Abstract
Background
The disease resistance phenotype is closely related to immunomodulatory function and immune tolerance and has far-reaching implications in animal husbandry and human health. Microbes play an important role in the initiation, prevention, and treatment of diseases, but the mechanisms of host–microbiota interactions in disease-resistant phenotypes are poorly understood. In this study, we hope to uncover and explain the role of microbes in intestinal diseases and their mechanisms of action to identify new potential treatments.
Methods
First, we established the colitis model of DSS in two breeds of sheep and then collected the samples for multi-omics testing including metagenes, metabolome, and transcriptome. Next, we made the fecal bacteria liquid from the four groups of sheep feces collected from H-CON, H-DSS, E-CON, and E-DSS to transplant the fecal bacteria into mice. H-CON feces were transplanted into mice named HH group and H-DSS feces were transplanted into mice named HD group and Roseburia bacteria treatment named HDR groups. E-CON feces were transplanted into mice named EH group and E-DSS feces were transplanted into mice in the ED group and Roseburia bacteria treatment named EDR groups. After successful modeling, samples were taken for multi-omics testing. Finally, colitis mice in HD group and ED group were administrated with Roseburia bacteria, and the treatment effect was evaluated by H&E, PAS, immunohistochemistry, and other experimental methods.
Results
The difference in disease resistance of sheep to DSS-induced colitis disease is mainly due to the increase in the abundance of Roseburia bacteria and the increase of bile acid secretion in the intestinal tract of Hu sheep in addition to the accumulation of potentially harmful bacteria in the intestine when the disease occurs, which makes the disease resistance of Hu sheep stronger under the same disease conditions. However, the enrichment of harmful microorganisms in East Friesian sheep activated the TNFα signalling pathway, which aggravated the intestinal injury, and then the treatment of FMT mice by culturing Roseburia bacteria found that Roseburia bacteria had a good curative effect on colitis.
Conclusion
Our study showed that in H-DSS-treated sheep, the intestinal barrier is stabilized with an increase in the abundance of beneficial microorganisms. Our data also suggest that Roseburia bacteria have a protective effect on the intestinal barrier of Hu sheep. Accumulating evidence suggests that host–microbiota interactions are associated with IBD disease progression.
Video Abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s40168-024-01932-8.
Keywords: Roseburia bacteria, Intestinal inflammation, Fecal microbiota transplantation (FMT), Gut microbiota
Introduction
Even within the same species, different microbial communities in the gut display enormous diversity, and the current data point to a relationship between host characteristics and gut bacteria [1]. The gut microbiota also undergoes compositional changes over the course of an individual’s life, as either the cause or consequence of changes in host health and disease status [2]. Host physiology can be altered at the cellular level by microbiome-induced cell signalling, proliferation and neurotransmitter biosynthesis, leading to mucosal and systemic alterations and thereby affecting homeostasis, barrier function, innate and adaptive immune responses, and metabolism [3]. The immune response to disease is an essential indicator of the host’s disease resistance phenotype, which differs depending on the microbiota composition [4]. The microbial communities and their metabolites and components are not only necessary for immune homeostasis but also influence the susceptibility of the host to many immune-mediated diseases and disorders [5]. Although disruption of the host’s intrinsic homeostasis and changes in the microbiota are what cause these diseases to develop, how the microbiota contributes to various disease-resistant phenotypes remains unknown. Given the significance and complexity of the microbiota in many species of sheep, there is growing interest in elucidating the microbiota’s composition and functional role to comprehend the processes that lead to disease resistance phenotypes [6]. In recent years, more and more researchers have focused on potentially beneficial bacteria. One of them is Roseburia bacteria, whose metabolites have been shown to prevent intestinal inflammation in 2002 as a gram-positive bacteria and obligate anaerobic, butyrate-producing bacterium which was first isolated from human feces [7]. However, current research has not applied Roseburia bacteria to cure diseases in domestic animals. In our study, Roseburia bacteria was firstly used as the primary therapeutic substance in DSS-induced acute colitis. The East Friesian sheep (EFS) is one of the most productive dairy sheep breeds that provide dairy products and byproducts needed for daily human life [8]. East Friesian sheep have been imported to many countries and are often used to improve the qualities of native sheep in various countries due to their good milk and meat production performance [9]. The Hu sheep is a famous sheep breed from the Taihu Plain in China that has the advantages of high prolificacy, year-round estrus, and fast growth. However, it is still difficult to develop their excellent traits in the advantages of two sheep [10]. Colitis is one of the most reasons. The costs associated with enteritis in the livestock sector, including deaths, lost productivity range from US$ 10 million to US$ 29 million each year, and the cost is increasing every year [11]. Therefore, the treatment and prevention of enteritis is very vital. A growing body of research shows differences in disease resistance between different species of sheep [12]. However, the mechanism of action of microorganisms is not clear. Although East Friesian sheep have many advantages. However, East Friesian sheep have poor disease resistance [13]. We hypothesize that East Friesian sheep may lack important host–microbiota interactions, resulting in in a less adaptive host under disease and inflammatory immunostimulant stress. DSS-induced colitis is a well-established model of acute colitis with ulceration that resembles ulcerative colitis in humans [14]. Ulcerative colitis, a type of IBD, can be viewed as an autoimmune disease that is strongly influenced by disruptions in host–microbiota homoeostasis [15]. The cause of colitis is most likely related to changes in microorganisms and metabolites in the host. This study provides information on the underlying fundamental link between the gut microbiome and host disease resistance phenotypes, which can aid in the identification of core gut microbial candidates associated with host health and disease resistance phenotypes [16]. Therefore, we used a DSS-induced colitis model to identify certain commensal bacteria and their metabolites in Hu sheep and East Friesian sheep that may affect the physiological adaptation of the host and thus susceptibility to IBD. Then, microbiomics, metabolomics, transcriptomics, and other multiomics methods were combined to explore the underlying mechanism of disease resistance.
Results
Different breeds of sheep exhibit different resistance phenotypes
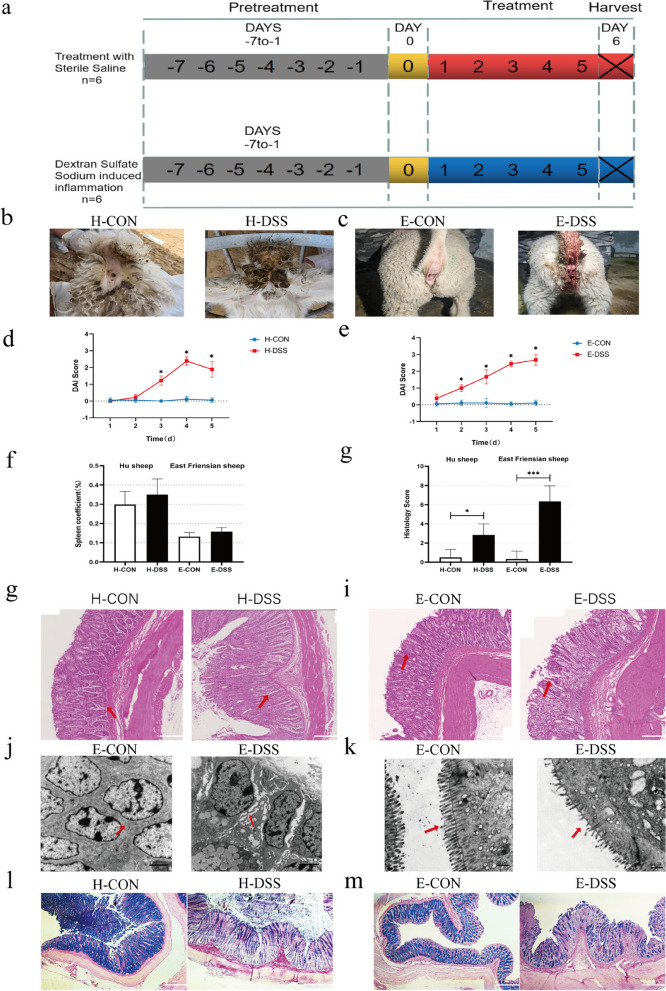
Hu sheep and East Friesian sheep exhibited different phenotypic disease resistance traits, which may be related to differences in the composition of their respective gut microbiota. To test this hypothesis, we first determined whether there is a significant phenotypic difference between Hu sheep and East Friesian sheep. Therefore, we compared the control Hu sheep with the model Hu sheep with acute colitis (H-CON and H-DSS) and the control East Friesian sheep with the model East Friesian sheep with acute colitis (E-CON and E-DSS). We selected 12 2-month-old male Hu sheep and 12 2-month-old male East Friesian sheep, and sheep of the same breed were randomly divided into two groups. After grouping, the sheep are first acclimatized to the environment for 7 days, during which all sheep eat and drink freely, and the initial body weight was measured on day 8. Afterwards, a 5-day DSS infusion was started, during which the control group was gavaged with saline. During the molding period, all the experiment sheep under ad libitum feeding and drinking. Finally, on the sixth day after gavage, samples were collected (Fig. 1a). We recorded diarrhea in Hu sheep and East Friesian sheep and compared with those in the control group; the Hu sheep and East Friesian sheep in the DSS group had obvious diarrhea symptoms (Fig. 1b, c). We used the disease activity index (DAI) to compare the differences in disease activity between the Hu sheep and East Friesian sheep control groups and the DSS-induced colitis model groups to more intuitively compare the differences in the resistance phenotypes between the two breeds of sheep. There was no significant change in the DAI values between the H-CON group and the E-CON group. However, the DAI values of the H-DSS and E-DSS groups increased significantly; interestingly, the DAI values of the H-DSS group showed a downwards trend in the later stage (Fig. 1d, e). The spleen, an immune organ that contains many lymphocytes, is an important indicator of immune phenotype [17]. The spleen coefficient of the DSS group was significantly greater than that of the control group (Fig. 1f). H&E staining of the colon tissues of Hu sheep and East Friesian sheep revealed that the colon inflammation and histopathology scores of the disease group were significantly greater than those of the control group, and those of the disease group of East Friesian sheep were significantly greater than those of the disease group of Hu sheep (Fig. 1g, i). Transmission electron microscopy imaging of the colon tissue of East Friesian sheep revealed that the intercellular connections, mitochondria, brush margins, and endoplasmic reticulum in the E-DSS group were significantly damaged compared with those in the E-CON group (Fig. 1j, k Extended Data Fig. 1a, b). The AB-PAS results showed that the number of goblet cells in the small intestinal epithelium of sheep in the DSS group was significantly lower than that in the control group (Fig. 1l, m). In addition, we also performed serum biochemical tests on samples from Hu sheep and East Friesian sheep and found that all the indicators showed different degrees of variation and that DSS-induced colonic inflammation also caused different degrees of damage to other parts of the body; this result indicated the successful establishment of our disease model (Extended Data Fig. 1c-t). These results show that in the disease model we established, the disease resistance phenotype of Hu sheep was significantly better than that of East Friesian sheep.
Fig. 1.
