Abstract
Digital twins in biomedical research, i.e. virtual replicas of biological entities such as cells, organs, or entire organisms, hold great potential to advance personalized healthcare. As all biological processes happen in space, there is a growing interest in modeling biological entities within their native context. Leveraging generative artificial intelligence (AI) and high-volume biomedical data profiled with spatial technologies, researchers can recreate spatially-resolved digital representations of a physical entity with high fidelity. In application to biomedical fields such as computational pathology, oncology, and cardiology, these generative digital twins (GDT) thus enable compelling in silico modeling for simulated interventions, facilitating the exploration of ‘what if’ causal scenarios for clinical diagnostics and treatments tailored to individual patients. Here, we outline recent advancements in this novel field and discuss the challenges and future research directions.
Keywords: Generative AI, Spatial omics, Multiplexed imaging, Digital twin
1. Introduction
Biomedical digital twins (DT), which commonly refer to high-fidelity digital representations of physical entities [38], [16], are designed to accurately mirror the biological, physiological, and pathological characteristics of the organisms they emulate. Over the past decade, substantial efforts have been dedicated to developing DTs [38] for biomedical applications. The progress of modeling comprehensive DTs has been continuously fueled and reshaped by the growing demand for personalized healthcare [34].
By analyzing detailed biomedical data—such as a patient's unique genetic profile, environmental factors, and lifestyle influences through methods of machine learning and artificial intelligence (AI), it becomes possible to accurately generate DTs with potential future clinical utility [12]. Personalized simulations that can predict disease progression [3], optimize treatment plans [14], and enhance patient outcomes [36] by tailoring interventions to the individual's specific biological and environmental context have significant potential in data-driven medicine. Further, these digital replicas allow for continuous monitoring and adjustment, ensuring that healthcare strategies remain adaptive and responsive to real-time changes in a patient's health status [70].
For biomarker and drug discovery [44], [5], [4], DTs can be created using comprehensive biomedical datasets to simulate drug effects and predict treatment efficacy. Future use of DTs and virtual patient cohorts could offer a possible pre-stage to randomized clinical trials (RCT), helping to reduce costs and increasing efficacy. In the field of medical diagnostics, DTs can enable the seamless integration of imaging data with genetic and molecular profiles to form virtual representations of disease states with great utility for medical research. Based on DT simulation, clinicians could potentially make more informed decisions regarding surgical planning, and targeted therapy selection, thereby improving the accuracy and effectiveness of personalized diagnostics [62]. As most biological processes occur within native spatial contexts (e.g., the tissue microenvironment, TME), the development of spatially resolved analytical and simulation techniques is a crucial advancement for understanding the complex interplay between cellular, molecular, and environmental factors. These techniques enable precise mapping and modeling of biological events in their native architecture, offering insights into tissue-specific responses, disease progression, and therapeutic interventions, thereby enhancing our ability to design targeted treatments.
This mini-review starts with a concise overview of pertinent approaches from the converging domains of generative AI and spatial technologies. By incorporating spatially profiled molecular and bioimage data into training generative AI models, we discuss existing methods that illustrate novel paths towards the construction of GDTs and describe future opportunities and challenges in the field.
1.1. The convergence of generative AI and spatial technologies
Generative AI, a novel technology that was initially developed for natural visual content creation [10], presents a promising analog for spatially resolved bioimage generation. Generative models demonstrate impressive proficiency in generating high-quality visual content [52], [42] through parallel GPU training. For instance, generative models have been employed in image generation for the study of rare diseases [1], graph learning, and molecular design [10]. These approaches can capture normal or disease phenotypes across various scales, from the cellular level to tissues, organs, and entire organisms. By leveraging these capabilities, generative AI has the potential to revolutionize how we visualize and understand complex biological structures and processes, ultimately enhancing our ability to diagnose and treat diseases more effectively.
The development of generative AI is paralleled by the evolution of highly resolved in situ methods for the co-profiling of morphological and molecular features of diagnostic tissue samples using cutting-edge spatial technologies in biomedical research [9]. The high-throughput quantification of spatial molecular data encompassing epigenomics, transcriptomics, and proteomics, alongside matched high-resolution bioimages stained with Hematoxylin and Eosin (H&E), (immuno)fluorescence (IF), imaging mass cytometry (IMC) etc. is not only poised to stimulate innovative biomedical discoveries, but offers critical proxies and paired training data in modeling the morpho-molecular causal associations.
1.2. Generative digital twins (GDT)
Similar to how text and image data pairs are utilized to train generative AI models, matched molecular and bioimage data can be effectively employed to train such models. In this mini-review, we introduce a novel concept termed Generative Digital Twins (GDT), i.e., generated (spatial) biological organisms by generative AI models trained with paired molecular and bioimage data. Profiling and intervening on input molecular data, these generative AI models enable in-silico biomedical investigations on causal “what if” scenarios of GDTs, in terms of visualizing and quantifying morphological transitions driven by spatial molecular changes. Such extensive causal investigations would otherwise be non-trivial or unethical to carry out using animal modeling. Given the virtual characteristics of GDTs, we can further circumvent the legal, ethical, and regulatory risks for implementing high-throughput biological interventions, which may also help reduce the need for interventions on laboratory animals according to the 3R principle (Refine, Reduce, Replace).
2. Generative AI in vision
In the field of computer vision, Generative Adversarial Nets (GAN) [21] and diffusion-based models [28] are recognized as leading methodologies among various generative models. Characterized by the adversarial training framework where a generator and discriminator engage in an iterative process, GANs have proven to be effective and efficient for image synthesis. As competitive alternatives, diffusion-based models represent a subset of probabilistic generative models that introduce noise to data and then learn to reverse this process for sample generation. In recent studies, these models have demonstrated superior performance in various generation tasks and thus challenged the dominance of GANS. By feeding text prompts to GAN- or diffusion-based models [32], [52], researchers can create realistic visual data for a wide range of real-world use cases, accommodating both unconditional and input-conditional generation.
