey et al., 2004). Raw scores from individual tests were converted to demographically adjusted T-scores which were then converted to domain and global T-scores. Individual test T-scores were also converted to deficit scores, ranging from 0 (T-score>39, no impairment) to 5 (T-score<20, severe impairment) and used to derive domain deficit scores (DDS) and a global deficit score (GDS), the latter which reflects the number and severity of NC deficits across the test battery, with a cutoff of GDS>0.5 used to classify NCI (Norman et al., 2011).
All participants also completed a modified version of the Lawton & Brody Activities of Daily Living (ADL) Scale, a self-report questionnaire assessing their current level of everyday functioning. The ADL Scale includes 13 items detailing their level of independent functioning across various domains of functioning (e.g., financial and medication management (Lawton and Brody, 1969). Participants are asked to rate their best and current level of independence across each domain. The total score is the total number of activities for which there is a need for increased assistance, with a range of 0 (no need for increased assistance) to 13 (increased dependence in all activities). Participants were classified as dependent in their ADLs if they reported increased dependence on two or more areas of functioning (Heaton et al., 2004).
Peripheral blood was collected to isolate leukocytes and plasma, which were stored in liquid nitrogen and −80 °C, respectively, until the assays were used. RT-PCR measured CSF and plasma viral loads in CLIA-certified laboratory. CD4 and CD8 T-lymphocyte counts were measured by flow cytometry for PWH. Nadir CD4 levels were taken from medical records and study-obtained values.
Peripheral Leukocytes specimens – For assays, archived specimens were thawed in fetal bovine serum, washed and viability was counted using disposable hemocytometers (Bulldog Bio, Portsmouth, NH). Cell numbers were adjusted for flow cytometry and for digital transcriptome.
Flow Cytometry - Washed cells with adjusted concentrations were resuspended in HBSS without phenol red, containing 2% fetal bovine serum and 0.2% sodium azide, and stained with pre-defined concentrations of subset and activation-specific antibodies. The antibodies used in staining were: CD4 (clone RPA-T4), CD8 (clone SK1), CD11b (clone M1/70), CD14 (clone M5E2), and CD16 (clone B73.1), all from BioLegend (San Diego, CA). We also used an antibody against MRP8/14 (clone Mac387, BioRad, Herculer, CA). Following incubation, cells were washed and resuspended in 4% Paraformaldehyde. Tubes were kept at 4 °C and protected from light until acquisition using a CytoFlex S benchtop platform (Beckman Coulter, Indianapolis, IN), and then analyzed using FlowJo software (FlowJo LLC, Ashland, OR).
RNA extraction – The RNA purification from pellets was performed using Nucleospin RNA Mini kit with DNA separation column (Macherey-Nagel, Bethlehem, PA). RNA quality and levels were monitored in a NanoDrop spectrophotometer ND-2000 (Thermo Scientific, Waltham, MA), and used to adjust concentrations for subsequent assays.
Digital Multiplex Gene Expression– The nCounter gene expression assays (NanoString Technologies, Seattle, WA) were performed using a custom-made combination of Neuropathology and Neuroinflammation NanoString panels. Briefly, panel code-set probes were hybridized with 150 ng of total RNA per specimen or total cell pellets over 18hr at 65 °C in a SimplyAmp Thermocycler (Applied Biosystems, Waltham, MA). Hybridized RNA was diluted in water and loaded into nCounter SPRINT cartridge (NanoString) in nCounter SPRINT Profiler, and then RNA-conjugated probes were counted via NanoString Sprint Profiler technology, following manufacturer’s protocols. Results from each panel were merged into a data file. Normalization was performed for each sample by dividing each sample’s raw count profiles by the geometric mean of 8 reference genes in nSolver, as previously described (Danaher et al., 2017). Genes that did not show signal in any specimen were excluded from subsequent systems analysis.
Statistical analysis – Gene expression scores were calculated as the log2 normalized expression of each transcript. Variance due to noise for each score was estimated in a linear mixed model. Ward’s linkage analysis was used to define hierarchical gene clusters to isolate biomarkers, followed by Analysis of Variance. Fitted value models were used to determine variable and intercept effects. Pairwise comparisons and false discovery rate (FDR) adjustments were calculated for each group. Multiple comparisons were performed using pairwise Tukey HSD, with Tukey-Kramer adjustment, or Bonferroni. Predictor screening was used to determine specific effects of individual variables, named HIV and METH, and their interaction. Mixed models were used to identify interactive effects of comorbidities and clinical parameters on gene expression, as well as genotype. All statistical analysis was performed in JMP Pro 15.2.0 software (SAS Institute Inc., Cary, NC, USA) and Prism 9.0 (GraphPad, Boston, MA).
Systems analysis and Visualization tools – Gene interactions were identified in Cytoscape 3.9.0 platform (www.cytoscape.org, 2020) (Shannon et al., 2003) using local search features in GeneMANIA plugin (Franz et al., 2018; Montojo et al., 2010, 2014a, 2014b; Warde-Farley et al., 2010) (www.genemania.org), with Homo sapiens sources from Reactome (Haw et al., 2011; Stein, 2004) (www.reactome.org, 2020) and BioGRID_ORGANISM (Oughtred et al., 2021; Stark et al., 2006) (https://thebiogrid.org, 2020), for identification of genes associated by pathway, protein interactions and genetic interactions, for visualization of broad patterns linked to group assignment. The analysis of processes and pathway annotations was then performed in DAVID Bioinformatics Resources 6.8 (https://david.ncifcrf.gov, 2020). Genes listed in significantly increased and decreased pathways were then placed back into Cytoscape 3.9.0 for visualization of narrow clusters of interest using GeneMANIA. Transcription factor regulation was predicted using ChEA3 (Keenan et al., 2019) using a 54 input gene set most likely upregulated in the HIV + METH + group as a whole, compared to HIV-METH+. The top 10 transcription factors ranked based on the highest number of overlapping targets (Supplementary Table 3) were visualized using iRegulon in Cytoscape 3.9.0 with merged predicted regulated targets, using only genes upregulated in PBLs from HIV + METH + subjects exhibiting COMT Met/Met genotype and not in Val/Met or Val/Val as input. (Basova et al., 2021; Tjitro et al., 2018).
3. Results
Peripheral leukocyte transcripts as markers of HIV, Meth, and their interaction with NCI were tested using a targeted digital multiplex custom analysis. The construction of a panel of 784 transcripts was guided by the knowledge of chronic inflammation and viral persistency linked to NCI in PWH, confounders (Mediouni et al., 2015) introduced by Meth due to the activation of both overlapping as well as non-redundant pathways leading to inflammation and potentially affecting the response to the virus in the brain (Basova et al., 2020, 2022). In addition, the construction of a biomarker panel was also guided by reports that both HIV and Meth, independently and together, lead to disorders of the dopaminergic system (Baek et al., 2020; Basova et al., 2018, 2023a; Calderon et al., 2017; Gaskill et al., 2013).
The specimens were selected from individuals assigned as males at birth, with similar age, education, and race/ethnicity distribution across groups (Supplementary Table 1), in order to decrease the number of variables and confounders, and increase power. The HIV + groups (HIV+/METH- and HIV+/METH+) did not differ in CD4 nadir, or in plasma or CSF viral load (Supplementary Table 1).
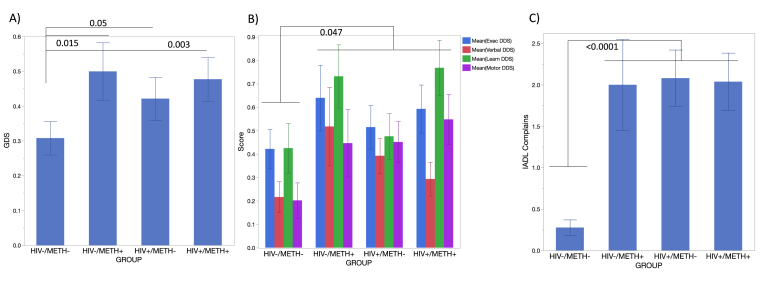
As expected, HIV + groups exhibited significantly lower values of current CD4+ T cells, and higher CD8+ T cells (Supplementary Figure 1A upper and lower panels, respectively). The percentage of CD11b + monocytes was significantly decreased in the blood of HIV+/METH + group compared to HIV-/METH- controls, but no differences were observed between other groups (Supplementary Figure 1B, upper panel). However, the percentage of activated CD11b + monocytes, expressing CD14 and CD16high, was significantly increased in HIV-/METH + subjects (Supplementary Figure 1B, middle panel), suggesting a higher availability of circulating pro-inflammatory monocytes linked to Meth use. Interestingly, in HIV+/METH- and in HIV+/METH + subjects, the percentage of this pro-inflammatory subset in the blood was as low as in negative controls. This could be due to higher sequestration into tissues. Importantly, both HIV and Meth, and the interaction between Meth use and HIV, were associated with worse global neurocognitive deficits, determined by Global deficit scores (GDS), and with domain-specific scores for Executive, Verbal, Learning, and Motor Mean domain deficit scores (DDS), and number of self-reported complaints associated with Instrumental Activities of Daily Living (IADL), as indicated in Fig. 1. The reasons for such effects, however, may differ between HIV and Meth use, making it critical to examine other additional molecular markers with relevance to neuroinflammation in the context of substance use on peripheral leukocytes. Signatures of the response of leukocytes to neurotransmitters that Meth enhances, may offer biomarkers that can distinguish the mechanisms leading to deficits in these groups.
Fig. 1.
Cognitive Assessments. A) Global Deficit Score (GDS), B) Mean Executive, Verbal, Learning, and Motor function domain deficit scores (DDS), C) Instrumental Activities of Daily Living (IADL) Complaints. P values in the figure for assigned comparisons. P values from adjusted all pairwise Tukey HSD multiple comparisons as indicated.
Our previous studies on PWH have indicated that disturbances in gene networks assigned to biological processes able to identify comorbidities and stratify individuals that are users of cannabis (Basova LAL et al., 2022), using analytical strategies that were similar to what was used here to sort effects of Meth chronic use detectable in PWH individuals with cognitive deficits. For that, the cohort was divided in groups based on HIV infection (H, + or -), and Meth use (M, + or -), as in H-M-, H + M-, H-M+, H + M+. The specimens were peripheral blood leukocytes (PBL) from PWH and controls. Co-morbidities and polysubstance use were additional factors considered in the analysis of DA signatures in human PBLs, as previously described (Basova LAL et al., 2022; Rogers et al., 2023).
All genes were normalized to 8 housekeepers prior to input. Using Gene Network Analysis (GNA) strategies, we found genes and biological processes in HIV + Meth users that matched DA signatures previously identified in vitro. The analysis of transcripts positively or negatively affected by HIV, Meth or their interactions in comparison to uninfected non-Meth users, indicated that functional annotations of interest to the cohort were significantly affected, including Neuroactive factors and receptors and dopaminergic response (p = 4.2 E−36), amphetamine addiction (p = 2.4 E −29) and Host-virus interactions (p = 1.2 E −11), as shown in Supplementary Table 2. To identify biomarker candidates, we used two strategies to segregate HIV in the context of Meth: 1) Hierarchical clustering, patterns and predictor screening on transcriptional values normalized by mean expression levels, and B) Pairwise comparisons of internally normalized values with a focus on upregulated transcripts. We sought to identify candidates with consistently significance and overlapping hits in both approaches. Additional candidates were further analyzed to consider other comorbidities and genotypes. Visualizations were facilitated by Cytoscape 3.9.0.
