Abstract
Objectives
This article describes the design and evaluation of MS Pattern Explorer, a novel visual tool that uses interactive machine learning to analyze fitness wearables’ data. Applied to a clinical study of multiple sclerosis (MS) patients, the tool addresses key challenges: managing activity signals, accelerating insight generation, and rapidly contextualizing identified patterns. By analyzing sensor measurements, it aims to enhance understanding of MS symptomatology and improve the broader problem of clinical exploratory sensor data analysis.
Materials and Methods
Following a user-centered design approach, we learned that clinicians have 3 priorities for generating insights for the Barka-MS study data: exploration and search for, and contextualization of, sequences and patterns in patient sleep and activity. We compute meaningful sequences for patients using clustering and proximity search, displaying these with an interactive visual interface composed of coordinated views. Our evaluation posed both closed and open-ended tasks to participants, utilizing a scoring system to gauge the tool’s usability, and effectiveness in supporting insight generation across 15 clinicians, data scientists, and non-experts.
Results and Discussion
We present MS Pattern Explorer, a visual analytics system that helps clinicians better address complex data-centric challenges by facilitating the understanding of activity patterns. It enables innovative analysis that leads to rapid insight generation and contextualization of temporal activity data, both within and between patients of a cohort. Our evaluation results indicate consistent performance across participant groups and effective support for insight generation in MS patient fitness tracker data. Our implementation offers broad applicability in clinical research, allowing for potential expansion into cohort-wide comparisons or studies of other chronic conditions.
Conclusion
MS Pattern Explorer successfully reduces the signal overload clinicians currently experience with activity data, introducing novel opportunities for data exploration, sense-making, and hypothesis generation.
Keywords: understanding patient experience, sensor data exploration, interactive machine learning, multiple sclerosis, data visualization
Introduction
The surge in digital health data, especially activity sensor data, complements traditional health records by offering detailed insights into patient wellness and disease expression, with potential for improved prevention and disease (self-)management. Consumer sensors like smartwatches allow for non-intrusive monitoring, providing high-resolution, multivariate data. This data supports various analytical approaches, including machine learning on lifestyle factors, potentially improving chronic disease management1–4 and personalized treatment options, thereby enhancing care quality.5
However, the complexity and granularity of continuously measured activity data pose challenges in finding meaningful patterns. Human input in the analysis, a core aspect of visual analytics (VA), can improve understanding by extracting meaningful and actionable signals on health-status changes6,7 (Figure 1). Despite existing VA solutions in healthcare, few address chronic disease management through consumer-grade sensor data. Monitoring factors like activity and sleep quality can prove invaluable to understanding chronic disease management.
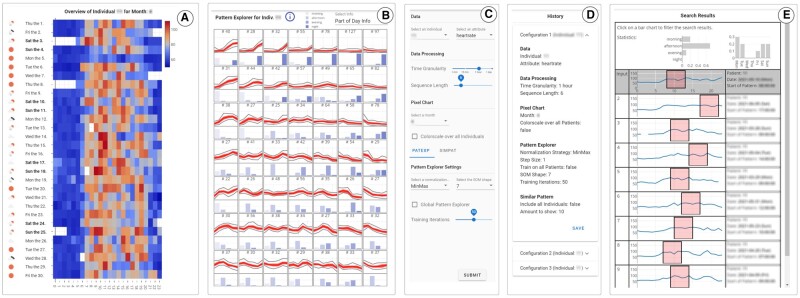
Figure 1.
The MS Pattern Explorer interface. Experts identify activity time series in the Temporal Overview (A) for a selected sensor and specific PwMS, at multiple temporal granularity levels. To explore available activity patterns of interest, experts can cluster sequences by their similarity in the Pattern Explorer View (B), and discovered patterns of interest can be contextualized through explanatory metadata visualizations. Data, model, and visualization parameters can be adjusted with the Configuration Panel (C). Provenance information on previous analysis configurations is tracked in the History View (D). Users can search for similar, user-specified activity sequences in the Similar Pattern View (E). To enable comparative between-PwMS analyses, MS Pattern Explorer automatically scales the visual encodings to accommodate for multiple PwMS.
This study addresses multiple sclerosis (MS) management, a chronic, neurodegenerative disease with highly variable symptoms, like fatigue and vision problems.8,9 There is no cure for MS; treatment aims to reduce relapses and slow progression. Physical activity is crucial for maintaining abilities and may slow disease progression.3,9,10 We refer to those affected as “person(s) with MS” or PwMS, and to MS domain experts as experts.
Background
Current analysis practices of MS domain experts
In MS research, analyzing PwMS physical activity data identifies challenges in maintaining physical activity, including motivation, fatigue, and accessibility. Remote sensors help sustain activity levels by providing motivation and useful data like heart rate and step count. The long-term goal of remote sensing is to guide healthcare providers in managing MS symptoms by leveraging undiscovered knowledge from sensor data across various timeframes and symptomology.
Analytical methods for time series data include regression trees, local dynamic complexity for detecting significant changes,11–14 ARIMA models for pattern analysis,15 and Bayesian structural time series modeling to assess rehabilitation effects, accounting for variables like age and disease duration.16 Addressing data quality, methods like the Kalman Filter are used for imputing missing data.15,17 This analytical process requires domain expertise in data analysis and a deep understanding of data characteristics. Optimized for data exploration support, our grid-based visualizations aid clinicians in such hypothesis testing. These visualizations can highlight variations in step count or sleep habits, indicating challenges PwMS face in maintaining stable activity levels after in-clinic rehabilitation, and thus making them more interpretable and actionable to healthcare providers.