In clinical diagnostics, generative AI has also shown exciting applications for algorithmic modeling of disease states [1]. By efficiently generating numerous (rare) disease images or virtual staining of high-plex biomarkers, they have the potential to substantially impact biomedical research. However, directly applying existing methods to high-resolution biomedical data (e.g., whole slice images (WSI) in digital pathology) remains challenging due to an exponential increase in computational complexity. For instance, 96-128 A100 GPUs were used to train a large GigaGAN [21] to synthesize natural images at a megapixel resolution. At the present time, the hardware requirements for generating WSIs at a gigapixel resolution can be computationally prohibitive, rendering the use of such models costly in biomedical research. To resolve the memory bottleneck inherent in generating high-resolution images, researchers frequently use patch-based generative models as a cost-effective and scalable solution. After training with a large amount of cropped patch-wise data, the methods summarized below can generate (arbitrarily) large images at inference, successfully circumventing the hardware constraints.
2.1. Patch-based GAN models
The synthesis of high-resolution images by stitching together small generated image patches is a longstanding technique in computer vision and graphics [19]. Within the GAN framework, Single-image GAN (SinGAN [55]) was first introduced to learn internal statistics from randomly cropped patches of a single natural image. Given its pyramid convolutional architecture, SinGAN has unlocked the generation of new images with an arbitrary size and aspect ratio, while maintaining the global and local structures of the training image. Motivated to solve the computational constraints, Lin et al. proposed conditional coordinate GAN (COCO-GAN [40]) to generate complete natural images. Trained with cropped and incomplete image patches, COCO-GAN employed the spatial coordinate as condition and can produce state-of-the-art full images during inference. To extend image generation beyond the boundary and resolution of learned images, InfinityGAN [41] decomposed global appearances into local structures and textures, both of which are instantiated using a neural implicit function, denoted structure synthesizer, and a padding-free generator, termed texture synthesizer. At inference time, InfinityGAN achieves arbitrary-sized image generation without stitching artifacts. Following the seminal StyleGAN framework [33], the ‘aligning latent and image spaces’ method (ALIS [57]) improved the infinite high-resolution images with diverse and complex content that smoothly transition from one to another. Similar to coordinate-conditioned studies, this was made possible using synchronous interpolations and coordinate-aware latent codes. Subsequently, Any-resolution GAN (Anyres-GAN [13]) aimed to synthesize patches at continuous scales to match the distribution of real patches. Trained on multi-resolution image datasets, the unconditional generator learns to generate image patches at continuous scales to match the distribution of real patches. At test time, Anyres-GAN can generate images at high resolutions with the desired sampling rate.
2.2. Patch-based diffusion models
Derived from the denoising diffusion model (DDM), the single image DDM (SinDDM) [35], acts as the successor of SinGAN, aimed to match the single image distribution through the training of its randomly cropped patches. Utilizing a fully convolutional denoiser with residual connections, SinDDM is capable of generating image samples of any desired dimensions that closely resemble the training image. To reduce the training time and improve data efficiency, Patch diffusion [61] encoded the trained patch location into the coordinate channel, while patch sizes are randomized to encode the cross-region dependency across multiple scales. Such modifications resulted in the competitive generation quality in terms of outstanding Fréchet inception score (FID), along with improved training efficiency. To reduce the computational cost of diffusion models, Patchdiffusion [43] suggested to simply reshaping the image into non-overlapping patches, however this patch-based model cannot guarantee the seamless stitching of neighboring patch images to complete a high-resolution image generation. More recently, another patch diffusion model (Path-DM) [18] implemented a novel collage strategy of the learned representations, yielding high-resolution generation results with reduced memory footprints. In this approach, smooth transitions between neighboring image tiles are imposed through the generation from concatenated latent representations.
Similar to standard generative models, these patch-based approaches can effectively generate desired images conditioned on textual input representations. Nevertheless, it remains non-trivial to apply the aforementioned methods for creating GDTs. For example, though it is theoretically feasible for SinGAN and SinDDM to generate images with any desired resolution, studies [64] suggest that the generation quality deteriorated drastically at the scale factor of 4. As shown in Table 1 that summarizes the existing patch-based models, many of which are additionally required to be conditioned on spatial coordinates. Such a design choice is not well-suited for biomedical applications, because the arrangement of biological structures is not governed by a rigid coordinate system. Instead, it is shaped by the complex interplay of genetic, epigenetic, gene expression and protein expression variability, which collectively determine the phenotype of a biological entity [23]. Therefore, seamlessly integrating these advancements into the construction of GDTs remains an open area that requires further exploration.
Table 1.
Summary table of patch-based and biomedical generative models. Top: The patch-based generative models built upon the GAN methodology; Middle: The patch-based generative models built upon DDM methodology; Bottom; Miscellaneous generative models for biomedical applications.
| GAN-based model | Backbone | Patch resolution | Conditional input | Patch → large image generation | Summary |
|---|---|---|---|---|---|
| SinGAN [55] | Conv2d blocks | coarse-to-fine | N/A | Input scaling | Adversarial training with a single image. |
| Coco-GAN [40] | Residual blocks | 64 × 64 | Coordinate | Feature merging | Synthesize large images by small patches conditioned on local coordinates and latent vectors. |
| ALIS [57] | StyleGAN2 | 256 × 256 | Coordinate | Latent code interpolation | Interpolation between the learned latent codes with regard to a 2d coordinate system. |
| InfiniteGAN [41] | StyleGAN2 | 101 × 101 | Coordinate | Padding-free convolution | Introducing a structure synthesizer and padding-free generator. |
| Anyres-GAN [13] | StyleGAN3 | 64 × 64 | Coordinate | Increasing coordinate sampling rate | Two-stage training paradigm that learns the global information and then learns the patch details. |
| Diffusion-based model | |||||
| SinDDM [35] | Residual blocks | coarse-to-fine | learned text codes | Input scaling | Similar to SingGAN, the application of single-image training paradigm. |
| Patch diffusion [61] | UNet | random | learned text codes | N/A | Reduce the training memory footprint via patch-wise operations and position embedding concatenation. |
| Patchdiffusion [43] | UNet | 64 × 64 | learned text codes | N/A | Reshaping the image into non-overlapping grid patches. |
| Path-dm [18] | UNet | 64 × 64 | learned text codes and coordinate | Feature collage | Performing the latent representation collage for both the training and inference. |
| Biomedical generative model | |||||
| Phenexplain [37] | StyleGAN2 | 128 × 128,256 × 256 | Drug perturbation label | N/A | Condition on drug perturbation with different concentration levels. |
| GILEA [66] | StyleGAN2 | 64 × 64 | N/A | N/A | Perturbation on real constructed cells by GAN inversion. |
| Grid-shift diffusion [24] | UNet | coarse-to-fine | N/A | Grid-shift sampling | Synthesis of WSIs by grid-shift technique. |
| RNA-cdm [11] | UNet | 256 × 256 | 1-D mRNA vector | N/A | Synthesis of WSI tiles using bulk RNA-sequence. |
| SST-editing [67] | StyleGAN2 | 128 × 128 | 1-D mRNA vector | N/A | Reconstruction of cellular morphological images using sub-cellular gene expression patterns. |
| IST-editing [64] | StyleGAN2 | 133 × 133 | 3-D mRNA array | Padding-free convolution and overlapped spatial mRNA array | Reconstruction of the gigapixel mouse pup using sub-cellular gene expression patterns. |
3. Spatial technologies
Emerging spatial technologies, which profile molecular data in space, are driving groundbreaking biomedical discoveries [39]. The rapid development and application of these technologies have been extensively reviewed in recent literature [39], [9], [45], underscoring their transformative impact across various domains of biological and medical sciences. Leveraging spatial omics technologies [49], [25], [30], [68], alongside multiplexed bioimaging including (immuno)fluorescence (IF) [6], [51] and imaging mass cytometry (IMC) [29], [48], researchers can simultaneously co-profile tens of thousands of data points on gene and protein expression in a spatial context. Spatial co-profiling enabled the unprecedented analysis of molecular landscapes within tissue samples, providing novel insights into the spatial organization and functional interactions of cellular components. Furthermore, the availability of fully integrated datasets holds great potential to uncover novel causal associations linking molecular-guided changes to morphology at the scale of cell, tissue, organ, and an entire organism [64].
