Abstract
Microbial cell factories (MCFs) are extensively used to produce a wide array of bioproducts, such as bioenergy, biochemical, food, nutrients, and pharmaceuticals, and have been regarded as the “chips” of biomanufacturing that will fuel the emerging bioeconomy era. Biotechnology advances have led to the screening, investigation, and engineering of an increasing number of microorganisms as diverse MCFs, which are the workhorses of biomanufacturing and help develop the bioeconomy. This review briefly summarizes the progress and strategies in the development of robust and efficient MCFs for sustainable and economic biomanufacturing. First, a comprehensive understanding of microbial chassis cells, including accurate genome sequences and corresponding annotations; metabolic and regulatory networks governing substances, energy, physiology, and information; and their similarity and uniqueness compared with those of other microorganisms, is needed. Moreover, the development and application of effective and efficient tools is crucial for engineering both model and nonmodel microbial chassis cells into efficient MCFs, including the identification and characterization of biological parts, as well as the design, synthesis, assembly, editing, and regulation of genes, circuits, and pathways. This review also highlights the necessity of integrating automation and artificial intelligence (AI) with biotechnology to facilitate the development of future customized artificial synthetic MCFs to expedite the industrialization process of biomanufacturing and the bioeconomy.
Introduction
Biomanufacturing refers to the use of a biological system composed of enzymes, microorganisms, or more advanced biological cells to manufacture diverse products with economic value, such as biofuels, biochemicals, nutrients, natural products, or other value-added products from renewable feedstocks, such as lignocellulosic biomass, via biotechnology [1]. MCFs are the “chips” of biomanufacturing. The design and construction of efficient and robust MCFs have the potential to meet the needs of environmental protection, carbon neutralization, and global economic development toward the coming sustainable bioeconomy. The concept of metabolic engineering was first officially suggested in 1991 with the development of molecular biology and genetic engineering tools to design metabolic pathways in organisms [2]. The development of next-generation sequencing (NGS) and spectrometry technology has enabled a systems biology approach for the deep understanding of microbial metabolic networks and regulation, which has been applied to the construction of higher-level MCFs [3]. The emergence of synthetic biology in combination with the accumulation of systems biology datasets and the development of automation and big data analysis technology have taken MCFs into a design or synthetic era. The integration of artificial intelligence (AI), automation, and synthetic biology with systems biology has led to the thorough understanding and modification of many chassis cells to enable the rational design and modification of both model and nonmodel chasses into robust and efficient cell factories capable of producing natural and nonnatural products of interest [4,5].
Although bioproducts such as biofuels, bioplastics, and biomedicines have been produced industrially via microbial fermentation, challenges persist regarding production scalability and cost. Therefore, the use of low-cost biomass feedstock for platform biochemical production and the development of MCFs with improved production performance and robustness are crucial to address the industrialization problems. To increase the production capacity of MCFs to meet industrial requirements, it is essential to reprogram and optimize model and nonmodel chasses to develop robust and efficient cell factories for industrial applications.
The accumulation of omics datasets and advancements in cutting-edge technologies have made it possible to analyze and interpret the metabolic pathways, regulatory networks, and mechanisms of microbial chassis cells in detail. Multidimensional rational or semirational design of microbial chassis cells can be performed by combining the design-build-test-learn (DBTL) strategy with synthetic biology tools. This involves exploring and designing biological parts, circuits, and modules to construct metabolic and regulatory pathways and then optimizing them for rational design and industrial development of diverse bioproducts by integrating spatiotemporal regulation [6].
In this review, we highlight the importance of understanding microbial chassis cells and provide a timeline-based overview of the advancements and strategies employed in the development of MCFs. We divide the chronology of the MCFs into 5 distinct periods and provide a detailed overview of the characteristics of each period (Fig. 1). The discovery and development of natural cell factories, as well as the creation of future artificial cell factories, are discussed. In addition, we summarize the advancements and strategies in the construction of robust and efficient MCFs with balanced and optimized metabolism in terms of substance, energy, physiology, and information. We also highlight the importance and necessity of synthetic biology strategies and corresponding enabling technologies in the construction and optimization of MCFs. Moreover, this review highlights the necessity of integrating automation and information technology (IT) with biotechnology to facilitate the development of future customized artificial synthetic MCFs that will help accelerate the advancement of biomanufacturing and the bioeconomy.
Fig. 1.
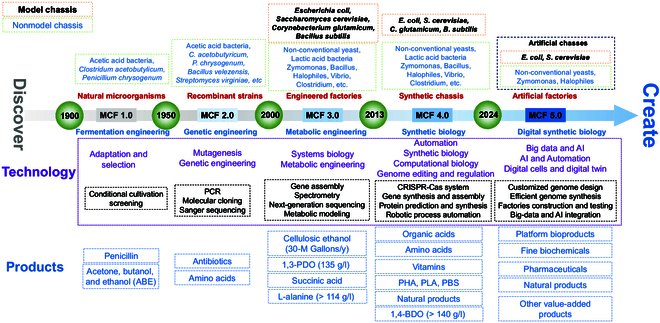
Development of MCFs from discovery to creation.
Development of Industrial Chassis Cells
The isolation and discovery of natural MCFs was followed by the generation of cell factories with superior performance through mutagenesis and genetic engineering. With advances in biotechnology, rational methods such as metabolic engineering have been applied to modify chassis cells to achieve more desirable characteristics. The emergence and development of synthetic biology have facilitated the establishment of technology platforms. It can be used not only for better understanding model and nonmodel microorganisms but also for guiding the rational design, synthesis, assembly, and construction of robust and efficient cell factories. Moreover, the accumulation of biological data and the continuous development of AI will guide the development of digital biology, which involves the application of digital methods to study and quantify cell factories from genes, circuits, and pathways to ecosystems [7]. Even in the development of future cell factories, information sharing between different species, such as some model and nonmodel chasses, can be achieved through digital synthetic biology, ultimately leading to the development of synthetic artificial chasses (Fig. 1). The representative products, including foods, fuels, chemicals, and medicines synthesized from different generations of MCF, are summarized in Table 1. However, few related reports about digital cells as a future development trend exist.
Table 1.
Summary of representative products synthesized from different generations of MCFs
| Class | Host | Target chemical | Strategy description | Titer (g/l) | Reference |
|---|---|---|---|---|---|
| MCF 1.0 | P. chrysogenum | Penicillin | Strain selection and isolation | 0.06 | [14] |
| C. acetobutylicum | Butanol | Strain selection and isolation | 10–12 | [157] | |
| MCF 2.0 | P. chrysogenum | Penicillin | X-ray mutagenesis | 0.3 | [14] |
| MCF 3.0 | E. coli | l-Alanine | Heterologous alanine dehydrogenase integration, competitive pathway deletion | 114 | [35] |
| E. coli | 1,3-Propanediol | Pathway design and optimization, cofactor supply | 130 | [158] | |
| S. cerevisiae | Artemisinic acid | Metabolic regulation, optimization of key genes expression levels, competitive pathway deletion | 0.1 | [106] | |
| C. glutamicum | l-Valine | Cofactor balance, competitive pathway deletion | 150 | [159] | |
| B. subtilis | Riboflavin | Mutagenesis of zwf and gnd | 15.7 | [160] | |
| C. acetobutylicum | Butanol | Overexpression of groESL | 17 | [157] | |
| MCF 4.0 | E. coli | 1,4-Butanodiol | Pathway design, energy supply, cell growth, redox balance | 18 | [39] |
| S. cerevisiae | Cannabinoids | Enzyme mining, copy number optimization | / | [107] | |
| Y. lipolytica | Succinic acid | Reconfiguration of the reductive TCA, adaptive laboratory evolution | 111.9 | [66] | |
| H. bluephagenesis | Polyhydroxyalkanoates (PHA) | Plasmid copy number optimization, cell morphology regulation | 80 | [100] | |
| Z. mobilis | Polyhydroxybutyrate (PHB) | Copy number optimization, cofactor supply, self-flocculation, C/N ratio | 74% DCW | [87] | |
| B. licheniformis | 2-Phenylethanol | Central metabolic pathway and phenylpyruvate pathway reconstruction, competitive pathway deletion, glucose transport system modulation | 6.24 | [161] | |
| Rhodotorula toruloides | Fatty acid ethyl esters | Overexpression of wax ester synthase genes | 9.97 | [162] |
Isolation and Discovery of Natural Industrial Microorganisms
Five thousand years ago, the discovery of the sour liquid produced by the natural fermentation of fruits opened the way to the understanding and use of vinegar. Vinegar plays an important role in the culinary, health, beverage, and chemical industries as well as other fields. Its production uses specialized aerobic acetic acid bacteria (AAB) to express ethanol dehydrogenase and acetaldehyde dehydrogenase, which oxidize the alcohol produced from grain or fruit fermentation to acetic acid. Acetobacter, the first AAB discovered, has industrial importance in edible vinegar production in the food industry. With the ongoing advancements in biotechnology, an increasing variety of AAB have been isolated [8] and widely applied in the production of cellulose, pigments, indole acetic acid, and ascorbic acid [9–11].
In the 20th century, energy shortages led to the rapid development of biobutanol, which is an important chemical feedstock and biofuel. Clostridium acetobutylicum has been extensively studied as an important strain for classical natural butanol production [12]. C. acetobutylicum can effectively undergo continuous industrial fermentation for acetone, butanol, and ethanol production at a ratio of 6:3:1 by using renewable biomass resources such as starch and molasses [13]. However, the high cost limits the development of butanol from microbial cells; thus, breeding high-proportion butanol strains and developing new fermentation processes are pivotal challenges.
The discovery of penicillin opened a new path for the use of antibiotics to treat infectious diseases, which marked a new era of antibiotics and drugs for humanity. The use of Penicillium chrysogenum made industrial fermentation production of penicillin a reality. However, the low titer of penicillin (~5 mg/l) led to an unaffordable price due to limitations in upstream strain engineering, fermentation optimization, and downstream processing for production isolation and purification [14]. Since the discovery of penicillin, more than 20,000 biologically active antibiotic drugs have been discovered in microorganisms [15]. For example, Cephalosporium acremonium was identified as a cephalosporin producer and actinomycetes as cephamycin, clavam, and carbapenem producers and, along with unicellular bacteria, as monocyclic β-lactam producers.
In addition, an increasing number of natural producers of biochemicals have been isolated and discovered, such as Anaerobiospirillum succiniciproducens and Mannheimia succiniciproducens for succinic acid [16,17], Rhodococcus opacus and Yarrowia lipolytica for lipids, fatty acids, and their derivatives [18,19], Corynebacterium glutamicum for amino acids [20], Zymomonas mobilis and Saccharomyces cerevisiae for ethanol [21,22], Lactobacillus for lactic acid [23], and Klebsiella pneumoniae for 1,3-propanediol (1,3-PDO) [24]. These naturally acquired strains have become the first generation of industrial microbial cell factories (MCF 1.0). However, it cannot completely meet the needs of industrial applications in terms of product titer, rate, and yield. The development of microbial chassis cells with better characteristics is critical.
Random Mutagenesis and Natural Selection
Owing to the limited information and genetic tools available for microorganisms selected from natural environments, random mutagenesis, which includes physical, chemical, and biological mutagenesis, was the primary method used to improve the features of these strains. The development of cell factories through mutagenesis and selection marked the second generation of industrial microbial cell factories (MCF 2.0).
Physical and chemical mutagenesis
Physical mutagenesis mainly uses physical factors, including ultraviolet (UV) light, x-rays, α-rays, γ-rays, and heavy ion irradiation, to induce mutations (Fig. 2). Recently, a novel mutagenesis tool, atmospheric and room temperature plasma (ARTP), which is a powerful mutagenesis tool to improve the yield of biochemicals, was developed. The advent of ARTP has dramatically enhanced the quality and characteristics of products derived from bacteria and fungi that are difficult to modify, such as antibiotics, organic acids, amino acids, vitamins, and steroids [25].
Fig. 2.
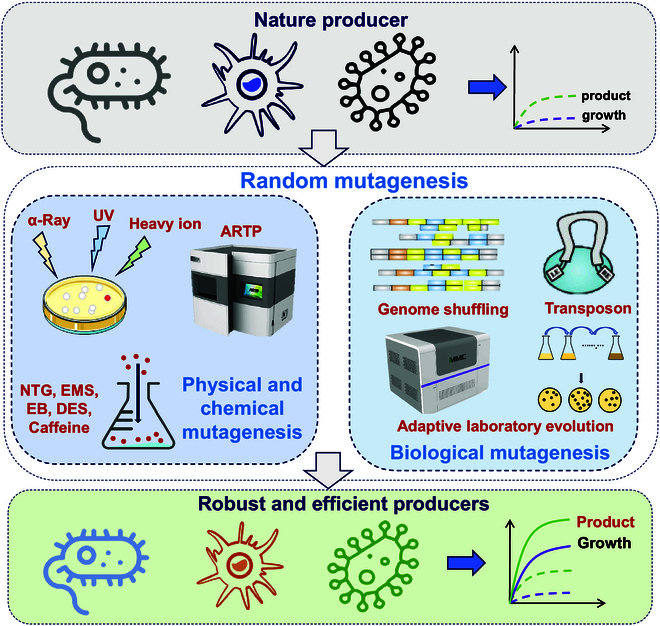
Strategies of random mutagenesis for natural MCF evolution
The mutants obtained through mutagenesis are listed in Table 2. Multiple mutants were acquired through mutagenesis, with enhanced bacteriocin production ranging from 103.48% to 551% [25], and an l-serine producer with a titer of 34.78 g/l was obtained via ARTP mutagenesis [26]. Chemical mutagenesis primarily uses chemical substances such as alkylating agents, base analogs, frameshift mutagens, deamination agents, and hydroxylation agents to obtain mutations. Although chemical mutagenesis provides high mutation efficiency, its use of toxic mutagens poses risks to human health and the environment. Therefore, the use of physical or biological mutagens will likely be the main trend in the future.
