Highlights
-
•
Avian influenza virus (AIV) spread to pinnipeds caused a remarkable mortality.
-
•
Phylogenetic analyses of AIV strains isolated from pinniped species was performed.
-
•
AIV strains isolated from pinnipeds bear characteristics of a highly pathogenic form.
-
•
Different evolutionary histories of different influenza virus genes were observed.
-
•
Amino acid substitutions that confer advantages for infecting mammals were found.
Keywords: Avian influenza, HPIAV, H5N1, Clade 2.3.4.4b, Seals, Sea lions, Fur seals
Abstract
Highly pathogenic influenza A virus (HPIAV) H5N1 within the genetic clade 2.3.4.4b has emerged in wild birds in different regions of the world, leading to the death of >70 million birds. When these strains spread to pinniped species a remarkable mortality has also been observed. A detailed genetic characterization of HPIAV isolated from pinnipeds is essential to understand the potential spread of these viruses to other mammalian species, including humans. To gain insight into these matters a detailed phylogenetic analysis of HPIAV H5N1 2.3.4.4b strains isolated from pinniped species was performed. The results of these studies revealed multiple transmission events from birds to pinnipeds in all world regions. Different evolutionary histories of different genes of HPIAV H5N1 2.3.4.4b strains gave rise to the viruses infecting pinnipeds in different regions of the world. European strains isolated from pinnipeds represent a completely different genetic lineage from strains isolated from South American ones. All strains isolated from pinnipeds bear characteristics of a highly pathogenic form for of avian influenza in poultry. Amino acid substitutions, previously shown to confer an adaptive advantage for infecting mammals, were observed in different genes in all pinniped species studied.
1. Introduction
Influenza A viruses (IAVs) are negative sense, single-stranded RNA viruses belonging to the family Orthomyxoviridae. IAV genomes comprise eight genomic RNA segments that encode for at least twelve viral proteins (Neumann et al., 2004).
Viral nomenclature is based on combinations of the two surface glycoproteins of the virus, hemagglutinin (HA) and neuraminidase (NA). Nineteen HA (H1–H19) and nine NA (N1–N9) subtypes are currently circulating in wild aquatic birds (Suttie et al., 2019). Avian Influenza A viruses (AIVs) are sporadically transmitted from waterfowl to domestic avian species. These viruses typically circulate in poultry flocks as low pathogenicity avian influenza virus (LPAIV), causing little to no apparent illness; however, some subtypes, such as H5 and H7, have the potential to mutate into high pathogenicity avian influenza virus (HPAIV) which can cause high mortality rates in domestic avian species (Kaplan and Webby, 2013). Since the 1990s, AIV H5 strains have shown a distinct evolutionary pattern by accumulating mutations and reassortment with other AIV subtypes and have evolved into 10 genetic clades (0–9) and several subclades based on the phylogenetic analysis of its HA gene segment (Sonnberg et al., 2013). In late 2020, HPAIV H5N1 within clade 2.3.4.4b emerged in wild birds and were detected in Africa, Asia, Europe and the Americas, leading to the death of over 70 million birds (Ouoba et al., 2022; Nagy et al., 2022; Cui et al., 2022; Bruno et al., 2023. These H5N1 2.3.4.4b viruses have recently spread to other host species, including wild mammals, raising concern about virus adaptation for persistent transmission in mammalian hosts (EFSA et al., 2022). Recently, HPIAV H5N1 2.3.4.4b strains spread to marine mammal populations of Europe, North and South America (Puryear et al., 2023; Mirolo et al., 2023; Rimondi et al., 2024), where a remarkable and significant cases surge (Leguia et al., 2023) as well as mortality was found (Ulloa et al., 2023). The virus reached South America in October 2022 and rapidly spread southwards through the Pacific Ocean (OFFLU, 2023; Plaza et al., 2024). It arrived to Uruguay at the end of August 2023, causing a great mortality (>2700 animals) of both, South American sea lions and South American fur seals.
In this study, we tested 18 wild dead marine mammals collected from the Uruguayan Atlantic coast in late 2023 for Influenza A virus (IAV), an obtained complete genome sequences from all segments of IAV from a sea lion (Otaria flavescens), and confirmed the presence of HPIAV H5N1 2.3.4.4b viruses in this sample. A detailed genetic characterization of HPIAV H5N1 2.3.4.b isolated from pinnipeds in all regions of the world will be critical to understand the potential spread of these viruses to other mammalian species, including humans.
In order to understand the evolution, adaptation and pandemic potential of HPIAV H5N1 2.3.4.4b strains in pinnipeds worldwide a detailed phylogenetic analysis was performed.
2. Materials and methods
2.1. Sample collection
On November 23th, 2023, 18 fresh oropharyngeal and cloacal swabs were obtained from wild dead marine mammals of varying species from the Uruguayan coast. The samples were placed in transport media composed of phosphate-buffered saline (PBS, pH 7.4) supplemented with 100 U/μL of penicillin and 100 U/μL of streptomycin and immediately shipped at 4̊ C to the laboratory for further analysis. Once arrived, total RNA was extracted from the samples were stored at -80̊ C.
2.2. RNA extraction
QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany) was used for total RNA extraction from samples, following the manufacturer's directions.
2.3. Molecular detection of avian influenza virus
5 μl of extracted ARN was first used to screen for IAV by quantitative reverse transcription polymerase chain reaction (qRT-PCR) for the M gene (Spackman et al., 2002). Each 20 μl was set up in 0.1 ml MicroAmp Fast PCR tubes with 1X TaqMan Fast Virus 1-Step Master Mix (Life Technologies, USA), 10 pmol of each primer, and 5 pmol of the probe. All M gene-positive samples were then tested by qRT-PCR for the H5 (Wang et al., 2002), with the same reaction setup used for the M gene. PCRs were performed on a QuantStudio™ 5 Real-Time PCR thermocycler (Applied Biosystems Inc., USA).
2.4. Full genome amplification and sequence of IAV
Positive samples for the M gene were amplified using one-step RT-PCR with two sets of primers that recognize the conserved 3′ and 5′ segment ends to simultaneously amplify all influenza genomic segments (available at: www.dx.doi.org/10.17504/protocols.io.n2bvj655wlk5/v1).
2.5. Sequencing of amplified PCR products
Sequencing of the amplified segments was performed according to Williams et al. (2023). Libraries were loaded into a FLO-MIN106D R9.4.1 flowcell and sequenced on the GridION X5 sequencing platform (ONT). Base calling and demultiplexing were performed with Guppy 6.3.9 (Oxford Nanopore Technologies, 2003a) using the super accuracy mode. Consensus genomes were generated using the Epi2Me workflow for Flu (Oxford Nanopore Technologies, 2003b) with default parameters.
Complete, high quality nucleotide sequences from HA, NA, M, PB1, NP, NS1 IAV genome segments obtained in this work were deposited in GISAID (EPI_ISL_19354810). Genome sequences for PA and PB2 segments, in which stretches of N were found, can be found under the same accession number.
