Abstract
An outbreak of OXA-23-producing carbapenem-resistant Acinetobacter baumannii amongst ICU-patients with COVID-19 likely occurred by transmission through inanimate surfaces, potentially facilitated by a contaminated positioning pillow shared between patients. Subsequent rapid spread may have been caused by exposure to respiratory secretions contaminating healthcare worker’s gloves and gowns during prone positioning.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13756-024-01485-3.
Introduction
Challenges in adhering to infection prevention and control guidance and escalation of antimicrobial usage during the COVID-19 pandemic led to a surge of hospital-acquired multidrug-resistant bacteria, including carbapenem-resistant Acinetobacter baumannii [1, 2].
In this report, we reveal potential reservoirs and transmission routes implicated in an outbreak of OXA-23-producing carbapenem-resistant A. baumannii (CRAB) among intensive care unit (ICU) patients with COVID-19.
Methods
Setting
The University Hospital Basel (USB) is a tertiary care center in the northwestern part of Switzerland with over 40,000 hospital admissions annually. During the COVID-19 pandemic, patients with SARS-CoV-2 infection were frequently repatriated from institutions abroad and transferred between various Swiss hospitals based on the availability of hospital resources. At the USB, COVID-19 patients needing intensive care, were cohorted in a dedicated area of the ICU.
In September 2021, a bronchial sample from one patient in this dedicated ICU cohort revealed OXA-23-producing CRAB. Subsequent outbreak investigation included environmental and patient screening, collection of epidemiological and clinical data and performance of detailed infection prevention and control audits. All CRAB derived from patients and environmental samples were analyzed using next generation sequencing (NGS) to investigate genetic relationship. As a quality assessment project, the Ethics Commission of Northwestern and Central Switzerland (EKNZ) confirmed that no approval was required (EKNZ-Request-2023-00647) so that the need to provide a consent to participate is waived.
Audits
Control audits were performed by the infection prevention and control team and had two main focuses. First, the handling of personal protective equipment (PPE) by health care workers (HCW), especially during prone positioning of the patients in the ICU was audited. Secondly, the cleaning and disinfection procedures performed by the environmental services staff were looked at to uncover any potential breach.
Screening
Upon detection of the outbreak, all patients hospitalized on the entire ICU were screened for colonization with CRAB. The following body sites were screened to assess colonization: wounds, catheter insertion sites, rectum, urine, tracheal secretion for ventilated patients and nasopharyngeal swabs for non-ventilated patients. Swabing was performed using eSwab® (Copan). Thereafter cross-sectional screenings of all patients in the COVID-19 cohort were performed on a weekly basis.
Due to the regular exchange of patients between the COVID-19 cohorts of the ICU and the general ward, all patients hospitalized in the COVID-19 cohort of the general ward were screened likewise. Patients were screened for colonization with CRAB of the rectum, urine, wounds, and puncture sites, as well as the respiratory tract if mechanically ventilated.
For outbreak investigation purposes, environmental screening swabs of high-touch areas were obtained to identify potential transmission sources.
Microbiological analysis
Search of bacterial growth from clinical specimens was performed using different agar media according to standard bacteriological procedures. Samples for screening for CRAB from patients as well as environmental samples were plated onto selective chromogenic agar plates (chromID® CARBA SMART agar, bioMérieux, Marcy-l’Étoile, France). Colorless bacterial colonies were identified by MALDI-TOF mass spectrometry (Bruker Daltonics, Bremen, Germany). The Vitek 2® system (bioMérieux, Marcy-l’Étoile, France) was used for antimicrobial susceptibility testing of all isolates. Molecular detection of CRAB was performed by eazyplex® SuperBug complete A kit (AmplexDiagnostics, Gars-Bahnhof, Germany) using LAMP technology on a Genie II instrument (AmplexDiagnostics) detecting all relevant carbapenemase genes of A. baumannii.
Next generation sequencing (NGS)
All CRAB strains were whole genome sequenced. Bacterial DNA was extracted with the EZ1 DNA Tissue kit (QIAGEN, Hilden, Germany) in the EZ1 advanced XL workstation (QIAGEN), according to manufacturer’s recommendations. Genomic libraries were prepared using the Illumina DNA Prep kit (Illumina, San Diego) and whole genome sequencing was performed on the NextSeq500 platform (Illumina, San Diego) (read length 2 × 150 bp). Assemblies (unicycler v0.3.0b) were analyzed in Ridom Seqsphere + v7.7.5 using the published core genome MLST scheme [3, 4]. Whole Genome Sequencing read data can be accessed at NCBI sequence read archive https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA977488 [Reviewer Link to be removed post acceptance: https://dataview.ncbi.nlm.nih.gov/object/PRJNA977488?reviewer=mnmrqcmui7n6nv0dqgrm4lbum3].