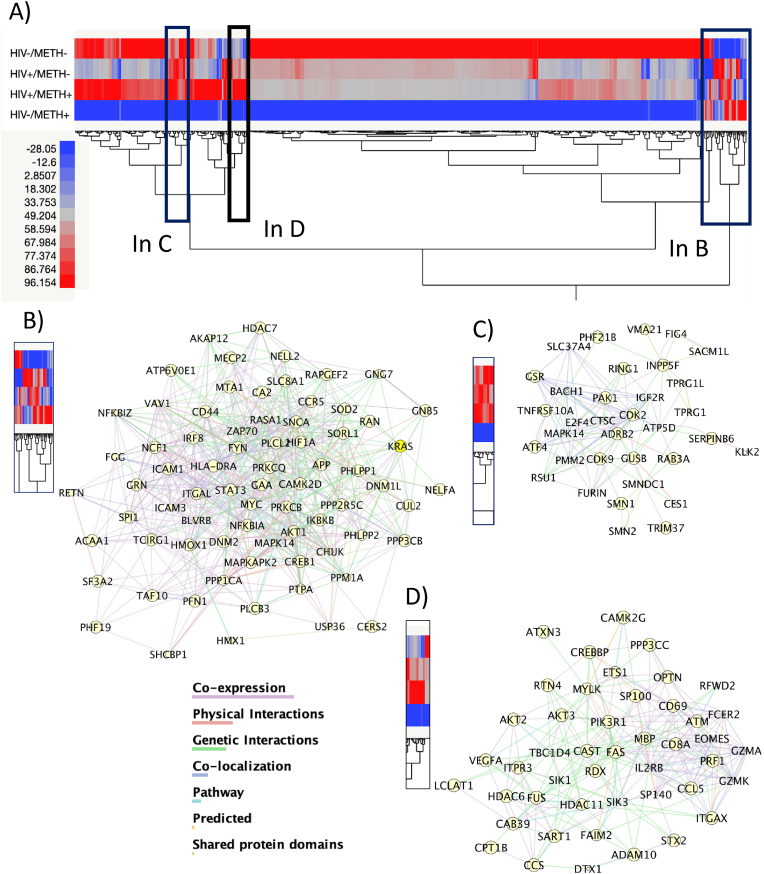
3.1. Hierarchical clustering (Fig. 2)
Fig. 2.
Hierarchical clustering and identification of biomarker patterns-associated interactions. Transcriptional values were normalized to housekeeping genes and then by the overall expression levels used in a Ward’s linkage analysis. Patterns were inspected visually, and gene clusters were gated for isolating sets of biomarkers. Interactions between gated genes were predicted in Genemania and visualized in Cytoscape 3.9.0. A) Heat maps showing 2-way dendrograms, with hierarchical gene clusters by group (HIV-METH-, HIV + METH-, HIV + METH+ and HIV-METH+). Rectangles indicate gated genes by patterns, magnified as follows. B) Genes with scrambled patterns that differ between groups, and their interactions. C) Genes transcriptionally suppressed in HIV-METH+ and their interactions. D) Genes upregulated in HIV + METH + compared to other groups, and their interactions.
In the first approach, the examination of means and error sum of squares in hierarchical clustering was visualized in heat maps showing 2-way dendrograms, with hierarchical gene clusters by group (HIV-METH-, HIV + METH-, HIV + METH+ and HIV-METH+) (Fig. 2). Interestingly, we observed a strong pattern of gene transcription suppression in leukocytes from HIV-METH + subjects (Fig. 2A), as seen by the color blue indicative of decrease in relation to the average expression levels, which was not observed in HIV + METH + peripheral leukocyte specimens. By inspecting clustering patterns, we selected clustered areas where normalized gene expression indicated a systematic difference in one non-control group of interest compared to the others (Fig. 2B–D), and which may more likely hold potential biomarkers and form clusters connected by genetic, physical, or pathway interactions, as well as co-expression and co-localization, suggestive of orchestrated behaviors in response to variables. For instance, the cluster in Fig. 3B indicated genes more likely to be increased by Meth, and potentially able to distinguish between HIV+ and HIV- Meth users. Interestingly, these proximal genes interact based on physical, genetic and pathway interactions, as well as due to co-expression, suggesting potential underlying orchestrated processes. Likewise, gated clusters suppressed by Meth in HIV- subjects but detectable in other groups (Fig. 3C) were found to interact by similar means, potentially contributing to converging cellular outcomes. Importantly, this strategy allowed the identification of genes increased in HIV + METH + subjects but not in other groups (Fig. 2D). The genes in Fig. 2D were linked to Neurodegeneration (KW_Diseases, p = 0.002), and with KEGG Pathway functional annotations in Apoptosis (p = 2.6E-9), Proteoglycans (p = 1.8E-6), HIV infection (p = 3.2E-5), VEGF signaling (p = 2.7E-5) and Cellular senescence (p = 1.5E-3), and due to their upregulation may be potential biomarkers of HIV in the context of Meth.
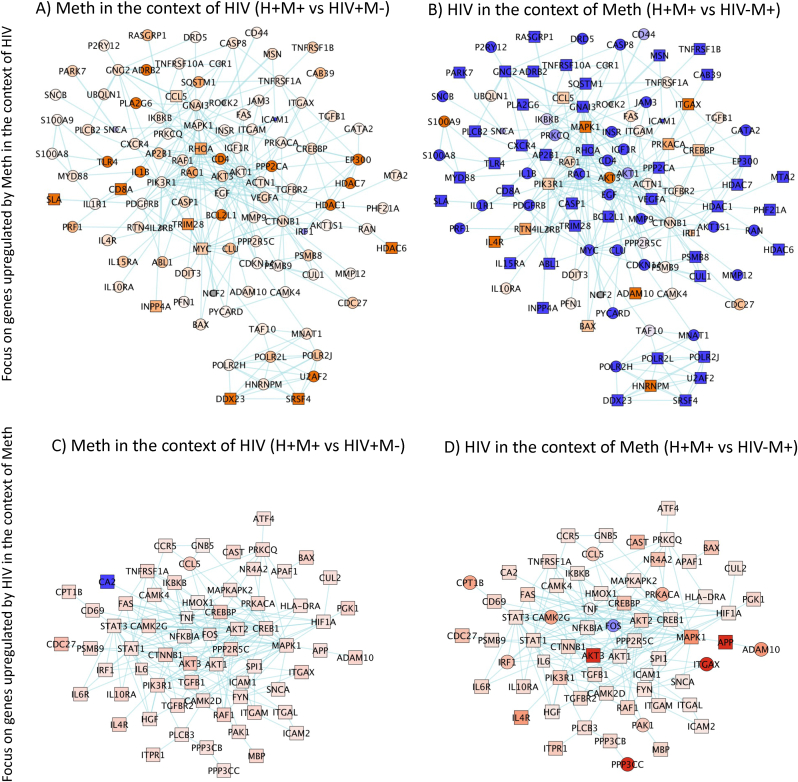
Fig. 3.
Gene Network Analysis (GNA) visualization of differences in the expression of DA-induced signatures detectable in human peripheral blood cells. DA signatures were validated by a Nanostring transcript panel. GNA strategies identified pathway-based clusters (blue connectors) visualized for the identification of nodes with biomarker value in H-M-, H + M-, H-M+, H+M+ subject specimens, n = 52–53/group. The 100 genes most upregulated by Meth (A, B) in the context of HIV, were visualized in A) HIV + METH + vs HIV + METH- and B) in HIV + METH + vs HIV-METH + comparisons. The 100 genes most upregulated by HIV (C, D) in the context of Meth, were visualized in C) HIV + METH + vs HIV + METH- and D) in HIV + METH + vs HIV-METH + comparisons. Shades of blue – downregulated, shades of orange – upregulated. Square shapes indicate p < 0.05. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2. Upregulated gene networks with a focus on context (Fig. 3)
We conducted gene network analyses and visualized differences in the expression of DA-induced signatures detectable in human peripheral blood cells. Fig. 3A–D shows results of comparisons with a focus on context; METH effects in the context of HIV (Fig. 3A and B), HIV effects in the context of METH (Fig. 3C and D). In this approach, transcriptional values were only internally normalized with housekeeping genes but not further normalized for average gene expression in the whole experiment, to enhance effects in individual genes.
Overall, the genes that were positively or negatively changed in HIV-Meth + subjects compared to HIV-METH- were linked to pathways in neurodegeneration (p = 6.0E-9) and dopaminergic synapse processes (p = 4.7E-5), including AKT3, FOS, G-protein subunits, ARRB2, CREBBP, CAL1 and COMT. Pathways on host-virus interactions (p = 9.8E-11), inflammation (p = 2.2E-6), and leukocyte adhesion (p = 4.2E-2) overlapped in genes involved in dopaminergic effects identified in pathway-based gene networks.
Fig. 3 shows the comparison between HIV + Meth+ and HIV + Meth-groups, which revealed transcripts with strong biomarker potential, due to their contribution to their distinctive transcriptional property behaviors indicating targeted effects of Meth in the context of HIV. For visualization, pathway-based gene networks were designed from the 100 genes most upregulated by Meth in the context of HIV (from the comparison between H+M+ vs H + M-, Fig. 3A and B), and by HIV in the context of Meth (from the comparison between H+M+ vs H-M+, Fig. 3C and D). Of these, 55 genes were significantly upregulated by Meth in the context of HIV, or H+M+ compared to H + M-, as indicated by square shapes, and were used in further analyses for effects of genotype in section 5 of results. The Cytoscape 3.9.0 software and GeneMania were applied, where colors indicated fold-change (shades of orange indicate increase, and shades of blue decrease), and shape of the nodes indicated statistical significance (circles p > 0.05 and squares p < 0.05 in the indicated comparisons). Hits with better biomarker potential for further testing were selected from these patterns, where genes significantly upregulated by Meth in the context of HIV (stronger orange and square shapes in Fig. 3A) were prioritized. Overlapping with Meth alone (>30% overlap) were genes annotated to neurodegeneration (p = 0.012), dopaminergic synapse (p = 1.8E-4), and inflammation (p = 0.0022). Host-virus interactions (p = 4.2E-11), autophagy and apoptosis (p = 6.5E-7 and 5.3E-3, respectively), splicing and positive regulation of transcription (p = 2.2E-8), and AGE-RAGE signaling pathway (p = 1.6E-7), were significantly perturbed by Meth in the context of HIV. Specific gene signatures upregulated by Meth in the context of HIV were CD8A, SLA, TRIM28, RhoA, INPP4A, HDCA6 and genes involved in RNA metabolism and splicing, such as SRSF4 and the DEAD-box gene DDX23. These biomarkers were efficient at distinguishing HIV + Meth users from the other groups. The increase in CD8A suggests a higher number of cytotoxic CD8 T cells, which were confirmed by flow cytometry (not shown). The increase in SLA (Src-like adaptor) detectable in humans, suggests enhanced adaptive immune responses, confirming our previous findings in the macaque model of HIV and Meth (Najera et al., 2016)). In addition, HDAC6 (Histone deacetylase 6) and TRIM28 (Tripartite Motif-containing Protein 28) are involved in silencing gene transcription, including HIV (Ait-Ammar et al., 2021).
