Abstract
It has been hypothesized that the major immediate-early (MIE) enhancer of cytomegalovirus (CMV) is important in determining virus tropism and latency because of its essential role in initiating the cascade of early gene expression necessary for virus replication. Although rat CMV (RCMV) and murine CMV (MCMV) exhibit extreme species specificity in vivo, they differ in their ability to replicate in tissue culture. MCMV can replicate in a rat embryo fibroblast (REF) cell line while RCMV does not grow in murine fibroblasts. The tropism is not due to a block in virus entry into the cell. We have constructed a recombinant RCMV in which the RCMV MIE enhancer has been replaced with that of MCMV. Growth of the recombinant virus in tissue culture remains restricted to rat cells, suggesting that other viral and/or host factors are more important in determining in vitro tropism. Unlike findings using recombinant MCMV in which the human CMV (HCMV) MIE enhancer substitutes for the native one (A. Angulo, M. Messerle, U. H. Koszinowski, and P. Ghazal, J. Virol. 72:8502–8509, 1998), infection with our recombinant virus at a low multiplicity of infection resulted in a substantial decrease in virus replication. This occurred despite comparable or increased MIE transcription from the recombinant virus. In vivo experiments showed that the recombinant virus replicates normally in the spleen during acute infection. Notably, the recombinant virus appears to be deficient in spreading to the salivary gland, suggesting a role for the MIE enhancer in tropism for certain tissues involved in virus dissemination. Four months after infection, recombinant virus with the foreign MIE enhancer was reactivated from spleen explants.
The cytomegaloviruses (CMVs) are betaherpesviruses characterized by extreme species specificity. The initial infection (often asymptomatic) leads to the development of latent states in multiple tissues, and during periods of immunosuppression, the virus can reactivate and cause severe and sometimes fatal disease. The transcription of CMV major immediate-early (MIE) genes is regulated by strong enhancers located upstream of the MIE promoter. The MIE proteins are made immediately after infection, and the MIE RNA can be transcribed under conditions that inhibit protein synthesis. This suggests that cellular and/or virion proteins are important for the activation of viral MIE genes. Herpes simplex virus MIE gene expression is enhanced by the virion protein VP16 (5). Similarly, the virion protein UL82 of human CMV (HCMV) can increase transcription of reporter genes regulated by the MIE enhancer (19).
Although the genetic architecture of the MIE loci for all the CMVs studied in detail is similar, the organizations of their respective MIE enhancers are quite different. The HCMV and the simian CMV (SCMV) enhancers share many of the same potential transcription binding sites, but the locations and numbers of these sites are different (7, 10). The murine CMV (MCMV) MIE enhancer spans approximately 700 bp containing six consensus binding sites for NF-κB and seven consensus binding sites for AP1 (11). The Maastricht rat CMV (RCMV), in contrast, contains no NF-κB sites and has more AP-1 sites than MCMV (6). Unlike the MIE enhancers mentioned above, the English RCMV enhancer possesses many fewer recognizable transcription factor binding sites or repeat sequences (29). Other than a TATA box, the only obvious consensus binding sites in the RCMV MIE enhancer are three CCAAT box transcription factor sites, one NF-κB site, and a few AP-1 binding sites. Despite the relative scarcity of known transcription factor binding sites, the English RCMV MIE enhancer activity compares well to those of HCMV and MCMV in transient-transfection assays of human, mouse, monkey, and rat cells (29).
Regulation of IE expression is thought to be important in whether an infection will be abortive or lytic or become latent. Whether an enhancer is activated or repressed could be due to the state of differentiation of the infected cell. For example, in vitro infection of human monocytes with HCMV leads to an abortive infection with little, if any, expression of IE proteins. Differentiation of the infected monocytes leads to abundant expression of IE proteins and a permissive infection (15, 17, 24). The change in the state of differentiation could reflect changes in the type or quantity of repressor and/or activator proteins, and these proteins could affect the MIE enhancer. Increased expression from the MIE enhancer can also be due to a response to external stimuli. For example, both HCMV and MCMV MIE enhancers contain retinoic acid response elements and the addition of physiological levels of retinoic acid leads to increased enhancer activity (1, 2).
The restricted tropism of the CMVs is a hallmark of these viruses. We have attempted unsuccessfully to infect several strains of mice (A/J, BALB/c, C57BL, C3H) as well as nude and SCID mice with RCMV (including direct salivary gland inoculation). Likewise, we have been unable to infect rats with MCMV. RCMV will not replicate in mouse embryo fibroblasts (MEF) or NIH 3T3 cells, although MCMV will grow to high titers in a line of rat embryo fibroblasts (REF). Generally, the species specificities exhibited by these viruses appear to be inherent properties of the viruses. Because the CMV MIE enhancer influences expression of the IE proteins, it could be that some of the characteristics of RCMV and MCMV in vitro and in vivo replication are due to virus-specific elements or their arrangement in the enhancer. Using MIE enhancer deletions and swaps, evidence against the MIE enhancer playing a decisive role in determining organ and species specificity for MCMV has been recently presented by two groups (4, 13). We have constructed similar enhancer-substituted recombinants of the English RCMV in order to determine the role of the MIE enhancer in RCMV biology. Our experiments are in general agreement with those reported for MCMV in that the MIE enhancer is not the sole determinant of viral tropism. However, we did observe phenotypic changes dependent on the source of the MIE enhancer. In addition, we show that in vitro reactivation from latency is not dependent on the presence of the native MIE enhancer. Continued studies with this recombinant virus will aid in understanding the role of the MIE enhancer in the pathogenesis of RCMV infection.
MATERIALS AND METHODS
Virus and cell cultures.
RCMV obtained from J. Hamilton (Duke University) was originally isolated and described by Priscott and Tyrrell (28). Virus was propagated in a REF cell line as previously described (8). We called this the English RCMV isolate to distinguish it from the Maastricht isolate, which appears to be a virus distinct from ours (6). REF, MEF, and NIH 3T3 cells were grown in minimum essential medium supplemented with 10% fetal calf serum, 2 mM glutamine, and gentamicin (20 μg/ml). An expression cassette containing the Cre recombinase gene (pBS185) was cotransfected with pSV2neo (Clontech, Palo Alto, Calif.) in REF cells, and a cell line was selected with G418 (400 μg/ml). This line expresses the Cre recombinase and is useful for recombining DNA between appropriately oriented loxP sites within viruses that can infect these cells.
