Abstract
Development of safe and effective gene transfer systems is critical to the success of gene therapy protocols for human diseases. Currently, several primate lentivirus-based gene transfer systems, such as those based on human and simian immunodeficiency viruses (HIV/SIV), are being tested; however, their use in humans raises safety concerns, such as the generation of replication-competent viruses through recombination with related endogenous retroviruses or retrovirus-like elements. Due to the greater phylogenetic distance from primate lentiviruses, feline immunodeficiency virus (FIV) is becoming the lentivirus of choice for human gene transfer systems. However, the safety of FIV-based vector systems has not been tested experimentally. Since lentiviruses such as HIV-1 and SIV have been shown to cross-package their RNA genomes, we tested the ability of FIV RNA to get cross-packaged into primate lentivirus particles such as HIV-1 and SIV, as well as a nonlentiviral retrovirus such as Mason-Pfizer monkey virus (MPMV), and vice versa. Our results reveal that FIV RNA can be cross-packaged by primate lentivirus particles such as HIV-1 and SIV and vice versa; however, a nonlentivirus particle such as MPMV is unable to package FIV RNA. Interestingly, FIV particles can package MPMV RNA but cannot propagate the vector RNA further for other steps of the retrovirus life cycle. These findings reveal that diverse retroviruses are functionally more similar than originally thought and suggest that upon coinfection of the same host, cross- or copackaging may allow distinct retroviruses to generate chimeric variants with unknown pathogenic potential.
Development of safe and effective gene transfer systems that allow the stable transduction of both dividing and nondividing cells is a desirable characteristic of gene therapy protocols for human diseases (13, 25, 26, 30, 42). Towards this end, lentivirus vectors are becoming one of the most promising vehicles due to their ability to (i) integrate the therapeutic gene into the host genome, ensuring its stable long-term expression; (ii) transduce nondividing cells, such as brain, liver, muscle, and hematopoietic cells, the main targets of gene therapy; (iii) encode large coding capacities, thereby allowing the expression of most genes of choice; and (iv) transduce only the therapeutic gene and not their own genes, thus avoiding host humoral and cellular responses to virus antigens that can eliminate the transduced cells (26).
Currently, several primate lentivirus-based gene transfer systems, such as human immunodeficiency virus type 1(HIV-1), type 2 (HIV-2), and simian immunodeficiency virus (SIV), are being tested; however, their use in humans is not very promising due to safety concerns such as the generation of replication-competent virus through recombination with related primate viruses. A gene transfer system based on nonprimate lentiviruses, such as feline immunodeficiency virus (FIV), which causes an immune deficiency syndrome similar to human AIDS in cats (8, 29), may circumvent such concerns. FIV is a lentivirus isolated from 2 to 20% of domestic and wild cats worldwide suffering from feline AIDS (reviewed in reference 29). FIV encodes only three accessory/regulatory genes, of which vif and rev are functionally equivalent to similar HIV-1 genes but tat has weak transactivation activity and is not functionally equivalent to HIV-1 tat since it does not interact with a TAR-like element (5, 36, 41). Phylogenetic comparisons and the absence of serological reactivity with other primate lentiviruses suggest that FIV is only distantly related to HIV-1 or SIV (3, 8, 27, 28, 40). Several features make FIV vectors excellent candidates for human gene therapy: (i) primates are not the natural host of FIV and despite prevalent human exposure to the virus, no human infection or seroconversion in humans has been reported (8, 18, 29, 44); (ii) FIV can infect and replicate in nondividing cells (13, 30, 42); and (iii) unlike primate AIDS models, feline AIDS models are much cheaper and are readily available due to shorter breeding cycles in which safety issues can be easily addressed (reviewed in reference 22).
Despite the aforementioned characteristics that make FIV suitable for gene delivery into humans, no studies exist that have directly tested the safety of FIV vectors in human cells either in vitro or in vivo. This is due primarily to the highly restricted tropism of FIV for feline cells attributed to its envelope. Additionally, many other steps in the FIV life cycle (nuclear import, integration, RNA export, etc.) require cellular factors for completion, which may not interact with FIV cis-acting elements when from human cells, resulting in further restriction to expression. This can be observed by the low transcriptional activity of the FIV long terminal repeat (LTR) (11, 23, 24, 36) and suboptimal Rev function in human cells (41). These restrictions could be overcome by the use of feline producer cells. However, use of feline producer cells for human gene transfer studies could introduce other hazards, such as transmission into humans of feline infectious agents like RD114, an endogenous feline retrovirus that can replicate in human cells (39). As an alternative to the use of feline producer cells in human gene transfer studies, various laboratories have successfully manipulated the FIV 5′ LTR by replacing its U3 (promoter) region with the human cytomegalovirus (hCMV) immediate-early promoter to allow its expression in human cells (13, 30).
The safety issues concerning the use of lentivirus vectors for human gene therapy have been highlighted by the fact that primate lentiviruses can cross-package each other's genomic RNAs. Efficient packaging of retroviral RNA entails specific recognition of a cis-acting RNA element (packaging signal, Ψ), located near the 5′ end of the retroviral RNA, by components of the Gag precursor protein. We have shown that SIV RNA can be efficiently cross-packaged by proteins of its close relative HIV-1 (31). Conversely, White et al. have shown that HIV-1 RNA can be cross-packaged by SIV particles as well (43). In addition to the observation that retroviral RNAs containing the Ψ of closely related viruses can be recognized by heterologous viral proteins and packaged, it has been clearly demonstrated that cross-packaging can occur in evolutionarily related yet molecularly different retroviruses. For example, among type C retroviruses, spleen necrosis virus (SNV) has been shown to cross-package murine leukemia virus (MLV) vector RNAs (9, 45, 46). The safety issues pertaining to cross-packaging of heterologous RNAs have been further compounded by the recent demonstration that infection with wild-type virus of previously transduced cells by HIV-1-based vectors can lead to competition for efficient packaging, mobilization, and recombination of the HIV-1 transfer vector (5).
In light of these concerns, we attempted to test the safety of FIV vectors by examining the ability of FIV genomic RNA to get cross-packaged into other primate lentivirus particles such as HIV-1 and SIV. Our results demonstrate that FIV is promiscuous in its ability to get cross-packaged into other lentivirus particles such as HIV-1 and SIV and vice versa. However, it cannot be packaged by a nonlentivirus particle such as Mason-Pfizer monkey virus (MPMV).
MATERIALS AND METHODS
Numbering system.
Nucleotide designations for the HIV-1NL4-3, SIVmac239, and FIVPetaluma (34TF10) strains and MPMV are based on GenBank accession numbers M19921 (1), M33262 (16), M25381 (40), and M12349 (35), respectively.
Plasmid construction. (i) HIV-1 packaging construct and transfer vectors.
The HIV-1 packaging construct, CMVΔR8.2 (Fig. 1A), was kindly provided by Didier Trono (Salk Institute, La Jolla, Calif.) and has been described previously (25). MB58 is an HIV-1 transfer vector (Fig. 1A) which was derived from HIV-GFP (43), kindly provided by Vicente Planelles (University of Rochester, Rochester, N.Y.). MB58 was constructed by replacing the XhoI-to-NotI gfp-containing fragment of HIV-GFP with the simian virus 40 (SV40)-expressed hygromycin gene cassette (SV-Hygr).
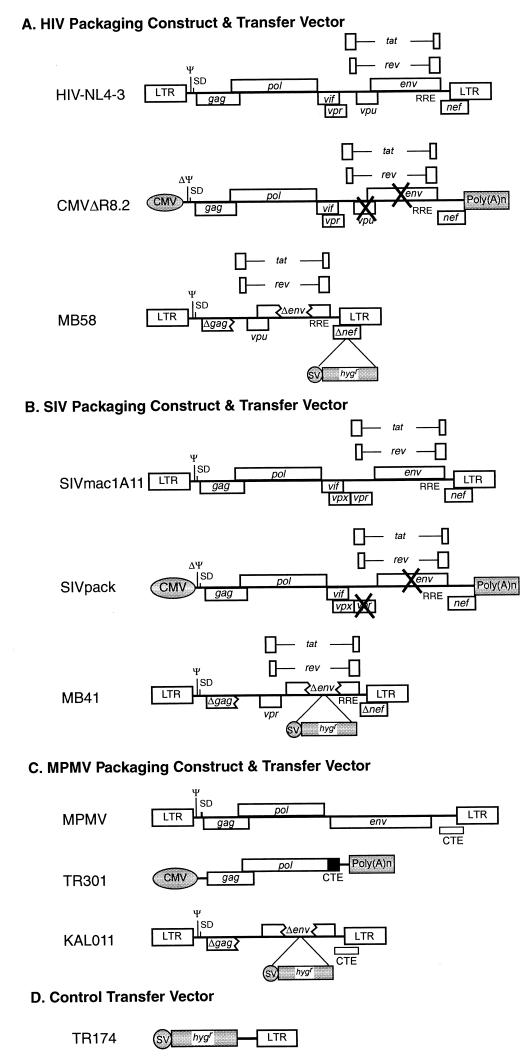
FIG. 1.