2.6. Sequences used in the analyses
The Global Initiative on Sharing Avian Influenza Data (GISAID) database was used throughout these studies. Available and comparable sequences of 57 HPIAV H5N1 2.3.4.4b strains isolated from pinnipeds in Europe, North- and South America, as well as Antarctica were used throughout these studies (representing the 85 % of strains isolated from pinnipeds for whom the complete segments genome sequences were known and available in GISAID database). For comparison purposes 72 HPIAV H5N1 isolated from birds and 2 from otters from these same regions were also obtained from GISAID. Only available strains for whom complete HA, NA, M, NP, PA, PB1, PB2 and NS1 gene sequences are known, were used throughout these studies. For strains names, date of isolation, host and accession numbers, see Supplementary Material Table 1 (for originating, submitting laboratories and authors of the sequences see Supplementary Material Table 2). Assignment to H5 2.3.4.4b genetic clade was done using FluSurver (available at http://flusurver.bii.a-star.edu.sg). All strains used throughout these studies were assigned to 2.3.4.4b clade.
Table 1.
Key amino acid analysis of HPIA H5N1 2.3.4.4b strains isolated from different species and regions.
 |
Table 2.
Amino acid polymorphisms found in HPIA H5N1 strains isolated from birds in different regions of the world in relation to A/Seal/Uruguay/H5/2023.
 |
2.7. Sequence alignment
Sequences were aligned using MAFFT version 7 program (Katoh et al., 2019). Only the coding regions of each segment were included in the alignment.
2.8. Bayesian Markov chain Monte Carlo analysis
Population dynamics and structure can influence the shape of a phylogenetic tree depicting the population history of an RNA virus population (Pannell, 2003). For these reasons, a structured coalescent model was used in the Bayesian Markov Chain Monte Carlo (MCMC) analyses of this work as implemented in the BEAST package v.2.5.2 (Bouckaert et al., 2019). This model is capable of overcoming bias in the inference of the genealogy obtained in the analyses and represent a suitable approach in the study of IAV populations (Vaughan et al., 2014). The best nucleotide evolutionary model was determined using jModelTest 2 software (Darriba et al., 2012) from the IQ-TREE web server (Trifinopoulos et al., 2016). Statistical uncertainty in the data was reflected by the 95 % highest probability density (HPD) values. Results were examined using the TRACER v1.6 program (available at: http://beast.bio.ed.ac.uk/Tracer). Convergence was assessed by effective sample sizes (ESS) above 200. Maximum clade credibility trees were generated by means of the use of the Tree Annotator program from the BEAST package. Visualization of the annotated trees were done using the FigTree program v1.4.2 (available at: http://tree.bio.ed.ac.uk).
2.9. Genotypes
Genotypes were classified using the eight segments of each assigned sample according to the scheme described in Youk et al. (2023) and confirmed by phylogenetic analysis (see Supplementary Material Fig. 1). HPIAV H5N1 2.3.4.4b strains isolated from pinnipeds in Europe belongs to genotype A3, while strains isolated in the East of North America belong to genotype A2. Strains isolated in the West of North America as well as strains isolated in South America belong to genotype B3.2 (for constellations found, see Supplementary Material Table 3).
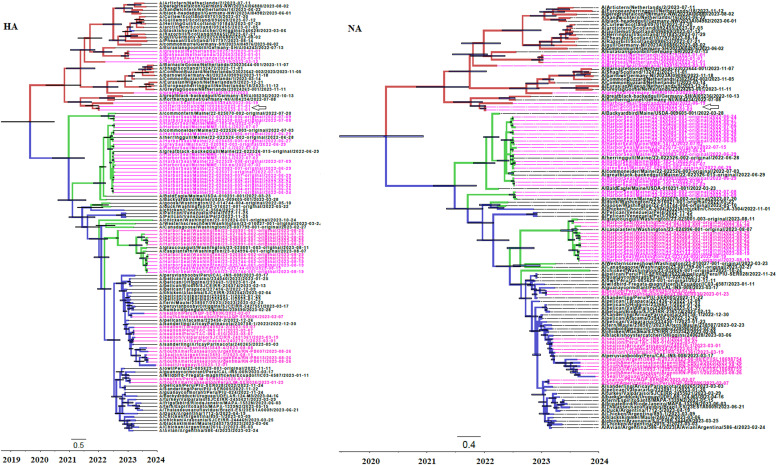
Fig. 1.
Bayesian MCMC phylogenetic tree analysis of HA and NA genes from HPIAV H5N1 2.3.4.4b strains. Maximum clade credibility trees are shown. Trees are rooted to the Most Recent Common Ancestor (MRCA). Time to the MRCA is shown in years at the bottom of the figure. Bar at the bottom of the trees denote time in years. Strains in the trees are shown by name followed by date of isolation. Clusters of strains isolated in Europe, North and South America are shown in red, green and blue, respectively. Node bars indicate the 95 % credibility values of the node heights. Strains isolated from birds are shown in black, while strains isolated from seals are shown in magenta. Strains isolated from otters are indicated by an arrow in magenta in both phylogenetic trees.
2.10. In silico translation
In order to observe the amino acid substitutions found in the proteins of the strains included in these analyses, the nucleotide sequences of each genome segment were translated in silico using software from the MEGA 11 program (Tamura et al., 2021).
3. Results
3.1. HPIAV H5N1 2.3.4.4b circulates in sea lions (Otaria flavescens) of Uruguay by the end of 2023
The analysis of 18 swabs samples obtained from dead sea lions in the Atlantic coast of Uruguay by November 23, 2023, revealed one positive sample for IAV. RNA samples from this specimen were directly amplified by PCR and the sequences were obtained as described in Materials & Methods section, without previous passage in cell cultures. Further analysis revealed that this IAV strain was a H5N1 2.3.4.4b strain. Full-genome IAV genome sequences were obtained from this sample and included in the phylogenetic analyses performed in these studies.
3.2. Phylogeographic analysis of HPIAV H5N1 2.3.4.4b genes circulating in birds and pinnipeds
HPIAV strains isolated from pinnipeds (for whom the complete gene sequences of the 8 IAV segments are known) have been isolated in Europe, North and South America. In order to study the degree of genetic variability and evolution of HPIAV in pinnipeds, a structured coalescent Bayesian MCMC analysis of all gene segments of strains isolated from seals and sea lions of these three regions of the world, as well as comparable gene sequences isolated from birds in these regions was performed.
