Abstract
Chemokines induce chemotaxis, cell migration, and inflammatory responses. We report the identification of an interleukin-8 (IL-8) homolog, termed vIL-8, encoded within the genome of Marek's disease virus (MDV). The 134-amino-acid vIL-8 shares closest homology to mammalian and avian IL-8, molecules representing the prototype CXC chemokine. The gene for vIL-8 consists of three exons which map to the BamHI-L fragment within the repeats flanking the unique long region of the MDV genome. A 0.7-kb transcript encoding vIL-8 was detected in an n-butyrate-treated, MDV-transformed T-lymphoblastoid cell line, MSB-1. This induction is essentially abolished by cycloheximide and herpesvirus DNA polymerase inhibitor phosphonoacetate, indicating that vIL-8 is expressed with true late (γ2) kinetics. Baculovirus-expressed vIL-8 was found to be secreted into the medium and shown to be functional as a chemoattractant for chicken peripheral blood mononuclear cells but not for heterophils. To characterize the function of vIL-8 with respect to MDV infection in vivo, a recombinant MDV was constructed with a deletion of all three exons and a soluble-modified green fluorescent protein (smGFP) expression cassette inserted at the site of deletion. In two in vivo experiments, the vIL-8 deletion mutant (RB1BvIL-8ΔsmGFP) showed a decreased level of lytic infection in comparison to its parent virus, an equal-passage-level parent virus, and to another recombinant MDV containing the insertion of a GFP expression cassette at the nonessential US2 gene. RB1BvIL-8ΔsmGFP retained oncogenicity, albeit at a greatly reduced level. Nonetheless, we have been able to establish a lymphoblastoid cell line from an RB1BvIL-8ΔsmGFP-induced ovarian lymphoma (MDCC-UA20) and verify the presence of a latent MDV genome lacking vIL-8. Taken together, these data describe the identification and characterization of a chemokine homolog encoded within the MDV genome that is dispensable for transformation but may affect the level of MDV in vivo lytic infection.
Marek's disease (MD) is a contagious, lymphoproliferative disease of domestic chickens in which mononuclear infiltration, demyelination of peripheral nerves, and T-cell lymphomas are common features (52). The etiologic agent of MD is a lymphotropic herpesvirus, MD virus (MDV), which can elicit rapid-onset T-cell lymphomas in chickens within several weeks of infection (reviewed in references 12 and 56). The severity of the symptoms usually correlates with the extent of viral replication in target cells. While the exact route of infection is not completely understood, cytolytic infection of B cells, followed by the latent infection of T cells, is thought to be among the crucial steps. During this initial process, a productive infection requires the successful recruitment of the host target cells, without eliciting strong host immune responses. The virus enters a latent state in T cells within 1 to 2 weeks postinfection and thereafter induces malignant transformation in this cell type. Vaccines that target this initial stage and diminish the virulence of incoming MDV have been developed. Despite this success, sporadic outbreaks of MD, due to newly evolved strains of MDV with increased virulence, have been reported (67). Knowledge of the pathways by which MDV infects and responds to cells of the host immune system is crucial to our understanding of the tumor biology of MD and the development of improved control strategies.
A number of gammaherpesviruses express genes related to cellular chemokines (47). Chemokines are collectively known as a family of structurally related cytokines that induce chemotaxis, or directed migration, of leukocyte subsets to the sites invaded by microbes (1). Thus, chemokines play important roles in cellular inflammation, both in the protective responses to the intruding pathogens and in the pathological processes associated with infection (55). Chemokines have been categorized into four subfamilies according to their protein structure, namely, CXC (or α), CC (or β), CXXXC (CX3C) fractalkine, and lymphotactin (1). The basis for this structural classification is the spacing of four invariable cysteines in all CXC and CC chemokines. The first two cysteines of CXC (CXXXC) chemokines are separated by 1 (3) intervening amino acid residue(s), while those in the CC subfamily are adjacent. Lymphotactin, on the other hand, is distinguished by possessing only 2, rather than 4, cysteines (29).
Structural analysis of interleukin 8 (IL-8), the prototype CXC chemokine, helped to define the critical residues that interact with the receptor to activate signaling pathways in the leukocytes that they recruit (15). IL-8 was originally identified as a monocyte-derived factor that attracts neutrophils in Boyden chamber assays (60, 66, 70). IL-8 recruits neutrophils in vivo by promoting cell adhesion and transendothelial migration abilities (2, 62). The recruited target cells become activated and exhibit biochemical properties, such as degranulation, oxidative burst, and calcium influx, leading to the enhancement of killing (5, 6). Viral infections are often accompanied by the upregulation of cellular IL-8 or other related chemokines (26, 41–43, 64).
The IL-8 homolog of chickens, 9E3/CEF4, was originally identified as an upregulated gene in Rous sarcoma virus-transformed cells (9, 64). This chemokine and a homologous chemokine, K60, were recently cloned from an activated chicken macrophage cell line cDNA library (28, 61) and show homology to IL-8 and growth-regulated protein alpha (Gro-α) (28, 38, 61). 9E3/CEF4, like IL-8, is greatly upregulated at sites of wounding (37) and appears to mitigate the healing cascade (36). Overproduction of IL-8 can be part of the host defense system but has been implicated also in inflammatory disorders, such as psoriasis (11) and rheumatoid arthritis (48). In some instances, IL-8-like chemokines, such as 9E3/CEF4, display strong angiogenic properties with a consequence of stimulating tumor cell growth (37) as well as stimulating growth of chick embyro fibroblasts (CEF) in culture (7).
We report here the identification and characterization of a CXC chemokine encoded within the repeated sequences flanking the unique long (UL) region of the MDV genome. This chemokine has been termed vIL-8 because of its high level of homology to cellular IL-8 families, including chicken 9E3/CEF4 and K60 (61). We have found that the MDV gene encoding vIL-8 is expressed with true late kinetics, its expression being abolished in both cycloheximide (CHX) and phosphonoacetic acid (PAA)-treated cells. Using a baculovirus expression system, we have found that vIL-8 is secreted and functions as a chemoattractant for chicken mononuclear cells but not for heterophils. In addition, to explore the function of vIL-8 during the course of MDV infection, both copies of the vIL-8 genes in the RB1B genome were replaced with a soluble-modified green fluorescent protein (smGFP) expression cassette, yielding a vIL-8 null mutant, termed RB1BvIL-8ΔsmGFP. In this paper, we describe the growth kinetics of RB1BvIL-8ΔsmGFP in cell culture, the oncogenicity of RB1BvIL-8ΔsmGFP in chickens, and the establishment of a T-lymphoblastoid cell line derived from tumors induced by RB1BvIL-8ΔsmGFP infection.
MATERIALS AND METHODS
Cells and viruses.
MSB-1 cells (3) were grown at 37°C in RPMI 1640 supplemented with 10% fetal bovine serum in the presence of 5% CO2. Virus replication was induced by the treatment of log-phase cells with 3 mM n-butyrate for 48 h or various time periods as noted. For MDV propagation, secondary CEF were prepared essentially as described (40) using specific-pathogen-free (SPF), 10-day-old embryonated eggs (Sunrise Inc., Catskill, N.Y.). MDV strain RB1B, a very virulent pathotype virus (58), was used for recombinant construction and was obtained from Robin W. Morgan, University of Delaware.
RACE.
The assays for rapid amplification of cDNA ends (RACE) were essentially identical to the procedures reported by Frohman et al. (19) with the following modifications. Specifically, polyadenylated [poly(A)] RNA was purified using oligo(dT) cellulose (type 7; Pharmacia, Piscataway, N.J.) spin columns. For first-strand synthesis, 2.5 μg of poly(A) RNA was primed with SS-dT oligonucleotides (Table 1), for 3′ RACE or dT20 for 5′ RACE, and reverse transcription (RT) was carried out using avian myeloblastosis virus-reverse transcriptase (Boehringer Mannheim, Indianapolis, Ind.) at 42°C for 1 h using the conditions specified by the manufacturer. The resulting first-strand cDNA was used directly as the DNA template in 3′ RACE. The DNA template for 5′ RACE was prepared by further treatment of the first strand cDNA with 60 U of terminal deoxynucleotidyltransferase (GIBCO-BRL, Gaithersburg, Md.) in the presence of 0.2 mM dATP for 15 min at 37°C. The second-strand synthesis was then primed with SS-dT primer and incubated with 120 U of T4 DNA polymerase; 24 U of Escherichia coli DNA ligase; and 5 U of RNase H in a buffer containing 0.2 mM dNTP, 100 mM KCl, 10 mM ammonium sulfate, 5 mM MgCl2, 0.15 mM β NAD, 20 mM Tris-HCl (pH 7.5), and 50 μg of bovine serum albumin/ml for 4 h at 15°C. Primers used in 5′ RACE were single stranded (SS) (Table 1). Primers used in 3′ RACE were SS and vIL-8F4 (Table 1). PCR conditions for denaturation, annealing, and polymerization were 93°C for 50 s, 56°C for 1 min, and 68°C for 3 min for 5′ RACE; and 93°C for 45 s, 60°C for 1 min, and 68°C for 4 min for 3′ RACE. High-fidelity Taq polymerase (Boehringer Mannheim) was used in all the RACE assays, and 30 cycles of amplification was applied to each reaction. PCR products were subsequently cloned into the pCR2.1-TOPO vector (Invitrogen, Carlsbad, Calif.).
TABLE 1.
Primers used in vIL-8 analysis
| Gene | Primer sequences | Product (bp) | Reference |
|---|---|---|---|
| Chicken β-actin | ACTF4: 5′-CCATGAAACTACCTTCAACTCCA-3′ | 628 (unspliced) | |
| ACTR4: 5′-GATTCATCGTACTCCTGCTTGCT-3′ | 274 (spliced) | 30 | |
| gC | gCF: 5′-TACTCTGCGATACGGAAG-3′ | 584 | 31 |
| gCR: 5′-GAGGAGTTGCAATGTTTCC-3′ | |||
| GFP | GFPF: 5′-CTTGTTGAATTAGATGGTGATGTT-3′ | 584 | 17 |
| GFPR: 5′-TTTCGAAAGGG-CAGATTGTG-3′ | |||
| ICP27 | ICP27F: 5′-GCAAGAAGGGCATCACCGAAGAA-3′ | 670 | 56 |
| ICP27R: 5′-ACGACGAATGCGCGATAACAAAAT-3′ | |||
| Meq | MEQF: 5′-CCCCTTCCCTGACGGCCTATCTGA-3′ | 421 | 27 |
| MEQR: 5′-CTGGGCGCAAAGTTCCTCCGTATC-3′ | |||
| RRL | RRLF: 5′-GCCGCCGTTGCTATTTCTCTA-3′ | 549 | 31 |
| RRLR: 5′-ATGACGCGTGATGCCAACAAC-3′ | |||
| vIL-8 (His tagged) | 5vIL-8NHE: 5′-GCACGCTAGCCGCCACCATGCAGGCGTTGTTGCTAG-3′ | 845 | This report |
| vIL-8HIS: 5′-CGGGATCCTAGTGATGATGATGGTGATGAAGACAGATATGGGA-3′ | |||
| vIL-8 (structural analysis) | vIL-8 no. 1: 5′-GGAGACCCAATAAC-AGGGAAATCGC-3′ | 939 | This report |
| vIL-8 no. 4: 5′-CGACAGACAGTTGTACACCTGCCTG-3′ | |||
| vIL-8 (RT-PCR) | vIL8F: 5′-TGCAGGCGTTGTTG-CTAGTATTGGTT-3′ | 699 (unspliced) | This report |
| vIL8R: 5′-ACAGGAGGTAGCAATTAATCAAAGAGA-3′ | 423 (spliced) | ||
| vIL-8 (SS) | 5′-CGTAGGTTACCGTATCGGATAG-3′ | See Fig. 2 | This report |
| vIL-8 (vIL-8B1) | 5′-GGTCCACACATACCTTCCTGTTCTTC-3′ | See Fig. 2 | This report |
| vIL-8 (SS-dT) | 5′-CGTAGGTTACCGTATCGGATAGCGGCCGCATTTTTTTTTTTTTTTTTT-3′ | See Fig. 2 | This report |
| vIL-8 (vIL-8F4) | 5′-GCAAGTGCGTGAAAGTCACTAATCG-3′ | See Fig. 2 | This report |
DNA sequence analysis.
