Abstract
Synthetic lethality (SL) is a gold mine of anticancer drug targets, exposing cancer-specific dependencies of cellular survival. To complement resource-intensive experimental screening, many machine learning methods for SL prediction have emerged recently. However, a comprehensive benchmarking is lacking. This study systematically benchmarks 12 recent machine learning methods for SL prediction, assessing their performance across diverse data splitting scenarios, negative sample ratios, and negative sampling techniques, on both classification and ranking tasks. We observe that all the methods can perform significantly better by improving data quality, e.g., excluding computationally derived SLs from training and sampling negative labels based on gene expression. Among the methods, SLMGAE performs the best. Furthermore, the methods have limitations in realistic scenarios such as cold-start independent tests and context-specific SLs. These results, together with source code and datasets made freely available, provide guidance for selecting suitable methods and developing more powerful techniques for SL virtual screening.
Subject terms: Machine learning, Data mining, Target identification, Cancer genetics, Genetic interaction
A comprehensive benchmarking of existing synthetic lethality (SL) prediction methods is lacking. Here, the authors compare 12 recently developed machine learning methods for SL prediction, assess their performance, and provide guidance on the selection of the most suitable method.
Introduction
The synthetic lethal (SL) interaction between genes was first discovered in Drosophila Melanogaster about a century ago1,2. SL occurs if mutations in two genes result in cell death, but a mutation in either gene alone does not. Based on this observation, Hartwell et al.3 and Kealin4 suggested that SL could be used to identify new targets for cancer therapy. In the context of cancer, where multiple genes are often mutated, identifying the SL partners of these genes and interfering with their function can lead to cancer cell death, but spare normal cells. PARP inhibitors (PARPi) are the first clinically approved drugs designed by exploiting SL5, which target the PARP proteins responsible for DNA damage repair for the treatment of tumors with BRCA1/2 mutations5–9. Clinical trials have shown that PARPi have promising treatment effect on lung, ovarian, breast, and prostate cancers9–12. Despite the success of PARPi, there are still few SL-based drugs that have passed the clinical trials so far, partly due to the lack of techniques to efficiently identify clinically relevant and robust SL gene pairs.
Many methods have been proposed for identifying potential SL gene pairs in the last decade. Various wet-lab experimental methods such as drug screening13, RNAi screening14, and CRISPR/Cas9 screening15 have been used to screen gene pairs with SL relationships16. However, due to the large number of pairwise gene combinations (~200 million in human cells)17, and considering the combinations of different genetic contexts (e.g., cancer types and cell lines), it is impractical to screen all potential SL pairs by these wet-lab methods. To reduce the search space of SL gene pairs, computational methods have been proposed. Statistical methods identify SL gene pairs based on hypotheses derived from specific biological knowledge. These methods are generally interpretable because they can reveal statistical patterns between a pair of genes18. So far, biologists have mainly relied on the statistical methods such as DAISY19, ISLE20, MiSL21, SiLi22, etc. Additionally, random forests (RF), a traditional machine learning method, are also frequently used23–25, probably because it is easier to understand than deep learning. However, the accuracy of these models largely depends on the reliability of the assumptions and feature extraction, and they are often hard to be scaled up. By contrast, deep learning methods can better capture the complex nonlinear relationships between input and output, enabling them to identify complex patterns in the data. However, deep learning methods have not been well received by the biological community, partly because biologists are more concerned with the accuracy and clinical relevance of computationally predicted SL gene pairs. Most of the deep learning methods for SL prediction so far are based on supervised or semi-supervised learning and therefore rely on the quality and quantity of labeled SL data, which are usually sparse and noisy. Furthermore, the inherent black-box nature of deep learning models makes it difficult to explain the prediction processes, hindering their practical application in real-world settings.
In this work, we conduct a comprehensive benchmarking of machine learning methods for SL prediction, providing guidelines for biologists on model utilization. Two recent reviews18,26 summarize data resources and computational methods associated with SL, but lack a systematic assessment of these methods’ performance. We address this gap by evaluating machine learning-based SL prediction methods on classification and ranking tasks across different scenarios, including the impact of negative sample quality from various sampling strategies. We compile a list of recently published traditional machine learning and deep learning methods for predicting SL interactions (see Supplementary Tables 14 and 15), from which we select three matrix factorization methods and nine deep learning methods for benchmarking (Table 1). To standardize input and output formats and to facilitate data processing and result aggregation, we use SL labels from SynLethDB27 and preprocess feature data from multiple other sources according to the model’s requirements, including knowledge graph (SynLethKG27), GO (Gene Ontology28), PPI (BioGRID29), and Pathway (KEGG30). To assess the generalizability and robustness of the models, we incorporate three data segmentation methods (DSMs) and four positive-to-negative ratios (PNRs) into our experimental design. Additionally, we investigate the impact of negative sample quality on the model performance by utilizing three negative sampling methods (NSMs). Finally, we also perform two prediction tasks (i.e., classification and ranking) to identify the most probable SL gene pairs. Benchmarking results indicate that integrating information from multiple data sources is beneficial for predicting results and improving the quality of training data, such as screening negative samples based on gene expression and removing SL calculated during the training process. Additionally, our study extends the SL prediction problem to a more realistic scenario, provides valuable insights into the performance of different AI approaches to SL prediction, and further provides some suggestions for future development of new methods.
Table 1.
List of supervised machine learning methods for SL prediction
| Model & Ref. | Year | Description |
|---|---|---|
| SL2MF31 | 2018 | SL2MF uses logistic matrix factorization to learn gene representations, which are then used to identify potential SL interactions. The authors design an importance weighting scheme to distinguish known and unknown SL pairs and combine PPI and GO information for the prediction. |
| GRSMF33 | 2019 | GRSMF is a method based on graph regularized self-representation matrix factorization (MF). It learns self-representation from known SL interactions and further integrates GO information to predict potential SL interactions. |
| CMFW32 | 2020 | CMFW is a collective matrix factorization-based method that integrates multiple heterogeneous data sources for SL prediction. |
| DDGCN34 | 2020 | DDGCN is the first graph neural network (GNN)-based method for SL prediction. It uses graph convolutional network (GCN) and known SL interaction matrix as features. The authors use coarse-grained node dropout and fine-grained edge dropout to address the issue of overfitting of GCNs on sparse graphs. |
| GCATSL35 | 2021 | GCATSL proposes a graph contextualized attention network to learn gene representations for SL prediction. The authors use data of GO and PPI to generate a set of feature graphs as model inputs and introduce attention mechanisms at the node and feature levels to capture the influence of neighbors and learn gene expression from different feature graphs. |
| SLMGAE36 | 2021 | SLMGAE is a method for predicting SL interactions by leveraging a multi-view graph autoencoder. The authors incorporate data from PPI and GO as supporting views, while utilizing the SL graph as the main view, and apply a graph autoencoder (GAE) to reconstruct these views. |
| MGE4SL37 | 2021 | MGE4SL is a method based on Multi-Graph Ensemble (MGE) to integrate biological knowledge from PPI, GO, and Pathway. It combines the embeddings of features with different neural networks. |
| KG4SL39 | 2021 | KG4SL is a novel model based on graphical neural networks (GNN), and the first method that utilizes knowledge graph (KG) for SL prediction. The integration of KG helps the model obtain more information. |
| PTGNN38 | 2021 | PTGNN is a pre-training method based on graph neural networks that can integrate various data sources and leverage the features obtained from graph-based reconstruction tasks to initialize models for downstream link prediction tasks. |
| PiLSL41 | 2022 | PiLSL is a graph neural network (GNN)-based method that predicts SL by learning the representation of pairwise interaction between two genes. |
| NSF4SL42 | 2022 | NSF4SL is a contrastive learning-based model for SL prediction that eliminates the need for negative samples. It frames the SL prediction task as a gene ranking problem and utilizes two interacting neural network branches to learn representations of SL-related genes, thereby capturing the characteristics of positive SL samples. |
| SLGNN40 | 2023 | SLGNN is a knowledge graph neural networks-based method for synthetic lethality prediction that models gene preferences in distinct relationships in a knowledge graph, providing better interpretability. |
Results
Benchmarking pipeline
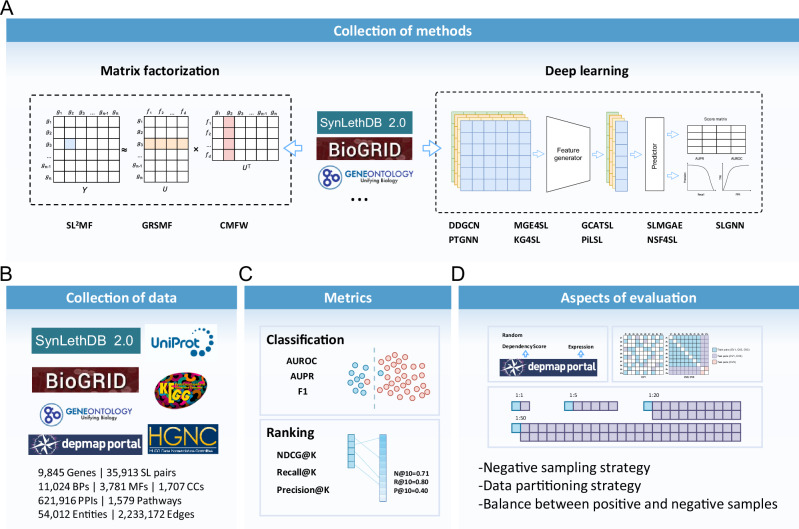
To evaluate machine learning methods for predicting SL interactions, we selected 12 methods published in recent years, including three matrix factorization-based methods (SL2MF31, CMFW32, and GRSMF33) and nine graph neural network-based methods (DDGCN34, GCATSL35, SLMGAE36, MGE4SL37, PTGNN38, KG4SL39, SLGNN40, PiLSL41, and NSF4SL42), see Table 1 for more details. The input data for these models varies, and in addition to SL labels, many kinds of data are used to predict SL, including GO28, PPI29, pathways30,43, and KG27, etc., and the detailed data requirements for each model are shown in Table 2. We believe that how many kinds of data inputs a model can accept is a function of the model’s own capabilities, and we focus on the model’s performance in various scenarios. To accomplish this, we designed 36 experimental scenarios, taking into account 3 different data splitting methods (DSMs), 4 positive and negative sample ratios (PNRs), and 3 negative sampling methods (NSMs) as shown in Fig. 1. In particular, these scenarios can be described as: (NSMN, CVi, 1:R), where N ∈ {Rand, Exp, Dep}; i ∈ {1, 2, 3}; R ∈ {1, 5, 20, 50} (see Methods for specific settings). After obtaining the results of all the methods in various experimental scenarios, we evaluated their performance for both classification and ranking tasks, and designed an overall score (see Methods) to better quantify their performance in Fig. 2. We also evaluated the scalability of all the models, including their computational efficiency and code quality. In addition, we evaluated the impact of labels from computational predictions on model performance in Supplementary Notes 2.1, and included a comparison between KR4SL44 and several deep learning methods on a dataset processed according to KR4SL in Supplementary Notes 2.2. For a more visual presentation of the benchmarking results, we provided figures and tables (Fig. 2, Table 3, Supplementary Figs. 3–20 and Supplementary Data 1) to show the results under various scenarios.