Table 2.
Examples of the application of physical and chemical mutagenesis to improve production
| Strain | Method | Product | Improvement (-fold) | Ref. |
|---|---|---|---|---|
| Actinobacillus succinogenes | UV + NTG | Succinic acid | 1.85 | [163] |
| Propionibacterium acidipropionici | UV | Propionic acid | 1.25 | [164] |
| Bacillus velezensis | DES + NTG | Surfactin | 3.99 | [165] |
| Streptomyces virginiae | UV | Virginiamycin | 11.6 | [166] |
| Cordyceps kyushuensis | UV + HNO2 | 3β,7α,15α-Trihydroxy-5-androsten-17-one | 9.6 | [167] |
| Actinosynnema pretiosum | ARTP | Ansericin | 1.93 | [168] |
Biological mutagenesis
Biological mutagenesis has led to innovations in mutation breeding. This not only accelerates mutagenesis efficiency but also ensures safety. This approach mainly includes techniques involving bacteriophages, plasmids, DNA transposon mutagenesis, and protoplast fusion, such as DNA and genome shuffling. Genome shuffling integrates traditional strain mutagenesis breeding methods with protoplast techniques, enabling the convergence of superior phenotypes from multiple parents into a single strain through multiple rounds of protoplast fusion. This approach accelerates the forward mutation process greatly in microbial cells. As a result, it has been widely applied to create strains for bioproducts with complicated pathways, such as the biofuels butanol and ethanol [27]; antibiotics such as doxorubicin [28], avilamycin [29], and nosipeptide [30]; and natural food preservatives such as ε-poly-l-lysine [31]. In addition, genome shuffling has also been used to improve other microbial traits, such as byproduct reduction. For example, genome shuffling was successfully used to reduce the glycerol byproduct in a mutant of industrial S. cerevisiae while increasing the tolerance to ethanol and inhibitory byproducts such as furfural and acetic acid in lignocellulosic hydrolysates [32,33].
Random mutagenesis plays an important role in the breeding of efficient MCFs. However, the mutations are highly random across the entire genome, leading to difficulty in obtaining the desired traits rapidly. Therefore, rational design and engineering of microbial chassis cells into efficient cell factories has become increasingly important.
Metabolic Engineering of MCFs
The success of the Human Genome Project and technological advances have led to a deeper understanding of microbial chassis cells at the genomic level. The accumulation of omics datasets and improvements in genetic tools have made genetic modifications in model microorganisms efficient for the production of diverse bioproducts, such as organic alcohols and acids, amino acids, organic amines, vitamins, natural products, and polyhydroxyalkanoates (PHAs). Research directions therefore shifted toward engineering model microorganisms, such as Escherichia coli, S. cerevisiae, C. glutamicum, and Bacillus subtilis, which have well-understood genetic backgrounds, abundant biological parts, and devices, and for which comprehensive genetic modification tools exist [4,34] (Table 3).
Table 3.
Characteristics of representative model and nonmodel microbial chassis cells
| Class | Strain | Growth | Safety | Genome (Mb) | Substrates | Genome manipulation tools | Products | Reference |
|---|---|---|---|---|---|---|---|---|
| Model microbes | E. coli | Facultative aerobic | Not GRAS | 4.64 | Glycerol, pentose, hexose, starch | Various tools | Alcohols, fatty acids, terpenoids | [68,124,169] |
| S. cerevisiae | Facultative aerobic | GRAS | 11.8, 16 chromosomes | Starch, sucrose, hexose | Various tools | Terpenoids, natural, products | [40,170] | |
| C. glutamicum | Facultative aerobic | GRAS | 3.28 | Sugars, alcohols, organic acids | HR, CRISPR-Cas9, CRISPR-Cpf1/dCpf1 | Alcohols, aminol acids, hyaluronan | [171,172] | |
| B. subtilis | Facultative aerobic | GRAS | 4.2 | Glucose, acetate, citrate | CRISPR-Cas9, CRISPR-dCas9-α/ω, CRISPR-Cpf1/dCpf1 | Enzymes, isobutanol, 2,3-BDO | [173–175] | |
| Nonmodel microbes | Y. lipolytica | Facultative aerobic | GRAS | 20.5, 6 chromosomes | Glucose, glycerol, sucrose, starch, inulin, cellobiose | NHEJ, ZFNs, TALENs, CRISPR-Cas9, (CRISPRi/CRISPRa) |
Lipid, FAAEs, terpenoids, alkanes | [98,176] |
| Rhodotorula toruloides | Facultative aerobic | GRAS | 20.2 | Glucose, xylose, cellobiose | CRISPR-Cas9, RNA interference | Lipid, biodiesel, carotenoids, enzyme | [162,177–179] | |
| Scheffersomyces stipitis | Facultative aerobic | GRAS | 15.4, 8 chromosomes | Glucose, fructose, xylose, mannose, galactose, cellobiose | CRISPR-Cas9 | Shikimic acid, ethanol, vitamin B9 | [180,181] | |
| Pichia pastoris | Facultative aerobic | GRAS | 9.6, 4 chromosomes | Glucose, glycerol, l-rhamnose, methanol | HR, CRISPR-Cas9 | Enzyme, terpenoids, alcohols | [48,49,182] | |
| Z. mobilis | Facultative anaerobic | GRAS | 2.2, 4 plasmids | Glucose, sucrose, fructose | HR, CRISPR-Cas9, CRISPR-Cas12a, endogenous type I-F CRISPR-Cas system | Ethanol, isobutanol, 2,3-BDO, PHB | [83,84,87,88] | |
| C. autoethanogenum | Obligate anaerobic | Not GRAS | 4.4 | CO, CO2 | Endogenous type I-B CRISPR cluster | Acetate, ethanol, 2,3-BDO; PHB | [183] | |
| C. acetobutylicum | Obligate anaerobic | Not GRAS | 4.1 | Glucose | CRISPR-Cas9/dCas9 | Acetone, ethanol, butanol | [184,185] | |
| H. bluephagenesis | Aerobic | Not GRAS | 4.09 | Glucose, butyrate | CRISPR-Cas9 CRISPRi CRISPR/AID | PHA, ectoine, l-threonine | [102,186] | |
| V. natriegens | Facultative anaerobic | GRAS | Chr: 3.24, Chr: 1.92 | Glucose, fructose, glycerol, citrate | CRISPR-Cas9 CRISPRi Cre-loxP system | PHA, alanine carotene melanin | [187] |
The model bacterial species E. coli is often used as a chassis to develop MCFs for the production of bulk chemicals such as l-alanine [35], 1,3-PDO [36], d-lactic acid [37], succinic acid [38], and 1,4-butanediol [39] because of its rapid growth and simple nutritional requirements. Owing to the lack of isoprene synthesis pathways required for the synthesis of terpene compounds, it is often used for the synthesis of simple terpene natural products through the 2-C-methyl-d-erythritol 4-phosphate (MEP) pathway. S. cerevisiae has a natural mevalonate (MVA) pathway, which leads to greater synthesis of precursor substances than does the MEP pathway [40]. Therefore, S. cerevisiae is commonly used for the synthesis of natural products [40]. C. glutamicum, a model microorganism for amino acid production, has also been engineered to produce natural products and nonessential amino acids. Owing to its strong secretion ability, B. subtilis is used to produce natural products including monosaccharide derivatives such as scyllo-inositol, terpenoids such as squalene, and active polysaccharides such as hyaluronic acid and chondroitin sulfate [41–44].
Therefore, these model microorganisms modified by metabolic engineering could be referred to as third-generation industrial microbial cell factories (MCF 3.0). This class not only possesses a series of technology platforms, including gene design, synthesis, editing, rich biological parts and devices, and massive databases but also establishes relatively mature commercial production infrastructures and business models, making them important chassis cells in biomanufacturing. However, it is still challenging to rapidly transform these model microorganisms into the desired MCFs for biomanufacturing.
Direction and strategies of metabolic engineering
Substance and energy metabolism are current primary modification targets for strain construction and improvement (Fig. 3). Substance metabolism modification can improve the utilization rate of substrates in cell factories, broaden their substrate utilization diversity, reduce the generation of redundant byproducts, and maximize the flow of carbon metabolism toward the target product. Energy metabolism modification can provide sufficient energy and cofactors for the chemical production process of cells, balancing the reducing power and promoting efficient production of target compounds.
Fig. 3.
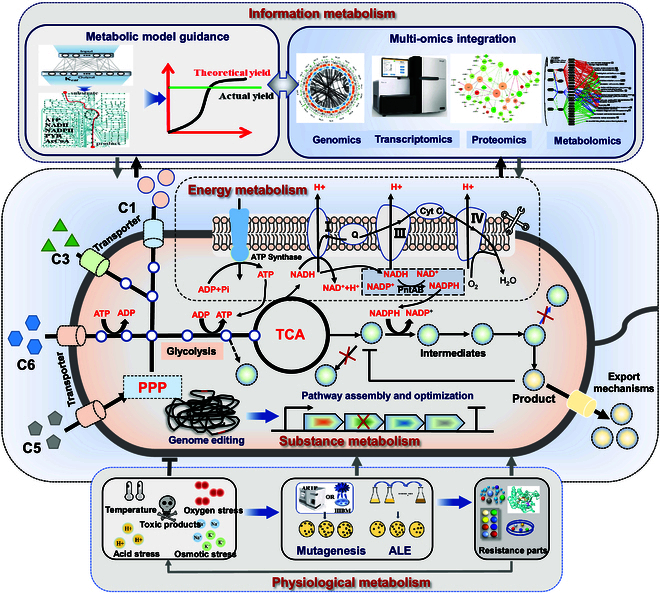
The direction and strategy of material, energy, physiology, and information metabolism optimization in MCFs.
Broadening substrate utilization
To actively respond to the impact of fluctuating raw material prices, developing microbial chassis cells with different substrate utilization capabilities is necessary. Lignocellulose is the most abundant renewable resource on Earth and the main raw feedstock for second-generation biorefining [45]. After pretreatment and enzymatic hydrolysis, the monosaccharides are released for utilization and fermentation by microorganisms. These monosaccharides include C5 sugars such as xylose and arabinose, as well as C6 sugars such as glucose, mannose, and galactose. However, the majority of microorganisms can use only specific sugars as carbon sources. Therefore, it is crucial to achieve economic biochemical production by coutilizing multiple carbon sources of lignocellulosic hydrolysates by introducing metabolic pathways for different carbon sources to expand the substrate utilization spectrum. Combining adaptive evolution after the introduction of substrate utilization pathways and transport systems, efficient substrate utilization cell factories have been obtained (Fig. 3). For example, the production of ethanol from xylose has been achieved by introducing exogenous xylose reductase and xylitol dehydrogenase or heterologous xylose isomerase pathways into yeast [46].
In addition to the use of lignocellulose as a raw material, C1 molecules such as CO2, methanol, and formate are also highly promising feedstocks [47]. At present, microorganisms that can utilize C1 molecules include those that use methyl compounds as the sole carbon source for the growth and metabolism of natural methyl nutrient microorganisms, as well as methanol-consuming strains such as E. coli and C. glutamicum obtained through metabolic engineering and adaptation. Through metabolic engineering of natural yeast, the synthesis of primary metabolites such as ethanol, isobutanol, and 2,3-butanediol (2,3-BDO), as well as secondary metabolites such as polyketones, terpenes, and fatty acid derivatives, has been achieved [48–52]. Carbon dioxide is the main raw material for third-generation biorefineries. Designing and developing microbial chassis cells to directly use CO2 to produce biofuels and biochemicals can greatly alleviate energy depletion and environmental problems [53]. The naturally occurring carbon sequestration pathway has been explored and applied to various model microorganisms, and autotrophic growth of E. coli has been achieved through continuous laboratory evolution by introducing RuBisCO carbon sequestration enzymes [54]. In addition, various artificial carbon sequestration pathways have been developed and applied [55].
Balancing cofactors and energy
Cofactors and adenosine triphosphate (ATP) are key regulatory factors in energy metabolism and are also important for maintaining cell growth and homeostasis [56]. Cells produce reducing power and ATP when metabolizing substrates such as glucose, and require them when synthesizing biochemicals. If the cofactor and ATP supply is imbalanced, the conversion rate and synthesis efficiency of the products will be affected.