Visual analytics in healthcare
Discussions with experts revealed a distinction between supporting single-patient analyses and multi-patient analyses, reflecting trends observed in related surveys.6,18,19 Single-patient methods, varying by data type, include LifeLines’ Gantt chart metaphor for medical event time series,20 Doccurate’s textual clinical history summaries,21 and VisuExplore’s multiple attribute visualizations.22 MS Pattern Explorer focuses on PwMS’ sensor data exploration and contextual analysis, like recent efforts in contextual analysis of Type 2 Diabetes by Philip et al.23 MS Pattern Explorer detects temporal patterns in activity data using a pixel-based visualization inspired by the approach of Gnaeus.24 MS Pattern Explorer is also inspired by single-to-multiple individual comparison cohort analysis approaches like CarePre25 and KAVAGait.26 At cohort-level, MS Pattern Explorer uses clustering and visual aggregation methods for the structured overview of time series data characteristics of PwMS.27–29 Finally, we use temporal attributes like the day of the week and weather conditions to find interesting relationships for the contextualization of time series patterns.30,31
Temporal exploration, search, and contextualization
Our approach for time series data exploration combines overviews and interactive techniques, influenced by best practices in dashboard design,32 human motion analysis,33 open research data,33,34 and exploratory search methods.33,34 We simplify data complexity through dimensionality reduction35 and clustering,42 using interactive visual queries inspired by query-by-example31 for effective pattern searching. This method streamlines searches,39 reduces the need for complete data familiarity,33,34 and adjusts for varying definitions of similarity.45 To contextualize patterns, we integrate metadata analysis with time series interpretation, using a Self-Organizing Map (SOM) algorithm41 for both clustering and mapping metadata to patterns, addressing our experts’ needs.36,45,46,48
Significance
This study acknowledges the significance of personal sensing devices in investigating not only physical activity but also other lifestyle factors like sleep quality and sleep-wake cycles. Working in collaboration with experts, we created MS Pattern Explorer, analyzing data from an observational MS study of smartwatches tracking the physical activity of 45 PwMS over a median of 7 weeks (see Figure 2).1,38 The study’s multivariate time series data presented analysis, visualization, and cognitive overload challenges concerning the merging and management of large data, and the identification of signals relevant to PwMS1 for insight and hypothesis generation. We identify and address 3 analysis challenges in experts’ existing workflows, concerning the exploration, search, and contextualization of time series activity data and metadata of PwMS. MS Pattern Explorer allows experts to personalize MS management for a given PwMS by providing comprehensive time-series sensor signals in one easy-to-use platform. Our iterative design approach included consultation with experts in clinical MS research and data science.
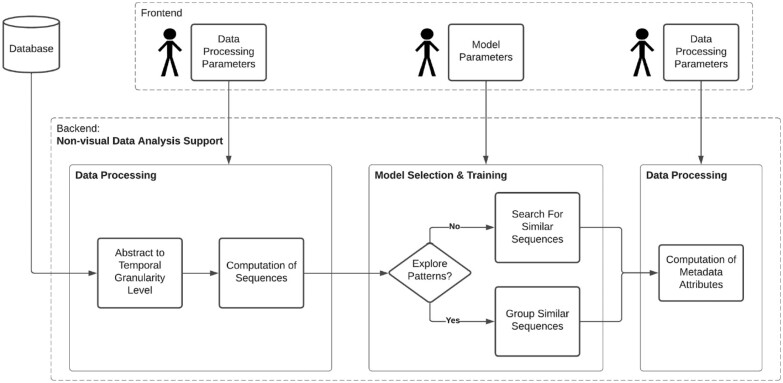
Figure 2:
Assembly of computational methods into a data science workflow. Data analysis support can be divided into the data processing area and the model selection and training area. Data processing activities occur before and after the model selection and training.
Objective
This study introduces the MS Pattern Explorer, a VA approach addressing challenges in analyzing smartwatch data from individuals with MS.1 Our collaboration with MS experts identifies 3 key analysis challenges in experts’ workflows with time series activity data:
Exploration: Exploring long time series at various granularities (minutes to days) lacks systematic support, making pattern discovery manual and slow for MS experts. Effective time series exploration and visual data abstractions would accelerate insight and hypothesis generation.
Search: Searching for specific time series sequences or patterns of user-defined lengths within or across PwMS is challenging, impeding the process of experts to assess the (re-)occurrence of search queries, to generalize findings, and to validate data-driven hypotheses. Meaningful query formulation and effective visual representation of search results would improve identification of significant activity patterns for individuals or cohorts.
Contextualization: Contextualizing time series patterns through auxiliary and explanatory metadata is currently a tedious manual process, challenging the experts to assess pattern frequency and significance. An example would be a pattern of high activity (step count or heart rate) surprisingly often happening Saturday morning, if the sun is shining. Effective visual analytics for detecting significant MS activity patterns would enhance experts’ sense-making abilities.
To address these challenges, MS Pattern Explorer uses multiple linked views, which together allow for enhanced discoverability and understanding of activity patterns of PwMS. We then study the usability of MS Pattern Explorer across 3 relevant user groups, to assess the accessibility of the tool to clinicians, data scientists, and patients themselves.
Methods
Setting
This study analyzed longitudinal activity measurement data collected as part of the “Barriers to physical activity in people with MS” (BarKA-MS) study.1 Participants were inpatients at the Valens Rehabilitation Centre, a clinic providing comprehensive inpatient and outpatient rehabilitation services, specializing in early neurological rehabilitation and internal and musculoskeletal rehabilitation. The primary objective of the BarKA-MS study was to assess physical activity levels in PwMS using an activity tracker during and after an inpatient rehabilitation stay. In addition, it sought to identify potential barriers to physical activity through standardized questionnaires and free-text assessments. Validation with research-grade sensor devices confirmed the suitability of the commercially available activity tracker to accurately measure several daily physical parameters, such as heart rate and step count.35,37,42 45 PwMS were equipped with a commercial activity tracker (“Fitbit Inspire HR”) and monitored during their rehabilitation stay as well as for 4 weeks after discharge (Appendix C—Figure S7).
Data characterization and terminology
We established a common understanding of data characteristics and usage of data-centric terminology between ourselves and our experts. The 2 activity data attributes considered (step count and heart rate) are univariate time series, with a temporal granularity up to 1 minute (heart rate). We defined 4 temporal granularity levels (1 minute, 15 minutes, 1 hour, 1 day), allowing experts to explore highly specific phenomena through analysis, such as inference on activity and sleep. A sequence in time series has a user-definable length and is used for interactive visual clustering and as a query in sequence searches. Its duration is determined by length and granularity, for example, “seven minutes”. Finally, we define patterns as generalizable, frequent temporal findings, represented by sets of similar sequences of equal length.