First, an analysis of virion surface HA and NA genes was performed (see Fig. 1). In both cases, strains in the trees are assigned to different clades according to their region of isolation (Europe, North and South America). HA and NA genes from strains isolated from birds and pinnipeds from the same region share a closer genetic relation among themselves and a more distant genetic relation with birds and pinnipeds of the other geographic regions analyzed (see Fig. 1). Interestingly, although all isolates included in these studies belong to 2.3.4.4b HPIAV clade, strains isolated from pinnipeds belong to different genetic lineages, even in the same geographic location, revealing multiple events of spillover to pinniped populations by different HPIAV strains (see Fig. 1, HA and NA trees). Strains isolated from otters in Europe share the same nodes with European strains isolated from seals (see Fig. 1, HA and NA trees).
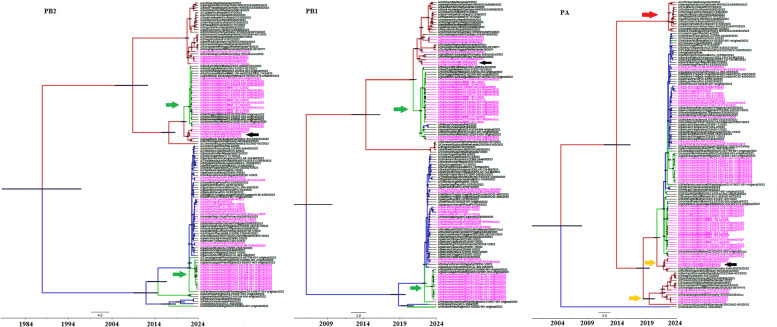
Then, the analysis of polymerase genes PB2 and PB1 revealed a completely different situation, where strains isolated from North American seals belong to two different genetic lineages (Fig. 2). Strains isolated in Maine (USA) have a closely related genetic relationship with strains isolated in Europe, while strains isolated in Washington are closely related to strains isolated in South America (see Fig. 2). Moreover, two different European genetic lineages have been found to circulate in European seals in these two genes (Fig. 2). Interestingly, strains isolated from otters in Scotland map in the same genetic lineage than strains isolated from Scottish seals. In the case of the polymerase PA gene, different genetic lineages are observed for strains isolated in Europe, one strictly composed of strains isolated from birds, while others were found composed of strains circulating in birds and seals (Fig. 2). These reveals that different PA genes were circulating in the bird population of Europe, but not all of them were passed to the seal populations (Fig. 2).
Fig. 2.
Bayesian MCMC phylogenetic tree analysis of PB2, PB1 and PA genes from HPIAV H5N1 2.3.4.4b strains. Maximum clade credibility trees are shown. North American clusters where strains isolated from seals were found are indicated by green arrows in PB2 and PB1 trees. Red arrow in the PA tree indicates a clade of strains only isolated from birds, while clades conformed by strains isolated from birds, seals and otters in Europe are indicated by orange arrows. Strains isolated from otters in Scotland are indicated by black arrows. The rest as same as Fig. 1.
The phylogenetic analysis of the M gene revealed that strains cluster according to their geographic location of isolation (Fig. 3), as was observed in HA and NA genes. In the case of NP and NS genes, strains isolated from seals in North America can be assigned to two completely different genetic lineages as previously observed for PB2 and PB1 genes (Fig. 3). Moreover, the analysis of NS1 gene revealed that strains isolated in Europe can be divided in two different clusters, one containing only strains isolated from birds and another conformed by strains isolated from birds, seals and otters, as observed in the case of PA gene (Fig. 3).
Fig. 3.
Bayesian MCMC phylogenetic tree analysis of M, NP and NS1 genes from HPIAV H5N1 2.3.4.4b strains. Maximum clade credibility trees are shown. The rest as same as Fig. 1, Fig. 2.
The results of these studies revealed a different evolutionary history of different genes in HPIAV H5N1 2.3.4.4b strains included in these studies.
3.3. Genetic characterization of HPIAV H5N1 2.3.4.4b isolated from pinnipeds
Since the phylogenetic analysis of HPIAV isolated from pinnipeds revealed that multiple genetic lineages coexist in time and in different geographic regions, it is important to study the genetic characteristics of those strains in order to understand the pandemic potential and the spread among different mammal hosts. To gain insight into the main genetic characteristics of HPIAV strains isolated from pinnipeds, a detailed analysis of key amino acid substitutions was done for HA, NA, M1, M2, NP, PA, PB1, PB2, NS1, and NS2/NEP genes of these strains.
The HA genes from all HPIAV strains isolated from pinnipeds and included in these studies bear multiple basic amino acids at their cleavage site (PLREKRRKR/GLF) (Table 1), characteristic of a high pathogenicity avian influenza virus (Isoda et al., 2022). Substitutions in the HA protein, which have been proven to increase in vitro binding to human-type receptor (i.e. S133A and S154N), have been also identified in almost all strains isolated from pinnipeds from seven different pinniped species (Table 1) (EFSA, 2024). On the other hand, amino acids Q222 and G224 also belonging to the receptor binding site have been found in all strains isolated from pinnipeds species (Table 1). These two sites have been shown to have an avian-like 2,3-sialic acid receptor binding preference (Mosaad et al., 2023). These substitutions were also present in all strains isolated from different avian species circulating in different continents of the world and included in these studies (Table 1).
All the NA sequences of HPIAV strains isolated from pinnipeds and avian species used in these studies were found without amino acid deletions in the NA stalk region (Table 1). This deletion has been previously associated to the high pathogenicity of H5N1 strains in ducks (Li et al., 2014).
Another important factor to consider is the potential of the H5N1 2.3.4.4b clade viruses to overcome the human/mammal antiviral response. In this regard, it has been
Previous studies have found that certain 2.3.4.4b clade viruses present a substitution in position 52 of NP (Y52/N/H) that allows the evasion of the antiviral activity of the human butyrophilin subfamily 3 member A3 (BTN3A3) protein (EFSA, 2024). This substitution was not found in strains isolated from pinnipeds from the west of North America, South America or Antarctica. Nevertheless, a substitution 52 in NP (Y52H) has been found in strains isolated from pinnipeds in Europe and the east of North America (Maine, USA) (Table 1).
Substitutions related to host adaptation in M protein, like N30D, I43M, T215A, which has been shown to increase the virulence of A/H5N1 subtype in mice and ducks were found in all strains isolated from pinnipeds (Nao et al., 2015; S. Fan et al., 2009) (see Table 1). In addition, a substitution in position 66 in accessory protein PB1-F2 (produced from a + 1 alternate reading frame of PB1 protein), also shown to increase virulence in A/H5N1 viruses in mice but not ducks, has been found in all strains isolated from pinnipeds (Schmolke et al., 2011) (Table 1).
Substitution D701N in the PB2 protein is known to play a prominent role in the adaptation of avian influenza A viruses to mammalian host. This substitution led to an increase in polymerase activity and replication efficiency in mammalian cells and in mouse pathogenicity (Czudai-Matwich et al., 2014). Interestingly, this substitution was found in pinniped species from South America, but not in pinnipeds isolated elsewhere (Table 1). A more in-depth analysis of the 229 available and comparable sequences from PB2 genes from strains isolated in the avian population of South America revealed that only 8 isolates have this substitution. This result suggest that they represented minor variants circulating in the avian population of South America (see Supplementary Material File 1). On the other hand, previous studies have shown that substitution E627K in PB2 increase viral polymerase activity in mammalian cells and is accompanied by improved viral replication at low temperatures (Hatta et al., 2007; Massin et al., 2001). This substitution was only found in strains isolated from pinnipeds in Scotland.