Schematic representation of HIV-1, SIV, and MPMV packaging constructs and transfer vectors used in the cross-packaging studies. (A) HIV-based packaging construct and transfer vector. (B) SIV-based packaging construct and transfer vector. (C) MPMV-based packaging construct and transfer vector. (D) Control vector without any packaging signal but containing the SV-Hygr cassette to measure any signs of retrofection. CMV, cytomegalovirus promoter/enhancer; SV, simian virus 40 early promoter; hygr, hygromycin B phosphotransferase gene; SD, splice donor; CTE, constitutive transport element from MPMV.
(ii) SIV packaging constructs and transfer vectors.
SIVpack (Fig. 1B) is a SIVmac1A11 (19)-derived packaging construct that was also kindly provided by V. Planelles and has been described before (43). MB41 is an SIVmac239-derived transfer vector that contains only the vpr, tat, and rev/RRE of SIV with an insertion of the SV-Hygr cassette at the NheI site at nucleotide (nt) 9000, replacing most of the gag/pol and env coding sequences (Fig. 1B).
(iii) FIV packaging construct and transfer vectors.
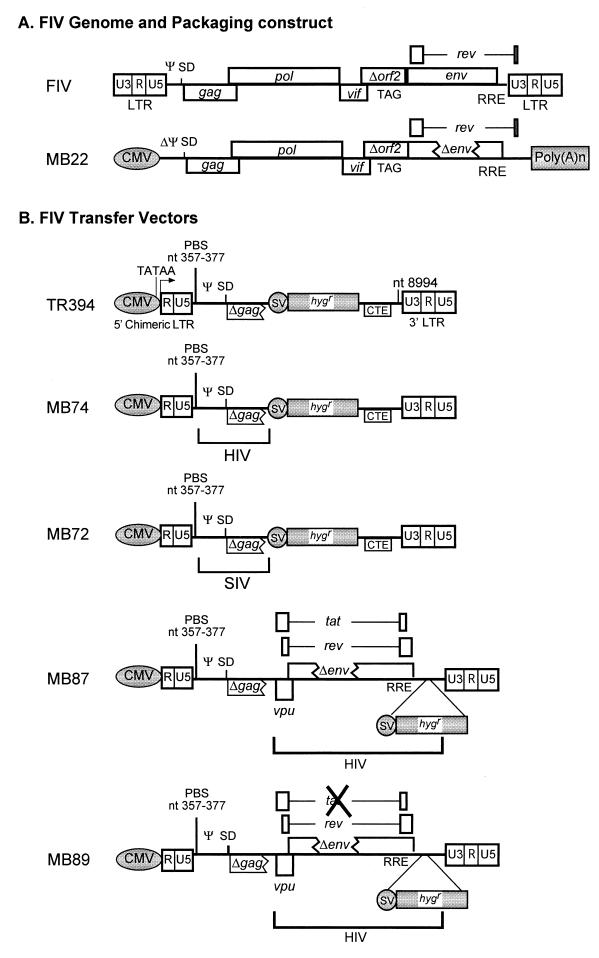
The FIV packaging construct, MB22 (Fig. 2A), uses the hCMV intron A promoter/enhancer (replacing the 5′ LTR) and bovine growth hormone (BGH) poly(A) sequences (replacing the 3′ LTR) to express all FIV genes from the Petaluma (34TF10) strain except env and orf2. Specifically, it contains FIV sequences from the SacI site (nt 503) in the 5′ untranslated region (UTR) to the BlpI site (nt 9204) in the beginning of the 3′ LTR with an 875-bp PflmI deletion in env. orf2 has a premature stop codon in 34TF10 (7). MB22 was constructed through several stages of cloning in which the ψ was deleted while maintaining the major splice donor. Such a design removed most of the cis-acting elements, thus minimizing the chances of recombination between the packaging construct and transfer vectors to generate replication-competent virus.
FIG. 2.
Schematic representation of the FIV packaging construct and transfer vectors used in the cross-packaging analysis of FIV RNA into HIV-1, SIV, and MPMV particles and vice versa. (A) FIV genome with open reading frames (ORFs) and FIV-based packaging construct. The U3 region of the FIV 5′ LTR was replaced with the CMV promoter to allow expression in human cells. (B) FIV-based transfer vectors. MB74, -72, -87, and -89 are chimeric derivatives of TR394 that contain portions of either HIV-1 or SIV sequences. MB89 contains a deletion in the Tat ORF, eliminating Tat expression. CMV, human cytomegalovirus promoter/enhancer; SV, simian virus 40 early promoter; hygr, hygromycin B phosphotransferase gene; SD, splice donor; CTE, constitutive transport element from MPMV.
TR394 (Fig. 2B), the FIV transfer vector, was constructed by PCR amplifying the two flanks of the FIV genome independently. The 5′ flank was amplified using oligonucleotides OTR399 (sense [S]; 5′ cccaagcttgagctcTGTGAAACTTCGAGGAGTCTC 3′; nt 203 to 223) and OTR400 (antisense [AS]; 5′ cctctAGAATACATATTTTCAGCT GCAGCAGC 3′; nt 960 to 934) (uppercase indicates viral and/or promoter sequences; lowercase indicates an artificially created restriction site), resulting in a 757-bp fragment containing FIV sequences from nt 203 in the 5′ U3 region to nt 960 in gag. The 3′ flank was amplified using oligonucleotides OTR401 (S; 5′ cctctagatatcGCCTTCAAGAGGATGATGACA 3′; nt 8994 to 9014) and OTR402 (AS; 5′ cccagatctcgagttaacTGCGAAGTTCTCGGCCCGGATTCC 3′; nt 9474 to 9451), resulting in a 480-bp fragment containing FIV sequences from nt 8994 in the 3′ UTR to nt 9474 at the end of the 3′ LTR. The two flanks of the virus were ligated into the cloning vector Rc/CMV (Invitrogen, Carlsbad, Calif.) followed by the insertion of the SV-Hygr cassette and MPMV constitutive transport element (CTE) (nt 8006 to 8240) to create TR394. FIV transfer vector MB87 (Fig. 2B) is a modification of TR394 containing an insertion of a fragment encoding the HIV-1 vpu, tat, and rev/RRE from HIV-GFP (43). MB89 (Fig. 2B) is a derivative of MB87 that contains a deletion of the HIV-1 tat ATG while maintaining vpu and rev/RRE.
MB72 and MB74 (Fig. 2B) are also derivatives of TR394 that maintain the FIV primer binding site (PBS) but replace the FIV ψ with that from either SIV or HIV-1, respectively. Using OTR405 (S; 5′ CGCGTTGACATTGATTAT 3′; nt 229 to 246 of Rc/CMV) and OTR460 (AS; 5′ ccccccatcgatgcggccgcGTTGCTGTAGAATCTCTCCTACCT 3′; FIV nt 377 to 357), the FIV 5′ UTR of TR394 was shortened to nt 377 and the FIV gag sequences were completely removed. MB72 was generated by inserting a PCR fragment after the FIV PBS containing the SIV ψ and necessary gag sequences (nt 1101 to 1940 of SIVmac239) using OTR465 (S; 5′aaaaaagcggccgcGGAGAGTGAGAGACTCCTGAG 3′; nt 1101 to 1121 immediately 3′ of SIVmac239 PBS) and OTR466 (AS; 5′ aaaaaagcggccgcCCTCCTCGTTTATAATATCTCTG 3′; nt 1940 to 1917 of SIVmac239), while MB74 (Fig. 2B) was created by cloning a PCR fragment containing the HIV-1 Ψ and relevant gag sequences after the FIV PBS using OTR462 (S; 5′ aaaaaagcggccgcAGCGAAAGTAAAGCCAGAGGA 3′; nt 659 to 679 3′ of the HIV-1 PBS) and OTR463 (AS; 5′ aaaaaagcggccgcCACTGGATGCAATCTATCCCA 3′; nt 1443 to 1423 in HIV-1 gag).
(iv) MPMV packaging construct and transfer vectors.
The MPMV packaging construct TR301 (Fig. 1C) expresses the gag/pol genes from the hCMV intron A promoter/enhancer, contains the MPMV CTE, and uses the BGH poly(A) sequences for transcript termination. The MPMV transfer vector, KAL011, contains cis-acting MPMV sequences from the 5′ LTR to nt 1171 of gag and, at the 3′ end, from nt 7181 to the end of the 3′ LTR with the insertion of the SV-Hygr cassette (Fig. 1C).
(v) Control vector for nonspecific RNA packaging.
To test the possibility of nonspecific packaging of RNA containing the SV-Hygr cassette through retrofection, we used TR174, which has been described before (31) and contains the SV-Hygr cassette with the 3′ SIV LTR providing the poly(A) sequence (Fig. 1D).
(vi) Envelope expression construct.
The vesicular stomatitis virus envelope protein G (VSV-G) expression construct, MD.G, was kindly provided by Didier Trono and has been described before (25).
All vectors were made through several stages of cloning, and further details of the design and construction of these vectors can be obtained from the authors upon request.