Table 2.
Data requirements of all models compared in this work
| Model | Data requirements | ||||
|---|---|---|---|---|---|
| SL | GO | PPI | KGa | Additional data | |
| SL2MF | ✓ | ✓ | ✓ | ||
| GRSMF | ✓ | ✓ | ✓ | ||
| CMFW | ✓ | ✓ | ✓ | ||
| DDGCN | ✓ | ||||
| SLMGAE | ✓ | ✓ | ✓ | ||
| MGE4SL | ✓ | ✓ | ✓ | Pathway, Protein-complex | |
| GCATSL | ✓ | ✓ | ✓ | ||
| PTGNN | ✓ | ✓ | ✓ | Protein sequence | |
| KG4SL | ✓ | ✓ | |||
| SLGNN | ✓ | ✓ | |||
| PiLSL | ✓ | ✓ | Gene expression | ||
| NSF4SL | ✓ | ✓ | |||
aKG includes 27 relationships, including GO and PPI.
Fig. 1. Workflow of the benchmarking study.
A A total of 12 methods are compared, including 3 matrix factorization-based methods and 9 deep learning methods. B We collected data from different databases to build a benchmark dataset. C This study compared the performance of the models in both classification and ranking tasks. D We also designed various experimental scenarios, including different negative sampling methods, positive-negative sample ratios, and data partitioning methods. The combinations of these scenarios constitute a task space ranging from easy to difficult tasks.
Fig. 2. Performance of the models.
A–C are the performance of the model under and NSMDep, respectively, where lighter colors indicate better performance. The figure contains five parts of information: a) A list of the 12 models. b) The overall scores of the models and the combined scores under the classification task and the ranking task only. c) and d) The performance of the models under the classification and ranking tasks, including six experimental scenarios consisting of 2 DSMs and 3 PNRs. e) The average time required for the models to complete one cross-validation.
Table 3.
The performance of the models under different DSMs and PNRs (on complete dataset)
| Models | (NSMRand, CVi, 1: 1), i = 1, 2, 3 | (NSMRand, CV1, 1: n), n = 5, 20, 50 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F1 score (Classification) | NDCG@10 (Ranking) | F1 score (Classification) | NDCG@10 (Ranking) | |||||||||
| CV1 | CV2 | CV3 | CV1 | CV2 | CV3 | 1:5 | 1:20 | 1:50 | 1:5 | 1:20 | 1:50 | |
| GRSMF | 0.849 | 0.750 | 0.677 | 0.284 | 0.104 | 0.000 | 0.812 | 0.770 | 0.713 | 0.294 | 0.319 | 0.334 |
| SL2MF | 0.766 | 0.667 | 0.667 | 0.280 | 0.005 | 0.000 | 0.738 | 0.703 | 0.664 | 0.281 | 0.281 | 0.281 |
| CMFW | 0.717 | 0.668 | 0.667 | 0.239 | 0.116 | 0.000 | 0.531 | 0.379 | 0.316 | 0.239 | 0.240 | 0.241 |
| SLMGAE | 0.883 | 0.779 | 0.738 | 0.270 | 0.101 | 0.039 | 0.855 | 0.809 | 0.757 | 0.309 | 0.336 | 0.351 |
| NSF4SL | 0.869 | 0.709 | 0.685 | 0.228 | 0.104 | 0.004 | 0.802 | 0.704 | 0.605 | 0.228 | 0.228 | 0.228 |
| PTGNN | 0.869 | 0.733 | 0.670 | 0.236 | 0.120 | 0.010 | 0.815 | 0.752 | 0.682 | 0.251 | 0.277 | 0.306 |
| PiLSL | 0.863 | 0.723 | 0.670 | - | - | - | 0.827 | 0.748 | - | - | - | - |
| KG4SL | 0.878 | 0.740 | 0.667 | 0.251 | 0.108 | 0.000 | 0.806 | 0.692 | 0.224 | 0.253 | 0.250 | 0.043 |
| SLGNN | 0.859 | 0.685 | 0.668 | 0.147 | 0.045 | 0.000 | 0.808 | 0.783 | 0.737 | 0.146 | 0.258 | 0.303 |
| DDGCN | 0.839 | 0.743 | 0.667 | 0.157 | 0.008 | 0.005 | 0.792 | 0.735 | 0.690 | 0.232 | 0.261 | 0.274 |
| GCATSL | 0.883 | 0.775 | 0.692 | 0.264 | 0.122 | 0.002 | 0.809 | 0.685 | 0.543 | 0.261 | 0.268 | 0.261 |
| MGE4SL | 0.697 | 0.670 | 0.668 | 0.003 | 0.004 | 0.004 | 0.335 | 0.098 | 0.039 | 0.042 | 0.014 | 0.003 |
The missing result ("-") is due to the inability of PiLSL to make batch predictions, which is required for a large number of gene pairs (> 800,000) when calculating the metrics. Bold formatting indicates the best-performing model in the given scenario, and underlined formatting indicates the second-best model.
Classification and ranking
For the problem of predicting SL interactions, most current methods still consider it as a classification problem, i.e., to determine whether a given gene pair has SL interaction. However, models with classification capabilities alone are insufficient for biologists, who need a curated list of genes that may have SL relationships with the genes they are familiar with. This list can empower biologists to conduct wet-lab experiments such as CRISPR-based screening. Among the evaluated methods, only NSF4SL originally regards this problem as a gene recommendation task, while other methods belong to the traditional discriminative models. To compute metrics for both tasks using these models, we adjusted the output layer, this modification ensures that every model produces a floating point score as its output.
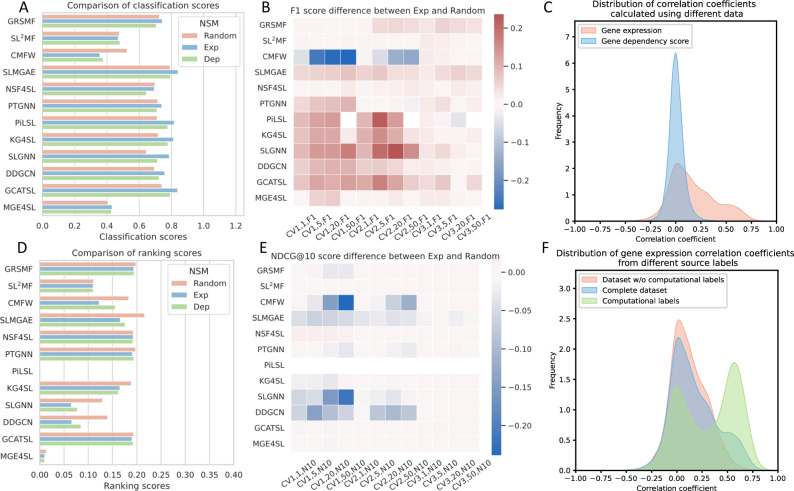
To assess the overall performance of the models in classification and ranking tasks, we employed separate Classification scores and Ranking scores (see “Methods”). Figure 2 presents these scores and the model’s performance across different scenarios and metrics. Based on the Classification scores, we found that when using negative samples filtered based on , the models usually had the best performance for the classification task. Among them, SLMGAE, GCATSL, and PiLSL performed the best with Classification scores of 0.842, 0.839, and 0.817, respectively. On the ranking task, the models performed slightly better under the scenario of NSMRand, and the top three methods were SLMGAE, GRSMF, and PTGNN, achieved Ranking scores of 0.216, 0.198, and 0.198, respectively. From these scores, SLMGAE is the model with the best overall performance.
In the following three sections, for the purpose of consistently assessing the performance of each model across classification and ranking tasks, we have designated a single metric for each task. Given our focus on the accurate classification of positive samples and the imbalance between positive and negative samples in experimental settings, we primarily employ F1 scores to gauge the models’ classification performance. Additionally, to appraise the model’s effectiveness in the ranking task, we mainly rely on the NDCG@10 metric, which takes into account the relevance and ranking of the genes in the SL prediction list.