Study design for the experiment using East Friesian sheep. a Control group of Hu sheep and the model group of Hu sheep with acute colitis (H-CON and H-DSS); control group of East Friesian sheep and the model group of East Friesian sheep with acute colitis (E-CON and E-DSS) (n = 6). b Phenotypic images of the control group and the DSS group. c Phenotypic diagram of the East Friesian sheep control group and the DSS group. d DAI of the Hu sheep control group and the DSS group (n = 6). e DAI of the East Friesian sheep control group and the DSS group (n = 6). f Spleen coefficient (n = 6). g Histology score. h H&E tissue sections of Hu sheep colon. i H&E tissue sections of East Friesian sheep colon. j, k Transmission electron microscopy tissue sections of East Friesian sheep colon. l AB-PAS tissue sections of Hu sheep colon. m AB-PAS tissue sections of East Friesian sheep colon (*P < 0.05, **P < 0.01, ***P < 0.001)
The colonic microbiota underwent dramatic remodelling in the control and DSS groups of the two sheep cultivars
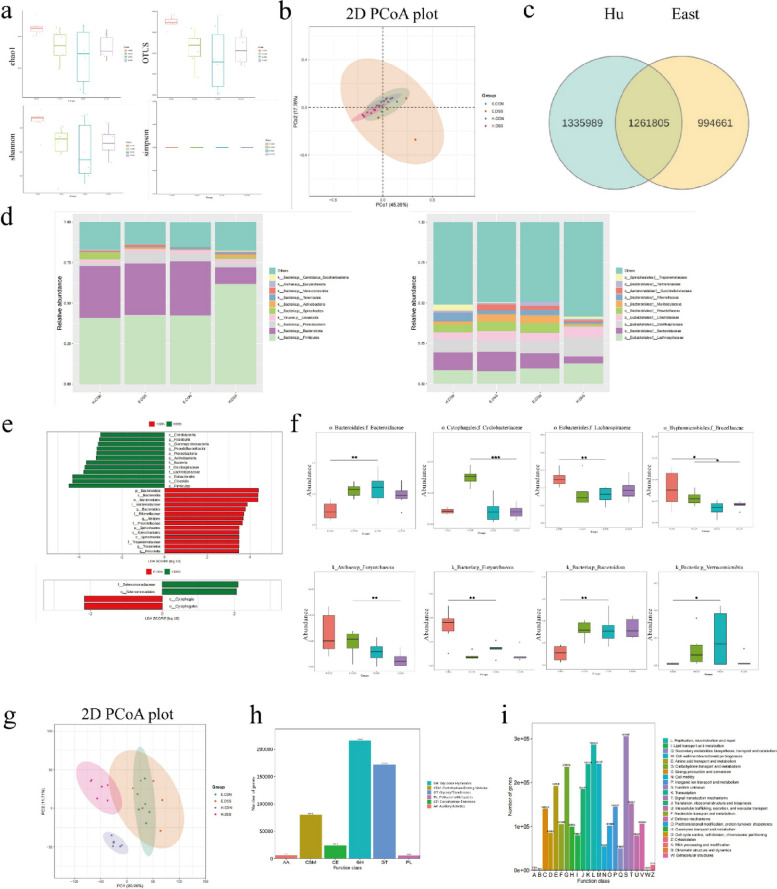
Based on the Hu sheep and East Friesian sheep phenotypes, we next determined the bacterial composition of the microbiota in the control and diseased groups of the two sheep breeds to elucidate the compositional differences between them. The richness index and indices such as the Shannon, Chao1, and other indices were used as measures of species diversity to evaluate α diversity. There were significant differences in α diversity between the H-DSS and E-DSS groups (Fig. 2a). However, there was no significant difference in β diversity between the disease group and the control group in the two breeds (Fig. 2b). We determined the total microbial community in the two types of sheep, including 1,335,989 microorganisms endemic to Hu sheep, 994,661 microorganisms endemic to East Friesian sheep, and 1,261,805 microorganisms common to both sheep (Fig. 2c). Our findings confirmed that sheep with DSS-induced colitis may experience alterations in the gut microbiota. To further understand the microbial differences between the control and diseased groups of these two breeds of sheep, we assessed the taxonomic composition of the colonic microbiota. Both sheep exhibited dramatic changes in bacterial abundance at both the phylum and family levels, with the abundances of the phyla Firmicutes, Proteobacteria, Actinobacteria, Tenericutes, and Candidatus_saccharibacteria increasing in the H-DSS group, while the abundances of Bacteroidota, Spirochaetes, and Verrucomicrobia decreased. In the E-DSS group, the abundances of Proteobacteria, Spirochaetes, Actinobacteria, and Verrucomicrobia increased, while those of Bacteroidota, Tenericutes, and Candidatus_saccharibacteria decreased. At the family level, Lachnospiraceae, Oscillospiraceae, Clostridiaceae, and Succinivibrionaceae abundance increased in the H-DSS group, while Bacteroidaceae, Prevotellaceae, Muribaculaceae, Rikenellaceae, and Treponemataceae abundance decreased. In the E-DSS group, the abundances of Bacteroidaceae, Succinivibrionaceae, and Treponemataceae increased, while those of Lachnospiraceae, Oscillospiraceae, Prevotellaceae, Muribaculaceae, Rikenellaceae, and Tannerellaceae decreased (Fig. 2d). The microbial abundance in the sheep after DSS induction changed significantly compared to that in the control group, but the microbial changes in the DSS group were significantly different in Hu sheep and East Friesian sheep. Interestingly, we found that the potential probiotics Lachnospiraceae, Oscillospiraceae, and Clostridiaceae were clustered in large numbers in the H-DSS group, while in the E-DSS group, the abundance of these probiotics decreased and potentially harmful bacteria such as Bacteroidaceae and Treponemataceae were clustered in large numbers. This phenomenon may be one of the reasons for the difference in the disease resistance phenotype between the two types of sheep. As harmful microorganisms are enriched in disease models, they may play a role in disease. However, potential beneficial microorganisms are also enriched in the intestinal tract of Hu sheep, and these potential beneficial microorganisms may play a certain role in the treatment and protection of the intestine [18]. Among these potentially beneficial bacteria, the most common are Lachnospiraceae and Oscillospiraceae. The probiotics Lachnospiraceae and Oscillospiraceae produce butyrate, which has a reparative and protective effect in the intestine [19]. Among the many microbial changes, there was the most obvious changes in Lachnospiraceae_Roseburia abundance, which increased in the H-DSS group and decreased in the E-DSS group; thus, the abundance showed the opposite trend in the two groups (Fig. 2e, f). Roseburia intestinalis is an anaerobic, gram-positive, slightly curved rod-shaped flagellated bacterium that produces butyrate in the colon. R. intestinalis has been shown to prevent intestinal inflammation and help maintain energy homeostasis via the production of metabolites [20]. The Hu sheep are highly resistant to intestinal diseases, which is likely because the butyrate secreted by Roseburia plays a role in repairing and protecting the intestine. In East Friesian sheep, on the one hand, intestinal lesions occurred due to the enrichment of harmful microorganisms, and on the other hand, the decrease in the abundance of potential beneficial microorganisms further aggravated the disease phenotype. Next, we analyzed microbial function, and the PCA results showed that there was a difference in microbial function between the H-DSS group and the H-CON group, while there was no significant difference between the E-DSS group and the E-CON group (Fig. 2g). Through the functional analysis of all microorganisms, we found that the main enriched pathways were lipid transport metabolism, amino acid transport metabolism, nucleotide transport metabolism, and carbohydrate transport metabolism (Fig. 2h, i).
Fig. 2.
Microbial results for Hu sheep and East Friesian sheep. a α diversity of Hu sheep and East Friesian sheep. b β diversity of Hu sheep and East Friesian sheep. c Venn diagram of Hu sheep and East Friesian sheep microorganisms. d Changes in microbial abundance at the phylum and genus levels. e Fold change diagram of differential microorganisms between Hu sheep and East Friesian sheep. f Box plot of microbial changes. g PCA diagram of microbial function. h, i Functional enrichment map of microbial genes (*P < 0.05, **P < 0.01)
Radical alteration of metabolite levels
Microorganisms are inextricably linked to their metabolites, and since the two breeds of sheep underwent dramatic microbial remodelling after DSS induction, we then evaluated the nontargeted metabolites of the two breeds of sheep. The PCA results showed significant changes in the metabolites of the colon contents of both breeds, which is clearly due to remodelling of the microorganism community (Extended Data Fig. 2a). There were 448 unique differentially abundant metabolites between E-DSS and E-CON, 217 unique differentially abundant metabolites between H-DSS and H-CON, and 26 identical differentially abundant metabolites found in both varieties (Extended Data Fig. 2b). Differences in these metabolites greatly affect the physiological state and function of the body [21]. Compared with those in the H-CON group, the differentially abundant metabolites in the H-DSS group were mainly heterocyclic compounds, amino acids and their metabolites, bile acids, organic acids and their derivatives, carbohydrates and their metabolites, etc. Compared with those in the E-CON group, the differentially abundant metabolites in the E-DSS group were mainly organic acids and their derivatives, hybrid compounds, amino acids and their metabolites, benzene, and its derivatives, etc. Among the differentially abundant metabolites, the levels of bile acids in the H-DSS group increased; bile acids play a very important role in the production of secondary bile acids and the regulation of host metabolism and immune system activity [22] (Extended Data Fig. 2c). In the E-DSS group, we found an increase in the proportion of benzene and its derivatives. Benzene and its derivatives can destroy the intestines, thereby aggravating inflammation [23]. The discovery of these two types of metabolites in the H-DSS group and the E-DSS group may be one of the reasons for the phenotypic differences in disease resistance between the two breeds (Extended Data Fig. 2d). There were 136 metabolites with upregulated expression and 107 metabolites with downregulated expression in the H-DSS group compared with the H-CON group. Compared with those in the E-CON group, there were 53 metabolites with upregulated expression and 421 metabolites with downregulated expression in the E-DSS group (Extended Data Fig. 2e, f). We selected the top 10 metabolites with upregulated and downregulated expression in the H-DSS group and the E-DSS group and found that most of them were different types of amino acids and nucleotides (Extended Data Fig. 2g, h). To determine the functions of the differentially abundant metabolites, we performed a KEGG enrichment assay. The KEGG enrichment results for H-CON vs. H-DSS and E-CON vs. E-DSS varied widely, but we noted that the differentially abundant metabolites in H-CON vs. H-DSS were mainly enriched in the secretion of primary cholic acid, which was consistent with our previous results (Extended Data Fig. 2i, j). The violin diagram shows this result intuitively (Extended Data Fig. 2k, l). To visualize the connections between microorganisms and metabolites, we performed a correlation analysis. The correlation analysis of microorganisms and metabolites in Hu sheep revealed that potentially beneficial bacteria, such as roses and rumen coccus, were positively correlated with potential beneficial metabolites, such as amino acids and short-chain fatty acids, while the correlation between microorganisms and metabolites in East Friesian sheep was not significant (Extended Data Fig. 3a, b).