Task characterization
We established a set of design requirements (R#) and user tasks (U#) through iterative co-design sessions with our clinical expert collaborators. These can be found in Tables 1 and 2, with a more detailed description in Appendix D. These requirements and tasks also guide the tool’s user study evaluation, mapping to specific analytical tasks evaluated (T#), as shown in Appendix B.
Table 1.
MS Pattern Explorer’s design requirements.
| Requirement | Description |
|---|---|
| R1 Attribute selection | Switch between heart rate and step count attributes interactively. |
| R2 Patient selection | Enable selection of a PwMS for detailed analysis. |
| R3 Granularity control | Adapt visual interfaces and algorithmic support to changes in time granularity. |
| R4 Sequence and pattern specification | Specify the length of temporal phenomena to limit search space and enhance search efficiency. |
| R5 Model control | Provide interactive control for ML clustering and retrieval. |
| R6 Within and between-PwMS analysis | Support workflows for both single and multiple PwMS analysis. |
| R7 Analysis history | Automatically track workflow history and enable return to different stages. |
Table 2.
MS Pattern Explorer’s user tasks.
| Task | Description |
|---|---|
| U1 Sequence analysis | Detailed longitudinal analysis of temporal sequences for selected PwMS, focusing on heart rate and step count. |
| U2 Pattern exploration | Explore patterns for selected MS activity attributes using clustering algorithms and interactive ML processes. |
| U3 Sequence search | Search for similar sequences based on a query, retrieving the k-most similar sequences. |
| U4 Temporal pattern localization | Localize pattern sequences along the temporal attribute to see distribution for a PwMS. |
| U5 Pattern contextualization | Relate frequent patterns to external metadata attributes like time of day, day of week, and weather conditions for context, to further support hypothesis generation. |
Data preparation
We derived metadata attributes to facilitate the contextualization of patterns in PwMS activity data, that is, the explanation of patterns with real-world context. An example would be a pattern that happens significantly more often during sunny weather. Four expert-identified, diversely sourced metadata attributes enrich pattern interpretation under various factors.
Computational analysis methods
Sequence computation
MS Pattern Explorer allows users to specify sequences and patterns of different lengths and granularities. This initiates on-the-fly sequence computation for the chosen time series attribute, using a sliding window approach to traverse PwMS activity data. The computed sequence vectors serve as the basis for our machine learning and information retrieval support (Figure 2). To enhance usability, sequences with missing values are excluded, and global min-max normalization is applied by default, with an optional user parameter for local normalization.
Machine learning support: self-organizing map (SOM)
To facilitate visual data exploration, we use a Self-Organizing Map (SOM) approach, due to its advantages in visual cluster analysis and pattern exploration, especially for unexplored data.41 SOM is a neural network trained by iterating over normalized sequences serving as data input vectors. We chose the Euclidean activation function as the similarity measure, which generalizes across all types of user-specified sequences (steps, heart-rate changes, sleep). The cells of the SOM are initialized using PCA dimensionality reduction.36 The SOM grid structure optimally utilizes display space, is effective for vector quantization, and represents patterns in a similarity-preserving way.43 The SOM’s grid size can be user-defined, with a default setup of 7 × 7 cells. Patterns identified by this approach are dynamic, emerging at runtime based on user behavior, rather than being precomputed on the input dataset.
Information retrieval support: k-nearest neighbor proximity search
A k-Nearest Neighbor (kNN) proximity search is used to facilitate search, providing the n most similar sequences, based on user input.44 Together with the experts, we decided for Euclidean distances applied to sequence vectors. The sequences with the smallest distances are presented as a list in the search results, either for within-PwMS or between-PwMS analysis.
Tool implementation
An overview of the MS Pattern Explorer’s implementation schematic can be seen in Figure 3.
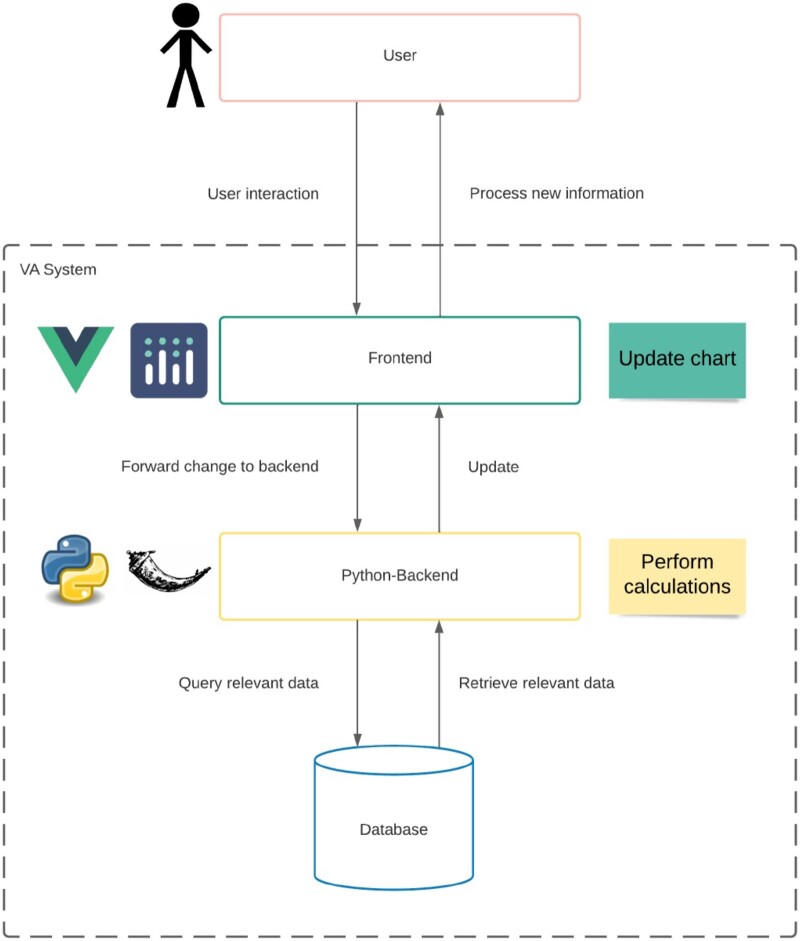
Figure 3:
The MS Pattern Explorer’s architecture. We use a Vue framework-based frontend with Plotly for visualizations, and a separate Python Flask Application for API call handling. User interaction involves frontend input, backend processing, and subsequent frontend updates with relevant data.