Plasmid constructs.
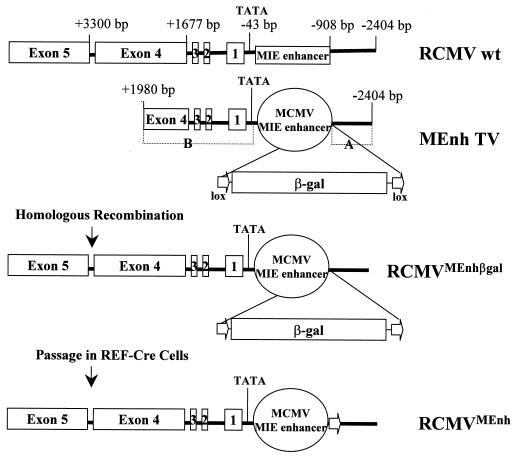
The RCMV KpnI I fragment, which spans the entire MIE enhancer and continues past exon 4 of IE1, was used as the basis for construction of our transfer vectors (Fig. 1). To form one flanking region (Fig. 1, MEnh TV, region A), an EcoRI-PstI fragment from bp −908 to −2404 (all RCMV positions are relative to the MIE cap site, which was set as +1) was cut from KpnI I and blunted, and KpnI linkers were ligated to both ends. This fragment was cloned into the KpnI I site of pSK (Stratagene, La Jolla, Calif.) to form pSKA. The other flanking region (Fig. 1, MEnh TV, region B) was constructed to contain the sequence from bp −49 (10 bp upstream of the TATA box) to bp +1980 and cloned into the plasmid containing the A flanking region described above. This was achieved by restricting RCMV KpnI I with FspI, which cuts in exon 4 of IE1 at bp +1980. The FspI site was changed to KpnI by blunting with Klenow and by the ligation of KpnI linkers. The fragment was then cut with HindIII, which cuts at bp +174, and the resulting 1,806-bp HindIII-KpnI fragment was cloned into pSK. PCR was used to amplify the region 5′ of the TATA box (bp −43) to the HindIII site (bp +174) in exon 1. The primer beginning at bp −49 contained an SpeI site, and the resulting SpeI-HindIII fragment (223 bp) was ligated into the plasmid containing the 1,806-bp HindIII-KpnI fragment to create pSKB. In order to move this flanking region (B) into the final transfer vector, the KpnI site was changed to SpeI prior to cutting it and ligating it into the SpeI site of pSKA. All PCR-amplified sequences and appropriate orientations of ligated fragments were confirmed by sequence data from the completed transfer vectors.
FIG. 1.
Generation of a recombinant RCMV (RCMVMEnh) that contains the MCMV MIE enhancer in place of the RCMV MIE enhancer. The DNA to be inserted includes the MCMV MIE enhancer from bp −31 to −835 relative to the MCMV IE1 cap site (therefore not including the MCMV TATA box) and the lox–β-Gal–lox expression cassette, which allows for identification of recombinant virus. The RCMVMEnh transfer vector (MEnh TV) contains the MCMV MIE enhancer, a β-Gal expression cassette with lox sequences at each end, flanked by two stretches of RCMV DNA (A and B), which allow for homologous recombination to occur. Region A contains the RCMV DNA sequence from bp −908 to −2404 relative to the cap site of RCMV IE1. Region B contains the RCMV DNA sequence from bp −49 to +1980 relative to the RCMV IE1 cap site and therefore contains the RCMV MIE TATA box-to-cap site region. Successful recombination results in a recombinant RCMV containing the MCMV MIE enhancer and the lox–β-Gal–lox cassette in place of the wt enhancer. Passage through the REF-Cre cell line recombines out the β-Gal expression cassette, leaving one lox site.
The MCMV MIE enhancer was amplified from pAMB25, which contains the entire MCMV MIE enhancer and coding regions and which was a gift from G. Keil (16). PstI-tailed primers from bp −835 to −31 relative to the MCMV IE1 cap site were used to isolate the 804-bp region of the MCMV MIE enhancer, and the sequence was verified and cloned along with a beta-galactosidase (β-Gal) expression cassette flanked by LoxP sites into the transfer vector.
To repair the MCMV enhancer RCMV recombinant virus (RCMVMEnh), the RCMV MIE enhancer from bp −47 to −908 was amplified using PstI-containing PCR primers and cloned into the PstI site in the transfer vector in place of the MCMV MIE enhancer to create the RCMV enhancer repair transfer vector.
The transfer vector for the generation of the wild-type (wt) β-Gal-expressing recombinant virus (RCMVβgal) contained the RCMV sequence from bp +178 bp to −4912 relative to the MIE cap site. The lox–β-Gal–lox expression cassette was cloned into the unique XhoI (blunted) site located at bp −1817 relative to the MIE cap site to create the appropriate transfer vector.
Transfection and recombinant virus isolation.
The methods of construction of the recombinant viruses are depicted schematically in Fig. 1. Subconfluent REF cells on six-well plates were cotransfected with transfer vector DNA (1 μg) and virion RCMV DNA (1 μg) by the calcium phosphate precipitation method as described previously (26). Supernatants from wells showing a cytopathic effect were frozen and screened for β-Gal-positive virus. For screening, REF cultures were infected and medium containing 0.9% methylcellulose was added to each well. The plates were incubated for 5 days at 37°C in 5% CO2, fixed, and stained for β-Gal-positive plaques as previously described (30). Recombinant virus was isolated by limiting-dilution cultures on REF cells in 96-well plates.
The repaired virus (RCMVrep) was generated by cotransfection of the RCMV enhancer repair transfer vector and virion DNA which had been extracted from the RCMVMEnh virus (β-Gal negative). The RCMVβgal virus was isolated following cotransfection with the lox–β-Gal–lox transfer vector and wt RCMV virion DNA.
Viral DNA analysis.
Virion DNA was prepared and used for Southern blot analysis as previously described (8). PCR-amplified MCMV and RCMV enhancers were used as probes. A nick translation system (Gibco-BRL, Gaithersburg, Md.) was used with [α-32P]dCTP (Amersham, Arlington Heights, Ill.) to label the probes. To sequence around the junctions of homologous recombination in recombinant and the repaired viruses, PCR-amplified products were cloned in pSK and sequenced using T3 and T7 primers and Sequenase v2.0 (Amersham).