The DNA sequence of each RACE insert was determined on both strands by a dideoxynucleotide chain termination method. The DNA sequences of RACE clones were compiled, aligned, and analyzed using MacVector (6.0) software. Deduced amino acid secondary structures were analyzed by Chou-Fasman, Kyte-Doolittle, or Robson-Garnier programs. Alignment of vIL-8 sequence with its mammalian homologs was performed by ClustalW profiling.
Kinetics of vIL-8 expression analysis. (i) Northern blot analysis.
Cells were lysed in TRIZOL solution (GIBCO-BRL), and total cellular RNA was isolated as specified by the manufacturer. Twenty micrograms of total RNA from each sample was separated by electrophoresis through a formaldehyde–1.2% agarose gel and blotted overnight to a nylon membrane (Nytran; Schleicher & Schuell) by standard procedures (4). The DNA probe used to detect the vIL-8 transcript was PCR amplified from the genomic DNA of MSB-1 cells, using primers vIL8B1 and vIL8F4. DNA probes for meq and chicken glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were from respective subclones as described previously (63).
(ii) RT-PCR analysis.
To firmly establish the kinetics of vIL-8 expression, DNA-free RNA was purified from 60-mm-diameter dishes of CEF infected with 10,000 PFU of RB1B-infected CEF in the presence or absence of 100 μg of CHX (Sigma)/ml or 100 μg of PAA (Sigma)/ml. CEF were plated for 2 h prior to the addition of CHX and PAA, and the inocula were thawed from frozen stocks, divided into equal aliquots, pelleted by centrifugation, and resuspended in growth medium containing the chemicals at the given concentrations. The infected cells were plated, and the infections were allowed to proceed for 24 h (37°C, 5% CO2). RNA was purified using RNAeasy (Qiagen Corp., Chatsworth, Calif.) and quantitated spectrophotometrically. To evaluate the kinetic class of MDV gene expression, primers were designed to amplify MDV genes of known kinetic class: immediate-early (IE), ICP27 (Table 1); early (E), the large subunit of ribonucleotide reductase (RRL) (Table 1); true late (L), gC (Table 1) as well as vIL-8 and Meq (Table 1). RT-PCR was performed using 500, 50, and 5 ng of RNA as template and the MasterAmp RT-PCR kit (Epicentre Technologies, Madison, Wis.). For RT-PCR, the following amplification conditions were used: 40 min at 60°C (RT step), 5 min at 95°C (initial denaturation), and 40 cycles of 95°C for 1 min, 55°C for 1 min, and 72°C for 2 min. The amplification terminated with a final elongation step of 7 min at 72°C. Amplified products were separated on a 1% agarose gel (1× Tris-borate-EDTA [TBE]), and images were acquired using the Gel-Doc system (Bio-Rad, Hercules, Calif.).
Plasmids.
An 18-bp sequence which encodes six histidines was fused to the 3′ end of the coding region of vIL-8 by PCR amplification using primers 5vIL-8NHE and vIL-8HIS (Table 1). The resulting PCR product was cloned into a baculovirus expression vector, pBlueBac2 (Invitrogen), as an 845-bp NheI-BamHI restriction fragment. The baculovirus-based expression construct, pBB2-vIL-8H, was confirmed by DNA sequence analysis.
For recombinant MDV construction, the BamHI-L fragment of the MDV genome (20) was used. The region encoding all three exons of vIL-8 was deleted from the BamHI-L fragment via excision of an 804-bp NcoI-ClaI fragment. An smGFP (17) expression cassette was constructed using dual expression vector pBKCMV (Stratagene, La Jolla, Calif.). Briefly, plasmid pBKCMV-smGFP has the open reading frame for smGFP fused in frame to the LacZ-alpha peptide in pBKCMV to yield a vector that expresses smGFP in E. coli (via the lac promoter) as well as in eucaryotic cells (via cytomegalovirus [CMV], i.e., promoter). This plasmid also contained a deletion of the simian virus 40 5′ splice donor/splice acceptor region downstream of smGFP. This region was removed as a 357-bp NotI-BclI fragment. The resulting smGFP expression cassette was contained within a 1,813-bp AseI-MluI fragment. This fragment was excised, blunt ended with the Klenow enzyme, and inserted into the blunt-ended NcoI-ClaI sites of the BamHI-L fragment with deletion of vIL-8. Recombinant plasmids were screened for smGFP expression and cassette orientation. Plasmid pYH6a has the smGFP expression cassette inserted in the same orientation as the original vIL-8 gene (see Fig. 7A).
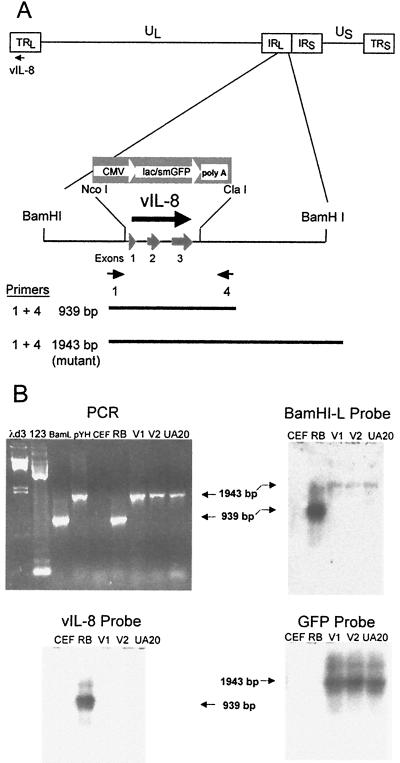
FIG. 7.
Structural characterization of RB1BvIL-8ΔsmGFP. (A) Schematic diagram of the MDV genome showing the region expressing the vIL-8 gene, the restriction sites used in the deletion of vIL-8, the smGFP expression cassette used in mutagenesis, and the location of PCR primers used in the structural characterization of the parental and mutant viruses. (B) An agarose gel showing the results of PCR amplification using primers 1 and 4 (shown in panel A) (upper left panel) and Southern blot hybridization analysis of these products using the BamHI-L fragment of the MDV genome (upper right panel), the vIL-8 cDNA (lower left panel), and the smGFP gene (lower right panel) as probes. The lanes are λd3, HindIII-digested lambda DNA molecular weight markers; 123, 123-bp DNA ladder markers (GIBCO-BRL); BamL, pUC19 containing the BamHI-L fragment of the MDV genome (20); pYH, transfer vector pYH6a; CEF, uninfected CEF DNA; RB, RB1B parental MDV-infected CEF DNA; V1, RB1BvIL-8ΔsmGFP clone 2-infected CEF DNA; V2, RB1BvIL-8ΔsmGFP reisolated from spleen cells of infected chickens; and UA20, DNA from cell line MDCC-UA20, established from an RB1BvIL-8ΔsmGFP-induced ovarian lymphoma.
Preparation of vIL-8–His fusion protein.
The baculovirus expression system was used to express recombinant vIL-8 protein. DNA encoding vIL-8 with six contiguous histidines tagged to its C terminus was inserted into baculovirus expression vector pBlueBac2. Insect Spodoptera frugiperda (Sf9) cells cotransfected with the expression vector and Bac-N-Blue linear AcMNPV DNA (Invitrogen) were used for generating high-titer recombinant baculovirus as described previously (69). High Five (Invitrogen) insect cells grown as suspension cultures in serum-free Express Five (GIBCO-BRL) medium were chosen for the production of recombinant vIL-8 protein. Briefly, cell-free supernatant was collected 72 h postinfection from cells infected with high-titer vIL-8–His recombinant viruses by centrifugation, concentrated 10-fold, and dialyzed into column buffer (50 mM Na phosphate, pH 8.0, containing 0.5 M NaCl and 20 mM imidazole) before being loaded onto a metal affinity column (Ni-resin; Novagen). After washing with 10 column volumes, proteins bound to the Ni-resin were eluted with a linear gradient of imidazole (20 to 500 mM) in column buffer. Fractions containing vIL-8–His (as judged by sodium dodecyl sulfate [SDS]–10 to 20% polyacrylamide gel electrophoresis [PAGE]) were pooled and dialyzed against phosphate-buffered saline. The resulting sample was concentrated eightfold using Centricon-3 filter units (Millipore) and subjected to gel filtration chromatography (Superose 12 column; Pharmacia). Fractions were collected every 2 min at a flow rate of 0.4 min/ml and were analyzed by SDS–10 to 20% PAGE followed by silver staining.
Chemotaxis assay.
For cell separation, 25 ml of fresh chicken whole blood was layered over an equal volume of 1-Step Polymorphs solution (Accurate Chemical and Scientific Co., Westbury, N.Y.) and centrifuged 30 min at 475 × g (20°C). Peripheral blood mononuclear cells (PBMC) and heterophils were separated into two distinct bands free from red blood cells after centrifugation and were collected and washed as specified by the manufacturer's instructions. Cells were resuspended in RPMI 1640 medium at a concentration of 106/ml. For the chemotaxis assay, a 12-well Costar Transwell (Corning Costar Co., Cambridge, Mass.) apparatus was used. Serial dilutions of purified vIL-8–His or recombinant human IL-8 (hIL-8) (R & D Systems, Minneapolis, Minn.) were made in RPMI 1640 medium, and 600 μl of each dilution was applied to the lower chamber of the assay wells and covered with a polycarbonate membrane with a 3-μm pore size for heterophils and 5-μm pore size for PBMC. A 100-μl cell suspension was then added to the top wells. The assay plates were incubated at 37o for 60 min for heterophils and for 40 min for PBMC. The cells in the lower chamber were then counted with a microscope (Nikon-TMS, Tokyo, Japan). Fibronectin (25 μg/ml [57 nM], Boehringer Mannheim) and medium alone were included in each experiment to serve as positive and negative controls, respectively.
Recombinant MDV construction.
RB1BvIL-8ΔsmGFP was constructed using RB1B-infected CEF DNA and the pYH6a transfer vector as described for lacZ-containing mutant MDVs (45) with minor modification. Essentially, 10 μg of RB1B-infected CEF DNA (RB1Bp12) was transfected with various concentrations of pYH6a (10, 50, 100, and 500 ng) via the calcium phosphate method optimized for MDV (40). At 6 days postinfection, fluorescent plaques were identified and passaged onto fresh CEF via scraping with a micropipette into 0.5 ml of 0.5% trypsin/EDTA (Sigma). After dissociation of the cells comprising the plaque (2 to 3 min), the trypsin was inactivated via the addition of 50 μl of filter-sterilized calf serum (GIBCO-BRL). The cells were then added to freshly plated secondary CEF on 60-mm-diameter gridded dishes (2 × 106 cells/dish) or diluted to 10 ml with complete medium (M199; 3% [vol/vol] calf serum; and penicillin, streptomycin, and neomycin [GIBCO-BRL]) and were spread across 96-well plates (100 μl of plaque suspension per well) that were plated with uninfected secondary CEF (1.25 × 104/well). Wells containing single, fluorescent plaques were then harvested via trypsin treatment and passaged again in 60-mm-diameter dishes and/or 96-well plates (as described above). This process was repeated until all formed plaques were fluorescent. At this point, stocks of mutant virus were made. The results presented here were obtained using one of four clones of RB1BvIL-8ΔsmGFP.