Tool evaluation
We conducted a 15-person user study with in-person and online interviews. Participants were equally distributed amongst 3 target user groups: data-scientists, clinicians working in the MS domain, and non-experts (members of neither of these). This classification allows for the assessment of the tool not only with the primary user group (clinicians), but also for the comparison with other relevant stakeholders in healthcare, in line with the design of similar studies.45 Participants were on average 34.1 years old (SD = 11.35) and 66.66% were male. None of the participants had seen MS Pattern Explorer before or performed analyses on the Barka-MS dataset and had varying degrees of experience with data analysis (data literacy). We asked the participants to answer a series of open (O) and closed (C) questions that comprehensively cover the tool’s feature set, to assess the intuitiveness of the tool in conducting exploratory data analysis across the 3 user groups. We grouped these questions under a set of analytical tasks, aiming for complete coverage of our characterized design requirements and user tasks (Tables 1 and 2, respectively). These questions can be found in Appendix B.
To accurately reflect task completion for closed questions, beyond binary assessment, participant responses were scored within the interval 0.0 (task failure) and 1.0 (task completion). We applied deductions of 0.2 or 0.5 points to the score if participants required minor usability hints or assistance. This allowed us to assess the degree to which our feature implementation was intuitive for our target user groups. Open question responses were evaluated based on the number of relevant insights identified, as in other literature on insight-based evaluation.33,46,47
The evaluation procedure included a brief introduction (5 minutes) to the research domain, followed by an overview of MS Pattern Explorer (5 minutes), where we deliberately only showed the different views without demonstrating any interaction techniques. Finally, the participants were asked to use MS Pattern Explorer to complete a set of usage tasks that comprehensively cover the tool’s interactive functionalities (20 minutes).
Results
MS Pattern Explorer tool
In this section, we refer to the requirements (R) and user tasks (U) identified in Tables 1 and 2 to clearly connect our user-centered development steps together.
Temporal overview
The Temporal Overview component (Figures 1A and 4) presents longitudinal time series data for one chosen attribute and PwMS (R1, R2, R3). Users can conduct detailed temporal sequence analysis at various granularities and identify multiple sequences through highlighting (U1). These sequences may represent previously explored patterns or search results (U2). Figure 4 captures absolute time, with the x-axis indicating finer granularities like days within a week (R3). This component adjusts to the user’s chosen temporal granularity (U4), accommodating different views, such as hours of a day (Figure 1). When set to 1 day granularity, the Temporal Overview displays the complete MS study period, as shown in Figure 4 for step count. The y-axis represents the week of the year, and the x-axis represents the day of the week. Users can switch to a between-PwMS comparison of sequences (R6), adjusting the scale of the color encoding of values to accommodate cohort-wide values, providing a global perspective useful for assessing PwMS activity variations. This feature is particularly valuable to experts when trying to understand the diverse activity levels among PwMS in the cohort (R6, U5) (Table 3).
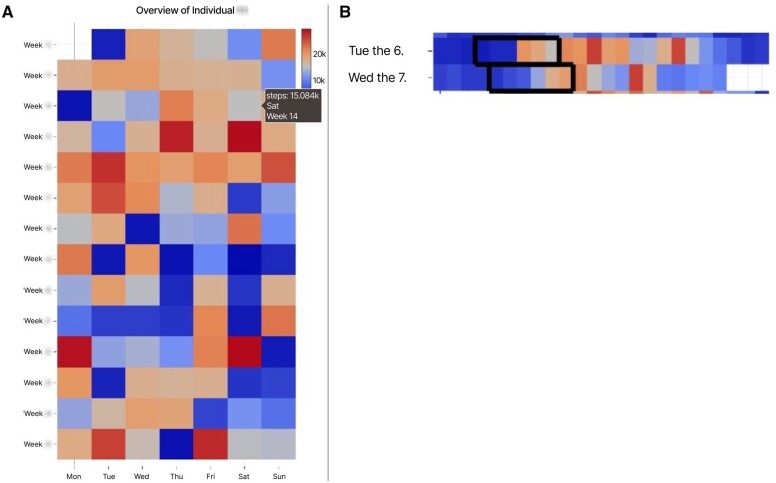
Figure 4.
(A) Temporal Overview of a PwMS’ weekly step counts; (B) exploring for PwMS activity patterns. In (B), 2 similar sequences occur on 2 consecutive days in the Temporal Overview (highlighted by the black rectangle marks).
Table 3.
Data and metadata attributes computation.
| Data attribute—type | Description | Contextual relevance | Source of data | Type and range of values |
|---|---|---|---|---|
| Part of the day—metadata | Splits the day into 4 segments | Helps detect energy loss in PwMS with fatigue during the day, a common symptom. | Computed values |
|
| Part of the week—metadata | Indicates on which weekdays specific patterns occur, applicable across all 4 granularity levels | Many PwMS are pursuing jobs or have family duties, leading to different physical activity patterns between weekdays and weekends. | Computed values | Categorical: Monday, Tuesday, Wednesday, Thursday, Friday, Saturday, Sunday |
| PwMS distribution—metadata | Anonymized PwMS IDs help identify if activity patterns are common to specific PwMS or are balanced in distribution across PwMS. | Identifies specific PwMS activity patterns, helping to manage relevance and cardinality. In the interface, the top 3 PwMS for each pattern are highlighted. | BarKA-MS39 | Categorical: Anonymized PwMS IDs |
| Weather condition—metadata | Weather data from a public API is integrated for multimodal analysis of PwMS sequences, while maintaining patient anonymity | Weather may impose additional barriers for physical activity, as heat can worsen specific MS symptoms. | Openweathermap Public API40 | Categorical: sunny, rainy, cloudy, snowy, etc. |
| Heart rate—data | Wearables-based measurement of PwMS heart rate | Approximates sleep and stress evaluations | BarKA-MS39 | Numerical |
| Steps—data | Wearables-based measurement of PwMS step count | Approximates level of activity, and indicates possible fatigue episode | BarkaMS39 | Numerical |
Exploration of activity patterns
MS Pattern Explorer enables effective time series exploration by implementing an interactive ML process, leveraging the Self-Organizing Maps (SOM) algorithm algorithm. 22 The clustering algorithm reveals frequent patterns of user-steerable length and model parameters (R5). Results of this exploratory cluster analysis are presented in the Pattern Explorer View (Figure 1B), a grid-based view with each cell containing one sequence pattern, where the variety and heterogeneity of patterns are easily identifiable by the clinicians (U5). An overview of the workflow for steering the exploratory analysis process can be seen in Figure 5.