Real-time quantitative PCR analysis.
Total-cell RNA was isolated at various times postinfection with wt virus, RCMVMEnh, and RCMVrep and was extracted with TRIzol (Gibco-BRL) by following the manufacturer's recommended procedures. Quantitation of MIE transcripts was performed with real-time PCR using the 7700 Sequence Detection System (PE Biosystems, Foster City, Calif.). This method is based on continuous optical monitoring of a fluorogenic probe (12, 14). Initial template concentration was derived from the cycle number (CT) at which the fluorescent signal crosses a threshold in the exponential phase of the PCR. The software default values were used in determining the threshold. Standard graphs of CT values obtained from serially diluted positive controls were used to derive values for unknowns. Each quantitation was done from plasmid standard curves with at least five values spanning 4 orders of magnitude or more. Each standard was determined in triplicate. The correlation coefficients calculated for each standard curve were as follows: 0.9737 for IE1, 0.9916 for IE2, and 0.9942 for GAPDH (glyceraldehyde-3-phosphate dehydrogenase).
Two separate primer-probe sets were generated for RCMV IE1 and RCMV IE2. The forward primer is common to both IE1 and IE2, being located in exon 2 of the RCMV MIE region. The reverse primer for IE1 is located in exon 4 and the reverse primer for IE2 is located in exon 5. Primer sequences were as follows: exon 2 forward, 5′-GATGAAGTGCGTGAGTCGGTAAATCAA-3′; IE1 exon 4, 5′-TTGGATGCATGTCGTGCGGATGTCT-3′; and IE2 exon 5, 5′-ATGGTCTCTCTGTTGATCCGGA ATATC-3′. The probes for IE1 and IE2 were designed to span the exon-exon border of exon 3 and exon 4 or exon 3 and exon 5, respectively, to eliminate concerns of DNA contamination. Probe sequences were as follows: IE1, 6FAM-CAGCCGTTCAAAGTCTTGTAATTGCCATCAAGACCGCG-TAMRA, and IE2, VIC-CAGCCGGGCCTAACGTGAGGCATATAGACATTGTTAC-TAMRA. PCR primers were synthesized by Life Technologies, Inc. (Gaithersburg, Md.), and fluorogenic probes were synthesized by PE Biosystems. Reverse transcriptase reactions used approximately 0.5 μg of RNA isolated using the TRIzol extraction method and Thermoscript Reverse Transcription kit (Gibco-BRL) in the presence of random hexamers. The reverse transcription reaction for each RNA sample was split into three separate PCRs for IE1, IE2, and GAPDH. The calculated copy number for each RNA sample was normalized to its concentration calculated by the GAPDH content. Each sample was tested in triplicate, and the mean of the three values is shown as the calculated copy number of the sample. Real-time PCRs were performed with universal master mix and universal cycling conditions (PE Biosystems).
In vitro infections.
REF cultures were infected with wt virus, RCMVMEnh, or RCMVrep at a high multiplicity of infection (MOI) of 10 PFU/ml or a low MOI of 0.01 PFU/ml. After a 1-h adsorption, the wells were washed three times with minimum essential medium. At various times after infection the plates were frozen and thawed, and the amount of infectious virus was determined by plaque assay on REF cells.
In vivo infections.
Six-week-old female Sprague-Dawley rats were infected intraperitoneally (i.p.) with mixed or individual virus pools. Spleens were harvested on days 2, 3, and 5, and salivary glands were harvested on day 16 postinfection. Following euthanasia with CO2, organs used for the detection of active virus infection were removed, minced, and sonicated, and titers were determined for REF cells in 24-well plates with and without methylcellulose overlays. Approximately one-half of the spleen and one-half of each salivary gland was used for sonication and titration. For the mixed-virus infections, the proportion of test virus to RCMVβgal was determined by staining first for β-Gal-positive virus and then staining with methylene blue.
To observe for latency and reactivation, rats were infected with either the mixed-virus pools or RCMVMEnh alone and spleens and salivary glands were harvested 120 days postinfection. One-third of each spleen and one-half of each salivary gland were tested for persistent virus as described above. One-tenth of each organ was frozen for DNA extraction and PCR analysis (see below). Fragments of spleen and salivary gland (1 to 2 mm3) from each rat were placed on REF cells in each well of a tc-6 plate (six fragments per well, approximately 5% of each organ per plate). Medium was changed every 2 to 3 days for up to 50 days or until virus was detected. Plates were examined daily for cytopathic effect, and twice weekly, supernatants were titrated for levels of virus in fresh REF cells. Plaques from isolated virus were stained for β-Gal and/or with methylene blue.
The sensitivity of our culture technique to detect infectious virus was determined by reconstruction experiments. Aliquots of 10 μl of medium containing 5, 50, or 500 PFU of wt RCMV was mixed with 500 μl of medium, salivary gland sonicate, or spleen sonicate, and titers in REF cultures were determined in the usual manner. Likewise, similar aliquots of virus were added to medium, salivary gland, or spleen before sonication and titration. Addition of virus to the sonicates or to the organs or medium prior to sonication resulted in a reduction of virus titer of approximately 50% in all cases when compared to the virus in the unsonicated medium (control). We estimate conservatively that we could detect at least 5 infectious units per organ.
Detection of RCMVMEnh and RCMVβgal by PCR.
DNA was extracted from spleen and salivary glands from rats 4 months postinfection. Approximately one-tenth of each organ was minced and lysed in TE9 (500 mM Tris-hydrochloride, pH 9.0, 20 mM EDTA, 10 mM NaCl), 0.5 mg of proteinase K (Sigma, St. Louis, Mo.)/ml, and 1% sodium dodecyl sulfate (Sigma) at 50°C for 1 h, passed through a 16-gauge needle four times, and incubated at 50°C another 2 h. The DNA was extracted multiple times in PC-9 (3 parts water-saturated phenol, 2 parts TE9, and 4 parts chloroform), followed by one extraction with chloroform, and precipitated in 0.3 M sodium acetate (pH 5.7) and 2.5 volumes of ethanol.