Generalizability to unseen genes
In this section, we utilized three different data splitting methods (DSMs) for cross-validation, namely CV1, CV2, and CV3 (see “Methods”), in the order of increasing difficulty. The performance of models given these DSMs reflects their ability to generalize from known to unknown SL relationships.
Among the three DSMs, CV1 is the most frequently used method for cross-validation. However, this method exclusively provides accurate predictions for genes present in the training set, lacking the ability to extend its predictive capabilities to genes unseen during training. The CV2 scenario can be characterized as a semi-cold start problem, i.e., one and only one gene in a gene pair is present in the training set. This scenario holds significant practical implications. Considering the existence of ~10,000 known genes involved in SL interactions, there is a substantial number of human genes remain unexplored. These genes, which have not yet received enough attention, likely include numerous novel SL partner genes of known primary genes mutated in cancers. CV3 is a complete cold-start problem, i.e., neither of the two genes is in the training set. Under CV3, the model must adeptly discern common patterns of SL relationships, to generalize to genes not encountered during training.
For the convenience of discussion, we fixed NSM to NSMRand and PNR to 1:1, i.e., our scenario is (NSMRand, CVi, 1:1), where i = 1, 2, 3. See Supplementary Data 1 for the complete results under all scenarios.
The left part of Table 3 shows the performance of the models under different DSMs while NSM and PNR are fixed to NSMRand and 1:1, respectively. From the table, it can be seen that, for the classification task, SLMGAE, GCATSL, and KG4SL performed better under the CV1 scenario, with F1 scores greater than 0.877. By contrast, CMFW and MGE4SL performed poorly, with F1 score less than 0.720. Under CV2, all the selected methods showed significant performance degradation compared to CV1. For example, the F1 scores of SLMGAE and GCATSL, still the top two methods, dropped to 0.779 and 0.775, respectively, while both of CMFW and MGE4SL decreased to less than 0.670. When the DSM is changed to CV3, only the F1 score of SLMGAE can still be above 0.730, while all the other methods drop below 0.700. For the ranking task, GRSMF, SL2MF, and SLMGAE exhibited better performance under the CV1 scenario with NDCG@10 greater than 0.270. However, under CV2, the NDCG@10 of almost all methods except for GCATSL and PTGNN are lower than 0.120. Lastly, when the DSM is CV3, the NDCG@10 scores of all the methods in this scenario become very low (lower than 0.010) except SLMGAE. Generally, SLMGAE, GCATSL, and GRSMF have good generalization capabilities. In addition, CV3 is a highly challenging scenario for all models, especially for the ranking task. However, it is worth mentioning that KR4SL has shown more promising performance compared to other methods in the CV3 scenario (see Supplementary Notes 2.2).
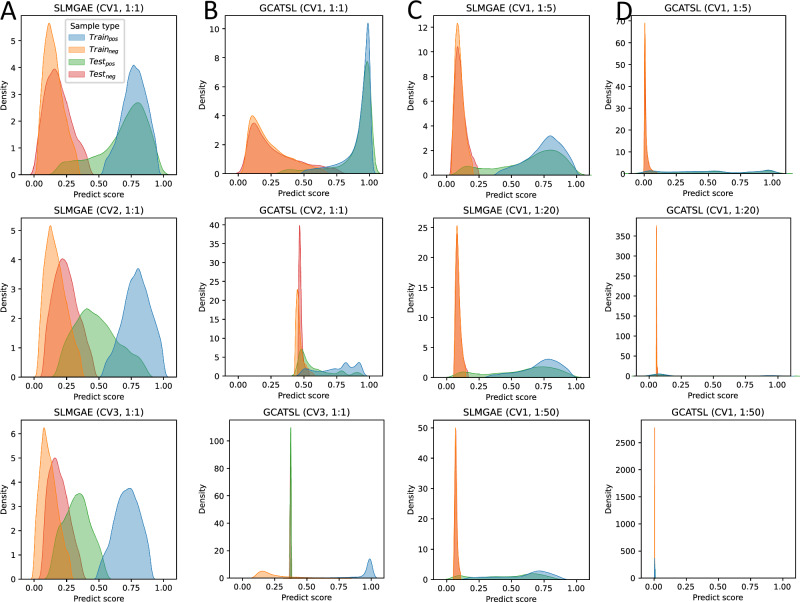
Moreover, Fig. 3A and B display the predicted score distributions of gene pairs in the training and testing sets for the SLMGAE and GCATSL models, respectively (see all methods in Supplementary Figs. 21–32). It is noteworthy that when the DSM is CV1, both models can differentiate between positive and negative samples effectively. As the challenge of generalization increases (in the case of CV2), there is a considerable change in the distribution of sample scores in the two models. In particular, for GCATSL, almost all negative sample scores in this model are concentrated around 0.5, while positive sample scores start to move towards the middle; for SLMGAE, only the positive sample distribution in the test set was significantly affected. When the task scenario becomes the most difficult CV3, the score distribution shift of the positive samples in the test set of the two models is more pronounced. For GCATSL, almost all sample scores in the test set are concentrated around 0.4, and the model cannot fail to have predictive ability in this scenario. Comparatively, SLMGAE is capable of distinguishing the samples in the test set with a relatively high degree of accuracy.
Fig. 3. Distribution of predicted scores.
A, B are the scores of gene pairs in the training and testing sets for the SLMGAE and GCATSL models under various data splitting methods (DSMs). C, D are the score of gene pairs in the training and testing sets for the SLMGAE and GCATSL models under various positive and negative sample ratios (PNRs). The results are obtained with NSMRand.
Robustness to increasing numbers of negative samples
In our study, so far, all negative samples used in training have been screened from unknown samples. As such, there could be false negative samples among the gene pairs categorized as negative. This situation could inadvertently introduce noise into the model’s training process. Furthermore, given the substantial disparity between the numbers of non-SL pairs and SL pairs, these models encounter the issue of imbalanced data. To assess the robustness of these models to noise stemming from negative samples, we conducted experiments by gradually increasing the number of negative samples. In our study, the number of negative samples is set to four levels: equal to the number of positive samples (1:1), five times the number of positive samples (1:5), twenty times the number of positive samples (1:20), and fifty times the number of positive samples (1:50). Notably, the 1:1 ratio corresponds to the conventional experimental configuration frequently adopted.
In this section, our experimental scenario for comparison is denoted as (NSMRand, CV1, 1:R) where R = 1, 5, 20, 50. From the right part of Table 3, it can be seen that as the number of negative samples increases, the models’ performance (F1 score) in classification tasks gradually decreases. This phenomenon is particularly pronounced for CMFW and MGE4SL. When the number of negative samples increases from one to five times that of positive samples (PNR is 1:5), the F1 scores of CMFW and MGE4SL drop dramatically from around 0.700 to 0.531 and 0.335, respectively. By contrast, several other methods, namely SLMGAE, PTGNN, PiLSL, KG4SL, and GCATSL, maintain their F1 scores above 0.800. When the number of negative samples is further increased to twenty times the number of positive samples (PNR is 1:20), only SLMGAE achieved an F1 score above 0.800, while CMFW and MGE4SL dropped to 0.379 and 0.098, respectively. Finally, when the number of negative samples is fifty times that of positive samples (PNR is 1:50), SLMGAE still outperformed other models with an F1 score of 0.757, followed by SLGNN with an F1 score of 0.737. Notably, compared with previous PNRs, KG4SL and GCATSL experienced a significant decline in their F1 scores, dropping to 0.224 and 0.543, respectively. On the other hand, in the context of ranking task, when PNR = 1:5, SLMGAE, GRSMF, PTGNN, and DDGCN exhibited a slight improvement in NDCG@10 than PNR is 1:1. At PNR = 1:20, the NDCG@10 for SLMGAE, GRSMF, PTGNN, and DDGCN continued to rise. Lastly, the NDCG@10 values for SLMGAE and GRSMF continue to increase to 0.351 and 0.334, respectively, when the PNR is changed to 1:50. Generally, SLMGAE and GRSMF have stronger robustness.
Figure 3C and D display the distribution of the scores for positive and negative samples predicted by SLMGAE and GCATSL across different PNRs. The figure shows the impact of the number of negative samples on the score of the given gene pair evaluated by the models. As the number of negative samples increases, an increasing number of positive samples in the testing set are assigned lower scores. Despite this effect, the majority of samples are still correctly classified by SLMGAE. But for GCATSL, when the number of negative samples is twenty times that of positive samples, i.e., PNR = 1:20, almost all samples are assigned very low scores. When the PNR becomes 1:50, almost all the scores given by GCATSL are concentrated in a very small score range (around 0), and the predictions of the model are no longer reliable. Furthermore, under the results of SLMGAE, it is evident that the distribution of negative samples becomes increasingly concentrated with a higher number of negative samples. And a notable phenomenon is observed in the previous results (Table 3), that certain models exhibit improved performance in the ranking task as the number of negative samples increases. We hypothesize that this improvement is attributed to the higher concentration of scores among negative samples, resulting in a greater number of positive samples achieving higher rankings. Consequently, the performance of some models under ranking tasks improves with increasing PNR.
Impact of negative sampling
Obtaining high-quality negative samples is crucial for the performance of the models. However, in the context of SL prediction, high-quality negative samples are scarce. Therefore, it is important to explore efficient and straightforward methods for obtaining high-quality negative samples from unknown samples. In this study, we evaluated three negative sampling approaches, namely , and NSMDep, which represents unconditional random negative sampling, negative sampling based on gene expression correlation, and negative sampling based on dependency score correlation, respectively (see Methods for details). Among these approaches, NSMRand has been widely used in existing SL prediction methods, and thus it will be used as the baseline for comparison. We denote the scenarios as (NSMN, CV1, 1:1), where N = Rand, Exp or Dep.