Intracellular ATP is regulated mainly by ATP-related enzyme metabolism and oxidative phosphorylation levels [57]. The activities of intracellular NADH [reduced form of nicotinamide adenine dinucleotide (oxidized form)], the electron transfer chain, and ATP synthase are the main sites for regulating intracellular ATP levels. To increase the ATP supply, metabolic engineering can be used by adding ATP energy substrates, controlling the generation and consumption of pH and ATP, and regulating the electron transfer chain. After the energy substrate citric acid was added to the fermentation system, the titer of poly-γ-glutamic acid (PGA) in B. licheniformis increased to 35 g/l [58].
The following 4 strategies can be implemented to optimize the supply of reducing power in the production process: regulating the synthesis and regeneration of the intracellular reducing power, changing the cofactor preference of key enzymes, consuming redundant cofactors, and introducing nonnatural cofactors. In regulating the synthesis and regeneration of intracellular cofactors, activating the combined expression of transhydrogenase and NAD (nicotinamide adenine dinucleotide) kinase can effectively increase intracellular NADPH (reduced form of nicotinamide adenine dinucleotide phosphate) levels and subsequently isobutanol production [59].
In addition, the Entner–Doudoroff (ED) pathway is often used as an alternative pathway to Embden–Meyerhof–Parnas (EMP) because of its efficient NADPH generation. For example, the ED metabolic pathway derived from Z. mobilis was introduced into E. coli, and the expression of related genes was regulated via ribosome binding site (RBS) libraries. The NADPH level in the recombinant E. coli was 25 times greater than that in the wild type, effectively increasing the production of terpenoids [60]. This strategy was also suitable for increasing isobutanol production in E. coli and C. glutamicum [61].
Improving robustness
MCFs are subjected to various physiological or physiochemical stress factors during fermentation using the inexpensive renewable resources, including changes in temperature, pH, oxygen, and osmotic pressure as well as changes in the concentrations of substrates, inhibitors, and toxic intermediates or byproducts. These stress factors can slow microbial metabolism and cell growth and sometimes even lead to the complete loss of production performance (Fig. 3).
Traditional mutagenesis and adaptive laboratory evolution (ALE) are effective strategies for improving strain robustness [62]. This method is widely used in both model and nonmodel chassis cells to improve the tolerance to toxic substances during the production process. The most common method is to increase the tolerance of chassis cells to toxic products to increase yield. For example, a serine-sensitive production strain lacking the l-serine degradation pathway was subjected to ALE to improve l-serine tolerance with gradually increasing concentrations from 3 to 100 g/l, and evolved strains with excellent growth performance at a titer of 50 g/l l-serine were isolated. These mutant strains presented improved serine production [63]. ALE can also be combined with genome sequencing and transcriptome analysis to further understand tolerance mechanisms at the genetic level [64,65]. In addition, ALE can effectively improve the metabolic disorders of microbial chassis cells. Through adaptive evolution of lipolytic yeast, ALE can effectively improve the metabolic disorders caused by the reductive tricarboxylic acid (TCA) cycle, thereby increasing cell growth ability and succinic acid production [66]. Therefore, ALE plays a crucial role in the development of industrial chassis cells.
Owing to the complicated screening process, prolonged cycles, and tedious workload involved in ALE, the pursuit of stress-resistant biological parts for rational design and engineering of microbial cells to increase their robustness has emerged as an alternative strategy to increase stress resistance (Fig. 3). With the accumulation of omics datasets, an increasing number of stress-resistant elements are being identified and applied for the construction of robust microorganisms. These stress-resistant elements mainly include genes related to cell walls and membranes, DNA repair, oxidative stress, compatible solutes, energy production and signal transduction, as well as efflux pumps, heat shock proteins, and global transcription factors [67], which can be used for genetic engineering to rationally modify strains for improved robustness.
For example, the tolerance of E. coli to organic alcohols and product yield were effectively improved by enhancing cell membrane biosynthesis and modifying membrane transport proteins [68–70]. In addition, heat shock proteins (GroELS, DnaK, DnaJ, HtpJ), DNA repair-related genes (RecA, UvrD), oxidative stress genes (Fpr, Gsh1, Glt1), and global transcription factors (CRP, RpoD) can also increase microbial tolerance to inhibitors [71–74]. Sulfur-containing amino acids are important essential amino acids, and their concentrations are closely related to microbial stress resistance. Hydrogen sulfide, an important intermediate product of sulfur metabolism, also plays an important role as an endogenous signaling molecule in response to reactive oxygen species (ROS) stress [75].
Rewiring the metabolic network
Owing to the varying number of genes involved in the biosynthetic pathway, the continual development of chassis strains that are proficient in producing high-yielding intermediate products reduces the complexity of pathway reconstruction greatly. This ensures an adequate supply of precursors, enabling the adoption of tailored strategies to optimize chassis cells, regulate metabolic balance, and ultimately facilitate the efficient construction of MCFs (Fig. 3). Genome-wide metabolic models were used to search for genetic targets for metabolic engineering, efficiently design a metabolic pathway reconstruction library, and combine with high-throughput biosensors to train different machine learning algorithms. A data generation cycle promoted the synthesis ability of aromatic amino acids in engineered yeast, resulting in 74% and 43% increases in tryptophan production titer and yield, respectively [76].
When reshaping the metabolic network of chassis cells, attention should be given to the interactions among various metabolic pathways to reduce the disturbance of common substances such as coenzymes and the energy supply caused by the introduction of biosynthesis pathways. Additionally, the introduction of heterologous pathways may have a negative influence on the physiological status and growth rate of chassis cells, as these heterologous products or intermediates may have certain toxicity toward the chassis cells.
Bioinformatics strategies are evolving with the continuous increase in high-throughput data such as whole-genome sequences of microorganisms, enabling comprehensive and systematic analysis, design, and regulation of microbial physiological metabolic functions. The emergence of the genome-scale metabolic network model (GSMM) based on genome sequence annotation and detailed biochemical information integration can help accurately predict and understand the mechanisms of microbial intracellular metabolism. At present, strain design and phenotype prediction analyses based on genome-scale metabolic models have been performed in various microorganisms, such as E. coli, B. subtilis, C. glutamicum, and S. cerevisiae. After years of revision and improvement, the accuracy of model predictions has improved. Through the combination of genome-wide metabolic modeling, genetic targets for strain modification can be predicted, and recombinant strains with improved titers, yields, and rates of the targeted products can be generated [77–79]. For example, the ability of engineering yeast to produce aromatic amino acids has been improved by combining high-throughput biosensors with the guidance of GSMM, leading to an increase in tryptophan production and yield of 74% and 43%, respectively [76].
Synthetic Biology-Guided Industrial Microbial Chassis Cell Modification
However, model microorganisms usually have innate disadvantages when using low-cost feedstocks, such as poor fermentation performance due to their low tolerance to toxic lignocellulosic hydrolysate, which limits the industrial application of model species in lignocellulosic biochemical production. With the development of systems and synthetic biology, more nonmodel microbial chassis cells with excellent industrial characteristics have been explored, such as Z. mobilis, Halomonas bluephagenesis, B. licheniformis, Clostridium autoethanogenum, C. acetobutylicum, Vibrio natriegens, and unconventional yeasts (Table 3), which can be used as chasses for economic biochemical production.
Synthetic biology provides powerful tools for designing, synthesizing, and reconstructing biological parts, circuits, and modules into functional synthetic cell factories. The breakthroughs in effective gene editing, synthesis, and assembly tools can facilitate the construction of robust and efficient industrial strains, and MCFs can be further improved to meet industrial needs.
Bioethanol is currently one of the most widely used bioenergy sources in the world. Its large-scale industrial production is achieved mainly by S. cerevisiae using grain crops such as corn or economic crops such as sugarcane. However, producing bioethanol from crops faces the challenges of competing with humans for food usage, which is not conducive to sustainable development. Therefore, the use of nonfood feedstocks for bioethanol production is a future trend. Z. mobilis is a nonmodel polyploid ethanologenic gram-negative bacterium with many industrial merits and unique physiological properties [80]; it is the only known microorganism that can utilize the ED pathway under anaerobic conditions [81] and has excellent characteristics compared with S. cerevisiae, such as a high sugar uptake rate, high ethanol yield, and ethanol tolerance [82]. DuPont has achieved the industrial production of 30 million gallons of cellulosic ethanol per year via C5-utilizing recombinant Z. mobilis [45]. In addition, efficient genome-editing tools based on heterologous clustered regularly interspaced short palindromic repeats (CRISPR)–CRISPR-associated protein 12a (Cas12a), endogenous type I-F CRISPR-Cas and associated repair pathways such as microhomology-mediated end joining (MMEJ), as well as a genome-wide iterative continuous editing (GW-ICE) system have been developed [83–86]. Many MCFs have been constructed for the production of acetaldehyde, lactate, l-alanine, l-serine, acetoin, 2,3-BDO, isobutanol, l-malate, succinic acid, poly-3-hydroxybutyrate (PHB), farnesene, and ethylene [21,87–97].
Succinic acid is a promising platform chemical used for the production of biomaterials such as 1,4-butanediol, γ-butyrolactone, tetrahydrofuran, and polybutylene succinate (PBS), which is also listed by the US Department of Energy as one of the 12 top platform chemicals. E. coli, in addition to some natural succinic acid-producing strains, is generally the most commonly used chassis cell for succinic acid production. Although these natural bacteria can achieve high yields and titers of succinic acid production, the fermentation process normally requires obligate anaerobic conditions with the addition of alkali to maintain neutral pH conditions, which increases costs in industrial production. Y. lipolytica has a strong ability to synthesize acetyl-CoA (coenzyme A), providing sufficient precursors for succinic acid production. High-efficiency succinic acid at low pH was achieved at a titer of 111.9 g/l by reconfiguring the reductive TCA cycle in Y. lipolytica [66]. Many important synthetic biology tools and components have been developed and applied in Y. lipolytica, such as DNA assembly technology, genome editing technology, and computational tools [98]. The development of these tools has further accelerated the construction of MCFs using Y. lipolytica as the chassis cell [98].
PHAs are polyesters synthesized by a wide range of bacteria and have been developed into various environmentally friendly plastic products [99]. H. bluephagenesis TD01 is an extreme microorganism that can accumulate ~80% (wt%) poly-β-hydroxybutyrate (PHB) naturally. Additionally, H. bluephagenesis has been engineered to produce 3-hydroxybutyrate, 3-hydroxypropionate, and 4-hydroxybutyrate [99–101] and other biochemicals, such as lysine, ectoine, cadaverine, and 5-aminolevulinic acid. Extremophiles such as halophilic bacteria provide a model and paradigm for the development of next-generation industrial biotechnology (NGIB) [102]. Lactic acid, another monomer of bioplastics, is not limited to natural microbial synthesis. It has been synthesized through synthetic biological methods in both model and nonmodel microorganisms [90].
Functional sugars are a type of carbohydrate that cannot be digested and absorbed by the human gastrointestinal tract and have special properties, such as sweetness flavor or the ability to promote probiotic growth. They mainly include functional oligosaccharides, sugar alcohols, and starch. These sugars can be used as functional food additives or raw materials alternatives to sucrose to meet the needs of special populations, such as those with diabetes and obesity. At present, its biosynthetic technology mainly includes enzymatic catalytic conversion technology and microbial fermentation technology. Food-grade enzyme expression systems were developed using B. subtilis and C. glutamicum as chassis strains, and various monosaccharides, dioxygenases, and branched-chain ketose compounds were generated by exploring different enzyme elements [103–105]. E. coli, S. cerevisiae, C. glutamicum, and B. subtilis were all designed and developed as chassis cells for human milk oligosaccharide (HMO) synthesis.
Artificial MCF synthesis of artemisinin is a landmark achievement of synthetic cell factories for biomedicine. Owing to their potential applications and great challenges, the construction of MCFs for natural products has attracted considerable attention. The high yield of terpenoid compounds in the cell factory when Y. lipolytica was used as the substrate increased the titer of squalene by a factor of 1,300 [106]. The biosynthesis of cannabidiol was achieved via the use of S. cerevisiae as the chassis cell [107]. The reconstruction and analysis of the biosynthesis pathway of Taxol have laid a solid foundation for elucidating its complete pathway [108,109].
The achievements in both nonmodel and model microorganisms led to the formation of fourth-generation industrial microbial cell factories (MCF 4.0), which can play important roles in bioenergy, biomaterials, bioplastics, biomedicines, food, and nutrients. Disruptive enabling technologies are the key to reprogramming biological genes, circuits, pathways, networks, and other components to continuously iterate and upgrade MCFs to accelerate the construction and application of efficient cell factories.
Enriching biological components and developing enabling technologies
Owing to the high complexity of cells, artificially implanted biological elements, circuits, or systems are influenced by the existing metabolic and regulatory pathways within the cells. Therefore, the continuous expansion of biological components and improvements in enabling technologies are needed. These include the excavation and identification of biological parts, circuits, and modules; the understanding of cell metabolism and regulatory networks among different chassis cells; the development of design theories and tools; and the improvement of high-throughput automatic assembly and testing methods for accelerating the development of microbial chassis cells and the construction of efficient MCFs (Fig. 4). Finally, enabling biotechnology tools need to be integrated with automation and AI for further applications.