Structural analysis of RB1B and RB1BvIL-8ΔsmGFP genomes.
To confirm the deletion of the vIL-8-encoding region from the MDV genome, PCR primers were designed that hybridize to sequences flanking the site of deletion. These primers are vIL-8 no. 1 and vIL-8 no. 4 (Table 1), which correspond to nucleotides (nt) 848 to 872 and the reverse complement of nt 1762 to 1786 of the BamHI-L fragment of the MDV genome, inclusive (31). The resulting PCR product is therefore 939 bp using the parental (RB1B) virus as template and 1948 bp using the mutant (RB1BvIL-8ΔsmGFP) virus as template, due to vIL-8 deletion and expression cassette insertion. To further confirm the structure of RB1BvIL-8ΔsmGFP, these PCR products were subjected to Southern blot hybridization analysis using the BamHI-L fragment, the deleted fragment (NcoI-ClaI), and GFP-specific probes. PCR-amplified products were separated via electrophoresis using a 1% agarose–1× TBE gel, transferred to nylon membranes (Nytran; Schleicher & Schuell) via capillary transfer (57), and subjected to UV cross-linking for immobilization (Stratalinker; Stratagene). The probes were labeled directly via cross-linking with alkaline phosphatase (Alk-Phos Direct; Amersham-Pharmacia). Prehybridization of blots, probe hybridization, and subsequent washes were performed according to the instructions of the manufacturer. After washing and substrate addition, blots were subject to autoluminography using X-ray film (X-OMAT-MR; Kodak, Rochester, N.Y.).
Expression analysis of RB1BvIL-8ΔsmGFP.
To correlate the deletion of the vIL-8 gene with a functional loss in gene expression, we performed RT-PCR analysis using total RNA purified from RB1B-, RB1BvIL-8ΔsmGFP-, and uninfected CEF (RNAeasy, Qiagen). The primers used were specific for (i) the chicken β-actin gene (exon IV→exon V, 628 bp [unspliced], 274 bp [spliced]), ACTF4 and ACTR4 (30); (ii) the smGFP gene (584-bp product), GFPF and GFPR (17); (iii) the MDV ICP27 gene (670-bp product), ICP27F and ICP27R (54); and (iv) the vIL-8 gene (exon I→exon III, 699 bp [unspliced], 423 bp [spliced]), vIL-8F and vIL-8R (Table 1). RT-PCR was performed using a MasterAmp RT-PCR kit (Epicentre Technologies) and serial dilutions of total RNAs (1 μg, 100 ng, or 10 ng per reaction). All primers were used at a concentration of 0.25 pmol per reaction. The program for the amplifications consisted of one cycle of 60°C for 40 min followed by one cycle of 95°C for 3 min and 30 cycles of 95°C for 1 min, 50°C for 1 min, and 72°C for 2 min. The reactions then had one final elongation step (72°C) for 7 min and were then held at 4°C until analyzed. For each gel, 10 μl of reaction products was separated on a 1% agarose gel in 1× TBE containing 0.5 μg of ethidium bromide per ml, as described previously (57). As a control for DNA contamination of RNA samples, chicken β-actin and vIL-8 primers were designed to span at least one intron. In addition, 20 ng of RB1B-infected CEF DNA was used as a control for vIL-8 gene amplification.
Characterization of RB1BvIL-8ΔsmGFP infection in vivo.
In order to characterize the function of the vIL-8 gene during infection, two independent in vivo studies were conducted. These studies were performed in the John Kirkpatrick Skeeles Poultry Health Laboratory, a biosecurity facility at the University of Arkansas. All work was performed at biosafety level 3 using animal rooms under negative pressure with high-efficiency particulate-filtered air entering and leaving each room. The birds were housed in glove box isolators and given a commercial starter feed and water ad libitum.
In vivo study 1.
This study employed SPF chickens (B15 × 7) provided as embryonated eggs by Richard L. Witter (USDA-ARS, Avian Disease and Oncology Laboratory, East Lansing, Mich.). For each virus-infected group, 25 1-day-old birds were injected with approximately 1,000 PFU of virus-infected CEF in 0.2 ml of medium via the intra-abdominal route. As mock-infected controls, 25 birds were injected with uninfected CEF. The titer was determined for each virus used via cocultivation of 10 and 100 μl of inoculum with secondary CEF. At 7, 14, 21, and 28 days postinoculation, three birds were removed from each treatment group for virus reisolation from peripheral blood leukocytes (PBL) and spleen cells. Cells were collected as previously described (45) and were cocultivated with freshly plated secondary CEF. Remaining chickens were maintained in isolation and monitored daily for the development of signs of MD (paralysis, depression, torticollis, etc.), and birds exhibiting severe signs were culled for necropsy and cell line establishment At 9 weeks postinoculation, all remaining birds were euthanatized and necropsied, and the number and site of MD-associated lesions were recorded.
In vivo study 2.
This study was performed essentially as described above, with the exception that commercially obtained SPF White Leghorn chickens were used (Sunrise Inc.). These chickens were obtained as embryonated eggs and hatched under SPF conditions at the John Kirkpatrick Skeeles Poultry Health Laboratory, University of Arkansas. Due to a poor hatch, the number of inoculated chickens was less than that used for in vivo study 1 (25 birds/group versus 35 birds/group).
Cell line establishment.
Lymphoblastoid cell lines were established from gross lymphomas essentially as described previously (18, 45), with the exception that a modified medium was used in the initial culturing of tumor cells (modified Iscove's medium, containing 10% fetal bovine serum, 10% chicken serum, 1× insulin/transferrin/selenium, 1× nonessential amino acids, 4 mM l-glutamine, 2 mM sodium pyruvate, 5 μM β-mercaptoethanol, 1× penicillin/streptomycin/neomycin, and 5× amphotericin B). All cell culture media and reagents were purchased from GIBCO-BRL.
Flow cytometry analysis of MDCC-UA20 cells.
For examination of the level of spontaneous smGFP expression in cell line MDCC-UA20, a FACSort flow cytometer was used (Becton-Dickinson, San Jose, Calif.). Ten thousand ungated cells were acquired per sample and subjected to analysis using CellQuest software (Becton-Dickinson). For comparison, reticuloendotheliosis virus-transformed cell line RECC-CU91 (53) and MDV-transformed cell lines MDCC-UA01 and UA04 (18) were also examined for GFP expression.
Nucleotide sequence accession number.
The cDNA sequence of vIL-8 described in this study has been deposited in GenBank under accession number AF065430.
RESULTS
Identification of vIL-8 transcript.
In the course of analyzing spliced variants of MDV oncoprotein Meq (27), we identified an alternatively spliced Meq-encoding cDNA which fused the bZIP domain of meq with sequences mapped in the BamHI-L fragment (L. F. Lee et al., unpublished). This variant is identical to a 677-bp cDNA first described by Peng et al. (51). For simplicity, this fusion transcript is referred to as MEQΔC-BamL in this paper. We noticed that the sequences derived from the BamHI-L region had a structure strikingly similar to those of CXC chemokines exemplified by IL-8 (see below). Yet, MEQΔC-BamL is unlikely to function as a chemokine because it lacks a signal peptide but instead is fused to a strong nuclear and nucleolar localization signal of meq (33). We hypothesized that MEQΔC-BamL was derived from an aberrant splicing product, in which the natural 5′ exon(s) of the IL-8-like chemokine was replaced by the single meq exon. Our supporting evidence for this was that Northern blot analysis of MSB-1 RNA, probed with the IL-8 portion of the MEQΔC-BamL sequence, revealed a single 0.7-kb transcript (Fig. 1). By contrast, the meq probe detected a series of higher-molecular-weight bands corresponding to differentially processed meq transcripts as previously described (27), none of which, however, comigrated with the 0.7-kb transcript.
FIG. 1.
Expression of vIL-8 is dramatically induced in MSB-1 cells treated with n-butyrate. For analysis, 4 × 106 MSB-1 cells were not treated (U) or treated with 3 mM n-butyrate (NB) for 48 h at 37°C to induce virus replication. Total RNA was purified from each sample and subjected to Northern blot hybridization analysis. Duplicate blots were prepared for different hybridizations. A distinct 0.7-kb transcript was identified when a DNA fragment containing vIL-8 sequences was used as a probe (vIL8). Without NB treatment, at least five transcripts could be identified using a probe encompassing the latent MDV EcoRI Q protein Meq (MEQ). As a comparison, chicken cellular rRNAs (28S and 18S) were stained with ethidium bromide to demonstrate the cellular transcription level in MSB-1 before and after NB treatment.
We examined several MDV-transformed lymphoblastoid cell lines as possible RNA sources for identifying IL-8 homolog-encoding transcripts. MSB-1, an MDV-transformed lymphoblastoid cell line (3) that does not express readily detectable levels of MEQΔC-BamL, was chosen for this purpose. We found that treatment of this cell line with n-butyrate, a chemical often used to induce the lytic replication of herpesviruses (13), appeared to differentially regulate the 0.7-kb and meq transcripts. Butyrate treatment increased the expression of the 0.7-kb transcript but appears to have downregulated the transcription of the meq transcripts (Fig. 1), although this effect may have been exacerbated by the overall decrease in RNA yield from the butyrate-treated cells (Fig. 1, ethidium bromide-stained lanes). These data do suggest, however, that the 0.7-kb transcript encodes an autonomous IL-8-like chemokine, distinct from the MEQΔC-BamL product. mRNA from n-butyrate-treated MSB-1 cells was therefore used as the source for RACE analysis of the putative vIL-8 transcript.
Molecular cloning of vIL-8.
The RT-PCR-based RACE approach was used to identify the ends of the vIL-8 cDNA, using primers derived from the known sequences of the IL-8 portion of MEQΔC-Bam-L (Fig. 2A and B). As shown in Fig. 2C, antisense primer vIL-8B1 produces a major 5′ RACE product of 0.3 kb. The nucleotide sequence of this product helped define the 5′ end of the cDNA and uncovered a putative translational initiation site of the open reading frame (see below). The sense primer vIL-8F4 produced a 3′ RACE product of 0.45 kb, in which a polyadenylation recognition signal near the 3′end was identified (see below).
FIG. 2.
Characterization of transcript encoding vIL-8 by RACE analysis. (A) Schematic drawing of the MDV genome and the location of vIL-8. Transcripts encompassing meq, meqΔC-BamL, and vIL-8 are depicted at the bottom of the diagram with exons in boxes and introns in solid lines. Abbreviations: IRL, long internal repeat; IRS, short internal repeat; TRL, long terminal repeat; TRS, short terminal repeat; UL, unique long sequence; US, unique short sequence. (B) Locations of the oligonucleotide primers used in RACE reactions. vIL-8F4 was used as a gene-specific primer in 3′ RACE. vIL-8B1 was used as a gene-specific primer in 5′ RACE. vIL-8FL-S and vIL-8FL-AS are primers derived from RACE results used to amplify the full length of the vIL-8 open reading frame. The empty arrows depict exons II and III present in the meqΔC-BamL and vIL-8 sequences. (C) Ethidium bromide-stained agarose gel (1%) demonstrates 0.3- and 0.45-kb PCR amplicons in 5′- and 3′ RACE reactions, respectively. Sizes are given on left in kilobases.