Transfections and infections of cells.
Cos cells were maintained at 37°C in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum from HyClone (Logan, Utah). HeLa CD4+ cells were maintained in DMEM supplemented with 7% calf serum from HyClone. Cos cells were transfected using 6 μl of Lipofectamine (Life Technologies, Gaithersburg, Md.) and 1 μg of each plasmid DNA (3 μg of total DNA per transfection) per well of a six-well plate using the manufacturer's suggested conditions. Virus stocks were harvested 72 h posttransfection and were subjected to low-speed centrifugation to remove cellular debris. Twelve to 18 h prior to infection, HeLa CD4+ cells were plated at 4.0 × 105 cells per 60-mm plate. A portion of the virus stock was used to infect HeLa CD4+ cells in the presence of 8 μg of DEAE-dextran (Pharmacia, Piscataway, N.J.)/ml. Selection for hygromycin resistance was initiated 48 h postinfection at 200 μg/ml (Calbiochem, La Jolla, Calif.). Ten days postinfection, hygromycin-resistant (Hygr) colonies were scored by either staining with 0.5% crystal violet in 50% methanol or expanding individual colonies for genomic DNA extraction.
Ultracentrifugation of virus particles.
Supernatants harvested from transfected cultures (9 ml) were clarified of cellular debris by low-speed centrifugation in a tabletop Sorvall centrifuge at 2,500 rpm for 5 min. Virus particles were pelleted by ultracentrifugation using a Sorvall TH641 swinging bucket rotor at 80,000 × g for 2 h at 4°C. Virus pellets were resuspended in 100 μl of TNE buffer (50 mM Tris-Cl, pH 7.4, 100 mM NaCl, 1 mM EDTA, pH 8.0) and were stored at −80°C until ready to be assayed. Twenty-five microliters of the virion pellet was used for protein extraction, while the remaining 75 μl was used to isolate virion RNA.
Western blot analysis.
Viral proteins were analyzed by Western blot analysis using the Enhanced Chemiluminescence kit (Amersham, Arlington Heights, Ill.) as described before (38). The anti-SIVmac251 p27 and anti-HIV-1-IIIB p24 monoclonal antibodies were commercially obtained (ABI, Columbia, Md.). The anti-MPMV Gag/Pol Pr78 polyserum was kindly provided by Eric Hunter (University of Alabama, Birmingham, Ala.). The FIV antibody is a polyclonal antiserum from a cat infected with the Petaluma strain of FIV and was kindly provided by Ellen Collisson (Texas A&M University, College Station, Tex.).
Viral and cellular RNA slot blot analysis.
Viral RNA was isolated from the pelleted virus particles of the transfected cells using the QiaAmp viral RNA kit (Qiagen, Valencia, Calif.) following the manufacturer's recommendations. Cellular RNA was isolated from transfected Cos cells 72 h posttransfection using the Qiagen RNeasy Total RNA kit according to the manufacturer's recommended protocol. Following RNA isolation, both viral and cellular RNAs were treated with RNase-free DNase (Life Technologies) to eliminate any contaminating DNA and were blotted using slot blot apparatus for analysis as described before (33, 34). The filters were hybridized with either a hygromycin gene-specific DNA probe or a β-actin cDNA probe (Clonetech, Palo Alto, Calif.) as described earlier (33).
Amplification of integrated proviral DNA.
In order to confirm that the Hygr colonies observed upon cross-packaging of FIV RNA into HIV-1 particles contained FIV proviral DNA and not contaminating plasmid DNA, individual Hygr colonies were expanded from the infected cells. Cellular lysates for PCR were prepared by incubating 2 × 106 cells at 65°C overnight with 200 μl of PCR lysis buffer (100 mM KCl, 10 mM Tris, pH 8, 2.5 mM MgCl2, 0.5% Tween 20, 0.5% Nonidet 40) containing 150 μg of proteinase K/ml. The reactions were boiled for 15 min to inactivate the proteinase K, were chilled on ice, and were centrifuged at 14,000 rpm for 20 min in the microcentrifuge to remove cellular debris.
To conduct nested PCR, the first round employed primers either within the CMV promoter, OTR457 (S; CGGGGTCATTAGTTCATAGCC; nt 274 to 294 of pRc/CMV), or the U3 region of FIV LTR, OTR455 (S; GGAACCCTGAAGAAATAGAAA; nt 14 to 34 of p34TF10), and FIV gag, OTR400 (AS; cctctagaATACATATTTTCAGCTGCAGCAGC; nt 960 to 934 of p34TF10). OTR400 contains an XbaI restriction endonuclease site for cloning purposes. The second round of PCR again employed primers either within the CMV promoter, OTR458 (S; CCATAGTAACGCCAATAGGGA; nt 394 to 414 of pRc/CMV) or the U3 region of FIV LTR, OTR456 (S; ACGTATAAGTTGTTCCATTGT; nt 161 to 181 of p34TF10), and FIV gag, OTR413 (AS; ccccccgcggccgcTCTTTCTTGTAAATCGCAAATAAC; nt 837 to 814 of p34TF10).
RESULTS
Design and construction of HIV-1-, SIV-, and FIV-based packaging systems.
Previously it was shown that SIV RNA can be efficiently packaged and propagated by HIV-1 particles (31). Similarly, HIV-1 RNA can be packaged and propagated by SIV protein (43). Based on these and other studies, we established a number of three-plasmid trans-complementation assays consisting of a series of packaging and transfer vectors to study the ability of FIV RNA to be cross-packaged into lentivirus (HIV-1 and SIV) or nonlentivirus (MPMV) particles. The design of these vectors was based on the premise that successful cross-packaging of FIV RNA into heterologous virus particles and its subsequent transduction would require that cis-acting sequences on FIV RNA be recognized by HIV-1, SIV, or MPMV proteins. These sequences would include the packaging signal (ψ) required for efficient incorporation of viral RNA into the assembling virion, the PBS and the polypurine tract (PPT) required for successful reverse transcription, and the attachment (att) sites required for the final step of integration into the transduced host cell. To allow propagation of these vectors into a wide variety of cells, we utilized the VSV-G protein as the envelope of choice.
CMVΔR8.2 and SIVpack are HIV-1 and SIV packaging constructs, respectively, that have been described before (25, 43). They contain the entire coding region of their respective viruses except for vpu and env genes in the case of HIV-1 (Fig. 1A), and vpr and env genes in the case of SIV (Fig. 1B). MB22 and TR301 are FIV and MPMV packaging constructs, respectively. MB22 expresses all FIV genes except env and orf2 (Fig. 2A), while TR301 expresses only Gag/Pol (Fig. 1C). These packaging constructs utilize the hCMV promoter to express the structural proteins, while transcription termination and polyadenylation are achieved by poly(A) sequences either from SV40 or BGH to minimize the generation of replication-competent virus via homologous recombination between the packaging construct and the transfer vector. Since the genomic RNA expressed from these vectors lacks Ψ, these constructs can only express the viral structural proteins, but not package their genomic RNAs in assembling virus particles.
Next, a series of transfer vectors were constructed expressing the hygromycin B phosphotransferase gene from the SV40 internal promoter/enhancer as a marker to assess successful transduction into target cells. MB58 is an HIV-1 transfer vector that contains all the cis-acting sequences necessary for its packaging into HIV-1 particles, including PBS, Ψ, PPT, att sites, and the first ∼300 bp of HIV-1 gag (since the packaging signal may extend into gag) (Fig. 1A). MB58 uses its own LTRs for promotion and termination functions of the genomic RNA, and in addition contains HIV-1 vpu, tat, rev, and RRE (Fig. 1A). MB41 (Fig. 1B) is an SIV vector similar to MB58. It differs from MB58 only in that it contains vpr instead of vpu in the background of SIV sequences. TR394 is an FIV transfer vector. Since FIV is not efficiently expressed in human cells, the U3 region of the 5′ LTR was substituted with the hCMV promoter to generate a chimeric 5′ LTR, while the 3′ LTR was maintained without any modifications. In addition, MPMV CTE, important for efficient nuclear export of genomic unspliced RNA (4, 33), was inserted downstream of the SV-Hygr cassette of TR394 to facilitate the nucleocytoplasmic transport of genomic RNA. Finally, TR174 is a control transfer vector for assessing nonspecific RNA packaging (Fig. 1D) (31).
FIV RNA can be cross-packaged and propagated by HIV-1 and SIV particles.
To determine whether FIV RNA could be cross-packaged and propagated by primate lentivirus particles, we tested the ability of an FIV transfer vector, TR394, to be cross-packaged by either HIV-1 (CMVΔR8.2) or SIV (SIVpack) packaging constructs using the three-plasmid trans complementation assays. Towards this end, either homologous or heterologous transfer and packaging vectors were cotransfected along with the env expression plasmid into African green monkey kidney Cos cells and the virus was harvested 72 h posttransfection, was processed for virion RNA and proteins, and was used to infect human HeLa CD4+ cells. Finally, the infected cultures were selected with medium containing hygromycin B to identify successfully transduced cells by the appearance of Hygr colonies 10 days postinfection.