The network with the most upregulated genes by HIV in the context of Meth (Fig. 3D) did not have power to distinguish the group comparisons (Fig. 4C). Yet, there were important genes, such as the transcription factors CREBBP and PRKACA, suggestive of signaling downstream GPCRs, as well as RANTES/CCL5, which affects inflammation and viral entry. As individual signatures, CREBBP and CCL5, along with other strong genes (Fig. 5) found by this and the previous approach were more likely to be upregulated in H+M+ specimens.
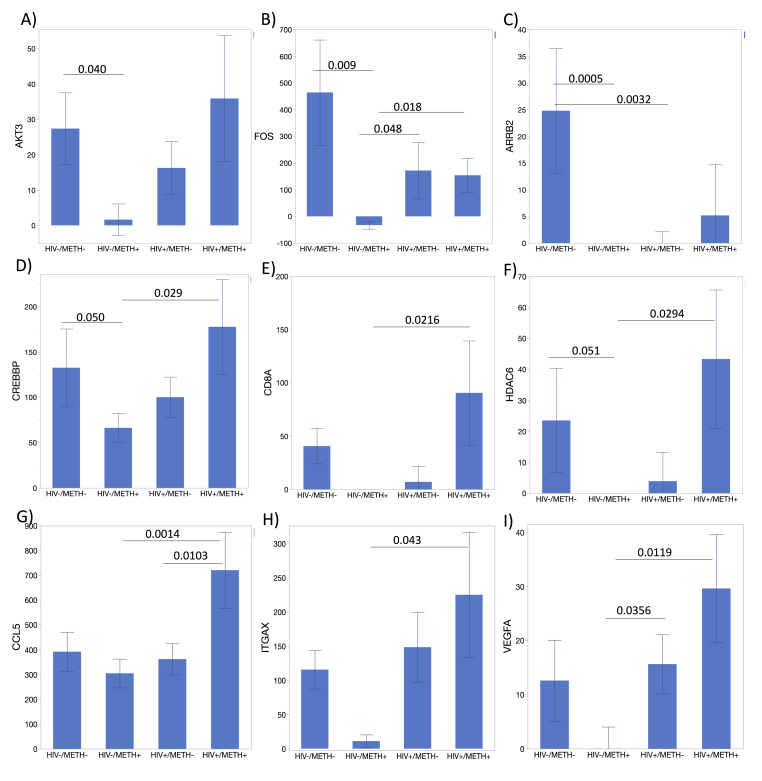
Fig. 4.
Individual transcripts with ability to distinguish the HIV/METH groups. Average counts per group for genes that are suppressed or unchanged in uninfected Meth users and show significance in ANOVA and one or more post-hoc group comparisons. A) AKT3, B) FOS, C) ARRB2, D) CREBBP, E) CD8A, F) HDAC6, G) CCL5, H) ITGAX, I) VEGFA. All transcripts were internally normalized to the average of 8 housekeeping genes. All genes in this figure had p < 0.05 in one-way ANOVA. Bonferroni’s post-hoc tests indicated in the figure for assigned comparisons, if significant.
Fig. 5.
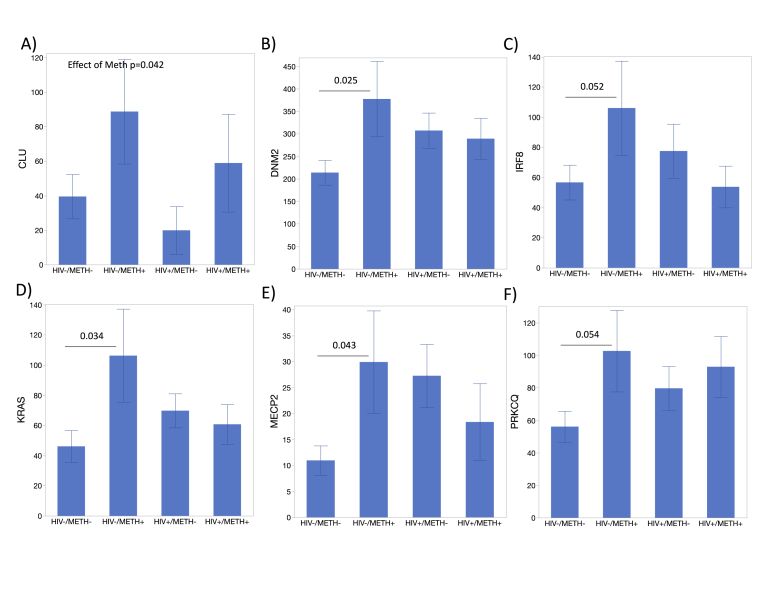
Individual transcripts upregulated in uninfected Meth users. Average counts per group for genes that are suppressed or unchanged in uninfected Meth users and show significance in ANOVA and in one or more post-hoc group comparisons. A) CLU, B) DNM2, C) IRF8, D) KRAS, E) MECP2, F) PRKCQ. All transcripts were internally normalized to the average of 8 housekeeping genes. All genes in this figure had p < 0.05 in one-way ANOVA. Bonferroni’s post-hoc tests indicated in the figure for assigned comparisons, if significant.
3.3. Overlapping genes between approaches and their numerical value
Within the expression patterns identified in the hierarchical clustering approach (Fig. 3), we selected genes that were also identified in gene network analysis (Fig. 4), and which showed statistical significance between groups, indicating potential biomarker value for identifying subjects in the groups, by HIV or METH status, or by interactions.
Overall, the majority of the individual transcriptional biomarkers replicated the main observations in Fig. 2A, showing significant suppression in uninfected Meth users compared to other groups (Fig. 4). These include transcripts also identified in GNA, such as AKT3 (Fig. 4A), FOS (Fig. 4B), and ARRB2 (Fig. 4C). On the other hand, some markers that were significantly suppressed in uninfected Meth users were also significantly upregulated in HIV + Meth users. These included CREBBP (Fig. 4D), CD8A (Fig. 4E), HDAC6 (Fig. 4F), ITGAX (Fig. 4H) and VEGFA (Fig. 4I). CCL5 expression raised special attention because it was significant and consistently upregulated in specimens from the HIV + METH + group compared to all the other groups, and it differed from other genes because it was not suppressed in uninfected Meth users (Fig. 4G), indicating a good biomarker value.
Despite the overall decrease in gene expression in the HIV-METH + group, upregulation in isolated genes identified in hierarchical clustering and in gene networks, proved useful to distinguish that group (Fig. 5). This was the case for CLU (Fig. 5A), DNM2 (Fig. 5B), IRF8 (Fig. 5C), KRAS (Fig. 5D), MECP2 (Fig. 5E), and PRKCQ (Fig. 5F). Together, the genes in Fig. 4, Fig. 5 can be important markers of METH use disorder alone and in the context of HIV.
3.4. Signature links with outcome
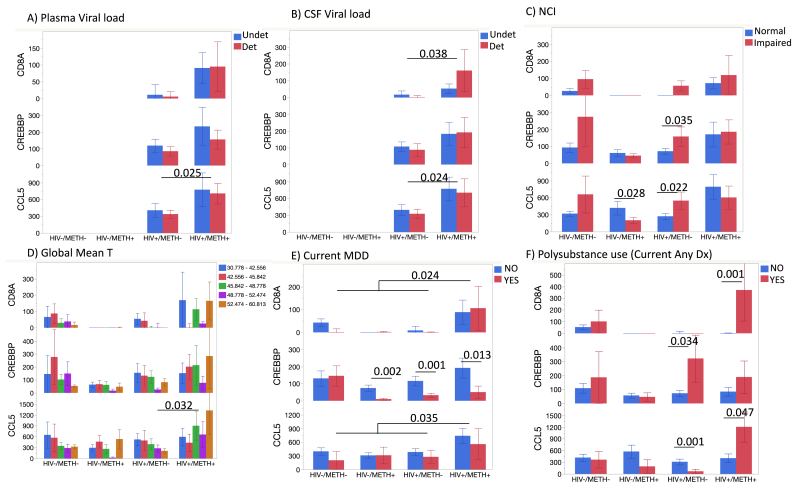
Among the signatures identified in both analysis approaches (hierarchical clustering and gene network analysis) as most elevated in HIV+/METH + individuals, we selected three genes for further examination in respect to important clinical variables. These genes were CD8A, CREBBP and CCL5, which were considered outstanding due to their overall highest expression levels, which may be beneficial to the development of detection assays for use clinical settings, and their ability to segregate groups. We examined whether their expression within groups could distinguish important outcomes, such as ability to control virus in plasma and CSF, as well as neurocognitive deficits or neuropsychiatric comorbidities (Fig. 6). Although there was an overall higher expression of these genes in HIV + Meth users compared to other groups, neither CD8A, CREBBP or CCL5 could distinguish between individuals with detectable vs. undetectable plasma or CSF viral load (Fig. 6A and B). NCI (as measured by GDS≥0.5) was distinguished in uninfected individuals by CREBBP, but not in HIV+ and/or METH + groups (Fig. 6C). CCL5 showed significant differences between NC normal and impaired HIV- Meth users and in HIV + non-users, but not in other groups. When examining Global Mean T scores, higher expression of CCL5 ocurred in more impaired individuals in both HIV+/METH- and HIV-/METH + groups, while in the HIV+/METH + group, CCL5 was higher in individuals with better performance (Fig. 6D). Decreased levels of CREBBP distinguished individuals with current Major depressive disorder (MDD) in all HIV+ and METH + groups (Fig. 6E). On the other hand, these genes were elevated in HIV + Meth users and also using any other substance (Fig. 6F).
Fig. 6.
Strongest overlapping signatures and links with cohort outcomes. Levels of CD8A, CREBBP and CCL5 transcripts were analyzed in respect to clinical variables of interest to the infection and to neurocognitive or psychiatric outcomes. A) Plasma viral load undetectable (Undet) and Detectable (Det); B) CSF Viral load (Undet/Det); C) Practice effect-corrected GDS values (GDS_pe = Normal/Impaired); D) Global Mean T scores stratified by numerical impairment levels, being higher values more impaired; p value from mixed models; E) Current Major Depressive Disorder (MDD); F) Polysubstance use (Current Any Diagnosis). Values represent Mean (Standard Error). Significant p values from multiple comparisons are indicated.