Results
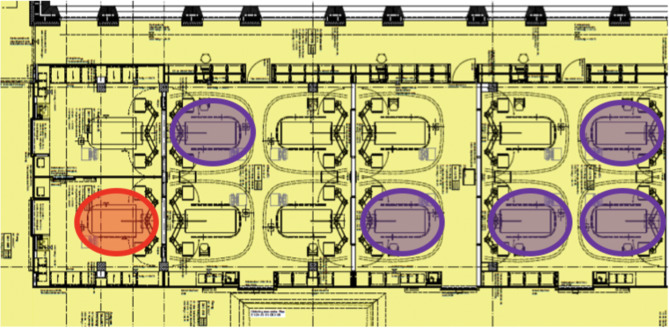
From July to October 2021, OXA-23-producing CRAB was recovered from six patients hospitalized in the COVID-19 cohort-ward of the ICU of the USB. The index patient had been repatriated from Serbia in July 2021 due to COVID-19 and placed under contact and droplet precautions in a single room during the entire hospital stay. OXA-23-producing CRAB was detected in a screening sample collected at admission from the urine and, during the course of hospitalization, from clinical respiratory tract specimens. Seventy-six days after the index patient’s death, massive counts of OXA-23-producing CRAB were recovered from a clinical respiratory tract specimen of a second COVID-19 patient and subsequent screening revealed CRAB colonization of the respiratory tract in four additional COVID-19 patients in neighboring rooms within six days. Figure 1 illustrates the floor plan and the location of all affected patients within the ICU COVID-19 cohort. Detailed patient characteristics, sites of colonization, need for antibiotic treatment due to infection with OXA-23-producing CRAB and outcome are summarized in supplementary Table 1. In addition, supplementary Table 2 details the results of antimicrobial resistance testing.
Fig. 1.
Floor plan of the COVID-19 cohort within the intensive care unit. Red circle: index patient with OXA-23-producing Acinetobacter baumannii in 07/2021; purple circles: patients with detection of OXA-23-producing A. baumannii throughout the cohort within six days, two months later
Audits revealed that prone positioning was performed by dedicated teams without consistent change of gloves and gowns between patients within the cohort. Contamination risk of gloves and gowns is high, due to close body contact, and contact with respiratory secretions, especially while stabilizing the tracheal tube during prone positioning. Some of the positioning pillows removed during repositioning were stored in non-patient-specific areas without immediate disinfection, resulting in potential contamination of surfaces.
A total of 100 environmental samples were collected from high-touch surfaces and equipment shared between patients within the cohort (supplementary Table 3). Among those, OXA-23-producing A. baumannii was recovered from three samples collected from a cuff pressure manometer, a drawer handle in proximity of the colonized patients, as well as from a reprocessed defective positioning pillow for the head used for prone positioning. Air samples taken in proximity of patients colonized with CRAB using an MAS-100 NT air sampling device (MBV, Stäfa, Switzerland) remained negative.
The genomes of all CRAB isolates recovered from clinical and environmental specimens were identified as belonging to Pasteur sequence type 2 and considered highly related as they differed by a maximum of one core genome allele (indicating a change of some sort, i.e. single or multiple single nucleotide polymorphisms) based in just one allele on NGS analyses. This sequence type had not been identified at our hospital within the previous four years.
After recognition of the outbreak, patients positive with OXA-23-producing CRAB were cohorted within the COVID-19 cohort. Reinforced infection prevention and control measures were introduced, such as establishing gloves and gown exchange for HCW between patients cohorted within the COVID-19 cohort after intense patient contact. When possible, care of patients colonized with CRAB was provided by a dedicated team to minimize staff turnover between rooms.
Repeated cleaning and disinfection, including UVC irradiation of the entire ICU COVID-19 cohort and H2O2 disinfection of mobile devices from affected rooms was performed. Materials such as positioning pillows for prone positioning were disposed of in case of visible damage. Washable materials were washed at a minimum of 60 °C. Surfaces in the COVID-19 cohort and dedicated OXA-23-producing CRAB cohort were disinfected twice daily. Cuff pressure manometers were replaced by disposable devices.