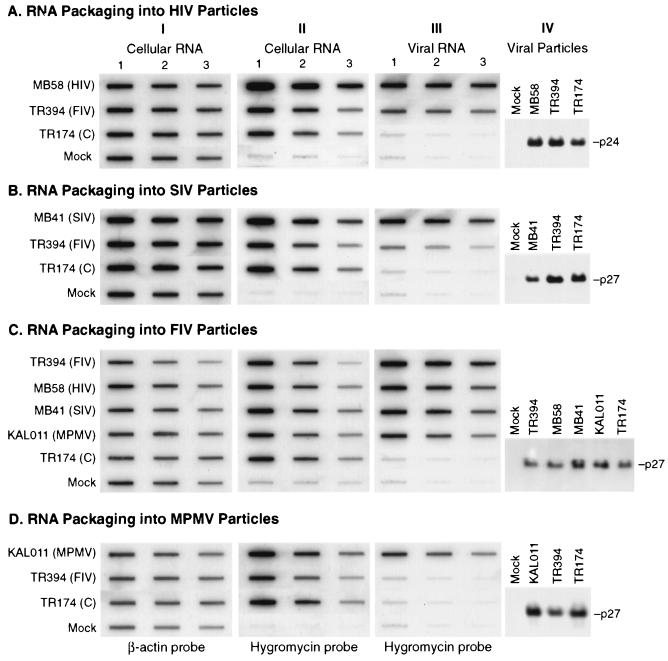
First, we analyzed the relative levels of virions produced in each of the transfected cultures followed by a direct analysis of the presence or absence of the homologous or heterologous RNAs in the virus particles. Western blot analysis of purified virions produced by HIV-1 and SIV packaging constructs revealed that similar amounts of virus particles were produced in each of the transfected cultures (Fig. 3A and B, panels IV). Slot blot analysis of RNA packaged by these particles revealed that both HIV-1 and SIV proteins were capable of packaging FIV transfer vector (TR394) RNA but with a lower efficiency than homologous HIV-1 (MB58) and SIV (MB41) vector RNAs (Fig. 3A and B, panels III). This is despite the fact that the intracellular transfer vector RNAs for each of the viruses were stable and were expressed efficiently (Fig. 3A and B, panels I and II). Together, these results reveal that FIV RNA can be detectably cross-packaged by both HIV-1 and SIV particles.
FIG. 3.
Packaging of retroviral RNAs into HIV-1 (A), SIV (B), FIV (C), and MPMV (D) particles. The transfer vector used in each trans- complementation assay is indicated to the left of panel I or at the top of panel IV. Panels I to III are slot blot analyses of cellular and viral RNAs hybridized with 32P-labeled probes that anneal either to a housekeeping gene, the β-actin (panel I), or the hygromycin gene in the transfer vector (panels II and III). Whole-cell or viral RNAs were isolated from transiently transfected Cos cells in the three-plasmid trans-complementation assay and were DNase treated, diluted, and subjected to slot blot analysis. Panel I represents a 1:2 dilution of 1 μg of cellular RNA (1, 0.5, and 0.25 μg), while panel II represents a 1:5 dilution of 5 μg of cellular RNA (5, 1, and 0.2 μg) blotted. Panel III contains a 1:2 dilution of purified virion RNA harvested from a 3/4 portion of 9 ml of virus-containing medium from the transfected cultures. Panel IV shows the Western blot analyses of the remaining 1/4 portion of the purified virus particles harvested from each transfection in the corresponding trans-complementation assays using appropriate antisera (see Materials and Methods for details). TR174 (C), control transfer vector that does not contain the 5′ LTR, PBS, or the packaging signal; Mock, cells transfected without DNA.
The in vivo transduction assay further revealed trans complementation of the HIV-1 vector (MB58) by the HIV-1 packaging construct (CMVΔR8.2) and the SIV vector (MB41) by the SIV packaging construct (SIVpack). This was evidenced by the transduction of the marker gene (hygromycin) to the target cells and the subsequent appearance of Hygr colonies (Table 1). In addition, RNA of the FIV transfer vector TR394 was also propagated by HIV-1 and SIV virus particles, though to a much lower efficiency than the homologous RNAs (83-fold lower compared to the homologous HIV-1 RNA and 166-fold lower compared to the homologous SIV RNAs) (Table 1). No Hygr colonies were observed when the control transfer vector, TR174, which lacks a packaging signal as well as other sequences necessary for virus replication and integration, was used as the transfer vector in the trans-complementation assays using HIV-1 or SIV particles (Table 1). This ensured that the colonies observed upon cross-packaging of FIV RNA into either HIV-1 or SIV particles were not due to retrofection (nonspecific packaging of random RNA and its subsequent transfer to target cells). Similarly, no Hygr colonies were observed in mock transfections without DNA. Together, these results demonstrate that FIV RNA can be cross-packaged and propagated by both HIV-1 and SIV proteins, although to a much lower efficiency.
TABLE 1.
Packaging of lentivirus and nonlentivirus transfer vectors into HIV, SIV, MPMV, and FIV particles produced with the VSV-G Env expression plasmida
| Transfer vector | Description of transfer vectors | Titers (CFU/ml)b
|
|||
|---|---|---|---|---|---|
| HIV proteins | SIV proteins | MPMV proteins | FIV proteins | ||
| TR394 | Chimeric 5′ FIV LTR,ccis-acting FIV sequencesd and MPMV CTE | 185 ± 17 | 185 ± 56 | 0 | 1,664 ± 42 |
| MB74 | Same as TR394 except that FIV ψ has been replaced with HIV ψ | 391 ± 51 | NDe | ND | 431 ± 175 |
| MB72 | Same as TR394 except that FIV ψ has been replaced with SIV ψ | ND | 659 ± 150 | ND | 343 ± 123 |
| MB87 | Same as TR394 with the insertion of HIV vpu, tat, rev, and RRE | ND | ND | ND | 7,200 ± 910 |
| MB89 | Same as TR394 with the insertion of HIV vpu, tat, rev, and RRE | ND | ND | ND | 2,427 ± 318 |
| MB58 | cis-acting HIV sequencesd and HIV vpu, tat, rev, and RRE | 15,387 ± 3,400 | ND | ND | 640 ± 97 |
| MB41 | cis-acting SIV sequencesd and SIV vpr, tat, rev, and RRE | ND | 30,700 ± 8,614 | ND | 96 ± 14 |
| KAL011 | cis-acting MPMV sequencesd and MPMV CTE | ND | ND | 32,320f | 0 |
| TR174 | SV-HYGRO cassette using 3′ SIV LTR as poly(A)sequences | 0 | 0 | 0 | 0 |
| MOCK | No DNA control | 0 | 0 | 0 | 0 |
No Hygr colonies were observed for any of the vectors when the transfections were performed without the VSV-G Env expression vector, MD.G, or with MD.G by itself, or when mock transfected.
Each value represents a mean of three experiments performed in duplicate except as explained in note f.
In the chimeric FIV 5′ LTR, the U3 has been replaced by the CMV promoter.
cis-acting sequences include PBS, PPT, ψ, att, 5′ LTR, and 3′ LTR.
ND, not done.
Value represents an average of two experiments performed in duplicate; the value for the third experiment was too numerous to count.
Individual Hygr colonies contain FIV proviral DNA.
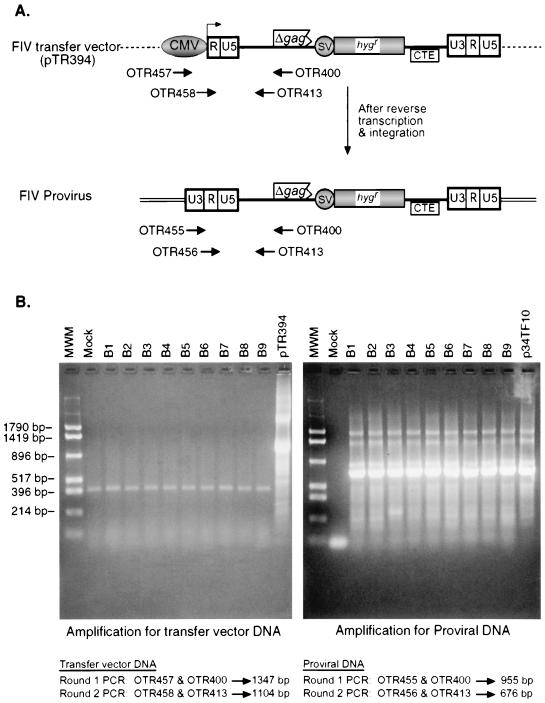
To ensure that the Hygr colonies observed in target cells were due to the successful transfer of FIV RNA by HIV-1 or SIV particles and not due to accidental transfer of the FIV transfer vector plasmid DNA from the transfected cells, individual Hygr colonies were expanded and analyzed by PCR. To distinguish between the two possibilities, primers were designed on the rationale that following delivery of the transfer vector RNA to the target cells and its subsequent reverse transcription, the U3 region of the 5′ LTR of the provirus will be provided by the U3 region of the 3′ LTR, resulting in a wild-type LTR, while the input plasmid DNA would maintain the 5′ chimeric LTR, as shown in Fig. 4A. Amplification of FIV-specific sequences from cell lysates of nine individual clones revealed that only the FIV proviral sequences were present in the individual Hygr colonies, while no contaminating transfer vector containing the 5′ chimeric LTR was detectable (Fig. 4B). These results demonstrate that the Hygr colonies observed upon cross-packaging of FIV RNA by either HIV-1 or SIV particles were due to the actual packaging and successful replication of FIV RNA and not because of carryover of contaminating FIV vector plasmid DNA from the transfected to the target cells.