3.5. Effects of comorbidities and COMT genotype
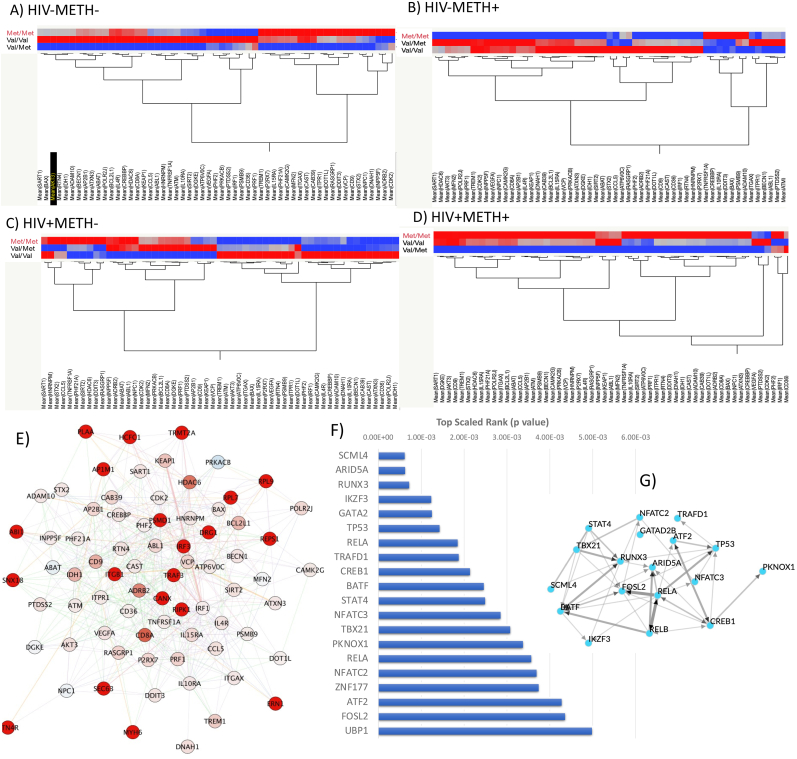
Val158met (rs4680) is a common single nucleotide polymorphism in the COMT gene which results in a change of one amino acid in the protein from a valine (Val) to a methionine (Met). The met allele is associated with approximately 40% lower enzymatic activity, leading to increased dopamine activity. Interestingly, the heterozygous genotype (Val/Met) is considered as the normal level of dopamine degradation or clinically non-actionable. Individuals with the Val/Val genotype display elevated COMT enzyme activity, thus reducing dopamine levels. Individuals with the Met/Met genotype have reduced COMT enzyme activity and thus elevated dopamine levels. We focused on a set of 55 genes that were significantly upregulated in METH in the context of HIV, as shown in Fig. 3A, and re-clustered these genes by COMT genotype normalized by the average expression within each group (Fig. 7). The goal of this approach was to test if any of the COMT genotypes could be responsible for driving the upregulation in HIV + METH users, and in relation to the attributed biological consequences of the COMT genotype. We found that while in METH-negative groups patterns are mixed for these genes (Fig. 7A and C), in METH users Met/Met genotype drives important distinctions. For instance, in HIV-METH+, Met/Met is associated with lower than average gene expression for the majority of the genes of interest (Fig. 7B). On the other hand, in HIV + METH + subjects, the Met/Met genotype was strongly linked to the upregulation of the same genes (Fig. 7D), suggesting that the genotype linked to higher DA levels in strongly influenced by METH and HIV interactions.
Fig. 7.
Hierarchical clustering of genes upregulated in the HIV + METH users in each group by COMT genotype, Visualization and Prediction of transcription factor usage regulated by COMT in HIV + METH users. A set of 55 genes upregulated in the HIV + METH + group was clustered by COMT genotype (Met/Met, Val/Met and Val/Val) using Ward method, with transcript counts normalized by individual groups' average. A) HIV-METH-, B) HIV-METH+, C) HIV + METH- and D) HIV + METH+. E) Network containing genes most strongly influenced by COMT genotype in HIV + METH+, linked by genetic interactions (green connectors) and physical interactions (red connectors). F) Prediction of transcription factor usage was performed using the genes in (D/E) as input in ChEA3 (://maayanlab.cloud/chea3), with p values assigned to the 20 highest ranked. G) Regulation studies and transcription factors' network based on their co-expression similarity constructed by Weighted Gene Co-expression Network Analysis (WGCNA) and Allegro Edge-Repulsive Strong Clustering in Cytoscape 3.9.0. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The examination of the upregulated patterns increased by HIV and METH, provided a restricted set of 55 genes that was used for making transcription factor usage predictions, which, while influenced by COMT Met/Met genotype, were potentially more strongly regulated by DA and modified by HIV (Fig. 7E). Using iRegulon in Cytoscape 3.9.0 these genes' input resulted in the prediction of transcription factors with overlapping and complementary regulatory actions. The 20 highest significance score transcription factors are indicated in Supplementary Table 3, with integrated scaled rank, the number of overlapping regulated genes among the ones found to be affected by COMT genotype, and database sources. The top 10 rank were SCML4, followed by ARID5A, RUNX3, IKZF3, GATA2, TP53, RELB, TRAFD1, CREB1 and STAT4. The prediction scores can be visualized in Fig. 7F. Fig. 7G displays a network scatter plot representing the 20 highest rank human transcription factors based on their co-expression similarity constructed by Weighted Gene Co-expression Network Analysis (WGCNA) over human TF expression data, and using Allegro Edge-Repulsive Strong Clustering.
The results suggest that the success of a panel of biomarkers for the identification of METH users in the context of HIV is attached to immune-dopaminergic interactions, influenced by individual genotypes in areas that influence levels and signaling of dopamine. The results also indicate that the pathways and mechanisms affected by the immune-modulatory effects of dopamine in the context of METH offers opportunities to single out HIV + METH users and the relevant outcomes in the population.
4. Discussion
A panel of markers that detect NCI in the context of Meth use might help monitor and manage disease, cognitive improvements, and infection in PWH who are drug users. A broad panel can also increase the power of discovery, by facilitating the identification of orchestrated effects on genes that are not evaluated as single biomarkers, but as network components linked by protein-protein or genetic interactions, co-expression, commons protein domains, or predicted interactions, and associated with the response of leukocytes, particularly innate immune HIV targets, to neurotransmitters that are abundant in Meth users' brains, such as dopamine (DA).
In this study, a transcriptional biomarker panel was used to investigate neuroimmune interactions in the context of HIV infection and METH use disorder in a human cohort. Our findings indicate that this panel can potentially distinguish lifetime METH use disorder in individuals with HIV. This is significant for monitoring the progression of HIV in METH users, especially those experiencing cognitive decline. This decline may be influenced by various factors, including the interaction between HIV and METH use, the impact on viral load control, and other health determinants that elevate classical inflammatory markers through distinct mechanisms. Such a panel is needed for monitoring progress in HIV + METH users, particularly the ones with cognitive decline, due to the confounder effects and interactions between the infection and the drug, decreased control of viral load, along with other health determinants increasing levels of classical inflammatory markers through potentially non-overlapping mechanisms.
We examined several biomarkers known to be associated with cognitive impairment in people with HIV (PWH). Notably, the increased proportion of CD14+CD16+ monocytes has been shown to be predictive of low neuropsychiatric deficits [30] and atrophy of brain structures (Kallianpur et al., 2020), a finding supported by transmigration of these inflammatory cell subsets under high dopamine conditions (Calderon et al., 2017), as seen in METH users. Other markers, such as soluble CD14 and soluble CD163 are found in monocytes, macrophages and activated microglia, and can predict CNS inflammation (Ancuta et al., 2008; Fischer-Smith et al., 2008a, 2008b) when elevated in plasma and CSF (Bryant et al., 2017; Burdo et al., 2010; Nowlin et al., 2015). However, these markers can increase in various conditions within the context of HIV (Burdo et al., 2011a, 2011b; Srinivasa et al., 2014; Walker et al., 2014), and not specifically due to substance use. Therefore, identifying markers of neuroHIV in drug users is crucial for tailoring treatment and care.
The study’s demographic focus on male individuals, with consistent age, education, and race, presents a limitation in terms of broader applicability but enhances the statistical power for this cohort. We observed expected differences in numbers of peripheral blood cells between groups, with HIV + individuals showing lower CD4 and higher CD8 cells irrespective of METH use. A notable finding was the lower percentage of CD11b + cells, suggesting evasion from the peripheral blood to tissues, including the brain, aligning with cognitive decline observations. This supports an inflammatory basis to neurocognitive impairment in PWH, particularly those using drugs (Mediouni et al., 2015; Nath et al., 2001).
We utilized digital transcriptome methods and various analytical strategies to explore whether the inflammatory cells in drug users differ from those in non-drug users and the specific mechanisms involved. Ward’s linkage method for hierarchical clustering of transcripts facilitated the identification of gene expression patterns, revealing signatures of neurodegeneration, apoptosis, and vascular adhesion in the peripheral blood cells of HIV + METH users.
We employed Gene Network Analysis (GNA) to further validate these findings to identify co-expression patterns among genes, revealing significant differences in a gene network related to neurodegeneration, dopaminergic synapse, and inflammation. This suggests that genes in dopaminergic pathways, important for neuro-immune functions, are potential biomarkers for inflammation in HIV + METH users.
The use of Ward’s linkage method for hierarchical clustering of transcripts allows an easy implementation and interpretation, applicable to small data sets such as the one used here (784 transcripts). Systems biology was used as an accessory strategy to identify the links between genes showing similar behaviors in hierarchical clustering, indicative of orchestrated changes, and confirming that signatures of neurodegeneration, apoptosis, and vascular adhesion can be identified in the peripheral blood cells of HIV + METH users compared to other groups.
An additional independent strategy was designed to further confirm identifying pathways relevant to the brain in blood cells. This included GNA to detect biological links among genes showing similar co-expression patterns between groups. There are multiple advantages to this method, given that the variability between human subjects, occurring due to background, co-morbidities, and genotype, may not show uniform differences in the expression of single markers but detect effects in pathways or biological processes relevant to HIV infection, inflammation, and the brain. By linking genes upregulated by METH in the context of HIV, or genes increased in H+M+ compared to H + M-, we found most significant differences in a single gene network with shared common elements in pathways linked to neurodegeneration, dopaminergic synapse, and inflammation, and supporting the idea that genes in dopaminergic pathways exerting important neuro-immune functions serve well as biomarkers of inflammation in HIV + METH users compared to non-METH users.