Nested PCR was used to detect RCMVMEnh DNA. Outer primers for RCMVMEnh were 5′-AGCCAAGTACACTGACTC-3′ (from −594 relative to the MCMV IE1 cap site) and 5′-CTGAGAACTGCGTTCCAC-3′ (from +26 relative to the cap site of RCMV IE1; antisense). Inner primer sequences were 5′-AACGCCATGTACTTTCCC-3′ (from −255 relative to the MCMV IE1 cap site) and 5′-AATTTCCAGGGGAAAACC-3′ (from −64 relative to the MCMV IE1 cap site; antisense), which should generate a 181-bp fragment. One microgram of DNA was used in the initial round of PCR, which consisted of one cycle of 94°C for 3 min followed by 30 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min. For the second round of PCR, 2 μl of DNA from the first round was used as template with the same PCR conditions as the first round except that the reaction mixture was amplified for 35 cycles. Both rounds were followed by a 7-min extension period at 72°C. PCR products (20 μl) were separated by electrophoresis on 2% agarose gels visualized by ethidium bromide staining. Southern blotting was performed by standard methods. Oligonucleotides representing sequences within the inner primer amplifications were labeled with 32P using T4 kinase (Gibco-BRL) by following the manufacturer's directions. The RCMVMEnh probe was 5′-CATAGCTGATTAATGGGA-3′ (from −177 relative to MCMV IE1 cap site). Hybridization was performed as previously described (8) except that the temperature and time for prehybridization and hybridization were 42°C and 2 h. Membranes were washed three times for 5 min at 42°C in 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate).
Direct salivary gland inoculation.
Rats were anesthetized with sodium pentobarbital, and the salivary glands were exposed. Approximately 105 PFU of either wt RCMV or RCMVMEnh was injected under direct visualization using a 30-gauge needle, and the wounds were clipped. Rats were sacrificed after 5 days and the salivary glands were processed and cultured for virus as described above.
RESULTS
Isolation and Southern blot analysis of the RCMVMEnh recombinant virus.
Cotransfection of MCMV enhancer transfer vector and RCMV virion DNA led to approximately 1 in 5,000 virus plaques being positive for β-Gal production. Virus expressing β-Gal (RCMVMEnhβgal) was isolated by multiple rounds of limiting-dilution cultures on 96-well plates. Successful isolation of recombinant viruses was confirmed by ethidium bromide visualization of restriction patterns on gels, Southern blottings, and negative PCR assays for wt virus (data not shown).The β-Gal expression cassette was removed by passage of the virus through a REF cell line expressing Cre recombinase, and the non-β-Gal virus (RCMVMEnh) was then isolated by limiting-dilution cultures.
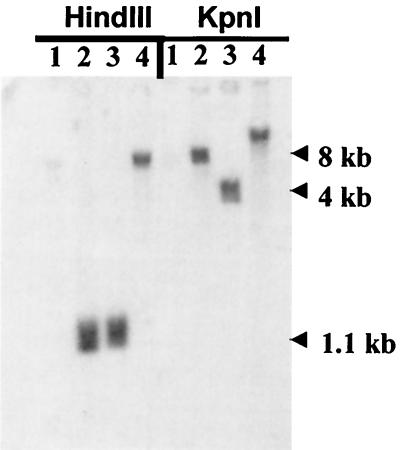
Southern blots of wt RCMV, wt MCMV, the RCMVMEnhβgal recombinant virus, and the non-β-Gal recombinant virus (RCMVMEnh) cut with HindIII or KpnI and probed with the MCMV MIE enhancer are shown in Fig. 2. Both recombinant viruses and wt MCMV were positive for the MCMV MIE enhancer. A new HindIII site provided by the loxP site directly 5′ to the MCMV enhancer resulted in bands of approximately 1.1 kbp following HindIII digestion of both recombinants. Because there was also a new KpnI site just 5′ to the β-Gal cassette, the difference in band size seen in the recombinants cut with KpnI and probed with the MCMV enhancer reflects the loss of the β-Gal cassette. The isolation of the repaired virus (RCMVrep) was confirmed by Southern blot analysis (data not shown). To ensure that homologous recombination and the excision of the β-Gal cassette occurred as expected in both RCMVMEnh and RCMVrep, DNA from around the recombination junctions at the 5′ end of the enhancers and around the FspI site in exon 4 was amplified, cloned, and sequenced and was found to be correct.
FIG. 2.
Southern blot analysis of recombinant RCMV. Shown is a Southern blot of wt RCMV (1), RCMVMEnhβgal (2), RCMVMEnh (3), and wt MCMV (4) virion DNA digested with HindIII or KpnI and probed with the MCMV enhancer.
In vitro virus replication.
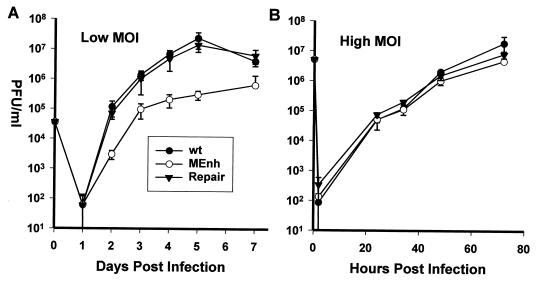
RCMV does not replicate in MEF or NIH 3T3 cells. The block is unlikely due to an inability to enter the cells and express MIE genes because by 4 h postinfection, MIE proteins are easily detected (data not shown). Therefore, it was not surprising that RCMVMEnh was unable to replicate in either type of murine cell (data not shown). Replication of wt RCMV, RCMVMEnh, and RCMVrep in REF cells was assayed after both a high MOI of 10 and a low MOI of 0.01. The average of three experiments is shown in Fig. 3. Although the only consistent difference at the high MOI was a 2- to 3-fold-lower yield of RCMVMEnh (Fig. 3B), a 10-fold difference in yield was consistently found in infections at low MOI (Fig. 3A).
FIG. 3.
One-step growth curves comparing wt virus, RCMVMEnh, and RCMVrep at low (A) and high (B) MOI of REF cells. Virus supernatants were collected postinfection at the times indicated, and titers were determined by standard plaque assay on REF cells. Data presented are averages of three experiments ± standard deviation.
Effect of the MCMV enhancer on MIE gene transcription.