FIG. 4.
The hygromycin-resistant colonies obtained from the successful transduction of the hygromycin gene by HIV-1 particles contain only the proviral FIV sequences and not FIV transfer vector sequences. (A) Schematic representation of the PCR amplification strategy used to distinguish between proviral FIV DNA from FIV transfer vector DNA. Upon reverse transcription and integration, the resulting FIV provirus should contain a wild-type 5′ LTR rather than the artificially introduced chimeric CMV/LTR. Two rounds of PCR were conducted to amplify the specific products; the first round utilized the outer primers, while the second round utilized the inner primers shown. Small black arrows, primers used in the PCR amplification procedure; CMV, cytomegalovirus promoter/enhancer; SV, simian virus 40 early promoter; hygr, hygromycin B phosphotransferase gene; SD, splice donor; CTE, constitutive transport element from MPMV. (B) Ethidium bromide-stained images of agarose gels containing the amplified nested PCR products. pTR394, transfer vector plasmid containing the chimeric CMV/FIV LTR; p34TF10, FIV molecular clone containing the wild-type FIV LTR. The sizes of the expected products from each round of PCR are shown below the agarose gels. Sizes of the molecular size markers are indicated to the left of the image.
Replacement of FIV packaging signal by that of either HIV-1 or SIV increases the packaging efficiency of FIV RNA by HIV-1 and SIV proteins.
The above in vivo packaging and trans- complementation assays suggested that the FIV packaging signal was being recognized by HIV-1 and SIV proteins. We further rationalized that by replacing the packaging signal of the FIV transfer vector, TR394, with that of HIV-1 or SIV, we should be able to improve the titers obtained by its packaging into HIV-1 or SIV particles. Therefore, the FIV packaging signal (but not the PBS) in TR394 was replaced with that from either HIV-1 or SIV, generating MB74 and MB72, respectively. MB74 and MB72 were tested in the trans-complementation assays using HIV-1 and SIV packaging constructs. As expected, both HIV-1 and SIV virus particles were able to package and propagate MB74 and MB72 with higher efficiency than TR394, as demonstrated by a twofold increase in viral titers in the case of HIV-1 (from 185 ± 17 to 391 ± 51 CFU/ml) and a 3.6-fold increase in viral titers in the case of SIV (from 185 ± 56 to 659 ± 150 CFU/ml) (Table 1). Although we did not look directly at incorporation of vector RNA in the virus particles, the fact that we substituted only the packaging signal of FIV with that of HIV-1 and no other cis-acting sequences (such as PBS, PPT, and att sites) leads us to believe that most of the rise in titers observed was due to increased packaging of the vector RNA and not due to other steps in the virus life cycle such as reverse transcription or integration.
FIV particles can package and propagate HIV-1 and SIV vector RNAs.
Since HIV-1 and SIV could cross-package and propagate FIV RNA, we tested the ability of FIV particles to reciprocally cross-package and propagate HIV-1 or SIV RNAs. Therefore, the FIV packaging construct, MB22, was tested in the trans-complementation assay with either FIV (TR394), HIV-1 (MB58), or SIV (MB41) transfer vectors. Once again, virions were purified from supernatants of the transfected cultures and were analyzed for the presence of either homologous or heterologous RNAs. Western blot analysis of purified virions using anti-FIV sera from infected cats revealed that similar amounts of virus particles were produced by each of the transfected cultures containing FIV, HIV-1, and SIV transfer vector RNAs (Fig. 3C, panel IV). Slot blot analysis of RNAs packaged by FIV particles revealed that both HIV-1 and SIV RNA could get cross-packaged into FIV particles at almost the same efficiency as the homologous FIV RNA (Fig. 3C, panel III) when normalized to the intracellular steady-state levels of transfer vector RNAs (Fig. 3C, panels I and II). These results demonstrate that both HIV-1 and SIV RNAs can be efficiently cross-packaged by FIV particles.
Analysis of the FIV trans-complementation assay revealed that, as expected, FIV particles produced by MB22 were able to efficiently propagate the homologous FIV vector RNA, as evidenced by the appearance of Hygr colonies (Table 1). In addition, FIV particles were able to cross-package and propagate both HIV-1 (MB58) and SIV (MB41) nucleic acids to the target cells. However, in contrast to the equal efficiency of packaging of HIV-1 and SIV RNAs by FIV particles observed in the slot blot analysis, the efficiency of heterologous vector RNA propagation by FIV particles was 2.6- and 17-fold lower for HIV-1 and SIV RNAs, respectively, than for the homologous vector RNA (640 ± 97 and 96 ± 14 CFU/ml for HIV-1 and SIV compared to 1,664 ± 42 CFU/ml for FIV) (Table 1). No Hygr was observed using the control transfer vector, TR174, or in mock transfections. These results demonstrate that both HIV-1 and SIV RNAs can be efficiently cross-packaged by FIV particles; however, FIV particles are less efficient in propagating HIV-1- and SIV-based transfer vector RNAs than the homologous RNA.
We next asked whether the cross-packaging of HIV-1 and SIV RNAs by FIV particles was due solely to the recognition of HIV-1 and SIV packaging signals by FIV proteins. Therefore, we tested the ability of our chimeric FIV transfer vectors, MB74 and MB72, which contained HIV-1 and SIV packaging signals, respectively, to be cross-packaged into FIV particles. These chimeric FIV transfer vectors were successfully packaged and propagated into FIV particles (Table 1), though to a lower efficiency than TR394 since TR394 contains the homologous FIV packaging signal while MB74 and MB72 carry heterologous packaging signals. Together, these results demonstrate that not only can HIV-1 and SIV packaging signals be recognized by FIV proteins but their nucleic acids can also be functionally propagated by FIV particles into the target cells.
Nonlentivirus particles cannot package FIV RNA.
The experiments described above clearly demonstrate that both primate and nonprimate lentiviruses (HIV-1, SIV, and FIV) were promiscuous in their ability to cross-package each other's RNAs. We next asked whether a nonlentiviral primate retrovirus such as MPMV could package and propagate a lentiviral RNA such as that from FIV. Therefore, we tested the ability of RNA from the FIV transfer vector, TR394, to be cross-packaged by MPMV particles in the MPMV trans-complementation assay. TR301 is the MPMV packaging construct that expresses MPMV gag/pol structural genes (Fig. 1C). Slot blot analysis of RNA from MPMV virions revealed that MPMV was unable to package FIV RNA (Fig. 3D, panel III) despite efficient expression of FIV transfer vector RNA intracellularly (Fig. 3D, panels I and II) and equal amounts of MPMV particle production in each transfection (Fig. 3D, panel IV). Finally, as expected from the results of the slot blot analysis, no Hygr colonies were obtained upon infection of the target cells with supernatants from cells cotransfected with the MPMV packaging construct (TR301) and FIV transfer vector, TR394 (Table 1), despite highly efficient transduction of the hygromycin gene by the homologous MPMV transfer vector (Table 1).
Conversely, we asked whether FIV proteins could cross-package MPMV transfer vector RNA. Therefore, we tested the ability of the FIV proteins to cross-package MPMV transfer vector (KAL011) RNA using FIV particles produced by MB22. Interestingly, slot blot analysis of RNA isolated from FIV particles revealed an almost equal level of packaged MPMV transfer vector (KAL011) RNA as the homologous FIV transfer vector (TR394) RNA (Fig. 3C, panel III). But surprisingly, a complete lack of Hygr colonies was observed upon infection of the target cells. These data reveal that FIV particles can efficiently cross-package MPMV RNA; however, this RNA cannot be propagated through the target cells, presumably due to blocks at other steps in the viral replication cycle such as reverse transcription and/or integration.
Altogether, these experiments reveal that while FIV is promiscuous in its ability to cross-package RNAs from other lentiviruses as well as nonlentiviruses, a nonlentivirus such as MPMV is more restrictive in its ability to cross-package heterologous RNAs.
Presence of HIV-1 tat on FIV vectors enhances vector titers.