Interestingly, genes like CCL5, CREBBP and HDAC6 which increased in HIV + METH users, intersect in immune pathways controlled by GPCRs, and regulate gene transcription, cell migration, inflammation, and neurotoxicity. The increase in CD8A transcripts matches the enrichment of CD8 cells in METH users, also suggesting anti-viral responses, although their specificity remains uncertain (Wang et al., 2018; Bartolotti and Lazarov, 2019; Gamo et al., 2008). We have previously shown that bystander CD8 cells are more harmful than virus-specific CD8 T cells with equal cytotoxic activity (Marcondes et al., 2008, 2015). Moreover, despite ART, virological failure is a common aspect of METH use (Ellis et al., 2003), with relative contribution of poor adherence but also a weakened immune competence (Massanella et al., 2015). Among the vascular endothelial growth factors, VEGFA is strongly associated with angiogenesis, and weakly associated with cell migration (Rapisarda and Melillo, 2012), but with potentially protective effects in cognitive function, despite enhancing inflammation via macrophage recruitment (Cao et al., 2004a, 2004b; Cursiefen et al., 2004). Interestingly, low VEGF plasma levels are a marker of amnestic mild cognitive impairment among older virally suppressed PWH (Serrano et al., 2021). Here, the VEGFA transcriptional upregulation in H + M + PBMCs did not distinguish differences in global cognitive scores (not shown), but correlated with lower CD11b + cells in the periphery. A decrease in peripheral leukocyte numbers despite the higher expression of activation markers may be suggestive of higher trafficking supporting tissue inflammation and may indicate higher disease severity, as reported in other CNS pathologies such as PD (Bhatia et al., 2021; Garre et al., 2017; Garre and Yang, 2018). This could be related to VEGFA, or be regulated independently of DA by factors linked to inflammation in the context of HIV infection (Garre et al., 2017; Garre and Yang, 2018). We have recently shown that the DA-regulated monocytic marker MRP8/14, which segregates METH users with detectable CSF viral load (Basova et al., 2023a), is a marker of redistribution into the CNS (Persidsky, 2015). New studies are necessary to measure VEGFA in plasma in the context of METH use, and respective to cognition, and how this relates to inflammatory cell trafficking.
The focus on a set of upregulated genes is important to biomarker discovery in HIV + METH users. Previous observations have indicated that METH causes suppression of gene transcription broadly (Basova et al., 2020, 2023b), with hubs of upregulation that vary between individuals, and often linked to inflammatory pathways (Basova et al., 2022; Bortell et al., 2015). These markers upregulated by METH in the context of HIV, are strong candidate biomarkers to track disease progress or improvements due to treatment, virological suppression or abstinence of METH.
Regarding CCL5, the highest expression patterns matched NC impairment. However, the highest levels of CCL5 transcripts were found in the HIV+/METH + groups, but in individuals with better cognitive performance. These individuals had undetectable CSF viral load (not shown), indicating that one potential explanation for this result relies on the ability of CCL5, which is produced by immune cells in the context of Meth (Najera et al., 2016; Bortell et al., 2015), to block viral entry via the CCR5 co-receptor (Cocchi et al., 1995), or enhance the anti-viral response (Fransen et al., 2000).
Genes that are decreased are also important, but may be diluted by the overall suppression of gene transcription caused by METH, which is observed in humans and confirmed in experimental models (Basova et al., 2020, 2023b). Some genes were upregulated in HIV-negative METH users, but not in HIV-positive METH users. These genes were CLU, DNM2, KRAS, MECP2, and PRKCQ, which are rather involved in neurological outcomes, dopaminergic disorders and substance use. For instance, CLU (clusterin), is a genetic risk association with Alzheimer’s disease, and may have implications in anxiety and in the development of such disease in METH users (Bettens et al., 2015). DNM2 encodes dynamin 2, a protein with structural and cytoskeleton functions, also increased by cocaine (Freeman et al., 2010). KRAS is an oncogene that is curiously responsive to METH addiction treatment (Li et al., 2014). MECP2 (Methyl-CpG-binding protein 2) is also interesting, as a transcriptional regulator that represses or activates the expression of target genes and playing a role in the regulation of drug addiction in the dopaminergic reward system (Bae et al., 2022). Similarly, PRKCQ is a protein kinase involved in dopaminergic neurotoxicity in the context of METH (Shin et al., 2019, 2021). Showing similar pattern, IRF8 has a link to inflammation and HIV control (Lacagnina et al., 2017; Assis et al., 2021).
Our observations highlight the importance of considering genes that show patterns of upregulation as well as those that are suppressed by METH use. Certain genes were upregulated in HIV-negative METH users but not in HIV-positive users, indicating their involvement in neurological outcomes and substance use disorders.
The results indicate that co-factors such as genotype may be critical and complementary in efforts to distinguish the susceptible population. For example, the enzyme COMT, which is involved in the metabolism of dopamine and other catecholamines (Barr et al., 2006; Panenka et al., 2013; Saloner et al., 2020), may account for disparities in the expression of transcriptional markers affected by dopamine in the context of METH. The stronger upregulation in biomarkers identified here was largely driven by individuals exhibiting the Met/Met COMT genotype, associated with higher DA levels. Interestingly, in HIV- subjects that are METH users, the same genotype was linked to a lower than average gene expression, indicating that the effects on DA levels in the context of METH are critically modified by HIV. This data further highlights the complexities of the underlying mechanisms of inflammation, despite the similar outcomes in therms of NCI between groups. It underlines the need for better biomarkers tailored to detect HIV in the context of METH and vice-versa. The effects of COMT were detected in relation to transcriptional patterns in leukocytes in the context of METH use. Genome Wide Association studies (GWAS) have identified links between COMT genotypes and bipolar disorder (Mullins et al., 2019), alcohol drinking (Feitosa et al., 2018), and chronic pain (Li et al., 2023), all potentially underlying or coexisting with substance use. However, the clustering of DA-regulated biomarkers is linked to the role of COMT as regulator of the levels of neurotransmitters that interact with immune cells (Barr et al., 2006; Panenka et al., 2013; Saloner et al., 2020). It remains to be examined if similar patterns in inflammatory markers may be detected in the disorders known to be linked to COMT genotypes from GWAS studies. Interestingly, the elevation of both CD8A and CCL5, which clustered in the HIV + METH + group, was strongly driven by concomitant alcohol use. Our observations lay the foundation for future studies that can better establish the connection of DA markers and COMT genotype, with concomitant alcohol/polysubstance use and METH.
The transcriptional patterns generated by COMT genotype in METH users allowed predictions of transcription factor usage that is affected by DA, and modified by HIV. SCML4 was the top ranked transcription factor predicted in correlation with effects of COMT genotype in METH and HIV. Interestingly, two of the top predicted transcription factors have been described in association with METH self-administration in rats (Khalid et al., 2023). In that model, SCLM4 was the fifth most downregulated gene, and RUNX3 was among the 25 most upregulated genes in dopaminergic regions of the brain following METH self administration (Khalid et al., 2023). IKZF3 is an IKAROS zink finger family protein acting on immune cell fitness and IL10 production (Ridley et al., 2020; Lazarian et al., 2021), but no reported links to substance use or dopamine. GATA-2, on the other hand, is expressed in dopaminergic areas of the brain with changes tightly linked to Parkinson’s disease (Scherzer et al., 2008). TP53 contributes to neurotoxic signaling via DA receptors (Lu et al., 2017). In a knock out mouse model, it has been demonstrated that TP53 plays a critical role in METH-mediated neurotoxicity and cell damage in dopaminergic terminal areas (Hirata and Cadet, 1997), with potentially long-term deleterious effects. RELB and RELA are components of the NFkB transcription complex, with important roles in neuro-inflammation, including in various dopaminergic pathologies (Perkins, 1997; Lanzillotta et al., 2015; Ghosh et al., 2007) and in HIV (Kiebala et al., 2010). TRAFD1 plays a role in regulating anti-viral responses (Sanada et al., 2008). CREB1 is strongly affected by DA (Konradi et al., 1993; Lee et al., 2010) and has been considered as a target in neurological disorders such as schizophrenia (Wang et al., 2018). These 10 highest scored transcription factors may be potential regulators of processes detectable in PBMCs and responsive to DA signaling and genotypes affecting DA levels in drug users, particularly the ones that are living with HIV.
There are limitations in our study that include a restricted inclusion criteria due to the small number of subjects per group, and the narrow hypothesis embedded in a 2 by 2 design. Within this design, MDD and polysubstance use, particularly alcohol, are potential confounders. In this context, CREBBP is a particularly interesting gene because it was able to distinguish individuals with current MDD in all HIV+ and METH + groups. Interestingly, variants of the CREBBP have been previously associated with MDD (Crisafulli et al., 2012), as well as changes in other components in the cAMP responsive pathway (Xiao et al., 2018; Blendy, 2006), and along with NFkB responsive inflammatory genes (Mellon et al., 2016). The overlap of substance use and depression, via CREB (Pandey et al., 2005) underlies the inflammatory character of these disorders (Chan et al., 2019; Enache et al., 2019), also present in HIV (Vasantiuppapokakorn et al., 2024). Anti-inflammatory treatments have been previously incorporated to anti-depressant trials with positive results (Kohler-Forsberg et al., 2019). Our results suggest that transcriptional levels of CREBBP can be a potential biomarker to track inflammatory processes linked to MDD in PWH that are Meth users.
Regarding the combinatorial role of METH and other drugs in variables such as CSF viral load, more studies need to be done. However, our approach improved the power of discovery of novel biomarkers that can identify uncontrolled CSF viral load and neuroinflammation in METH users by establishing an a priori focus on DA-regulated genes in leukocytes. Our results indicate that the power of group segregation by a biomarker panel can be enhanced by the knowledge of genotypes and transcription factor usage predictions. The effects of DA in cells of the CNS and in peripheral leukocytes have different implications to disease outcomes due to differences in cell targets expressing DA receptors (neurons, immune cells, microglia, vs. lymphocytes and monocytes). However, as the expression of molecular phenotypes in the two compartments overlaps in time, the peripheral blood cells influenced by DA offer a window to events occurring in immune cells of the brain.
The identification of biomarkers that can track progress in substance users is important but also a complex problem due the multiple mechanisms affected by the drug, or by the virus. Both HIV and METH act by different means to trigger inflammation in the brain and elsewhere, where distinctive underlying causes converge in one outcome. In conclusion, the identification of biomarkers is a crucial step in understanding and managing the complex interplay between HIV infection, METH use, and their combined impact on neuroimmune functions. Future research, particularly longitudinal studies and interventions, and examination of other genotypes, will be vital for advancing our ability to monitor, track, and treat this challenging population effectively.
Funding
This work was funded by NIH/NIDA R01DA047822 and R61DA059924 to MCGM; P30MH062512 to the HNRC.
Data availability statement
All data can be available upon request.
CRediT authorship contribution statement
Liana V. Basova: Visualization, Validation, Project administration, Investigation, Formal analysis, Data curation. Tera Riley: Visualization, Methodology. Donald Franklin: Resources, Project administration, Investigation, Conceptualization. Violaine Delorme-Walker: Supervision, Methodology, Formal analysis. Wei Ling Lim: Validation, Methodology. Igor Grant: Resources, Conceptualization. Scott L. Letendre: Writing – review & editing, Visualization, Validation, Conceptualization. Jennifer E. Iudicello: Writing – review & editing, Resources, Investigation, Formal analysis, Conceptualization. Mariana Cherner: Writing – review & editing, Resources, Investigation, Conceptualization. Ronald J. Ellis: Writing – review & editing, Resources, Investigation, Data curation, Conceptualization. Maria Cecilia Garibaldi Marcondes: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Software, Resources, Project administration, Methodology, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization.