NS segment encodes for at least two proteins, NS1 and NS2 (now known as Nuclear Export Protein, NEP). NS1 protein performs multiple functions that affect IAV replication and virulence (Marc, 2014). In addition to NS1, viral segment 8 encodes a 121 amino acid polypeptide from a spliced form of the segment mRNA transcript (NEP) which is involved in mediating the export of viral ribonucleoproteins (vRNPs) from the host cell nucleus and ensuring that the viral genomic segments are available for packaging on the cellular periphery (Paterson and Fodor, 2012).
Substitutions in NS1 decreasing the host antiviral response in chickens, ferrets, or mice, at positions 42, 149 and 205 have been found in all strains isolated from pinnipeds (Table 1) (Jiao et al., 2008; Imai et al., 2010). Previous studies have shown that a substitution in position 16 of NEP (M16I) leads to an enhancement of avian polymerase activity in mammalian cells (Mänz et al., 2012). This substitution was not found in all seven pinniped species studied (not shown). Recent studies revealed that NEP harbors a highly conserved SUMO-interacting motif (SIM). Disruption of the integrity of this SIM in NEP has deleterious effects on the replication and pathogenicity of AIVs in mammalian hosts, but not in avian hosts (Sun et al., 2023). Notably, SIM wild type motif (LLEVE) is highly conserved in avian isolates of H1-H16 IAVs. This motif was found in all strains isolated from pinnipeds (not shown).
While this work was in process, HPIAV H5N1 2.3.4.4b strains isolated from other three pinniped species is South America and Antarctica. This permitted to compare the substitutions found in key amino acids of these three species with the other pinniped species included in these studies (Table 1). Most of the amino acid substitutions were shared by all pinniped species isolated in South America and Antarctica, with the exception of the substitutions found in PB1-F2 and PB2 in strains isolated from pinnipeds in Antarctica (Table 1).
In order to observe if key amino acid substitutions found in pinniped species were previously circulating in the avian population, the substitutions found in strains isolated from pinniped species were compared with strains isolated from avian species in Europe, North and South America and Antarctica. The results of these studies revealed that the majority of the key amino acid substitutions were already circulating in the avian population of the different regions studied. These results revealed that HPIAV have evolved to acquire these substitutions in the avian population previous to expand in the pinniped species. Nevertheless, an analysis of amino acid substitutions found in the genome of strains isolated from avian species from Europe, North- and South America and Antarctica in relation to the genome of strain isolated from pinniped Otaria flavescens (A/Seal/Uruguay/H5/2023) revealed that although the pinniped specie share most of the sites with avian species isolated in South America, they are not identical, revealing that HPIAV strains continue to evolve in the seal population (see Table 2).
4. Discussion
There is an increasing concern of a new epidemic or pandemic in environments where IAV evolve and spread, since new strains may carry mutations of mammalian adaptation, show enhanced polymerase activity and replication in these species, increased virulence, increased binding to human-like receptors, as well as evasion of immune response (EFSA, 2024). For these reasons, a detailed genetic characterization of HPIAV H5N1 strains isolated in marine mammals like pinnipeds is mandatory to understand its emergence, spread, evolution and pandemic potential.
HPAIV H5 clade 2.3.4.4b infection of mammals was reported in 2021 and 2022 during the ongoing epizootic in birds in Eurasia and the Americas (Puryear et al., 2023). More recently, H5N1 clade 2.3.4.4b infection was confirmed in grey seals (Halichoerus grypus) from coastal waters of the Netherlands and Germany (Mirolo et al., 2023) while an unusual mass mortality events caused by epizootics of HPAIV H5N1 infection have been reported in harbor seals (Phoca vitulina) in North America (Puryear et al., 2023; Lair et al., 2024). By the beginning of 2023, HPIAV H5N1 of the clade 2.3.4.4b were detected in marine mammals of South America, caused the death of thousands of South American sea lions (Otaria flavescens/byronia) (Plaza et al., 2024) and a catastrophic mortality of southern elephant seals (Mirounga leonina) in Argentina (Campagna et al., 2024). The results of these studies revealed multiple events of transmission from birds to pinnipeds in all regions of the world and the capacity of different HPAIV H5N1 2.3.4.4b strains circulating in different regions of the world to infect as well as cause mortality in the seven different pinnipeds species studied (see Fig. 1 and Table 1).
Different evolutionary history of different genes of HPAIV H5N1 2.3.4.4b strains gave rise to the viruses infecting pinnipeds in different regions of the world. European strains isolated from seals constitute a completely different genetic lineage from strains isolated in South America in all genes (see Figs. 1 to 3). North American strains isolated from seals share a close genetic relation with South American strains in surface genes HA and NA, as well as in the M and PA genes. Nevertheless, internal genes like PB2, PB1, NP as well as NS1 gene revealed that two different genetic lineages gave rise to strains isolated in North America: strains isolated from pinnipeds in Maine (who share a close genetic relation with strains isolated from European pinnipeds) and strains isolated from pinnipeds in Washington (who share a close genetic relation with strains isolated in South American pinnipeds) (see Fig. 2, Fig. 3 and Table 1). This is in agreement with recent results revealing the presence of reassortment events in the evolution of H5N1 2.3.4.4b viruses (Cho et al., 2023; Marandino et al., 2023). Moreover, the results of these studies are in agreement with previous ones revealing that genotype A3 viruses can be traced back to H5N1 viruses from the Netherlands that dispersed throughout Europe, while evidence of reassortment involving up to five internal gene segments (PB2, PB1, PA, NP, and NS) were found between the A1 genotype viruses isolated from pinnipeds in North and South America (Youk et al., 2023). These viruses resulted to be genotype B3.2 is in agreement with very recent results (Rivetti et al., 2024) (for constellations found, see Supplementary Material Table 3).
These results are also in agreement with previous studies revealing the transatlantic spread of HPIAV H5N1 by migratory wild birds from Europe to North America (Alkie et al., 2023: Günther et al., 2022) that ultimately gave rise to the strains isolated from pinnipeds in the east of North America (Maine, USA), while other migratory birds with different movement patterns along the pacific (birds breeding in North America and Eurasia and wintering in southern Eurasia) ultimately gave rise to the strains isolated from pinnipeds in the west of North America (Washington, USA) (Peterson et al., 2007). Moreover, the results of these studies revealed an important role of the Pacific route in the spread of H5N1 viruses to the South American region in agreement with recent results (Rivetti et al., 2024) (see Supplementary Material Fig. 2).