Earlier, we had observed that the titer obtained from the packaging of RNA from the FIV transfer vector, TR394, was much lower when packaged by FIV particles (Table 1) than the titers of HIV-1- and SIV-based transfer vectors, MB58 and MB41, when packaged by their homologous virus particles (Table 1). MB58 and MB41 contain tat, rev, and RRE in addition to either vpu or vpr, respectively, as opposed to TR394, which does not contain any accessory genes. Therefore, we asked whether the accessory genes from HIV-1 could enhance titers obtained from FIV transfer vector transduction. To address this question, we created an FIV/HIV chimeric transfer vector, MB87 (Fig. 2B), which maintains all the necessary cis-acting sequences from FIV important for virus replication and, in addition, encodes a fragment containing the HIV-1 accessory genes vpu, tat, and rev/RRE, replacing MPMV CTE. Tests of MB87 in the FIV trans-complementation assay revealed that in the presence of HIV-1 accessory genes, the titer of the FIV transfer vector was enhanced fourfold (7,200 ± 910 CFU/ml) upon transduction when compared to TR394 (1,664 ± 42 CFU/ml) (Table 1).
Among the accessory genes of HIV-1 present in MB87, we suspected that tat might play a role in titer enhancement of FIV transfer vectors. We could exclude vpu since it has a cell-specific effect which is not apparent in our producer Cos cells, while we could exclude the rev/RRE regulatory system since MPMV CTE was already present in the vector and was providing analogous functions. Therefore, to determine the effects of Tat on titer enhancement, we decided to delete tat in the chimeric FIV/HIV transfer vector MB87 to create MB89 (Fig. 2B). Abrogation of tat expression in MB89 resulted in decreasing the vector titer to nearly the same level as that of TR394 when packaged and propagated by the homologous FIV proteins (2,427 ± 318 versus 1,664 ± 42 CFU/ml) (Table 1). These results clearly demonstrate that HIV-1 tat enhances the titers of FIV transfer vectors, probably by increasing the amount of genomic RNA available for encapsidation.
DISCUSSION
Currently, FIV-based gene transfer vectors are being studied for their potential to be used for human gene therapy in addition to vectors based on HIV and SIV. FIV, being a feline lentivirus, is more distantly related to HIV and its primate cousin, SIV (3, 8, 27, 29, 40); this evolutionary distance is thought to reduce the likelihood of recombination between FIV and primate lentiviruses that could potentially generate replication-competent variants. A number of studies have raised considerable safety concerns about the use of primate lentiviruses for human gene therapy, including the demonstration that HIV-1 is able to cross-package SIV (31) as well as HIV-2 RNA (14, 15, 21), while reciprocally, SIV can cross-package HIV-1 RNA (43). So far, however, no studies exist that have tested if HIV-1 or SIV can cross-package the genomes of nonprimate lentiviruses such as FIV. In this study, we attempted to address the safety of FIV-based gene transfer systems by analyzing the ability of HIV-1 and SIV particles to cross-package FIV transfer vector RNA and vice versa. Our data demonstrate that FIV is highly promiscuous in its ability to get cross-packaged and propagated by primate lentiviruses such as HIV-1 or SIV but not by a nonlentivirus such as MPMV. Reciprocally, FIV particles can package HIV-1 and SIV RNAs. Interestingly, FIV particles can package MPMV RNA but cannot propagate it further, unlike MPMV particles that cannot package FIV RNA. Finally, we demonstrate that cross-packaging between FIV and primate lentiviruses is due to the recognition of viral packaging signals by the heterologous proteins. Together, these results suggest that there may be more functional similarities between divergent lentiviruses than has been appreciated before and raise important questions about the design and use of FIV-based vectors for human gene therapy.
FIV RNA was readily cross-packaged and propagated by both HIV-1 and SIV particles; however, the efficiency was much lower than that observed for the homologous RNAs in the trans-complementation assays (Table 1). The same was true when HIV-1 and SIV RNAs were cross-packaged by FIV particles (Table 1). Successful transduction of the hygromycin gene in our in vivo packaging assay entailed that, in addition to RNA packaging, reverse transcription and integration must also have taken place. Since we used VSV-G as the envelope protein to pseudotype all our viruses, this eliminated variation in viral entry into the target cells. Therefore, the difference in titers observed could be attributed to differences in either RNA packaging, reverse transcription, and/or integration efficiencies. Slot blot analysis of the amount of viral RNA packaged in the particles provided a direct estimate of the packaging efficiency (Fig. 3). These data revealed that HIV-1 and SIV particles cross-packaged FIV RNA less efficiently than the homologous RNA (Fig. 3A and B, panel III), thus contributing to the lower efficiency of vector transduction observed (Table 1). However, FIV particles were able to package either the homologous FIV RNA or the heterologous HIV-1, SIV, or MPMV RNAs with very similar efficiencies (Fig. 3C, panel III). Yet, their ability to functionally propagate the heterologous RNA was much attenuated in the case of HIV-1 or SIV or was completely abrogated as in the case of MPMV (Table 1). The block in subsequent transduction steps of the hygromycin gene on MPMV RNA could be at either the reverse transcription or integration steps since, being heterologous, the FIV reverse transcriptase and/or integrase may not be able to mediate these steps efficiently using MPMV genomic RNA as the substrate.
Although we observed reciprocity in cross-packaging among lentiviruses, this was not the case with the nonlentivirus MPMV. A direct examination of the RNA content of virions revealed that whereas FIV particles were able to package MPMV RNA, MPMV particles, on the other hand, were unable to package FIV RNA, revealing a more stringent requirement of MPMV for RNA packaging compared to lentiviruses. Nonreciprocal packaging has also been observed in other type C retroviruses where the RNA genome from MLV could be packaged by the proteins of SNV (9, 45, 46) but MLV proteins could not package SNV RNA (6). Unlike the case of FIV particles cross-packaging MPMV RNA, however, the restriction in this case was at the level of the inability of MLV proteins to recognize the SNV packaging signal (6).
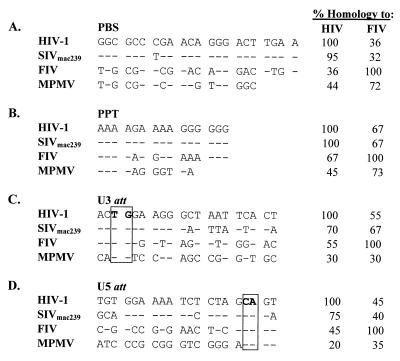
The fact that lentiviruses can not only cross-package but also propagate each other's genomes reveals that they must be promiscuous in their ability to reverse transcribe and integrate the heterologous RNAs. This process, therefore, must involve either (i) recognition of each other's PBS and PPT sequences by the reverse transcriptase and/or (ii) recognition of each other's att sites by the integrase of the packaging virus. We have compared these cis-acting sequences among HIV-1, SIV, FIV, and MPMV. HIV-1 and SIV are nearly identical in their PBS and PPT (Fig. 5A and B); therefore, the difference in transduction efficiencies observed during cross-packaging by these viruses probably lies at the level of integration efficiency. Interestingly, although FIV PBS has only about 36 and 32% sequence homology to HIV-1 and SIV PBS, respectively, compared to 72% to MPMV PBS, FIV proteins were able to both package and propagate HIV-1 and SIV RNAs, when they could only package but not propagate MPMV RNA. This difference could be due to the fact that (i) MPMV PBS and PPT are shorter than those of FIV, HIV-1, and SIV and, therefore, there may be better compatibility of FIV reverse transcriptase for HIV-1 and SIV PBS and/or PPT compared to MPMV; and/or (ii) the FIV integrase may recognize HIV-1 and SIV att sites better than MPMV att sites. Further comparison of the att sites among the four viruses indicates that, as expected, the HIV-1 and SIV U3 and U5 att sites were the most homologous to each other, followed by FIV, while MPMV att sites were the least homologous among the four viruses (Fig. 5C and D). The observed similarities in the att sites follows the relative titers obtained in our in vivo transduction assays, where FIV could propagate the homologous FIV RNA most efficiently, followed by HIV-1 and SIV RNAs, while it could not propagate MPMV RNA at all (Table 1). This observation is in agreement with the studies of Masuda and colleagues that revealed that HIV-1 RNA substituted with FIV att sites was integrated with 70% efficiency compared to wild-type HIV-1, while HIV-1 RNA substituted with att sites from the more distantly related murine leukemia virus was integrated with less than 0.5% of the efficiency of the wild type (20).
FIG. 5.
Comparison of retroviral cis-acting sequences important during reverse transcription and integration, including the following: (A) PBS, primer binding site; (B) PPT, polypurine tract; (C) U3 att, 3′ attachment site in the U3 region of the 3′ LTR; and (D) U5 att, 5′ attachment site in the U5 region of the 5′ LTR. The canonical TG dinucleotide in the U3 att sequences and CA dinucleotide in the U5 att sequences are boxed. All sequences are shown in comparison to HIV-1, where “–” indicates homology with HIV-1, while differences are shown by the actual nucleotide. Columns to the right indicate the percent homology of each viral sequence to either HIV-1 or FIV with homology to self indicated as 100%.