Declaration of competing interest
The authors of the manuscript “Identifying Methamphetamine Use Predictors in HIV Infection: Immune-Dopaminergic Signatures in Peripheral Leukocytes and the Role of COMT Genotype” have no conflicts of interest to declare.
Acknowledgements
The authors thank Christine Auciello and Krista Scrivner for administrative assistance, and Dr. Howard Fox (University of Nebraska Medical Center) for discussions during the conceptualization of this study. We thank Jennifer Marquie-Beck (Director of Research Operations at UCSD/HNRP) and Dr. Marcus Kaul (University of California Riverside School of Medicine, Division of Biomedical Sciences) for participating in discussions and providing important insights.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbih.2024.100873.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
We have submitted all raw and processed data to NCBI Gene Expression Omnibus (GEO) and it is available with accession number GSE273630.
References
- Ait-Ammar A., Bellefroid M., Daouad F., et al. Inhibition of HIV-1 gene transcription by KAP1 in myeloid lineage. Sci. Rep. 2021;11(1):2692. doi: 10.1038/s41598-021-82164-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ancuta P., Kamat A., Kunstman K.J., et al. Microbial translocation is associated with increased monocyte activation and dementia in AIDS patients. PLoS One. 2008;3(6) doi: 10.1371/journal.pone.0002516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assis M.A., Carranza P.G., Ambrosio E. A “Drug-Dependent” immune system can compromise protection against infection: the relationships between psychostimulants and HIV. Viruses. 2021;13(5) doi: 10.3390/v13050722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson J.H., Higgins J.A., Vigil O., et al. Psychiatric context of acute/early HIV infection. The NIMH multisite acute HIV infection study: IV. AIDS Behav. 2009;13(6):1061–1067. doi: 10.1007/s10461-009-9585-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae J., Ahn S., Cho D.W., Kim H.S., Han S.C., Im H.I. Claustral MeCP2 regulates methamphetamine-induced conditioned place preference in cynomolgus monkey. Exp. Neurobiol. 2022;31(6):390–400. doi: 10.5607/en22034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek E.J., Kim H., Basova L.A., et al. Sex differences and Tat expression affect dopaminergic receptor expression and response to antioxidant treatment in methamphetamine-sensitized HIV Tat transgenic mice. Neuropharmacology. 2020;178 doi: 10.1016/j.neuropharm.2020.108245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr A.M., Panenka W.J., MacEwan G.W., et al. The need for speed: an update on methamphetamine addiction. J. Psychiatry Neurosci. 2006;31(5):301–313. [PMC free article] [PubMed] [Google Scholar]
- Bartolotti N., Lazarov O. CREB signals as PBMC-based biomarkers of cognitive dysfunction: a novel perspective of the brain-immune axis. Brain Behav. Immun. 2019;78:9–20. doi: 10.1016/j.bbi.2019.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basova L., Najera J.A., Bortell N., et al. Dopamine and its receptors play a role in the modulation of CCR5 expression in innate immune cells following exposure to Methamphetamine: implications to HIV infection. PLoS One. 2018;13(6) doi: 10.1371/journal.pone.0199861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basova L.V., Kesby J.P., Kaul M., Semenova S., Marcondes M.C.G. Systems biology analysis of the antagonizing effects of HIV-1 tat expression in the brain over transcriptional changes caused by methamphetamine sensitization. Viruses. 2020;12(4) doi: 10.3390/v12040426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basova L., Lindsey A., McGovern A.M., Ellis R.J., Marcondes M.C.G. Detection of H3K4me3 identifies neuroHIV signatures, genomic effects of methamphetamine and addiction pathways in postmortem HIV+ brain specimens that are not amenable to transcriptome analysis. Viruses. 2021;13(4) doi: 10.3390/v13040544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basova L.V., Vien W., Bortell N., Najera J.A., Marcondes M.C.G. Methamphetamine signals transcription of IL1beta and TNFalpha in a reactive oxygen species-dependent manner and interacts with HIV-1 Tat to decrease antioxidant defense mechanisms. Front. Cell. Neurosci. 2022;16 doi: 10.3389/fncel.2022.911060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basova L.V., Lindsey A., McGovern A., et al. MRP8/14 is a molecular signature triggered by dopamine in HIV latent myeloid targets that increases HIV transcription and distinguishes HIV+ methamphetamine users with detectable CSF viral load and brain pathology. Viruses. 2023;15(6):1363. doi: 10.3390/v15061363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basova L.V., Lindsey A., McGovern A., et al. MRP8/14 is a molecular signature triggered by dopamine in HIV latent myeloid targets that increases HIV transcription and distinguishes HIV+ methamphetamine users with detectable CSF viral load and brain pathology. Viruses. 2023;15(6) doi: 10.3390/v15061363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basova LAL S.E., Milner R., Ellis R.J., Cherner M., Iudicello J., Marcondes M.C.G. Polygenic networks in peripheral leukocytes indicate patterns associated with HIV infection and context-dependent effects of cannabis use. Brain Behav. Immun. - Health. 2022;20 doi: 10.1016/j.bbih.2022.100414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettens K., Vermeulen S., Van Cauwenberghe C., et al. Reduced secreted clusterin as a mechanism for Alzheimer-associated CLU mutations. Mol. Neurodegener. 2015;10:30. doi: 10.1186/s13024-015-0024-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia D., Grozdanov V., Ruf W.P., et al. T-cell dysregulation is associated with disease severity in Parkinson’s Disease. J. Neuroinflammation. 2021;18(1):250. doi: 10.1186/s12974-021-02296-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blendy J.A. The role of CREB in depression and antidepressant treatment. Biol. Psychiatr. 2006;59(12):1144–1150. doi: 10.1016/j.biopsych.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Bortell N., Morsey B., Basova L., Fox H.S., Marcondes M.C. Phenotypic changes in the brain of SIV-infected macaques exposed to methamphetamine parallel macrophage activation patterns induced by the common gamma-chain cytokine system. Front. Microbiol. 2015;6:900. doi: 10.3389/fmicb.2015.00900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant A.K., Moore D.J., Burdo T.H., et al. Plasma soluble CD163 is associated with postmortem brain pathology in human immunodeficiency virus infection. AIDS. 2017;31(7):973–979. doi: 10.1097/qad.0000000000001425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdo T.H., Soulas C., Orzechowski K., et al. Increased monocyte turnover from bone marrow correlates with severity of SIV encephalitis and CD163 levels in plasma. PLoS Pathog. 2010;6(4) doi: 10.1371/journal.ppat.1000842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdo T.H., Lentz M.R., Autissier P., et al. Soluble CD163 made by monocyte/macrophages is a novel marker of HIV activity in early and chronic infection prior to and after anti-retroviral therapy. J. Infect. Dis. 2011;204(1):154–163. doi: 10.1093/infdis/jir214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdo T.H., Lo J., Abbara S., et al. Soluble CD163, a novel marker of activated macrophages, is elevated and associated with noncalcified coronary plaque in HIV-infected patients. J. Infect. Dis. 2011;204(8):1227–1236. doi: 10.1093/infdis/jir520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderon T.M., Williams D.W., Lopez L., et al. Dopamine increases CD14(+)CD16(+) monocyte transmigration across the blood brain barrier: implications for substance abuse and HIV neuropathogenesis. J. Neuroimmune Pharmacol. 2017;12(2):353–370. doi: 10.1007/s11481-017-9726-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L., Jiao X., Zuzga D.S., et al. VEGF links hippocampal activity with neurogenesis, learning and memory. Nat. Genet. 2004;36(8):827–835. doi: 10.1038/ng1395. [DOI] [PubMed] [Google Scholar]
- Cao R., Eriksson A., Kubo H., Alitalo K., Cao Y., Thyberg J. Comparative evaluation of FGF-2-, VEGF-A-, and VEGF-C-induced angiogenesis, lymphangiogenesis, vascular fenestrations, and permeability. Circ. Res. 2004;94(5):664–670. doi: 10.1161/01.RES.0000118600.91698.BB. [DOI] [PubMed] [Google Scholar]
- Carey C.L., Woods S.P., Gonzalez R., et al. Predictive validity of global deficit scores in detecting neuropsychological impairment in HIV infection. J. Clin. Exp. Neuropsychol. 2004;26(3):307–319. doi: 10.1080/13803390490510031. [DOI] [PubMed] [Google Scholar]
- Carrico A.W., Cherenack E.M., Flentje A., et al. A positive affect intervention alters leukocyte DNA methylation in sexual minority men with HIV who use methamphetamine. Brain Behav. Immun. 2024;120:151–158. doi: 10.1016/j.bbi.2024.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattie J.E., Woods S.P., Iudicello J.E., Posada C., Grant I., Group T. Elevated neurobehavioral symptoms are associated with everyday functioning problems in chronic methamphetamine users. J. Neuropsychiatr. Clin. Neurosci. Summer. 2012;24(3):331–339. doi: 10.1176/appi.neuropsych.11080192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.L., Cathomas F., Russo S.J. Central and peripheral inflammation link metabolic syndrome and major depressive disorder. Physiology. 2019;34(2):123–133. doi: 10.1152/physiol.00047.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherenack E.M., Chavez J.V., Martinez C., et al. Stimulant use, HIV, and immune dysregulation among sexual minority men. Drug Alcohol Depend. 2023;251 doi: 10.1016/j.drugalcdep.2023.110942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocchi F., DeVico A.L., Garzino-Demo A., Arya S.K., Gallo R.C., Lusso P. Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells. Science. 1995;270(5243):1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- Coley J.S., Calderon T.M., Gaskill P.J., Eugenin E.A., Berman J.W. Dopamine increases CD14+CD16+ monocyte migration and adhesion in the context of substance abuse and HIV neuropathogenesis. PLoS One. 2015;10(2) doi: 10.1371/journal.pone.0117450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisafulli C., Shim D.S., Andrisano C., et al. Case-control association study of 14 variants of CREB1, CREBBP and CREM on diagnosis and treatment outcome in major depressive disorder and bipolar disorder. Psychiatr. Res. 2012;198(1):39–46. doi: 10.1016/j.psychres.2011.08.022. [DOI] [PubMed] [Google Scholar]
- Cursiefen C., Chen L., Borges L.P., et al. VEGF-A stimulates lymphangiogenesis and hemangiogenesis in inflammatory neovascularization via macrophage recruitment. J. Clin. Invest. 2004;113(7):1040–1050. doi: 10.1172/JCI20465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danaher P., Warren S., Dennis L., et al. Gene expression markers of tumor infiltrating leukocytes. J. Immunother. Cancer. 2017;5:18. doi: 10.1186/s40425-017-0215-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis R.J., Childers M.E., Cherner M., et al. Increased human immunodeficiency virus loads in active methamphetamine users are explained by reduced effectiveness of antiretroviral therapy. J. Infect. Dis. 2003;188(12):1820–1826. doi: 10.1086/379894. [DOI] [PubMed] [Google Scholar]