Host restriction limits cross-species transmission of AIV from migratory aquatic birds to mammals (Long et al., 2019). These viruses must overcome multiple host range barriers to effectively infect and spread among mammals in both structural and non-structural proteins (Sun et al., 2023). Most of the H5N1 2.3.4.4b strains isolated from pinnipeds bear HA receptor binding domain sites that increase binding to human-type receptor, like the substitutions found in positions 133 and 154, as well as avian-like type receptors sites at positions 222 and 224) (Table 1). These substitutions were already present in strains circulating in the avian population and might represent an evolutionary advantage acquired by the virus to infect mammals and facilitate the spillover to pinnipeds and other mammals as recently reported (Erdelyan et al., 2024). Nevertheless, more studies will be needed to address the role of these particular substitutions in the adaptation to mammalian hosts.
A major species barrier known in the restriction of avian virus to adapt to mammals reside in the PB2 gene (Sun et al., 2023). Substitution D701N in the PB2 protein is known to play a prominent role in the adaptation of avian influenza A viruses to mammalian hosts. This substitution led to an increase in polymerase activity and replication efficiency in mammalian cells and in mouse pathogenicity (Czudai-Matwich et al., 2014). Interestingly, this substitution was found in different pinniped species of South America, but not in pinnipeds isolated elsewhere (Table 1). An analysis of 229 available and comparable PB2 sequences from strains isolated from avian species in South America revealed that only 8 isolates have N at position 701 (see Supplementary Material File 1). These results revealed that this substitution was present in the avian population of South America as a minor variant. Importantly, this study revealed that the evolution of HPAIV permits the selection of minor variants that bear substitutions that may allow them to better adapt to other host and environments (see Table 1). Moreover, whereas most AIV isolates contain glutamic acid (E) at position 627 of PB2. This position is frequently mutated to lysine (K) in human-derived isolates, including H5N1 isolates that cause a high morbidity in humans (Mänz et al., 2012). This substitution was only found in strains isolated from pinnipeds in Scotland, also revealing that minor variants might be selected to adapt to other hosts.
Recent studies have shown that NS2 acts as a cofactor in AIV adaptation to mammalian hosts (Sun et al., 2023). These studies revealed that a conserved SUMO-interacting motif (SIM) in NS2 is required for its avian polymerase-enhancing properties and disrupting SIM impairs AIV replication and pathogenicity in mammalian hosts, but not in avian hosts (Sun et al., 2023). All strains isolated from the seven pinniped species studies bears a characteristic SIM wild type motif (LLEVE) in NS2 (not shown) revealing that the selection of strains bearing this motif may also represent an evolutionary advance to the adaptation to a mammalian host, like pinnipeds. Therefore, adaptation to new mammalian hosts may be determined not only by substitutions in structural proteins but in non-structural proteins as well.
As HPIAV continues to evolve, probably acquiring amino acid substitutions that permits the virus to adapt to different host and environments the pandemic potential of these findings is particularly concerning. During 2023, at least 52,000 pinnipeds died in South America, likely due to the rapid spread of HPAIV H5N1 2.3.4.4b strains. In Peru, 10,457 sea lions were recorded dead. In Chile, >20,000 pinnipeds died, including sea lions, fur seals, and elephant seals (OFFLU 2023). Argentina reported over 18,000 deaths, including a mortality rate of 97.4 % among Southern elephant seal pups born in the 2023 breeding season (Campagna et al. 2023). In Uruguay, between September and December 2023, at least 2713 pinnipeds died (Szteren and Franco-Trecu, in preparation). Finally, in southern Brazil, approximately 1000 pinnipeds were reported dead (de Carvalho-Araujo et al., 2024). This highlights the urgent need for continued surveillance and research to understand the mechanisms behind these substitutions and their implications for both animal and human health.
The results of this work revealed that in addition of having some receptor binding sites that bind preferentially to mammalian-like receptors and a high-pathogenic characteristic cleavage site of HA protein, strains isolated from pinnipeds have substitutions in several internal genes that may provide an evolutionary advantage to adapt to mammalian hosts like pinnipeds (see Table 1). Nevertheless, these valuable findings need to be handled with care, since changes in biological characteristics for specific point mutations and motifs may sometimes only be associated with specific hosts. On the other hand, the results of this work highlight the capacity of HPIAV H5N1 2.3.4.4b strains to acquire amino acid substitutions that permit them a rapid spillover to different host species and its epidemic/pandemic potential (OFFLU, 2023; Campagna et al., 2024). Although key substitutions among strains circulating in avian and pinniped populations in a specific geographic area can be shared, strains isolated from pinnipeds are not necessarily identical to the avian ones (see Table 1, Table 2). This fact reveals that HPIAV continues to evolve in both avian and pinniped population acquiring new substitutions that might have a phenotypic effect. Moreover, substitutions present in minor variants in the avian reservoir in a particular region, can be found in different pinniped species of that particular region. Although these studies used the complete segment genomic sequences of 57 strains isolated from pinnipeds (representing the 85 % of the ones available at the time this work was performed) as new sequences became available it is possible that new amino acid substitutions will be found in key sites of HPIAV genome. More studies will be needed to address the possible effect of these new substitutions in the evolution and pathology of HPIAV.
5. Conclusion
The results of these studies revealed multiple transmission events from birds to pinnipeds in all regions of the world. All strains isolated from seven different pinniped species, isolated from different regions of the world, bears characteristics of a high pathogenic form. Most of the key substitutions found in the proteins of HPIAV strains infecting pinnipeds were already present in the avian population in relation to the geographic area of isolation. These results revealed the selection of HPIAV strains circulating in the avian population are capable of infecting both avian and mammals’ populations. Interestingly, even minor variants carrying key substitutions in non-structural proteins may be selected in the adaptation process to other hosts. Strains isolated from European pinnipeds represent a completely different genetic lineage from strains isolated from pinniped in America. The Pacific route played an important route of spread from the west of North America to South American for birds and pinnipeds. Amino acid substitutions found in the HA receptor binding sites of avian and pinniped species permits to bind to human as well as avian type receptors. Amino acid substitutions, previously shown to confer an adaptive advantage for infecting mammals, were observed in different genes of the seven different pinniped species studied. Enhanced monitoring and preventive measures are crucial to mitigate the risks associated with the spillover of HPIAV to pinnipeds and other mammal host species.
Ethics statement
No live animals were involved in these studies. Samples obtained in Uruguay were from wild dead South American sea lions (Otaria flavescens).
Funding sources
This work was supported by Agencia Nacional de Investigación e Innovación and PEDECIBA, Uruguay. We acknowledge Comisión Sectorial de Investigación Científica, Universidad de la República, Uruguay, for support through Grupos I + D grant.
Author statement
The reason of this letter is to let you know that Mercedes Paz, Valentina Franco-Trecu, Diana Szteren, Alicia Costábile, Cecilia Portela, Alfredo Bruno, Gonzalo Moratorio, Pilar Moreno and Juan Cristina, we are the authors of the manuscript “Understanding the emergence of highly pathogenic Avian Influenza A virus H5N1 in pinnipeds: an evolutionary approach”.