To ensure that these two RACE products were derived from the same transcript, primers vIL-8FL-S and vIL-8FL-AS, which are located at the ends of the 5′ and 3′ RACE products, respectively, were used to amplify the region that expresses the entire open reading frame of vIL-8 (Fig. 2B). An RT-PCR product of 454 bp was obtained, and the complete sequence of this amplicon (capital letters) superimposed on the BamHI-L genomic sequences is depicted in Fig. 3A. This alignment reveals three exons in the cDNA: the first exon is 80 bp long, with an initiation methionine located at nt 17 to 19 (the beginning of the cDNA is hereby set as nt 1). Exons II and III are identical to those found in meq–BamHI-L (51), which we recognized as the IL-8-like domains. The entire cDNA thus encodes a 134-amino-acid polypeptide with the hallmarks of a CXC chemokine: (i) exon I contains 21 amino acids; (ii) exons II and III contain 4 cysteines (circled) which are positionally conserved in all CXC chemokines; (iii) the overall amino acid sequences share significant homology with the IL-8 family of chemokines (see below). In addition, a consensus TATA box, TATATAA, is found at −32 nt and, as described above, a noncanonical polyadenylation sequence, ATTAAA, is located 15 nt upstream of the 3′ end and adjacent to the translational stop of vIL-8 exon III. These results suggest that vIL-8 has its own transcriptional unit in the MDV genome and that the previously described MEQΔC-BamL transcript may represent an aberrant or alternatively spliced product.
FIG. 3.
Fine structure of vIL-8 cDNA. (A) The nucleotide sequence of a full-length cDNA compared to its genomic counterpart. cDNA sequences are in capitals and blocked by lines while consensus splice donor/splice acceptor in introns are in bold. Putative TATA box (TATATAA) and polyadenylation recognition (ATTAAA) sequences are underlined. Deduced amino acid sequences are shown beneath the nucleotide sequences. The 17 hydrophobic residues of the first exon are in boldface. Four cysteines conserved in all CXC chemokines are circled. (B) Alignment of amino acid sequences of vIL-8 and various mammalian and avian IL-8 homologs. The four conserved cysteines and the signature ELR motif are indicated on top of the alignment. Similar (shaded in light gray) and identical (shaded in dark gray) amino acids are indicated. The abbreviation used and GenBank accession number for each sequence are rhIL8 (rhesus macaque IL-8), P51495; huIL8 (hIL-8), P10145; ovIL8 (ovine IL-8), P36925; doIL8 (dog IL-8), JN0841; rabIL8 (rabbit IL-8), P19874; 9E3/CEF4 (CEF protein 9E3), M16199; moGro (mouse Gro), P12850; moMIP2 (murine macrophage inflammatory protein 2), P10889; moBLC (Mus musculus B-lymphocyte chemoattractant), AF044196; and ratPF4 (rat PF4), P06765.
Comparison of vIL-8 with cellular IL-8 genes.
The vIL-8 amino acid sequence was compared to sequences in the present database using BlastSearch. Sequences with the highest homology scores were those from IL-8 and its related CXC chemokines, such as Gro and platelet factor 4 (PF4) (Fig. 3B), all of which are implicated in the chemotaxis and inflammatory responses (5). Noticeably, chicken 9E3/CEF4 (9, 64) and K60 (61), postulated to be the avian orthologs of IL-8 (7), were also identified in this analysis. The overall amino acid sequence identities between vIL-8 and human IL-8, vIL-8 and 9E3/CEF4, and vIL-8 and mouse Gro are 23, 22, and 20%, respectively. Despite the significant similarities with viral and mammalian IL-8s, there is an interesting difference: the 3-amino-acid motif ELR (glutamate-leucine-arginine), which immediately precedes the first CXC motif and is thought to be the determinant of the neutrophil specificity, is conspicuously absent. In place of this ELR motif is a DKR (asparate-lysine-arginine) motif. There are other IL-8-like chemokines that do not have ELR motifs. such as PF4 and the recently discovered B-lymphocyte chemoattractant (22, 32). In both cases, the primary target cells are not neutrophils. This raises an intriguing question about the target cell specificity of vIL-8.
Expression kinetics of vIL-8.
To understand the role of vIL-8 in the MDV life cycle, we first determined the kinetics of vIL-8 expression. Initially, log-phase MSB-1 cells were treated with n-butyrate for different time periods, in combination with herpesvirus DNA polymerase inhibitor PAA (21). Total RNAs from each sample were subjected to Northern blot analysis using the vIL-8 cDNA fragment as a probe. As mentioned before, there was a basal level of vIL-8 expressed in untreated MSB-1 cells, presumably due to a small population of cells undergoing reactivation (Fig. 4A, lane 1). The induction follows relatively slow kinetics and was evident only after 36 h of treatment. Furthermore, this induction was largely inhibited by PAA treatment (Fig. 4A, lane 6), suggesting that vIL-8 is a lytic transcript with true late (γ2) kinetics (21).
FIG. 4.
Expression kinetics of vIL-8 in MSB-1 cells. (A) MSB-1 cells were treated with n-butyrate (3 mM) for 0 to 48 h, as indicated. For the 36-h time point, duplicate cell cultures were prepared for the treatment of cells with PAA (100 μM). Total RNAs were extracted at the end of chemical treatment, and concentrations were determined. Twenty micrograms of RNA from each sample was loaded to each lane, and duplicate filters were prepared for vIL-8 hybridization (upper gel) and GAPDH hybridization (lower gel) (33). (B) RNA was purified from RB1B-infected CEF that were not treated (left lanes), treated with 100 μg of CHX/ml (center lanes), or treated with 100 μg of PAA/ml (right lanes) and was used as the template for RT-PCR. For each RNA treatment group, three concentrations of RNA were used (500, 50 and 5 ng, left to right, respectively). RT-PCR was performed as described in the text, and the entire reaction products (total, 25 μl) were loaded on a 1% agarose gel. The products for RT-PCR were ICP27, 670 bp; RRL, 549 bp; gC, 584 bp; vIL-8, 699 bp (unspliced) and 423 bp (spliced); and Meq, 421 bp (Table 1). Arrows indicate the RT-PCR products of the appropriate size. Far right lanes contain molecular size markers (Hyper-Ladder; Bioline). (C) Ethidium bromide-stained agarose gel of 1 μg of each RNA used for RT-PCR analysis in panel B, demonstrating that the RNAs used were not degraded. Mrk, Hyper-Ladder DNA size markers in base pairs. Un, untreated.
To firmly establish the kinetics of vIL-8 expression during MDV infection, CEF were infected with MDV (RB1B) in the presence and absence of CHX and PAA (Fig. 4B). As controls for IE, early (E), and true late (L, γ2) genes, primers were designed to amplify MDV ICP27, RRL, and gC gene products. In addition, primers specific for the meq gene were used to establish the kinetics of Meq expression during lytic infection. Our results (Fig. 4B) clearly demonstrate that vIL-8 is expressed with true late kinetics, as its expression is blocked by both CHX and PAA. A problem with kinetic analysis of MDV gene expression is the carryover of transcripts of all kinetic classes present in the inoculum. This carryover does allow for some background expression of RRL, gC, and vIL-8 genes in the presence of CHX and/or PAA (Fig. 4B). This level of expression is minor, however, and RRL expression is largely blocked by CHX but not by PAA, and gC and vIL-8 expression is largely inhibited by CHX and PAA. Aiding in this assessment of expression is the use of 3 orders of magnitude (500, 50, and 5 ng) of template, which allows clear distinction among the three kinetic classes.
Interestingly, we determined that meq is apparently expressed as an IE gene during lytic infection. These data, as well as our Northern blot results for meq expression, must be viewed from the standpoint that both techniques do not provide information regarding the sense or antisense nature of the transcripts detected. This is a particularly important consideration for this region of the genome, as both sense and antisense transcripts have been detected during latent and lytic infection (50).
In any event, our data do indicate that vIL-8 is expressed as a true late gene and therefore is likely to be involved in the replication (lytic) phase of MDV. Given its similarity to cellular chemokines, one possible role of vIL-8 is the recruitment of target cells for MDV infection. As a first step to test this hypothesis and to demonstrate that vIL-8 is biologically active, we chose the baculovirus expression system to obtain recombinant vIL-8 for chemotaxis studies.
Expression and purification of recombinant MDV vIL-8.
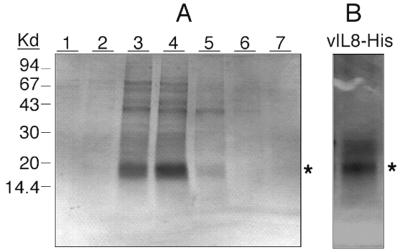
To facilitate the purification of baculovirus-expressed protein, the vIL-8 open reading frame was tagged with six contiguous histidines at its C terminus. Cell culture supernatant from insect cells infected with vIL-8–His recombinant baculoviruses was partially purified by passing through an Ni-nitrilotriacetic acid affinity column followed by gel filtration chromatography (Fig. 5). A protein band with an apparent molecular mass of 18 to 20 kDa in SDS-PAGE was eluted from the affinity column (Fig. 5A, fractions 3 and 4). After gel filtration, the pooled fraction (42- to 44-min fractions) contained primarily the vIL-8–His protein (Fig. 5B), which was used for subsequent studies. Further verification that the 18-kDa band was indeed vIL-8 came from the observations that a monoclonal antibody against the C-terminal six histidines and rabbit antisera against two synthetic peptides derived from the deduced vIL-8 open reading frame also showed immunoreactivity to this protein (data not shown). The elution behavior of this protein in gel filtration (corresponding to an apparent molecular mass of 12 to 15 kDa) suggests that the majority of the recombinant vIL-8–His exists in a monomer form. In addition, the fact that the recombinant vIL-8–His was isolated from the supernatant suggests that the signal sequence is functional, resulting in the secretion of the vIL-8 protein.
FIG. 5.
Purification of six-His-tagged vIL-8 produced by baculovirus expression system. (A) Purification of recombinant vIL-8–His by Ni-affinity column chromatography. Cell-free supernatants collected from High Five insect cells infected with vIL-8–His recombinant baculovirus were prepared and loaded onto a Ni column as described in Materials and Methods. Bound vIL-8–His was eluted by a 20 to 500 mM imidazole gradient. Twenty microliters of each elution fraction was loaded onto an SDS–10 to 20% PAGE gel, separated electrophoretically, and stained with Coomassie blue. Kd, kilodaltons. (B) Silver staining demonstrates the purity of vIL-8–His after gel filtration. Elution fractions 3 and 4 of the Ni column were pooled, concentrated eightfold by Centricon-3 spin concentration, and further purified by gel filtration chromatography (Superose 12). Fractions were collected every 2 min at a flow rate of 0.4 min/ml. Purity of the eluent with the highest chemotaxis activity (42- to 44-min fractions) was analyzed by SDS–10 to 20% PAGE followed by silver staining. A major protein band with a size of approximately 20 kDa is indicated with an asterisk.
MDV vIL-8 as a chemoattractant for chicken PBMC.