- Enache D., Pariante C.M., Mondelli V. Markers of central inflammation in major depressive disorder: a systematic review and meta-analysis of studies examining cerebrospinal fluid, positron emission tomography and post-mortem brain tissue. Brain Behav. Immun. 2019;81:24–40. doi: 10.1016/j.bbi.2019.06.015. [DOI] [PubMed] [Google Scholar]
- Feitosa M.F., Kraja A.T., Chasman D.I., et al. Novel genetic associations for blood pressure identified via gene-alcohol interaction in up to 570K individuals across multiple ancestries. PLoS One. 2018;13(6) doi: 10.1371/journal.pone.0198166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer-Smith T., Bell C., Croul S., Lewis M., Rappaport J. Monocyte/macrophage trafficking in acquired immunodeficiency syndrome encephalitis: lessons from human and nonhuman primate studies. J. Neurovirol. 2008;14(4):318–326. doi: 10.1080/13550280802132857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer-Smith T., Tedaldi E.M., Rappaport J. CD163/CD16 coexpression by circulating monocytes/macrophages in HIV: potential biomarkers for HIV infection and AIDS progression. AIDS Res. Hum. Retrovir. 2008;24(3):417–421. doi: 10.1089/aid.2007.0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitting S., Booze R.M., Mactutus C.F. Neonatal intrahippocampal glycoprotein 120 injection: the role of dopaminergic alterations in prepulse inhibition in adult rats. J. Pharmacol. Exp. Therapeut. 2006;318(3):1352–1358. doi: 10.1124/jpet.106.105742. [DOI] [PubMed] [Google Scholar]
- Fransen S., Copeland K.F., Smieja M., Smaill F., Rosenthal K.L. RANTES production by T cells and CD8-mediated inhibition of human immunodeficiency virus gene expression before initiation of potent antiretroviral therapy predict sustained suppression of viral replication. J. Infect. Dis. 2000;181(2):505–512. doi: 10.1086/315270. [DOI] [PubMed] [Google Scholar]
- Franz M., Rodriguez H., Lopes C., et al. GeneMANIA update 2018. Nucleic Acids Res. 2018;46(W1):W60–W64. doi: 10.1093/nar/gky311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman W.M., Lull M.E., Patel K.M., et al. Gene expression changes in the medial prefrontal cortex and nucleus accumbens following abstinence from cocaine self-administration. BMC Neurosci. 2010;11:29. doi: 10.1186/1471-2202-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamo K., Kiryu-Seo S., Konishi H., et al. G-protein-coupled receptor screen reveals a role for chemokine receptor CCR5 in suppressing microglial neurotoxicity. J. Neurosci. 2008;28(46):11980–11988. doi: 10.1523/JNEUROSCI.2920-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garre J.M., Yang G. Contributions of monocytes to nervous system disorders. J. Mol. Med. (Berl.) 2018;96(9):873–883. doi: 10.1007/s00109-018-1672-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garre J.M., Silva H.M., Lafaille J.J., Yang G. CX3CR1(+) monocytes modulate learning and learning-dependent dendritic spine remodeling via TNF-alpha. Nat. Med. 2017;23(6):714–722. doi: 10.1038/nm.4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaskill P.J., Calderon T.M., Luers A.J., Eugenin E.A., Javitch J.A., Berman J.W. Human immunodeficiency virus (HIV) infection of human macrophages is increased by dopamine: a bridge between HIV-associated neurologic disorders and drug abuse. Am. J. Pathol. 2009;175(3):1148–1159. doi: 10.2353/ajpath.2009.081067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaskill P.J., Carvallo L., Eugenin E.A., Berman J.W. Characterization and function of the human macrophage dopaminergic system: implications for CNS disease and drug abuse. J. Neuroinflammation. 2012;9:203. doi: 10.1186/1742-2094-9-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaskill P.J., Calderon T.M., Coley J.S., Berman J.W. Drug induced increases in CNS dopamine alter monocyte, macrophage and T cell functions: implications for HAND. J. Neuroimmune Pharmacol. 2013;8(3):621–642. doi: 10.1007/s11481-013-9443-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A., Roy A., Liu X., et al. Selective inhibition of NF-kappaB activation prevents dopaminergic neuronal loss in a mouse model of Parkinson’s disease. Proc. Natl. Acad. Sci. U. S. A. 2007;104(47):18754–18759. doi: 10.1073/pnas.0704908104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grelotti DJM J., Delorme-Walker V., Iudicello J., Ellis R., Grant I., Letendre S., Marcondes M.C.G., Cherner M. On the issue of treating HIV in people with syndemic mental and substance use disorders. Nature Mental Health. 2024 doi: 10.1038/s44220-024-00283-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halkitis P.N., Parsons J.T., Stirratt M.J. A double epidemic: crystal methamphetamine drug use in relation to HIV transmission among gay men. J. Homosex. 2001;41(2):17–35. doi: 10.1300/J082v41n02_02. [DOI] [PubMed] [Google Scholar]
- Haw R.A., Croft D., Yung C.K., et al. The reactome BioMart. Database. 2011;2011:bar031. doi: 10.1093/database/bar031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton R.K., Marcotte T.D., Mindt M.R., et al. The impact of HIV-associated neuropsychological impairment on everyday functioning. J. Int. Neuropsychol. Soc. 2004;10(3):317–331. doi: 10.1017/S1355617704102130. [DOI] [PubMed] [Google Scholar]
- Hirata H., Cadet J.L. p53-knockout mice are protected against the long-term effects of methamphetamine on dopaminergic terminals and cell bodies. J. Neurochem. 1997;69(2):780–790. doi: 10.1046/j.1471-4159.1997.69020780.x. [DOI] [PubMed] [Google Scholar]
- Kallianpur K.J., Jahanshad N., Sailasuta N., et al. Regional brain volumetric changes despite 2 years of treatment initiated during acute HIV infection. AIDS. 2020;34(3):415–426. doi: 10.1097/QAD.0000000000002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan A.B., Torre D., Lachmann A., et al. ChEA3: transcription factor enrichment analysis by orthogonal omics integration. Nucleic Acids Res. 2019;47(W1):W212–W224. doi: 10.1093/nar/gkz446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesby J.P., Najera J.A., Romoli B., et al. HIV-1 TAT protein enhances sensitization to methamphetamine by affecting dopaminergic function. Brain Behav. Immun. 2017;65:210–221. doi: 10.1016/j.bbi.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesby J.P., Chang A., Najera J.A., Marcondes M.C.G., Semenova S. Brain reward function after chronic and binge methamphetamine regimens in mice expressing the HIV-1 TAT protein. Curr. HIV Res. 2019;17(2):126–133. doi: 10.2174/1570162X17666190703165408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler R.C., Ustun T.B. The world mental health (WMH) survey initiative version of the world health organization (WHO) composite international diagnostic Interview (CIDI) Int. J. Methods Psychiatr. Res. 2004;13(2):93–121. doi: 10.1002/mpr.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalid Elhadi APD., Cadet Jean Lud. Modeling methamphetamine use disorder and relapse in animals: short- and long-term epigenetic, transcriptional., and biochemical consequences in the rat brain. Neurosci. Biobehav. Rev. 2023;155 doi: 10.1016/j.neubiorev.2023.105440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiebala M., Polesskaya O., Yao Z., Perry S.W., Maggirwar S.B. Nuclear factor-kappa B family member RelB inhibits human immunodeficiency virus-1 Tat-induced tumor necrosis factor-alpha production. PLoS One. 2010;5(7) doi: 10.1371/journal.pone.0011875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler-Forsberg O., C N.L., Hjorthoj C., Nordentoft M., Mors O., Benros M.E. Efficacy of anti-inflammatory treatment on major depressive disorder or depressive symptoms: meta-analysis of clinical trials. Acta Psychiatr. Scand. 2019;139(5):404–419. doi: 10.1111/acps.13016. [DOI] [PubMed] [Google Scholar]
- Konradi C., Kobierski L.A., Nguyen T.V., Heckers S., Hyman S.E. The cAMP-response-element-binding protein interacts, but Fos protein does not interact, with the proenkephalin enhancer in rat striatum. Proc. Natl. Acad. Sci. U. S. A. 1993;90(15):7005–7009. doi: 10.1073/pnas.90.15.7005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacagnina M.J., Rivera P.D., Bilbo S.D. Glial and neuroimmune mechanisms as critical modulators of drug use and abuse. Neuropsychopharmacology. 2017;42(1):156–177. doi: 10.1038/npp.2016.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzillotta A., Porrini V., Bellucci A., et al. NF-kappaB in innate neuroprotection and age-related neurodegenerative diseases. Front. Neurol. 2015;6:98. doi: 10.3389/fneur.2015.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawton M.P., Brody E.M. Assessment of older people: self-maintaining and instrumental activities of daily living. Gerontologist. Autumn. 1969;9(3):179–186. [PubMed] [Google Scholar]
- Lazarian G., Yin S., Ten Hacken E., et al. A hotspot mutation in transcription factor IKZF3 drives B cell neoplasia via transcriptional dysregulation. Cancer Cell. 2021;39(3):380–393 e8. doi: 10.1016/j.ccell.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.G., Cho H.Y., Park S.W., Seo M.K., Kim Y.H. Effects of olanzapine on brain-derived neurotrophic factor gene promoter activity in SH-SY5Y neuroblastoma cells. Prog. Neuro-Psychopharmacol. Biol. Psychiatry. 2010;34(6):1001–1006. doi: 10.1016/j.pnpbp.2010.05.013. [DOI] [PubMed] [Google Scholar]