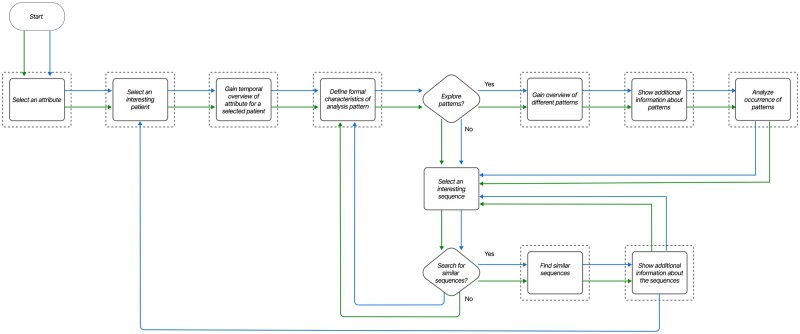
Figure 5:
Experts’ workflow when performing exploratory data analysis. First, experts get an overview of the patient, and define the formal characteristics of a pattern. Then, experts explore new patterns, or search for similar sequences. We support 2 main workflows: within-patient analysis (bottom arrows, in green), and across-patient analysis (top arrows, in blue).
The pattern of every cell is encoded with a thick red line, which allows for gaining an overview of the variety of patterns (U1, U2). Thin black lines represent the heterogeneity of sequences distributed in every pattern/cell. At the top of every cell, the sequence count is shown (eg, #25), that is, the number of sequences that the clustering algorithm has assigned to a particular cell. Figure 1B shows 6-hour heart rate patterns for a PwMS, with upward trends at the top left of the SOM, low heart rate patterns at the top right, and diverse patterns with high heart rates located at the bottom.
The linking between the Pattern Explorer View (B) and the Temporal Overview (A) allows users to analyze the distribution of temporal occurrences of a pattern (U2, U4). A click on a cluster cell selects the patter, linking it to the Temporal Overview where these pattern sequences are highlighted. This can, for example, be seen in Figure 4B, where sequences with upward trends in the time series are highlighted.
Users are in control of the normalization strategy, the grid dimensions, and the number of training iterations (R5). While designed for simplicity with standard SOM parameters,22,23 it also offers adjustable settings for more tailored analysis, catering to both individual and group-level (between-PwMS) explorations (R6).
Searching for PwMS activity sequences
MS Pattern Explorer enables effective search for temporal activity patterns with the Similar Pattern View (Figure 1E), aiding clinicians in finding sequences similar to a predefined query using a proximity search algorithm (R4, U3). This algorithm retrieves and ranks the k-most similar sequences, displayed together with the query sequence always at the top. Users select a query sequence from the Temporal Overview (A), with user-specified data processing parameters (U5). The number of sequences shown in the Similar Pattern View is adjustable.
Each list element shows the retrieved sequence within its temporal attribute context (blue line chart) using a rectangle metaphor with a bright-red background to highlight the sequence.
Each sequence is accompanied by relevant metadata like the PwMS ID and sequence start date, maintaining data privacy. An interactive bar chart on the right of every list element shows attributes like the time of day or day of the week, for contextualization and filtering (U2, U5). This functionality enables users to drill down into specific data subsets, such as sequences occurring on Thursday afternoons, blurred in Figure 1E for data privacy reasons.
In the between-PwMS analysis mode, experts can search and compare sequences across multiple PwMS to identify patterns and similarities (R6, U4), simplifying the selection of interesting PwMS for further analysis (R2). The interface allows for easy navigation between different PwMS analyses and supports iterative searches, where selecting a sequence from the result list can trigger a new search based on that sequence (R7). Appendix A—Case Study 2 exemplifies these results.
Contextualizing identified PwMS activity patterns
MS Pattern Explorer aids users in contextualizing sequence patterns, whether found through pattern exploration or sequence search. The Pattern Explorer View (B) offers a bar chart at the bottom of each SOM cell, showing the distribution of selected metadata attributes, with a color legend explaining these attributes. An example includes the contextualization of heart rate patterns with time-of-day information, like morning or evening (U4). This tool is particularly useful in between-PwMS analyses, helping experts determine if patterns are common across multiple PwMS or specific to individuals (R6). Additionally, the Similar Pattern View (E) offers contextualization through bar charts indicating metadata attribute occurrences, like time of day or day of the week, aiding in understanding search results (U1, U4, U5). For instance, it can show sequences primarily occurring in the afternoon, but not on Wednesdays or at night (Figure 4B). Appendix A—Case Study 1 exemplifies these results.