After implementation of these measures no further cases were detected in weekly cross-sectional screenings of patients hospitalized in the COVID-19 cohort of the ICU or on the general ward until December 2021 and the outbreak was declared terminated. Among 175 patients identified as having had either direct contact with one of the patients colonized or with the dedicated COVID-19 cohort on the ICU during the outbreak, 80 patients were able to be screened, all of whom tested negative for CRAB.
Discussion
We report an outbreak of genetically highly related OXA-23-producing CRAB among six patients within a COVID-19 ICU cohort. We hypothesize that transmission of a strain of OXA-23-producing CRAB from the index patient to the first contact patient occurred through inanimate surfaces, potentially facilitated by a contaminated positioning pillow shared between patients within the COVID-19 cohort. Subsequent rapid spread may have been caused by exposure to respiratory secretions contaminating HCW’s gloves and gowns during prone positioning. The latter is supported by initial colonization of the respiratory tract with high bacterial loads.
During the COVID-19 pandemic, numerous outbreaks with multidrug resistant bacteria, especially CRAB were reported worldwide [5–8]. In Italy, for example, incidence of CRAB increased from 5.1/100,000 ICU patient days pre-pandemic to 26.4/100,000 during pandemic [9]. Our outbreak and others support the importance of contaminated surfaces and equipment in facilitating ongoing transmission.
A. baumannii has been shown to be able to survive for prolonged periods on dry surfaces [10] and Gottesmann and colleagues [5] showed that inconsistent cleaning and disinfection of the surrounding patient area (medication room) led to an outbreak with CRAB in their COVID-19 dedicated hospital, shortly after reopening the COVID-19 dedicated wards. Low adherence to proper use of PPE, especially in times of high workload and low staffing and breaches in environmental cleaning, as experienced during the COVID-19 pandemic, have all been implicated as facilitating the spread of multidrug-resistant organisms [11, 12].
The outbreak in our institution was terminated by reinforcement of the cleaning and disinfection procedures of the patient environment and equipment shared within the cohort as well as introduction of consistent changes of PPE when moving between patients and within the cohort. We acknowledge the limitation of not having performed systematic skin screening to detect colonization with A. baumannii and has recently been shown to be the most sensitive site for its detection [13]. Colonization of the skin was, however, accounted for to a limited degree during our outbreak by screening catheter insertion sites and wounds.
Conclusion
Positioning pillows shared between patients may serve as reservoirs for multidrug-resistant organisms and are challenging to reprocess by standard cleaning and disinfection regimens. Prone positioning with exposure to respiratory secretions may constitute a potential transmission-route within COVID-19-cohorts. Initial colonization of the respiratory tract with high bacterial loads, suggests transmission by contact within proximity of the patient’s airway. Screening of the respiratory tract, in addition to performance of skin sampling by established methods, may be considered during outbreak investigations involving mechanically ventilated patients.
Screening of the respiratory tract, in addition to performance of skin sampling by established methods, may be considered during outbreak investigations involving mechanically ventilated patients.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Magdalena Schneider, Elisabeth Schultheiss, Clarisse Straub, Rosa-Maria Vesco, Daniel Gander, Valerie Courtet and Vincent Hartmann (USB) for excellent technical assistance. Bioinformatic analyses were partially performed at sciCORE (http://scicore.unibas.ch), the scientific computing center at the University of Basel.
Author contributions
SZ collected, analyzed and interpreted the data and wrote the first draft of the manuscript.SK, MvR, AP, RK collected, analyzed and interpreted the data and revised the manuscript.AE, HSS, PS, DG analyzed and interpreted the data and revised the manuscript.SB, SM, HP interpreted the data and revised the manuscript.STS conceptualized the study analyzed and interpreted the data and revised the manuscript.
Funding
This study was supported by the University Hospital Basel and the University Basel.
Data availability
Whole Genome Sequencing read data can be accessed at NCBI secuence read archive https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA977488 [Reviewer Link to be removed post acceptance: https://dataview.ncbi.nlm.nih.gov/object/PRJNA977488?reviewer=mnmrqcmui7n6nv0dqgrm4lbum3].