Based on the findings from the previous two subsections, certain characteristics regarding the model’s generalizability and robustness in the context of NSMRand can be observed. Here, we investigated two additional negative sampling methods ( and NSMDep). Our observations revealed that models utilizing negative samples from demonstrate improved classification performance compared to NSMRand. On the other hand, models employing negative samples from NSMDep do not show significant performance differences relative to NSMRand (see Supplementary Data 1). The results of the classification and ranking tasks are presented in Fig. 4A and D. The majority of models demonstrate a marked improvement in the classification task when using , with GCATSL’s Classification score increasing from 0.709 to 0.808. Other models such as SLMGAE, GRSMF, KG4SL, DDGCN, and PiLSL all achieved Classification scores above 0.720. On the other hand, SL2MF and CMFW experienced a decrease in performance. In the ranking task, the performance of NSMDep was better than , but not as good as NSMRand. CMFW, DDGCN, and SLMGAE experienced a considerable decrease in their Ranking scores when using either NSMDep or , while the other models did not demonstrate a significant variation.
Fig. 4. Model performance under various negative sampling methods and analysis of known sample correlations.
A, D show the overall score comparison of the models on the classification and ranking tasks under all three negative sampling methods (NSMs). B, E illustrates the differences between classification and ranking tasks using and NSMRand in different scenarios, as measured by the F1 score and NDCG@10 metrics. Red and blue signify an increase and a decrease, respectively, with darker shades indicating a larger difference. C illustrates the distribution of correlation coefficients between known SL pairs of genes under different data sources. F illustrates the distribution of correlation coefficients between gene expression levels of SL pairs from different label sources.
We also assessed the impact of negative sampling on the generalization and robustness of the model. As shown in Fig. 4B and E, the negative samples obtained through have a small impact on the performance of matrix factorization-based methods, except for the CMFW model, which has a significant decrease in performance in classification tasks compared to NSMRand. On the contrary, for deep learning-based methods, the negative samples obtained through improve the classification task performance of the model in various scenarios, especially for the CV1 and CV2 scenarios. It is noteworthy that the performance of the NSF4SL model is not affected by the quality of negative samples, as it “does not use negative samples" (i.e., does not use negative samples at all) during the training process. For ranking tasks, except for CMFW and SLGNN, the quality of negative sample has a relatively small impact on most models.
Furthermore, we investigated the potential reasons underlying the different impacts of the negative sampling method. As shown in Fig. 4C, the distribution of gene expression correlation scores for known SL gene pairs exhibits a bias towards positive scores. Note that selects gene pairs with negative correlation coefficient of gene expression. It is possible that, distinguishes the distribution of positive and negative samples in advance, reducing the difficulty of classification, it is able to because improve the performance of the models in the classification task. By contrast, there is a symmetric distribution of correlation based on dependency scores, hampering the model’s ability to learn more effective features from the negative samples selected by NSMDep.
Impact of computationally derived labels on performance
Comparing Tables 3 and 4, it is evident that the performance of all the models across various DSMs generally improves significantly after excluding SL labels predicted by computational methods from the dataset (The complete results can be found in Supplementary Data 2). Of note, NSF4SL is elevated to the second best (i.e., runner-up) on the classification task, with F1 scores increasing by 0.108 and 0.135 in the CV1 and CV2 scenarios, respectively. Nevertheless, SLMGAE remains the best-performing model overall (see Supplementary Table 4). An explanation for the observed improvement in performance might be that, by filtering out the computationally derived SL labels, we have also reduced the noise in the model’s training data. Specifically for NSF4SL, which only relies on positive samples during training, the improvement in positive sample quality enables NSF4SL to achieve notably better ranking on dataset without computationally derived labels compared with complete dataset. We performed correlation analysis on gene expression within each of the three datasets: the complete dataset, the dataset excluding computationally predicted labels, and the dataset of solely computationally predicted labels. Results revealed that computationally predicted SL gene pairs predominantly clustered around the correlation coefficients of 0 and 0.5 (Fig. 4F).
Table 4.
The performance of the models under different DSMs and PNRs (on dataset without computationally derived labels)
| Models | (NSMRand, CVi, 1: 1), i = 1, 2, 3 | (NSMRand, CV1, 1: n), n = 5, 20, 50 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F1 score (Classification) | NDCG@10 (Ranking) | F1 score (Classification) | NDCG@10 (Ranking) | |||||||||
| CV1 | CV2 | CV3 | CV1 | CV2 | CV3 | 1:5 | 1:20 | 1:50 | 1:5 | 1:20 | 1:50 | |
| GRSMF | 0.955 | 0.807 | 0.687 | 0.459 | 0.180 | 0.000 | 0.907 | 0.829 | 0.780 | 0.462 | 0.478 | 0.524 |
| SL2MF | 0.870 | 0.667 | 0.667 | 0.449 | 0.009 | 0.000 | 0.850 | 0.801 | 0.743 | 0.449 | 0.449 | 0.449 |
| CMFW | 0.761 | 0.671 | 0.667 | 0.388 | 0.201 | 0.000 | 0.582 | 0.425 | 0.353 | 0.388 | 0.389 | 0.389 |
| SLMGAE | 0.965 | 0.835 | 0.761 | 0.491 | 0.198 | 0.028 | 0.945 | 0.903 | 0.855 | 0.531 | 0.570 | 0.586 |
| NSF4SL | 0.977 | 0.844 | 0.734 | 0.465 | 0.195 | 0.026 | 0.939 | 0.852 | 0.737 | 0.465 | 0.465 | 0.465 |
| PTGNN | 0.957 | 0.792 | 0.672 | 0.404 | 0.215 | 0.009 | 0.917 | 0.837 | 0.759 | 0.462 | 0.489 | 0.512 |
| PiLSL | 0.962 | 0.799 | 0.678 | - | - | - | 0.931 | 0.840 | - | - | - | - |
| KG4SL | 0.966 | 0.816 | 0.693 | 0.404 | 0.196 | 0.000 | 0.911 | 0.793 | 0.452 | 0.405 | 0.411 | 0.320 |
| SLGNN | 0.963 | 0.732 | 0.667 | 0.310 | 0.099 | 0.000 | 0.945 | 0.897 | 0.717 | 0.372 | 0.432 | 0.345 |
| DDGCN | 0.902 | 0.790 | 0.671 | 0.236 | 0.038 | 0.005 | 0.881 | 0.823 | 0.767 | 0.333 | 0.375 | 0.408 |
| GCATSL | 0.970 | 0.839 | 0.708 | 0.430 | 0.208 | 0.001 | 0.926 | 0.739 | 0.610 | 0.425 | 0.424 | 0.432 |
| MGE4SL | 0.721 | 0.677 | 0.672 | 0.004 | 0.005 | 0.007 | 0.470 | 0.183 | 0.039 | 0.101 | 0.017 | 0.004 |
The missing result ("-") is due to the inability of PiLSL to make batch predictions, which is required for a large number of gene pairs (> 800,000) when calculating the metrics. Bold formatting indicates the best-performing model in the given scenario, and underlined formatting indicates the second-best model.
Comparison with context-specific methods
In recent years, increasing attention has been paid to the prediction of context-specific SLs, leading to the development of methods such as MVGCN-iSL45 and ELISL46. To further assess the performance of machine learning methods in the context-specific settings, we benchmarked 7 models on cancer cell-line-specific data, including ELISL and MVGCN-iSL, along with the following 5 models which demonstrated superior performance among the 12 methods in the previous benchmarking study: SLMGAE, NSF4SL, KG4SL, PTGNN, and GCATSL. These 7 models were tested on 4 cancer cell lines: 293T (KIRC)47, Jurkat (LAML)48, OVCAR8 (OV)49, and HeLa (CESC)47,50. From the results, it is evident that SLMGAE performed particularly well on 293T and OVCAR8. NSF4SL performed the best on HeLa. MVGCN-iSL showed consistently competitive performance across all the cancer types except for 293T. ELISL’s overall performance was good but not highly competitive across the four cancer types. Detailed results can be found in Supplementary Notes 2.3.
Discussion
Here, we present a comprehensive benchmarking study of 12 machine-learning methods for predicting SL interactions. We constructed a dataset from multiple sources and evaluated all the methods on this dataset. Our results demonstrate that the predictive capabilities of these methods vary under different experimental settings. Specifically, among the matrix factorization-based methods, GRSMF exhibited superior accuracy and stability compared to the other two methods. Among the deep learning methods, SLMGAE showed the most competitive performance overall, although other methods such as NSF4SL are also promising. In our experimental settings, the deep learning methods have outperformed the matrix factorization methods overall, but the latter have their own strengths. This benchmarking framework can be used to evaluate new models, providing a platform for consistent model training and testing to facilitate future development of AI techniques in this field.