All authors had agreed that Dr. Juan Cristina will be the corresponding author. All authors have read and approved the final version of this revised version of manuscript. The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper. The funding institutions has no role in the decision of publishing of this manuscript. No artificial intelligence was used in any aspect of this work.
CRediT authorship contribution statement
Mercedes Paz: Investigation, Formal analysis. Valentina Franco-Trecu: Investigation, Formal analysis. Diana Szteren: Investigation, Formal analysis. Alicia Costábile: Formal analysis, Data curation. Cecilia Portela: Investigation, Formal analysis. Alfredo Bruno: Writing – review & editing, Formal analysis. Gonzalo Moratorio: Writing – original draft, Funding acquisition. Pilar Moreno: Writing – review & editing, Funding acquisition. Juan Cristina: Writing – review & editing, Writing – original draft, Validation, Supervision, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper. The funding institutions has no role in the decision of publishing of this manuscript.
Acknowledgments
We acknowledge CSIC, Universidad de la República and Institute Pasteur – Montevideo, for encouragement and support. We gratefully acknowledge all data contributors (i.e., the authors and their originating laboratories responsible for obtaining the specimens, and their submitting laboratories for generating the genetic sequences and metadata) for sharing via the GISAID Initiative, on which this research is based. For originating and submitting laboratories as well as authors, see Supplementary Material Table 2. We acknowledge anonymous reviewers for their useful comments to improve the quality of this work.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.virusres.2024.199472.
Contributor Information
Mercedes Paz, Email: mpaz@pasteur.edu.uy.
Valentina Franco-Trecu, Email: vfranco-trecu@fcien.edu.uy.
Diana Szteren, Email: diana@fcien.edu.uy.
Alicia Costábile, Email: acostabile@fcien.edu.uy.
Cecilia Portela, Email: cportela@pasteur.edu.uy.
Pilar Moreno, Email: pmoreno@fcien.edu.uy.
Juan Cristina, Email: cristina@cin.edu.uy.
Appendix. Supplementary materials
Data availability
Data will be made available on request.
References
- Alkie T.N., Byrne A.M.P., Jones M., Mollett B., Bourque L., Lung O., James J., Yason C., Banyard A., Sullivan D., Signore A., Lang A., Sullivan D., Baker M., Dawe B., Brown I., Berhane Y. Recurring trans-atlantic incursion of clade 2.3.4.4b H5N1 viruses by long distance migratory birds from Northern Europe to Canada in 2022/2023. Viruses. 2023;15:1836. doi: 10.3390/v15091836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert R., Vaughan T.G., Barido-Sottani J., Duchene S., Fourment M., Gavryushkina A., et al. BEAST 2.5: an advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2019;15 doi: 10.1371/journal.pcbi.1006650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno A., Alfaro-Núñez A., de Mora D., Armas R., Olmedo M., Garcés J., Vaca M.S., De la Torre E., Jarrin D., Burbano L., Salas J., Imbacuan C., Chanatasig J., Barrionuevo M., Galante M.C., Salas V., Goñi N., Cristina J., Domingues C.S., Oliveira-Montesino L., Gomes-Cardoso F., Reischak D., Garcia-Bereguiain M.A. Phylogenetic analysis reveals that the H5N1 avian influenza A outbreak in poultry in Ecuador in November 2022 is associated with the highly pathogenic clade 2.3.4.4b. Int. J. Infect. Dis. 2023;133:27–30. doi: 10.1016/j.ijid.2023.04.403. [DOI] [PubMed] [Google Scholar]
- Campagna C., Uhart M., Falabella V., Campagna J., Zavattieri V., Vanstreels R.E., Lewis M.N. Catastrophic mortality of southern elephant seals caused by H5N1 avian influenza. Marine Mam. Sci. 2024;40:322–325. doi: 10.1111/mms.13101. [DOI] [Google Scholar]
- Cho A., Si Y., Kim D., Seo Y., Lee D., Kim D., Seo Y., Lee D., Kim D., Lee D., Son Y., Jeong H., Song C., Lee D. Novel avian influenza A(H5N6) virus in wild birds, South Korea. Emerg. Infect. Dis. 2023;30:1285–1288. doi: 10.3201/eid3006.240192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui P., Shi J., Wang C., Zhang Y., Xing X., Kong H., Yan C., Zeng X., Liu L., Tian G., Li C., Deng G., Chen H. Global dissemination of H5N1 influenza viruses bearing the clade 2.3.4.4b HA gene and biologic analysis of the ones detected in China. Emerg. Microbe. Infect. 2022;11:1693–1704. doi: 10.1080/22221751.2022.2088407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czudai-Matwich V., Otte A., Matrosovich M., Gabriel G., Klenk H.D. PB2 mutations D701N and S714R promote adaptation of an influenza H5N1 virus to a mammalian host. J. Virol. 2014;88:8735–8742. doi: 10.1128/JVI.00422-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D., Taboada G.L., Doallo R., Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat. Method. 2012;30:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Carvalho-Araujo A., Cho A.Y., Silva L.M.N., Corrêa T.C., de Souza G.C., Albuquerque A.S., Macagnan E., Kolesnikvoas C.K.M., Meurer R., Vieira J.V., Lemos G.G., Barreto A.S., Dick J.L., Groch K.R., de Castilho P.V., Amgarten D., Malta F., Miller M., Dorlass E.G., Palameta S., Lee S.H., Arns C.W., Durigon E.L., Pinho J.R.R., Lee D.H., Ferreira H.L. Mortality in sea lions is associated with the introduction of the H5N1 clade 2.3.4.4b virus in Brazil October 2023: whole genome sequencing and phylogenetic analysis. BMC Vet. Res. 2024;20:285. doi: 10.1186/s12917-024-04137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan S., Deng G., Song J., Tian G., Suo Y., Jiang Y., Guan Y., Bu Z., Kawaoka Y., Chen H. Two amino acid residues in the matrix protein M1 contribute to the virulence difference of H5N1 avian influenza viruses in mice. Virology. 2009;384:28–32. doi: 10.1016/j.virol.2008.11.044. [DOI] [PubMed] [Google Scholar]