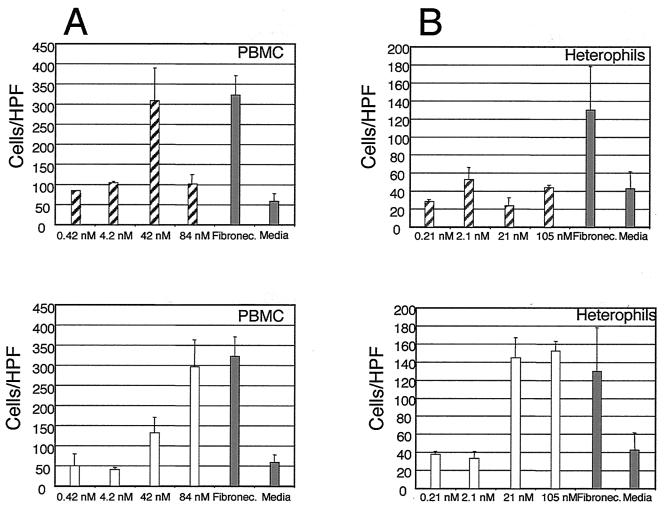
A major function of chemokines is chemotaxis. Thus, we first tested whether vIL-8 is able to recruit chicken PBL. Freshly prepared chicken blood samples were fractionated through a Percoll gradient, which gave rise to two distinct bands, namely, PBMC and heterophils. The PBMC fraction contained monocytes, thrombocytes, and lymphocytes, whereas the heterophil fraction was principally the chicken equivalent of human neutrophils. Chemotaxis assays were conducted using a modification of the method described by Boyden (10). The purified vIL-8 protein fractions were assayed for chemotactic activity by placing the protein to be tested in the medium, which was separated from the target cells by a membrane (3- to 5-μm thickness). The number of cells migrating through the membrane in response to the protein was counted, and the results are shown in Fig. 6. MDV vIL-8 exhibited a strong chemotactic activity toward chick PBMC, with the maximal response elicited at a concentration of 42 nM (Fig. 6A, upper panel). At a higher concentration (84 nM), the activity decreased, consistent with previous reports that receptors are downmodulated after full occupancy (8, 35). Recombinant hIL-8 also elicited a positive response (Fig. 6A, lower panel), although a higher concentration of ligand was required. In contrast, when chicken heterophils were used as targets, hIL-8 exhibited a strong effect at 21 nM, while vIL-8 failed to “chemoattract” the heterophils at all concentrations tested (Fig. 6B). These data suggest that MDV vIL-8 is biologically active and functions like a chemokine. They also reveal an interesting difference in target cell specificity between vIL-8 and hIL-8. We have recently received clones of the chicken IL-8 orthologs 9E3/CEF4 and K60 (62; a gift from Peter Staeheli, University of Freiburg) and plan to investigate the possible interactions of vIL-8 with native chicken IL-8 signaling and chemotaxis.
FIG. 6.
Chemotaxis assays demonstrate that MDV vIL-8 is a functional chicken PBMC chemoattractant. The chemotactic activity of vIL-8 toward chicken PBMC (A) or heterophils (B) was assayed using Costar Transwell plates. For panel A, chemotaxis was performed using 5-μm-pore-size Transwell plates with the indicated concentrations of vIL-8 at 37°C for 40 min. For panel B, a 3-μm-pore-size Transwell plate was used for heterophils and the incubation was increased to 1 h. In each assay, a positive control consisting of medium with 15 μg of fibronectin and a negative control containing medium alone were included. Results were averaged from at least three independent experiments. ░⃞, control; ▨, vIL-8; □, hIL-8. HTP, high-power field.
The MDV vIL-8 gene is nonessential for replication in cell culture.
Given the location of vIL-8 in the MDV genome, i.e., within the repeats flanking the UL region downstream of the meq gene, and the difference in expression patterns between meq and the vIL-8 gene, we felt that vIL-8 is likely to be important in MDV infection. Most genes expressed within herpesvirus repeats are involved in the regulation of gene expression (39), and this region is of particular interest in the MDV genome, as this region is unique to serotype 1 (oncogenic) MDVs (25, 44).
To examine the role of vIL-8 during MDV infection, therefore, we constructed a deletion mutant having both copies of the vIL-8 coding sequences (long terminal repeat [TRL] and long internal repeat [IRL]) removed and replaced with a smGFP expression cassette (Fig. 7A). The smGFP expression cassette was inserted between NcoI and ClaI sites in the BamHI-L fragment, resulting in the deletion of all three exons encoding vIL-8. Mutant viruses constructed using this mutated BamHI-L fragment were examined structurally for deletion of both of the vIL-8 coding regions. Of four clones constructed, three were found to have deleted both copies of the vIL-8 region (data not shown). Clone 2, designated RB1BvIL-8ΔsmGFP, was examined further via PCR, Southern blot hybridization, and RT-PCR analysis (Fig. 7B and 8). Using primers that flank the site of deletion, PCR products of 939 and 1,948 bp were observed when parent virus (RB1B)- and deletion mutant (RB1BvIL-8ΔsmGFP)-infected CEF DNA were used as templates, respectively. Moreover, Southern blot hybridization analysis of these PCR products demonstrated that the vIL-8 coding regions were, in fact, absent from the genome of RB1BvIL-8ΔsmGFP, RB1BvIL-8ΔsmGFP reisolated from chickens (spleen cells) during in vivo infection, and from a cell line established from an RB1BvIL-8ΔsmGFP-induced tumor (MDCC-UA20) (Fig. 7B).
FIG. 8.
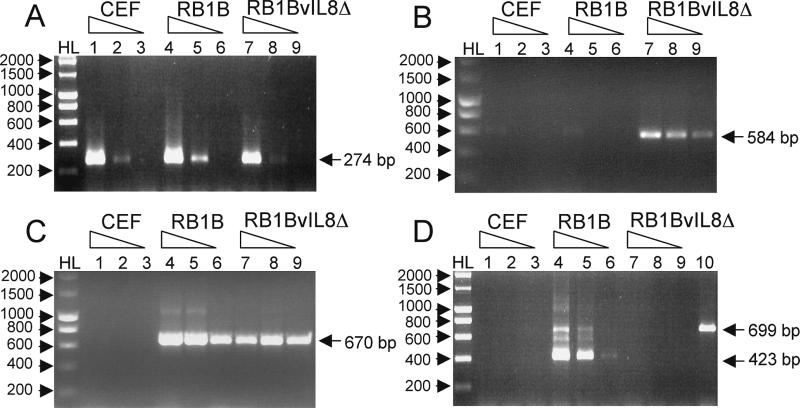
Expression analysis of uninfected and RB1B- and RB1BvIL-8ΔsmGFP-infected CEF RNAs. The expression of cellular and virus gene products was examined using RT-PCR and primers specific for chicken β-actin (exon IV→exon V) (A); the smGFP gene (B); the MDV ICP27 gene (C); and the MDV vIL-8 gene (exon I→exon III) (D). Each picture shows the ethidium bromide-stained 1% agarose gel of 10 μl of each reaction product (total reaction volume, 50 μl). The templates used were lanes 1 to 3, 1 μg, 100 ng, and 10 ng of CEF total RNA, respectively; lanes 4 to 6, 1 μg, 100 ng, and 10 ng of RB1B-infected CEF total RNA, respectively; lanes 7 to 9, 1 μg, 100 ng, and 10 ng of RB1BvIL-8ΔsmGFP-infected CEF total RNA, respectively. Lane 10 in panel D represents 20 ng of RB1B-infected CEF DNA as a control for DNA contamination. HL denotes DNA Hyper-Ladder molecular size standard (Bioline). Sizes of specific RT-PCR products and molecular size standards are identified with arrows.
To demonstrate a functional absence of vIL-8 expression associated with deletion of these regions from the MDV genome, we performed RT-PCR analysis using primers specific for cellular actin (cellular and DNA contamination control), smGFP (RB1BvIL-8ΔsmGFP control), ICP27 (for MDV infection control), and vIL-8 expression (Fig. 8). To compare relative levels of expression among these genes, we selected primers that generated small products (200 to 700 bp) and used RNA template concentrations spanning 3 orders of magnitude (1 μg, 100 ng, and 10 ng). Results using chicken β-actin primers indicate that comparable levels of RNA are present in all samples and that this RNA is devoid of DNA contamination (Fig. 8A). The primers specific for smGFP amplification demonstrate that this gene is expressed in abundance in infected cells and is readily detectable in 10 ng of total RNA (Fig. 8B). The level of smGFP expression, however, appeared lower than that of ICP27, an IE gene (Fig. 8C). The level of ICP27 expression, in fact, was comparable among RB1B- and RB1BvIL-8ΔsmGFP-infected CEF RNAs, demonstrating that cells were infected to comparable levels. The high abundance of ICP27 message may be explained by the fact that the MDV ICP27 gene is downstream of glycoprotein K (gK), with no transcriptional terminator between the translational stop of gK and the initiation codon of ICP27 (56). Consequently, our ICP27 primers detect ICP27-specific transcripts as well as gK read-through transcripts. For our intents and purposes, however, these RT-PCR products demonstrate comparable levels of MDV infection between the parental (Fig. 8, lanes 4 to 6) and recombinant (Fig. 8, lanes 7 to 9) infected CEF.
Using primers spanning exons I and III of vIL-8, we found readily detectable levels of vIL-8 expression in RB1B- but not RB1BvIL-8ΔsmGFP-infected CEF RNA, correlating a functional loss of expression with deletion of both copies of the gene from the genome (Fig. 8D). The level of vIL-8 expression appeared less than the expression of ICP27, and unspliced and single-spliced forms of the vIL-8 transcripts were clearly visible. These products are unprocessed and partially processed RNAs, as RT-PCR analysis using the primers employed in structural analysis (Fig. 7A), which contained one primer downstream of the putative poly(A) site, failed to amplify any products from RB1B- or RB1BvIL-8ΔsmGFP-infected CEF RNAs (data not shown).
In vivo replication of RB1BvIL-8ΔsmGFP.
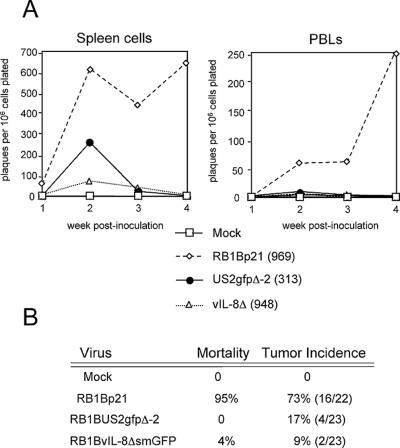
To examine the effect of vIL-8 deletion on the ability of MDV to replicate and induce pathology in chickens, we performed two in vivo experiments (Fig. 9 and 10). In in vivo experiment 1, RB1BvIL-8ΔsmGFP replicated to lower levels than an equal-passage level parent virus (RB1Bp21) or a recombinant RB1B having a GFP expression cassette inserted at the nonessential US2 gene (RB1BUS2gfpΔ-2) (18) (Fig. 9). This impairment in replication was particularly evident in reisolation attempts from PBL, from which RB1BvIL-8ΔsmGFP could be reisolated only at weeks 2 and 3 postinoculation. Given that the US2gfp recombinant was inoculated at one-third the dosage of RB1BvIL-8ΔsmGFP (313 PFU/bird vs. 948 PFU/bird, respectively), it does not appear that the decrease in virus replication for RB1BvIL-8ΔsmGFP in vivo can be attributed to GFP expression. Despite this significant growth impairment, RB1BvIL-8ΔsmGFP was able to induce tumors in two birds (Fig. 9B) and a cell line was established from one of these tumors (MDCC-UA20). The ability to establish a T-cell line using RB1BvIL-8ΔsmGFP indicates that (i) the vIL-8 gene is nonessential for transformation and that (ii) even high levels of smGFP expression are tolerated by many chicken cell types.
FIG. 9.