Fig. 4.
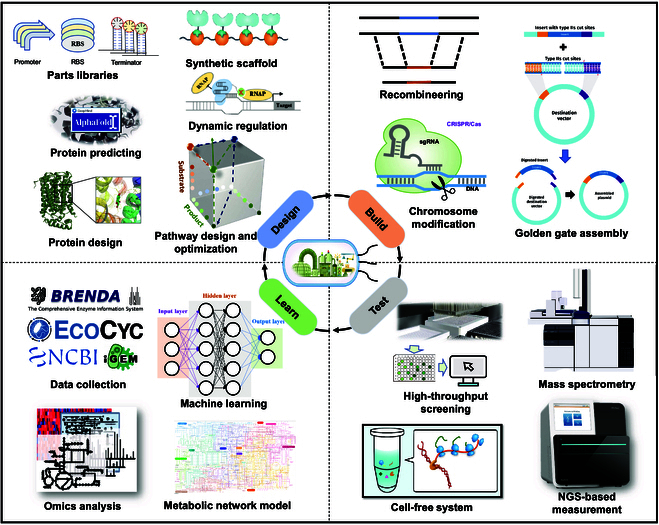
Enabling technologies and applications of synthetic biology for the construction and optimization of MCF 4.0.
Characterization and applications of biological parts
Biological parts are one of the fundamental elements in synthetic biology and constitute the cornerstone of synthetic biology. Standard biological parts include DNA sequences such as promoters, terminators, transcription units, plasmids, conjugation transfer elements, transposons, and protein coding regions, as well as RNA sequences such as RBSs and protein domains. The excavation, identification, and modification of biological components is an important research direction in the field of synthetic biology.
An increasing number of biological parts, including promoters and RBSs, have been applied to regulate metabolic network flux, increase target product yield, and reduce byproduct generation. It has become an inevitable trend to build a promoter library with a range of strengths, as is the case for yeast and E. coli [110]. In addition, artificial RBS and untranslated region sequences with different translation levels can be predicted and designed using computational tools [111].
For the convenience of different levels of design, researchers have developed corresponding analysis tools for different nonstandard biological components, such as Softberry software and online analysis websites for promoter prediction analysis and the Database for Prokaryotic OpeRons (DOOR) for prokaryotic operator prediction analysis [112,113]. In addition, a series of biological element algorithms (Algorithm) tools, metabolic pathway constructions, and protein expression processes (Workflows) have been developed. These include the RBS Library Calculator for predicting translation rates and controlling expression levels; the RBS Library Calculator for predicting expression changes, optimizing expression levels, and designing libraries; the Operator Calculator for predicting the expression of operons and multiple proteins; the ELSA Calculator for regulating multiple genes; the Syth Success Calculator for optimizing DNA synthesis; the NRP Calculator for mining and designing nonrepetitive elements; and the Riboswitch Calculator for predicting riboswitches and designing RNA sensors [114,115].
Gene circuits
With an increasing number of bioparts being excavated and characterized, redesigning and constructing bioparts enables artificially designed biomolecular pathways to regulate metabolic networks in MCFs, which has enormous potential in biological manufacturing. Gene circuits comprise 2 basic types: transcriptional gene regulatory circuits and protein-based signaling circuits.
Transcriptional gene regulatory circuits control gene expression through transcription. The widely used transcriptional gene regulatory circuits are T7 RNAP, LacI, and TetR. The T7 expression system established on the basis of the specific and efficient recognition between T7 RNAP and the PT7 promoter is currently one of the most powerful expression systems for exogenous proteins in E. coli and has been widely used for protein expression [116,117]. The TetR-derived promoter has also been used for the production of biochemicals, such as isobutanol and acetoin [88,93]. Promoter–transcription factor circuits have also been used for the dynamic regulation of MCFs. Transcription factors can bind to specific metabolites and promoter region DNA, thereby activating or inhibiting downstream gene expression. Acyl-CoA responsive dynamic sensor regulatory system (DSRS) was developed to regulate the expression of fatty acid ethyl ester production genes, resulting in a 3-fold increase in fatty acid ethyl ester production to 1.5 g/l [118].
With in-depth research on the quorum-sensing (QS) mechanism of microorganisms, dynamic regulation based on QS has also been used to decouple cell growth and production processes. By regulating the expression levels of the cell density-dependent genes luxI and luxR and changing the switching time of trigger gene expression, this system has been applied to the production of 1,4-butanediol and bisabolene and the redirection of the TCA cycle to isopropanol through trigger switching [119–121].
Developing and optimizing genome-editing strategies
For industrial applications of MCFs, the chromosomal expression of biosynthetic genes is often favored over plasmid-based expression because of the instability of plasmids. Zinc-finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) constitute a powerful class of tools for genetic modifications in the past 10 years, and with the elucidation of the mechanisms of the CRISPR-Cas system, various CRISPR-based technologies have been developed as powerful genome editing tools for MCF construction [122].
The CRISPR-Cas system originates from the immune system of microorganisms and has recently attracted great interest as a gene editing tool because of its precision and rapid characteristics compared with those of conventional tools. For model microorganism applications, CRISPR-Cas9 was first introduced into E. coli for gene editing [123] and then optimized to achieve simultaneous editing of 3 genes [124]. To increase the curing and editing efficiency of plasmid editing, a Rock-Paper-Scissors strategy, a robust and fast iterative genome-editing strategy, was designed and applied in E. coli and K. pneumoniae [125]. Additionally, a mature CRISPR-Cas editing system was constructed and applied in other model microorganisms (Table 3).
As important industrial chassis cells for cellulosic ethanol production, gene editing systems based on exogenous CRISPR-Cas12a and endogenous I-F-type CRISPR-Cas systems have been successfully developed in Z. mobilis, as well as CRISPRi technology based on dCas9 and the guide RNA (gRNA) design network tool CRISpy-pop [83,84,126], providing diverse genetic tools for genome editing and modification and for successfully constructing efficient cell factories for d-lactic acid and acetoin production [90,93]. Transposon mutation, homologous recombination, the CRISPR-Cas9 editing system, and a multigene editing system have been established in V. natriegens [127,128], which has accelerated the process of metabolic engineering and genome simplification optimization and achieved the production of recombinant proteins, melanin, carotenoids, and violets [129–134]. In addition, genome-editing systems have been established in some nonmodel microorganisms, and the synthesis of biochemicals has been achieved, accelerating the industrial application of nonmodel microbial chassis cells [135,136].
Genome minimization
Genome minimization is an important strategy for the rational construction of excellent chassis cells. Cell energy consumption is reduced by knocking out redundant genes, allowing more energy to be used for target product production [137]. Various microorganisms have demonstrated excellent characteristics through genome minimization, such as increased biomass, genetic stability, transformation efficiency, and product yield [137]. For example, 663 kb of genomic DNA was removed from E. coli MDS42, resulting in a 2-order increase in transformation efficiency compared with that of the parental strain [138]. After 1,670 kb of genomic DNA was deleted, the E. coli DGF-298 genome was more stable, with a higher growth rate and cell density. From the perspective of biological product synthesis, the simplified B. subtilis BSK814G2 genome lacks 814.4 kb of genomic DNA, but the cells accumulate 115.2 mg/l guanosine (~4.4 times greater than that of the original strain). The minimization of B. subtilis BSK756T3 resulted in a 5.2-fold increase in the concentration of intracellular 151.2 mg/l thymidine [139]. In addition, after 7.7% of the genomic DNA in Pseudomonas mendocina NK-01 was deleted, the production of PHA and alginate oligosaccharides increased by 114.8% and 27.8%, respectively [139]. Therefore, through genome minimization, a microbial chassis with excellent traits and high yield can be obtained.
High-throughput screening
High-throughput screening can be used to obtain desired mutants from large variant libraries quickly and efficiently. With the development of molecular biology and gene editing technology, genetic engineering methods continue to be iterated and innovated, resulting in different mutant libraries. Combining traditional microplate-based screening methods with automated equipment can reduce labor intensity and improve screening efficiency to 103 to 105 colonies per day, and has been successfully used in various industrial microbial strain screening methods [140,141].
In addition, fluorescent markers, mass spectrometry, and single-cell Raman spectroscopy (SCRS) have also been introduced on the basis of their unique characteristics. SCRS technology can recognize panoramic information at the level of living single cells and nonlabeled states, thereby distinguishing complex functional phenotypes. It has advantages such as being fast and economic, can be coupled with downstream omics research, and is considered a new single-cell phenotype recognition technology [142]. Flow cytometry screening, such as fluorescence-activated cell sorting (FACS), can analyze and sort individual cells on the basis of specific criteria. However, the application of FACS is limited to analyzing fluorescence signals related to intracellular target products or metabolites bound to membranes and cannot be used for identifying cells that overproduce secreted metabolites and secreted extracellular enzymes. Mass spectrometry is highly sensitive and does not require specific labeling, which can provide key technical support for high-throughput screening of MCFs without epigenetic traits [143].
Compared with the above screening methods, droplet microfluidics, which mainly involve the formation of monodisperse microdroplets through the shearing of 2 incompatible fluids, can monitor quantitative single-cell metabolite concentrations with increased screening efficiency. More precise operations such as droplet incubation, fusion, division, detection, and sorting in the chip can be achieved. To better achieve high-throughput and automated continuous cultivation of microbial chassis cells to obtain mutant strains with excellent characteristics, a microbial microdroplet culture system (MMC) was developed by combining microfluidic technology and photoelectric sensing and automation technology for parallel cultivation and growth curve monitoring of microbial droplets. This accelerated the evolution of numerous microbial chassis cells, greatly improving the efficiency of obtaining excellent phenotype samples, such as substrate utilization ability, solvent tolerance, and robustness of microbial substrate cells [144,145].
Artificial intelligence
Computer-aided design (CAD) of biomolecules with specific functions greatly improves the ability and quality of biological parts and systems. The rational design of biological parts, circuits, pathways, and systems can be achieved through AI or computational biology to achieve better biological expression and higher production efficiency. For example, AI algorithms can be used to design and optimize promoter sequences, thereby increasing the expression level of target genes and reducing side effects. A machine learning model for predicting promoter strength in E. coli was designed on the basis of artificial neural networks and support vector machines [146].
Moreover, computational biology methods can also be used to simulate and predict the expression levels and functions of enzymes in different environments. Design methods and tools such as FireProt, GRAPE-WEB, PROSS, Funclib, ABACUS2, the Swiss Model, ConSurf, and ROSIE have been developed based on the selective modification and stability enhancement of enzymes, greatly improving the efficiency of enzyme design [147–153]. With the application of AI, an increasing number of key enzymes and metabolic pathways in metabolic networks have been identified by analyzing large amounts of metabolic datasets. Computational biology methods can also be used to simulate and predict the performance of metabolic pathways under different conditions, thereby helping to optimize the design of metabolic engineering and better guiding the construction and development of efficient MCFs.
Creation of Customized Artificial Cell Factories
The development of a knowledge base of microbial cells and the technological advancements in AI and automation have made rational design, synthesis, and testing of chassis cells more intelligent, greatly reducing the trial-and-error frequency of industrial chassis cells during their development process. In addition, the rapid accumulation of omics data and AI has led to the efficient discovery and analysis of many genes of unknown function in the genome. Customized industrial chassis cells can be obtained by modifying genotype-associated phenotypes, which can also be referred to as fifth-generation industrial microbial cell factories (MCF 5.0).
The development of sequencing technology and the rapid growth of biological data resource models have brought new understanding and development opportunities to the design of MCFs. The customized design of MCFs can be based on massive amounts of data resources and adopt biological design automation. The automated design of cell factories can be completed through a series of algorithms to predict and screen biosynthetic pathways, the design of regulatory components and pathways, and the adaptability of pathways and the chassis. On the basis of data- and knowledge-driven approaches, intelligent design and automated high-throughput construction testing are assisted to further develop the ability to synthesize biobased products via the use of diverse chassis cells, such as those of microorganisms, animals, and plants. This enables comprehensive development from the local modification of genetic components to new creations, spanning from the local optimization of genetic networks to overall adaptation, local genome replacement to de novo design and synthesis, and the passive observation of cellular functions for precise control [154]. The efficient and intelligent creation of enzymes and cell factories promotes the green and sustainable development of the bioeconomy, especially cell factories, which are regarded as “chips” that are directly related to research and industrial development in biological manufacturing.
Future Perspectives of Industrial Digital MCFs
Achievements have been made in constructing cell factories to synthesize biochemicals using model or nonmodel microbial chassis cells by combining systems biology, synthetic biology, and metabolic engineering technologies. However, the majority of target product syntheses have not yet met the requirements of industrial production at present, and there is an urgent need to achieve breakthroughs in yield and productivity. Therefore, it is necessary to further explore and study the key biological components in the biosynthetic pathway of biochemicals, design and reconstruct the metabolic mode of chassis cells, optimize the adaptability of synthesis pathways and metabolic networks, and combine process engineering technology with intelligent control strategies to support fermentation processes. In the era of systems and synthetic biology, the constantly evolving biotechnology and rapidly developing IT will provide abundant tools and resources for the construction and optimization of efficient MCFs in biological manufacturing.