RNA-seq analysis was used to explore the mechanism underlying the disease resistance phenotype in the two sheep breeds
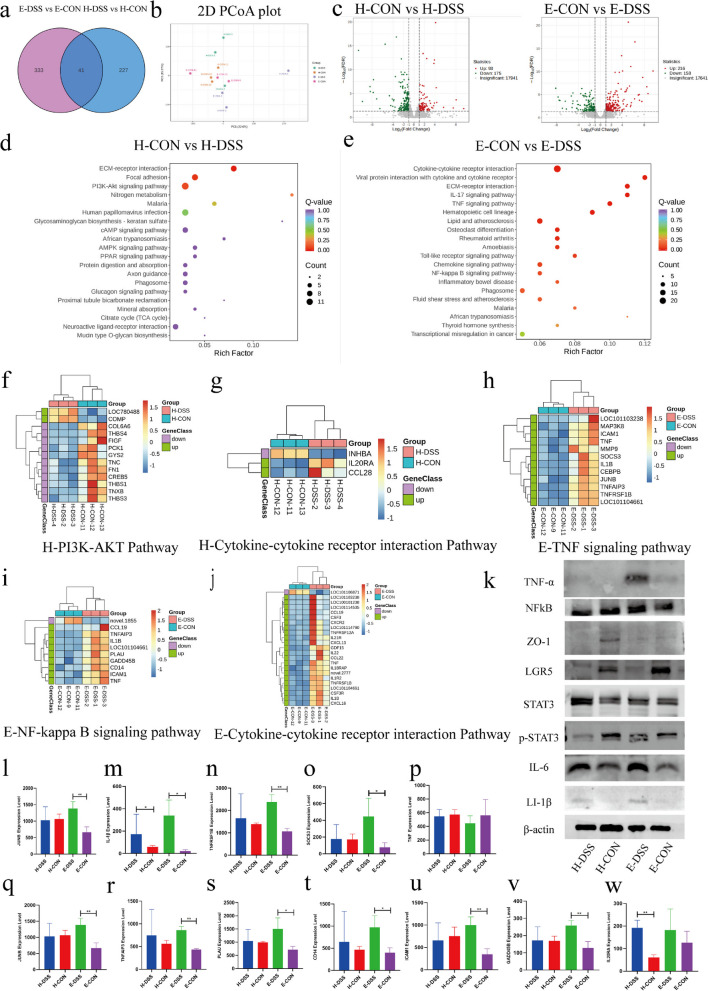
To explore the underlying role of changes in the gut microbiota in host colon disease resistance, we used RNA-seq analysis to quantify the gene expression profiles in the colon of these two varieties of sheep. There were 227 (differentially expressed genes) DEGs specific to the H-CON vs. H-DSS group, 333 DEGs specific to the E-CON vs. E-DSS group, and 41 DEGs common in both groups (Fig. 3a). The PCA plot revealed that there were significant differences between genes in the H-DSS group and those in the E-DSS group (Fig. 3b). Figure 3c shows the volcano plots that were generated to visualize the distribution in gene expression between control and diseased Hu sheep and East Friesian sheep. In total, 93 genes had upregulated expression and 175 genes had downregulated expression in H-DSS sheep compared with H-CON sheep, and 216 genes had upregulated expression and 158 genes had downregulated expression in E-DSS sheep compared with E-CON sheep (Fig. 3c). To better understand the genetic differences between microbial and host interactions in control and diseased Hu sheep and East Friesian sheep, KEGG enrichment analysis was performed. ECM-receptor interaction, focal adhesion, and the PI3K-Akt signalling pathway were enriched in DEGs between H-DSS sheep and H-CON sheep (Fig. 3d). In E-DSS sheep, KEGG analysis revealed that the DEGs in the E-DSS sheep when compared with E-CON sheep were enriched in the cytokine‒cytokine receptor interaction pathway, the IL-17 signalling pathway, the TNF signalling pathway, the Toll-like receptor signalling pathway, and the NF-kappa B signalling pathway (Fig. 3e). Some signalling pathways, such as the TNF signalling pathway, the IL-17 signalling pathway, and the NF-kappa B signalling pathway, showed enrichment of DEGS in the E-DSS group but not DEGs in the H-DSS group. The genes in these pathways exhibited substantially upregulated expression in E-DSS sheep, which indicated that inflammation was exacerbated in East Friesian sheep. Next, we generated a heatmap of the signalling pathways and found that key genes, such as MAP3K8, TNF, IL1β, JUNB, PLAU, CD14, and other proinflammatory genes, had significantly upregulated expression in the E-DSS-enriched signalling pathway (Fig. 3f–j). By determining the RNA and protein levels of these key genes in these enrichment pathways, we found that the relative expression levels in the E-DSS group were significantly greater than those in the H-DSS group (Fig. 3k–w). Based on these results, we hypothesize that the reason why the disease resistance phenotype of Hu sheep is better than that of East Friesian sheep is that the intestinal microbiota of Hu sheep changes after DSS induction; the abundance of beneficial bacteria, such as rose and rumen coccus, increased, and the total bile content of metabolites increased, which protected the intestine. Compared with H-CON sheep, East Friesian sheep showed the opposite result; due to the enrichment of the potentially harmful bacteria Bacteroidaceae and Treponemataceae in the intestine and the enrichment of benzene and its derivatives, colon inflammation was aggravated, and the TNF signalling pathway, the NF-kappa B signalling pathway, and the IL-17 signalling pathway were activated, which further aggravated intestinal disease in the E-DSS group.
Fig. 3.
RNA-seq analysis was used to explore the mechanism underlying the disease resistance phenotype in the two sheep breeds. a Venn diagram of differentially expressed genes. b PCA map of differentially expressed genes. c Volcano plot of differentially expressed genes. d KEGG enrichment map of differentially expressed genes in Hu sheep. e KEGG enrichment map of differentially expressed genes in East Friesian sheep. f Gene heatmap of the PI3K-AKT pathway in Hu sheep. g Gene heatmap of the cytokine‒cytokine signalling pathway in Hu sheep. h Gene heatmap of the TNF signalling pathway in East Friesian sheep. i Gene heatmap of the NF-kappa B signalling pathway in East Friesian sheep. j Gene heatmap of the cytokine‒cytokine signalling pathway in East Friesian sheep. k Western blotting was used to validate the immune status and intestinal structure in sheep. l–w Relative RNA expression of differentially differentiated genes. The data are presented as the mean ± SEM (n = 3 mice per group) (*P < 0.05, **P < 0.01)
Remodelling of the gut microbiome in mice
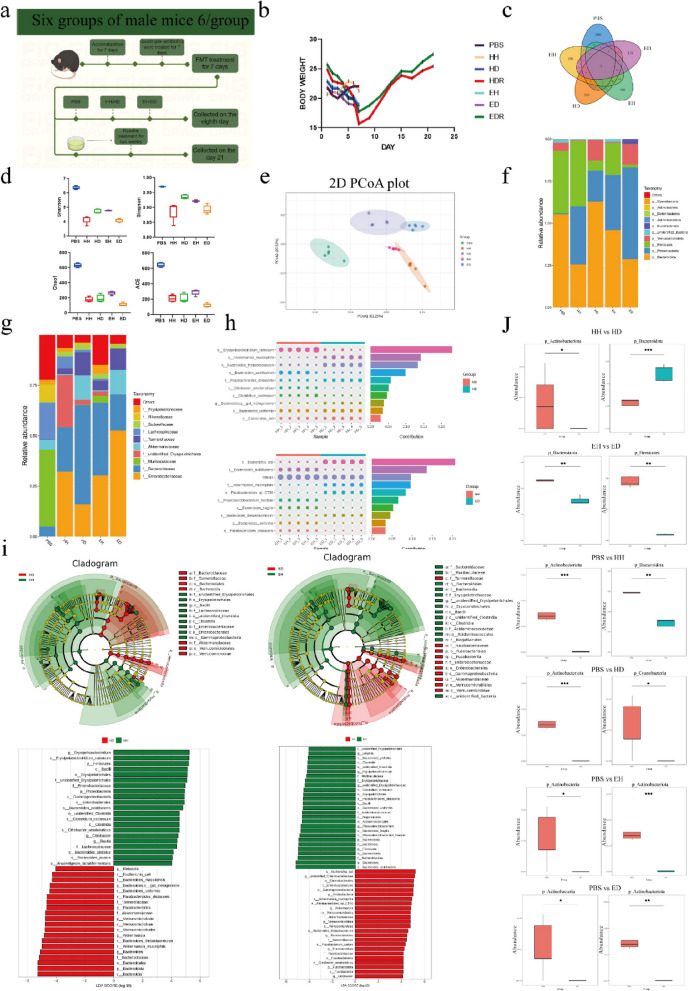
To validate our results, we performed fecal microbiota transplantation (FMT) from mice. FMT is an attractive strategy to correct microbial dysbiosis in patients with diarrhea-predominant irritable bowel syndrome [24]. The vaccination period was 1 week, with each mouse receiving 200 μL of fecal bacterial solution per day [25]. The control, HH, HD, EH, ED, HDR, and EDR groups were included. All groups except the control group were given 1 g/ml fecal microbiota solution from Hu sheep and East Friesian sheep for seven consecutive days [26]. The samples from the control, HH, HD, EH, and ED groups were collected on the eighth day after 7 days of FMT, and samples from the HDR group and the EDR group were subjected to intragastric Roseburia treatment beginning on the eighth day. The duration of the treatment was 2 weeks, and the samples were collected after 2 weeks (Fig. 4a). The results of weight change showed that the weight of rats in the PBS group was slightly greater than the initial body weight after FMT, the weights of the rats in the HD group and the ED group showed a significant downwards trend, and the weight of rats in the EH group also decreased but was greater than that of rats in the ED group. However, rats in the HDR group and the EDR group treated with Roseburia bacteria exhibited significant weight recovery after treatment; their body weights were even greater than their initial body weight (Fig. 4b). We then evaluated the colon contents of mice via 16S sequencing. Venn diagram analysis revealed that the intestinal microbiota of mice after FMT changed significantly compared with that of the PBS group (Fig. 4c). We used Chao1, Shannon, Simpson, and ACE indices to reflect the α diversity of samples, and significant changes in the α diversity were detected mice the underwent FMT compared to those in the control group. There were also significant differences between the HH and HD groups and between the EH and ED groups (Fig. 4d). These differences were also demonstrated by the PCoA results, which indicated successful FMT in mice; the results revealed differences in the gut microbiota between the different groups of sheep (Fig. 4e). Proteobacteria, Actinobacteria, Verrucomicrobia, Bacteroidetes, Actinobacteria, Fusobacteriota, Euryarchaeota, and Deferribacteres were the top 10 genera with the greatest changes in abundance at the posterior-door level in the FMT treatment group (Fig. 4f). We also examined differences in the abundance of bacteria at the phylum level between groups (Fig. 4j). There were changes in Enterobacteriaceae, Bacteroidaceae, Muribaculaceae, Unidentified Erysipelotrichales, Akkermansiaceae, Tannereuaceae, Lachnospiraceae, Sutterellaceae, Rikenellaceae, and Erysipelotrichaceae abundance at the family level (Fig. 4g-h). To identify specific bacterial genera that were characteristic of the two breeds, linear discriminant analysis effect size (LEFSe) was used to further evaluate the differences in bacterial composition between animals (Fig. 4i). We found similar results in both types of sheep, with a reduction in Lachnospiraceae abundance in the HD group compared to the HH group, and a large decrease in Lachnospiraceae abundance in the ED group compared to the EH group; these results are consistent with the sheep microbiome results (Fig. 4 j).
Fig. 4.
Remodelling of the mouse gut microbiome. a Experimental treatment and grouping were performed as follows: PBS, normal control group (not treated); HH, H-CON feces gavage for 7 days; HD, H-DSS feces gavage for 7 days; HDR, H-DSS feces gavage for 7 days; Roseburia gavage for 14 days; EH, E-CON feces gavage for 7 days; ED, E-DSS feces gavage for 7 days; and EDR, E-DSS feces gavage for 7 days; Roseburia gavage for 14 days. b Weight change (n = 6). c Venn diagram of differential microorganisms in FMT mice. d α diversity of mice. eβ diversity of mice; f, g Changes in microbial abundance at the phylum and genus levels. h Fold change diagram of differential microorganisms between HH and HD, EH, and ED. i Fold change plot of differential microorganisms in LEfSe mice. j Box diagram of differentially abundant microbes in mice (*P < 0.05, **P < 0.01)
Changes in metabolites in mice after FMT
The metabolite profiles of mice also underwent dramatic remodelling in the different groups of sheep, as was shown via metabolite analysis (Extended Data Fig. 4a). There were 2470 metabolites with upregulated expression and 1266 metabolites with downregulated expression in the HD group compared with the HH group. Compared with those in the EH group, there were 2637 metabolites with upregulated expression and 1163 metabolites with downregulated expression in the ED group (Extended Data Fig. 4b, c). The Venn diagram results showed that the types of metabolites changed dramatically in all five groups (Extended Data Fig. 4d). Among these changes, the changes in the HH vs. HD group involved bile acids, while benzene and its substituted derivatives were more abundant in the EH vs. ED group. This finding is also in line with previous results obtained from these sheep (Extended Data Fig. 4e, f). We also measured the level of the top 49 metabolites (Extended Data Fig. 4g, h), and to determine the function of the differentially abundant metabolites, we performed KEGG enrichment analysis (Extended Data Fig. 4i, j). Notably, in the HH vs. HD comparison, bile secretion was also the main enriched pathway. As an important factor in regulating the composition of intestinal microbes, bile continuously affects the host, and the main component of bile that exerts different effects is bile acid [27].