The increased titer observed with FIV/HIV chimeric transfer vector MB87, which contains HIV-1 tat, as opposed to MB89, which lacks tat, when packaged in the homologous FIV particles could be attributed to the trans-activating effects of HIV-1 Tat on the hCMV promoter. HIV-1 Tat, in addition to trans-activating its own LTR, has been reported to trans-activate other viral promoters such as the JC virus late promoter (38) and the human and murine CMV (mCMV) promoters (10, 17). Either direct or indirect binding of Tat to the negative regulatory region upstream of the enhancer region in the mCMV promoter is thought to allow Tat to interact with cellular factors that bind both upstream and downstream regulatory elements, resulting in activation of gene expression (17). The hCMV promoter is used in our FIV transfer vectors to substitute the FIV U3 region due to its weak promoter activity in primate cells (reviewed in reference 13). At present, we do not know if FIV Tat would also trans-activate the hCMV promoter since our FIV packaging construct, MB22, did not express Tat due to the presence of a premature stop codon in orf2 (7). Johnston and colleagues did not observe any effects of FIV Tat on the transduction efficiencies of their CMV-containing FIV transfer vectors when packaged either in the presence or absence of Tat, suggesting that FIV Tat, if at all, may be only a weak trans-activator of the CMV promoter (13). Therefore, we attribute the increase (∼threefold) in the titers of MB87 to the action of HIV-1 Tat on the hCMV promoter.
The fact that only a low level of trans-activation was observed by HIV-1 Tat (∼threefold) compared to what is normally observed (∼100-fold) could be due to the presence of the SV40 origin of replication in our transfer vectors and that of the SV40 large T antigen in our producer Cos cells. Under these conditions, Srinivasakumar and colleagues have observed only partial trans-activation of the HIV-1 LTR by Tat (37). Finally, the slight increase in titers (∼1.5-fold) observed in MB89 (in which tat was mutated) compared to TR394 could be due to the use of the rev/RRE regulatory system in MB89 versus the MPMV CTE that was used in TR394. The MPMV CTE has been shown to function less efficiently in heterologous genomic systems (32).
The reciprocity in packaging between FIV and the primate lentiviruses raises concerns for the proposed safety of lentivirus vectors for human gene transfer studies. These concerns gain further significance from the recent observation that certain strains of FIV are able to establish a low but sustained level of productive infection in human peripheral blood mononuclear cells, as well as in immortalized human cell lines following cell-free infection (12). A low level of FIV replication in human cells combined with our findings that FIV particles can cross-package HIV-1/SIV RNA and vice versa suggests possibilities of recombination between these virus sequences as well as endogenous retroviruses upon coinfection of the same individual. This study further supports concerns regarding the transplantation of organs from nonhuman primates and other animals such as swine into humans (2). Such cross-species transplantation can result in the introduction of foreign retroviruses into the human recipient, allowing such foreign retroviruses and human endogenous retroviruses and/or retrovirus-like elements to interact to generate recombinants with unknown pathogenic potential (46).
Genetic recombination between different retroviruses has previously been documented (46, 47). The RNA genomes of MLV and SNV can not only copackage in vivo but also recombine during minus-strand DNA synthesis (46, 47). The ability of FIV and the primate lentiviruses to copackage and recombine has not yet been examined. However, in this study we have shown that chimeric transfer vectors between FIV and primate lentiviruses such as HIV-1 or SIV (MB72 and -74, packaging signal chimeras; MB87 and -89, accessory gene chimeras; Fig. 2B) were stable. Furthermore, FIV proteins could cross-package and propagate these chimeric vectors efficiently in target cells (Table 1). It would be interesting to see if HIV/FIV chimeric viruses can replicate in a feline model, since such a model may allow the study of determinants of HIV-1 replication and pathogenesis in vivo.
ACKNOWLEDGMENTS
We thank Didier Trono (Salk Institute, La Jolla, Calif.) for providing CMVΔR8.2 and MD.G, Vicente Planelles (University of Rochester, Rochester, N.Y.) for providing HIV-GFP and SIV pack, Eric Hunter (University of Alabama, Birmingham, Ala.) for providing anti-MPMV Gag/Pol antibodies, and Ellen Collisson (Texas A&M University, College Station, Tex.) for providing FIV immune serum. The FIV molecular clone, p34TF10, was obtained from the AIDS Research and Reference Reagent Program of the National Institutes of Health. We thank Farah Mustafa (The University of Texas at Austin, Austin, Tex.), A. Srinivasan (Thomas Jefferson University, Philadelphia, Pa.), and Antonito Panganiban (University of New Mexico, Albuquerque, N. Mex.) for stimulating discussions.
This work was supported by funds from the University Cancer Foundation at the University of Texas M.D. Anderson Cancer Center and in part by grants from the American Heart Association (AHA 9950182N) and the Texas Board of Higher Education (ATP 003657-0096-1997).
REFERENCES
- 1.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bach F H, Robson S C, Winkler H, Ferran C, Stuhlmeier K M, Wrighton C M, Hancock W W. Barriers to xenotransplantation. Nat Med. 1995;1:869–873. doi: 10.1038/nm0995-869. [DOI] [PubMed] [Google Scholar]
- 3.Bachmann M H, Mathiason-Dubard C, Learn G H, Rodrigo A G, Sodora D L, Mazzetti P, Hoover E A, Mullins J I. Genetic diversity of feline immunodeficiency virus: dual infection, recombination, and distinct evolutionary rates among envelope sequence clades. J Virol. 1997;71:4241–4253. doi: 10.1128/jvi.71.6.4241-4253.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bray M, Prasad S, Dubay J W, Hunter E, Jeang K-T, Rekosh D, Hammarskjold M J. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc Natl Acad Sci USA. 1994;4:1256–1260. doi: 10.1073/pnas.91.4.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bukovsky A A, Song J, Naldini L. Interaction of human immunodeficiency virus-derived vectors with wild-type virus in transduced cells. J Virol. 1999;73:7087–7092. doi: 10.1128/jvi.73.8.7087-7092.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Certo J L, Shook B F, Yin P D, Snider J T, Hu W-S. Nonreciprocal pseudotyping: murine leukemia virus proteins cannot efficiently package spleen necrosis virus-based vector RNA. J Virol. 1998;72:5408–5413. doi: 10.1128/jvi.72.7.5408-5413.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Parseval A, Elder J H. Demonstration that orf2 encodes the feline immunodeficiency virus transactivating (Tat) protein and characterization of a unique gene product with partial Rev activity. J Virol. 1999;73:608–617. doi: 10.1128/jvi.73.1.608-617.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elder J H, Phillips T R. Molecular properties of feline immunodeficiency virus (FIV) Infect Agents Dis. 1993;2:361–374. [PubMed] [Google Scholar]
- 9.Embertson J E, Temin H M. Lack of competition results in efficient packaging of heterologous murine retroviral RNAs and reticuloendotheliosis virus encapsidation-minus RNAs by the reticuloendotheliosis virus helper cell line. J Virol. 1987;61:2675–2683. doi: 10.1128/jvi.61.9.2675-2683.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ho W-Z, Harouse J M, Rando R F, Gonczol E, Srinivasan A, Plotkin S A. Reciprocal enhancement of gene expression and viral replication between human cytomegalovirus and human immunodeficiency virus type 1. J Gen Virol. 1990;71:97–103. doi: 10.1099/0022-1317-71-1-97. [DOI] [PubMed] [Google Scholar]
- 11.Ikeda Y, Tomonaga K, Kawaguchi Y, Komoto M, Inoshima Y, Tohya Y, Miyazawa T, Kai C, Mikami T. Feline immunodeficiency virus can infect a human cell line (MOLT-4) but establishes a state of latency in the cells. J Gen Virol. 1996;77:1623–1630. doi: 10.1099/0022-1317-77-8-1623. [DOI] [PubMed] [Google Scholar]
- 12.Johnston J, Power C. Productive infection of human peripheral blood mononuclear cells by feline immunodeficiency virus: implications for vector development. J Virol. 1999;73:2491–2498. doi: 10.1128/jvi.73.3.2491-2498.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnston J C, Gasmi M, Lim L E, Leland E, Elder J H, Yee J-K, Jolly D J, Campbell K P, Davidson B L, Sauter S. Minimum requirements for efficient transduction of dividing and nondividing cells by feline immunodeficiency virus vectors. J Virol. 1999;73:4991–5000. doi: 10.1128/jvi.73.6.4991-5000.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaye J F, Lever A M L. Human immunodeficiency virus types 1 and 2 differ in the predominant mechanism used for selection of genomic RNA for encapsidation. J Virol. 1999;73:3023–3031. doi: 10.1128/jvi.73.4.3023-3031.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaye J F, Lever A M L. Nonreciprocal packaging of human immunodeficiency virus type 1 and type 2 RNA: a possible role for the p2 domain of Gag in RNA encapsidation. J Virol. 1998;72:5877–5885. doi: 10.1128/jvi.72.7.5877-5885.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kestler H W, III, Kodama T, Ringler D, Marthas M, Pedersen N, Lackner A, Regier D, Sehgal P, Daniel M, King N, Desrosiers R C. Induction of AIDS in rhesus monkeys by molecularly cloned simian immunodeficiency virus. Science. 1990;248:1109–1112. doi: 10.1126/science.2160735. [DOI] [PubMed] [Google Scholar]