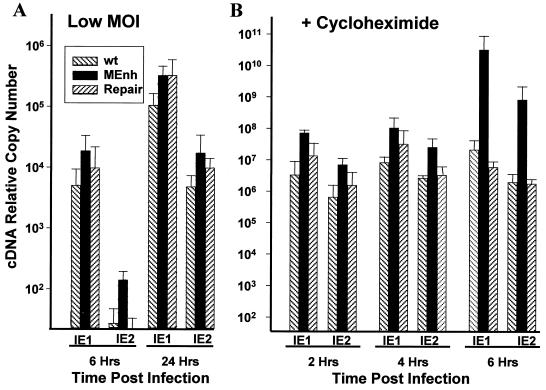
The decreased virus production of RCMVMEnh after infection at a low MOI might be due to a deficiency in MIE transcription. We therefore examined MIE transcription using quantitative PCR to determine transcription levels of IE1 (exon 4) and IE2 (exon 5). After infection at a low MOI, which resulted in a log lower virus production for RCMVMEnh compared to the wt and repaired viruses, comparable expression of IE1 and IE2 transcripts was found for all three viruses at 6 and 24 h (Fig. 4A). Similar MIE expression results were found for infections at high MOI and when dot blottings instead of the quantitative PCR assays were used (data not shown). In all experiments and at all time points, the expression of IE1 and IE2 from the RCMVMEnh virus was at least equal to and usually higher than that from the wt or repaired virus.
FIG. 4.
Real-time quantitative PCR analysis of MIE transcription. Total RNA of cells infected with wt virus, RCMVMEnh (MEnh), or RCMVrep (repair) was extracted postinfection as indicated. Following reverse transcription, primers specific for exon 2 and either exon 4 (IE1) or exon 5 (IE2) were used in PCRs with internal exon 4- or exon 5-specific fluorescent probes in a quantitative PCR sequence detector. Results were normalized to those of GAPDH. Cells were infected at a low MOI (A) or at a high MOI in the presence of cycloheximide (B).
We also examined transcription under IE conditions (with cycloheximide present). A representative experiment after infection at a high MOI is shown in Fig. 4B. Transcription of IE1 and IE2 in the presence of CH was comparable for all three viruses at 2 and 4 h postinfection, but at 6 h, the production of both transcripts was 2 to 3 logs higher for RCMVMEnh.
In vivo acute infections.
We next determined the in vivo effects of the enhancer switch. Because of a wide variability of virus titers in organs between individual rats, in some experiments we mixed the test virus with RCMVβgal, which served as an internal control for infection. A similarly wide variation among the organ titers of individual mice after MCMV infection has also been noted (13). wt virus, RCMVMEnh, or RCMVrep (2 × 106 PFU) was mixed with RCMVβgal (107 PFU) and injected i.p. Fivefold more RCMVβgal was used in mixing experiments because, from previous experience, we found that this virus does not replicate as well as wt virus in vivo. We first compared the viruses in acute infections of the spleen. Three rats per group were infected with the mixed-virus pools (the test virus plus RCMVβgal), and then the animals were sacrificed and spleen homogenates were assayed for virus 1, 3, and 5 days after infection. As seen in Fig. 5A, by day 3, the percentages of the three viruses compared to those of RCMVβgal were approximately equal. By day 5 postinfection, most of the virus was cleared from the spleen, in that only 2 of 9 rats had detectable virus; one had wt virus only and one had RCMVMEnh only (data not shown). To rule out a complementation effect that RCMVβgal might have had on the replication of RCMVMEnh, rats (four per group) were infected with wt virus, RCMVMEnh, or RCMVrep alone and spleens were harvested 2 days later. Spleens from all four rats infected with RCMVMEnh virus were positive for virus at titers similar to those of rats infected with wt virus or RCMVrep alone (data not shown).
FIG. 5.
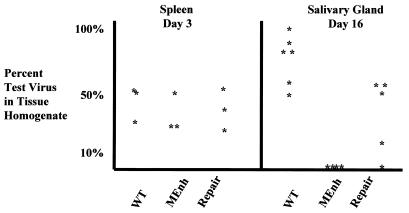
Acute in vivo infections. Because of the variability in organ titers of mice and rats following inoculation with identical viral pools, a mixture of wt virus, RCMVMEnh, and RCMVrep with RCMVβgal was used to assess differences in in vivo replication. RCMVβgal served as an internal control. Rats were inoculated with the virus pools, and the organs were harvested at the times indicated. Results are expressed as the percent of wt virus, RCMVMEnh, or RCMVrep (non-blue plaques) in the mixture, the remainder being RCMVβgal (blue plaques). Thus, an asterisk at 10% indicates that of the virus isolated from that organ, 10% was wt, RCMVMEnh, or RCMVrep and 90% was RCMVβgal. Not all rats were positive for virus. Spleens were harvested from rats (three per group) 3 days postinfection, and salivary glands were harvested from rats (six per group) 6 days postinfection.
We next compared wt virus, RCMVMEnh, and RCMVrep mixed with RCMVβgal for the ability to reach and replicate in the salivary gland after i.p. infection. Like MCMV, RCMV reaches a peak virus titer in the salivary gland approximately 2 weeks after i.p. infection. Therefore, rats (six per test virus) were infected with each mixed-virus pool and sacrificed at day 16 postinfection, and submaxillary salivary glands were removed and assayed for virus. As seen in Fig. 5B, salivary glands of five out of six rats infected with wt virus plus RCMVβgal had increased titers of wt virus while the salivary gland from one rat had equal titers of both viruses. Three out of five salivary glands from rats infected with RCMVrep plus RCMVβgal showed equal titers of both viruses, and the virus titer from the salivary gland from one rat was 16% RCMVrep and 84% RCMVβgal. One salivary gland from the RCMVrep plus RCMVβgal group yielded only RCMVβgal, which was at a very low titer, and the salivary gland from one rat was negative for any virus. In contrast to wt and repaired viruses, in each of the four rats positive for virus following infection with the RCMVMEnh plus RCMVβgal mixture, less than 5% of the virus isolated was RCMVMEnh. Twelve rats per group were then infected with either wt virus, repaired virus, or RCMVMEnh alone, and the salivary glands were harvested 16 days later and virus titers were determined. The salivary glands from all 12 rats infected with wt virus were positive for virus and 9 of the 12 rats infected with repaired virus were positive. However, no virus was isolated from the salivary glands of the 12 rats infected with RCMVMEnh (data not shown). Direct inoculation of the salivary gland with RCMVMEnh alone resulted in replication of the virus when assayed 5 days later (data not shown), suggesting that there is no inherent limitation of growth by this virus in the salivary gland.