The construction of an efficient cell factory integrates disciplines such as stoichiometry, computer simulation, and biotechnology, especially with the current development of synthetic biotechnology, making it possible to transform living organisms to serve humanity. In the design and assembly process of cell factories, it is necessary not only to evaluate the feasibility of metabolic pathways through stoichiometry but also to optimize and design more efficient synthesis pathways and components through computer simulation and other tools. On this basis, biotechnology is used to assemble and fine tune the synthesis module, ensuring a sufficient supply of precursors in the metabolic pathway, smooth metabolic flow, cofactor balance, and feedback inhibition release, thereby obtaining an efficient cell factory for synthesizing target chemicals. With the advancement of synthetic biotechnology, many expression elements and regulatory methods have been developed, and these tools will gradually be applied to the construction and optimization of artificial synthetic cell factories.
AI is playing an increasingly important role in synthetic biology. In the past few years, AI has achieved great success in protein design. With their powerful feature extraction, data statistics, and function fitting capabilities, advanced AI models learn basic features and interaction relationships from existing protein structure and sequence data and fit generalized function models for application in various protein design tasks. Taking AlphaFold as an example, combining advanced machine learning techniques and biophysical knowledge enables AlphaFold to improve the accuracy of protein structure prediction, thereby accelerating the progress of biological science [155]. The predictive accuracy of the powerful protein–protein interactions of AlphaFold3 also represents a major step forward in predicting the structure of biomolecules [156]. The prediction and development of proteins by AI can be further applied to the development and optimization of efficient MCFs, promoting the development of cell factories.
With the further development and cross-integration of automation, IT, and biotechnology, various technologies are expected to become increasingly mature. The understanding of chassis cells is gradually increasing, the library of biological components is constantly expanding, and the design, construction, optimization, and adaptation of artificial metabolic pathways are becoming more convenient and practical. Fermentation process optimization and control are becoming increasingly intelligent, which will lead to breakthroughs in the efficient, green, and low-cost synthesis of customized artificial cell factories for economic biochemical production and commercialization.
Conclusion
The discovery and creation of cell factories is a long process, from initially requiring a large number of human resources to accelerating the iteration and updating of MCFs with the emergence of efficient and effective tools. The development of MCFs is undergoing diverse processes, and the application and development of model and nonmodel microorganisms have promoted the industrial application of MCFs. The emergence of synthetic biology is a revolutionary subversion of MCFs. However, in the era of AI, the construction and optimization of MCFs not only require advanced biotechnology methods but also rely on digital technologies with stronger simulation and computing capabilities to reduce time and material costs. With the support of multiple biotechnologies and AI, MCF 5.0 will demonstrate better performance and a more intelligent form.
Acknowledgments
Funding: This work was supported by the National Key Research and Development Program of China (2022YFA0911800), the International Science and Technology Cooperation Base of Hubei Province (SH2318), and the Innovation Base for Introducing Talents of Discipline of Hubei Province (2019BJH021).
Author contributions: S.Y. conceived the idea with inputs from all the authors. X.Y. and Q.H. wrote the draft. X.Y., Q.H., B.G., and S.Y. revised the manuscript. All the authors read and approved the final manuscript.
Competing interests: The authors declare that S.Y. is the founder of Wuhan ZymoBiotech Inc.
References
- 1.Xie D. Continuous biomanufacturing with microbes—Upstream progresses and challenges. Curr Opin Biotechnol. 2022;78: Article 102793. [DOI] [PubMed] [Google Scholar]
- 2.Bailey JE. Toward a science of metabolic engineering. Science. 1991;252(5013):1668–1675. [DOI] [PubMed] [Google Scholar]
- 3.Zhang XE, Liu C, Dai J, Yuan Y, Gao C, Feng Y, Wu B, Wei P, You C, Wang X, et al. Enabling technology and core theory of synthetic biology. Sci China Life Sci. 2023;66(8):1742–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho JS, Kim GB, Eun H, Moon CW, Lee SY. Designing microbial cell factories for the production of chemicals. JACS Au. 2022;2(8):1781–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi KR, Jang WD, Yang D, Cho JS, Park D, Lee SY. Systems metabolic engineering strategies: Integrating systems and synthetic biology with metabolic engineering. Trends Biotechnol. 2019;37(8):817–837. [DOI] [PubMed] [Google Scholar]
- 6.Carbonell P, Jervis AJ, Robinson CJ, Yan C, Dunstan M, Swainston N, Vinaixa M, Hollywood KA, Currin A, Rattray NJW, et al. An automated design-build-test-learn pipeline for enhanced microbial production of fine chemicals. Commun Biol. 2018;1:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morris RW, Bean CA, Farber GK, Gallahan D, Jakobsson E, Liu Y, Lyster PM, Peng GCY, Roberts FS, Twery M, et al. Digital biology: An emerging and promising discipline. Trends Biotechnol. 2005;23(3):113–117. [DOI] [PubMed] [Google Scholar]
- 8.Yamada Y, Yukphan P. Genera and species in acetic acid bacteria. Int J Food Microbiol. 2008;125(1):15–24. [DOI] [PubMed] [Google Scholar]
- 9.Bastián F, Cohen A, Piccoli P, Luna V, Bottini R, Baraldi R, Bottini R. Production of indole-3-acetic acid and gibberellins A1 and A3 by Acetobacter diazotrophicus and Herbaspirillum seropedicae in chemically-defined culture media. Plant Growth Regul. 1998;24:7–11. [Google Scholar]
- 10.Fuentes-Ramírez LE, Bustillos-Cristales R, Tapia-Hernández A, Jiménez-Salgado T, Wang ET, Martínez-Romero E, Caballero-Mellado J. Novel nitrogen-fixing acetic acid bacteria, Gluconacetobacter johannae sp. nov. and Gluconacetobacter azotocaptans sp. nov., associated with coffee plants. Int J Syst Evol Microbiol. 2001;51(4):1305–1314. [DOI] [PubMed] [Google Scholar]
- 11.Nguyen VT, Flanagan B, Gidley MJ, Dykes GA. Characterization of cellulose production by a Gluconacetobacter xylinus strain from Kombucha. Curr Microbiol. 2008;57(5):449–453. [DOI] [PubMed] [Google Scholar]
- 12.Kim S, Jang YS, Ha SC, Ahn JW, Kim EJ, Hong Lim J, Cho C, Shin Ryu Y, Kuk Lee S, Lee SY, et al. Redox-switch regulatory mechanism of thiolase from Clostridium acetobutylicum. Nat Commun. 2015;6(5):449–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones DT, Woods DR. Acetone-butanol fermentation revisited. Microbiol Rev. 1986;50:484–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Demain AL, Elander RP. The beta-lactam antibiotics: Past, present, and future. Anton Leeuw Int J G. 1999;75(1-2):5–19. [DOI] [PubMed] [Google Scholar]
- 15.Bérdy J. Bioactive microbial metabolites. J Antibiot. 2005;58:1–26. [DOI] [PubMed] [Google Scholar]
- 16.Lee JW, Yi J, Kim TY, Choi S, Ahn JH, Song H, Lee MH, Lee SY. Homo-succinic acid production by metabolically engineered Mannheimia succiniciproducens. Metab Eng. 2016;38:409–417. [DOI] [PubMed] [Google Scholar]
- 17.Lee PC, Lee SY, Hong SH, Chang HN, Park SC. Biological conversion of wood hydrolysate to succinic acid by Anaerobiospirillum succiniciproducens. Biotechnol Lett. 2003;25(2):111–114. [DOI] [PubMed] [Google Scholar]
- 18.Qiao K, Wasylenko TM, Zhou K, Xu P, Stephanopoulos G. Lipid production in Yarrowia lipolytica is maximized by engineering cytosolic redox metabolism. Nat Biotechnol. 2017;35(2):173–177. [DOI] [PubMed] [Google Scholar]
- 19.Kurosawa K, Plassmeier J, Kalinowski J, Rückert C, Sinskey AJ. Engineering l-arabinose metabolism in triacylglycerol-producing Rhodococcus opacus for lignocellulosic fuel production. Metab Eng. 2015;30:89–95. [DOI] [PubMed] [Google Scholar]
- 20.Eggeling L, Bott M. Handbook of Corynebacterium glutamicum. Boca Raton (FL): CRC Press; 2005.
- 21.Yang S, Fei Q, Zhang Y, Contreras LM, Utturkar SM, Brown SD, Himmel ME, Zhang M. Zymomonas mobilis as a model system for production of biofuels and biochemicals. Microb Biotechnol. 2016;9:699–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jansen MLA, Bracher JM, Papapetridis I, Verhoeven MD, Bruijn H, Waal PP, AJA Van Maris, Klaassen P, Pronk JT. Saccharomyces cerevisiae strains for second-generation ethanol production: From academic exploration to industrial implementation. FEMS Yeast Res. 2017;17(5):fox044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song AA-L, In LLA, Lim SHE, Rahim RA. A review on Lactococcus lactis: From food to factory. Microb Cell Factories. 2017;16(1):55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun Y, Shen JT, Yan L, Zhou JJ, Jiang LL, Chen Y, Yuan JL, Feng EM, Xiu ZL. Advances in bioconversion of glycerol to 1,3-propanediol: Prospects and challenges. Process Biochem. 2018;71:134–146. [Google Scholar]
- 25.Zhang Q, Miao R, Feng R, Yan J, Wang T, Gan Y, Zhao J, Lin J, Gan B. Application of atmospheric and room-temperature plasma (ARTP) to microbial breeding. Curr Issues Mol Biol. 2023;45(8):6466–6484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X, Zhang X, Xu G, Zhang X, Shi J, Xu Z. Integration of ARTP mutagenesis with biosensor-mediated high-throughput screening to improve l-serine yield in Corynebacterium glutamicum. Appl Microbiol Biotechnol. 2018;102:5939–5951. [DOI] [PubMed] [Google Scholar]
- 27.Gao X, Zhao H, Zhang G, He K, Jin Y. Genome shuffling of Clostridium acetobutylicum CICC 8012 for improved production of acetone-butanol-ethanol (ABE). Curr Microbiol. 2012;65(2):128–132. [DOI] [PubMed] [Google Scholar]
- 28.Zhang J, Wang X, Diao J, He H, Zhang Y, Xiang W. Streptomycin resistance-aided genome shuffling to improve doramectin productivity of Streptomyces avermitilis NEAU1069. J Ind Microbiol Biotechnol. 2013;40(8):877–889. [DOI] [PubMed] [Google Scholar]
- 29.Lv XA, Jin YY, Li YD, Zhang H, Liang XL. Genome shuffling of Streptomyces viridochromogenes for improved production of avilamycin. Appl Microbiol Biotechnol. 2013;97(2):641–648. [DOI] [PubMed] [Google Scholar]
- 30.Wang Q, Zhang D, Li Y, Zhang F, Wang C, Liang X. Genome shuffling and ribosome engineering of Streptomyces actuosus for high-yield nosiheptide production. Appl Biochem Biotechnol. 2014;173(6):1553–1563. [DOI] [PubMed] [Google Scholar]
- 31.Wang L, Chen X, Wu G, Zeng X, Ren X, Li S, Tang L, Mao Z. Genome shuffling and gentamicin-resistance to improve ε-poly-L-lysine productivity of Streptomyces albulus W-156. Appl Biochem Biotechnol. 2016;180(6):1601–1617. [DOI] [PubMed] [Google Scholar]
- 32.Wang PM, Zheng DQ, Liu TZ, Tao XL, Feng MG, Min H, Jiang XH, Wu XC. The combination of glycerol metabolic engineering and drug resistance marker-aided genome shuffling to improve very-high-gravity fermentation performances of industrial Saccharomyces cerevisiae. Bioresour Technol. 2012;108:203–210. [DOI] [PubMed] [Google Scholar]
- 33.Cheng C, Almario MP, Kao KC. Genome shuffling to generate recombinant yeasts for tolerance to inhibitors present in lignocellulosic hydrolysates. Biotechnol Lett. 2015;37(11):2193–2200. [DOI] [PubMed] [Google Scholar]
- 34.Yilmaz S, Nyerges A, Oost J, Church GM, Claassens NJ. Towards next-generation cell factories by rational genome-scale engineering. Nat Catal. 2022;5(9):751–765. [Google Scholar]