Our evaluation showed that despite using only SL data, DDGCN can achieve performance comparable to models that rely on multiple external data sources (e.g., PTGNN), possibly due to its unique design of dual dropout. In addition, although both SLMGAE and MGE4SL incorporate multiple graph views and attention mechanisms, SLMGAE demonstrates far superior performance compared to MGE4SL. Further analysis of the two methods reveals that SLMGAE optimizes the model through three distinct objectives corresponding to SL, PPI, and GO, respectively. It distinguishes the main view from supporting views and reconstructs multiple graphs for different views via graph auto-encoders (GAEs). By contrast, MGE4SL optimizes the model by fusing all the information using a cross-entropy loss, which may introduce noise. The auto encoder has been shown51 to reduce information loss caused by Laplacian smoothing52. This may be one of the reasons why SLMGAE performs so well. KG4SL and GCATSL exhibit noticeable performance degradation when the PNR changes from 1:20 to 1:50, i.e., with an overwhelming proportion of negative samples, indicating the models’ inability to discriminate between sparse positive and abundant negative samples. Furthermore, models that integrate different types of data, such as GO and PPI, usually yield better results than DDGCN. However, if done inappropriately, such integration could be counterproductive, as in the case of MGE4SL. We also observed that, although KG4SL utilizes multiple relationships contained in the knowledge graph, its performance lags behind that of SLMGAE in both classification and ranking tasks. This suggests that it is challenging to incorporate an excess of diverse supplementary information into a predictive model.
Based on our study, we identified several issues in exploring SL prediction that deserve attention. Firstly, most models do not consider the realistic imbalance between the numbers of SL and non-SL gene pairs. Theoretically, SL interactions constitute only a small fraction of all gene-gene interactions. As a result, there are much more non-SL gene pairs than SL gene pairs. The models should be able to handle highly imbalanced data in order to make accurate predictions in real-world applications. Secondly, the authors of most current methods have primarily used the CV1 data splitting approach, which has significant limitations for predicting new potential SL gene pairs. This scenario focuses on genes with known SL relationships, often resulting in a “streetlight effect” where other potentially significant SL gene pairs tend to be overlooked. Moreover, within the currently known gene sets, the number of SL gene pairs may have reached saturation, necessitating exploration of new scenarios. The CV2 and CV3 scenarios are designed to uncover overlooked new SL interactions. Compared to CV1, the CV2 scenario evaluates a model’s ability to generalize patterns learned from training data to genes that have not yet participated in the model training process. However, most methods have mediocre performance in the real-world scenarios of CV2, possibly because most of the benchmarked methods were tailored for the CV1 scenario. Unsurprisingly, these methods perform even worse in the CV3 scenario, because CV3 represents a complete cold-start problem, where both genes in the test pairs are unseen during the training. Such a lack of prior exposure means that these models cannot leverage any previously learned patterns or relationships associated with the new genes in the test set, severely limiting their predictive ability. Additionally, the complexity and sparsity of SL relationships exacerbate this issue, making these models struggle to generalize effectively from known data to unseen gene pairs. KR4SL44 has shown better performance in the CV3 scenario compared to other benchmarked methods (see Supplementary Notes 2.2). This may be attributed to the graph reasoning mechanism of KR4SL, which allows it to learn deeper graph features corresponding to SL relationships, thus achieving better generalization capability. Thirdly, the lack of high-quality negative samples can affect the evaluation of the model. Generally, when evaluating the performance of a discriminant model, it is necessary to test the model with both positive and negative samples of known labels. However, in the context of SL prediction, obtaining high-quality negative samples is often a daunting task. The false negative samples included in the test dataset could potentially skew the evaluation metrics. In recent years, with the development of CRISPR technology, there have been works53–55 using combinatorial CRISPR/Cas956 to screen SL gene pairs, and the data generated from these experiments can provide reliable negative samples. But for a larger space of gene pairs, the wet-lab method is still labor-intensive. Finally, it is essential to incorporate multiple data sources, such as GO and PPI, to enhance the generalizability of the model. Additional data related to genes and genetic interactions can provide features for a complete set of genes and provide information for predicting new SL interactions in CV2 and CV3 scenarios. Additionally, as a valuable data source, KG contains information on multiple aspects of a gene and is organized in a graph format that can be more interpretable. However, the current methods for predicting SL interactions (e.g., KG4SL, PiLSL, NSF4SL, SLGNN, and KR4SL) are still rudimentary in harnessing KG’s potential.
We recommend several future directions to explore in the field of SL prediction by machine learning. Firstly, predicting context-specific (e.g., cancer-specific or cell-line-specific) SL interactions is key to developing cancer precision medicine. SL interactions are context-specific, meaning that the SL relationship between a pair of genes may only occur in one type of cancer (or cell line) but not in another. ELISL46 and MVGCN-iSL45 are methods published in recent years for predicting context-specific SL interactions. We conducted experiments on these and several other methods (see Supplementary Notes 2.3) and found that current approaches are still unable to adequately address this issue. Additionally, the cross-cell-line prediction task may also offer a potential solution to this problem. We conducted a simple experiment on this task (see Supplementary Notes 2.5). Based on our results, the task of predicting cross-cell-line SLs requires models to possess robust generalization capabilities across cell lines, which is currently challenging. The models need to effectively integrate multi-omics data from different cell lines, to capture disparities and commonalities among the cell lines. Secondly, SL interactions can be used to guide drug repositioning, by identifying new targets of approved drugs or discovering novel combination therapies. Zhang et al. developed SLKG57, which provides a computational platform for designing SL-based tumor therapy, and they demonstrated that SLKG could help identify promising repurposed drugs and drug combinations. Furthermore, computationally inferred SL interactions may be used to predict drug responses at patient level, indicating the SLs are clinically relevant. For instance, ISLE20 mines TCGA cohort to identify the most likely clinically relevant SL interactions (cSLi) from a given candidate set of experimentally SLi. Moreover, it has been shown that cSLi can successfully predict patients’ drug treatment response and provide patient stratification signatures. Thirdly, it is urgent to develop deep learning methods that can predict clinically relevant SLs based on personalized data. Although statistical methods such as SLIdR58 and ISLE have attempted to address this issue, deep learning models have not yet demonstrated their ability to recognize clinically relevant SL interactions. In order to fill the gap between deep learning models and clinical applications, it is necessary to make better use of in vivo or in vitro data. Fourthly, the concept of SL needs extension and refinement. Several classes of SL, such as synthetic dosage lethality (SDL)59,60 and collateral lethality61,62, have been proposed by researchers to capture the inherent complexity of the SL concept. Furthermore, Li et al.63 categorized SLs into two types: non-conditional/original and conditional SL. Conditional SL refers to synthetic lethal interactions that occur under specific conditions, such as genetic background, hypoxia, high levels of reactive oxygen species (ROS), and exposure to DNA-damaging agents and radiation. The distinction between the various SL definitions is often overlooked in the current literature of computational SL prediction. Fifthly, the lack of high-quality negative samples poses a challenge for researchers in SL prediction. While NSF4SL42 alleviates this problem by using a contrastive learning framework to dispense with the need of using negative samples, most classification models still rely on them. Last but not the least, most current machine learning models lack interpretability, making it difficult to assess the reliability of their predictions. Although performing well in terms of accurate and sensitive prediction, these models often do not incorporate underlying biological mechanisms. Improving the interpretability of the machine learning models can make them more practically useful and informative for users, including experimental biologists. The methods of PiLSL41, PTGNN38, and KR4SL44 can provide some degree of interpretability by uncovering the prediction process and providing evidence about the biological mechanisms of SL, e.g., by highlighting edges or paths with higher attention weights than the rest of a graph. More powerful techniques of interpretability analysis, however, remain to be designed in the future.
Methods
Data
For this benchmarking study, we obtained synthetic lethality (SL) label data from the database of SynLethDB 2.0, which contains a knowledge graph named SynLethKG27. In addition, we collected other data used by some models studied here including protein-protein interaction (PPI), gene ontology (GO), pathways, protein complexes, and protein sequences. For details on the model data requirements, see Table 2.
To ensure consistency, we standardized the gene names in the data using the HUGO Gene Nomenclature Committee (HGNC, http://www.genenames.org/)64 and the Ensembl Database65.
Synthetic lethality labels
After filtering, we finally obtained a dataset consisting of 9845 unique genes and 35,913 positive SL gene pairs from SynLethDB. Among the 35,913 pairs, 26,591 were identified through CRISPR, RNAi, and text mining, whereas the rest 9322 pairs were predicted using different computational methods. Specifically, we used the data on K562 cell line collected from Horlbeck et al.17 as the independent test set while employing the SynLethDB dataset used in the benchmarking study as the training set. The results can be found in Supplementary Notes 2.4. We used GI score < −3 to screen positive samples and matched the gene set in the obtained dataset with the genes in our benchmarking study. Finally, we obtained 1292 positive SL gene pairs as our independent test dataset. For negative samples, we ultimately screened 82,553 gene pairs with a GI score of > −3 and the gene set from our benchmarking study, from which negative samples were selected based on the GI score in descending order. We collected 54,012 entities and 2,233,172 edges from the knowledge graph of SynLethKG. To facilitate integration, all the data were aligned by gene names. Next, we describe our way of pre-processing prior knowledge.
Protein-protein interaction (PPI)
We extracted PPI data from the BioGRID database29 to construct a PPI network, where the nodes represent a subset of proteins from 9845 genes while the edges represent the interactions between the nodes. 621,916 PPIs were extracted for our experiment. Additionally, we reimplemented the FSweight algorithm66 on the PPI network to calculate the functional similarity matrix of proteins as input for SL2MF. We also collected 3403 protein complexes data related to the benchmarking test dataset from the CORUM67 database as input for MGE4SL.
Gene ontology (GO)
We collected GO data from the Gene Ontology database28, which consists of Gene Ontology Annotation (GOA) and GO terms. We collected 16,512 GO terms from the database, including 11,024 Biological Process (BP) terms, 3781 Molecular Function (MF) terms, and 1707 Cellular Component (CC) terms. To compute the functional similarity between genes and the semantic similarity between GO terms, we employed the R package GOSemSim68.