- Günther A., Krone O., Svansson V., Pohlmann A., King J., Hallgrimsson G.T., Skarphéðinsson K.H., Sigurðardóttir H., Jónsson S.R., Beer M., Brugger B., Harder T. Iceland as stepping stone for spread of highly pathogenic avian influenza virus between Europe and North America. Emerg. Infect. Dis. 2022;28:2383–2388. doi: 10.3201/eid2812.221086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatta M., Hatta Y., Kim J.H., Watanabe S., Shinya K., Nguyen T., Lien P.S., Le Q.M., Kawaoka Y. Growth of H5N1 influenza A viruses in the upper respiratory tracts of mice. PLoS Pathog. 2007;3:1374–1379. doi: 10.1371/journal.ppat.0030133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai H., Shinya K., Takano R., Kiso M., Muramoto Y., Sakabe S., Murakami S., Ito M., Yamada S., Le M.T., Nidom C.A., Sakai-Tagawa Y., Takahashi K., Omori Y., Noda T., Shimojima M., Kakugawa S., Goto H., Iwatsuki-Horimoto K., Horimoto T., Kawaoka Y. The HA and NS genes of human H5N1 influenza A virus contribute to high virulence in ferrets. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isoda N., Onuma M., Hiono T., Sobolev I., Lim H.Y., Nabeshima K., Honjyo H., Yokoyama M., Shestopalov A., Sakoda Y. Detection of new H5N1 high pathogenicity avian influenza viruses in winter 2021–2022 in the far east, which are genetically close to those in Europe. Viruses. 2022;4:2168. doi: 10.3390/v14102168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao P., Tian G., Li Y., Deng G., Jiang Y., Liu C., Liu W., Bu Z., Kawaoka Y., Chen H. A single-amino-acid substitution in the NS1 protein changes the pathogenicity of H5N1 avian influenza viruses in mice. J. Virol. 2008;82:1146–1154. doi: 10.1128/JVI.01698-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan B.S., Webby R.J. The avian and mammalian host range of highly pathogenic avian H5N1 influenza. Virus Res. 2013;178:3–11. doi: 10.1016/j.virusres.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Rozewicki J., Yamada K.D. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinformat. 2019;4:1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lair S., Quesnel L., Signore A.V., Delnatte P., Embury-Hyatt C., Nadeau M.S., Lung O., Ferrell S.T., Michaud R., Berhane Y. Outbreak of highly pathogenic avian influenza A(H5N1) virus in seals, St. Lawrence Estuary, Quebec, Canada. Emerg. Infect. Dis. 2024;30:1133–1143. doi: 10.3201/eid3006.231033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leguia M., Garcia-Glaessner A., Muñoz-Saavedra B., Juarez D., Barrera P., Calvo-Mac C., Jara J., Silva W., Ploog K., Amaro L., Colchao-Claux P., Johnson C.K., Uhart M.M., Nelson M.I., Lescano J. Highly pathogenic avian influenza A (H5N1) in marine mammals and seabirds in Peru. Nat. Commun. 2023;14:5489. doi: 10.1038/s41467-023-41182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Chen S., Zhang X., Fu Q., Zhang Z., Shi S., Zhu Y., Gu M., Peng D., Liu X. A 20-amino-acid deletion in the neuraminidase stalk and a five-amino-acid deletion in the NS1 protein both contribute to the pathogenicity of H5N1 avian influenza viruses in mallard ducks. PLoS ONE. 2014;9:e95539. doi: 10.1371/journal.pone.0095539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J.S., Mistry B., Haslam S.M., Barclay W.S. Host and viral determinants of influenza A virus species specificity. Nat. Rev. Microbiol. 2019;17:67–81. doi: 10.1038/s41579-018-0115-z. [DOI] [PubMed] [Google Scholar]
- Marandino A., Tomas G., Panzera Y., Leizagoyen C., Perez R., Bassetti L., Negro R., Rodríguez S., Perez R. Spreading of the high-pathogenicity avian influenza (H5N1) virus of clade 2.3.4.4b into Uruguay. Viruses. 2023;15:1906. doi: 10.3390/v15091906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mänz B., Brunotte L., Reuther P., Schwemmle M. Adaptive mutations in NEP compensate for defective H5N1 RNA replication in cultured human cells. Nat. Commun. 2012;3(802) doi: 10.1038/ncomms1804. [DOI] [PubMed] [Google Scholar]
- Marc D. Influenza virus non-structural protein NS1: interferon antagonism and beyond. J. Gen. Virol. 2014;95:2594–2611. doi: 10.1099/vir.0.069542-0. [DOI] [PubMed] [Google Scholar]
- Massin P., van der Werf S., Naffakh N. Residue 627 of PB2 is a determinant of cold sensitivity in RNA replication of avian influenza viruses. J. Virol. 2001;75:5398–5404. doi: 10.1128/JVI.75.11.5398-5404.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirolo M., Pohlmann A., Ahrens A.K., Kühl B., Rubio-Garcìa A., Kramer K., Meinfelder U., Rosenberger T., Morito H.L., Beer M., Ludlow M., Wohlsein P., Baumgärtner W., Harder T., Osterhaus A. Highly pathogenic avian influenza A virus (HPAIV) H5N1 infection in two European grey seals (Halichoerus grypus) with encephalitis. Emerg. Microbe. Infect. 2023;12 doi: 10.1080/22221751.2023.2257810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosaad Z., Elhusseiny M., Zanaty A., Fathy M.M., Hagag N.M., Mady W.H., Said D., Elsayed M.M., Erfan A.M., Rabie N., et al. Emergence of highly pathogenic avian influenza A virus (H5N1) of clade 2.3.4.4b in Egypt, 2021–2022. Pathogens. 2023;12(90) doi: 10.3390/pathogens12010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A., Černíková L., Stará M. A new clade 2.3.4.4b H5N1 highly pathogenic avian influenza genotype detected in Europe in 2021. Arch. Virol. 2022;167:1455–1459. doi: 10.1007/s00705-022-05442-6. [DOI] [PubMed] [Google Scholar]
- Nao N., Kajihara M., Manzoor R., Maruyama J., Yoshida R., Muramatsu M., Miyamoto H., Igarashi M., Eguchi N., Sato M., Kondoh T., Okamatsu M., Sakoda Y., Kida H., Takada A. A single amino acid in the M1 protein responsible for the different pathogenic potentials of H5N1 highly pathogenic avian influenza virus strains. PLoS ONE. 2015;10 doi: 10.1371/journal.pone.0137989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann G., Brownlee G.G., Fodor E., Kawaoka Y. Orthomyxovirus replication, transcription, and polyadenylation. Curr. Top. Microbiol. Immunol. 2004;283:121–143. doi: 10.1007/978-3-662-06099-5_4. [DOI] [PubMed] [Google Scholar]
- Ouoba L.B., Habibata-Zerbo L., Zecchin B., Barbierato G., Hamidou-Ouandaogo S., Palumbo E., Giussani E., Bortolami A., Niang M., Traore-Kam A., Terregino C., Guitti-Kindo M., Angot A., Guigma D., Barro N., Fusaro A., Monne I. Emergence of a reassortant 2.3.4.4b highly pathogenic H5N1 avian influenza virus containing H9N2 PA gene in Burkina Faso, West Africa, in 2021. Viruses. 2022;14:1901. doi: 10.3390/v14091901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OFFLU, WHAH, FAO, 2023. Continued Expansion of High Pathogenicity Avian Influenza H5 in Wildlife in South America and Incursion into the Antarctic region. https://www.offlu.org/wp-content/uploads/2023/12/OFFLU-wildlife-statement-no.-II.pdf (accessed 1 December 2023).