- 35.Liu P, Xu H, Zhang X. Metabolic engineering of microorganisms for L-alanine production. J Ind Microbiol Biotechnol. 2022;49(2):kuab057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee JH, Lama S, Kim JR, Park SH. Production of 1,3-propanediol from glucose by recombinant Escherichia coli BL21(DE3). Biotechnol Bioprocess Eng. 2018;23(2):250–258. [Google Scholar]
- 37.Zhou S, Causey TB, Hasona A, Shanmugam KT, Ingram LO. Production of optically pure D-lactic acid in mineral salts medium by metabolically engineered Escherichia coli W3110. Appl Environ Microbiol. 2003;69(1):399–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu X, Tan Z, Xu H, Chen J, Tang J, Zhang X. Metabolic evolution of two reducing equivalent-conserving pathways for high-yield succinate production in Escherichia coli. Metab Eng. 2014;24:87–96. [DOI] [PubMed] [Google Scholar]
- 39.Yim H, Haselbeck R, Niu W, Pujol-Baxley C, Burgard A, Boldt J, Khandurina J, Trawick JD, Osterhout RE, Stephen R, et al. Metabolic engineering of Escherichia coli for direct production of 1,4-butanediol. Nat Chem Biol. 2011;7(7):445–452. [DOI] [PubMed] [Google Scholar]
- 40.Bian G, Deng Z, Liu T. Strategies for terpenoid overproduction and new terpenoid discovery. Curr Opin Biotechnol. 2017;48:234–241. [DOI] [PubMed] [Google Scholar]
- 41.Tanaka K, Natsume A, Ishikawa S, Takenaka S, Yoshida KI. A new-generation of Bacillus subtilis cell factory for further elevated scyllo-inositol production. Microb Cell Factories. 2017;16(1):67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Song Y, Guan Z, van Merkerk R, Pramastya H, Abdallah II, Setroikromo R, Quax WJ. Production of squalene in Bacillus subtilis by squalene synthase screening and metabolic engineering. J Agric Food Chem. 2020;68:4447–4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang S, Wang Y, Wei C, Liu Q, Jin X, du G, Chen J, Kang Z. A new sRNA-mediated posttranscriptional regulation system for Bacillus subtilis. Biotechnol Bioeng. 2018;115(12):2986–2995. [DOI] [PubMed] [Google Scholar]
- 44.Jin P, Zhang L, Yuan P, Kang Z, Du G, Chen J. Efficient biosynthesis of polysaccharides chondroitin and heparosan by metabolically engineered Bacillus subtilis. Carbohydr Polym. 2016;140:424–432. [DOI] [PubMed] [Google Scholar]
- 45.Zhao X, Liu C, Bai F. Making the biochemical conversion of lignocellulose more robust. Trends Biotechnol. 2024;42(4):418–430. [DOI] [PubMed] [Google Scholar]
- 46.Chen S, Xu Z, Ding B, Zhang Y, Liu S, Cai C, Li M, Dale BE, Jin M. Big data mining, rational modification, and ancestral sequence reconstruction inferred multiple xylose isomerases for biorefinery. Sci Adv. 2023;9(5):eadd8835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhong W, Li H, Wang Y. Design and construction of artificial biological systems for one-carbon utilization. Biodes Res. 2023;5:0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meesapyodsuk D, Chen Y, Ng SH, Chen J, Qiu X. Metabolic engineering of Pichia pastoris to produce ricinoleic acid, a hydroxy fatty acid of industrial importance. J Lipid Res. 2015;56:2102–2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dong C, Qiao J, Wang X, Sun W, Chen L, Li S, Wu K, Ma L, Liu Y. Engineering Pichia pastoris with surface-display minicellulosomes for carboxymethyl cellulose hydrolysis and ethanol production. Biotechnol Biofuels. 2020;13:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Siripong W, Wolf P, Kusumoputri TP, Downes JJ, Kocharin K, Tanapongpipat S, Runguphan W. Metabolic engineering of Pichia pastoris for production of isobutanol and isobutyl acetate. Biotechnol Biofuels. 2018;11:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang Z, Zhang Z. Production of (2R, 3R)-2,3-butanediol using engineered Pichia pastoris: Strain construction, characterization and fermentation. Biotechnol Biofuels. 2018;11:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gao J, Li Y, Yu W, Zhou YJ. Rescuing yeast from cell death enables overproduction of fatty acids from sole methanol. Nat Metab. 2022;4(7):932–943. [DOI] [PubMed] [Google Scholar]
- 53.Liu Z, Wang K, Chen Y, Tan T, Nielsen J. Third-generation biorefineries as the means to produce fuels and chemicals from CO2. Nat Catal. 2020;3(3):274–288. [Google Scholar]
- 54.Gleizer S, Ben-Nissan R, Bar-On YM, Antonovsky N, Noor E, Zohar Y, Jona G, Krieger E, Shamshoum M, Bar-Even A, et al. Conversion of Escherichia coli to generate all biomass carbon from CO2. Cell. 2019;179(6):1255–1263 e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Santos Correa S, Schultz J, Lauersen KJ, Soares RA. Natural carbon fixation and advances in synthetic engineering for redesigning and creating new fixation pathways. J Adv Res. 2023;47:75–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kok S, Kozak BU, Pronk JT, Maris AJ. Energy coupling in Saccharomyces cerevisiae: Selected opportunities for metabolic engineering. FEMS Yeast Res. 2012;12(4):387–397. [DOI] [PubMed] [Google Scholar]
- 57.Hara KY, Kondo A. ATP regulation in bioproduction. Microb Cell Factories. 2015;14:198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yoon SH, Hwan Do J, Yup Lee S, Nam CH. Production of poly-γ-glutamic acid by fed-batch culture of Bacillus licheniformis. Biotechnol Lett. 2000;22(7):585–588. [Google Scholar]
- 59.Shi A, Zhu X, Lu J, Zhang X, Ma Y. Activating transhydrogenase and NAD kinase in combination for improving isobutanol production. Metab Eng. 2013;16:1–10. [DOI] [PubMed] [Google Scholar]
- 60.Ng CY, Farasat I, Maranas CD, Salis HM. Rational design of a synthetic Entner-Doudoroff pathway for improved and controllable NADPH regeneration. Metab Eng. 2015;29:86–96. [DOI] [PubMed] [Google Scholar]
- 61.Liang S, Chen H, Liu J, Wen J. Rational design of a synthetic Entner-Doudoroff pathway for enhancing glucose transformation to isobutanol in Escherichia coli. J Ind Microbiol Biotechnol. 2018;45(3):187–199. [DOI] [PubMed] [Google Scholar]
- 62.Qureshi AS, Zhang J, Bao J. High ethanol fermentation performance of the dry dilute acid pretreated corn stover by an evolutionarily adapted Saccharomyces cerevisiae strain. Bioresour Technol. 2015;189:399–404. [DOI] [PubMed] [Google Scholar]
- 63.Mundhada H, Seoane JM, Schneider K, Koza A, Christensen HB, Klein T, Phaneuf PV, Herrgard M, Feist AM, Nielsen AT. Increased production of L-serine in Escherichia coli through adaptive laboratory evolution. Metab Eng. 2017;39:141–150. [DOI] [PubMed] [Google Scholar]
- 64.Yang Q, Yang Y, Tang Y, Wang X, Chen Y, Shen W, Zhan Y, Gao J, Wu B, He M, et al. Development and characterization of acidic-pH-tolerant mutants of Zymomonas mobilis through adaptation and next-generation sequencing-based genome resequencing and RNA-Seq. Biotechnol Biofuels. 2020;13(1):144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Horinouchi T, Sakai A, Kotani H, Tanabe K, Furusawa C. Improvement of isopropanol tolerance of Escherichia coli using adaptive laboratory evolution and omics technologies. J Biotechnol. 2017;255:47–56. [DOI] [PubMed] [Google Scholar]
- 66.Cui Z, Zhong Y, Sun Z, Jiang Z, Deng J, Wang Q, Nielsen J, Hou J, Qi Q. Reconfiguration of the reductive TCA cycle enables high-level succinic acid production by Yarrowia lipolytica. Nat Commun. 2023;14(1):8480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Deparis Q, Claes A, Foulquie-Moreno MR, Thevelein JM. Engineering tolerance to industrially relevant stress factors in yeast cell factories. FEMS Yeast Res. 2017;17(4): Article fox036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tan Z, Yoon JM, Nielsen DR, Shanks JV, Jarboe LR. Membrane engineering via trans unsaturated fatty acids production improves Escherichia coli robustness and production of biorenewables. Metab Eng. 2016;35:105–113. [DOI] [PubMed] [Google Scholar]
- 69.Yuan Y, Bi C, Nicolaou SA, Zingaro KA, Ralston M, Papoutsakis ET. Overexpression of the Lactobacillus plantarum peptidoglycan biosynthesis murA2 gene increases the tolerance of Escherichia coli to alcohols and enhances ethanol production. Appl Microbiol Biotechnol. 2014;98(19):8399–8411. [DOI] [PubMed] [Google Scholar]
- 70.Tan Z, Khakbaz P, Chen Y, Lombardo J, Yoon JM, Shanks JV, Klauda JB, Jarboe LR. Engineering Escherichia coli membrane phospholipid head distribution improves tolerance and production of biorenewables. Metab Eng. 2017;44:1–12. [DOI] [PubMed] [Google Scholar]
- 71.Suo Y, Luo S, Zhang Y, Liao Z, Wang J. Enhanced butyric acid tolerance and production by class I heat shock protein-overproducing Clostridium tyrobutyricum ATCC 25755. J Ind Microbiol Biotechnol. 2017;44(8):1145–1156. [DOI] [PubMed] [Google Scholar]
- 72.Abdullah Al M, Sugimoto S, Higashi C, Matsumoto S, Sonomoto K. Improvement of multiple-stress tolerance and lactic acid production in Lactococcus lactis NZ9000 under conditions of thermal stress by heterologous expression of Escherichia coli dnaK. Appl Environ Microbiol. 2010;76(13):4277–4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu C, Zhang J, Du G, Chen J. Heterologous expression of Lactobacillus casei RecO improved the multiple-stress tolerance and lactic acid production in Lactococcus lactis NZ9000 during salt stress. Bioresour Technol. 2013;143:238–241. [DOI] [PubMed] [Google Scholar]
- 74.Luo J, Song Z, Ning J, Cheng Y, Wang Y, Cui F, Shen Y, Wang M. The ethanol-induced global alteration in Arthrobacter simplex and its mutants with enhanced ethanol tolerance. Appl Microbiol Biotechnol. 2018;102:9331–9350. [DOI] [PubMed] [Google Scholar]
- 75.Yan X, Wang X, Yang Y, Wang Z, Zhang H, Li Y, He Q, Li M, Yang S. Cysteine supplementation enhanced inhibitor tolerance of Zymomonas mobilis for economic lignocellulosic bioethanol production. Bioresour Technol. 2022;349: Article 126878. [DOI] [PubMed] [Google Scholar]
- 76.Zhang J, Petersen SD, Radivojevic T, Ramirez A, Pérez-Manríquez A, Abeliuk E, Sánchez BJ, Costello Z, Chen Y, Fero MJ, et al. Combining mechanistic and machine learning models for predictive engineering and optimization of tryptophan metabolism. Nat Commun. 2020;11:4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lee SJ, Lee DY, Kim TY, Kim BH, Lee J, Lee SY. Metabolic engineering of Escherichia coli for enhanced production of succinic acid, based on genome comparison and in silico gene knockout simulation. Appl Environ Microbiol. 2005;71:7880–7887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bodor Z, Tompos L, Nechifor AC, Bodor K. In silico analysis of 1,4-butanediol heterologous pathway impact on Escherichia coli metabolism. Rev Chim. 2019;70:3448–3455. [Google Scholar]
- 79.Bro C, Regenberg B, Forster J, Nielsen J. In silico aided metabolic engineering of Saccharomyces cerevisiae for improved bioethanol production. Metab Eng. 2006;8:102–111. [DOI] [PubMed] [Google Scholar]
- 80.Fuchino K, Wasser D, Soppa J. Genome copy number quantification revealed that the ethanologenic alpha-proteobacterium Zymomonas mobilis is polyploid. Front Microbiol. 2021;12: Article 705895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang X, He Q, Yang Y, Wang J, Haning K, Hu Y, Wu B, He M, Zhang Y, Bao J, et al. Advances and prospects in metabolic engineering of Zymomonas mobilis. Metab Eng. 2018;50:57–73. [DOI] [PubMed] [Google Scholar]
- 82.Xia J, Yang Y, Liu C, Yang S, Bai F. Engineering Zymomonas mobilis for robust cellulosic ethanol production. Trends Biotechnol. 2019;37:960–972. [DOI] [PubMed] [Google Scholar]
- 83.Zheng Y, Han J, Wang B, Hu X, Li R, Shen W, Ma X, Ma L, Yi L, Yang S, et al. Characterization and repurposing of the endogenous type I-F CRISPR-Cas system of Zymomonas mobilis for genome engineering. Nucleic Acids Res. 2019;47(21):11461–11475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shen W, Zhang J, Geng B, Qiu M, Hu M, Yang Q, Bao W, Xiao Y, Zheng Y, Peng W, et al. Establishment and application of a CRISPR-Cas12a assisted genome-editing system in Zymomonas mobilis. Microb Cell Factories. 2019;18(1):162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang X, Wu B, Sui X, Zhang Z, Liu T, Li Y, Hu G, He M, Peng N. CRISPR-mediated host genomic DNA damage is efficiently repaired through microhomology-mediated end joining in Zymomonas mobilis. J Genet Genomics. 2021;48(2):115–122. [DOI] [PubMed] [Google Scholar]