Persistence and latency.
Rats (four per group) were infected with virus mixtures containing either wt virus plus RCMVβgal, RCMVMEnh plus RCMVβgal, or RCMVrep plus RCMVβgal and were evaluated 120 days after infection for the presence of persistent infectious virus in the spleen, the presence of viral DNA, and the ability to reactivate virus. No persistent virus could be found in the spleens of any of the rats at 120 days postinfection. Three out of four rats infected with wt virus plus RCMVβgal produced virus after the explantation of their spleens: two rats produced mixed infections and one rat produced only RCMVβgal (Table 1). Similarly, three of four rats infected with RCMVrep plus RCMVβgal produced virus from explanted spleen cultures: one had a mixed infection, one was infected with RCMVβgal only, and one was infected with RCMVrep only. Two of four rats that received RCMVMEnh plus RCMVβgal produced both viruses from explanted spleens.
TABLE 1.
Virus isolation from spleen explant cultures at 120 days
| Viruses (no. of rats infected) | Spleen explant cultures
|
|
|---|---|---|
| No. positive | Virus isolated (no. of cultures) | |
| Wt, RCMVβgal (4) | 3 | Mixed (2), RCMVβgal (1) |
| RCMVMEnh, RCMVβgal (4) | 2 | Mixed (2) |
| RCMVrep, RCMVβgal (4) | 3 | Mixed (2), RCMVβgal (1), RCMVrep (1) |
| RCMVMEnh (6) | 4 | RCMVMEnh (4) |
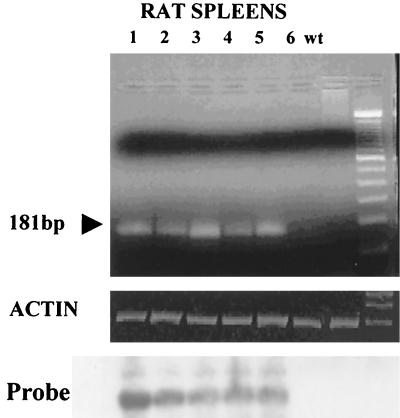
To rule out complementation of RCMVMEnh due to the presence of RCMVβgal, six rats were infected with RCMVMEnh alone and spleens and salivary glands were tested for persistent virus and explanted for reactivation at 120 days postinfection. No persistent infectious virus was isolated from any of the spleens or salivary glands. DNA extracted from the spleens of five of the six rats infected with RCMVMEnh alone were positive by PCR (Fig. 6), and four of these rats reactivated RCMVMEnh. In contrast, only one of six salivary glands from these rats was positive by PCR for RCMVMEnh (data not shown). All salivary glands were negative for persisting virus, and none of them reactivated the virus.
FIG. 6.
PCR of spleen DNA from six rats infected with RCMVMEnh only. Approximately 120 days postinfection, DNA from spleens was used for PCR with primers specific for RCMVMEnh virus. The Southern blot was probed with an internal oligonucleotide.
DISCUSSION
It is a long-held view that the MIE locus plays a critical role in the life cycle of CMVs. The basic genetic organization of this region is similar in the different species of CMV, with at least two principle proteins (IE1 and IE2) being produced by differential splicing. Loss of the exon 4 product (IE1) appears nonlethal although MIE transcription and viral replication are diminished in infections at low MOI with these mutants of HCMV (13) and MCMV (4). IE2 is essential for replication of both viruses (3, 21).
Control of the MIE products resides in the MIE enhancer/promoter regions, which vary considerably among the CMV species (6, 7, 10, 11, 29). The presence and arrangement of specific binding sites have evolved over millions of years for each CMV species and should reflect unique requirements for specific tissues or purposes (lytic infection, latency, or reactivation needs). Surprisingly, recent experiments suggest that little more than a minimal promoter is sufficient for MCMV MIE expression and viral replication in tissue culture (4). Furthermore, an MIE enhancer swap from HCMV for that in MCMV resulted in no observable phenotype changes in vitro. After infection of mice, such recombinants grew normally in mouse liver but less well in extrahepatic tissues (13). However, it is not possible to observe for phenotype changes in all infected tissues under the various conditions faced by the host, and reactivation from latency was not examined.
We had parallel experiments under way utilizing RCMV as the test virus. We constructed an RCMV recombinant in which we replaced the native MIE enhancer with that from MCMV. The two previously reported sets of MIE enhancer-swap MCMV recombinants differed from each other in that in one set the replacement began 20 bp 5′ of the TATA box and thus retained the native promoter and a short adjacent upstream sequence (4) while the other set replaced the native promoter beginning five bases 5′ of the cap site and thus replaced the native TATA box and surrounding sequences (13). Viruses in the latter set were thus missing any native cis-acting repressor sequence (crs), if such a sequence in MCMV (23) was present in the same relative position as in HCMV (20, 27). Even so, there appeared to be no differences in these viruses in MIE transcription characteristics or virus production in tissue culture, making the role of IE2 (HCMV) and, IE3 (MCMV) autoregulation unclear. Our recombinant virus replaced the native MIE enhancer 10 bases 5′ of the TATA box and thus retained any putative crs elements, if located, as in HCMV. We have not found evidence for RCMV IE2-mediated autoregulation despite extensive transfection experiments (unpublished data).
Unlike the findings for the HCMV MIE enhancer swap recombinants of MCMV, our recombinant RCMV, in which the MIE enhancer was replaced with that from MCMV, exhibits a diminished capacity to replicate in vitro and an altered organ tropism in vivo. Using real-time quantitative PCR, we found that the presence of the MCMV enhancer led to increased transcription of IE1 and IE2 RNA, but only in the presence of cycloheximide. The presence of virion proteins or extant cellular transcription factors able to interact with binding sites in the enhancer are important determinants of initial transcription and would be operative in the absence of protein synthesis. The larger number and variety of transcription factor binding sites in the MCMV MIE might allow for more efficient initiation of transcription under these circumstances. In the absence of cycloheximide, the transcription of IE1 and IE2 was the same regardless of the source of the MIE enhancer during infections at both low and high MOI.