3.1. Spatial omics
One of the initial attempts at achieving spatial molecular profiling was based on sequencing methods [54], [59]. Solutions such as the Visium [49] and Stereo-seq [15] platforms subsequently emerged as widely-used alternatives enabling the in-situ profiling of RNA expression through the adaptation of next-generation sequencing (NGS) technologies. To achieve the seamless generation of GDTs using spatial molecular data, sequencing-based methods are nonetheless sub-optimal, primarily due to their limited spot-wise spatial resolution and the lack of molecular readouts in gaps between adjacent spots.
In contrast, in situ hybridization (ISH)-based approaches have demonstrated strong capabilities in detecting high-plexed gene expression at sub-cellular resolution, exemplified by newly released platforms such as CosMx [25], Xenium [30] and MERFISH [68] (Please see also Table 2 for reference). Specifically, a recent report [26] has showcased the successful in situ characterization of up to 6000 transcriptomes using CosMx technology, while maintaining the sub-cellular resolution for individual mRNA readouts. This further substantiates the potential of ISH-based technologies in profiling high-quality molecular data for training path-based generative models.
Table 2.
The overview of widely-used spatial technologies.
| Methods | Company | Imaging/Sequencing technology | Resolution | N_readouts | (Processed) Representation |
|---|---|---|---|---|---|
| Imaging mass cytometry (IMC) [2] | Standard BioTools | Multiplexed mass spectrometry (MMS) | Sub-cellular | <50 protein markers | learned latent representation |
| Multiplexed ion beam imaging (MIBI) [48] | Ionpath | MMS | Sub-cellular | <50 protein markers | learned latent representation |
| Co-detection by indexing (CODEX) [6] | Akoya Biosciences | Cyclic immunofluorescence (IF) | Diffraction-limited | Up to 60 protein markers | learned latent representation |
| COMET [51] | Lunaphore | Cyclic IF | Diffraction-limited | Up to 40 protein markers | learned latent representation |
| GeoMX DSP [27] | Nanostring | Light-based dissection | <10 microns | Whole transcriptome and >570 proteins | Imputed readouts between the spot gap |
| Visium [49] | 10x | Spatial barcoding | 55 microns in diameter | Whole transcriptome | Imputed readouts between the spot gap |
| Visum HD [46] | 10x | Spatial barcoding | 2 × 2 microns in square | Whole transcriptome | Processed readouts through bin aggregation |
| Stereo-seq [15] | BGI | Spatial barcoding | 220 nm in diameter | Whole transcriptome | Processed readouts through bin aggregation |
| CosMx [25] | Nanostring | Multiplexed in situ hybridization (ISH) | Sub-cellular | Up to 6000 RNAs and 64 proteins | Raw readouts |
| Xenium [30] | 10x | Multiplexed ISH | Sub-cellular | Up to 5000 RNAs | Raw readouts |
| MERFISH [68] | Vizgen | Multiplexed ISH | Sub-cellular | Up to 1000 RNAs | Raw readouts |
3.2. Multiplexed imaging
At the same time, multiplexed bioimaging has become increasingly available and standardized, enabling the application to clinical samples at scale. For instance, imaging mass cytometry (IMC) [2], [58], which simultaneously profiles up to 50 protein biomarkers at sub-cellular resolution in human or animal tissues, demonstrates potential in understanding cellular identity, function and spatial distribution patterns at scale. Similar applications are also achievable by IF-based methods such as e.g., Codex [6], Vectra Multispectral Imaging [20] and COMET [51]. Given the critical role of the mRNA-to-protein translation process in spatial biology, there is considerable potential for technological synergies between spatially resolved sequencing approaches (DNA, RNA) and multiplexed imaging or mass spectrometry solutions (protein) in the future development of GDTs. Integrating these technologies could deliver new insights into the spatial analysis of mRNA-to-protein translation occurring in biological systems.
3.3. Spatial training data for generative AI
Derived from the data processing pipeline used for single-cell sequencing, a prevalent approach in handling spatial transcriptomic (ST) data is the summarized gene-versus-cell table [47]. Concretely, the spatial gene-level readouts (the number of RNA transcripts detectable for a given gene) are often reduced to a one-dimensional column associated with individual cells. Since the precise spatial coordinates are excluded from the summarized gene table, this data formality does not fully reflect the spatial localization of gene expression patterns and is not ideal for establishing a high-resolution GDT. Instead, a multi-channel gene expression ‘image’, which preserves the exact spatial organization of transcripts, is more favorable to seamlessly construct a large-scale virtual organism. To improve the data availability as well as reduce the computational cost, it is necessary to collect patch-wise image data with low pixel resolutions from the whole tissue sections for training generative models.