Pathway
We retrieved pathway data from multiple databases, including KEGG30 and Reactome43. A total of 409 KEGG pathways and 1170 Reactome pathways related to the genes in our experiment were collected. We obtained the genes involved in the same pathway and used them to construct a symmetrical mask matrix. In the matrix, a value of 1 is assigned when two genes are in the same pathway, and 0 otherwise. These pathway data are used as inputs for MGE4SL.
Dependency score and gene expression
We collected the data of gene expression and gene dependency scores in different cell lines from the DepMap database69. Using these data, we designed two new negative sampling methods to evaluate the impact of negative samples on the models’ performance.
Experimental scenarios
Different proportions of negative samples
To evaluate the performance of synthetic lethality (SL) prediction methods, it is crucial to use realistic scenarios for training and testing. In most existing methods, the number of negative samples is set equal to the number of positive samples, i.e., a positive-negative ratio (PNR) of 1:1. However, in real data, there are much more non-SL gene pairs than SL gene pairs. Therefore, we tested four different PNRs in our experiments, i.e., 1:1, 1:5, 1:20, and 1:50.
Data split methods
Suppose H = {ga, gb, . . . gn} is the set of all human genes, and PSL = {(gi, gj), (gk, gl), . . . } is the set of all known human SL gene pairs. The human gene set H can be divided into two sets, Hseen and Hunseen, which represent currently known genes with SL interactions and the rest genes, respectively. Clearly Hseen ∪ Hunseen = H and . For the machine learning models in this benchmarking study, there are generally three situations encountered in the actual prediction of whether a pair of genes (ga, gb) have SL interaction: (1). {(ga, gb)∣ga ∈ Hseen, gb ∈ Hseen}, (2). {(ga, gb)∣ga ∈ Hseen, gb ∈ Hunseen}, and (3). {(ga, gb)∣ga ∈ Hunseen, gb ∈ Hunseen}.
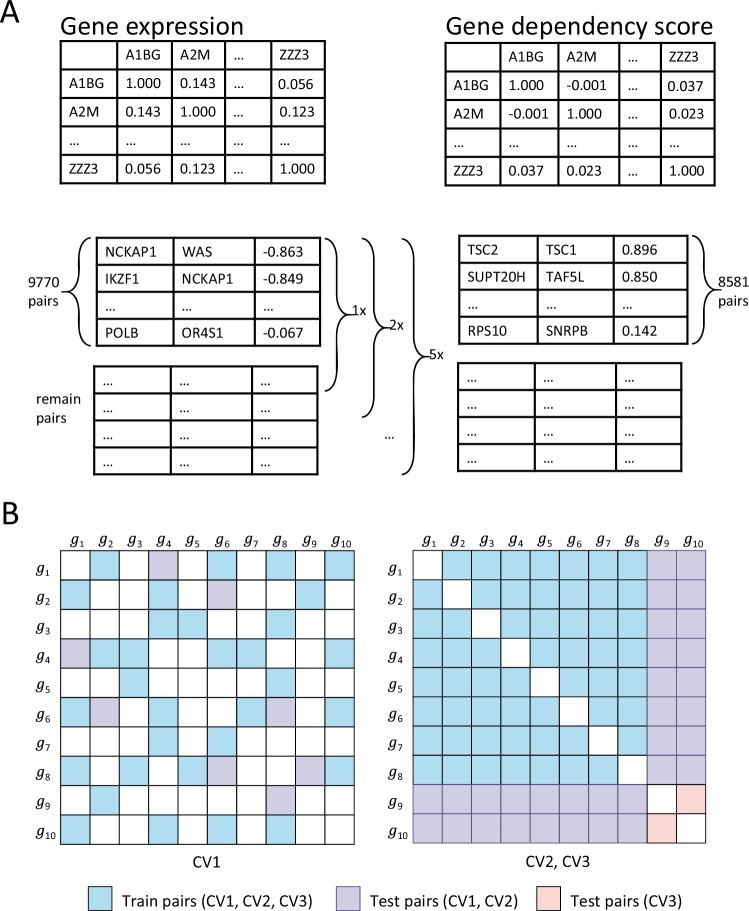
We simulated these possible scenarios through three data split methods (DSMs), namely CV1, CV2, and CV3 (Fig. 5B). The settings are as follows:
CV1: we split the data into training and testing sets by SL pairs, where both genes of a tested pair may have occurred in some other gene pairs in the training set.
CV2: we split the data by genes, where only one gene of a tested pair is present in the training set.
CV3: we split the data by genes, where neither of a tested pair of genes is present in the training set.
Fig. 5. Schematic diagram of negative sampling strategy and data splitting method.
A depicts the negative sampling steps based on gene expression and gene dependency scores. B illustrates the different data partitioning methods: the left matrix shows an example when DSM = CV1, with the blue and purple areas representing the randomly sampled training and test samples, respectively. For DSM = CV2 or CV3, their training samples are both drawn from the blue area, while the purple and orange represent the test sample regions for CV2 and CV3 respectively.
Negative sampling methods
To train the deep learning models for SL prediction, a sufficient number of gene pairs are required, including negative samples. However, non-SL gene pairs are rarely known, which makes it difficult to satisfy the requirements of deep learning models. Therefore, negative sampling is often needed to obtain negative SL data for learning. A common strategy is to randomly select gene pairs from unknown samples as negative samples, which may include false negatives.
To address this issue, we designed two new negative sampling methods (NSMs) based on the DepMap database69. The DepMap database includes gene expression data, gene mutation data, gene dependency scores data, etc. of many cell lines. The gene dependency scores were assessed through the utilization of CRISPR technology, which involves the examination of cellular activity after the single knockout of a specific gene, and a higher gene dependency score indicates lower cell activity. From DepMap database, we obtained gene expression data and gene dependency scores obtained by CRISPR knockout experiments. We have designed two new negative sampling methods (NSMs) using these data: and NSMDep.
The is based on the correlation of expression between two genes. For each pair of genes, we calculated the correlation coefficient of expression () between the genes across the cell lines. From Fig. 4C, we observe that known SL gene pairs tend to have positive correlations of gene expression, i.e., > 0. Therefore, our sampling step is as follows:
We arranged all gene pairs in ascending order of their correlation scores;
To ensure that each gene appears in the negative samples, we first traverse the gene pairs sequentially from the beginning to find the smallest set of gene pairs that can contain all genes;
From the remaining samples, we extract them in order (ascending order in and descending order in NSMDep) and stop when the quantity reaches one, five, twenty, and fifty times the number of positive samples.
Similarly, NSMDep is based on the correlation of the gene dependency score. For each pair of genes, we calculated the correlation coefficient of the dependency score (corrDep) between the two genes. We found that the corrDep of the pairs of known SL genes were distributed mainly in the range [−0.2, 0.2]. Therefore, we first take absolute values for all corrDep and use a sampling step similar to , but unlike , in the first step of the sampling, we rank all gene pairs according to their correlation scores from the highest to the lowest (Fig. 5A shows the specific negative sampling steps).
Evaluation metrics
To comprehensively evaluate the performance of the models, we utilized six metrics. For the classification task, we used three metrics: area under the receiver operating characteristic curve (AUROC), area under the precision-recall curve (AUPR), and F1 score. These metrics are commonly used for binary classification. For the gene ranking task, we employed three metrics: normalized discounted cumulative gain (NDCG@K), Recall@K, and Precision@K. NDCG@K measures whether the known SL gene pairs are in a higher position in the predicted list of a model, while Recall@K and Precision@K are used to evaluate the model’s ability to measure its coverage of relevant content and accuracy in returning the top-K results, respectively. The definitions of these metrics are as follows:
Area Under the Receiver Operating Characteristic Curve (AUROC): AUROC measures the model’s ability to classify samples at different thresholds. It is calculated as the area under the receiver operating characteristic curve, which is a curve plotted with false positive rate on the x-axis and true positive rate on the y-axis. The value of AUROC ranges between 0 and 1.
Area Under the Precision-Recall Curve (AUPR): AUPR is a performance metric used to evaluate binary classifiers, which measures the average precision across different recall levels. Like AUROC, AUPR can be used to evaluate the performance of classifiers in the presence of imbalanced classes or uneven sample distributions. AUPR is a more sensitive metric than AUROC, particularly for classification of imbalanced data.
F1 score: The F1 score is a metric for evaluating the overall effectiveness of a binary classification model by considering both precision and recall. Combining precision and recall into a single value, it provides a balanced measure of the effectiveness of the model.
- Normalized discounted cumulative gain (NDCG@K): NDCG@K can be used to evaluate the ability of a model in ranking candidate SL partners for a gene gi. NDCG@K is calculated as NDCG@K = DCG@K/IDCG@K, where IDCG@K is the maximum DCG@K value among the top-K predictions, and DCG@K is calculated as:
where denotes all known genes that have SL relationships with gene gi, Gi(j) is the j-th gene on the list of predicted SL partners for gene gi, and is the indicator function.1 - Recall@K: Recall@K measures the proportion of correctly identified hits among the top K predicted SL partners to the total known SL partners for gene gi.
2 - Precision@K: Precision@K represents the proportion of correctly identified SL partners among the top K predicted SL partners of gene gi.