- 86.Binan G, Yalun W, Xinyan W, Yongfu Y, Peng Z, Yunhaon C, Xuan Z, Chenguang L, Fengwu B, Ping X, et al. Efficient genome-editing tools to engineer the recalcitrant non-model industrial microorganism Zymomonas mobilis. Trends Biotechnol. 2024. [DOI] [PubMed] [Google Scholar]
- 87.Li Y, Wang Y, Wang R, Yan X, Wang J, Wang X, Chen S, Bai F, He Q, Yang S. Metabolic engineering of Zymomonas mobilis for continuous co-production of bioethanol and poly-3-hydroxybutyrate (PHB). Green Chem. 2022;24(6):2588–2601. [Google Scholar]
- 88.Qiu M, Shen W, Yan X, He Q, Cai D, Chen S, Wei H, Knoshaug EP, Zhang M, Himmel ME, et al. Metabolic engineering of Zymomonas mobilis for anaerobic isobutanol production. Biotechnol Biofuels. 2020;13:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vriesekoop F, Pamment NB. Acetaldehyde stimulation of the growth of Zymomonas mobilis subjected to ethanol and other environmental stresses: Effect of other metabolic electron acceptors and evidence for a mechanism. Fermentation. 2021;7(2):80. [Google Scholar]
- 90.Hu M, Bao W, Peng Q, Hu W, Yang X, Xiang Y, Yan X, Li M, Xu P, He Q, et al. Metabolic engineering of Zymomonas mobilis for co-production of D-lactic acid and ethanol using waste feedstocks of molasses and corncob residue hydrolysate. Front Bioeng Biotechnol. 2023;11:1135484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Uhlenbusch I, Sahm H, Sprenger GA. Expression of an L-alanine dehydrogenase gene in Zymomonas mobilis and excretion of L-alanine. Appl Environ Microbiol. 1991;57:1360–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang Z, Wang X, Yan X, Yi H, He S, Zhang H, Zhou X, He Q, Yang S. Metabolic engineering of an industrial bacterium Zymomonas mobilis for anaerobic l-serine production. Synth Syst Biotechnol. 2024;9(2):349–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bao W, Shen W, Peng Q, Du J, Yang S. Metabolic engineering of Zymomonas mobilis for acetoin production by carbon redistribution and cofactor balance. Fermentation. 2023;9:113. [Google Scholar]
- 94.Yang S, Mohagheghi A, Franden MA, Chou YC, Chen X, Dowe N, Himmel ME, Zhang M. Metabolic engineering of Zymomonas mobilis for 2,3-butanediol production from lignocellulosic biomass sugars. Biotechnol Biofuels. 2016;9(1):189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Khandelwal R, Srivastava P, Bisaria VS. Expression of Escherichia coli malic enzyme gene in Zymomonas mobilis for production of malic acid. J Biotechnol. 2022;351:23–29. [DOI] [PubMed] [Google Scholar]
- 96.Lee KY, Park JM, Kim TY, Yun H, Lee SY. The genome-scale metabolic network analysis of Zymomonas mobilis ZM4 explains physiological features and suggests ethanol and succinic acid production strategies. Microb Cell Factories. 2010;9:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.He Y, Wu B, Xia W, Zhao KY, Qin Y, Tan Q, Yu QH, Liu PT, Hu GQ, He MX. Metabolic engineering of Zymomonas moblis for ethylene production from straw hydrolysate. Appl Microbiol Biotechnol. 2021;105(4):1709–1720. [DOI] [PubMed] [Google Scholar]
- 98.Larroude M, Rossignol T, Nicaud J-M, Ledesma-Amaro R. Synthetic biology tools for engineering Yarrowia lipolytica. Biotechnol Adv. 2018;36(8):2150–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tao W, Lv L, Chen G-Q. Engineering Halomonas species TD01 for enhanced polyhydroxyalkanoates synthesis via CRISPRi. Microb Cell Factories. 2017;16(1):48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tan D, Wu Q, Chen J-C, Chen G-Q. Engineering Halomonas TD01 for the low-cost production of polyhydroxyalkanoates. Metab Eng. 2014;26:34–47. [DOI] [PubMed] [Google Scholar]
- 101.Kazak Sarilmiser H, Ates O, Ozdemir G, Arga KY, Toksoy OE. Effective stimulating factors for microbial levan production by Halomonas smyrnensis AAD6T. J Biosci Bioeng. 2015;119(4):455–463. [DOI] [PubMed] [Google Scholar]
- 102.Ye J, Lin Y, Yi X, Yu Z, Liu X, Chen G. Synthetic biology of extremophiles: A new wave of biomanufacturing. Trends Biotechnol. 2023;41(3):342–357. [DOI] [PubMed] [Google Scholar]
- 103.Yang J, Zhu Y, Li J, Men Y, Sun Y, Ma Y. Biosynthesis of rare ketoses through constructing a recombination pathway in an engineered Corynebacterium glutamicum. Biotechnol Bioeng. 2015;112:168–180. [DOI] [PubMed] [Google Scholar]
- 104.Yang J, Li J, Men Y, Zhu Y, Zhang Y, Sun Y, Ma Y. Biosynthesis of l-sorbose and l-psicose based on C-C bond formation catalyzed by aldolases in an engineered Corynebacterium glutamicum strain. Appl Environ Microbiol. 2015;81(13):4284–4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li J, Yang J, Men Y, Zeng Y, Zhu Y, Dong C, Sun Y, Ma Y. Biosynthesis of 2-deoxysugars using whole-cell catalyst expressing 2-deoxy-D-ribose 5-phosphate aldolase. Appl Microbiol Biotechnol. 2015;99(19):7963–7972. [DOI] [PubMed] [Google Scholar]
- 106.Ro DK, Paradise EM, Ouellet M, Fisher KJ, Newman KL, Ndungu JM, Ho KA, Eachus RA, Ham TS, Kirby J, et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440(7086):940–943. [DOI] [PubMed] [Google Scholar]
- 107.Luo X, Reiter MA, d’Espaux L, Wong J, Denby CM, Lechner A, Zhang Y, Grzybowski AT, Harth S, Lin W, et al. Complete biosynthesis of cannabinoids and their unnatural analogues in yeast. Nature. 2019;567:123–126. [DOI] [PubMed] [Google Scholar]
- 108.Yang C, Wang Y, Su Z, Xiong L, Wang P, Lei W, Yan X, Ma D, Zhao G, Zhou Z. Biosynthesis of the highly oxygenated tetracyclic core skeleton of Taxol. Nat Commun. 2024;15(1):2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jiang B, Gao L, Wang H, Sun Y, Zhang X, Ke H, Liu S, Ma P, Liao Q, Wang Y, et al. Characterization and heterologous reconstitution of Taxus biosynthetic enzymes leading to baccatin III. Science. 2024;383(6683):622–629. [DOI] [PubMed] [Google Scholar]
- 110.Wang Y, Wang H, Wei L, Li S, Liu L, Wang X. Synthetic promoter design in Escherichia coli based on a deep generative network. Nucleic Acids Res. 2020;48(12):6403–6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Reeve B, Hargest T, Gilbert C, Ellis T. Predicting translation initiation rates for designing synthetic biology. Front Bioeng Biotechnol. 2014;2:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mao F, Dam P, Chou J, Olman V, Xu Y. DOOR: A database for prokaryotic operons. Nucleic Acids Res. 2009;37(suppl_1):D459–D463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mao X, Ma Q, Zhou C, Chen X, Zhang H, Yang J, Mao F, Lai W, Xu Y. DOOR 2.0: Presenting operons and their functions through dynamic and integrated views. Nucleic Acids Res. 2014;42(D1):D654–D659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Espah Borujeni A, Cetnar D, Farasat I, Smith A, Lundgren N, Salis HM. Precise quantification of translation inhibition by mRNA structures that overlap with the ribosomal footprint in N-terminal coding sequences. Nucleic Acids Res. 2017;45(9):5437–5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Salis HM. The ribosome binding site calculator. Methods Enzymol. 2011;498:19–42. [DOI] [PubMed] [Google Scholar]
- 116.Landick R. Active-site dynamics in RNA polymerases. Cell. 2004;116(3):351–353. [DOI] [PubMed] [Google Scholar]
- 117.Studier FW, Moffatt BA. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189(1):113–130. [DOI] [PubMed] [Google Scholar]
- 118.Zhang F, Carothers JM, Keasling JD. Design of a dynamic sensor-regulator system for production of chemicals and fuels derived from fatty acids. Nat Biotechnol. 2012;30(4):354–359. [DOI] [PubMed] [Google Scholar]
- 119.Soma Y, Hanai T. Self-induced metabolic state switching by a tunable cell density sensor for microbial isopropanol production. Metab Eng. 2015;30:7–15. [DOI] [PubMed] [Google Scholar]
- 120.Liu H, Lu T. Autonomous production of 1,4-butanediol via a de novo biosynthesis pathway in engineered Escherichia coli. Metab Eng. 2015;29:135–141. [DOI] [PubMed] [Google Scholar]
- 121.Kim EM, Woo HM, Tian T, Yilmaz S, Javidpour P, Keasling JD, Lee TS. Autonomous control of metabolic state by a quorum sensing (QS)-mediated regulator for bisabolene production in engineered E. coli. Metab Eng. 2017;44:325–336. [DOI] [PubMed] [Google Scholar]
- 122.Liu L, Helal SE, Peng N. CRISPR-Cas-based engineering of probiotics. Biodes Res. 2023;5:0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31(3):233–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Li Y, Lin Z, Huang C, Zhang Y, Wang Z, Tang YJ, Chen T, Zhao X. Metabolic engineering of Escherichia coli using CRISPR-Cas9 meditated genome editing. Metab Eng. 2015;31:13–21. [DOI] [PubMed] [Google Scholar]
- 125.Wang J, Sui X, Ding Y, Fu Y, Feng X, Liu M, Zhang Y, Xian M, Zhao G. A fast and robust iterative genome-editing method based on a rock-paper-scissors strategy. Nucleic Acids Res. 2021;49(12): Article e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Stoneman HR, Wrobel RL, Place M, Graham M, Krause DJ, de Chiara M, Liti G, Schacherer J, Landick R, Gasch AP, et al. CRISpy-Pop: A web tool for designing CRISPR/Cas9-driven genetic modifications in diverse populations. G3 (Bethesda). 2020;10(11):4287–4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Dalia TN, Yoon SH, Galli E, Barre FX, Waters CM, Dalia AB. Enhancing multiplex genome editing by natural transformation (MuGENT) via inactivation of ssDNA exonucleases. Nucleic Acids Res. 2017;45(12):7527–7537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Dalia TN, Hayes CA, Stolyar S, Marx CJ, McKinlay JB, Dalia AB. Multiplex genome editing by natural transformation (MuGENT) for synthetic biology in Vibrio natriegens. ACS Synth Biol. 2017;6:1650–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hoffart E, Grenz S, Lange J, Nitschel R, Müller F, Schwentner A, Feith A, Lenfers-Lücker M, Takors R, Blombach B. High substrate uptake rates empower Vibrio natriegens as production host for industrial biotechnology. Appl Environ Microbiol. 2017;83(12): Article e01614-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kim JY, Oh JJ, Jeon MS, Kim GH, Choi YE. Improvement of Euglena gracilis paramylon production through a cocultivation strategy with the Indole-3-acetic acid-producing bacterium Vibrio natriegens. Appl Environ Microbiol. 2019;85(19): Article e01548-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Fernández-Llamosas H, Castro L, Blázquez ML, Díaz E, Carmona M. Speeding up bioproduction of selenium nanoparticles by using Vibrio natriegens as microbial factory. Sci Rep. 2017;7(1):16046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wang Z, Tschirhart T, Schultzhaus Z, Kelly EE, Chen A, Oh E, Nag O, Glaser ER, Kim E, Lloyd PF, et al. Melanin produced by the fast-growing marine bacterium Vibrio natriegens through heterologous biosynthesis: Characterization and application. Appl Environ Microbiol. 2020;86(5): Article e02749-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ellis GA, Tschirhart T, Spangler J, Walper SA, Medintz IL, Vora GJ. Exploiting the feedstock flexibility of the emergent synthetic biology chassis Vibrio natriegens for engineered natural product production. Mar Drugs. 2019;17(12):679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Becker W, Wimberger F, Zangger K. Vibrio natriegens: An alternative expression system for the high-yield production of isotopically labeled proteins. Biochemistry. 2019;58(25):2799–2803. [DOI] [PubMed] [Google Scholar]
- 135.Biggs BW, Bedore SR, Arvay E, Huang S, Subramanian H, McIntyre EA, Duscent-Maitland CV, Neidle EL, Tyo KEJ. Development of a genetic toolset for the highly engineerable and metabolically versatile Acinetobacter baylyi ADP1. Nucleic Acids Res. 2020;48(9):5169–5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Huang H, Chai C, Li N, Rowe P, Minton NP, Yang S, Jiang W, Gu Y. CRISPR/Cas9-based efficient genome editing in Clostridium ljungdahlii, an autotrophic gas-fermenting bacterium. ACS Synth Biol. 2016;5:1355–1361. [DOI] [PubMed] [Google Scholar]