Analogous to the text-image data pairs used for training generative AI models, we take spatial molecular readouts (arrays) as the input and a ‘prompt’. These molecular prompts are then utilized to construct biologically plausible high-resolution representations of physical entities, i.e., GDTs. For effective modeling of morpho-molecular causal associations, the combination of ST data and optionally prepared (H&E-stained) WSI from the same tissue section becomes essential to facilitate the generative process. In exploring the mRNA-to-protein causal linkage, we can approach these biomedical questions using synergistic spatial ST data and multiplexed bioimages. By feeding edited gene expression patterns to the constructed GDT, we can simulate and assess multi-scale phenotypic transitions within the virtual system.
4. Towards GDT of an entire organism
Given the costly and rapidly evolving nature of spatial technologies, not until recently did we gain access to large-scale standardized datasets including spatial molecular data [31]. For those pioneer methods that did not train on spatially-resolved molecular data, we nevertheless discuss their insights and contributions relevant to this novel research field (see also Table 1).
To reveal invisible and subtle phenotypic effects induced by genetic, chemical, or disease-based perturbations, Lamiable et al. [37] suggested new variants of conditional-GAN (Phenexplain) to traverse the latent representations between two trained conditions. Depending on perturbation type and drug concentration level, such latent traversals enable the simulation of smooth transitions from untreated to treated cells that can be applied to a wide variety of drug candidates for high-throughput in silico screening. Using multiplexed cell datasets, Phenexplain has shown the effective generation of otherwise invisible cell phenotypes, where the quantitative differences between real and generated phenotypes can be measured by well-established statistical tests.
Subsequently, Wu and Koelzer introduced GAN Inversion-enabled Latent Eigenvalue Analysis (GILEA) for cell-level phenome profiling and editing [66]. Trained on large-scale SARS-CoV-2 datasets RxRx19 [17] stained with the multiplexed fluorescence Cell Painting protocol [8], this study developed a two-stage GAN Inversion model to reconstruct digital replicas of a comprehensive in vitro cell culture dataset including stimulation conditions with over 1800 drug compounds at up to 8 different concentrations. By quantifying the sorted eigenvalue difference of latent representations used in the reconstruction (a simple alternative to the FID score [65]), GILEA measures drug efficacy in restoring the phenoprint of SARS-CoV-2 infected cells to the normal condition. More importantly, this study demonstrated that latent representations can be indirectly modulated through their largest principal components, leading to plausible phenotypic transitions that provide biomedical interpretations of drug effect identification. While these studies investigated the generation of cell culture images, the adaptation of these findings to the tissue, organ, and organism system requires further experimental evidence.
Meanwhile, the generation of virtual whole slide images (WSI) at a gigapixel scale has been explored under the DDM framework (Grid-shift diffusion [24]). This approach employs a coarse-to-fine sampling scheme to synthesize high-resolution WSIs. To address and mitigate stitching artifacts, Grid-shift diffusion introduces a computationally efficient grid-shift technique applied at each diffusion step. This technique ensures that boundary pixels between adjacent patches are seamlessly transferred, resulting in a more coherent image with reduced artifacts. However, due to limitations in the available datasets, these methods are unable to incorporate spatial molecular data into the generative process, which limits their potential for molecularly guided bioimage generation.
This issue has been partially addressed in the study [11] in experiments on well-curated TCGA datasets. Trained on pairwise image tiles extracted from WSIs and associated (bulk)RNA-sequencing data, Carrillo-Perez et al. utilized RNA-guided cascade diffusion models (RNA-cdm [11]) to synthesize digital tumor samples. Though the proposed RNA-cdm can alleviate the issue of biomedical data scarcity and facilitate data augmentation for downstream prediction tasks, the bulk RNA expression profile is unlikely to fully capture the spatial variability embedded in gigapixel cancer WSIs. Therefore, it becomes critical to generate local ground truth of gene expression readouts to address the heterogeneity present in cancer tissue.
Leveraging advanced spatial transcriptomic profiling at a sub-cellular resolution, In Silico Spatial Transcriptomic editing (SST-editing [67]) was introduced to generate cellular digital twins stained with IF biomarkers such as DAPI and B2M. Given a center-cropped tumor or normal liver cell, SST-editing enables the faithful reconstruction of a cell phenotype when taking its spatial gene expression table as the input. The reconstruction accuracy can be quantitatively assessed using well-established Peak Signal-to-Noise Ratio (PSNR) and Structural Similarity Index Measure (SSIM). By modifying the sample covariance matrix of tumor cell collections to resemble that of normal cells (or vice versa), SST-editing can reverse the tumor cellular morphology to the normal one, demonstrating the in silico editability of the proposed method. Importantly, this study was conducted at the cellular level, and the spatial resolution of generated digital cells is less than pixels. This highlights the need for innovative approaches that can be scaled up to encompass tissue, organ, and even whole-organism levels.
To extend model applicability to large-sized organisms, Wu et al. [63] developed Infinite Spatial Transcriptomic editing (IST-editing) to reconstruct a gigapixel digital twin of an entire mouse pup. This was accomplished by utilizing padding-free StyleConv layers and incorporating overlapping gene expression images between adjacent tiles, which guarantees the seamless generation of high-resolution digital representations. Further, IST-editing showcased biologically plausible interventions through multi-scale gene expression editing on mammalian organs, such the colon and liver regions. Last but not least, initial experiments on the 3D mouse brain atlas [69], which profiles a comprehensive 500-plexed gene profile and cell morphology across the entire mouse brain, provided proof-of-concept for constructing a digital twin of a whole mouse brain in 3D space, as shown in Fig. 1. These advancements thus pave a promising path towards developing GDTs of an entire organism.
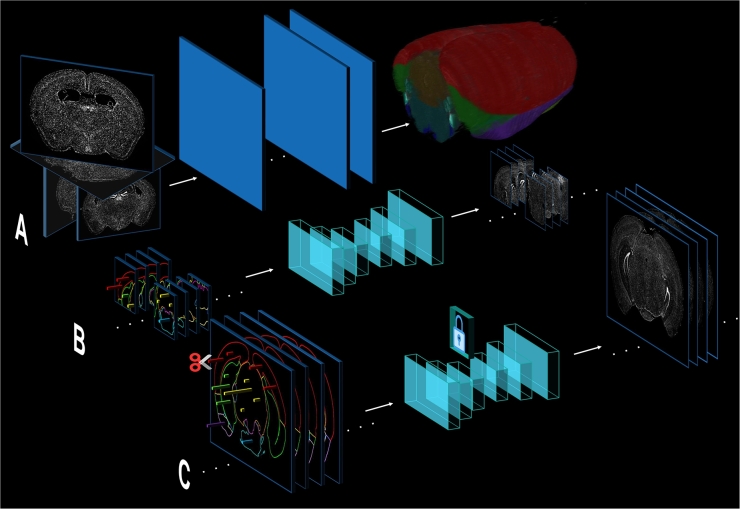
Fig. 1.