Comparison of RB1B and RB1BvIL-8ΔsmGFP, in vivo experiment 1. (A) Graphs of virus reisolation data for spleen cells and PBL from chickens (B15 × 7) inoculated with mock-infected CEF (Mock); equal-passage-level parent virus-infected CEF (RB1Bp21); RB1BUS2gfpΔ-2-infected CEF (US2gfpΔ-2); and RB1BvIL-8ΔsmGFP-infected CEF (vIL-8Δ). The numbers in parentheses indicate PFU inoculated per chicken. Each time point represents the mean plaque number counted on triplicate 60-mm-diameter dishes of CEF cocultivated with spleen cells or PBL at 6 days postplating. The y axis denotes plaques formed on CEF monolayers per 106 cell plated. (B) Summary of virus-specific mortality and tumor incidence data for this trial. Numbers in parentheses indicate the actual number of birds containing tumors divided by the number of birds left in treatment groups after virus reisolation attempts (i.e., after 4 weeks postinoculation).
FIG. 10.
Comparison of RB1B and RB1BvIL-8ΔsmGFP, in vivo experiment 2. (A) Graphic representations of virus reisolation data for spleen cells and PBL from commercial SPF chickens inoculated with mock-infected CEF (Mock); parent virus (RB1B)-infected CEF (RB1Bp12); and RB1BvIL-8ΔsmGFP-infected CEF (vIL-8Δ). Each time point represents the mean plaque number counted on triplicate 60-mm-diameter dishes of CEF cocultivated with spleen cells or PBL at 6 days postplating. The numbers in parentheses indicate PFU inoculated per chicken. The y axis denote plaques formed on CEF monolayers per 106 cell plated. (B) Summary of virus-specific mortality and tumor incidence data for this trial. Numbers in parentheses indicate the actual number of birds containing tumors divided by the number of birds left in treatment groups after virus reisolation attempts (i.e., after 4 weeks postinoculation).
In vivo experiment 1 employed B15 × 7 chickens, which are genetically defined and highly susceptible to MDV pathology, particularly to neurological lesions (68). To assess the ability of RB1BvIL-8ΔsmGFP to replicate in genetically outbred, commercial SPF chickens, we performed an additional in vivo experiment (Fig. 10). During in vivo study 2, the reisolation levels of RB1BvIL-8ΔsmGFP from spleen cells and PBL were again an order of magnitude less than the levels of parent virus (RB1Bp12). In this study, RB1BvIL-8ΔsmGFP induced one tumor, indicating that this virus retained oncogenicity unrelated to the genetic susceptibility of the chickens infected.
Taken together, these results demonstrate that the vIL-8 gene is nonessential for MDV replication in cell culture, during in vivo infection, and moreover, for transformation of chicken T cells. As hypothesized, given the timing of expression of vIL-8, the function of vIL-8 is likely to be associated with lytic infection, perhaps affecting target cell access or recruitment. Our current lack of a rescue mutant, however, limits our conclusions with regard to specific deficits in replication due to the possibility of MDV spontaneous mutation when passaged repeatedly in cell culture.
The smGFP cassette inserted at the vIL-8 loci is constitutively expressed in the cell line MDCC-UA20.
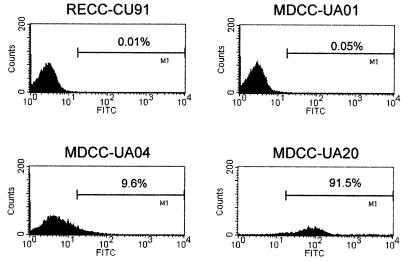
Having excised an RB1BvIL-8ΔsmGFP-induced ovarian lymphoma during in vivo experiment 1, we noted that, like other GFP- and Lac-expressing MDVs, the marker gene was apparently repressed in situ and in the initial stages of cell culture (18; data not shown). Upon culturing these cells and after cell line establishment, we noted, via microscopic examination, spontaneous smGFP expression in a low but consistent number of cells (data not shown). We reported this observation previously for an RB1BUS2gfpΔ-1-transformed lymphoblastoid cell line (18) and several lac-containing MDV-transformed cell lines (46). By flow cytometry, however, we determined that the UA20 cell line constitutively expressed detectable levels of smGFP (Fig. 11) compared to non-MDV-transformed (CU91), parental RB1B-transformed (UA01), and RB1BUS2gfpΔ-1-transformed (UA04) cell lines. These data suggest that there is constitutive expression of the smGFP cassette in the context of the latent MDV genome. This expression could be due to (i) copy number of the expression cassette, (ii) the location of the cassette within the repeats versus unique sequences of the MDV genome, or (iii) the location of the cassette downstream of the meq gene, a well-characterized latent gene (27), resulting in read-through transcripts from meq in the transformed cells.
FIG. 11.
Spontaneous expression of smGFP in MDCC-UA20 cells. Histogram plots of flow cytometry data are shown depicting the number and fluorescence intensity of GFP-expressing lymphoblastoid cells. The x axis denotes arbitrary fluorescence units (log scale) for the FL-1 (fluorescein isothiocyanate [FITC], GFP) channel, and the y axis denotes cell number. For each plot, 104 cells were acquired. The cell lines used were RECC-CU91, a REV-transformed T-lymphoblastoid cell line (53); MDCC-UA01, an RB1B-transformed T-lymphoblastoid cell line (18); MDCC-UA04, an RB1BUS2gfpΔ-1-transformed T-lymphoblastoid cell line (18); and MDCC-UA20, an RB1BvIL-8ΔsmGFP-transformed T-lymphoblastoid cell line.
DISCUSSION
We describe here the identification and cloning of a novel chemokine encoded by MDV, termed vIL-8. vIL-8 mRNA is transcribed from the IRL region of the MDV genome via two splicings. Deduced amino acid sequence of vIL-8 cDNA reveals a significant similarity with mammalian IL-8 families, including a prototypical signal peptide and four positionally conserved cysteines. Baculovirus-expressed vIL-8 was shown to be functional as a secreted protein and as a chemoattractant for chicken mononuclear cells.
The alignment of cDNA and genomic sequences identifies three exons for the 0.7-kb vIL-8 mRNA. The N-terminal 21 amino acids encoded by exon I are rich in hydrophobic residues (80%) and likely to serve as a signal peptide (16). Exons II and III constitute the secreted portion of vIL-8. The highly conserved motifs and spacing classify vIL-8 as a member of the IL-8-like CXC family. Consequently, vIL-8 is the first reported CXC chemokine encoded by an alphaherpesvirus (34). Recently, CMV has also been shown to encode Gro-α/IL-8-like chemokines (UL146, UL147) (49). Aside from these examples, most other viruses encode homologs of either CC chemokines or CXC chemokine receptors. While the overall structure of vIL-8 is similar to those of CXC chemokines, vIL-8 carries an unusually long C-terminal extension. Whether and how this long C terminus modulates the binding to receptors or to polyglycans remains to be determined.
The crucial residues involved in binding of CXC chemokines to receptors have been mapped to the N terminus. In particular, the three residues right before the CXC motif are deemed to be essential (15, 23). Mutations or deletions of them render the molecules incapable of recruiting target cells. All IL-8-like molecules that target neutrophils carry the tri-amino acids ELR in these positions (5). Chemokine PF4, which carries DLS (or DLQ) in these positions, fails to attract neutrophils. Addition of ELR residues to an N-terminally truncated PF4 increases its chemotactic potential for neutrophils (14). In vIL-8, DKR, a putative hybrid motif of ELR and DLS, is present in this region. Interestingly, the UL147 gene product of CMV (vCXC-2) contains a DRR motif at these positions, suggesting a possible similarity of function between this betaherpesvirus-borne CXC chemokine and MDV-borne vIL-8 (49). Thus, it is difficult to predict whether vIL-8 has a tropism toward neutrophils. Our chemotaxis experiments suggest that vIL-8, unlike hIL-8, is more potent as a chemoattractant for chicken mononuclear cells than for heterophils. We hoped that this comparison could be extended to chicken IL-8 homologs (62). Unfortunately, we were unable to obtain purified chicken IL-8 homologs for this study and plan to construct expression vectors for both 9E3/CEF4 and the recently discovered K60 genes to investigate the possible interactions of vIL-8 with native chicken IL-8 signaling and chemotaxis. In addition, future experiments are required to determine more precisely which cell types in the chicken mononuclear cell fraction are the authentic targets for vIL-8.
While this is the first IL-8-like molecule found to be encoded by an alphaherpesvirus, the association of cellular IL-8 expression with viral infection has long been documented. Cultures of corneal keratinocytes infected with herpes simplex virus type 1 were shown to synthesize at the rate of more than 30 ng per million cells of IL-8 during an 18-h incubation (43). Similarly, the induction of IL-8 synthesis could be demonstrated in hepatocytes infected with hepatitis B virus, monocytes infected with human CMV, fibroblasts infected with human herpesvirus 6, or leukemia cells infected with human T-cell leukemia virus I (22, 36, 42). IL-8 is known as an important mediator of inflammation which recruits and activates neutrophils to sites of infection, often inducing some of the symptoms associated with virus infections (1, 5). An IL-8-activated neutrophil is characterized by enhanced migration, phagocytosis, superoxide generation, and granule release (6). It has also been recognized that signals transduced by the interaction of IL-8 with IL-8R, a G-protein-coupled receptor, are responsible for the activation of neutrophils. Recently, the observation that members of the p21-activated kinase family of kinases are transiently activated in neutrophils stimulated with IL-8 further elucidated the molecular process of IL-8 signaling (24). Taken together, the overproduction of cellular IL-8 in viral infections could be a reactive response of the host cells, aimed at reducing viral load.
If the function of cellular IL-8 is to curtail the production of viruses at entry into a host, then why does MDV encode an IL-8-like gene? Obviously, a great deal has yet to be learned before this question can be answered, and we can only speculate on some of the plausible reasons. First, vIL-8 may be involved in recruiting and/or activating phagocytic cells in the lungs (macrophage, dendritic cells, and heterophils), the primary site of MDV entry (12). This recruitment or activation may aid in the early transmission of MDV to the primary lymphoid organs. A similar dissemination function has been postulated for vCXC-1 (the UL146 gene product) encoded by CMV (49).
Second, as suggested by this study, the target of MDV vIL-8 may not be neutrophils but rather B or T cells. It is known that MDV infection in vivo is comprised of an early cytolytic infection of B cells, followed by a latent infection of T cells. The recruitment of B and T cells is therefore crucial to the initial establishment of infection. Expression of vIL-8 may help recruit proper target cells for MDV. Third, vIL-8 may serve as a decoy for chicken IL-8. We have found that baculovirus-expressed vIL-8 does not elicit calcium mobilization in either heterophils or mononuclear cells (data not shown). Cellular IL-8 is known to form dimers when present at high concentrations (6). Therefore, it is possible that vIL-8 may antagonize host IL-8 by either competing for binding to the receptor or the formation of an inactive dimer. The absence of calcium mobilization as a result of vIL-8-binding may be due, however, to the absence of additional protein modifications not catalyzed in insect cells, and the above observation must therefore be viewed with this caveat.
A recently discovered CC-like chemokine scavenger, called the M3 protein, encoded by murine gammaherpesvirus 68, shows remarkably broad cellular receptor usage, binding to certain CC, CXC, and CX3C receptors (49, 65) and inhibiting the signaling of these receptors. Therefore, vIL-8 may preferentially attract target cells while blocking the attraction of other leukocytes.
An additional possibility is that vIL-8 may augment MDV gene expression and enhance viral replication. It has been demonstrated that the exposure of human fibroblasts to IL-8 augments both infectious virus production and replication of CMVs at least eightfold (41). This is understandable, since the signals responding to IL-8 stimulation, such as p21-activated kinase phosphorylation and calcium mobilization, are also those which activate transcription factors. If B or T cells carry receptors that interact with vIL-8, the IL-8 signals may act upon viral or cellular promoters that transcribe genes conducive for viral replications. In this regard, it is noteworthy that the newly discovered B-lymphocyte chemoattractant chemokine, which shares significant homology with vIL-8, binds a receptor specific for B cells, known as BLR (22, 29).