User-centered evaluation results of MS Pattern Explorer
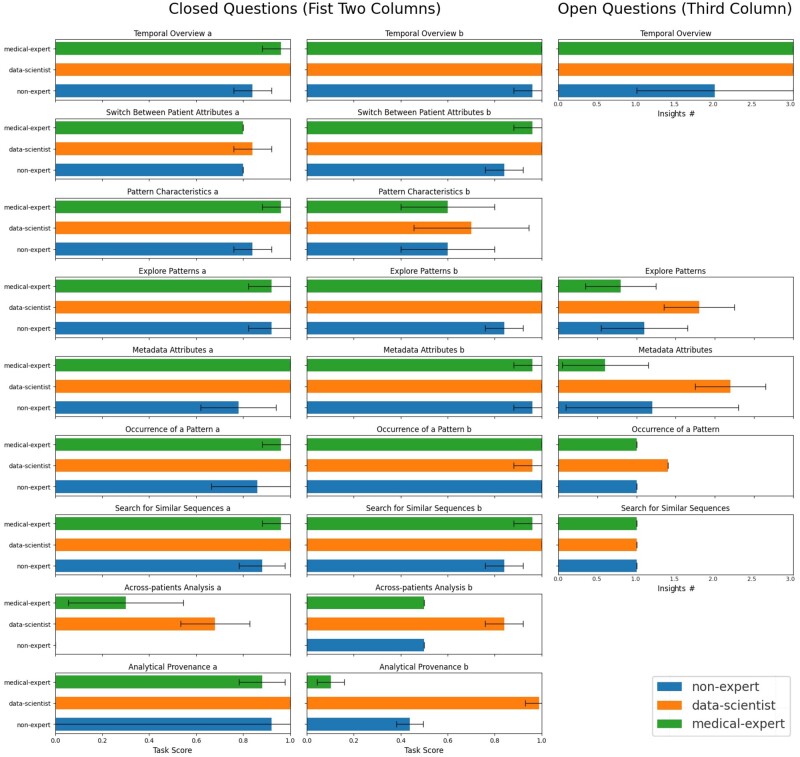
In this section, we refer to the analysis tasks (T) identified in Appendix B to connect our user-centered development to our evaluation strategy. We observed distinct performance patterns among data scientists, medical experts, and non-experts in handling both closed and open questions. Data scientists consistently outperformed both medical experts and non-experts, with a notably lower variability in scores for open questions among participants of this group (Figure 6). Medical experts had a marginal advantage over non-experts in closed questions, but their performances were comparable in open questions. When interacting with the Temporal Overview of a PwMS (T1), data scientists mainly focused on data-specific insight like missing values, while non-experts observed general patterns (eg, morning activity), or related the time series to medical events (eg, physiotherapy). The identified patterns were assessed qualitatively through discussions with experts, in line with other studies on biomarker identification via fitness trackers.35,39,48
Figure 6:
Results of the user study. From left to right: Column 1 and 2: Task scores for the closed questions across user groups, represented as colored bars. Each question charted shows the average score of a user group’s participant, with error bars indicating the standard error. Participants generally had the most difficulty solving across-patients analysis, with no considerable performance differences between user groups. Column 3: Task score for the open questions across user groups, represented as colored standard error bars with a capped global maximum of 3 insights. Overall, data scientists made more insights than both non-experts and medical experts.
The biggest performance gap can be observed in questions T3–T5 (see Appendix B), where participants defined and explored patterns and gained additional knowledge through metadata attributes. Regarding the tool’s objectives (exploration, search, and contextualization of patterns in temporal activity data), data scientists scored higher in most tasks, especially in pattern identification and metadata interpretation. However, there are no considerable performance differences between user groups.
Discussion
Our results indicate that although domain experts using MS Pattern Explorer were sometimes overwhelmed by the parametrization-dependent analyses, they were able to interpret high-level patterns with ease. The tool provided simple, interpretable information usually inaccessible to them. We discuss the implications, possible extensions, and limitations of our work below:
Implications for clinician workflow
MS Pattern Explorer represents a meaningful development in medical informatics. Clinicians often hesitate to use wearable device data due to concerns about time investment, data volume, and the challenge of extracting meaningful insights.48 Our tool aims to address these issues by making data more accessible and useful, offering a new range of analytical possibilities. It seeks to bridge the gap between data science and frontline healthcare by providing visual analyses that could facilitate the integration of wearable technology into routine care. The history functionality standardizes analyses, making them repeatable across time and patients, thus enhancing comparability. Additionally, clinicians can use predefined analysis sets to discuss physical activity and fatigue with patients, addressing a need identified by expert neurologists. By addressing these gaps, our tool aims to support clinicians in leveraging the potential of wearables, to enhance patient care.
Interactive data labeling
MS Pattern Explorer enables users to explore and compare time series patterns identified via precomputed features and a pretrained model. Our approach could benefit from the incorporation of additional knowledge from experts or PwMS, facilitated by interactive data labeling performed while using the tool. Future work may include allowing PwMS to comment on their wellbeing, like existing approaches in healthcare.49–51
Transferability to other domains
MS Pattern Explorer uses consumer sensing devices, making the analyzed signals generic. Feedback from medical experts suggests our approach could also be useful in other domains, such as long-COVID management.3 However, involving relevant stakeholders in selecting signals, processing data, creating metadata, and designing the system is crucial. MS Pattern Explorer cannot be assumed to work as-is in a new disease context. Overall, the innovation of MS Pattern Explorer lies in the creation of its analysis and visualization toolbox, and the testing of its stakeholder activation strategy.
Performance gaps by user group
In questions T3-T5 (see Appendix B), we suspect the following reasons for a performance gap: (1) Non-experts and medical experts lacked exploratory data analysis experience, making it difficult to find interesting patterns. (2) Their limited analytical background made interpreting clustering results hard. (3) They found a visualization of 7 × 7 clusters (default parameter settings) overwhelming. (4) Metadata attributes needed further explanation before being understood and valued. (5) While most data scientists were able to perform between-PwMS analyses (T8) with minimal guidance, others encountered difficulties steering multiple parameters simultaneously. This suggests a need for further development iterations, or a training program for using the tool. Additionally, medical experts showed little interest in between-PwMS analysis, preferring the use of our approach in consultations with single patients.
Scalability
MS Pattern Explorer depends on the user-defined data granularity and sequence length, and so the computation of sequences and model results happens at runtime. To address problems occurring for larger data sizes, we store every configuration in a database, enabling quick lookups of precomputed results. However, the memory required for storing these values, and the computational complexity for larger data remain limiting factors.
Conclusions
We present MS Pattern Explorer, a VA system for the exploration, search, and contextualization of temporal activity data of PwMS. Designed with expert collaboration, it leverages interactive clustering and coordinated, interactive views. Our tool effectively aids exploratory sensor data analysis and insight generation for PwMS, with potential for its extension into cohort comparisons and applications to other chronic diseases.
Supplementary Material
Acknowledgments
We are most grateful to the participants of the BarKA-MS study who dedicated their time to support this research, the Valens Rehabilitation Center for making this research possible, and the University of Zürich Digital Society Initiative for their generous support of the authors’ work.
Contributor Information
Gabriela Morgenshtern, Institute for Informatics, University of Zürich, 8050 Zürich, Switzerland; Digital Society Initiative, University of Zürich, 8001 Zürich, Switzerland.