RNA-seq analysis was used to explore the mechanism underlying the disease resistance phenotype in mice
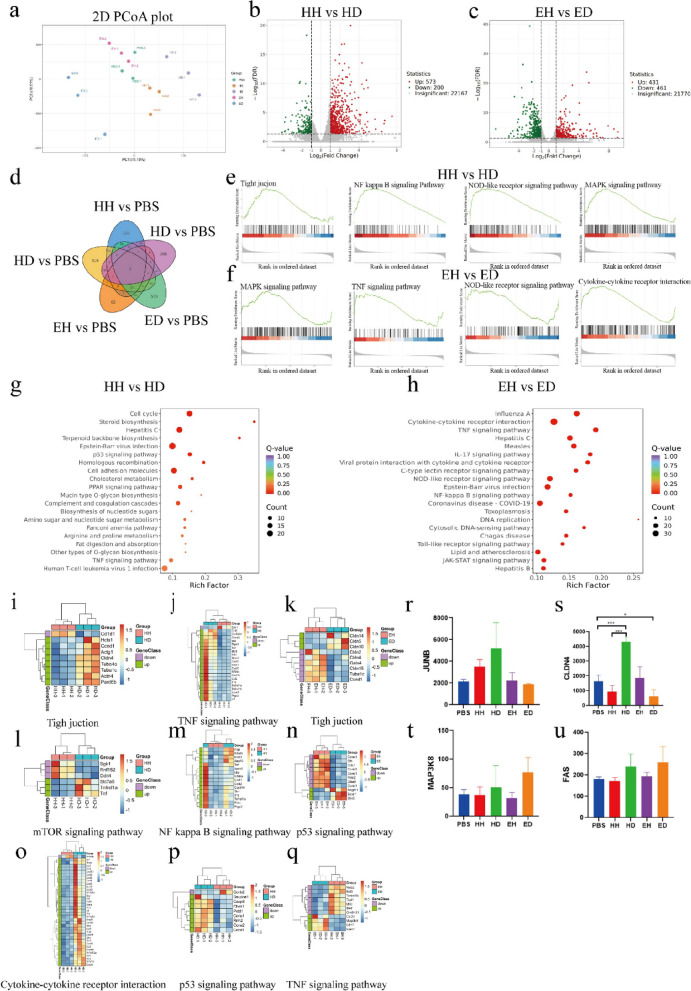
To validate our previous results, we performed RNA-seq using samples from five additional groups of mice and found significant genetic differences between each group, as shown in the PCA plot and Venn diagrams (Fig. 5a, d). In total, 573 genes had upregulated expression and 200 genes had downregulated expression in the HD group compared with those in the HH group, and 431 genes had upregulated expression and 461 genes had downregulated expression in the ED group compared with those in the EH group (Fig. 5b, c). The GSEA results revealed significant changes in proteins involved in tight junctions, the NF-kappa B signalling pathway, the MAPK signalling pathway, and the NOD-like receptor signalling pathway in the HH vs. HD comparison; there were significant changes in DEGS involved in the MAPK signalling pathway, the TNF signalling pathway, the NOD-like receptor signalling pathway, and the cytokine‒cytokine receptor interaction pathway in the EH vs. ED comparison (Fig. 5e, f). To comprehensively determine the changes in DEGs between the two groups of mice, we performed KEGG enrichment analysis; the results showed that the DEGs in the HH vs. HD group were enriched mainly in the cell cycle, the p53 signalling pathway, and the PPAR signalling pathway (Fig. 5g). In the EH vs. ED comparison, the DEGs were enriched in the TNF signalling pathway, the IL17 signalling pathway, the NF-kappa B signalling pathway, and the JAK-STAT signalling pathway (Fig. 5h). We performed a heatmap analysis of the pathways and quantified the key differential genes (Fig. 5i-u). By comparing the results of the two different breeds of sheep, we found that DEGs of both groups were enriched in the TNF signalling pathway and the NF-kappa B signalling pathway. Tumor necrosis factor (TNF) is a major mediator of apoptosis, inflammation, and immunity and has been implicated in the pathogenesis of a wide spectrum of human diseases, including sepsis, diabetes, cancer, osteoporosis, multiple sclerosis, rheumatoid arthritis, and inflammatory bowel diseases [28]. TNF works through two receptors, TNFR1 (also known as p55) and TNFR2 (also known as p75). TNFR1 is expressed in most tissues, and cross-linking with TNF produces a classic proinflammatory response. The most classic response is the activation of the NF-κB and c-Jun pathways, and the other pathways also activate MAPK; this activation results in the expression of various classical proinflammatory cytokines, such as IL-1, IL-6, and GM-CSF [29]. When TNF binds to TNFR2, the intracellular domain recruits the existing cytoplasmic TRAF2-cIAP1-cIAP2 complex. cIAP has ubiquitin ligase activity that inhibits the functions of caspases and other apoptosis-inducing factors, thereby initiating classical and noncanonical NF-κB activation [30]. These results suggest that the TNF-type immune response is an important driver of inflammatory autoimmune diseases and is dominant in East Friesian sheep. Taken together, these results strongly suggest that East Friesian sheep are more susceptible to inflammatory stimuli, which can induce sustained inflammation.
Fig. 5.
RNA-seq analysis was used to explore the mechanism underlying the disease resistance phenotype in mice. a PCA map of DEGs. b Volcano plot of HH and HD DEGs. c Volcano plot of EH and ED DEGs. d Venn diagram of DEGs. d GSEA diagram of HH and HD DEGs. f GSEA diagram of EH and ED DEGs. g KEGG enrichment map of HH and HD DEGs. h KEGG enrichment map of EH and ED DEGs. i Gene heatmap of the tight junction pathway in HH and HD. j Gene heatmap of the TNF signalling pathway in HH and HD. k Gene heatmap of the tight junction pathway in EH and ED. l Gene heatmap of the mTOR signalling pathway in HH and HD. m Gene heatmap of the NF-kappa B signalling pathway in HH and HD. n Gene heatmap of the p53 signalling pathway in EH and ED. o Gene heatmap of the cytokine‒cytokine receptor interaction in HH and HD. p Gene heatmap of the p53 signalling pathway in HH and HD. q Gene heatmap of the TNF signalling pathway in EH and ED. r Relative expression of JUNB. s The data are presented as the mean ± SEM (n = 6 mice per group). *P < 0.05, **P < 0.01, *** P < 0.001 were determined by one-way ANOVA with Bonferroni’s multiple comparisons test
Therapeutic effect of Roseburia bacteria on DSS-induced colitis in Hu sheep and East Friesian sheep
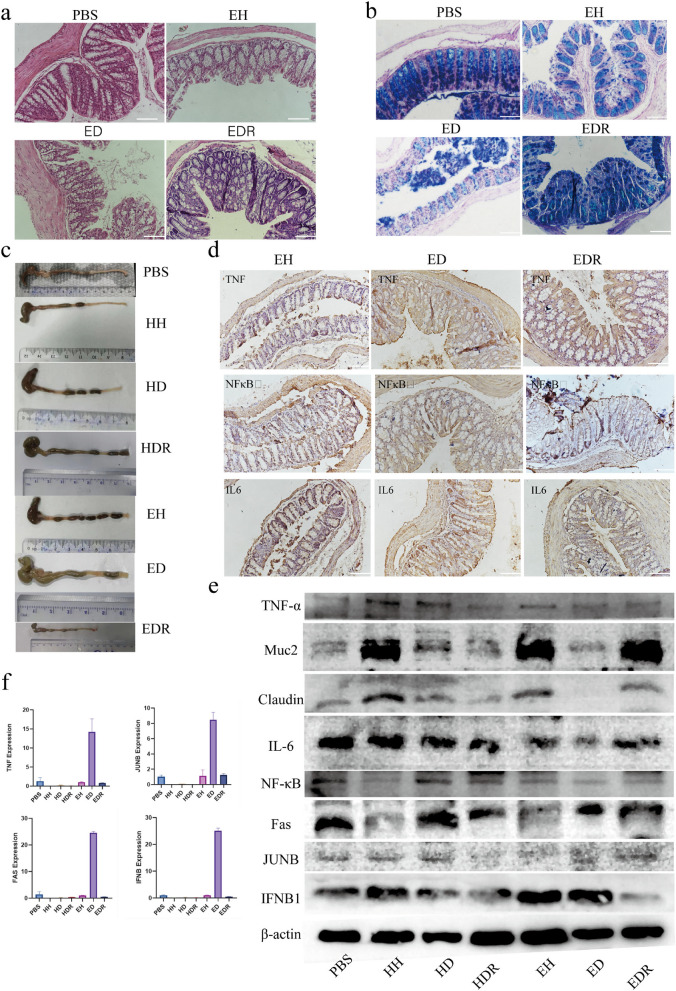
To explore whether Roseburia plays a role in DSS-induced colitis in Hu sheep and East Friesian sheep, FMT transplant mouse experiments were used in this study, and samples from HDR and EDR sheep were given to mice via FMT for 2 weeks. According to the results of our experiments, we determined that the phenotype of resistance to intestinal diseases in Hu sheep is better than that of East Friesian sheep because the beneficial bacteria, including Roseburia, are enriched in the intestinal tract of Hu sheep, and the secretion of bile and bile acids is greater. These two changes play a protective and reparative role in the gut of sheep. However, the enrichment of harmful microorganisms and metabolites in East Friesian sheep activated the TNFα signalling pathway, which increased the level of downstream proinflammatory factors, thereby damaging the intestine. However, the increase in beneficial microorganisms and beneficial metabolites in the intestinal tract of Hu sheep was not found in East Friesian sheep, which led to further aggravation of inflammation in the intestinal tract. In summary, we found that Roseburia bacteria are essential for protection against DSS-induced acute colitis, and to verify the effect of Roseburia as a potential probiotic in the intestine, we performed FMT via gavage to mice. After 2 weeks of treatment, we observed significant weight gain in the treated mice (Fig. 4b). Further histological analysis revealed severe body injury, intestinal mucosal damage, and crypt loss in the HD and ED groups. Roseburia increased the height of the colonic crypt and the width of the muscle layer (Fig. 6a, Extended Data Fig. 5a), suggesting that Roseburia significantly reduced cell infiltration and mucosal damage in mice with colitis. PAS staining revealed a decrease in the number of goblet cells in the HD and ED groups, while Roseburia treatment increased the number of mucin-producing goblet cells (Fig. 6b, Extended Data Fig. 5b). We observed a significant reduction in colon shortening in the HD and ED groups after Roseburia treatment (Fig. 6c). These results demonstrated that the TNFα signalling pathway was activated in both the DSS-induced colitis model group and the ED group of mice. To verify the therapeutic effect of Roseburia, we performed immunohistochemistry using samples from the two treatment groups, HDR and EDR. We found that the levels of TNFα, NFκB, and the inflammatory factor IL6, which were previously increased in the HD and ED groups, significantly decreased (Fig. 6d, Extended Data Fig. 5c). We performed a novel quantitative analysis of the PAS and immunohistochemistry results and found that the number of TNFα-, NFκB-, and IL6-positive cells significantly decreased after treatment (Extended Data Fig. 5d). Finally, we validated the therapeutic effects of Roseburia at the protein and RNA levels. We observed a significant increase in the level of the tight junction marker Claudin after treatment compared to that in the disease group. The TNFα and NFκB signalling pathways were significantly inhibited, the level of the inflammatory cytokine IL6 was significantly reduced, and the production of key factors in the TNF signalling pathway, such as Fas, JUNB, and Ifnb, was inhibited (Fig. 6e, f). This result suggests that Roseburia bacteria have a therapeutic effect in the gut of mice with colitis, which confirms our findings about the underlying sources of differences in disease resistance between the two breeds of sheep.