3 - To evaluate the overall performance of a model, we calculate its performance under classification and ranking tasks separately and combine them with equal weights to obtain an indicator that reflects the overall performance of the model, i.e., Overall = (Classification score + Ranking score)/2. The classification and ranking scores are calculated as follows, respectively:
4 5 6
In the context of predicting new SL relationships, the CV2 scenario is more realistic and prevalent, and thus it holds greater significance. For most models, CV3 is overly challenging. Therefore, when calculating the integrative classification and ranking scores, we set the weights for CV1, CV2, and CV3 scenarios to 40%, 50%, and 10%, respectively.7
Model selection and implementation
In this study, we benchmarked 12 in-silicon methods for synthetic lethality prediction, including three matrix factorization-based methods and nine deep learning-based methods (Table 1). Among these, PTGNN and NSF4SL use self-supervised learning, while the other methods are supervised or semi-supervised learning depending on specific scenarios. The details of these methods can be found in the Supplementary Methods. For all the methods, their implementation details are as follows:
GRSMF33: due to the lack of executable code in the code repository of the method itself, the code version we used is GRSMF implemented in GCATSL https://github.com/lichenbiostat/GCATSL/tree/master/baseline%20methods/GRSMF. We set num_nodes to 9845, which is the number of genes in our data.
SL2MF31: we used the code of SL2MF from https://github.com/stephenliu0423/SL2MF. The num_nodes was set to 9845.
CMFW32: we used the code of CMFW from https://github.com/lianyh/CMF-W.
SLMGAE36: we used the code of SLMGAE from https://github.com/DiNg1011/SLMGAE. We used the default settings.
NSF4SL42: we used the code of NSF4SL from https://github.com/JieZheng-ShanghaiTech/NSF4SL. The settings aug_ratio = 0.1 and train_ratio = 1 were used.
PTGNN38: we used the code of PTGNN from https://github.com/longyahui/PT-GNN. We have limited the maximum length of protein sequences to 600 and redesigned the word dictionary based on the original paper.
PiLSL41: we used the code of PiLSL from https://github.com/JieZheng-ShanghaiTech/PiLSL. We set the following parameters: –hop 3, –batch_size 512. When calculating the metrics for the ranking task, we need to calculate the scores of all gene pairs, which are about 50 million. PiLSL is a pair by pair prediction approach that demands significant time to obtain all the necessary scores. Therefore, we only considered the performance of the model under the classification task when the PNRs of 1:1, 1:5, and 1:20.
KG4SL39: we used the code of KG4SL from https://github.com/JieZheng-ShanghaiTech/KG4SL. The default parameters are used for the experiment.
SLGNN40: we used the code of SLGNN from https://github.com/zy972014452/SLGNN. The default parameters are used for the experiment.
DDGCN34: we used the code of DDGCN from https://github.com/CXX1113/Dual-DropoutGCN. We set dropout = 0.5 and lr = 0.01, which are the default parameter settings.
GCATSL35: we used the code of GCATSL from https://github.com/lichenbiostat/GCATSL, The default parameters are used for the experiment.
MGE4SL37: we used the code of MGE4SL from https://github.com/JieZheng-ShanghaiTech/MGE4SL, The default parameters are used for the experiment.
Computational resource
The experiments were conducted on a workstation equipped with 4 Intel(R) Xeon(R) Gold 6242 CPUs @ 2.80 GHz, with a total of 64 cores and 22,528 KB cache, along with 503 GB of memory. The system was also equipped with three Tesla V100s 32 GB GPUs, providing a total of 96 GB of GPU memory. The operating system used was Linux Ubuntu 20.04.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Information
Source data
Acknowledgements
We thank Weifan Mao, Zhen Yue, Siyu Tao, and Yang Yang for their assistance in the preliminary study and revision of this paper.
Author contributions
J.Z., M.W., and Y.F. conceived this idea, J.Z. and M.W. designed and guided the project, Y.F., Y.L., Q.L., M.W., and J.Z. participated in manuscript writing, Y.F. completed experiments and result analysis, Y.F. designed charts, H.W., and Y.O. assisted in literature review.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
Source data are provided with this paper. All the data used in our research comes from publicly available sources, including SL labels from SynLethDB 2.0 https://synlethdb.sist.shanghaitech.edu.cn/#/download. The Entrez IDs of the genes come from the NCBI database https://www.ncbi.nlm.nih.gov/gene/. Ensemble ID from https://asia.ensembl.org/index.html. The PPI data come from the data released by BioGRID on June 25, 2022, and the download link is https://downloads.thebiogrid.org/File/BioGRID/Release-Archive/BIOGRID-4.4.211/BIOGRID-ALL-4.4.211.tab.zip. GO annotation and GO term data are respectively from http://geneontology.org/gene-associations/goa_human.gaf.gz and http://geneontology.org/docs/download-ontology/#go_obo_and_owl. The gene expression data and gene dependency score data of the cell lines are from DepMap Public 22Q4 https://figshare.com/articles/dataset/DepMap_22Q4_Public/21637199/2. Pathway data are from KEGG database https://www.kegg.jp/kegg-bin/download_htext?htext=hsa00001&format=json&filedir=kegg/brite/hsa. Pathway data are from Reactome database released on Sep 15, 2022 https://download.reactome.org/82/databases/gk_current.sql.gz. The protein complexes data are from the CORUM database, released on Sep 9, 2022 https://mips.helmholtz-muenchen.de/fastapi-corum/public/file/download_archived_file?version=4.0. The protein sequence data are from the UniProt70 database, released on July 22, 2022 https://www.uniprot.org/help/downloads. All processed training data in this study are publicly available in the Zenodo repository (10.5281/zenodo.13691648)71 with unrestricted access. Source data are provided with this paper.
Code availability
The custom code for integrating the models is available on GitHub at https://github.com/JieZheng-ShanghaiTech/SL_benchmark, while the data is provided in the Zenodo repository at 10.5281/zenodo.1369164871 with unrestricted access.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
2/13/2025
A Correction to this paper has been published: 10.1038/s41467-025-56368-x
Contributor Information
Min Wu, Email: wumin@i2r.a-star.edu.sg.
Jie Zheng, Email: zhengjie@shanghaitech.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-52900-7.
References
- 1.Bridges, C. B. The origin of variations in sexual and sex-limited characters. Am. Nat.56, 51–63 (1922). [Google Scholar]
- 2.Dobzhansky, T. Genetics of natural populations. XIII. Recombination and variability in populations of Drosophila pseudoobscura. Genetics31, 269–290 (1946). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hartwell, L. H., Szankasi, P., Roberts, C. J., Murray, A. W. & Friend, S. H. Integrating genetic approaches into the discovery of anticancer drugs. Science278, 1064–1068 (1997). [DOI] [PubMed] [Google Scholar]
- 4.Kaelin, W. G. Choosing anticancer drug targets in the postgenomic era. J. Clin. Investig.104, 1503–1506 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lord, C. J. & Ashworth, A. PARP inhibitors: synthetic lethality in the clinic. Science355, 1152–1158 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Satoh, M. S. & Lindahl, T. Role of poly(ADP-ribose) formation in DNA repair. Nature356, 356–358 (1992). [DOI] [PubMed] [Google Scholar]
- 7.De Vos, M., Schreiber, V. & Dantzer, F. The diverse roles and clinical relevance of PARPs in DNA damage repair: Current state of the art. Biochem. Pharmacol.84, 137–146 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Krishnakumar, R. & Kraus, W. L. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol. Cell39, 8–24 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bryant, H. E. et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase (vol 434, pg 913, 2005). Nature447, 346–346 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Farago, A. F. et al. Combination olaparib and temozolomide in relapsed small cell lung cancer. Cancer Discov.9, 1372–1387 (2019). [DOI] [PMC free article] [PubMed]
- 11.Moore, K. et al. Maintenance olaparib in patients with newly diagnosed advanced ovarian cancer. N. Engl. J. Med.379, 2495–2505 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Fong, P. C. et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N. Engl. J. Med.361, 123–134 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Liu, L. et al. Synthetic lethality-based identification of targets for anticancer drugs in the human signaling network. Sci. Rep.8, 8440 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Setten, R. L., Rossi, J. J. & Han, S.-P. The current state and future directions of RNAi-based therapeutics. Nat. Rev. Drug Discov.18, 421–446 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Behan, F. M. et al. Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens. Nature568, 511–516 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Topatana, W. et al. Advances in synthetic lethality for cancer therapy: cellular mechanism and clinical translation. J. Hematol. Oncol.13, 1–22 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horlbeck, M. A. et al. Mapping the genetic landscape of human cells. Cell174, 953–967.e22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang, J. et al. Computational methods, databases and tools for synthetic lethality prediction. Brief. Bioinform.23, bbac106 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jerby-Arnon, L. et al. Predicting cancer-specific vulnerability via data-driven detection of synthetic lethality. Cell158, 1199–1209 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Lee, J. S. et al. Harnessing synthetic lethality to predict the response to cancer treatment. Nat. Commun.9, 2546 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sinha, S. et al. Systematic discovery of mutation-specific synthetic lethals by mining pan-cancer human primary tumor data. Nat. Commun.8, 15580 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang, C. et al. Mapping the landscape of synthetic lethal interactions in liver cancer. Theranostics11, 9038–9053 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Kegel, B., Quinn, N., Thompson, N. A., Adams, D. J. & Ryan, C. J. Comprehensive prediction of robust synthetic lethality between paralog pairs in cancer cell lines. Cell Syst.12, 1144–+ (2021). [DOI] [PubMed] [Google Scholar]