Despite normal transcription kinetics, virus production in tissue culture by the RCMVMEnh recombinant was diminished, particularly during infection at low MOI. This result differs from those presented by Angulo et al. using MCMV with the HCMV MIE enhancer (4). This disparity could be the result of sequences unique to the RCMV MIE enhancer which were not complemented for by the MCMV enhancer. We also have preliminary data that suggest an RCMV recombinant which has the HCMV MIE enhancer replacing the RCMV enhancer also does not replicate as well as wt virus at low MOI (unpublished observations).
The relationship of MIE expression and viral growth is not clear. In an extensive study of HCMV recombinants with deletions in the MIE enhancer, Meier and Pruessner (22) found that deletion of the distal portion of the enhancer resulted in diminished virus production and that this correlated with decreased MIE transcription. However, our finding of equivalent MIE transcription regardless of which enhancer was present, even at low MOI when recombinant virus growth was diminished, indicates an uncoupling of the MIE expression and virus growth under some conditions. This is seen in the extreme when infection of nonpermissive cells allows for MIE expression without subsequent production of infectious virus (18). Our results suggest that there may be a threshold effect in permissive cells above which the amount of MIE transcription does not ultimately determine virus production. The manner in which the MIE enhancer may influence virus production other than through its regulation of MIE transcription is unclear but may include maintenance of an open chromatin structure that facilitates replication (9, 25).
An important aspect of CMV biology addressed by MIE enhancer swap experiments concerns the role of the enhancer in virus tropism and species specificity. In tissue culture, MCMV can replicate in both MEF and REF cells while RCMV does not replicate in MEF cells. The presence of the MCMV MIE enhancer in the RCMV recombinant virus did not confer the capacity to the virus to replicate in mouse cells. This was not surprising since RCMV can enter mouse cells and express IE1 and IE2, indicating that the MIE genes are probably not involved in this replication block. These results are consistent with the findings obtained after switching the HCMV MIE enhancer for that of MCMV, in which viral tropism was basically unchanged (1, 13). Furthermore, we were unable to detect infectious virus after infection of BALB/c mice with RCMVMEnh (unpublished observations).
The substitution of the MCMV enhancer for that of the RCMV MIE enhancer affected the ability of the recombinant virus to replicate in vivo. While replication of the recombinant virus in the spleen early after i.p. infection was similar to that of wt and the repaired viruses, much less recombinant virus was present in the salivary glands after i.p. infection. However, intrasalivary gland inoculation led to growth of RCMVMEnh comparable to that of wt and repaired virus, implying a deficiency in the spread from initial or secondary infection sites to the salivary gland. This suggests that the RCMV MIE enhancer may play a role in vivo in viral spread and/or tissue tropism.
The only instance where active RCMVMEnh replication could be detected in the salivary gland after i.p. inoculation was when the recombinant virus was infected with RCMVβgal, suggesting a complementation effect allowing for RCMVMEnh replication. This complementation was probably not important for initial replication of RCMVMEnh in the spleen because when infected alone, RCMVMEnh appeared to replicate to normal titers in the spleen. It is possible that secondary sites important for spread to the salivary gland (e.g., circulating mononuclear cells) are not readily infected by RCMVMEnh and require complementation by wt RCMVβgal virus. Experiments are under way using in situ PCR to identify such sites where complementation may occur.
At 120 days postinfection, the spleens from the two of four rats with a mixed infection and five of six rats infected with RCMVMEnh alone showed detectable RCMVMEnh DNA by PCR and, in most instances, reactivated the virus. Thus, the native MIE enhancer is not required for replication, establishment of latency, or reactivation of the virus from the spleen.
Results presented here suggest that the RCMV MIE enhancer can play an important role in the tissue tropism and, thus, pathogenesis of in vivo RCMV infection. This differs from the conclusions in MIE enhancer swap experiments with MCMV. Furthermore, RCMV reactivation from the spleen may occur when a foreign MIE enhancer is present. However, current definitions of latency are based on the finding of no infectious virus in an organ and the production of infectious virus after explantation, immunosuppression, or some other “reactivating” manipulation. Operationally, the definition of latency thus depends on the sensitivity of the assay for persisting infectious virus. Until molecular markers that uniquely characterize the CMV latent state are identified, definitive statements cannot be made concerning latency experiments in this or other studies. It may be that the differences seen among the CMV enhancers reflect differences in the sites of replication, latency, and mechanism of reactivation in each species.
ACKNOWLEDGMENTS
We thank Gary Hayward, John Nicholas, and M. Stanley for helpful discussions.