The phenotype exhibited by the deletion mutant virus, RB1BvIL-8ΔsmGFP, in vivo does provide some insight into the possible function of vIL-8. This mutant showed marked growth impairment in spleen cells and PBL, despite wild-type replication in cell culture (data not shown). If vIL-8 were required for enhanced viral gene expression during the lytic infection of fibroblasts, cells that normally express IL-8 receptor (59), then ablation of this gene would be likely to affect ability of this virus to replicate in fibroblasts. Our results suggest that vIL-8 is likelier to be involved in leukocyte target recruitment or immunoevasion, as RB1BvIL-8ΔsmGFP established a poor lytic infection and was largely cleared from infected chickens by 4 weeks postinoculation. Despite this poor initial infection, however, RB1BvIL-8ΔsmGFP retained the ability to transform T cells, suggesting that its abilities to establish and maintain latency were not abrogated. Our conclusions regarding the phenotype of RB1BvIL-8ΔsmGFP must be viewed with the caveat that a rescue mutant has not yet been generated. From RB1BvIL-8ΔsmGFP-infected chickens, we have isolated viruses that have deleted the smGFP expression cassette, and one of these has been plaque purified and passaged in vivo (data not shown). This virus, termed NF-1, has deletions of vIL-8, the smGFP cassette, and additional flanking sequences. We plan to use this virus for the reinsertion of the vIL-8 gene at another locus to confirm our observations regarding the function of vIL-8 in MDV in vivo infection.
The constitutive expression of smGFP in the RB1BvIL-8ΔsmGFP-transformed cell line UA20 is interesting, considering previous findings with heterologous promoters inserted within the MDV genome (18, 48). This constitutive expression likely reflects a difference in structure and/or expression of the IRL/TRL during MDV latent infection, since a construct containing another GFP (S65T; Clontech, Palo Alto, Calif.) expressed using the same promoter (CMV) inserted at the US2 locus was repressed in the tumor and is largely repressed in a derived cell line (18) (Fig. 11).
In summary, we have identified an IL-8-like molecule encoded by MDV. This gene is expressed as a true late gene during lytic infection, suggesting its involvement in viral replication. We have further shown that baculovirus-expressed vIL-8 is functionally active as a secreted protein that attracts chicken PBMC. A recombinant MDV having both copies of the vIL-8 gene deleted from the genome failed to express vIL-8 transcripts, replicated to wild-type levels in cell culture, yet showed marked growth impairment in vivo. Despite this growth impairment, the vIL-8 deletion virus was capable of inducing tumors, demonstrating that this gene is nonessential for transformation of chicken T cells. A cell line derived from a vIL-8 deletion mutant-induced tumor showed constitutive expression of a marker gene, suggesting that these regions of the genome (the repeats flanking the UL region) have the capacity to be transcriptionally active in MDV cell lines. The present study provides a basic framework to understand how this viral chemokine (virokine) functions in MDV replication and in cell-type specific virus-host interactions.
ACKNOWLEDGMENTS
M.S.P. and S.-F.L. contributed equally to this work.
We thank Juinn-Lin Liu for providing the MEQ fragment used in Northern blot analysis. We are grateful to Narayan Rath (University of Arkansas) for the helpful suggestions for the separation of chicken heterophils from lymphocytes. We also thank Cindy Cisar (University of Arkansas) for β-actin, smGFP, ICP27, RRL and gC primers, and critical review of the manuscript.
This research was supported by grants to H.-J.K. from the U.S. Department of Agriculture (93-37024-9340), the National Cancer Institute (CA46613), and the Council for Tobacco Research (4034) and to M.S.P. from the U.S. Department of Agriculture (97-35204-5067).
Footnotes
Arkansas Agricultural Experiment Station manuscript no. 00108.
REFERENCES
- 1.Adams D H, Lloyd A R. Chemokines: leucocyte recruitment and activation cytokines. Lancet. 1997;349:490–495. doi: 10.1016/s0140-6736(96)07524-1. [DOI] [PubMed] [Google Scholar]
- 2.Adams D H, Shaw S. Leucocyte-endothelial interactions and regulation of leucocyte migration. Lancet. 1994;343:831–836. doi: 10.1016/s0140-6736(94)92029-x. [DOI] [PubMed] [Google Scholar]
- 3.Akiyama Y, Kato S, Iwa N. Continuous cell culture from lymphoma of Marek's disease. Biken J. 1973;16:177–179. [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1995. [Google Scholar]
- 5.Baggiolini M. Chemokines and leukocyte traffic. Nature. 1998;392:565–568. doi: 10.1038/33340. [DOI] [PubMed] [Google Scholar]
- 6.Baggiolini M, Dewald B, Moser B. Interleukin-8 and related chemotactic cytokines—CXC and CC chemokines. Adv Immunol. 1994;55:97–179. [PubMed] [Google Scholar]
- 7.Barker K A, Hampe A, Stoeckle M Y, Hanafusa H. Transformation-associated cytokine 9E3/CEF4 is chemotactic for chicken peripheral blood mononuclear cells. J Virol. 1993;67:3528–3533. doi: 10.1128/jvi.67.6.3528-3533.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Becker E L, Showell H J, Naccache P H, Freer R J, Walenga R W, Shaafi R I. Chemotactic factors: locomotory hormones. In: Karnovsky M L, Bolis L, editors. Phagocytosis, past and future. New York, N.Y: Academic Press, Inc.; 1982. pp. 87–103. [Google Scholar]
- 9.Bedard P A, Alcorta D, Simmons D L, Luk K C, Erikson R L. Constitutive expression of a gene encoding a polypeptide homologous to biologically active human platelet protein in Rous sarcoma virus-transformed fibroblasts. Proc Natl Acad Sci USA. 1987;84:6715–6719. doi: 10.1073/pnas.84.19.6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyden S V. The chemotactic effect of mixtures of antibody and antigen on polymorphonuclear leucocytes. J Exp Med. 1962;115:453–466. doi: 10.1084/jem.115.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruch-Gerharz D, Fehsel K, Suschek C, Michel G, Ruzicka T, Kolb-Bachofen V. A proinflammatory activity of interleukin 8 in human skin: expression of the inducible nitric oxide synthase in psoriatic lesions and cultured keratinocytes. J Exp Med. 1996;184:2007–2012. doi: 10.1084/jem.184.5.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calnek B W. Marek's disease—a model for herpesvirus oncology. Crit Rev Microbiol. 1986;12:293–320. doi: 10.3109/10408418509104432. [DOI] [PubMed] [Google Scholar]
- 13.Chen W Y, Bailey E C, McCune S L, Dong J Y, Townes T M. Reactivation of silenced, virally transduced genes by inhibitors of histone deacetylase. Proc Natl Acad Sci USA. 1997;94:5798–5803. doi: 10.1073/pnas.94.11.5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clark-Lewis I, Dewald B, Geiser T, Moser B, Baggiolini M. Platelet factor 4 binds to interleukin 8 receptors and activates neutrophils when its N terminus is modified with Glu-Leu-Arg. Proc Natl Acad Sci USA. 1993;90:3574–3577. doi: 10.1073/pnas.90.8.3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clark-Lewis I, Schumacher C, Baggiolini M, Moser B. Structure-activity relationships of interleukin-8 determined using chemically synthesized analogs. Critical role of NH2-terminal residues and evidence for uncoupling of neutrophil chemotaxis, exocytosis, and receptor binding activities. J Biol Chem. 1991;266:23128–23134. [PubMed] [Google Scholar]
- 16.Dalbey R E, Von Heijne G. Signal peptidases in prokaryotes and eukaryotes—a new protease family. Trends Biochem Sci. 1992;17:474–478. doi: 10.1016/0968-0004(92)90492-r. . (Erratum, 18:25, 1993.) [DOI] [PubMed] [Google Scholar]
- 17.Davis S J, Vierstra R D. Soluble, highly fluorescent variants of green fluorescent protein (GFP) for use in higher plants. Plant Mol Biol. 1998;36:521–528. doi: 10.1023/a:1005991617182. [DOI] [PubMed] [Google Scholar]
- 18.Dienglewicz R L, Parcells M S. Establishment of a lymphoblastoid cell line using a mutant MDV containing a green fluorescent protein expression cassette. Acta Virol. 1999;43:106–112. [PubMed] [Google Scholar]
- 19.Frohman A M, Dush M K, Martin G R. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fukuchi K, Sudo M, Lee Y S, Tanaka A, Nonoyama M. Structure of Marek's disease virus DNA: detailed restriction enzyme map. J Virol. 1984;51:102–109. doi: 10.1128/jvi.51.1.102-109.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furman P A, Coen D M, St. Clair M H, Schaffer P A. Acyclovir-resistant mutants of herpes simplex virus type 1 express altered DNA polymerase or reduced acyclovir phosphorylating activities. J Virol. 1981;40:936–941. doi: 10.1128/jvi.40.3.936-941.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gunn D M, Ngo V N, Ansel K M, Ekland E H, Cyster J G, Williams L T. A B-cell-homing chemokine made in lymphoid follicles activates Burkitt's lymphoma receptor-1. Nature. 1998;391:799–803. doi: 10.1038/35876. [DOI] [PubMed] [Google Scholar]
- 23.Hebert C A, Vitangcol R V, Baker J B. Scanning mutagenesis of interleukin-8 identifies a cluster of residues required for receptor binding. J Biol Chem. 1991;266:18989–18994. [PubMed] [Google Scholar]
- 24.Huang R, Lian J P, Robinson D, Badwey J A. Neutrophils stimulated with a variety of chemoattractants exhibit rapid activation of p21-activated kinases (Paks): separate signals are required for activation and inactivation of Paks. Mol Cell Biol. 1998;18:7130–7138. doi: 10.1128/mcb.18.12.7130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Igarashi T, Takahashi M, Donovan J, Jessip J, Smith M, Hirai K, Tanaka A, Nonoyama M. Restriction enzyme map of herpesvirus of turkey DNA and its collinear relationship with Marek's disease virus DNA. Virology. 1987;157:351–358. doi: 10.1016/0042-6822(87)90277-7. [DOI] [PubMed] [Google Scholar]
- 26.Inagi R, Guntapong R, Nakao M, Ishino Y, Kawanishi K, Isegawa Y, Yamanishi K. Human herpesvirus 6 induces IL-8 gene expression in human hepatoma cell line, Hep G2. J Med Virol. 1996;49:34–40. doi: 10.1002/(SICI)1096-9071(199605)49:1<34::AID-JMV6>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 27.Jones D, Lee L, Liu J L, Kung H J, Tillotson J K. Marek disease virus encodes a basic-leucine zipper gene resembling the fos/jun oncogenes that is highly expressed in lymphoblastoid tumors. Proc Natl Acad Sci USA. 1992;89:4042–4046. doi: 10.1073/pnas.89.9.4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaiser P, Hughes S, Bumstead N. The chicken 9E3/CEF4 CXC chemokine is the avian orthologue of IL8 and maps to chicken chromosome 4 syntenic with genes flanking the mammalian chemokine cluster. Immunogenetics. 1999;49:673–684. doi: 10.1007/s002510050664. [DOI] [PubMed] [Google Scholar]