Conceptual illustration of the algorithmic paradigm for constructing a generative digital twin (GDT). A. The image registration of consecutive slide sections profiled for an entire organism. Here, we illustrate the example of registering coronal sections of the mouse brain atlas. B. The training of generative AI models using patch-wise paired data, which consists of spatial gene expression arrays and associated cellular morphological bioimages. C. The construction of the entire digital brain atlas using spatial gene expression as the input. On top of that, selected molecular-level interventions can be performed and drive associated morphological transitions.
5. Outlook and future directions
Developing robust and intervenable GDTs with low ethical risks presents a promising and inexpensive alternative for testing disease therapeutics. This methodology extrapolates previously unseen physiological and pathological states of the generated virtual entity, empowering researchers to conduct counterfactual predictions on the whole biological system. Importantly, this once science-fiction vision is becoming increasingly attainable thanks to the advancement and convergence of patch-based generative AI and spatial technologies. Researchers now have access to highly detailed spatial omics data, e.g., the mouse brain atlas [69]. Utilizing patch-based models trained on paired gene expression and bioimage data cropped from the atlas, it is then feasible to reconstruct a 3D digital mouse brain with affordable hardware budgets. Built upon attention blocks [60], this interaction-through-construction methodology provides a powerful framework for deciphering complex gene-gene interactions within stratified anatomical structures and neural layers. In addition, we gain valuable insights into the causal association between spatial genetic organization and cellular morphology, through the lens of quantifiable neuron phenotypic transitions driven by gene expression-guided editing. As promising as the long-term vision for GDTs may appear, several challenges should be tackled on the way to deliver a reliable in silico system.
5.1. Data storage and processing at unprecedented scales
Trained on cropped biomedical data, patch-based AI approaches indeed resolve the computational bottlenecks within the highly parallel GPU training paradigm. However, this improvement in computational efficiency does not mitigate the storage and processing challenges associated with raw omic data from which patch-wise training samples are extracted. This is simply because of the large-scale and high-resolution nature of the biomedical data with up to a gigapixel resolution [64]. If we extend the organism profiling to the 3D space, by preparing consecutive slices of tissue sections, the resulting raw data resolution can be up to petavoxel, as exemplified by both mouse brain atlas [69] and the fragment of human cerebral cortex studies [56]. At unprecedented scales of data volume, selecting the appropriate data format—such as tables, arrays, or tensors—as well as the choice of storage hardware and its placement within the computational cluster, becomes crucial for enhancing workflow efficiency.
Another distinctive challenge is the high-sparsity and high-plex nature of gene expression readouts. While existing tools can handle sparse data with a limited number of channels, they will immediately exhaust CPU and GPU memory resources when processing spatial gene expression arrays with up to 20000 channels. This limitation underscores the need for developing novel, open-source sparse matrix tools designed to efficiently manage and process such molecular data.
5.2. Aligning in silico discoveries to real biological processes
Complementary to in vivo experiments, generative modeling can provide insightful predictions about how complex biological components interact through the construction of GDTs and unlock the extrapolation capability of virtual biological systems among novel and unseen experimental conditions. Additionally, the ability to simulate interventions [67], [64] on specific gene groups or well-established pathways within GDTs represents an important advantage of algorithmic approaches. Such simulations provide a powerful tool for understanding and manipulating biological processes in ways that may be ethically or legally prohibitive in traditional in vivo settings.
Reliable biological simulation. Nonetheless, special care must be taken when interpreting results derived from GDTs. Despite significant advances in multi-scale spatial profiling technologies, how to fully model a biological system with all its necessary functional and organizational details remains unclear [50]. As discussed in the previous section, the number of spatially resolved protein readouts in many studies is still limited to less than 100, which significantly lags behind the amount of over 100'000 proteins [53] that perform functions in the human body. Second, many biological processes and interactions remain unknown or only partially characterized, leading to gaps in our domain knowledge.
Trustworthy hypothesis validation. The complexity of spatial interpretations generated by computational modeling and especially any biological conclusions drawn from this simulated data require thorough external validation. To ensure reliability and biological relevance, in silico findings must be carefully compared against empirical data. Further, new ways of measuring the accuracy of generated outputs may be required for application in the biomedical research space. Instead of using widely used measurements in calibrating modern generic AI models [7], biomedical-tailored metrics may need to be proposed for more specialized applications such as assessing the construction fidelity of GDTs. Such challenges, for instance, have been investigated in the IST-editing study [64] using nuclear morphometrics. By quantitatively assessing differences in nuclear size and count between GDTs driven by edited gene expressions and the ground truth, the authors rigorously evaluated construction quality through biologically interpretable metrics. These specialized metrics would then provide a more accurate evaluation of the models' performance in capturing the intricate details of biological systems. When analyzing emerging phenomena within virtual systems, close collaboration between computational scientists and biomedical domain experts is critical to ensure that the models are grounded in biological reality and that the insights gained are both scientifically valid and clinically relevant.
5.3. Developing dynamic GDTs using spatial-temporal technologies
Biology occurs not only in spatial dimensions but also over time. Given the wide range of cellular organizations obtained by multi-omic technologies, continuous patterns can be discerned through pseudo-time trajectory analysis [22] or multi-section analysis at different developmental stages [15]. To construct dynamic GDTs, time- and spatial-resolved measurements can be utilized to train temporal generative AI models, which have proven effective in diverse video generation tasks [42].
However, pseudo-temporal analyses unavoidably rely on static snapshots of different samples at various stages of development or disease progression. These snapshots, while informative, do not directly track the changes within the same individual over time, potentially missing transient or rapid events. In the meantime, these analyses typically infer temporal trajectories by averaging data across a population of cells or tissues, which can obscure individual variability and unique responses. This averaging effect might overlook rare or stochastic events that are crucial to understanding biological processes. To develop robust and dynamic GDTs, true spatial-temporal technologies need to be advanced, and continuous temporal data need to be generated for individual biological objects to facilitate the complete modeling of a physical system. Alongside progressing profiling technologies, more efficient data management strategies are required to handle the exponentially growing data volume with up to exa- and zetta-resolutions. Despite these challenges, we stay optimistic and enthusiastic about the future of this nascent research domain.