REFERENCES
- 1.Angulo A, Chandraratna R A S, LeBlanc J F, Ghazal P. Ligand induction of retinoic acid receptors alters an acute infection by murine cytomegalovirus. J Virol. 1998;72:4589–4600. doi: 10.1128/jvi.72.6.4589-4600.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angulo A, Ghazal P. Regulation of human cytomegalovirus by retinoic acid. Scand J Infect Dis. 1995;99:113–115. [PubMed] [Google Scholar]
- 3.Angulo A, Ghazal P, Messerle M. The major immediate-early gene ie3 of mouse cytomegalovirus is essential for viral growth. J Virol. 2000;74:11129–11136. doi: 10.1128/jvi.74.23.11129-11136.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Angulo A, Messerle M, Koszinowski U H, Ghazal P. Enhancer requirement for murine cytomegalovirus growth and genetic complementation by the human cytomegalovirus enhancer. J Virol. 1998;72:8502–8509. doi: 10.1128/jvi.72.11.8502-8509.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Batterson W, Roizman B. Characterization of the herpes simplex virion-associated factor responsible for the induction of alpha genes. J Virol. 1983;46:371–377. doi: 10.1128/jvi.46.2.371-377.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beisser P S, Kaptein S J, Beuken E, Bruggeman C A, Vink C. The Maastricht strain and English strain of rat cytomegalovirus represent different betaherpesvirus species rather than strains. Virology. 1998;246:341–351. doi: 10.1006/viro.1998.9196. [DOI] [PubMed] [Google Scholar]
- 7.Boshart M, Weber F, Jahn G, Dorsch-Hasler K F B, Schaffner W. A very strong enhancer is located upstream of an immediate early gene of human cytomegalovirus. Cell. 1985;41:521–530. doi: 10.1016/s0092-8674(85)80025-8. [DOI] [PubMed] [Google Scholar]
- 8.Burns W H, Barbour G M, Sandford G R. Molecular cloning and mapping of rat cytomegalovirus DNA. Virology. 1988;166:140–148. doi: 10.1016/0042-6822(88)90155-9. [DOI] [PubMed] [Google Scholar]
- 9.Cereghini S, Yaniv M. Assembly of transfected DNA into chromatin: structural changes in the origin-promoter-enhancer region upon replication. EMBO J. 1984;3:1243–1253. doi: 10.1002/j.1460-2075.1984.tb01959.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang Y N, Jeang K T, Lietman T, Hayward G S. Structural organization of the spliced immediate-early gene complex that encodes the major acidic nuclear (IE1) and trans-activator (IE2) proteins of African green monkey cytomegalovirus. J Biomed Sci. 1995;2:105–130. doi: 10.1007/BF02253062. [DOI] [PubMed] [Google Scholar]
- 11.Dorsch-Hasler K, Keil G M, Weber F, Jasin M, Schaffner W, Koszinowski U H. A long and complex enhancer activates transcription of the gene coding for the highly abundant immediate early mRNA in murine cytomegalovirus. Proc Natl Acad Sci USA. 1985;82:8325–8329. doi: 10.1073/pnas.82.24.8325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibson E E, Heid C A, Williams P M. A novel method for real time quantitative RT-PCR. Genome Res. 1996;6:995–1001. doi: 10.1101/gr.6.10.995. [DOI] [PubMed] [Google Scholar]
- 13.Grzimek N K A, Podlech J, Steffens H-P, Holtappels R, Schmalz S, Reddehase M J. In vivo replication of recombinant murine cytomegalovirus driven by the paralogous major immediate-early promoter-enhancer of human cytomegalovirus. J Virol. 2000;73:5043–5055. doi: 10.1128/jvi.73.6.5043-5055.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heid C A, Stevens J, Livak K J, Williams P M. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 15.Ibanez C E, Schrier R, Ghazal P, Nelson J A. Human cytomegalovirus productively infects primary differentiated macrophages. J Virol. 1991;65:6581–6588. doi: 10.1128/jvi.65.12.6581-6588.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keil G M, Ebeling-Keil A, Koszinowski U H. Immediate-early genes of murine cytomegalovirus: location, transcripts, and translation products. J Virol. 1987;61:526–533. doi: 10.1128/jvi.61.2.526-533.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kondo K, Kaneshima H, Mocarski E S. Human cytomegalovirus latent infection of granulocyte-macrophage progenitors. Proc Natl Acad Sci USA. 1994;91:11879–11883. doi: 10.1073/pnas.91.25.11879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lafemina R L, Hayward G S. Differences in cell-type-specific blocks to immediate early gene expression and DNA replication of human, simian and murine cytomegalovirus. J Gen Virol. 1988;69:355–374. doi: 10.1099/0022-1317-69-2-355. [DOI] [PubMed] [Google Scholar]
- 19.Liu B, Stinski M F. Human cytomegalovirus contains a tegument protein that enhances transcription from promoters with upstream ATF and AP-1 cis-acting elements. J Virol. 1992;66:4434–4444. doi: 10.1128/jvi.66.7.4434-4444.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Macias M P, Stinski M F. An in vitro system for human cytomegalovirus immediate early 2 protein (IE2)-mediated site-dependent repression of transcription and direct binding of IE2 to the major immediate early promoter. Proc Natl Acad Sci USA. 1993;90:707–711. doi: 10.1073/pnas.90.2.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marchini A, Liu H, Zhu H. Human cytomegalovirus with IE-2 (UL122) deleted fails to express early lytic genes. J Virol. 2001;75:1870–1878. doi: 10.1128/JVI.75.4.1870-1878.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meier J L, Pruessner J A. The human cytomegalovirus major immediate-early distal enhancer region is required for efficient viral replication and immediate-early gene expression. J Virol. 2000;74:1602–1613. doi: 10.1128/jvi.74.4.1602-1613.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Messerle M, Buhler B, Keil G M, Koszinowski U H. Structural organization, expression, and functional characterization of the murine cytomegalovirus immediate-early gene 3. J Virol. 1992;66:27–36. doi: 10.1128/jvi.66.1.27-36.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minton E J, Tysoe C, Sinclair J H, Sissons J G. Human cytomegalovirus infection of the monocyte/macrophage lineage in bone marrow. J Virol. 1994;68:4017–4021. doi: 10.1128/jvi.68.6.4017-4021.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nelson J A, Groudine M. Transcriptional regulation of the human cytomegalovirus major immediate-early gene is associated with induction of DNase I-hypersensitive sites. Mol Cell Biol. 1986;6:452–461. doi: 10.1128/mcb.6.2.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Hare P, Hayward G S. Expression of recombinant genes containing herpes simplex virus delayed-early and immediate-early regulatory regions and trans activation by herpesvirus infection. J Virol. 1984;52:522–531. doi: 10.1128/jvi.52.2.522-531.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pizzorno M C, Hayward G S. The IE2 gene products of human cytomegalovirus specifically down-regulate expression from the major immediate-early promoter through a target sequence located near the cap site. J Virol. 1990;64:6154–6165. doi: 10.1128/jvi.64.12.6154-6165.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Priscott P K, Tyrrell D A J. The isolation and partial characterisation of a cytomegalovirus from the brown rat, Rattus norvegiec. Arch Virol. 1982;73:145–160. doi: 10.1007/BF01314723. [DOI] [PubMed] [Google Scholar]
- 29.Sandford G R, Burns W H. Rat cytomegalovirus has a unique immediate early gene enhancer. Virology. 1996;222:310–317. doi: 10.1006/viro.1996.0428. [DOI] [PubMed] [Google Scholar]
- 30.Voigt S, Sandford G R, Ding L, Burns W H. Identification and characterization of a spliced C-type lectin-like gene encoded by rat cytomegalovirus. J Virol. 2001;75:603–611. doi: 10.1128/JVI.75.2.603-611.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]