- Li M.D., Wang J., Niu T., et al. Transcriptome profiling and pathway analysis of genes expressed differentially in participants with or without a positive response to topiramate treatment for methamphetamine addiction. BMC Med. Genom. 2014;7:65. doi: 10.1186/s12920-014-0065-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.J., Richter E.I., Okafor C.N., et al. Social genomics of methamphetamine use, HIV viral load, and social adversity. Ann. Behav. Med. 2022;56(9):900–908. doi: 10.1093/abm/kaab096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Brimmers A., van Boekel R.L.M., Vissers K.C.P., Coenen M.J.H. A systematic review of genome-wide association studies for pain, nociception, neuropathy, and pain treatment responses. Pain. 2023;164(9):1891–1911. doi: 10.1097/j.pain.0000000000002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T., Kim P., Luo Y. Tp53 gene mediates distinct dopaminergic neuronal damage in different dopaminergic neurotoxicant models. Neural Regen. Res. 2017;12(9):1413–1417. doi: 10.4103/1673-5374.215243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcondes M.C., Sopper S., Sauermann U., et al. CD4 deficits and disease course acceleration can be driven by a collapse of the CD8 response in rhesus macaques infected with simian immunodeficiency virus. AIDS. 2008;22(12):1441–1452. doi: 10.1097/QAD.0b013e3283052fb5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcondes M.C., Morsey B., Emanuel K., Lamberty B.G., Flynn C.T., Fox H.S. CD8+ T cells maintain suppression of simian immunodeficiency virus in the central nervous system. J. Infect. Dis. 2015;211(1):40–44. doi: 10.1093/infdis/jiu401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massanella M., Gianella S., Schrier R., et al. Methamphetamine use in HIV-infected individuals affects T-cell function and viral outcome during suppressive antiretroviral therapy. Sci. Rep. 2015;5 doi: 10.1038/srep13179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mediouni S., Marcondes M.C., Miller C., McLaughlin J.P., Valente S.T. The cross-talk of HIV-1 Tat and methamphetamine in HIV-associated neurocognitive disorders. Front. Microbiol. 2015;6:1164. doi: 10.3389/fmicb.2015.01164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellon S.H., Wolkowitz O.M., Schonemann M.D., et al. Alterations in leukocyte transcriptional control pathway activity associated with major depressive disorder and antidepressant treatment. Transl. Psychiatry. 2016;6(5) doi: 10.1038/tp.2016.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montojo J., Zuberi K., Rodriguez H., et al. GeneMANIA Cytoscape plugin: fast gene function predictions on the desktop. Bioinformatics. 2010;26(22):2927–2928. doi: 10.1093/bioinformatics/btq562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montojo J., Zuberi K., Rodriguez H., Bader G.D., Morris Q. GeneMANIA: fast gene network construction and function prediction for Cytoscape. F1000Res. 2014;3:153. doi: 10.12688/f1000research.4572.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montojo J., Zuberi K., Shao Q., Bader G.D., Morris Q. Network Assessor: an automated method for quantitative assessment of a network’s potential for gene function prediction. Front. Genet. 2014;5:123. doi: 10.3389/fgene.2014.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran L.M., Aksenov M.Y., Booze R.M., Webb K.M., Mactutus C.F. Adolescent HIV-1 transgenic rats: evidence for dopaminergic alterations in behavior and neurochemistry revealed by methamphetamine challenge. Curr. HIV Res. 2012;10(5):415–424. doi: 10.2174/157016212802138788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran L.M., Booze R.M., Webb K.M., Mactutus C.F. Neurobehavioral alterations in HIV-1 transgenic rats: evidence for dopaminergic dysfunction. Exp. Neurol. 2013;239:139–147. doi: 10.1016/j.expneurol.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins N., Bigdeli T.B., Borglum A.D., et al. GWAS of suicide attempt in psychiatric disorders and association with major depression polygenic risk scores. Am. J. Psychiatr. 2019;176(8):651–660. doi: 10.1176/appi.ajp.2019.18080957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najera J.A., Bustamante E.A., Bortell N., et al. Methamphetamine abuse affects gene expression in brain-derived microglia of SIV-infected macaques to enhance inflammation and promote virus targets. BMC Immunol. 2016;17(1):7. doi: 10.1186/s12865-016-0145-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath A., Anderson C., Jones M., et al. Neurotoxicity and dysfunction of dopaminergic systems associated with AIDS dementia. J. Psychopharmacol. 2000;14(3):222–227. doi: 10.1177/026988110001400305. [DOI] [PubMed] [Google Scholar]
- Nath A., Maragos W.F., Avison M.J., Schmitt F.A., Berger J.R. Acceleration of HIV dementia with methamphetamine and cocaine. J. Neurovirol. 2001;7(1):66–71. doi: 10.1080/135502801300069737. [DOI] [PubMed] [Google Scholar]
- Norman M.A., Moore D.J., Taylor M., et al. Demographically corrected norms for african Americans and caucasians on the hopkins verbal learning test-revised, brief visuospatial memory test-revised, stroop color and word test, and Wisconsin card sorting test 64-card version. J. Clin. Exp. Neuropsychol. 2011;33(7):793–804. doi: 10.1080/13803395.2011.559157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowlin B.T., Burdo T.H., Midkiff C.C., Salemi M., Alvarez X., Williams K.C. SIV encephalitis lesions are composed of CD163(+) macrophages present in the central nervous system during early SIV infection and SIV-positive macrophages recruited terminally with AIDS. Am. J. Pathol. 2015;185(6):1649–1665. doi: 10.1016/j.ajpath.2015.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oughtred R., Rust J., Chang C., et al. The BioGRID database: a comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci. 2021;30(1):187–200. doi: 10.1002/pro.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S.C., Chartoff E.H., Carlezon W.A., Jr., et al. CREB gene transcription factors: role in molecular mechanisms of alcohol and drug addiction. Alcohol Clin. Exp. Res. 2005;29(2):176–184. doi: 10.1097/01.alc.0000153550.31168.1d. [DOI] [PubMed] [Google Scholar]
- Panenka W.J., Procyshyn R.M., Lecomte T., et al. Methamphetamine use: a comprehensive review of molecular, preclinical and clinical findings. Drug Alcohol Depend. 2013;129(3):167–179. doi: 10.1016/j.drugalcdep.2012.11.016. [DOI] [PubMed] [Google Scholar]
- Perkins N.D. Achieving transcriptional specificity with NF-kappa B. Int. J. Biochem. Cell Biol. 1997;29(12):1433–1448. doi: 10.1016/s1357-2725(97)00088-5. [DOI] [PubMed] [Google Scholar]
- Persidsky Y. Insights into end-organ injury in HIV infection: dynamics of monocyte trafficking to the brain in SIV encephalitis. Am. J. Pathol. 2015;185(6):1548–1551. doi: 10.1016/j.ajpath.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapisarda A., Melillo G. Role of the VEGF/VEGFR axis in cancer biology and therapy. Adv. Cancer Res. 2012;114:237–267. doi: 10.1016/B978-0-12-386503-8.00006-5. [DOI] [PubMed] [Google Scholar]
- Ridley M.L., Fleskens V., Roberts C.A., et al. IKZF3/Aiolos is associated with but not sufficient for the expression of IL-10 by CD4(+) T cells. J. Immunol. 2020;204(11):2940–2948. doi: 10.4049/jimmunol.1901283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J.M., Iudicello J.E., Marcondes M.C.G., et al. The combined effects of cannabis, methamphetamine, and HIV on neurocognition. Viruses. 2023;15(3) doi: 10.3390/v15030674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross E.J., Williams R.S., Viamonte M., et al. Overamped: stimulant use and HIV pathogenesis. Curr. HIV AIDS Rep. 2023;20(6):321–332. doi: 10.1007/s11904-023-00672-y. [DOI] [PubMed] [Google Scholar]
- Saloner R., Paolillo E.W., Umlauf A., et al. Conditional effects of lifetime alcohol consumption on methamphetamine-associated neurocognitive performance. J. Int. Neuropsychol. Soc. 2019;25(8):787–799. doi: 10.1017/S1355617719000493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saloner R., Cherner M., Sundermann E.E., et al. COMT val158met genotype alters the effects of methamphetamine dependence on dopamine and dopamine-related executive function: preliminary findings. Psychiatr. Res. 2020;292 doi: 10.1016/j.psychres.2020.113269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanada T., Takaesu G., Mashima R., Yoshida R., Kobayashi T., Yoshimura A. FLN29 deficiency reveals its negative regulatory role in the Toll-like receptor (TLR) and retinoic acid-inducible gene I (RIG-I)-like helicase signaling pathway. J. Biol. Chem. 2008;283(49):33858–33864. doi: 10.1074/jbc.M806923200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherzer C.R., Grass J.A., Liao Z., et al. GATA transcription factors directly regulate the Parkinson’s disease-linked gene alpha-synuclein. Proc. Natl. Acad. Sci. U. S. A. 2008;105(31):10907–10912. doi: 10.1073/pnas.0802437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple S.J., Patterson T.L., Grant I. The context of sexual risk behavior among heterosexual methamphetamine users. Addict. Behav. 2004;29(4):807–810. doi: 10.1016/j.addbeh.2004.02.013. [DOI] [PubMed] [Google Scholar]
- Serrano V.B., Montoya J.L., Campbell L.M., et al. The relationship between vascular endothelial growth factor (VEGF) and amnestic mild cognitive impairment among older adults living with HIV. J. Neurovirol. 2021;27(6):885–894. doi: 10.1007/s13365-021-01001-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin E.J., Jeong J.H., Sharma G., et al. Protein kinase Cdelta mediates methamphetamine-induced dopaminergic neurotoxicity in mice via activation of microsomal epoxide hydrolase. Food Chem. Toxicol. 2019;133 doi: 10.1016/j.fct.2019.110761. [DOI] [PubMed] [Google Scholar]
- Shin E.J., Jeong J.H., Hwang Y., et al. Methamphetamine-induced dopaminergic neurotoxicity as a model of Parkinson’s disease. Arch Pharm. Res. (Seoul) 2021;44(7):668–688. doi: 10.1007/s12272-021-01341-7. [DOI] [PubMed] [Google Scholar]
- Srinivasa S., Fitch K.V., Petrow E., et al. Soluble CD163 is associated with shortened telomere length in HIV-infected patients. J. Acquir. Immune Defic. Syndr. 2014;67(4):414–418. doi: 10.1097/QAI.0000000000000329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark C., Breitkreutz B.J., Reguly T., Boucher L., Breitkreutz A., Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34(Database issue):D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein L.D. Using the reactome database. Curr. Protoc. Bioinformat. 2004 doi: 10.1002/0471250953.bi0807s7. Chapter 8:Unit 8 7. [DOI] [PubMed] [Google Scholar]
- Tjitro R., Campbell L.A., Basova L., et al. Modeling the function of TATA box binding protein in transcriptional changes induced by HIV-1 tat in innate immune cells and the effect of methamphetamine exposure. Front. Immunol. 2018;9:3110. doi: 10.3389/fimmu.2018.03110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasantiuppapokakorn M., Pattanasin S., Kittiyaowamarn R., et al. Depressive symptoms and HIV among a cohort of adolescent young men and transgender women who have sex with men, Bangkok and Nakhon Sawan, Thailand, 2017-2019. AIDS Care. 2024;36(7):974–982. doi: 10.1080/09540121.2024.2354866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J.A., Sulciner M.L., Nowicki K.D., Miller A.D., Burdo T.H., Williams K.C. Elevated numbers of CD163+ macrophages in hearts of simian immunodeficiency virus-infected monkeys correlate with cardiac pathology and fibrosis. AIDS Res. Hum. Retrovir. 2014;30(7):685–694. doi: 10.1089/AID.2013.0268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Xu J., Lazarovici P., Quirion R., Zheng W. cAMP response element-binding protein (CREB): a possible signaling molecule link in the pathophysiology of schizophrenia. Front. Mol. Neurosci. 2018;11:255. doi: 10.3389/fnmol.2018.00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warde-Farley D., Donaldson S.L., Comes O., et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38(Web Server issue):W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X., Zhang C., Grigoroiu-Serbanescu M., et al. The cAMP responsive element-binding (CREB)-1 gene increases risk of major psychiatric disorders. Mol. Psychiatr. 2018;23(9):1957–1967. doi: 10.1038/mp.2017.243. [DOI] [PubMed] [Google Scholar]
- Zauli G., Secchiero P., Rodella L., et al. HIV-1 Tat-mediated inhibition of the tyrosine hydroxylase gene expression in dopaminergic neuronal cells. J. Biol. Chem. 2000;275(6):4159–4165. doi: 10.1074/jbc.275.6.4159. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data can be available upon request.
We have submitted all raw and processed data to NCBI Gene Expression Omnibus (GEO) and it is available with accession number GSE273630.