CRediT authorship contribution statement
Jiqing Wu: Writing – review & editing, Writing – original draft, Conceptualization. Viktor H. Koelzer: Writing – review & editing, Supervision, Project administration, Funding acquisition, Conceptualization.
Declaration of Competing Interest
J.W. declares no competing interests. V.H.K. reports being an invited speaker for Sharing Progress in Cancer Care (SPCC) and Indica Labs; advisory board of Takeda; sponsored research agreements with Roche and IAG, all unrelated to the current study. V.H.K. is a participant in several patent applications on the assessment of cancer immunotherapy biomarkers by digital pathology, a patent application on multimodal deep learning for the prediction of recurrence risk in cancer patients, and a patent application on predicting the efficacy of cancer treatment using deep learning.
References
- 1.Alajaji S.A., Khoury Z.H., Elgharib M., Saeed M., Ahmed A.R., Khan M.B., et al. Generative adversarial networks in digital histopathology: current applications, limitations, ethical considerations, and future directions. Mod Pathol. 2023;100369 doi: 10.1016/j.modpat.2023.100369. [DOI] [PubMed] [Google Scholar]
- 2.Ali H.R., Jackson H.W., Zanotelli V.R., Danenberg E., Fischer J.R., Bardwell H., et al. Imaging mass cytometry and multiplatform genomics define the phenogenomic landscape of breast cancer. Nat Cancer. 2020;1:163–175. doi: 10.1038/s43018-020-0026-6. [DOI] [PubMed] [Google Scholar]
- 3.Allen A., Siefkas A., Pellegrini E., Burdick H., Barnes G., Calvert J., et al. A digital twins machine learning model for forecasting disease progression in stroke patients. Appl Sci. 2021;11:5576. [Google Scholar]
- 4.Auriemma Citarella A., Di Biasi L., De Marco F., Tortora G. Entail: yet another amyloid fibrils classifier. BMC Bioinform. 2022;23:517. doi: 10.1186/s12859-022-05070-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Auriemma Citarella A., Di Biasi L., Risi M., Tortora G. Snarer: new molecular descriptors for snare proteins classification. BMC Bioinform. 2022;23:148. doi: 10.1186/s12859-022-04677-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Black S., Phillips D., Hickey J.W., Kennedy-Darling J., Venkataraaman V.G., Samusik N., et al. Codex multiplexed tissue imaging with dna-conjugated antibodies. Nat Protoc. 2021;16:3802–3835. doi: 10.1038/s41596-021-00556-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borji A. Pros and cons of gan evaluation measures. Comput Vis Image Underst. 2019;179:41–65. [Google Scholar]
- 8.Bray M.A., Singh S., Han H., Davis C.T., Borgeson B., Hartland C., et al. Cell painting, a high-content image-based assay for morphological profiling using multiplexed fluorescent dyes. Nat Protoc. 2016;11:1757–1774. doi: 10.1038/nprot.2016.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bressan D., Battistoni G., Hannon G.J. The dawn of spatial omics. Science. 2023;381 doi: 10.1126/science.abq4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao H., Tan C., Gao Z., Xu Y., Chen G., Heng P.A., et al. A survey on generative diffusion models. IEEE Trans Knowl Data Eng. 2024 [Google Scholar]
- 11.Carrillo-Perez F., Pizurica M., Zheng Y., Nandi T.N., Madduri R., Shen J., et al. Generation of synthetic whole-slide image tiles of tumours from rna-sequencing data via cascaded diffusion models. Nat Biomed Eng. 2024:1–13. doi: 10.1038/s41551-024-01193-8. [DOI] [PubMed] [Google Scholar]
- 12.Cen S., Gebregziabher M., Moazami S., Azevedo C.J., Pelletier D. Toward precision medicine using a “digital twin” approach: modeling the onset of disease-specific brain atrophy in individuals with multiple sclerosis. Sci Rep. 2023;13 doi: 10.1038/s41598-023-43618-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chai L., Gharbi M., Shechtman E., Isola P., Zhang R. European conference on computer vision. Springer; 2022. Any-resolution training for high-resolution image synthesis; pp. 170–188. [Google Scholar]
- 14.Chaudhuri A., Pash G., Hormuth D.A., Lorenzo G., Kapteyn M., Wu C., et al. Predictive digital twin for optimizing patient-specific radiotherapy regimens under uncertainty in high-grade gliomas. Front Artif Intell. 2023;6 doi: 10.3389/frai.2023.1222612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen A., Liao S., Cheng M., Ma K., Wu L., Lai Y., et al. Spatiotemporal transcriptomic atlas of mouse organogenesis using dna nanoball-patterned arrays. Cell. 2022;185:1777–1792. doi: 10.1016/j.cell.2022.04.003. [DOI] [PubMed] [Google Scholar]
- 16.Corral-Acero J., Margara F., Marciniak M., Rodero C., Loncaric F., Feng Y., et al. The ‘digital twin'to enable the vision of precision cardiology. Eur Heart J. 2020;41:4556–4564. doi: 10.1093/eurheartj/ehaa159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cuccarese MF, Earnshaw BA, Heiser K, Fogelson B, Davis CT, McLean PF, et al. Functional immune mapping with deep-learning enabled phenomics applied to immunomodulatory and covid-19 drug discovery. 2020. bioRxiv.