- Oxford Nanopore Technologies, 2003a. https://community.nanoporetech.com/docs/prepare/library_prep_protocols/Guppy-protocol/v/gpb_2003_v1_revax_14dec2018/guppy-software-overview (accessed 1 December 2023).
- Oxford Nanopore Technologies, 2003b. https://labs.epi2me.io/workflows/wf-flu/ (accessed 1 December 2023).
- Pannell J.R. Coalescence in a metapopulation with recurrent local extinction and recolonization. Evolut. (N.Y.) 2003;57:949–961. doi: 10.1111/j.0014-3820.2003.tb00307.x. [DOI] [PubMed] [Google Scholar]
- Peterson A.T., Benz B.W., Papes M. Highly pathogenic H5N1 avian influenza: entry pathways into North America via bird migration. PLoS ONE. 2007;2:e261. doi: 10.1371/journal.pone.0000261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson D., Fodor E. Emerging roles for the influenza A virus nuclear export protein (NEP) PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1003019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaza P.I., Gamarra-Toledo V., Rodríguez-Euguí J., Rosciano N., Lambertucci S.A. Pacific and Atlantic sea lion mortality caused by highly pathogenic avian influenza A(H5N1) in South America. Travel. Med. Infect. Dis. 2024;59 doi: 10.1016/j.tmaid.2024.102712. [DOI] [PubMed] [Google Scholar]
- Puryear W., Sawatzki K., Hill N., Foss A., Stone J.J., Doughty L., Walk D., Gilbert K., Murray M., Cox E., Patel P., Mertz Z., Ellis S., Taylor J., Fauquier D., Smith A., DiGiovanni R.A.Jr, van de Guchte A., Gonzalez-Reiche A.S., Khalil Z., van Bakel H., Torchetti M.K., Lantz K., Lenoch J.B., Runstadler J. Highly pathogenic avian influenza A(H5N1) virus outbreak in New England Seals, United States (2023) Emerg. Infect. Dis. 2023;29:786–791. doi: 10.3201/eid2904.221538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimondi A., Vanstreels R.E.T., Olivera V., Donini A., Lauriente M.M., Uhart M.M. Highly pathogenic avian influenza A(H5N1) viruses from multispecies outbreak, Argentina, August 2023. Emerg. Infect. Dis. 2024;30:812–814. doi: 10.3201/eid3004.231725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivetti A.V., Jr., Reischak D., de Oliveira C.H.S., Otaka J.N.P., Domingues C.S., Freitas T.L., Cardoso F.G., Montesino L.O., da Silva A.L.S., Camillo S.C.A., Malta F., Amgarten D., Goés-Neto A., Aguiar E.R.G.R., de Almeida I.G., Pinto C.A., Fonseca A.A., Jr., Camargos M.F. Phylodynamics of avian influenza A(H5N1) viruses from outbreaks in Brazil. Virus Res. 2024;347 doi: 10.1016/j.virusres.2024.199415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmolke M., Manicassamy B., Pena L., Sutton T., Hai R., Varga Z.T., Hale B.G., Steel J., Pérez D.R., García-Sastre A. Differential contribution of PB1-F2 to the virulence of highly pathogenic H5N1 influenza A virus in mammalian and avian species. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnberg S., Webby R.J., Webster R.G. Natural history of highly pathogenic avian influenza H5N1. Virus Res. 2013;178:63–77. doi: 10.1016/j.virusres.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spackman E., Senne D.A., Myers T.J., Bulaga L.L., Garber L.P., Perdue M.L., Lohman K., Daum L.T., Suarez D.L. Development of a real-time reverse transcriptase PCR assay for type A influenza virus and the avian H5 and H7 hemagglutinin subtypes. J. Clin. Microbiol. 2002;40:3256–3260. doi: 10.1128/JCM.40.9.3256-3260.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Kong H., Yu M., Zhang Z., Zhang H., Na L., Qu Y., Zhang Y., Chen H., Wang X. The SUMO-interacting motif in NS2 promotes adaptation of avian influenza virus to mammals. Sci. Adv. 2023;9 doi: 10.1126/sciadv.adg5175. eadg5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttie A., Deng Y.M., Greenhill A.R., Dussart P., Horwood P.F., Karlsson E.A. Inventory of molecular markers affecting biological characteristics of avian influenza A viruses. Virus Gene. 2019;55:739–768. doi: 10.1007/s11262-019-01700-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021;38:3022–3027. doi: 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifinopoulos J., Nguyen L.T., von Haeseler A., Minh B.Q. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucl. Acid. Res. 2016;44:W232–W235. doi: 10.1093/nar/gkw256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulloa M., Fernández A., Ariyama N., Colom-Rivero A., Rivera C., Nuñez P., Sanhueza P., Johow M., Araya H., Torres J.C., Gomez P., Muñoz G., Agüero B., Alegría R., Medina R., Neira V., Sierra E. Mass mortality event in South American sea lions (Otaria flavescens) correlated to highly pathogenic avian influenza (HPAIV) H5N1 outbreak in Chile. Vet. Q. 2023;43:1–10. doi: 10.1080/01652176.2023.2265173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan T.G., Kühnert D., Popinga A., Welch D., Drummond A.J. Efficient Bayesian inference under the structured coalescent. Bioinformatics. 2014;30:2272–2279. doi: 10.1093/bioinformatics/btu201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Li Y., Zhang F., Jiang N., Zhuang Q., Hou G., Jiang L., Yu J., Yu X., Liu H., Zhao C., Yuan L., Huang B., Wang K. Reverse transcription recombinase-aided amplification assay for H5 subtype avian influenza virus. Virol. J. 2002;19(129) doi: 10.1186/s12985-022-01807-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams T.G.S., Snell L.B., Alder C., Charalampous T., Alcolea-Medina A., Sehmi J.K., Al-Yaakoubi N., Humayun G., Miah S., Lackenby A., Zambon M., Batra R., Douthwaite S., Edgeworth J.D., Nebbia G. Feasibility and clinical utility of local rapid Nanopore influenza A virus whole genome sequencing for integrated outbreak management, genotypic resistance detection and timely surveillance. Microb. Genom. 2023;9 doi: 10.1099/mgen.0.001083. mgen001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youk S., Torchetti M.K., Lantz K., Lenoch J.B., Killian M.L., Leyson C., Bevins S.N., Dilione K., Ip H.S., Stallknecht D.E., Poulson R.L., Suarez D.L., Swayne D.E., Pantin-Jackwood M.J. H5N1 highly pathogenic avian influenza clade 2.3.4.4b in wild and domestic birds: introductions into the United States and reassortments, December 2021-April 2022. Virology. 2023;587 doi: 10.1016/j.virol.2023.109860. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.