Yves Rutishauser, Institute for Informatics, University of Zürich, 8050 Zürich, Switzerland.
Christina Haag, Institute for Implementation Science, University of Zürich, 8006 Zürich, Switzerland.
Viktor von Wyl, Digital Society Initiative, University of Zürich, 8001 Zürich, Switzerland; Institute for Implementation Science, University of Zürich, 8006 Zürich, Switzerland.
Jürgen Bernard, Institute for Informatics, University of Zürich, 8050 Zürich, Switzerland; Digital Society Initiative, University of Zürich, 8001 Zürich, Switzerland.
Author contributions
Jürgen Bernard and Viktor von Wyl are the guarantors of this article. Tool development: Yves Rutishauser. Manuscript preparation: Gabriela Morgenshtern and Jürgen Bernard. Writing support: Christina Haag and Viktor von Wyl. Logistics and administration: Gabriela Morgenshtern and Jürgen Bernard.
Supplementary material
Supplementary material is available at Journal of the American Medical Informatics Association online.
Funding
None declared.
Conflicts of interest
None declared.
Data availability
Data are available on request.
References
- 1. Sieber C, Haag C, Polhemus A, et al. Feasibility and scalability of a fitness tracker study: results from a longitudinal analysis of persons with multiple sclerosis. Front Digit Health. 2023;5(1):1006932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Block VJ, Zhao C, Hollenbach JA, et al. Validation of a consumer-grade activity monitor for continuous daily activity monitoring in individuals with multiple sclerosis. Mult Scler J Exp Transl Clin. 2019;5(4):2055217319888660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gadaleta M, Radin JM, Baca-Motes K, et al. Passive detection of COVID-19 with wearable sensors and explainable machine learning algorithms. NPJ Digit Med. 2021;4(1):166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mohr DC, Zhang M, Schueller SM.. Personal sensing: understanding mental health using ubiquitous sensors and machine learning. Annu Rev Clin Psychol. 2017;13(1):23–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Seals A, Pilloni G, Kim J, et al. ‘are they doing better in the clinic or at home?’: Understanding clinicians’ needs when visualizing wearable sensor data used in remote gait assessments for people with multiple sclerosis. CHI Conference on Human Factors in Computing Systems. New York, NY, USA: ACM; 2022. 10.1145/3491102.3501989 [DOI]
- 6. Preim B, Lawonn K.. A survey of visual analytics for public health. Comput Graph Forum. 2020;39(1):543–580. [Google Scholar]
- 7. Rind A. Interactive information visualization to explore and query electronic health records. FNT Human–Comput Interact. 2013;5(3):207–298. [Google Scholar]
- 8. Engelhardt B, Comabella M, Chan A.. Multiple sclerosis: immunopathological heterogeneity and its implications. Eur J Immunol. 2022;52(6):869–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McGinley MP, Goldschmidt CH, Rae-Grant AD.. Diagnosis and treatment of multiple sclerosis: a review. JAMA. 2021;325(8):765–779. [DOI] [PubMed] [Google Scholar]
- 10. Amatya B, Khan F, Galea M.. Rehabilitation for people with multiple sclerosis: an overview of cochrane reviews. Cochrane Database Syst Rev. 2019;1(1):CD012732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chevance G, Baretta D, Heino M, et al. Characterizing and predicting person-specific, day-to-day, fluctuations in walking behavior. PLoS One. 2021;16(5):e0251659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schiepek G, Strunk G.. The identification of critical fluctuations and phase transitions in short term and coarse-grained time series-a method for the real-time monitoring of human change processes. Biol Cybern. 2010;102(3):197–207. [DOI] [PubMed] [Google Scholar]
- 13. Olthof M, Hasselman F, Strunk G, et al. Critical fluctuations as an early-warning signal for sudden gains and losses in patients receiving psychotherapy for mood disorders. Clin Psychol Sci. 2020;8(1):25–35. [Google Scholar]
- 14. Therneau T, Atkinson B, Ripley B. Rpart: Recursive partitioning and regression trees. R Package Version 4.1. CRAN R package repository; 2021. https://cran.r-project.org/web/packages/rpart/
- 15. Turicchi J, O'Driscoll R, Finlayson G, et al. Data imputation and body weight variability calculation using linear and nonlinear methods in data collected from digital smart scales: simulation and validation study. JMIR Mhealth Uhealth. 2020;8(9):e17977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu J, Spakowicz DJ, Ash GI, et al. Bayesian structural time series for biomedical sensor data: a flexible modeling framework for evaluating interventions. PLoS Comput Biol. 2021;17(8):e1009303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gomez V, Maravall A.. Estimation, prediction, and interpolation for nonstationary series with the Kalman filter. J Am Stat Assoc. 1994;89(426):611. [Google Scholar]
- 18. Guo Y, Guo S, Jin Z, et al. Survey on visual analysis of event sequence data. IEEE Trans Vis Comput Graph. 2022;28(12):5091–5112. [DOI] [PubMed] [Google Scholar]
- 19. Canali S, Schiaffonati V, Aliverti A.. Challenges and recommendations for wearable devices in digital health: data quality, interoperability, health equity, fairness. PLOS Digit Health. 2022;1(10):e0000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Plaisant C, Mushlin R, Snyder A, et al. LifeLines: using visualization to enhance navigation and analysis of patient records. Proc AMIA Symp. 1998:76–80. [PMC free article] [PubMed] [Google Scholar]
- 21. Sultanum N, Singh D, Brudno M, et al. Doccurate: a curation-based approach for clinical text visualization. IEEE Trans Vis Comput Graph. 2018;25(1):142-151. 10.1109/TVCG.2018.2864905 [DOI] [PubMed] [Google Scholar]
- 22. Rind A, Miksch S, Aigner W, Turic T, Pohl M. VisuExplore: gaining new medical insights from visual exploration. Proceedings of the 1st International Workshop on Interactive Systems in Healthcare (WISH@CHI2010). 2010. 10.1007/978-3-642-23768-3_24 [DOI]