Fig. 6.
Therapeutic effect of Roseburia bacteria on DSS-induced colitis. a H&E tissue sections of mice. b AB-PAS tissue sections of mice. c Mouse colon length. d Mouse immunohistochemistry results. e Western blotting was used to validate the immune status and intestinal structure in mice. f Relative RNA expression
Conclusions
Here, the effects of different disease-resistant varieties of sheep on host–microbiota interactions were investigated through microbiota sequencing, metabolomics, transcriptomics, and other analyses. Our study showed that in H-DSS-treated sheep, the intestinal barrier is stabilized with an increase in the abundance of beneficial microorganisms. Our data also suggest that Roseburia bacteria have a protective effect on the intestinal barrier of Hu sheep. Accumulating evidence suggests that host–microbiota interactions are associated with IBD disease progression. Therefore, the results of this study may be useful in the identification of key gut microbes that are relevant to host health and may support a shift in thinking about anti-inflammatory strategies from enhancing immunity to immunosuppression.
Discussion
Within the gut, the colon is home to the densest and most metabolically active microbial community [31]. Although colitogenic pathobionts promote IBD development, commensal bacteria are also crucial for reducing IBD susceptibility [32]. FMT revealed that mice transplanted with feces from East Friesian sheep were more likely to develop DSS-induced colitis than mice transplanted with feces from Hu sheep [33]. The microbiota may play a major role in gut health, including in the maturation of host immune responses, protection against enteric pathogen proliferation and response to or the modification of specific drugs [34]. In the present study, diseased Hu sheep exhibited reduced injury in terms of weight loss, DAI, and limited crypt loss; this result provides evidence that the microbiota is associated with disease resistance. Zhao et al.’s study reported intestinal disease resistance of pigs; due to the different microbial composition in the host intestine, the resistance to intestinal diseases and the repair ability of damaged intestine in Min pigs were stronger than those of Yorkshire pigs under the same disease model conditions, and there were also significant differences in microbial β diversity in two different breeds of pigs [35]. Important host–microbiota interactions may be critical to host physiology and disease phenotypes [36]. We integrated microbiota sequencing, metabolomics, and transcriptomics to study host–microbiota crosstalk mechanisms that contribute to disease resistance and used FMT to validate these mechanisms in mice.
Despite scientific efforts in recent decades, the etiology and pathogenesis of the two major inflammatory bowel diseases, namely, Crohn’s disease and ulcerative colitis, remain unclear [37]. According to the results of multiple studies, the development of either disease is the result of an exaggerated or insufficiently suppressed immune response to an undefined luminal antigen, likely derived from the microbial flora [38]. This inflammatory process leads to mucosal damage and therefore further disruption of epithelial barrier function, resulting in an increased influx of bacteria into the intestine and further accelerating the inflammatory process [37]. In contrast to East Friesian sheep, Hu–DSS sheep displayed an increase in Firmicutes and major reductions in Bacteroidetes and Spirochaetes abundances [39]. At the genus level, more specific shifts were clearly observed. Our data suggest that Bacteroidetes and Proteobacteria were enriched in diseased sheep of both cultivars. Nevertheless, the extent and severity of these changes were less pronounced in Hu sheep than in East Friesian sheep. Palmla et al. found that the number of proteobacteria strains isolated from IBD patient samples with virulent properties increased, and the number of bacteria in this group was positively correlated with IBD recurrence [40]. Bacteroides increased in patients who had previously undergone surgical resection was reported in the study of Clooney et al. due to its involvement in mucin metabolism and its role in the destruction of the protective mucus layer [41]. An increase in these bacteria is common in IBD-associated dysbiosis, suggesting that these bacteria may be potentially harmful microorganisms and play a role in the pathogenesis of IBD [42]. In our study, downregulation of ZO-1 expression was observed in E-DSS sheep; this result is consistent with our inference about alteration of the intestinal barrier versus the role of microorganisms. Metabolites produced by the gut microbiota have been shown to influence the development of colorectal disease [43]. The concentration of butyrate, one of the essential metabolites of the human body, is inversely correlated with the incidence of colorectal disease [44]. Existing studies have shown that butyrate is positively correlated with the production of short-chain fatty acids in the body [45]. As an indispensable beneficial substance in the human body, short-chain fatty acids play an irreplaceable role in the prevention of intestinal diseases and the repair of damaged intestines [46]. Therefore, it is important for microorganisms that produced short-chain fatty acids to change in the host. In this study, we revealed for the first time that the phenotypic difference between Hu sheep and East Friesian sheep was due to microbial remodelling caused by IBD, which affected the species and abundance of their metabolites; we also identified the key potentially beneficial microbe Roseburia. Lachnospiraceae are among the main producers of SCFAs [47]. SCFAs, especially butyrate, have beneficial effects in terms of regulating intestinal immune function and inhibiting intestinal inflammation [48]. The depletion of these bacteria in East Friesian sheep may influence the reduction in SCFA concentrations and amplify intestinal inflammation [49]. These findings were reinforced by the inflammatory cytokine levels and mucosal barrier function. Remarkably, the abundances of many potentially beneficial microbes, such as Lactobacillus species and bacteria of the genus Ruminococcaceae belonging to Firmicutes, were increased in Hu-DSS-treated sheep. In addition, since Lachnospiraceae is considered a potential probiotic that is negatively correlated with inflammation and metabolism-related diseases [50], the higher abundance of Lachnospiraceae in Hu–DSS sheep suggests that Hu sheep may have disease defenses.
The gut microbiota can produce a variety of metabolites through anaerobic fermentation, including exogenous undigested dietary components that reach the colon, as well as endogenous compounds produced by microorganisms and hosts. Microbial metabolites can enter and interact with host cells, thereby influencing immune and inflammatory responses [51]. By pairing metabolomics with microbiota sequencing analysis, a strong correlation between the colonic microbiota and metabolites was found; these analyses also showed changes in the levels of bile acids and other metabolites. Previous studies have shown that specific classes of metabolites, particularly bile acids, are involved in the pathogenesis of IBD [34]. We observed a decrease in the concentration of bile acids and the inhibition of metabolism in East Friesian sheep. Hu sheep showed a very different trend from that of East Friesian sheep. In Hu sheep, the level of bile and bile acid metabolism was elevated, which is associated with alterations in the local microbiota and reduced mucosal inflammation. Overall, the results of the present study indicated that the beneficial gut microbes in Hu sheep alleviated injury in the colon and altered the levels of metabolites, based on the correlation between the gut microbiota and metabolites.
To further investigate the potential link among the gut microbiota, the host immune response, and intestinal barrier function in these two sheep breeds, RNA-seq analysis was performed using samples from the colons of Hu sheep and East Friesian sheep. KEGG analysis revealed that Hu sheep and East Friesian sheep had adequate immune function, including immune responses, inflammatory responses, and antibacterial humoral immune responses mediated by antimicrobial peptides, which indicated that they responded to exogenous stimuli. Although the responses were similar, the colonic inflammatory response was more active in E-DSS sheep than in H-DSS sheep. Specifically, several signalling pathways and downstream inflammation-related pathways, such as Toll-like receptor binding, the Nod-like receptor signalling pathway, the NIK/NF-κB pathway, the I-κB kinase/NF-κB signalling pathway, and the proinflammatory cytokine secretion pathway, were enriched in DEGS in East Friesian sheep but not in DEGS in Hu sheep. In E-DSS sheep, with the increase in potentially harmful microbes, such as Oscillospiraceae [52], microbial ligand signals are received in response to intestinal barrier damage and thus activate TNF signalling and downstream inflammation-related pathways. A reduction in bile acid may further exacerbate intestinal barrier dysfunction. In E-DSS-treated sheep, enrichment of Bacteroidetes and Spirochetes may increase the levels of benzene and its derivatives, thereby disrupting intestinal barrier function. Moreover, the increase in bile production may be due to an increase in several beneficial microorganisms that help maintain colon ZO1 levels [53]. In our study, it was found that there was a significant recovery of intestinal mucin in mice after Roseburia treatment, and the quantitative of markers related to tight junctions increased significantly, while inflammatory factors decreased significantly, suggesting that Roseburia may play a role in protecting and repairing the intestinal barrier damaged. These data suggest that in this IBD model, adjustment of the microbial community structure and the subsequent recovery of metabolites in Hu sheep may protect the intestinal barrier, while the increase in potentially harmful bacteria in E-DSS and the sharp decrease in potentially beneficial bacteria, including Lachnospiraceae, in Hu sheep activated the TNF signalling pathway, thereby promoting the secretion of inflammatory factors, further aggravating intestinal damage, and ultimately leading to differences in the disease resistance phenotype between the two sheep. In the study of Engevik et al., it was also confirmed that microorganisms are involved in the development of inflammation. This study proves that the increase of potentially harmful microorganisms activates the TNF signalling pathway and promotes the secretion of inflammatory factors, causing damage to advocacy [54]. In our study, for the first time, we used multiomics to show that the mechanism of disease resistance is closely related to the increase in Roseburia in sheep, verified the reason for the poor disease resistance of East Friesian sheep, and verified the results by FMT; although Roseburia was a key potential probiotic that was identified in this study, the reason for the opposite trend in the abundance of Roseburia bacteria in the gut of the two types of sheep is unknown. Additionally, due to time and experimental conditions, we have not yet analyzed the causes of metabolite changes related to Roseburia [55]. We do not know what the relationship between potentially beneficial and harmful bacteria is in the gut or what impact these relationships have on the host [56], which may be the direction of future research. However, in our study, it can be determined that Roseburia bacteria are beneficial bacteria for the intestinal disease resistance in two sheep, and Roseburia bacteria showed a very good therapeutic effect in the colitis. The results suggest that Roseburia bacteria may be a promising venue for the treatment of sheep colitis.
Methods
Animals and experimental design
Twelve Hu sheep (2-month-old male) and 12 East Friesian sheep (2-month-old male) were selected from a commercial farm. The weight range was 15 kg ± 3 kg, and all the sheep were in the same physiological state and individually housed in stainless steel metabolism crates with automatic troughs and drinking water nipples for free access to water. Animals received the same diet. After 7 days of acclimation, the sheep of each breed were randomized into two treatments administered via gavage for 5 days: CON (control, sterile saline) and DSS (2 g/kg). All sheep were anesthetized and euthanized on the 14th day after modeling. Samples were taken from all mice after being sacrificed using the cervical dislocation. Samples were collected from each sheep immediately after slaughter. Colon tissue was rinsed and stored immediately at − 80 °C until further analysis. Colon contents were collected and stored at − 80 °C until microbiota and metabolite analyses.