- 18.Ding Z., Zhang M., Wu J., Tu Z. The twelfth international conference on learning representations. 2023. Patched denoising diffusion models for high-resolution image synthesis. [Google Scholar]
- 19.Efros A.A., Leung T.K. Proceedings of the seventh IEEE international conference on computer vision, IEEE. 1999. Texture synthesis by non-parametric sampling; pp. 1033–1038. [Google Scholar]
- 20.Frei A.L., McGuigan A., Sinha R.R., Jabbar F., Gneo L., Tomasevic T., et al. Multiplex analysis of intratumoural immune infiltrate and prognosis in patients with stage ii–iii colorectal cancer from the scot and quasar 2 trials: a retrospective analysis. Lancet Oncol. 2024;25:198–211. doi: 10.1016/S1470-2045(23)00560-0. [DOI] [PubMed] [Google Scholar]
- 21.Goodfellow I., Pouget-Abadie J., Mirza M., Xu B., Warde-Farley D., Ozair S., et al. Generative adversarial nets. Adv Neural Inf Process Syst. 2014;27 [Google Scholar]
- 22.Haghverdi L., Büttner M., Wolf F.A., Buettner F., Theis F.J. Diffusion pseudotime robustly reconstructs lineage branching. Nat Methods. 2016;13:845–848. doi: 10.1038/nmeth.3971. [DOI] [PubMed] [Google Scholar]
- 23.Haniffa M., Taylor D., Linnarsson S., Aronow B.J., Bader G.D., Barker R.A., et al. A roadmap for the human developmental cell atlas. Nature. 2021;597:196–205. doi: 10.1038/s41586-021-03620-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harb R., Pock T., Müller H. Proceedings of the IEEE/CVF winter conference on applications of computer vision. 2024. Diffusion-based generation of histopathological whole slide images at a gigapixel scale; pp. 5131–5140. [Google Scholar]
- 25.He S., Bhatt R., Brown C., Brown E.A., Buhr D.L., Chantranuvatana K., et al. High-plex imaging of rna and proteins at subcellular resolution in fixed tissue by spatial molecular imaging. Nat Biotechnol. 2022;40:1794–1806. doi: 10.1038/s41587-022-01483-z. [DOI] [PubMed] [Google Scholar]
- 26.He S., Patrick M., Reeves J.W., Danaher P., Preciado J., Phan J., et al. Path to the holy grail of spatial biology: spatial single-cell whole transcriptomes using 6000-plex spatial molecular imaging on ffpe tissue. Cancer Res. 2023;83:5637. [Google Scholar]
- 27.Hernandez S., Lazcano R., Serrano A., Powell S., Kostousov L., Mehta J., et al. Challenges and opportunities for immunoprofiling using a spatial high-plex technology: the nanostring geomx® digital spatial profiler. Front Oncol. 2022;12 doi: 10.3389/fonc.2022.890410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ho J., Jain A., Abbeel P. Denoising diffusion probabilistic models. Adv Neural Inf Process Syst. 2020;33:6840–6851. [Google Scholar]
- 29.Irmisch A., Bonilla X., Chevrier S., Lehmann K.V., Singer F., Toussaint N.C., et al. The tumor profiler study: integrated, multi-omic, functional tumor profiling for clinical decision support. Cancer Cell. 2021;39:288–293. doi: 10.1016/j.ccell.2021.01.004. [DOI] [PubMed] [Google Scholar]
- 30.Janesick A., Shelansky R., Gottscho A.D., Wagner F., Williams S.R., Rouault M., et al. High resolution mapping of the tumor microenvironment using integrated single-cell, spatial and in situ analysis. Nat Commun. 2023;14:8353. doi: 10.1038/s41467-023-43458-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jaume G., Doucet P., Song A.H., Lu M.Y., Almagro-Pérez C., Wagner S.J., et al. Hest-1k: a dataset for spatial transcriptomics and histology image analysis. 2024. arXiv:2406.16192 arXiv preprint.
- 32.Kang M., Zhu J.Y., Zhang R., Park J., Shechtman E., Paris S., et al. Proceedings of the IEEE/CVF conference on computer vision and pattern recognition. 2023. Scaling up gans for text-to-image synthesis; pp. 10124–10134. [Google Scholar]
- 33.Karras T., Laine S., Aittala M., Hellsten J., Lehtinen J., Aila T. Proceedings of the IEEE/CVF conference on computer vision and pattern recognition. 2020. Analyzing and improving the image quality of stylegan; pp. 8110–8119. [Google Scholar]
- 34.Katsoulakis E., Wang Q., Wu H., Shahriyari L., Fletcher R., Liu J., et al. Digital twins for health: a scoping review. npj Digit Med. 2024;7:77. doi: 10.1038/s41746-024-01073-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kulikov V., Yadin S., Kleiner M., Michaeli T. International conference on machine learning, PMLR. 2023. Sinddm: a single image denoising diffusion model; pp. 17920–17930. [Google Scholar]
- 36.Lal A., Li G., Cubro E., Chalmers S., Li H., Herasevich V., et al. Development and verification of a digital twin patient model to predict specific treatment response during the first 24 hours of sepsis. Crit Care Explor. 2020;2 doi: 10.1097/CCE.0000000000000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lamiable A., Champetier T., Leonardi F., Cohen E., Sommer P., Hardy D., et al. Revealing invisible cell phenotypes with conditional generative modeling. Nat Commun. 2023;14:6386. doi: 10.1038/s41467-023-42124-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laubenbacher R., Mehrad B., Shmulevich I., Trayanova N. Digital twins in medicine. Nat Comput Sci. 2024;4:184–191. doi: 10.1038/s43588-024-00607-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lewis S.M., Asselin-Labat M.L., Nguyen Q., Berthelet J., Tan X., Wimmer V.C., et al. Spatial omics and multiplexed imaging to explore cancer biology. Nat Methods. 2021;18:997–1012. doi: 10.1038/s41592-021-01203-6. [DOI] [PubMed] [Google Scholar]
- 40.Lin C.H., Chang C.C., Chen Y.S., Juan D.C., Wei W., Chen H.T. Proceedings of the IEEE/CVF international conference on computer vision. 2019. Coco-gan: generation by parts via conditional coordinating; pp. 4512–4521. [Google Scholar]
- 41.Lin C.H., Lee H.Y., Cheng Y.C., Tulyakov S., Yang M.H. Infinitygan: towards infinite-pixel image synthesis. 2021. arXiv:2104.03963 arXiv preprint.
- 42.Liu Y., Zhang K., Li Y., Yan Z., Gao C., Chen R., et al. Sora: a review on background, technology, limitations, and opportunities of large vision models. 2024. arXiv:2402.17177 arXiv preprint.
- 43.Luhman T., Luhman E. Improving diffusion model efficiency through patching. 2022. arXiv:2207.04316 arXiv preprint.