- 29.Kelner G S, Kennedy J, Bacon K B, Kleyensteuber S, Largaespada D A, Jenkins N A, Copeland N G, Bazan J F, Moore K W, Schall T J, et al. Lymphotactin: a cytokine that represents a new class of chemokine. Science. 1994;266:1395–1399. doi: 10.1126/science.7973732. [DOI] [PubMed] [Google Scholar]
- 30.Kost T A, Theodorakis N, Hughes S H. The nucleotide sequence of the chick cytoplasmic beta-actin gene. Nucleic Acids Res. 1983;11:8287–8301. doi: 10.1093/nar/11.23.8287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee L F, Wu P, Sui D, Ren D, Kamil J, Kung H J, Witter R L. The complete unique long sequence and the overall genomic organization of the GA strain of Marek's disease virus. Proc Natl Acad Sci USA. 2000;97:6091–6096. doi: 10.1073/pnas.97.11.6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Legler D F, Loetscher M, Roos R S, Clark-Lewis I, Baggiolini M, Moser B. B cell-attracting chemokine 1, a human CXC chemokine expressed in lymphoid tissues, selectively attracts B lymphocytes via BLR1/CXCR5. J Exp Med. 1998;187:655–660. doi: 10.1084/jem.187.4.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu J L, Lee L F, Ye Y, Qian Z, Kung H J. Nucleolar and nuclear localization properties of a herpesvirus bZIP oncoprotein, MEQ. J Virol. 1997;71:3188–3196. doi: 10.1128/jvi.71.4.3188-3196.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu J L, Lin S F, Xia L, Brunovskis P, Li D, Davidson I, Lee L F, Kung H J. MEQ and V-IL8: cellular genes in disguise? Acta Virol. 1999;43:94–101. [PubMed] [Google Scholar]
- 35.Luscinskas F W, Kiely J M, Ding H, Obin M S, Hebert C A, Baker J B, Gimbrone M A., Jr In vitro inhibitory effect of IL-8 and other chemoattractants on neutrophil-endothelial adhesive interactions. J Immunol. 1992;149:2163–2171. [PubMed] [Google Scholar]
- 36.Martins-Green M, Feugate J E. The 9E3/CEF4 gene product is a chemotactic and angiogenic factor that can initiate the wound-healing cascade in vivo. Cytokine. 1998;10:522–535. doi: 10.1006/cyto.1997.0311. [DOI] [PubMed] [Google Scholar]
- 37.Martins-Green M, Hanafusa H. The 9E3/CEF4 gene and its product the chicken chemotactic and angiogenic factor (cCAF): potential roles in wound healing and tumor development. Cytokine Growth Factor Rev. 1997;8:221–232. doi: 10.1016/s1359-6101(97)00016-6. [DOI] [PubMed] [Google Scholar]
- 38.Martins-Green M, Tilley C, Schwarz R, Hatier C, Bissell M J. Wound-factor-induced and cell cycle phase-dependent expression of 9E3/CEF4, the avian gro gene. Cell Regul. 1991;2:739–752. doi: 10.1091/mbc.2.9.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McGeoch D J. The genomes of the human herpesviruses: contents, relationships, and evolution. Annu Rev Microbiol. 1989;43:235–265. doi: 10.1146/annurev.mi.43.100189.001315. [DOI] [PubMed] [Google Scholar]
- 40.Morgan R W, Cantello J L, McDermott C H. Transfection of chicken embryo fibroblasts with Marek's disease virus DNA. Avian Dis. 1990;34:345–351. [PubMed] [Google Scholar]
- 41.Murayama T, Kuno K, Jisaki F, Obuchi M, Sakamuro D, Furukawa T, Mukaida N, Matsushima K. Enhancement of human cytomegalovirus replication in a human lung fibroblast cell line by interleukin-8. J Virol. 1994;68:7582–7585. doi: 10.1128/jvi.68.11.7582-7585.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murayama T, Ohara Y, Obuchi M, Khabar K S A, Higashi H, Mukaida N, Matsushima K. Human cytomegalovirus induces interleukin-8 production by a human monocytic cell line, THP-1, through acting concurrently on AP-1- and NF-κB-binding sites of the interleukin-8 gene. J Virol. 1997;71:5692–5695. doi: 10.1128/jvi.71.7.5692-5695.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oakes J E, Monteiro C A, Cubitt C L, Lausch R N. Induction of interleukin-8 gene expression is associated with herpes simplex virus infection of human corneal keratocytes but not human corneal epithelial cells. J Virol. 1993;67:4777–4784. doi: 10.1128/jvi.67.8.4777-4784.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ono M, Katsuragi-Iwanaga R, Kitazawa T, Kamiya N, Horimoto T, Niikura M, Kai C, Hirai K, Mikami T. The restriction endonuclease map of Marek's disease virus (MDV) serotype 2 and collinear relationship among three serotypes of MDV. Virology. 1992;191:459–463. doi: 10.1016/0042-6822(92)90210-g. [DOI] [PubMed] [Google Scholar]
- 45.Parcells S M, Anderson A S, Morgan T W. Retention of oncogenicity by a Marek's disease virus mutant lacking six unique short region genes. J Virol. 1995;69:7888–7898. doi: 10.1128/jvi.69.12.7888-7898.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parcells S M, Dienglewicz R L, Anderson A S, Morgan R W. Recombinant Marek's disease virus (MDV)-derived lymphoblastoid cell lines: regulation of a marker gene within the context of the MDV genome. J Virol. 1999;73:1362–1373. doi: 10.1128/jvi.73.2.1362-1373.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pease J E, Murphy P M. Microbial corruption of the chemokine system: an expanding paradigm. Semin Immunol. 1998;10:169–178. doi: 10.1006/smim.1998.0129. [DOI] [PubMed] [Google Scholar]
- 48.Peichl P, Ceska M, Effenberger F, Haberhauer G, Broell H, Lindley I J. Presence of NAP-1/IL-8 in synovial fluids indicates a possible pathogenic role in rheumatoid arthritis. Scand J Immunol. 1991;34:333–339. doi: 10.1111/j.1365-3083.1991.tb01554.x. [DOI] [PubMed] [Google Scholar]
- 49.Penfold E M, Dairaghi D J, Duke G M, Saederup N, Mocarski E S, Kemble G W, Schall T J. Cytomegalovirus encodes a potent alpha chemokine. Proc Natl Acad Sci USA. 1999;96:9839–9844. doi: 10.1073/pnas.96.17.9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peng Q, Shirazi Y. Isolation and characterization of Marek's disease virus (MDV) cDNAs from a MDV-transformed lymphoblastoid cell line: identification of an open reading frame antisense to the MDV Eco-Q protein (Meq) Virology. 1996;221:368–374. doi: 10.1006/viro.1996.0388. [DOI] [PubMed] [Google Scholar]
- 51.Peng Q, Zeng M, Bhuiyan Z A, Ubukata E, Tanaka A, Nonoyama M, Shirazi Y. Isolation and characterization of Marek's disease virus (MDV) cDNAs mapping to the BamHI-I2, BamHI-Q2, and BamHI-L fragments of the MDV genome from lymphoblastoid cells transformed and persistently infected with MDV. Virology. 1995;213:590–599. doi: 10.1006/viro.1995.0031. [DOI] [PubMed] [Google Scholar]
- 52.Powell P C, Payne L N. Marek's disease. In: McFerran J B, McBulty M S, editors. Virus infections of birds. Vol. 4. New York, N.Y: Elsevier Science; 1993. pp. 37–75. [Google Scholar]
- 53.Pratt W D, Morgan R W, Schat K A. Characterization of reticuloendotheliosis virus-transformed avian T-lymphoblastoid cell lines infected with Marek's disease virus. J Virol. 1992;66:7239–7244. doi: 10.1128/jvi.66.12.7239-7244.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ren D, Lee L F, Coussens P M. Identification and characterization of Marek's disease virus genes homologous to ICP27 and glycoprotein K of herpes simplex virus-1. Virology. 1994;204:242–250. doi: 10.1006/viro.1994.1528. [DOI] [PubMed] [Google Scholar]
- 55.Rollins B J. Chemokines. Blood. 1997;90:909–928. [PubMed] [Google Scholar]
- 56.Ross N L. T-cell transformation by Marek's disease virus. Trends Microbiol. 1999;7:22–29. doi: 10.1016/s0966-842x(98)01427-9. [DOI] [PubMed] [Google Scholar]
- 57.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 58.Schat K A, Calnek B W, Fabricant J. Characterization of two highly oncogenic strains of Marek's disease virus. Avian Pathol. 1982;11:593–605. doi: 10.1080/03079458208436134. [DOI] [PubMed] [Google Scholar]
- 59.Schonbeck U, Brandt E, Petersen F, Flad H D, Loppnow H. IL-8 specifically binds to endothelial but not to smooth muscle cells. J Immunol. 1995;154:2375–2383. [PubMed] [Google Scholar]
- 60.Schroder J M, Mrowietz U, Morita E, Christophers E. Purification and partial biochemical characterization of a human monocyte-derived, neutrophil-activating peptide that lacks interleukin 1 activity. J Immunol. 1987;139:3474–3483. [PubMed] [Google Scholar]
- 61.Sick C, Schneider K, Staeheli P, Weining K C. Novel chicken CXC and CC chemokines. Cytokine. 2000;12:181–186. doi: 10.1006/cyto.1999.0543. [DOI] [PubMed] [Google Scholar]
- 62.Springer T A. Traffic signals for lymphocyte recirculation and leukocyte emigration: the multistep paradigm. Cell. 1994;76:301–314. doi: 10.1016/0092-8674(94)90337-9. [DOI] [PubMed] [Google Scholar]
- 63.Stone E M, Rothblum K N, Alevy M C, Kuo T M, Schwartz R J. Complete sequence of the chicken glyceraldehyde-3-phosphate dehydrogenase gene. Proc Natl Acad Sci USA. 1985;82:1628–1632. doi: 10.1073/pnas.82.6.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sugano S, Stoeckle M Y, Hanafusa H. Transformation by Rous sarcoma virus induces a novel gene with homology to a mitogenic platelet protein. Cell. 1987;49:321–328. doi: 10.1016/0092-8674(87)90284-4. [DOI] [PubMed] [Google Scholar]
- 65.van Berkel V, Barrett J, Tiffany H L, Fremont D H, Murphy P M, McFadden G, Speck S H, Virgin H I. Identification of a gammaherpesvirus selective chemokine binding protein that inhibits chemokine action. J Virol. 2000;74:6741–6747. doi: 10.1128/jvi.74.15.6741-6747.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Walz A, Peveri P, Aschauer H, Baggiolini M. Purification and amino acid sequencing of NAF, a novel neutrophil-activating factor produced by monocytes. Biochem Biophys Res Commun. 1987;149:755–761. doi: 10.1016/0006-291x(87)90432-3. [DOI] [PubMed] [Google Scholar]
- 67.Witter R L. Increased virulence of Marek's disease virus field isolates. Avian Dis. 1997;41:149–163. [PubMed] [Google Scholar]
- 68.Witter R L, Gimeno I M, Reed W M, Bacon L D. An acute form of transient paralysis induced by highly virulent strains of Marek's disease virus. Avian Dis. 1999;43:704–720. [PubMed] [Google Scholar]
- 69.Wu Z, Eaton S F, Laue T M, Johnson K W, Sana T R, Ciardelli T L. Coiled-coil molecular recognition: directed solution assembly of receptor ectodomains. Protein Eng. 1994;7:1137–44. doi: 10.1093/protein/7.9.1137. [DOI] [PubMed] [Google Scholar]
- 70.Yoshimura T, Matsushima K, Tanaka S, Robinson E A, Appella E, Oppenheim J J, Leonard E J. Purification of a human monocyte-derived neutrophil chemotactic factor that has peptide sequence similarity to other host defense cytokines. Proc Natl Acad Sci USA. 1987;84:9233–9237. doi: 10.1073/pnas.84.24.9233. [DOI] [PMC free article] [PubMed] [Google Scholar]