Declarations
Competing interests
S. Tschudin-Sutter serves as editor of the journal Antimicrobial Resistance and Infection Control.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.CDC, COVID-19:. U.S. Impact on Antimicrobial Resistance, Special Report 2022. Atlanta, GA: U.S. Department of Health and Human Services, CDC; 2022. https://www.cdc.gov/drugresistance/covid19.html.
- 2.Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022;399(10325):629–655. doi: 10.1016/S0140-6736(21)02724-0. Epub 2022 Jan 19. Erratum in: Lancet. 2022;400(10358):1102. PMID: 35065702; PMCID: PMC8841637. [DOI] [PMC free article] [PubMed]
- 3.Higgins PG, Prior K, Harmsen D, Seifert H. Development and evaluation of a core genome multilocus typing scheme for whole-genome sequence-based typing of Acinetobacter baumannii. PLoS ONE. 2017;12(6):e0179228. 10.1371/journal.pone.0179228. PMID: 28594944; PMCID: PMC5464626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13(6):e1005595. 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gottesman T, Fedorowsky R, Yerushalmi R, Lellouche J, Nutman A. An outbreak of carbapenem-resistant Acinetobacter baumannii in a COVID-19 dedicated hospital. Infect Prev Pract. 2021;3(1):100113. 10.1016/j.infpip.2021.100113. Epub 2021 Jan 9. PMID: 34316574; PMCID: PMC7794049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Russo A, Gavaruzzi F, Ceccarelli G, Borrazzo C, Oliva A, Alessandri F, Magnanimi E, Pugliese F, Venditti M. Multidrug-resistant Acinetobacter baumannii infections in COVID-19 patients hospitalized in intensive care unit. Infection. 2022;50(1):83–92. 10.1007/s15010-021-01643-4. Epub 2021 Jun 27. PMID: 34176088; PMCID: PMC8236000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shinohara D, Dos Santos Saalfeld S, Martinez H, Altafini D, Costa B, Fedrigo N, Tognim M. Outbreak of endemic carbapenem-resistant Acinetobacter baumannii in a coronavirus disease 2019 (COVID-19)–specific intensive care unit. Infect Control Hosp Epidemiol. 2022;43(6):815–7. 10.1017/ice.2021.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perez S, Innes GK, Walters MS, Mehr J, Arias J, Greeley R, Chew D. Increase in Hospital-Acquired Carbapenem-Resistant Acinetobacter baumannii infection and colonization in an Acute Care Hospital during a Surge in COVID-19 admissions - New Jersey, February-July 2020. MMWR Morb Mortal Wkly Rep. 2020;69(48):1827–31. 10.15585/mmwr.mm6948e1. PMID: 33270611; PMCID: PMC7714028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pascale R, Bussini L, Gaibani P, Bovo F, Fornaro G, Lombardo D, Giannella M. Carbapenem-resistant bacteria in an intensive care unit during the coronavirus disease 2019 (COVID-19) pandemic: a multicenter before-and-after cross-sectional study. Infect Control Hosp Epidemiol. 2022;43(4):461–6. 10.1017/ice.2021.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Green, et al. Bacterial hydrophilins promote pathogen dissication tolerance. Cell Host Microbe. 2022. 10.1016/j.chom.2022.03.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thoma R, Seneghini M, Seiffert SN, et al. The challenge of preventing and containing outbreaks of multidrug-resistant organisms and Candida Auris during the coronavirus disease 2019 pandemic: report of a carbapenem-resistant Acinetobacter baumannii outbreak and a systematic review of the literature. Antimicrob Resist Infect Control. 2022;11:12. 10.1186/s13756-022-01052-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Segala FV, Bavaro DF, Di Gennaro F, Salvati F, Marotta C, Saracino A, Murri R, Fantoni M. Impact of SARS-CoV-2 epidemic on Antimicrobial Resistance: A literature review. Viruses. 2021;13(11):2110. 10.3390/v13112110. PMID: 34834917; PMCID: PMC8624326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nutman A, Levi GD, Keren-Paz A, et al. Active surveillance for carbapenem-resistant Acinetobacter baumannii (CRAB) carriage. Microbiol Spectr. 2023;11(6):e0314623. 10.1128/spectrum.03146-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Whole Genome Sequencing read data can be accessed at NCBI secuence read archive https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA977488 [Reviewer Link to be removed post acceptance: https://dataview.ncbi.nlm.nih.gov/object/PRJNA977488?reviewer=mnmrqcmui7n6nv0dqgrm4lbum3].