- 17.Kim Y, Risser R. TAR-independent transactivation of the murine cytomegalovirus major immediate-early promoter by the tat protein. J Virol. 1993;67:239–248. doi: 10.1128/jvi.67.1.239-248.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lutz H. Feline retroviruses: a brief review. Vet Microbiol. 1990;23:131–146. doi: 10.1016/0378-1135(90)90143-j. [DOI] [PubMed] [Google Scholar]
- 19.Marthas M L, Banapour B, Sutjipto S, Siegel M E, Marx P A, Gardner M B, Pedersen N C, Luciw P A. Rhesus macaques inoculated with molecularly cloned simian immunodeficiency virus. J Med Primatol. 1989;18:311–319. [PubMed] [Google Scholar]
- 20.Masuda T, Kuroda M J, Harada S. Specific and independent recognition of U3 and U5 att sites by human immunodeficiency virus type 1 integrase in vivo. J Virol. 1998;72:8396–8402. doi: 10.1128/jvi.72.10.8396-8402.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCann E M, Lever A M. Location of cis-acting signals important for RNA encapsidation in the leader sequence of human immunodeficiency virus type 2. J Virol. 1997;71:4133–4137. doi: 10.1128/jvi.71.5.4133-4137.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller R J, Cairns S, Bridges S, Sarver N. Human immunodeficiency virus and AIDS: insights from animal lentiviruses. J Virol. 2000;74:7187–7195. doi: 10.1128/jvi.74.16.7187-7195.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miyazawa T, Kawaguchi Y, Kohmoto M, Tomonaga K, Mikami T. Comparative functional analysis of the various lentivirus long terminal repeats in human colon carcinoma cell line (SW 480 cells) and feline renal cell line (CRFK cells) J Vet Med Sci. 1994;56:895–899. doi: 10.1292/jvms.56.895. [DOI] [PubMed] [Google Scholar]
- 24.Miyazawa T, Kawaguchi Y, Kohmoto M, Sakuragi J, Adachi A, Fukasawa M, Mikami T. Production of feline immunodeficiency virus in feline and non-feline non-lymphoid cell lines by transfection of an infectious molecular clone. J Gen Virol. 1992;73:1543–1546. doi: 10.1099/0022-1317-73-6-1543. [DOI] [PubMed] [Google Scholar]
- 25.Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage F H, Verma I M, Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 26.Naldini L, Verma I M. Lentiviral vectors. In: Friedmann T, editor. The development of human gene therapy. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1999. pp. 47–60. [Google Scholar]
- 27.Olmsted R A, Barnes A K, Yamamoto J K, Hirsch V M, Purcell R H, Johnson P R. Molecular cloning of feline immunodeficiency virus. Proc Natl Acad Sci USA. 1989;86:2448–2452. doi: 10.1073/pnas.86.7.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olmsted R A, Langley R, Roelke M E, Goeken R M, Adger-Johnson D, Goff J P, Albert J P, Packer C, Laurenson M K, Caro T M, et al. Worldwide prevalence of lentivirus infection in wild feline species: epidemiologic and phylogenetic aspects. J Virol. 1992;66:6008–6018. doi: 10.1128/jvi.66.10.6008-6018.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pedersen N C. The feline immunodeficiency virus. In: Levy J A, editor. The Retroviridae. Vol. 2. New York, N.Y: Plenum Press; 1993. pp. 181–228. [Google Scholar]
- 30.Poeschla E M, Wong-Staal F, Looney D J. Efficient transduction of nondividing human cells by feline immunodeficiency virus lentiviral vectors. Nat Med. 1998;4:354–357. doi: 10.1038/nm0398-354. [DOI] [PubMed] [Google Scholar]
- 31.Rizvi T A, Panganiban A. Simian immunodeficiency virus RNA is efficiently encapsidated by human immunodeficiency virus type 1 particles. J Virol. 1993;67:2681–2688. doi: 10.1128/jvi.67.5.2681-2688.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rizvi T A, Schmidt R D, Lew K A, Keeling M E. Rev/RRE-independent Mason-Pfizer monkey virus-constitutive transport element-dependent propagation of SIVmac239 vectors using a single round of replication assay. Virology. 1996;222:457–463. doi: 10.1006/viro.1996.0444. [DOI] [PubMed] [Google Scholar]
- 33.Rizvi T A, Lew K A, Murphy E C, Jr, Schmidt R D. Role of Mason-Pfizer monkey virus constitutive transport element in the propagation of MPMV vectors by genetic complementation using homologous/heterologous env genes. Virology. 1996;224:517–532. doi: 10.1006/viro.1996.0558. [DOI] [PubMed] [Google Scholar]
- 34.Rizvi T A, Schmidt R D, Lew K A. Mason-Pfizer monkey virus (MPMV) constitutive transport element (CTE) functions in a position-dependent manner. Virology. 1997;236:118–129. doi: 10.1006/viro.1997.8728. [DOI] [PubMed] [Google Scholar]
- 35.Sonigo P, Barker C S, Hunter E, Hobson S-W. Nucleotide sequences of Mason-Pfizer monkey virus: an immunosuppressive D-type retrovirus. Cell. 1986;45:375–385. doi: 10.1016/0092-8674(86)90323-5. [DOI] [PubMed] [Google Scholar]
- 36.Sparger E E, Shacklett B L, Gregg L-R, Barry P A, Pedersen N C, Elder J H, Luciw P A. Regulation of gene expression directed by the long terminal repeat of the feline immunodeficiency virus. Virology. 1992;187:165–177. doi: 10.1016/0042-6822(92)90305-9. [DOI] [PubMed] [Google Scholar]
- 37.Srinivasakumar N, Chazal N, Maria C-H, Prasad S, Hammarskjold M L, Rekosh D. The effect of viral regulatory protein expression on gene delivery by human immunodeficiency virus type 1 vectors produced in stable packaging cell lines. J Virol. 1997;71:5841–5848. doi: 10.1128/jvi.71.8.5841-5848.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tada H, Rappaport J, Lashgari M, Amini S, Wong-Staal F, Khalili K. Trans-activation of the JC virus late promoter by the tat protein of type 1 human immunodeficiency virus in glial cells. Proc Natl Acad Sci USA. 1990;87:3479–3483. doi: 10.1073/pnas.87.9.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takeuchi Y, Cosset F C, Lachmann P J, Okada H, Weiss R A, Collins M K L. Type C retrovirus inactivation by human complement is determined by both the viral genome and the producer cell. J Virol. 1994;68:8001–8007. doi: 10.1128/jvi.68.12.8001-8007.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Talbott R T, Sparger E E, Lovelace K M, Fitch W M, Pedersen N C, Luciw P A, Elder J H. Nucleotide sequence and genomic organization of feline immunodeficiency virus. Proc Natl Acad Sci USA. 1989;86:5743–5747. doi: 10.1073/pnas.86.15.5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tomonaga K, Miyazawa T, Kawaguchi Y, Kohmoto M, Inoshima Y, Mikami T. Comparison of the Rev transactivation of feline immunodeficiency virus in feline and non-feline cell lines. J Vet Med Sci. 1994;56:199–201. doi: 10.1292/jvms.56.199. [DOI] [PubMed] [Google Scholar]
- 42.Wang G, Slepushkin V, Zabner J, Keshavjee S, Johnston J C, Sauter S L, Jolly D J, Dubensky T W, Davidson B L, McCray P B. Feline immunodeficiency virus vectors persistently transduce nondividing airway epithelia and correct the cystic fibrosis defect. J Clin Investig. 1999;104:55–62. doi: 10.1172/JCI8390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.White S M, Renda M, Nam N-Y, Klimatcheva E, Zhu Y, Fisk J, Halterman M, Rimel B, Federoff H, Pandya S, Rosenblatt J D, Planelles V. Lentivirus vectors using human and simian immunodeficiency virus elements. J Virol. 1999;73:2832–2840. doi: 10.1128/jvi.73.4.2832-2840.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamamoto J K, Hansen H, Ho E W, Morishita T Y, Okuda T, Sawa T R, Nakamura R M, Pedersen N C. Epidemiologic and clinical aspects of feline immunodeficiency virus infection in cats from the continental United States and Canada and possible mode of transmission. J Am Vet Med Assoc. 1989;194:213–220. [PubMed] [Google Scholar]
- 45.Yang S, Temin H M. A double hairpin structure is necessary for the efficient encapsidation of spleen necrosis virus retroviral RNA. EMBO J. 1994;13:713–726. doi: 10.1002/j.1460-2075.1994.tb06311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yin P H, Hu W-S. RNAs from genetically distinct retroviruses can copackage and exchange genetic information in vivo. J Virol. 1997;71:6237–6242. doi: 10.1128/jvi.71.8.6237-6242.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yin P H, Pathak V K, Rowan A E, Teufel II R J, Hu W-S. Utilization of nonhomologous minus-strand DNA transfer to generate recombinant retroviruses. J Virol. 1997;71:2487–2494. doi: 10.1128/jvi.71.3.2487-2494.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]