We used 8-week-old male C57BL/6JNifdc male mice (20g ± 3g) without specific pathogens provided by Weitong Lihua Experimental Animal Technology Co., Ltd. (Beijing, China) in this study. The mice were maintained under standard temperature, humidity, and light conditions (room temperature 22 °C, humidity 55–60%, and light/dark cycles for 12 h) and were allowed free access to food and water. Commercial mouse chow and water were autoclaved prior to use. To determine the role of microbes in the FMT model, the mice were administered an antibiotic cocktail (vancomycin 0.5, ampicillin 0.5, metronidazole 0.5, and neomycin sulfate 1 g L − 1; ABX) for 7 days before FMT induction [25]. After 1 week of acclimation, the mice were divided into 7 groups (n = 8), which were PBS group, HH group, HD group, HDR group, EH group, ED group, and EDR group. Among them, the PBS group was given PBS by gavage in the control group, the HH group was given healthy Hu sheep fecal liquid, the HD group was given fecal liquid from sheep treated with DSS, the HDR group was also given fecal liquid of Hu sheep treated with DSS and Roseburia bacteria, the EH group was given healthy East Friesian sheep fecal liquid, the ED group was given fecal liquid from East Friesian sheep treated with DSS, and the EDR group was also given fecal liquid of East Friesian sheep treated with DSS and Roseburia bacteria. All but the PBS group required antibiotic therapy. The prepared fecal bacterial solution was transferred to recipient mice in the experimental group through the oral forced feeding method. The vaccination period was 1 week, with each mouse receiving 200 μL of fecal bacterial solution per day.
Disease activity index (DAI) and histological activity index (HAI) scoring criteria
The body weight and DAI were recorded for each group of mice. The length of the colon was measured when the mice were euthanized. All animal procedures were approved by the Institutional Animal Care and Use Committee of Inner Mongolia University. The DAI was calculated as (body weight loss score + fecal trait score + fecal blood score)/3.
DAI scoring criteria
| Score | Weight loss% | Stool consistency | Blood in stool |
|---|---|---|---|
| 0 | 0 | Normal | Normal |
| 1 | 1–5 | Soft | Normal |
| 2 | 5–10 | Fluid | Occult blood |
| 3 | 10–20 | Fluid | Occult blood |
| 4 | ≥ 20 | Diarrhea | Gross bleeding |
Histological scoring criteria
After sampling, histopathological analysis was performed on each sample, and each individual selected the most complete section for H&E staining. Each slide selects three fields of view for histopathological scoring, and all pathology scores are done by the same technician to ensure the impartiality of the results.
| Score | Inflammatory cell infiltration | Depth of invasion | Crypt injury | Pathological range (%) |
|---|---|---|---|---|
| 0 | None | None | None | 0 |
| 1 | ± | Mucosa layer | 1/3 | 1–25 |
| 2 | + | Mucosa and submucosa | 2/3 | 26–50 |
| 3 | + + | Full | Full | 51–75 |
| 4 | + + + | Full | Full | 76–100 |
Organ indices
The spleen and colon were collected and evaluated by the organ index formula: organ index (%) = (organ weight/body weight) × 100%.
H&E staining
The severity of colonic lesions was scored macroscopically and histologically. Colon samples were fixed in 4% paraformaldehyde, embedded in paraffin, sectioned, and stained with hematoxylin and eosin (H&E) for histological examination to determine the severity of inflammation and the extent of mucosal and crypt damage. Images were captured with a 100 × magnification imaging system.
AB-PAS staining
Deparaffinization to water: xylene I, 5 min; xylene II, 5 min; xylene III, 5 min; absolute ethanol, 1 min; 95% ethanol, 1 min; 75% ethanol, 1 min; and distilled water wash for 5 min. Add Alixin blue staining solution dropwise for 10–20 min and wash three times with distilled water for 1–2 min each time. Put it in the oxidant for 5 min. Rinse with tap water and soak in distilled water two times. Dip into Schiff staining solution for 10–20 min. Pour off the Schiff staining solution and rinse with running water for 10 min. Hematoxylin staining solution stains the nucleus for 1–2 min and washes with water. Acidic differentiation solution differentiates for 2–5 s and washes with water. Return to blue with Scott bluing solution for 3 min and wash with water for 3 min. Dehydration and transparency: 75% ethanol for 1 min, 95% ethanol for 1 min, absolute ethanol for 1 min, xylene three times, 1–2 min each time, and neutral gum mounting.
Transmission electron microscope
Colon tissue sections were cut into ultra-thin sections at a thickness of 60–68 nm. A JEM-1010 Transmission Electron Microscope (Institute of Food Science and Technology CAAS, Beijing, China) was used for electron micrographs.
Preparation of the donor fecal bacterial solution
Donor East Friesian sheep and Hu sheep feces are collected and mice are subjected to FMT for consecutive 7 days. The feces were collected and stored in sterile 5-mL centrifuge tubes, with a portion stored in − 80 °C ultralow temperature refrigerators for microbial detection. Fungal solutions were prepared according to previously reported methods [57]. The remaining two groups of fecal samples were mixed separately in sterile PBS solution (1 g/5 mL). After stirring evenly, the solution was filtered through double-layered sterile gauze. After centrifugation (2000 r/min, 5 min), the supernatant was discarded, and the cells were resuspended in the same volume of sterile PBS to obtain a fecal bacterial solution. One milliliter of sterile glycerol (100%) was added to every 10 mL of fecal bacterial solution, after which the solution was distributed into a 15-mL centrifuge tube and stored at − 80 °C [26].
Immunohistochemistry
Heat-mediated antigen retrieval was performed using 0.01 M citrate buffer (pH 6.0) or 1 mM EDTA (pH 8.0) for 20 min in a microwave for immunohistochemistry analysis. After cooling to room temperature, the sections were immersed in 3% H2O2 for 10 min, blocked with blocking solution at RT for 1 h, and incubated with primary antibodies overnight at 4 °C. The sections were then immunostained by the ABC peroxidase method (Vector Laboratories) with diaminobenzidine (DAB) as the substrate and hematoxylin as the counterstain. Images were captured using a Nikon inverted fluorescence microscope (Nikon, SMZ7457).
Western blotting
Total protein was extracted using lysis buffer (Solarbio, R0030) containing 0.1 M PMSF (Solarbio, P0100) and 10 g/L phosphatase inhibitor (Thermo Scientific, A32957) on ice. After centrifugation at 13,000 × g for 10 min at 4 °C, the protein concentration in the supernatant was determined using a Pierce BCA Protein Assay Kit (Thermo Scientific, 23,227). Proteins were separated by 10–12% SDS–PAGE, transferred to a nitrocellulose membrane (BIO-RAD, 1620177), blocked with 5% skim milk (BD, 232100) for 1 h at room temperature, and then incubated with primary antibody overnight at 4 °C and then with secondary antibody for 1 h at room temperature. The signals were visualized with Pierce ECL Western blotting Substrate (Thermo Scientific, 32,209) and detected with an E-BLOT contact nondestructive quantitative WB imaging system (TouchImager, S2303063).
Quantitative RT-PCR analysis
Total RNA was isolated using the RNeasy Mini Kit (Qiagen,74104) according to the manufacturer’s protocol. cDNA was synthesized using the GoScript Reverse Transcription System (Promega, A5001). Real-time PCR was performed with the KAPA SYBR FAST qPCR Kit (KAPA Biosystems, KR0389) on a LightCycler 96 instrument (Roche Molecular Systems) with at least three biological replicates, and all results were similar. Relative transcript levels were assessed using the 2-ΔΔCt method, and GAPDH served as an endogenous control. The primer pairs used in this study are described in Extended Data Table 2.
RNA-seq analysis
RNA-seq was performed using intact colon tissue samples from different treatment groups. Total RNA was extracted using TRIzol reagent following the manufacturer’s procedure. The cDNA library construction and bioinformatics analysis were used to construct cDNA libraries, which were sequenced on an Illumina NovaSeq6000 (California, USA). Gene expression levels were estimated by calculating fragment values. Exons per thousand nucleotides were mapped to one million reads (FPKM). We selected genes with a fold change > 2 and a P value < 0.05. Then, we used Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis to determine enrichment of upregulated and downregulated DEGs in specific pathways. The data are presented as reads per kilobase million.
Culture of Roseburia
Volatile fatty acid solution
A volatile fatty acid solution was obtained by mixing 1.9 mL of acetic acid, 0.7 mL of propionic acid, 90 μL of isobutyric acid, 100 μL of valeric acid, and 100 μL of isovaleric acid, filtered and sterilized, and stored at 4 °C.
Vitamin solution
Two milligrams biotin, 2 mg folic acid, 10 mg pyridoxine hydrochloride, 2 mg thiamine hydrochloride × 2H2O, 2 mg vitamin B2, 12 mg niacin, 5 mg D-calcium pantothenate, 5 mg vitamin B12, 5 mg p-p-aminobenzoic acid, and 5 mg lipoic acid were dissolved in 1000 mL water, filtered and sterilized, and stored at 4 °C.
YCFA medium preparation
Weigh the basal medium (except sodium bicarbonate, heme chloride, and cysteine) and dissolve it in water
Boil for 10 min, add sodium bicarbonate, heme chloride, and cysteine, and adjust pH to 6.7–6.8.
-
(2)
Add 100 μL of vitamin solution and 2.89 mL of volatile per 10 mL of medium under sterile conditions.
Hair fatty acid solution
YCFA medium can be purchased from the German Microbial Culture Collection Center DSMZ, or you can purchase the domestic R. intestinalis culture and treatment R. intestinalis was anaerobically cultured at 37 °C using YCFA medium, after growth to mid-log, the bacterial solution was collected, 8000 × g, centrifuged at 4 °C for 10 min, the supernatant was discarded, and resuspended in sterile PBS at a final concentration of 10 [9] CFU/mL. Dosage for mice, 2 × 10 [8] CFU of R. intestinalis per day.
Extraction and PCR amplification of genomic DNA from the gut microbiota
16 S rRNA gene sequencing of fecal samples was performed by Wuhan Maiwei Metabolic Biotechnology Co., Ltd. The results were analyzed using the Maiwei Cloud platform. The samples were extracted via the CTAB method, after which the purity and concentration of the DNA were determined by agarose gel electrophoresis. An appropriate amount of sample DNA was transferred to a centrifuge tube and diluted to 1 ng /μL with sterile water. Using diluted genomic DNA as a template and based on the selection of sequencing regions, specific primers with Barcode were used, using the Phusion from New England Biolabs ® High Fidelity PCR Master Mix with GC Buffer and efficient high-fidelity enzymes for PCR to ensure amplification efficiency and accuracy. The 341F (CCTAYGGRBGCACAG) and 806R (GGACTACNNGGGTATCTAAT) primer binding sequences were used to amplify the V3–V4 region of the 16S rDNA gene in bacteria, after which an amplicon pool was prepared for amplification. Library construction and machine sequencing were done using TruSeq ® DNA PCR Free Sample Preparation Kit for library construction and Qubit (ThermoFisher Scientific)/Agilent Bioanalyzer 2100 (Agilent Technologies Inc. USA) to construct the library System/Q-PCR quantification. After the library is qualified, use NovaSeq6000 for machine sequencing.
Sequencing data processing
The data for each sample were split from the offline data based on the barcode sequence and PCR amplification primer sequence, and the barcode and primer sequences were compared. Using fastp (v0.22.0, https://github.com/OpenGene/fastp), the original reads were filtered to obtain high-quality reads via the following filtering method: automatically detect and remove joint sequences; remove reads with an N base quantity of 15 or more; remove reads with low-quality bases (mass value ≤ 20) accounting for more than 50%; delete those with an average mass less than 20 within the 4-base window interval; delete the polyG at the end; and delete reads with a length less than 150 bp. High-quality dual-end reads were obtained using FLASH (v1.2.11, http://ccb.jhu.edu/software/FLASH/) to obtain high-quality tag data (clean tags). The tag sequences were obtained through vsearch (v2.22.1); the chimeric sequences were compared and identified with the species annotation database (https://github.com/torognes/vsearch). Ultimately, the chimeric sequences were removed to obtain the final effective tags.