- 23. Philip NY, Razaak M, Chang J, et al. A data analytics suite for exploratory predictive, and visual analysis of type 2 diabetes. IEEE Access. 2022;10(1):13460-13471. [Google Scholar]
- 24. Federico P, Unger J, Amor-Amorós A, et al. Gnaeus: utilizing clinical guidelines for knowledge-assisted visualisation of EHR cohorts. EuroVA@EuroVis. 2015. 10.2312/eurova.20151108. [DOI]
- 25. Jin Z, Cui S, Guo S, et al. CarePre. ACM Trans Comput Healthcare. 2020;1(1):1–20. [Google Scholar]
- 26. Wagner M, Slijepcevic D, Horsak B, et al. KAVAGait: knowledge-assisted visual analytics for clinical gait analysis. IEEE Trans Vis Comput Graph. 2019;25(3):1528–1542. [DOI] [PubMed] [Google Scholar]
- 27. Belden JL, Wegier P, Patel J, et al. Designing a medication timeline for patients and physicians. J Am Med Inform Assoc. 2019;26(2):95–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bögl M, Aigner W, Filzmoser P, et al. Visual analytics for model selection in time series analysis. IEEE Trans Vis Comput Graph. 2013;19(12):2237–2246. [DOI] [PubMed] [Google Scholar]
- 29. Ahmad S, Sessler D, Kohlhammer J. Towards a Comprehensive Cohort Visualization of Patients with Inflammatory Bowel Disease. 2021 IEEE Workshop on Visual Analytics in Healthcare (VAHC), New Orleans, LA, USA. IEEE; 2021. 10.1109/VAHC53616.2021.00009 [DOI]
- 30. Bernard J, Ruppert T, Scherer M, et al. Guided discovery of interesting relationships between time series clusters and metadata properties. Proceedings of the 12th International Conference on Knowledge Management and Knowledge Technologies. New York, NY, USA: Association for Computing Machinery; 2012. 10.1145/2362456.2362485 [DOI]
- 31. Bernard J, Ruppert T, Scherer M, et al. Content-based layouts for exploratory metadata search in scientific research data. Proceedings of the 12th ACM/IEEE-CS joint conference on Digital Libraries. New York, NY, USA: ACM; 2012. 10.1145/2232817.2232844 [DOI]
- 32. Bach B, Freeman E, Abdul-Rahman A, et al. Dashboard design patterns. IEEE Trans Vis Comput Graph. 2023;29(1):342–352. [DOI] [PubMed] [Google Scholar]
- 33. Saraiya P, North C, Duca K.. An insight-based methodology for evaluating bioinformatics visualizations. IEEE Trans Vis Comput Graph. 2005;11(4):443–456. [DOI] [PubMed] [Google Scholar]
- 34. Bendeck A, Bromley D, Setlur V. SlopeSeeker: a search tool for exploring a dataset of quantifiable trends. Proceedings of the 29th International Conference on Intelligent User Interfaces, Greenville, SC, USA. ACM; 2024. 10.1145/3640543.3645208 [DOI]
- 35. Polhemus A, Sieber C, Haag C, et al. Non-equivalent, but still valid: establishing the construct validity of a consumer fitness tracker in persons with multiple sclerosis. PLoS Digit Health. 2023;2(1):e0000171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jolliffe IT. Principal component analysis for special types of data. Principal Component Analysis. Springer-Verlag; 2006:338–372. 10.1007/0-387-22440-8_13 [DOI] [Google Scholar]
- 37. Polhemus A, Haag C, Sieber C, et al. Methodological heterogeneity biases physical activity metrics derived from the Actigraph GT3X in multiple sclerosis: a rapid review and comparative study. Front Rehabil Sci. 2022. 10.3389/fresc.2022.989658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. von Wyl V. Barriers to Physical Activity in People With MS (BarKA-MS). ClinicalTrials.gov; 2021. https://clinicaltrials.gov/study/NCT04746807
- 39. Sieber C, Haag C, Polhemus A, et al. Exploring the major barriers to physical activity in persons with multiple sclerosis: observational longitudinal study. JMIR Rehabil Assist Technol. 2024. 10.2196/52733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weather conditions. Accessed June 25, 2024. https://openweathermap.org/weather-conditions
- 41. Kohonen T. Self-Organizing Maps. Springer; 1995. 10.1007/978-3-642-97610-0 [DOI] [Google Scholar]
- 42. Gotz D, Sun J, Cao N, et al. Visual cluster analysis in support of clinical decision intelligence. AMIA Annu Symp Proc. 2011;2011:481–490. [PMC free article] [PubMed] [Google Scholar]
- 43. Kohonen T. Essentials of the self-organizing map. Neural Netw. 2013;37(1):52–65. [DOI] [PubMed] [Google Scholar]
- 44. Altman NS. An introduction to kernel and nearest-neighbor nonparametric regression. Am Stat. 1992;46(3):175–185. [Google Scholar]
- 45. Bernard J, Sessler D, Kohlhammer J, et al. Using dashboard networks to visualize multiple patient histories: a design study on post-operative prostate cancer. IEEE Trans Vis Comput Graph. 2019;25(3):1615–1628. [DOI] [PubMed] [Google Scholar]
- 46. North C. Toward measuring visualization insight. IEEE Comput Graph Appl. 2006;26(3):6–9. [DOI] [PubMed] [Google Scholar]
- 47. Chang R, Ziemkiewicz C, Green TM, et al. Defining insight for visual analytics. IEEE Comput Graph Appl. 2009;29(2):14–17. [DOI] [PubMed] [Google Scholar]
- 48. Daniore P, Nittas V, Haag C, et al. From wearable sensor data to digital biomarker development: ten lessons learned and a framework proposal. NPJ Digit Med. 2024;7(1):161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bernard J, Sessler D, Bannach A, et al. A visual active learning system for the assessment of patient well-being in prostate cancer research. Proceedings of the 2015 Workshop on Visual Analytics in Healthcare. New York, NY, USA: ACM; 2015. 10.1145/2836034.2836035 [DOI]
- 50. Wang Q, Laramee RS.. EHR STAR: the state‐of‐the‐art in interactive EHR visualization. Comput Graph Forum. 2022;41(1):69–105. [Google Scholar]
- 51. Levy-Fix G, Kuperman GJ, Elhadad N. Machine learning and visualization in clinical decision support: current state and future directions, arXiv, arXiv:1906.02664, preprint: not peer reviewed. 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available on request.