- 137.Vickers CE, Blank LM, Krömer JO. Grand challenge commentary: Chassis cells for industrial biochemical production. Nat Chem Biol. 2010;6(12):875–877. [DOI] [PubMed] [Google Scholar]
- 138.Pósfai G, Plunkett G 3rd, Fehér T, Frisch D, Keil GM, Umenhoffer K, Kolisnychenko V, Stahl B, Sharma SS, Arruda M, et al. Emergent properties of reduced-genome Escherichia coli. Science. 2006;312:1044–1046. [DOI] [PubMed] [Google Scholar]
- 139.Li Y, Zhu X, Zhang X, Fu J, Wang Z, Chen T, Zhao X. Characterization of genome-reduced Bacillus subtilis strains and their application for the production of guanosine and thymidine. Microb Cell Factories. 2016;15:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zeng W, Du G, Chen J, Li J, Zhou J. A high-throughput screening procedure for enhancing α-ketoglutaric acid production in Yarrowia lipolytica by random mutagenesis. Process Biochem. 2015;50:1516–1522. [Google Scholar]
- 141.Zeng W, Guo L, Xu S, Chen J, Zhou J. High-throughput screening technology in industrial biotechnology. Trends Biotechnol. 2020;38(8):888–906. [DOI] [PubMed] [Google Scholar]
- 142.He Y, Wang X, Ma B, Xu J. Ramanome technology platform for label-free screening and sorting of microbial cell factories at single-cell resolution. Biotechnol Adv. 2019;37(6): Article 107388. [DOI] [PubMed] [Google Scholar]
- 143.Ali A, Abouleila Y, Shimizu Y, Hiyama E, Emara S, Mashaghi A, Hankemeier T. Single-cell metabolomics by mass spectrometry: Advances, challenges, and future applications. TrAC Trends Anal Chem. 2019;120: Article 115436. [Google Scholar]
- 144.Jian X, Guo X, Wang J, Tan ZL, Xing XH, Wang L, Zhang C. Microbial microdroplet culture system (MMC): An integrated platform for automated, high-throughput microbial cultivation and adaptive evolution. Biotechnol Bioeng. 2020;117(6):1724–1737. [DOI] [PubMed] [Google Scholar]
- 145.Lou J, Wang J, Yang Y, Yang Q, LI R, Hu M, He Q, du J, Wang X, Li M, et al. Development and characterization of efficient xylose utilization strains of Zymomonas mobilis. Biotechnol Biofuels. 2021;14(1):231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Meng H, Ma Y, Mai G, Wang Y, Liu C. Construction of precise support vector machine based models for predicting promoter strength. Quant Biol. 2017;5(1):90–98. [Google Scholar]
- 147.Musil M, Stourac J, Bendl J, Brezovsky J, Prokop Z, Zendulka J, Martinek T, Bednar D, Damborsky J. FireProt: Web server for automated design of thermostable proteins. Nucleic Acids Res. 2017;45(W1):W393–W399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Goldenzweig A, Goldsmith M, Hill SE, Gertman O, Laurino P, Ashani Y, Dym O, Unger T, Albeck S, Prilusky J, et al. Automated structure- and sequence-based design of proteins for high bacterial expression and stability. Mol Cell. 2018;70(2):380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Khersonsky O, Lipsh R, Avizemer Z, Ashani Y, Goldsmith M, Leader H, Dym O, Rogotner S, Trudeau DL, Prilusky J, et al. Automated design of efficient and functionally diverse enzyme repertoires. Mol Cell. 2018;72(1):178–186.e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Xiong P, Hu X, Huang B, Zhang J, Chen Q, Liu H. Increasing the efficiency and accuracy of the ABACUS protein sequence design method. Bioinformatics. 2020;36(1):136–144. [DOI] [PubMed] [Google Scholar]
- 151.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31(13):3381–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: Calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38(suppl_2):W529–W533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Moretti R, Lyskov S, Das R, Meiler J, Gray JJ. Web-accessible molecular modeling with Rosetta: The Rosetta online server that includes everyone (ROSIE). Protein Sci. 2018;27(1):259–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Shi Z, Liu P, Liao X, Mao Z, Zhang J, Wang Q, Sun J, Ma H, Ma Y. Data-driven synthetic cell factories development for industrial biomanufacturing. Biodes Res. 2022;2022: Article 9898461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596(7873):583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Abramson J, Adler J, Dunger J, Evans R, Green T, Pritzel A, Ronneberger O, Willmore L, Ballard AJ, Bambrick J, et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature. 2024;630:493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lee SY, Park JH, Jang SH, Nielsen LK, Kim J, Jung KS. Fermentative butanol production by Clostridia. Biotechnol Bioeng. 2008;101(2):209–228. [DOI] [PubMed] [Google Scholar]
- 158.Nakamura CE, Whited GM. Metabolic engineering for the microbial production of 1,3-propanediol. Curr Opin Biotechnol. 2003;14(5):454–459. [DOI] [PubMed] [Google Scholar]
- 159.Hasegawa S, Suda M, Uematsu K, Natsuma Y, Hiraga K, Jojima T, Inui M, Yukawa H. Engineering of Corynebacterium glutamicum for high-yield L-valine production under oxygen deprivation conditions. Appl Environ Microbiol. 2013;79:1250–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wang Z, Chen T, Ma X, Shen Z, Zhao X. Enhancement of riboflavin production with Bacillus subtilis by expression and site-directed mutagenesis of zwf and gnd gene from Corynebacterium glutamicum. Bioresour Technol. 2011;102(4):3934–3940. [DOI] [PubMed] [Google Scholar]
- 161.Zhan Y, Shi J, Xiao Y, Zhou F, Wang H, Xu H, Li Z, Yang S, Cai D, Chen S. Multilevel metabolic engineering of Bacillus licheniformis for de novo biosynthesis of 2-phenylethanol. Metab Eng. 2022;70:43–54. [DOI] [PubMed] [Google Scholar]
- 162.Zhang Y, Peng J, Zhao H, Shi S. Engineering oleaginous yeast Rhodotorula toruloides for overproduction of fatty acid ethyl esters. Biotechnol Biofuels. 2021;14(1):115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hu S, You Y, Xia F, Liu J, Dai W, Liu J, Wang Y. Genome shuffling improved acid-tolerance and succinic acid production of Actinobacillus succinogenes. Food Sci Biotechnol. 2019;28:817–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Luna-Flores CH, Palfreyman RW, Krömer JO, Nielsen LK, Marcellin E. Improved production of propionic acid using genome shuffling. Biotechnol J. 2017;12(2):1600120. [DOI] [PubMed] [Google Scholar]
- 165.Chen L, Chong XY, Zhang YY, Lv YY, Hu YS. Genome shuffling of bacillus velezensis for enhanced surfactin production and variation analysis. Curr Microbiol. 2020;77(1):71–78. [DOI] [PubMed] [Google Scholar]
- 166.Tong QQ, Zhou YH, Chen XS, Wu JY, Wei P, Yuan LX, Yao JM. Genome shuffling and ribosome engineering of Streptomyces virginiae for improved virginiamycin production. Bioprocess Biosyst Eng. 2018;41(5):729–738. [DOI] [PubMed] [Google Scholar]
- 167.Sun J, Li H, Ni Y, Zhang X, Shi J, Xu Z. Genome shuffling of colletotrichum lini for improving 3β,7α,15α-trihydroxy-5-androsten-17-one production from dehydroepiandrosterone. J Ind Microbiol Biotechnol. 2017;44(6):937–947. [DOI] [PubMed] [Google Scholar]
- 168.Li J, Guo S, Hua Q, Hu F. Improved AP-3 production through combined ARTP mutagenesis, fermentation optimization, and subsequent genome shuffling. Biotechnol Lett. 2021;43:1143–1154. [DOI] [PubMed] [Google Scholar]
- 169.Su B, Wu M, Zhang Z, Lin J, Yang L. Efficient production of xylitol from hemicellulosic hydrolysate using engineered Escherichia coli. Metab Eng. 2015;31:112–122. [DOI] [PubMed] [Google Scholar]
- 170.Ye Z, Shi B, Huang Y, Ma T, Xiang Z, Hu B, Kuang Z, Huang M, Lin X, Tian Z, et al. Revolution of vitamin E production by starting from microbial fermented farnesene to isophytol. Innovations. 2022;3(3): Article 100228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Sheng Q, Wu X, Xu X, Tan X, Li Z, Zhang B. Production of l-glutamate family amino acids in Corynebacterium glutamicum: Physiological mechanism, genetic modulation, and prospects. Synth Syst Biotechnol. 2021;6(4):302–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wang Y, Hu L, Huang H, Wang H, Zhang T, Chen J, du G, Kang Z. Eliminating the capsule-like layer to promote glucose uptake for hyaluronan production by engineered Corynebacterium glutamicum. Nat Commun. 2020;11(1):3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Guiziou S, Sauveplane V, Chang HJ, Clerté C, Declerck N, Jules M, Bonnet J. A part toolbox to tune genetic expression in Bacillus subtilis. Nucleic Acids Res. 2016;44(15):7495–7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Yu W, Jin K, Wu Y, Zhang Q, Liu Y, Li J, du G, Chen J, Lv X, Ledesma-Amaro R, et al. A pathway independent multi-modular ordered control system based on thermosensors and CRISPRi improves bioproduction in bacillus subtilis. Nucleic Acids Res. 2022;44(15):7495–7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Hao T, Han B, Ma H, Fu J, Wang H, Wang Z, Tang B, Chen T, Zhao X. In silico metabolic engineering of Bacillus subtilis for improved production of riboflavin, Egl-237, (R,R)-2,3-butanediol and isobutanol. Mol BioSyst. 2013;9(8):2034–2044. [DOI] [PubMed] [Google Scholar]
- 176.Tsigie YA, Wang CY, Truong CT, Ju YH. Lipid production from Yarrowia lipolytica Po1g grown in sugarcane bagasse hydrolysate. Bioresour Technol. 2011;102(19):9216–9222. [DOI] [PubMed] [Google Scholar]
- 177.Park YK, Nicaud JM, Ledesma-Amaro R. The engineering potential of Rhodosporidium toruloides as a workhorse for biotechnological applications. Trends Biotechnol. 2018;36:304–317. [DOI] [PubMed] [Google Scholar]
- 178.Otoupal PB, Ito M, Arkin AP, Magnuson JK, Gladden JM, Skerker JM. Multiplexed CRISPR-Cas9-based genome editing of Rhodosporidium toruloides. mSphere. 2019;4(2): Article e00099-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Wen Z, Zhang S, Odoh CK, Jin M, Zhao ZK. Rhodosporidium toruloides—A potential red yeast chassis for lipids and beyond. FEMS Yeast Res. 2020;20(5): Article foaa038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Kobayashi Y, Inokuma K, Matsuda M, Kondo A, Hasunuma T. Resveratrol production from several types of saccharide sources by a recombinant Scheffersomyces stipitis strain. Metab Eng Commun. 2021;13: Article e00188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Mastella L, Senatore V, Beltrani T, Branduardi P. Scheffersomyces stipitis ability to valorize different residual biomasses for vitamin B9 production. Microb Biotechnol. 2023;16(2):392–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Bumrungtham P, Promdonkoy P, Prabmark K, Bunterngsook B, Boonyapakron K, Tanapongpipat S, Champreda V, Runguphan W. Engineered production of isobutanol from sugarcane trash hydrolysates in Pichia pastoris. J Fungi. 2022;8(8):767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Bourgade B, Humphreys CM, Millard J, Minton NP, Islam MA. Design, analysis, and implementation of a novel biochemical pathway for ethylene glycol production in Clostridium autoethanogenum. ACS Synth Biol. 2022;11(5):1790–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Xu M, Zhao J, Yu L, Tang IC, Xue C, Yang ST. Engineering Clostridium acetobutylicum with a histidine kinase knockout for enhanced n-butanol tolerance and production. Appl Microbiol Biotechnol. 2015;99(5):1011–1022. [DOI] [PubMed] [Google Scholar]
- 185.Fu H, Yang ST, Wang M, Wang J, Tang IC. Butyric acid production from lignocellulosic biomass hydrolysates by engineered Clostridium tyrobutyricum overexpressing xylose catabolism genes for glucose and xylose co-utilization. Bioresour Technol. 2017;234:389–396. [DOI] [PubMed] [Google Scholar]
- 186.Du H, Zhao Y, Wu F, Ouyang P, Chen J, Jiang X, Ye J, Chen GQ. Engineering Halomonas bluephagenesis for L-threonine production. Metab Eng. 2020;60:119–127. [DOI] [PubMed] [Google Scholar]
- 187.Hoff J, Daniel B, Stukenberg D, Thuronyi BW, Waldminghaus T, Fritz G. Vibrio natriegens: An ultrafast-growing marine bacterium as emerging synthetic biology chassis. Environ Microbiol. 2020;22(10):4394–4408. [DOI] [PubMed] [Google Scholar]