Alpha diversity analysis
Using the photoseq (v1.40.0) and vegan (v2.6.2) packages of R software (v4.2.0), the Chao1, Shannon, Simpson, ACE, and PD-whole-tree indices were calculated. R software (v4.2.0) was used to plot dilution curves, rank abundance curves, and species accumulation curves, and intergroup difference analysis of alpha diversity indices was performed using R software. The analysis of intergroup differences in the alpha diversity index was conducted using both parametric and nonparametric tests.
Beta diversity analysis
Comparative analysis of diversity was conducted using the photoseq (v1.40.0) package of R software (v4.2.0) to calculate the UniFrac distance and construct a UPGMA sample clustering tree. R software (v4.2.0) was used to construct PCA, PCoA, and NMDS diagrams. PCA was performed with the R software stats package, while PCoA and NMDS analysis were performed with the R software photoseq (v1.40.0) package. R software was used for intergroup analysis of the differences in the beta diversity index, and parametric and nonparametric tests were conducted.
LEfSe was performed with LEfSe (v1.1.2) software, with a default LDA score filtering value of 3.6. Metastats analysis was performed with Mothur software to perform intergroup permutation tests at various classification levels (phylum, class, order, family, genus, and specifications) to obtain P values. Then, the Benjamin and Hochberg false discovery rate was used to correct the P values and obtain q values. ANOSIM, MRPP, and Adonis analyses were conducted using the ANOSIM function, mrpp function, and adonis function of the R vegan package, respectively. AMOVA was performed with the AMOVA function in motif software. Analysis of species with significant intergroup differences was performed using R software for intergroup t tests and plotting.
The collected samples were thawed on ice, and metabolites were extracted using 50% methanol buffer. Then, LC–MS analysis was performed on all samples according to the system manufacturer’s instructions, and metabolites eluted from the column were detected using a TripleTOF 6600 + high-resolution tandem mass spectrometer (SCIEX, USA). The metabolites were annotated using the online KEGG database by matching the precise molecular weight data (m/z) of the sample with data in the database.
Bioinformatics analysis
PCA
Unsupervised PCA (principal component analysis) was performed by the statistics function prcomp within R (www.r-project.org). The data were subjected to unit variance scaling before unsupervised PCA.
Hierarchical clustering analysis and Pearson correlation coefficients
The hierarchical cluster analysis (HCA) results for the samples and metabolites are presented as heatmaps with dendrograms, while Pearson correlation coefficients (PCCs) of samples were calculated by the cor function in R and are presented in heatmaps. Both HCA and PCC were carried out with the R package pheatmap. For HCA, normalized signal intensities of metabolites (unit variance scaling) were visualized as a color spectrum.
Differentially abundant metabolite selection
Significantly differentially abundant metabolites between groups were determined by the VIP, P value (P value < 0.05, Student’s t test) and absolute log2FC (fold change). VIP values were extracted from the OPLS-DA results, which also contained score plots and permutation plots that were generated using the R package MetaboAnalystR. The data were log2 transformed (log2) and mean centered before OPLS-DA. To avoid overfitting, a permutation test (200 permutations) was performed.
KEGG annotation and enrichment analysis
The identified metabolites were annotated using the KEGG compound database (http://www.kegg.jp/kegg/compound/), and the annotated metabolites were subsequently mapped to the KEGG pathway database (http://www.kegg.jp/kegg/pathway.html). Pathways with significantly differentially expressed metabolites were then subjected to metabolite set enrichment analysis (MSEA), and their significance was determined by hypergeometric test P values.
Statistical analysis
All the statistical analyses were conducted using GraphPad Prism, version 7.0 (GraphPad, La Jolla, CA) or the R package. At least three biologically independent experiments were performed unless stated otherwise. All the data are presented as the mean ± standard deviation (SD). The P value was obtained by unpaired two-tailed Student’s t test, and asterisks denote statistical significance (*P < 0.05; **P < 0.01; ***P < 0.001).
Supplementary Information
Additional file 1: Extended Data Figure legends. Extended Data Fig. 1 (a-b) Transmission electron microscopy images of colon tissue sections from East Friesian sheep; (c-t) biochemical analysis of Hu sheep and East Friesian sheep serum. Extended Data Fig. 2 (a) Metabolite PCA map; (b) Venn diagram of differentially abundant metabolites in Hu sheep and East Friesian sheep; (c) H-CON vs. H-DSS differentially abundant metabolite correlation network; (d) E-CON vs. E-DSS differentially abundant metabolite correlation network; (d) H-CON vs. H-DSS differentially abundant metabolite volcano plot; (d) E-CON vs. E-DSS differentially abundant metabolite volcano plot; (d) differential dynamic distribution of metabolite content in H-CON vs. H-DSS; (d) differential dynamic distribution of metabolite content in E-CON vs. E-DSS; (d) H-CON vs. H-DSS differentially abundant metabolite GO enrichment analysis; (d) E-CON vs. E-DSS differentially abundant metabolite GO enrichment analysis; (d) violin plot of H-CON vs. H-DSS differentially abundant metabolites; (d) violin plot of E-CON vs. E-DSS differentially abundant metabolites. Extended Data Fig. 3 (a) Microbial correlation plot of H-CON vs. H-DSS with the metabolite Spearman; (b) microbial correlation plot of E-CON vs. E-DSS with the metabolite Spearman. Extended Data Fig. 4 (a) Metabolite PCA map in mice; (b) HH vs. HD differentially abundant metabolite volcano plot; (c) EH vs. ED differentially abundant metabolite volcano plot; (d) Venn diagram of differentially abundant metabolites in mice; (e) HH vs. HD differentially abundant metabolite correlation network in mice; and (f) EH vs. ED differentially abundant metabolite correlation network in mice. (g) Violin plot of HH vs. HD differentially abundant metabolites in mice; (h) violin plot of EH vs. ED differentially abundant metabolites in mice; (i) HH vs. HD differentially abundant metabolite GO enrichment analysis in mice; and (j) EH vs. ED differentially abundant metabolite GO enrichment analysis in mice. Extended Data Fig. 5 (a) H & E tissue sections of mice; (b) B-PAS tissue sections of mice; (c) mouse immunohistochemistry results; and (d) PAS/Alcian blue staining, TNF-positive cells, NF-κB-positive cells, and IL6-positive cells. Extended Data Fig. 6 TNFα binds to the extracellular region of tumour necrosis factor receptor (TNFR1), which forms a trimer of TNFR1, recruits the intracellular signalling protein TRAF2/5, activates the NF-κB signalling pathway through TAK1, and ultimately promotes the related inflammatory factor Fas through CREB. Release of IL6, IL1β, JUNB, etc.
Acknowledgements
The authors thank Liming Liu (Inner Mongolia Shengle Biotechnology Co., Ltd, Hohhot, China ) for sheep administration. We thank Wuhan Metware Biotechnology Co., Ltd. for assisting in sequencing and bioinformatics analysis.
Authors’ contributions
Y.S.,S.B.,X.L designed research; S.Y.,R.D, performed research; S.Y.,R.D.,W.Y.,H.Z.,Y.X.,Y.L.,T.,H.B,Y.C,Y.Z.G,C analyzed data; S.Y.and Y.S. produced figures; S.Y.and Y.S. wrote the manuscript.
Funding
This work was supported by the Scientific and Technological Innovation 2030-major project funding (no. 2022ZD0401403); the Introduction Project of High-Level Talents in Inner Mongolia University Biology (no.10000–22120301/011); platform construction of biological breeding of dairy cattle and sheep (No.2023KJHZ0019); and 2024 Inner Mongolia University "Steed Plan" high-level talent funding(10000–23122101/021).
Data availability
The datasets supporting the conclusions of this article are available in the NCBI Sequence Read Archive (SRA) repository under accession number PRJNA1109790, PRJNA1109823, PRJNA1110381 and PRJNA1110339 (available on May 13, 2024).Our metabolome data has been uploaded to the NGDC database with PRJCA026594 (ngdc.cncb.ac.cn) (Release Date May 29 , 2024).
Declarations
Ethics approval and consent of participate
All animal experiments were performed in accordance with the National Research Council Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee at Inner Mongolia University, China. The approval number is NMGDX (Wu) 2022–0003.
Consent for publication
All authors have consented to publication.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Shuo Yan and Ruilin Du contributed equally to this work.
Contributor Information
Xihe Li, Email: lixh@imu.edu.cn.
Siqin Bao, Email: baosq@imu.edu.cn.
Yongli Song, Email: ylsong@imu.edu.cn.