- 44.Moingeon P., Chenel M., Rousseau C., Voisin E., Guedj M. Virtual patients, digital twins and causal disease models: paving the ground for in silico clinical trials. Drug Discov Today. 2023;28 doi: 10.1016/j.drudis.2023.103605. [DOI] [PubMed] [Google Scholar]
- 45.Moses L., Pachter L. Museum of spatial transcriptomics. Nat Methods. 2022;19:534–546. doi: 10.1038/s41592-022-01409-2. [DOI] [PubMed] [Google Scholar]
- 46.Oliveira MF, Romero JP, Chung M, Williams S, Gottscho AD, Gupta A, et al. Characterization of immune cell populations in the tumor microenvironment of colorectal cancer using high definition spatial profiling. 2024. bioRxiv, 2024–06.
- 47.Palla G., Spitzer H., Klein M., Fischer D., Schaar A.C., Kuemmerle L.B., et al. Squidpy: a scalable framework for spatial omics analysis. Nat Methods. 2022;19:171–178. doi: 10.1038/s41592-021-01358-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ptacek J., Locke D., Finck R., Cvijic M.E., Li Z., Tarolli J.G., et al. Multiplexed ion beam imaging (mibi) for characterization of the tumor microenvironment across tumor types. Lab Investig. 2020;100:1111–1123. doi: 10.1038/s41374-020-0417-4. [DOI] [PubMed] [Google Scholar]
- 49.Rao N., Clark S., Habern O. Bridging genomics and tissue pathology: 10x genomics explores new frontiers with the visium spatial gene expression solution. Genet Eng Biotechnol News. 2020;40:50–51. [Google Scholar]
- 50.Read M.N., Alden K., Timmis J., Andrews P.S. Strategies for calibrating models of biology. Brief Bioinform. 2020;21:24–35. doi: 10.1093/bib/bby092. [DOI] [PubMed] [Google Scholar]
- 51.Rivest F., Eroglu D., Pelz B., Kowal J., Kehren A., Navikas V., et al. Fully automated sequential immunofluorescence (seqif) for hyperplex spatial proteomics. Sci Rep. 2023;13 doi: 10.1038/s41598-023-43435-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rombach R., Blattmann A., Lorenz D., Esser P., Ommer B. Proceedings of the IEEE/CVF conference on computer vision and pattern recognition. 2022. High-resolution image synthesis with latent diffusion models; pp. 10684–10695. [Google Scholar]
- 53.Šali A. 100,000 protein structures for the biologist. Nat Struct Biol. 1998;5:1029–1032. doi: 10.1038/4136. [DOI] [PubMed] [Google Scholar]
- 54.Schena M., Shalon D., Davis R.W., Brown P.O. Quantitative monitoring of gene expression patterns with a complementary dna microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 55.Shaham T.R., Dekel T., Michaeli T. Proceedings of the IEEE/CVF international conference on computer vision. 2019. Singan: learning a generative model from a single natural image; pp. 4570–4580. [Google Scholar]
- 56.Shapson-Coe A., Januszewski M., Berger D.R., Pope A., Wu Y., Blakely T., et al. A petavoxel fragment of human cerebral cortex reconstructed at nanoscale resolution. Science. 2024;384 doi: 10.1126/science.adk4858. [DOI] [PubMed] [Google Scholar]
- 57.Skorokhodov I., Sotnikov G., Elhoseiny M. Proceedings of the IEEE/CVF international conference on computer vision. 2021. Aligning latent and image spaces to connect the unconnectable; pp. 14144–14153. [Google Scholar]
- 58.de Souza N., Zhao S., Bodenmiller B. Multiplex protein imaging in tumour biology. Nat Rev Cancer. 2024;24:171–191. doi: 10.1038/s41568-023-00657-4. [DOI] [PubMed] [Google Scholar]
- 59.Ståhl P.L., Salmén F., Vickovic S., Lundmark A., Navarro J.F., Magnusson J., et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 2016;353:78–82. doi: 10.1126/science.aaf2403. [DOI] [PubMed] [Google Scholar]
- 60.Vaswani A., Shazeer N., Parmar N., Uszkoreit J., Jones L., Gomez A.N., et al. Attention is all you need. Adv Neural Inf Process Syst. 2017;30 [Google Scholar]
- 61.Wang Z., Jiang Y., Zheng H., Wang P., He P., Wang Z., et al. Patch diffusion: faster and more data-efficient training of diffusion models. Adv Neural Inf Process Syst. 2024;36 [Google Scholar]
- 62.Wu C., Lorenzo G., Hormuth D.A., Lima E.A., Slavkova K.P., DiCarlo J.C., et al. Integrating mechanism-based modeling with biomedical imaging to build practical digital twins for clinical oncology. Biophys Rev. 2022;3 doi: 10.1063/5.0086789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu J, Berg I, Koelzer, Ist-editing V. Infinite spatial transcriptomic editing in a generated gigapixel mouse pup. In: Medical Imaging with Deep Learning.
- 64.Wu J., Berg I., Koelzer V. Ist-editing: infinite spatial transcriptomic editing in a generated gigapixel mouse pup. International conference on medical imaging with deep learning; PMLR; 2024. [Google Scholar]
- 65.Wu J., Koelzer V. NeurIPS 2022 workshop on distribution shifts: connecting methods and applications. 2022. Sorted eigenvalue comparison deig: a simple alternative to dfid. [Google Scholar]
- 66.Wu J., Koelzer V.H. Gilea: in silico phenome profiling and editing using gan inversion. Comput Biol Med. 2024;179 doi: 10.1016/j.compbiomed.2024.108825. [DOI] [PubMed] [Google Scholar]
- 67.Wu J., Koelzer V.H. Sst-editing: in silico spatial transcriptomic editing at single-cell resolution. Bioinformatics. 2024;40 doi: 10.1093/bioinformatics/btae077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xia C., Fan J., Emanuel G., Hao J., Zhuang X. Spatial transcriptome profiling by merfish reveals subcellular rna compartmentalization and cell cycle-dependent gene expression. Proc Natl Acad Sci. 2019;116:19490–19499. doi: 10.1073/pnas.1912459116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yao Z., van Velthoven C.T., Kunst M., Zhang M., McMillen D., Lee C., et al. A high-resolution transcriptomic and spatial atlas of cell types in the whole mouse brain. Nature. 2023;624:317–332. doi: 10.1038/s41586-023-06812-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhu Y.C., Wagg D., Cross E., Barthorpe R. Model validation and uncertainty quantification, volume 3: proceedings of the 38th IMAC, a conference and exposition on structural dynamics 2020. Springer; 2020. Real-time digital twin updating strategy based on structural health monitoring systems; pp. 55–64. [Google Scholar]