- 24.Benfatto, S. et al. Uncovering cancer vulnerabilities by machine learning prediction of synthetic lethality. Mol. Cancer20, 111 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, J. et al. Identification of synthetic lethality based on a functional network by using machine learning algorithms. J. Cell. Biochem.120, 405–416 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Tang, S. et al. Synthetic lethal gene pairs: experimental approaches and predictive models. Front. Genet.13, 961611 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang, J. et al. SynLethDB 2.0: A web-based knowledge graph database on synthetic lethality for novel anticancer drug discovery. Database.2022, baac030 (2022). [DOI] [PMC free article] [PubMed]
- 28.Ashburner, M. et al. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat. Genet.25, 25–9 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oughtred, R. et al. The BioGRID database: a comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci.30, 187–200 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M. & Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res.51, D587–D592 (2022). [DOI] [PMC free article] [PubMed]
- 31.Liu, Y., Wu, M., Liu, C., Li, X.-L. & Zheng, J. SL2MF: predicting synthetic lethality in human cancers via logistic matrix factorization. IEEE/ACM Trans. Comput. Biol. Bioinform.17, 748–757 (2020). [DOI] [PubMed] [Google Scholar]
- 32.Liany, H., Jeyasekharan, A. & Rajan, V. Predicting synthetic lethal interactions using heterogeneous data sources. Bioinformatics36, 2209–2216 (2020). [DOI] [PubMed] [Google Scholar]
- 33.Huang, J., Wu, M., Lu, F., Ou-Yang, L. & Zhu, Z. Predicting synthetic lethal interactions in human cancers using graph regularized self-representative matrix factorization. BMC Bioinform.20, 1–8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cai, R., Chen, X., Fang, Y., Wu, M. & Hao, Y. Dual-dropout graph convolutional network for predicting synthetic lethality in human cancers. Bioinformatics36, 4458–4465 (2020). [DOI] [PubMed] [Google Scholar]
- 35.Long, Y. et al. Graph contextualized attention network for predicting synthetic lethality in human cancers. Bioinformatics37, 2432–2440 (2021). [DOI] [PubMed] [Google Scholar]
- 36.Hao, Z. et al. Prediction of synthetic lethal interactions in human cancers using multi-view graph auto-encoder. IEEE J. Biomed. Health Inform.25, 4041–4051 (2021). [DOI] [PubMed] [Google Scholar]
- 37.Lai, M. et al. Predicting synthetic lethality in human cancers via multi-graph ensemble neural network (IEEE, 2021). [DOI] [PubMed]
- 38.Long, Y. et al. Pre-training graph neural networks for link prediction in biomedical networks. Bioinformatics38, 2254–2262 (2022). [DOI] [PubMed] [Google Scholar]
- 39.Wang, S. et al. KG4SL: knowledge graph neural network for synthetic lethality prediction in human cancers. Bioinformatics37, i418–i425 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu, Y., Zhou, Y., Liu, Y., Wang, X. & Li, J. SLGNN: synthetic lethality prediction in human cancers based on factor-aware knowledge graph neural network. Bioinformatics39, btad015 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu, X. et al. PiLSL: pairwise interaction learning-based graph neural network for synthetic lethality prediction in human cancers. Bioinformatics38, ii106–ii112 (2022). [DOI] [PubMed] [Google Scholar]
- 42.Wang, S. et al. NSF4SL: negative-sample-free contrastive learning for ranking synthetic lethal partner genes in human cancers. Bioinformatics38, ii13–ii19 (2022). [DOI] [PubMed] [Google Scholar]
- 43.Gillespie, M. et al. The reactome pathway knowledgebase 2022. Nucleic Acids Res.50, D687–D692 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, K., Wu, M., Liu, Y., Feng, Y. & Zheng, J. KR4SL: knowledge graph reasoning for explainable prediction of synthetic lethality. Bioinformatics39, i158–i167 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fan, K., Tang, S., Gökbağ, B., Cheng, L. & Li, L. Multi-view graph convolutional network for cancer cell-specific synthetic lethality prediction. Front. Genet. 13, 1103092 (2022). [DOI] [PMC free article] [PubMed]
- 46.Tepeli, Y. I., Seale, C. & Gonçalves, J. P. ELISL: early-late integrated synthetic lethality prediction in cancer. Bioinformatics40, btad764 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shen, J. P. et al. Combinatorial CRISPR-Cas9 screens for de novo mapping of genetic interactions. Nat. Methods14, 573–576 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Han, K. et al. Synergistic drug combinations for cancer identified in a CRISPR screen for pairwise genetic interactions. Nat. Biotechnol.35, 463–474 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Najm, F. J. et al. Orthologous CRISPR-Cas9 enzymes for combinatorial genetic screens. Nat. Biotechnol.36, 179–189 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao, D. et al. Combinatorial CRISPR-Cas9 metabolic screens reveal critical redox control points dependent on the KEAP1-NRF2 regulatory axis. Mol. Cell69, 699–708.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma, M., Na, S. & Wang, H. AEGCN: an autoencoder-constrained graph convolutional network. Neurocomputing432, 21–31 (2021). [Google Scholar]
- 52.Li, Q., Han, Z. & Wu, X.-m. Deeper insights into graph convolutional networks for semi-supervised learning. in Proc. of the AAAI Conference on Artificial Intelligence32 (2018).
- 53.Ito, T. et al. Paralog knockout profiling identifies DUSP4 and DUSP6 as a digenic dependence in MAPK pathway-driven cancers. Nat. Genet.53, 1664–1672 (2021). [DOI] [PubMed] [Google Scholar]
- 54.Parrish, P. C. R. et al. Discovery of synthetic lethal and tumor suppressor paralog pairs in the human genome. Cell Rep.36, 109597 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thompson, N. A. et al. Combinatorial CRISPR screen identifies fitness effects of gene paralogues. Nat. Commun.12, 1302 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vidigal, J. A. & Ventura, A. Rapid and efficient one-step generation of paired gRNA CRISPR-Cas9 libraries. Nat. Commun.6, 8083 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang, B. et al. The tumor therapy landscape of synthetic lethality. Nat. Commun.12, 1275 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Srivatsa, S. et al. Discovery of synthetic lethal interactions from large-scale pan-cancer perturbation screens. Nat. Commun.13, 7748 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reid, R. J. D. et al. A synthetic dosage lethal genetic interaction between CKS1B and PLK1 is conserved in yeast and human cancer cells. Genetics204, 807–819 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.O’Neil, N. J., Bailey, M. L. & Hieter, P. Synthetic lethality and cancer. Nat. Rev. Genet.18, 613–623 (2017). [DOI] [PubMed] [Google Scholar]
- 61.Muller, F. L., Aquilanti, E. A. & Depinho, R. A. Collateral lethality: a new therapeutic strategy in oncology. Trends Cancer1, 161–173 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dey, P. et al. Genomic deletion of malic enzyme 2 confers collateral lethality in pancreatic cancer. Nature542, 119–123 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li, S. et al. Development of synthetic lethality in cancer: molecular and cellular classification. Signal Transduct. Target. Ther.5, 241 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Seal, R. L. et al. Genenames.org: the HGNC resources in 2023. Nucleic Acids Res. 51, D1003–D1009 (2022). [DOI] [PMC free article] [PubMed]
- 65.Cunningham, F. et al. Ensembl 2022. Nucleic Acids Res.50, D988–D995 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chua, H. N., Sung, W.-K. & Wong, L. Exploiting indirect neighbours and topological weight to predict protein function from protein-protein interactions. Bioinformatics22, 1623–1630 (2006). [DOI] [PubMed] [Google Scholar]
- 67.Tsitsiridis, G. et al. CORUM: The comprehensive resource of mammalian protein complexes-2022. Nucleic Acids Res.51, D539–D545 (2022). [DOI] [PMC free article] [PubMed]
- 68.Yu, G. Gene ontology semantic similarity analysis using GOSemSim. in Methods in Molecular Biology2117, 207–215 (2020). [DOI] [PubMed]
- 69.Tsherniak, A. et al. Defining a cancer dependency map. Cell170, 564–576.e16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.The UniProt Consortium. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res.49, D480–D489 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Feng, Y. et al. Benchmarking machine learning methods for synthetic lethality prediction in cancer. Zenodo repository, https://zenodo.org/records/13691648 (2024). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Information
Data Availability Statement
Source data are provided with this paper. All the data used in our research comes from publicly available sources, including SL labels from SynLethDB 2.0 https://synlethdb.sist.shanghaitech.edu.cn/#/download. The Entrez IDs of the genes come from the NCBI database https://www.ncbi.nlm.nih.gov/gene/. Ensemble ID from https://asia.ensembl.org/index.html. The PPI data come from the data released by BioGRID on June 25, 2022, and the download link is https://downloads.thebiogrid.org/File/BioGRID/Release-Archive/BIOGRID-4.4.211/BIOGRID-ALL-4.4.211.tab.zip. GO annotation and GO term data are respectively from http://geneontology.org/gene-associations/goa_human.gaf.gz and http://geneontology.org/docs/download-ontology/#go_obo_and_owl. The gene expression data and gene dependency score data of the cell lines are from DepMap Public 22Q4 https://figshare.com/articles/dataset/DepMap_22Q4_Public/21637199/2. Pathway data are from KEGG database https://www.kegg.jp/kegg-bin/download_htext?htext=hsa00001&format=json&filedir=kegg/brite/hsa. Pathway data are from Reactome database released on Sep 15, 2022 https://download.reactome.org/82/databases/gk_current.sql.gz. The protein complexes data are from the CORUM database, released on Sep 9, 2022 https://mips.helmholtz-muenchen.de/fastapi-corum/public/file/download_archived_file?version=4.0. The protein sequence data are from the UniProt70 database, released on July 22, 2022 https://www.uniprot.org/help/downloads. All processed training data in this study are publicly available in the Zenodo repository (10.5281/zenodo.13691648)71 with unrestricted access. Source data are provided with this paper.
The custom code for integrating the models is available on GitHub at https://github.com/JieZheng-ShanghaiTech/SL_benchmark, while the data is provided in the Zenodo repository at 10.5281/zenodo.1369164871 with unrestricted access.