References
- 1.Zhernakova A, et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science. 2016;352:565–9. 10.1126/science.aad3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen L, et al. The long-term genetic stability and individual specificity of the human gut microbiome. Cell. 2021;184:2302-2315.e2312. 10.1016/j.cell.2021.03.024. [DOI] [PubMed] [Google Scholar]
- 3.Schirmer M, Garner A, Vlamakis H, Xavier RJ. Microbial genes and pathways in inflammatory bowel disease. Nat Rev Microbiol. 2019;17:497–511. 10.1038/s41579-019-0213-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao X, et al. Host-microbiota interaction-mediated resistance to inflammatory bowel disease in pigs. Microbiome. 2022;10. 10.1186/s40168-022-01303-1. [DOI] [PMC free article] [PubMed]
- 5.de Vos WM, Tilg H, Van Hul M, Cani PD. Gut microbiome and health: mechanistic insights. Gut. 2022;71:1020–32. 10.1136/gutjnl-2021-326789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mamun MAA, et al. The composition and stability of the faecal microbiota of Merino sheep. J Appl Microbiol. 2020;128:280–91. 10.1111/jam.14468. [DOI] [PubMed] [Google Scholar]
- 7.Kang X, et al. Roseburia intestinalis generated butyrate boosts anti-PD-1 efficacy in colorectal cancer by activating cytotoxic CD8(+) T cells. Gut. 2023;72:2112–22. 10.1136/gutjnl-2023-330291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li X, et al. Whole genome re-sequencing reveals artificial and natural selection for milk traits in East Friesian sheep. Front Vet Sci. 2022;9. 10.3389/fvets.2022.1034211. [DOI] [PMC free article] [PubMed]
- 9.Kominakis A, Hager-Theodorides AL, Saridaki A, Antonakos G, Tsiamis G. Genome-wide population structure and evolutionary history of the Frizarta dairy sheep. Animal. 2017;11:1680–8. 10.1017/s1751731117000428. [DOI] [PubMed] [Google Scholar]
- 10.Tüfekci H, Sejian V. Stress factors and their effects on productivity in sheep. Animals. 2023;13. 10.3390/ani13172769. [DOI] [PMC free article] [PubMed]
- 11.Kong LC, et al. Characterization of bacterial community changes and antibiotic resistance genes in lamb manure of different incidence. Sci Rep. 2019;9:10101. 10.1038/s41598-019-46604-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu Y, et al. ADDAGMA: A database for domestic animal gut microbiome atlas. Comput Struct Biotechnol J. 2022;20:891–8. 10.1016/j.csbj.2022.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X, et al. Whole genome re-sequencing reveals artificial and natural selection for milk traits in East Friesian sheep. Front Vet Sci. 2022;9: 1034211. 10.3389/fvets.2022.1034211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghia JE, et al. Serotonin has a key role in pathogenesis of experimental colitis. Gastroenterology. 2009;137:1649–60. 10.1053/j.gastro.2009.08.041. [DOI] [PubMed] [Google Scholar]
- 15.Pu Z, et al. Systematic understanding of the mechanism and effects of Arctigenin attenuates inflammation in dextran sulfate sodium-induced acute colitis through suppression of NLRP3 inflammasome by SIRT1. American journal of translational research. 2019;11:3992–4009. [PMC free article] [PubMed] [Google Scholar]
- 16.Vijay A, Valdes AM. Role of the gut microbiome in chronic diseases: a narrative review. Eur J Clin Nutr. 2022;76:489–501. 10.1038/s41430-021-00991-6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Lewis SM, Williams A, Eisenbarth SC. Structure and function of the immune system in the spleen. Science immunology4. 2019. 10.1126/sciimmunol.aau6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He C. et al. View from the Biological Property: Insight into the functional diversity and complexity of the gut mucus. Int J Mol Sci .2023;24.10.3390/ijms24044227. [DOI] [PMC free article] [PubMed]
- 19.Huang S, et al. Lachnospiraceae-derived butyrate mediates protection of high fermentable fiber against placental inflammation in gestational diabetes mellitus. Science advances. 2023;9:eadi733. 10.1126/sciadv.adi7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nie K, et al. Roseburia intestinalis: A beneficial gut organism from the discoveries in genus and species. Front Cell Infect Microbiol. 2021;11: 757718. 10.3389/fcimb.2021.757718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qi R, et al. The intestinal microbiota contributes to the growth and physiological state of muscle tissue in piglets. Sci Rep. 2021;11:11237. 10.1038/s41598-021-90881-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jia W, Li Y, Cheung KCP, Zheng X. Bile acid signaling in the regulation of whole body metabolic and immunological homeostasis. Science China Life sciences. 2023. 10.1007/s11427-023-2353-0. [DOI] [PubMed] [Google Scholar]
- 23.Zhang L, et al. Probiotics ameliorate benzene-induced systemic inflammation and hematopoietic toxicity by inhibiting Bacteroidaceae-mediated ferroptosis. The Science of the total environment. 2023;899: 165678. 10.1016/j.scitotenv.2023.165678. [DOI] [PubMed] [Google Scholar]
- 24.Singh P, et al. Effect of antibiotic pretreatment on bacterial engraftment after fecal microbiota transplant (FMT) in IBS-D. Gut Microbes. 2022;14: 2020067. 10.1080/19490976.2021.2020067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gou H, et al. Traditional medicine Pien Tze Huang suppresses colorectal tumorigenesis through restoring gut microbiota and metabolites. Gastroenterology. 2023;165:1404–19. 10.1053/j.gastro.2023.08.052. [DOI] [PubMed] [Google Scholar]
- 26.Cheng J, et al. Sheep fecal transplantation affects growth performance in mouse models by altering gut microbiota. J Animal Sci. 2022;100:skac303. 10.1093/jas/skac303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larabi AB, Masson HLP, Bäumler AJ. Bile acids as modulators of gut microbiota composition and function. Gut Microbes. 2023;15: 2172671. 10.1080/19490976.2023.2172671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen G, Goeddel DV. TNF-R1 signaling: a beautiful pathway. Science. 2002;296:1634–5. 10.1126/science.1071924. [DOI] [PubMed] [Google Scholar]
- 29.Webster JD, Vucic D. The balance of TNF mediated pathways regulates inflammatory cell death signaling in healthy and diseased tissues. Frontiers in cell and developmental biology. 2020;8:365. 10.3389/fcell.2020.00365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monaco C, Nanchahal J, Taylor P, Feldmann M. Anti-TNF therapy: past, present and future. Int Immunol. 2015;27:55–62. 10.1093/intimm/dxu102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sender R, Fuchs S, Milo R. Are we really vastly outnumbered? revisiting the ratio of bacterial to host cells in humans. Cell. 2016;164:337–40. 10.1016/j.cell.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 32.Kamada N, Seo SU, Chen GY, Núñez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013;13:321–35. 10.1038/nri3430. [DOI] [PubMed] [Google Scholar]
- 33.Burrello C, et al. Therapeutic faecal microbiota transplantation controls intestinal inflammation through IL10 secretion by immune cells. Nat Commun. 2018;9:5184. 10.1038/s41467-018-07359-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schirmer M, Garner A, Vlamakis H, Xavier RJ. Microbial genes and pathways in inflammatory bowel disease. Nat Rev Microbiol. 2019;17:497–511. 10.1038/s41579-019-0213-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao X, et al. Host-microbiota interaction-mediated resistance to inflammatory bowel disease in pigs. Microbiome. 2022;10:115. 10.1186/s40168-022-01303-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buffington SA, et al. Dissecting the contribution of host genetics and the microbiome in complex behaviors. Cell. 2021;184:1740-1756.e1716. 10.1016/j.cell.2021.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmidt C, Stallmach A. Etiology and pathogenesis of inflammatory bowel disease. Minerva Gastroenterol Dietol. 2005;51:127–45. [PubMed] [Google Scholar]
- 38.Ney LM, et al. Short chain fatty acids: key regulators of the local and systemic immune response in inflammatory diseases and infections. Open Biol. 2023;13: 230014. 10.1098/rsob.230014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rezvannejad E, Nanaei HA, Esmailizadeh A. Detection of candidate genes affecting milk production traits in sheep using whole-genome sequencing analysis. Vet Med Sci. 2022;8:1197–204. 10.1002/vms3.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palmela C, et al. Adherent-invasive Escherichia coli in inflammatory bowel disease. Gut. 2018;67:574–87. 10.1136/gutjnl-2017-314903. [DOI] [PubMed] [Google Scholar]
- 41.Clooney AG, et al. Ranking microbiome variance in inflammatory bowel disease: a large longitudinal intercontinental study. Gut. 2021;70:499–510. 10.1136/gutjnl-2020-321106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Santana PT, Rosas SLB, Ribeiro BE, Marinho Y, de Souza HSP. Dysbiosis in inflammatory bowel disease: pathogenic role and potential therapeutic targets. Int J Mol Sci. 2022;23:3464. 10.3390/ijms23073464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zitvogel L, Daillère R, Roberti MP, Routy B, Kroemer G. Anticancer effects of the microbiome and its products. Nat Rev Microbiol. 2017;15:465–78. 10.1038/nrmicro.2017.44. [DOI] [PubMed] [Google Scholar]
- 44.Wang T, et al. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012;6:320–9. 10.1038/ismej.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mann ER, Lam YK, Uhlig HH. Short-chain fatty acids: linking diet, the microbiome and immunity. Nat Rev Immunol. 2024;24:577–95. 10.1038/s41577-024-01014-8. [DOI] [PubMed] [Google Scholar]
- 46.Zhang D, et al. Short-chain fatty acids in diseases. Cell Commun Signal. 2023;21:212. 10.1186/s12964-023-01219-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mukherjee A, Lordan C, Ross RP, Cotter PD. Gut microbes from the phylogenetically diverse genus Eubacterium and their various contributions to gut health. Gut Microbes. 2020;12: 1802866. 10.1080/19490976.2020.1802866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun M, Wu W, Liu Z, Cong Y. Microbiota metabolite short chain fatty acids, GPCR, and inflammatory bowel diseases. J Gastroenterol. 2017;52:1–8. 10.1007/s00535-016-1242-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deleu S, Machiels K, Raes J, Verbeke K, Vermeire S. Short chain fatty acids and its producing organisms: an overlooked therapy for IBD? EBioMedicine. 2021;66: 103293. 10.1016/j.ebiom.2021.103293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McCarville JL, Chen GY, Cuevas VD, Troha K, Ayres JS. Microbiota metabolites in health and disease. Annu Rev Immunol. 2020;38:147–70. 10.1146/annurev-immunol-071219-125715. [DOI] [PubMed] [Google Scholar]
- 51.Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol. 2016;16:341–52. 10.1038/nri.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dennis VA, et al. Live Borrelia burgdorferi spirochetes elicit inflammatory mediators from human monocytes via the Toll-like receptor signaling pathway. Infect Immun. 2009;77:1238–45. 10.1128/iai.01078-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mohanty I, et al. The changing metabolic landscape of bile acids - keys to metabolism and immune regulation. Nat Rev Gastroenterol Hepatol. 2024. 10.1038/s41575-024-00914-3. [DOI] [PubMed] [Google Scholar]
- 54.Engevik MA, et al. Fusobacterium nucleatum secretes outer membrane vesicles and promotes intestinal inflammation. mBio. 2021;12. 10.1128/mBio.02706-20. [DOI] [PMC free article] [PubMed]
- 55.Sana TR, et al. Global mass spectrometry based metabolomics profiling of erythrocytes infected with Plasmodium falciparum. PLoS One. 2013;8: e60840. 10.1371/journal.pone.0060840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hills RD, Jr. et al. Gut microbiome: profound implications for diet and disease. Nutrients. 2019;11.10.3390/nu11071613 [DOI] [PMC free article] [PubMed]
- 57.Zhang W, et al. Fecal microbiota transplantation (fmt) alleviates experimental colitis in mice by gut microbiota regulation. J Microbiol Biotechnol. 2020;30:1132–41. 10.4014/jmb.2002.02044. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Extended Data Figure legends. Extended Data Fig. 1 (a-b) Transmission electron microscopy images of colon tissue sections from East Friesian sheep; (c-t) biochemical analysis of Hu sheep and East Friesian sheep serum. Extended Data Fig. 2 (a) Metabolite PCA map; (b) Venn diagram of differentially abundant metabolites in Hu sheep and East Friesian sheep; (c) H-CON vs. H-DSS differentially abundant metabolite correlation network; (d) E-CON vs. E-DSS differentially abundant metabolite correlation network; (d) H-CON vs. H-DSS differentially abundant metabolite volcano plot; (d) E-CON vs. E-DSS differentially abundant metabolite volcano plot; (d) differential dynamic distribution of metabolite content in H-CON vs. H-DSS; (d) differential dynamic distribution of metabolite content in E-CON vs. E-DSS; (d) H-CON vs. H-DSS differentially abundant metabolite GO enrichment analysis; (d) E-CON vs. E-DSS differentially abundant metabolite GO enrichment analysis; (d) violin plot of H-CON vs. H-DSS differentially abundant metabolites; (d) violin plot of E-CON vs. E-DSS differentially abundant metabolites. Extended Data Fig. 3 (a) Microbial correlation plot of H-CON vs. H-DSS with the metabolite Spearman; (b) microbial correlation plot of E-CON vs. E-DSS with the metabolite Spearman. Extended Data Fig. 4 (a) Metabolite PCA map in mice; (b) HH vs. HD differentially abundant metabolite volcano plot; (c) EH vs. ED differentially abundant metabolite volcano plot; (d) Venn diagram of differentially abundant metabolites in mice; (e) HH vs. HD differentially abundant metabolite correlation network in mice; and (f) EH vs. ED differentially abundant metabolite correlation network in mice. (g) Violin plot of HH vs. HD differentially abundant metabolites in mice; (h) violin plot of EH vs. ED differentially abundant metabolites in mice; (i) HH vs. HD differentially abundant metabolite GO enrichment analysis in mice; and (j) EH vs. ED differentially abundant metabolite GO enrichment analysis in mice. Extended Data Fig. 5 (a) H & E tissue sections of mice; (b) B-PAS tissue sections of mice; (c) mouse immunohistochemistry results; and (d) PAS/Alcian blue staining, TNF-positive cells, NF-κB-positive cells, and IL6-positive cells. Extended Data Fig. 6 TNFα binds to the extracellular region of tumour necrosis factor receptor (TNFR1), which forms a trimer of TNFR1, recruits the intracellular signalling protein TRAF2/5, activates the NF-κB signalling pathway through TAK1, and ultimately promotes the related inflammatory factor Fas through CREB. Release of IL6, IL1β, JUNB, etc.
Data Availability Statement
The datasets supporting the conclusions of this article are available in the NCBI Sequence Read Archive (SRA) repository under accession number PRJNA1109790, PRJNA1109823, PRJNA1110381 and PRJNA1110339 (available on May 13, 2024).Our metabolome data has been uploaded to the NGDC database with PRJCA026594 (ngdc.cncb.ac.cn) (Release Date May 29 , 2024).